Abstract
JavaProtein Dossier (JPD) is a new concept, database and visualization tool providing one of the largest collections of the physicochemical parameters describing proteins' structure, stability, function and interaction with other macromolecules. By collecting as many descriptors/parameters as possible within a single database, we can achieve a better use of the available data and information. Furthermore, data grouping allows us to generate different parameters with the potential to provide new insights into the sequence–structure–function relationship. In JPD, residue selection can be performed according to multiple criteria. JPD can simultaneously display and analyze all the physicochemical parameters of any pair of structures, using precalculated structural alignments, allowing direct parameter comparison at corresponding amino acid positions among homologous structures. In order to focus on the physicochemical (and consequently pharmacological) profile of proteins, visualization tools (showing the structure and structural parameters) also had to be optimized. Our response to this challenge was the use of Java technology with its exceptional level of interactivity. JPD is freely accessible (within the Gold Sting Suite) at http://sms.cbi.cnptia.embrapa.br, http://mirrors.rcsb.org/SMS, http://trantor.bioc.columbia.edu/SMS and http://www.es.embnet.org/SMS/ (Option: JavaProtein Dossier).
INTRODUCTION
One of the fundamental biochemical–biophysical problems is how to expose the intricate relationships among the forces that govern protein folding. Now, a general perception is emerging: the more parameters that can be collected under a common computational platform, the more relationships among those parameters that can be established. Such a newly established knowledge environment could consequently make possible more accurate prediction of these forces and their interdependence.
The disproportionate growth of the available data on one side, and the lack of available and appropriate tools that can establish adequate relationships among them on the other, are key sources of the noticeable ‘lag time’ before we can actually create applicable solutions based on databases generated by genome projects. Given the growing volume of the PDB (Protein Data Bank) (1) and the parallel trend in importance that protein structures have for understanding basic biological processes, the need to collect and then thoroughly describe structures is becoming ever more crucial.
It is clear that the human capacity for understanding and conceptualizing information is limited when faced with such large volumes of data. Therefore, the automation of data collection, analysis, summarization, trend discovery and characterization, as well as flagging the anomalies, is of crucial importance for further knowledge growth.
This area of automation is thriving on multidisciplinary effort involving information technology in general and database technology, artificial intelligence, statistics, high-performance computing and data visualization in particular. Data visualization is of particular interest in the field of bioinformatics, where we struggle to come up with the best visual display of measured and calculated quantities by means of the combined use of graphics and text.
Both statistical assessment and graphic representation are effective ways to describe, explore and summarize a very large set of numbers, making a combination of the two into a powerful instrument for reasoning about quantitative information (2). JPD (JavaProtein Dossier) communicates a large set of the physicochemical properties of proteins at a residue-by-residue level through a unique graphic interface.
At the same time JPD is a step toward compiling the most diversified and complete database of structure–function descriptors, which can and will be used as a platform for knowledge discovery. The Gold Sting release containing JPD currently offers a total of 125 numerical/textual descriptors for any given protein structure deposited in the PDB (3). This certainly places JPD among very few, if any, products available in this category with such a degree of completeness in terms of listed types of parameters, and definitely in terms of graphics quality.
An amino acid sequence (as a string of one-letter codes) can be mapped/annotated according to a number of parameters, shown in the rows underneath. Adequate display of the numerical value for any given parameter belonging to any amino acid in the sequence is achieved by appropriate color coding. This is the principal difference between some other software packages (4,5) and our Gold Sting component JPD.
JavaProtein Dossier INTRINSICS
JPD is completely integrated with our Sting Millennium (6) molecular sequence/structure viewer. All the parameters presented in JPD can be mapped on the three-dimensional (3D) structure by means of appropriate atom coloring. The Chime® molecular visualization plugin (MDL Inc.) is used to provide the molecular rendering. Other components of the Gold Sting Suite (to be described in detail elsewhere and which have undergone significant modifications from what they were in our earlier Sting Millennium Suite) are also fully integrated with the functionalities of JPD.
