Abstract
e2g is a web-based server which efficiently maps large expressed sequence tag (EST) and cDNA datasets to genomic DNA. It significantly extends the volume of data that can be mapped in reasonable time, and makes this improved efficiency available as a web service. Our server hosts large collections of EST sequences (e.g. 4.1 million mouse ESTs of 1.87 Gb) in precomputed indexed data structures for efficient sequence comparison. The user can upload a genomic DNA sequence of interest and rapidly compare this to the complete collection of ESTs on the server. This delivers a mapping of the ESTs on the genomic DNA. The e2g web interface provides a graphical overview of the mapping. Alignments of the mapped EST regions with parts of the genomic sequence are visualized. Zooming functions allow the user to interactively explore the results. Mapped sequences can be downloaded for further analysis. e2g is available on the Bielefeld University Bioinformatics Server at http://bibiserv.techfak.uni-bielefeld.de/e2g/.
INTRODUCTION
High-throughput cDNA and expressed sequence tag (EST) sequencing projects have generated a vast amount of data representing the transcribed portion of the organisms under study. As soon as (parts of) the sequence of the associated genome becomes available, the cDNA and ESTs are mapped to the genomic sequence e.g. to detect new genes, verify the exon–intron structure of predicted genes and determine splice variants.
Mapping ESTs or cDNAs to a genomic sequence is a standard task in molecular biology, and there are several tools available for this task [see, e.g. (1–7)]. Most of these tools were developed for small-scale tasks, where sensitivity was the main design goal. The essential step is usually a costly dynamic programming method with a running time quadratic to the size of the input. Therefore, some tools apply filtering methods first. These scan the ESTs in linear time to find regions containing highly conserved matches to the genomic sequence. Unfortunately, none of the existing tools can efficiently handle complete EST collections of vertebrates with millions of ESTs. This is because the fast filtering methods (if any) still have to scan the entire EST collection. Moreover, there are only a few tools available which provide a comprehensive graphical representation of the sometimes contradictory mappings of the ESTs or cDNAs to the genomic sequence. A good example of such a visualization tool is SpliceNest (8), which, however, only allows the visualization of static datasets.
e2g is a tool providing both efficient mapping of user-provided genomic sequences and convenient visualization. It uses an efficient index structure precomputed for the EST collection under consideration. The index structure allows the user to find highly conserved matches between the genomic sequence and the EST collection much more quickly than with a scanning-based method. The mapping of the ESTs or cDNAs is visualized as colored blocks (representing the length and direction of the matches) relative to the genomic sequence. The user can interactively explore the set of matches by zooming into regions of interest.
IMPLEMENTATION
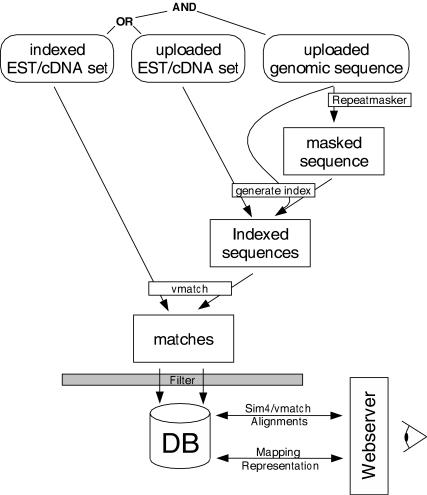
Figure 1 shows the data flow from sequence upload, via EST mapping, to output generation. e2g can be used in two different basic modes.
Figure 1.
Data flow in e2g. The user uploads a genomic sequence, which is optionally masked by RepeatMasker. Moreover, the user either chooses an EST collection with a precomputed index or uploads his/her own EST/cDNA set. The uploaded genomic sequence is matched against the index using Vmatch. Spurious matches are filtered and the remaining matches are stored in a database. The web server generates overviews of the mapping for the region chosen by the user. An alignment of the matching sequences, and a spliced alignment of a selected EST, are computed on demand by Vmatch and sim4, respectively.
The user uploads a genomic DNA sequence and chooses one of the EST collections available on the server. Currently, EST collections for Homo sapiens (5.4 × 106 ESTs) and Mus musculus (4.1 × 106 ESTs) and corresponding index structures are available.
The user uploads a genomic DNA sequence as well as the cDNA/EST collection to be mapped. In this case, the index structure for the collection is first computed by the server.
