Abstract
The MyHits web server (http://myhits.isb-sib.ch) is a new integrated service dedicated to the annotation of protein sequences and to the analysis of their domains and signatures. Guest users can use the system anonymously, with full access to (i) standard bioinformatics programs (e.g. PSI-BLAST, ClustalW, T-Coffee, Jalview); (ii) a large number of protein sequence databases, including standard (Swiss-Prot, TrEMBL) and locally developed databases (splice variants); (iii) databases of protein motifs (Prosite, Interpro); (iv) a precomputed list of matches (‘hits’) between the sequence and motif databases. All databases are updated on a weekly basis and the hit list is kept up to date incrementally. The MyHits server also includes a new collection of tools to generate graphical representations of pairwise and multiple sequence alignments including their annotated features. Free registration enables users to upload their own sequences and motifs to private databases. These are then made available through the same web interface and the same set of analytical tools. Registered users can manage their own sequences and annotations using only web tools and freeze their data in their private database for publication purposes.
INTRODUCTION
The Generalized Profile and HMM (hidden Markov model) technologies are the methods of preference to detect domains in protein sequences (1). Unfortunately, these methods require both good computer skills and powerful computing resources. Two different approaches exist currently to help researchers overcome these limitations.
On the one hand the PSI-BLAST tool (2) allows users to easily define their own potential protein domains and to use them to search the public databases. PSI-BLAST, despite being very fast and easy to use through a simple web interface, hides several steps that may be critical for the development of good protein domain descriptors. For example, it does not give access to the multiple alignment implicitly calculated between iterations. On the other hand, precomputed hit lists databases such as Pfam (3), SMART (4) and InterPro (5) provide fast access to already identified protein domains in the Swiss-Prot and TrEMBL protein databases (6). Following this lead, we previously developed the Hits database of precomputed matches including locally developed databases (7,8).
Normally, precomputed hit lists mine only the public databases, whereas users often possess unpublished sequences as well as structural or biochemical data that may shed new light on a protein region, domain or signature. This private information may prove essential in updating existing protein domain descriptors or in the definition of new ones, but it is currently difficult to integrate using web tools.
This creates the need for another generation of tools, which we have tried to assemble in our MyHits server.
ERGONOMICS OF THE WEB SITE
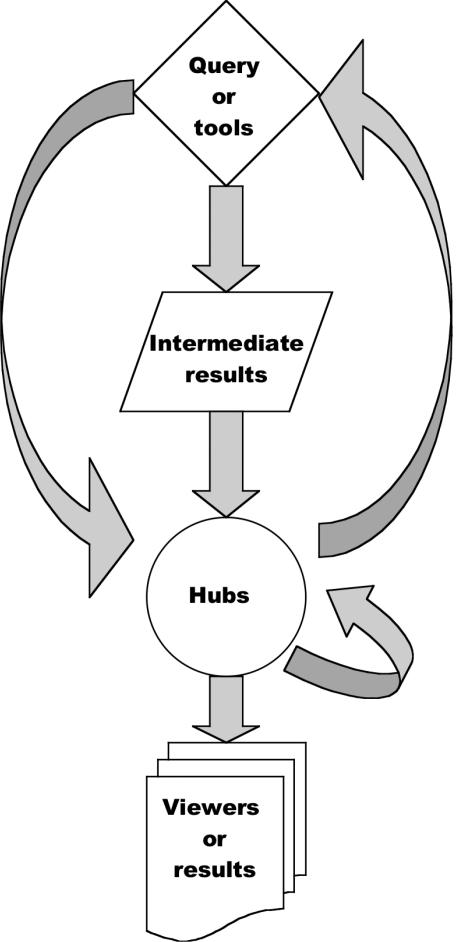
Conceiving a user interface means facing a necessary trade-off between the users learning curve and the versatility of the solution offered. We intend to fulfil the needs of the researcher with some education in bioinformatics, but not necessarily in computer science. MyHits is somewhat complex, but we feel its flexibility more than compensates for its complexity. To further reduce the learning curve, MyHits is extensively documented and illustrated with examples. Simple web applications typically have two stages: a query form and a results page. However, sometimes, users may wish to further explore their results in an alternative format, or using a specialized tool. MyHits allows the user to select among a choice of viewers. The choice is made on a special page that we call a hub. Each basic data type [e.g. protein, motif, multiple sequence alignment (MSA)] has its own hub. Any query that produces results of a given type links its results page to the corresponding hub. This means that two different query forms can lead to the same hub. In many cases, the results of a query can be fed into another query (e.g. a protein query produces a list of motifs, which can in turn be fed into the motif query). This is also done via the hub. Finally, the hub itself can serve as an entry point, if all users need is to view the data (i.e. no querying) (Figure 1).