The JPD package is implemented using Java™. However, a number of other programming languages are used for completing a variety of tasks: JavaScript is used to communicate between the Java Sequence window and the Chime plugin; the PERL language is used for processing web requests through a CGI; the C++ programming language is used for more intensive programming tasks in structure parameter calculations and their weekly updates.
JavaProtein Dossier MODES
JPD has two working modes, permitting its use with (i) a single PDB structure (Figure 1) and (ii) a structural alignment of two protein chains [previously aligned with the CE (7) software] (Figure 2). Three different windows allow the coordinated visualization of sequences, structures and their parameters: the Gold Sting sequence window, the Gold Sting Chime window and the JPD parameters window. The Gold Sting sequence window presents the resulting alignment of two sequences (using a structural, not sequence, alignment). The Gold Sting Chime window shows 3D representations of two superimposed molecular structures. Finally, the JPD parameters window shows the parameters of both chains, also following the structural alignment. In order to satisfy optimal structural alignment obtained by the CE algorithm, gaps are introduced when necessary.
Figure 1.
This is a snapshot of the JPD window showing all the available structural/functional parameters from the JPD_DB for the 3ktq.pdb file, chain A. This window is made up of the file menu area, the speed icons area and the parameter display area. The user can find detailed information on JPD basics and specific details on any area/parameter within the JPD Help pages. The content of the JPD-produced image in this figure shows a highly evolutionarily conserved region: Motif A (residues 605–618) of the Taq pol I protein. According to Patel and Loeb (21), a wide spectrum of residue substitutions was obtained for this particular region. However, except for the Asp_610, all other residues are mutable with no consequence on the wild-type activity. Only the Asp_610 has, in addition to low relative entropy, a high value for internal contacts energy (the red box in the Contacts Energy row). Consequently, one is able to find out what the factors are that influence the mutability without a change in activity, just by looking at this very simple picture.
Figure 2.
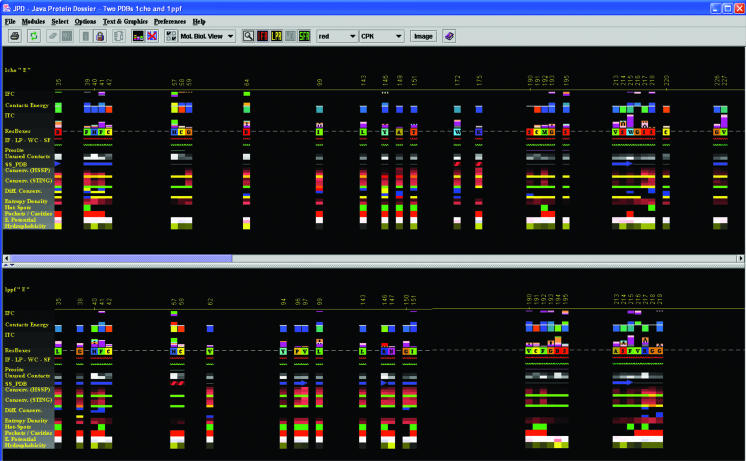
Structural alignment in JPD. This is one of the key features that JPD offers to users—the possibility to structurally align two proteins and consequently display ‘aligned’ structural parameters. In this case, we used two serino proteases: Chymotrypsin in complex with turkey ovomucoid third domain (1cho.pdb) and elastase in complex with the same inhibitor (1ppf.pdb). As a result of the alignment, Gold Sting shows, in the Chime window, two aligned structures and, in the Gold Sting sequence window, the sequences of two chains, aligned according to the structural alignment. This particular picture was obtained after clicking on the IFR icon in the JPD speed icons area. The displayed pattern shows only the residues at the interface area of the two aligned chains. After the user selects to view only the IFR region, JPD continues to preserve the alignment mode, so that the user can inspect the spatial position of these important ‘interfacing’ residues (of course in comparison with the homologous and aligned structures). This clearly offers a rapid insight into the mechanism of the specificity for certain substrates/inhibitors. In this specific case, we see two different enzymes binding to the same inhibitor. Both enzymes demonstrate a similar pattern of IFRs, yet variations between the two ensembles of IFRs should suggest the key elements responsible for the successful binding of the inhibitor to both enzyme chains.