In both modes, RepeatMasker (http://ftp.genome.washington.edu/RM/RepeatMasker.html) is optionally applied to the uploaded genomic sequence to mask organism-specific repeats. Simple repeats and low-complexity regions are always masked by default. Also, GenScan (9) is run to obtain an initial ab initio gene prediction. Furthermore, the user can upload a file containing a gene annotation of the corresponding genomic sequence. Currently, there is a 5 MB limitation on the size of uploaded data. In the following, we will assume the first mode because we expect it to be the standard mode.
The index structure is an enhanced suffix array (10) stored in several files. It provides rapid access to all substrings of the ESTs of the given collection. The enhanced suffix array is precomputed only once, and can be used many times for different genomic sequences. Given the enhanced suffix array, the software tool Vmatch (http://www.vmatch.de/) matches the genomic DNA against the enhanced suffix array to obtain exact matches. These are extended using the X-drop algorithm of (11). This gives highly conserved matches between ESTs and the genomic sequences. Sometimes there are a large number of spurious hits, typically caused either by DNA contaminations in the EST library or by repeats missed by RepeatMasker. Therefore, a match is discarded if there is no other match in the same EST. For convenient and fast access, the positions of the remaining matches are stored in a relational database. The matches represent a mapping of a subset of the ESTs to specific positions on the genomic sequence. The user can select an EST, and a spliced alignment for the EST is computed on the fly using sim4 (2). This allows for the detection of splice site signals. Running sim4 on a single EST and a small region of the genomic sequence does not add much to the running time of e2g. In the future, we will add other spliced alignment tools for different levels of sensitivity and speed.
All steps in the e2g dataflow (shown as rectangles in Figure 1) are implemented using web services technology (http://www.w3.org/TR/ws-arch/). e2g runs on a Sun Solaris compute server with eight 900 MHz UltraSparc III CPUs and 64 GB of RAM. The web interface and its underlying CGI framework are implemented as messaging services. This allows us to easily integrate more servers if necessary and develop standalone clients which are independent of the web interface and can be used in an automated way.
WEB INTERFACE
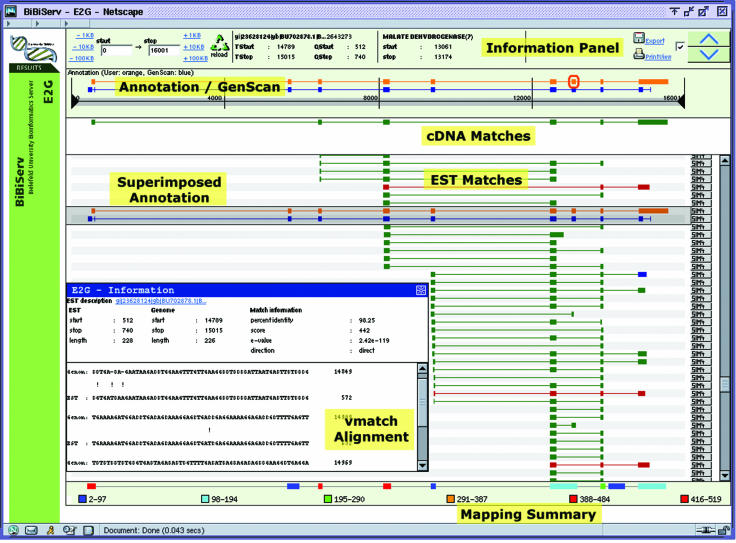
Figure 2 shows a screenshot of a graphical overview produced by e2g when uploading a 16.5 kb genomic sequence from M.musculus (Genbank GI: 28515921, bases 60 000–76 500) to compare it to 4.1 million ESTs from the same species. The overview is split into five panels, arranged from top to bottom:
Figure 2.
Screenshot of the e2g web interface showing the mapping of mouse ESTs/cDNAs to mouse genomic sequence (Genbank GI: 28515921, bases 60 000–76 500). The information is split into five panels: general information, gene annotation, cDNA matches, EST matches and mapping summary (from top to bottom). Forward matches are shown in green, reverse-complemented in red. The transparent gray-shaded box in the middle of the image contains the annotation uploaded by the user. It can be moved over matching ESTs to further inspect their exon/intron structure. The popup window shows the Vmatch alignment of the blue-highlighted exon in the EST match panel.
General information panel. The top of the window provides general information about the current task. The user can zoom into a region of interest within the submitted genomic sequence. The positions of the highlighted matches in the EST and the genomic sequence are displayed. This part of the overview also provides links to download the sequences or GI numbers of matching ESTs.