Figure 1.
Description of the MyHits hub system. This figure depicts the different routes leading from a query form to the results page.
HANDLING LARGE LISTS OF PROTEINS
A protein sequence may appear several times in different sequence databases because it has been sequenced and/or predicted more than once. Moreover, these different copies can bear distinct annotations that are related to the protein function in Swiss-Prot, or to the underlying exon location in trome (9), for example. Nowadays a simple database search can yield thousands of matches, pushing the manual screening of the results to the limits. Indeed, we believe that postprocessing tools for mining this information may offer new perspectives.
We provide a tool for the automated clustering of identical and highly similar protein sequences. Typically, this permits the extraction of a representative subset out of a large set of sequences. For example, we systematically postprocess the output of database search programs with a taxonomic filter and with a procedure to group the matched sequences with an identity level equal or superior to a user-defined threshold.
Graphical representation of the sequences and their attached features may speed up visual scanning of a large set of sequences. We provide access to several Java-based viewers [e.g. SEView (10), Dotlet (11), Jalview (12)], as well as access to a new graphical representation of sequence alignments including aligned feature annotations.
HANDLING MULTIPLE SEQUENCE ALIGNMENTS
We propose a collection of web-based tools to simplify the task of producing and improving multiple sequence alignments. As far as protein sequences are concerned, MSAs play a central role as the repositories of information. However, producing a high-quality MSA is very time-consuming, as it often needs to be edited by hand to make it compliant with available structural or biochemical information. Protein domain databases such as Pfam (3) and SMART (4) offer ‘seed’ MSAs of relatively good quality. Nevertheless, in our experience, these can be inadequate for particular applications. The list of proteins they encompass may miss some ‘important’ sequences for a particular problem (e.g. the one corresponding to the latest solved structure), or the MSA may cover a too divergent list of proteins, and restraining the MSA to a subfamily may improve it. The database search services we offer rely on two search engines, PSI-BLAST and pfsearch (13). Owing to its response time, the former is more adapted to interactive usage than the latter, but with somewhat inferior results, especially concerning alignment quality. The output of the two search methods was amended such that an MSA could be recovered in all cases. This MSA can be used as the start of a new database search. This mechanism, incidentally, permits the user to change the searched databases, to modify the search parameters and even to switch to another search engine at any iteration. We recommend using high-quality databases such as Swiss-Prot for ‘training’ an MSA with PSI-BLAST, and then to use it as the query to search databases of lower quality [e.g. trEST, trGEN, and trome (9)].
Another important aspect of the pivotal role played by the MSA is the possibility to realign the matched sequences using other programs between two searches. For that purpose we offer ClustalW (14) and T-Coffee (15) for automated alignment, as well as a modified version of the Jalview applet to manually edit the MSA and send it back to the hub. When alignment quality is critical, this strategy should always be envisaged as counteracting the general tendency of lowering alignment quality through successive search cycles.
We also provide a pattern search yielding an output that can similarly be recovered as an MSA.
PUBLIC AND PRIVATE DATABASES
A user has access to two different sorts of resources: on the one hand the public databases, and on the other unpublished private data (e.g. new sequences or additional biochemical evidence). A recurring request is the possibility to save working sets of sequences derived from public and private resources. Indeed, it would be useful to keep some sequences together and unmodified during a certain period of time (e.g. until a set of experiments has been completed or even until a paper has been published). Moreover, users might want to add personal annotations to their sequences or use their own predictors to automatically annotate their sequences.