JavaProtein Dossier STRUCTURAL/FUNCTIONAL PARAMETERS
We have added some extra features in order to build the new Gold Sting JPD Database (JPD_DB), based on the previously implemented SMS DB (Sting Milleneum Suite Database) [described earlier by Neshich et al. (6)]. The server side of Gold Sting is responsible for regularly taking updates from all relevant public domain databases used by Gold Sting: PDB, HSSP (Homology-derived Structures of Proteins) (8–10) and PROSITE (11). At the same time, the Gold Sting server is also responsible for calculating a number of macromolecular properties for each PDB structure: electrostatic potential is calculated using modified Delphi (12) software (details will be discussed elsewhere); curvature is calculated using the SurfRace (13) software; solvent accessible area for each protein chain and for the whole molecular complex is calculated using the Surfv (14) software, adapted to our own requirements; secondary structure identification is calculated according to DSSP (15) and STRIDE (16); intra- and interchain amino acid contacts as well as protein–DNA interaction are calculated using our own software, ‘contacts’; hydrophobicity is assigned according to Radzicka and Wolfenden (17); dihedral angles are calculated by our own ‘Ramachandran’ program; and PROSITE patterns are identified using the Ps_Scan (18) software. The ‘Sponge’ and ‘Density’ parameters are calculated using the double cubic lattice method (19) in combination with public library procedures such as those of the BALL's library (Biochemical Algorithms Library—http://voyager.bioinf.uni-sb.de/OK/BALL) and our own code (details to be described elsewhere). In addition, we used the Rate4Site (20) software to calculate the ‘Evolutionary Pressure’ parameter shown within the JPD Conservation row. Pockets are calculated using the ‘pocket’ program designed by Patrice Koehl (http://csb.stanford.edu/koehl/ProShape/) and adapted by us to our needs.
In Table 1 we list all available structural/functional parameters (in their order of appearance in the actual JPD window). An ‘X’ indicate the presence of a parameter in one or more of the three available default views which the user can select to display in the JPD window.
Table 1. List of parameters accessible in JPD.
| Parameter name | View all | Molecular biology view | Crystallography view |
|---|---|---|---|
| (1) IFR Contacts | X | X | X |
| (2) Density Energy for IFR Contacts | X | ||
| (3) IFR Contacts Energy | X | X | X |
| (4) Internal Contacts Energy | X | X | X |
| (5) Internal Contacts Energy—Sliding Window | X | ||
| (6) Density Energy for Internal Contacts | X | ||
| (7) Density Energy Internal for Contacts—Sliding Window | X | ||
| (8) Internal Contacts | X | X | X |
| (9) Sequence box | X | X | X |
| (10) IFR area + Extended IFR area | X | X | X |
| (11) Ligand Pocket AA | X | X | X |
| (12) Water Contacting AA | X | X | |
| (13) Surface + Bearly Surface AA | X | X | X |
| (14) Prosite | X | X | X |
| (15) Unused Contacts Energy | X | X | |
| (16) Unused Contacts | X | X | |
| (17) SS_PDB | X | X | X |
| (18) SS_DSSP | X | X | |
| (19) SS_Stride | X | X | |
| (20) Multiple Occupancy | X | X | |
| (21) Temperature Factor @ Ca | X | X | |
| (22) Temperature Factor @ LHA | X | X | |
| (23) Temperature Factor Mean | X | X | |
| (24) Temperature Factor Max | X | X | |
| (25) Conservation—Evolutionary Pressure | X | X | X |
| (26) Conservation—Relative Entropy 100 | X | X | |
| (27) Conservation—Relative Entropy | X | X | X |
| (28) Conservation—Reliability | X | X | |
| (29) STING SH2Qs Conservation—Evolutionary Pressure | X | X | X |
| (30) STING SH2Qs Conservation—Relative Entropy 100 | X | X | |
| (31) STING SH2Qs Conservation—Relative Entropy | X | X | X |
| (32) STING SH2Qs Conservation—Reliability | X | X | |
| (33) Diff. (HSSP- SH2Qs) Conservation—Evolutionary Pressure | X | X | |
| (34) Diff. (HSSP- SH2Qs) Conservation—Relative Entropy 100 | X | X | |
| (35) Diff. (HSSP- SH2Qs) Conservation—Relative Entropy | X | X | |
| (36) Diff. (HSSP- SH2Qs) Conservation—Reliability | X | X | |
| (37) Entropy Density @ IFR area | X | X | X |