Annotation panel. The second section of the window shows gene predictions for the genomic sequence, as uploaded by the user (orange colored) and delivered by GenScan (blue colored). If the prediction refers to the forward strand, then the exons are shown above the line representing the genome, otherwise below.
cDNA mapping panel. cDNA matches on the genomic sequence are shown as colored blocks. Forward matches are shown in green, reverse-complemented matches are shown in red.
EST mapping panel. EST matches on the genomic sequence are shown in the same way as cDNA match. The two kinds of match are separated since cDNAs are usually of higher quality, and thus matches to the genomic sequence are more reliable.
Mapping summary panel. The bottom panel provides a summary of all matches, shown as colored boxes. The color code represents the coverage of a region, i.e. the relative number of matches in the region. For example, in Figure 2, regions with high coverage are represented by red boxes and regions with low coverage by blue boxes.
The GenScan and uploaded annotation from the annotation panel can be superimposed on the cDNA and EST matches by dragging a transparent image over the lower part of the window. For web browsers that do not support JavaScript mouse events, the up/down buttons in the upper right corner of the window provide the same functionality. The transparent image conveniently allows the user to compare the gene prediction with the matches found.
By clicking on a match, an alignment (computed by Vmatch) between this individual region of the EST and the genomic sequence is shown in a popup window. The alignment is supplemented by additional information such as positions in the genomic sequence and in the EST, scores, identity values and E-values (Figure 2, bottom). Additionally, whenever the button on the right is clicked, sim4 is run to produce a spliced alignment over the whole EST sequence.
PERFORMANCE EVALUATION
For our performance experiments, we mapped all ESTs from M.musculus (total length 1.87 Gb) to a 16.5 kb genomic sequence (same as above) from the same species. Analyses were run on a Sun-UltraSparc III CPU (900 MHz). To show the limits of a scanning-based approach we first applied sim4 to this dataset. Since sim4 cannot handle files >2 GB, we split the 2.4 GB file containing the mouse ESTs into two files. The total running time of sim4 was 3.5 h, which is, of course, too long for a web service application.
e2g delivers a mapping (without any spliced alignment) for the same data size in much less time, using a precomputed index structure. With the default parameter setting, all 3889 matches of length ≥30, containing exact seeds of length ≥20, and with identity ≥98% are computed in 30 s. This is about the same time as required for storing the match positions in the database and generating the graphical overview.
Acknowledgments
ACKNOWLEDGEMENTS
We acknowledge the fruitful discussions with Carsten Drepper and Phillip Hahn, who have inspired us to develop e2g. Thanks to Ute Willhöft for valuable suggestions to improve previous versions of this manuscript.
REFERENCES
- 1.Mott R. (1997) EST_GENOME: a program to align spliced DNA sequences to unspliced genomic DNA. CABIOS, 13, 477–478. [DOI] [PubMed] [Google Scholar]
- 2.Florea L., Hartzell,G., Zhang,Z., Rubin,G.M. and Miller,W. (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res., 8, 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Usuka J., Zhu,W. and Brendel,V. (2000) Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics, 16, 203–211. [DOI] [PubMed] [Google Scholar]
- 4.Gemund C., Ramu,C., Altenberg-Greulich,B. and Gibson,T.J. (2001) Gene2EST: a BLAST2 server for searching expressed sequence tag (EST) databases with eukaryotic gene-sized queries. Nucleic Acids Res., 29, 1272–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Val C., Glatting,K.H. and Suhai,S. (2003) cDNA2Genome: a tool for mapping and annotating cDNAs. BMC Bioinformatics, 4, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee B.T.K., Tan,T.W. and Ranganathan,S. (2003) MGAlignIt: a web service for the alignment of mRNA/EST and genomic sequences. Nucleic Acids Res., 31, 3533–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coward E., Haas,S.A. and Vingron,M. (2002) SpliceNest: visualization of gene structure and alternative splicing based on EST clusters. Trends Genet., 18, 53–55. [Google Scholar]
- 9.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–91. [DOI] [PubMed] [Google Scholar]
- 10.Abouelhoda M.I., Kurtz,S. and Ohlebusch,E. (2004) Replacing suffix trees with enhanced suffix arrays. J. Discrete Algor., 2, 53–86. [Google Scholar]
- 11.Zhang Z., Schwartz,S., Wagner,L. and Miller,W. (2000) A greedy algorithm for aligning DNA sequences. J. Comput. Biol., 7, 203–214. [DOI] [PubMed] [Google Scholar]