With these goals in mind, MyHits offers services to two types of users: the guest user and the registered user. A guest user has access to all public databases and to all tools, but cannot save private sequences and motifs within the system. MyHits is populated by default with a list of publicly available databases of sequences and motifs. These databases (Tables 1 and 2) are available to all users for browsing, searching, scanning, and so on. They are automatically and incrementally updated on a weekly basis.
Table 1. Motif databases available in MyHits.
| Code | Status | Description |
|---|---|---|
| mypat | Private | User's own patterns |
| pat | Public | Prosite patterns |
| prf | Public | Prosite profiles |
| ipr | Public | InterPro motif entries |
Table 2. Sequence databases available in MyHits.
| Automatic hit lists calculations |
||||||
|---|---|---|---|---|---|---|
| Code | Status | Description | mypat | pat | prf | ipr |
| mypep | Private | User's own protein sequences | ✓+ | ✓+ | ✓+ | − |
| sw | Public | Swiss-Prot current release | − | ✓ | ✓ | ✓* |
| sw_var | Public | Splice variants reconstructed from Swiss-Prot annotations | − | ✓ | ✓ | ✓* |
| tr | Public | TrEMBL current release | − | ✓ | ✓ | ✓* |
| tr_var | Public | Splice variants reconstructed from TrEMBL annotations | − | ✓ | ✓ | ✓* |
| tn | Public | TrEMBL new sequences | − | ✓ | ✓ | − |
| to | Public | trome sequences | − | ✓ | ✓ | − |
| te | Public | trEST sequences | − | ✓ | ✓ | − |
| tg | Public | trGEN sequences | − | ✓ | ✓ | − |
An exhaustive calculation of hit lists is performed between public sequence databases and public motif databases (shown by a ‘✓’). By default, the private sequence database (mypep) is calculated against the private motif database (mypat) and the public motif databases (‘✓+’), but not the reverse (‘−’). The InterPro motifs (ipr) and hit lists are derived directly from the official distribution (no calculation is done locally) (‘✓*’).
Guest users can register for free to become registered users, with the following advantages. Currently the registered user can access a private database of sequences (mypep) and one of patterns (mypat). The registered user can also store multiple alignments in progress in a private database (mymsa). We plan to add other types of motif databases (myprf, myhmm) in the future. Each user database has a standard limit in terms of the number of entries and the size of sequences. These numbers can be increased upon request, depending on the availability of resources.
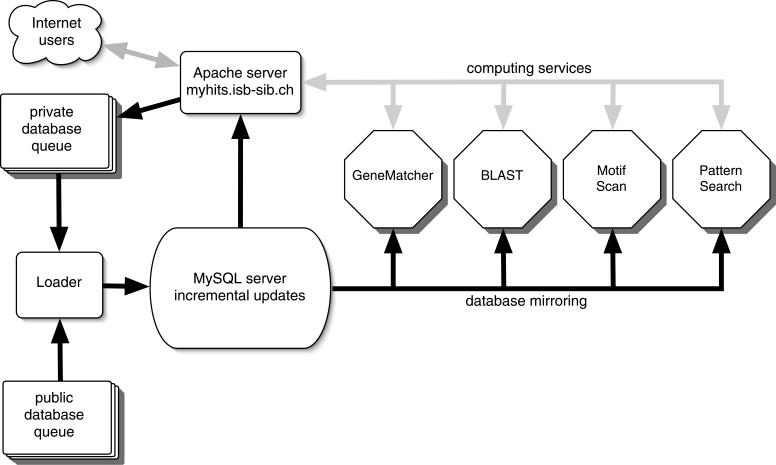
We have developed an asynchronous incremental updating process that is the technical heart of the new MyHits system (to be described elsewhere). Based on a relational database, MyHits lowers the cost of updates by using internally non-redundant sequence databases. The mirroring of the sequences on search engines is done automatically as well (Figure 2). Both the mypep and mypat databases are automatically added to our queuing system every time a user modifies an entry. The full update and mirroring steps of the new entries can take from several minutes to hours depending on the load of the server. During this process the old version of the user database is still available.