| (38) Entropy Density Internal | X | X | X |
| (39) Entropy Density Sliding Window | X | ||
| (40) Hot Spots | X | X | |
| (41) Accessibility in Complex | X | X | |
| (42) Accessibility in Isolation | X | X | |
| (43) Relative Accessibility | X | X | |
| (44) Dihedral Angles | X | X | |
| (45) Pockets/Cavities in Complex | X | X | X |
| (46) Pockets/Cavities in Isolation | X | X | X |
| (47) Electrostatic Potential @ Ca | X | X | |
| (48) Electrostatic Potential @ LHA | X | X | |
| (49) Electrostatic Potential @ Surface | X | X | X |
| (50) Electrostatic Potential Atom Average | X | X | X |
| (51) Hydrophobicity | X | X | X |
| (52) Curvature | X | ||
| (53) Distance from N-terminal | X | ||
| (54) Distance from C-terminal | X | ||
| (55) Distance from center of gravity | X | ||
| (56) Density at IFR area | X | X | |
| (57) Density Internal | X | X | |
| (58) Density Internal Sliding Window | X | X | |
| (59) Sponge at IFR area | X | X | |
| (60) Sponge Internal | X | X | |
| (61) Sponge Internal Sliding Window | X | X | |
| COLUMN TOTALS | 61 | 32 | 41 |
Many of the structural/functional parameters are calculated using a variety of default conditions (the variable volume size for a probing sphere, the atom at which the center of the probing sphere is placed or the variable size for a sliding window), consequently bringing the total number of available numerical values for structural/functional parameters reported by JPD to 125. Detailed description of all the parameters mentioned and the procedures used to obtain them is given in the JPD Help pages.
JavaProtein Dossier USER INTERFACE
JPD displays a selection of structural parameters (Figure 1) based on a choice of one among the three possible default views: ‘Molecular Biology view’, ‘Crystallography view’ and ‘View all’. An image generated by JPD is actually a user interface: an interactive window for accessing and graphically representing stored data. This window can be split to accommodate the display of the parameters belonging to two protein chains (Figure 2). In addition, in this window there is a menu and a tool bar with icons for the most used tasks. All these components of the main JPD window are described in detail in JPD Help pages.
The JPD structural parameters interactive window provides a graphic summary of several important structural characteristics for a chosen protein. The pivotal position among all the structural parameters is given to the amino acid sequence cartoon, labeled as ResBoxes. This row is accompanied by two histograms representing the intrachain atomic contacts (ITC) and the interchain atomic contacts (IFC). Above the ITC and below the IFC histograms, there is a row showing the corresponding contacts energy. The interface area (IF), water contacting (WC), ligand pocket forming (LP) and surface forming (SF) residues are indicated by wavy lines immediately underneath the sequence. Below the sequence cartoon there is a row labeled as Prosite, where a solid line indicates the regions of the protein sequence which contain a Prosite pattern. The next three rows show the secondary structure of the molecule using three different secondary structure indicators (PDB, DSSP, STRIDE). The following five parameters displayed for each amino acid and annotated with color-coded scales represent respectively: temperature factor, sequence conservation [two types: HSSP and Sequences Homolog to the Query (Structure-having Sequence) (SH2Qs) in a multiple alignment (relative sequence entropy), hot spots (hydrophobic patches at the molecular surface), solvent accessibility of the protein chain in isolation and in the complex with any other protein chain present in PDB file, and dihedral angles. In addition, seven more parameters are calculated with the aim of identifying relevant regions of proteins such as active sites and regions of protein recognition and interaction: pockets/cavities, electrostatic potential, hydrophobicity, curvature, distance from the N- and C-terminals, density and sponge. The numerical value of any structural parameter is easily obtained on screen by placing the cursor above the corresponding area.