Figure 2.
Schematic representation of the MyHits server. All user interactions are performed via the web using a browser. Users can access the different tools and public databases (grey arrows). In addition, registered users can manage their private databases (upload or edit their own sequences directly from the browser). Public, as well as the private databases which have been modified, are included in the incremental update process and the hit lists are then calculated taking into account the new sequences and motifs (dark arrows).
MyHits delivers a unique set of services, providing answers to some of the current limitations of the tools dedicated to protein domain identification and to some of the recurring end-user needs such as the ability to transfer results from one application to another in a very convenient and easy way with a simple mouse click. Thanks to users' feedback during the courses organized by the Swiss EMBnet node (http://www.ch.embnet.org) or obtained through its helpdesk (helpdesk@mail.ch.embnet.org) (16), as well as by continuous contact with Prosite and Swiss-Prot annotators, the MyHits server will improve continuously in the future.
REFERENCES
- 1.Hofmann K. (2000) Sensitive protein comparisons with profiles and hidden Markov models. Brief. Bioinformatics, 1, 167–178. [DOI] [PubMed] [Google Scholar]
- 2.Schaffer A.A., Aravind,L., Madden,T.L., Shavirin,S., Spouge,J.L., Wolf,Y.I., Koonin,E.V. and Altschul,S.F. (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res., 29, 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman A., Coin,L., Durbin,R., Finn,R.D., Hollich,V., Griffiths-Jones,S., Khanna,A., Marshall,M., Moxon,S., Sonnhammer,E.L. et al. (2004) The Pfam protein families database. Nucleic Acids Res., 32, D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultz J., Copley,R.R., Doerks,T., Ponting,C.P. and Bork,P. (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res., 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulder N.J., Apweiler,R., Attwood,T.K., Bairoch,A., Barrell,D., Bateman,A., Binns,D., Biswas,M., Bradley,P., Bork,P. et al. (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res., 31, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O'Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jongeneel C.V. (2000) Searching the expressed sequence tag (EST) databases: panning for genes. Brief. Bioinformatics, 1, 76–92. [DOI] [PubMed] [Google Scholar]
- 8.Pagni M., Iseli,C., Junier,T., Falquet,L., Jongeneel,V. and Bucher,P. (2001) trEST, trGEN and Hits: access to databases of predicted protein sequences. Nucleic Acids Res., 29, 148–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sperisen P., Iseli,C., Pagni,M., Stevenson,B.J., Bucher,P. and Jongeneel,C.V. (2004) trome, trEST and trGEN: databases of predicted protein sequences. Nucleic Acids Res., 32, D509–D511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Junier T. and Bucher,P. (1998) SEView: a Java applet for browsing molecular sequence data. In Silico Biol., 1, 13–20. [PubMed] [Google Scholar]
- 11.Junier T. and Pagni,M. (2000) Dotlet: diagonal plots in a web browser. Bioinformatics, 16, 178–179. [DOI] [PubMed] [Google Scholar]
- 12.Clamp M., Cuff,J., Searle,S.M. and Barton,G.J. (2004) The Jalview Java alignment editor. Bioinformatics, 20, 426–427. [DOI] [PubMed] [Google Scholar]
- 13.Sigrist C.J., Cerutti,L., Hulo,N., Gattiker,A., Falquet,L., Pagni,M., Bairoch,A. and Bucher,P. (2002) PROSITE: a documented database using patterns and profiles as motif descriptors. Brief. Bioinformatics, 3, 265–274. [DOI] [PubMed] [Google Scholar]
- 14.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Notredame C., Higgins,D.G. and Heringa,J. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol., 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 16.Falquet L., Bordoli,L., Ioannidis,V., Pagni,M. and Jongeneel,C.V. (2003) Swiss EMBnet node web server. Nucleic Acids Res., 31, 3782–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]