Owing to the limited space and different scope of this paper, we neither mention all the parameters present in JPD nor describe a procedure for calculating each one. However, the user is able to find such information in the extensive JPD Help pages.
JPD allows the user to make very informed decisions about the possible role of specific amino acids in defining the function of a protein. It also helps in deciphering what effect a specific mutation will possibly have on the structure and function of a protein, specifically by observing the changes in intra- and interface contact signatures, the sum of the energy values for established contacts, the electrostatic potential at the surface and the conservation.
Selection of amino acids can be made with any combination of conditions, permitting powerful identification of functionally/structurally important regions or sites. For example, the user can select residues located at the interface between two chains, having a negative electrostatic potential at the surface they form, which are preserved in terms of evolution. (For detailed examples, see the JPD help manual: JPD Select Residues option).
AN EXAMPLE JavaProtein Dossier APPLICATION
In Figures 1, 2 and 3, we show snapshots produced by JPD during a session that analyzed several different proteins. The figure legends describe both the details of the proteins (structures) analyzed and the potential benefits the user may derive from using JPD. The user can infer many valuable conclusions about how important any specific amino acid is for protein stability, for protein function and for binding to inhibitor/substrate. The biological meaning of these exercises is specifically emphasized.
Figure 3.
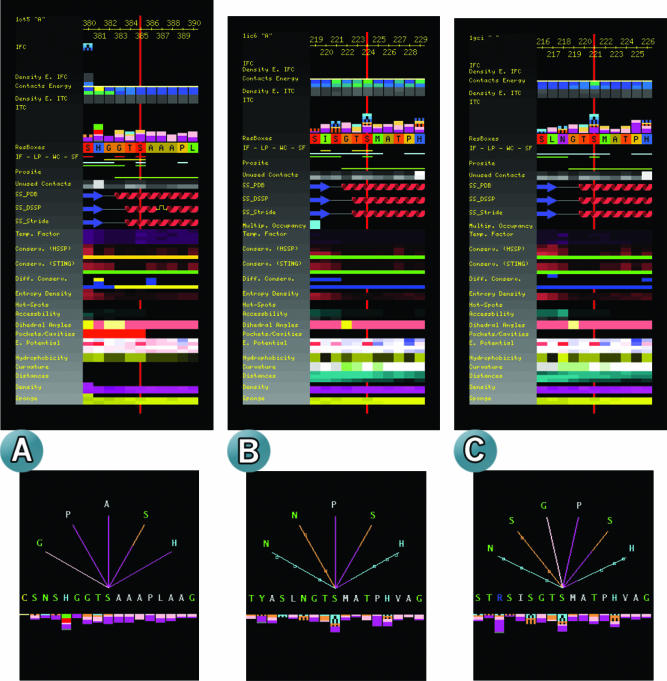
A comparative study of the evolutionarily preserved catalytic Serine in three Pro-hormone convertases. The three structures analyzed here (1ot5.pdb, 1gci.pdb and 1ic6.pdb) belong to the conserved family of eukaryotic serine proteases responsible for certain functions in the secretory pathway. The endoproteases are responsible for the process of maturation of a variety of precursors which are then transformed into peptides and proteins that perform some clinically relevant functions (22). (A) Data on Ser_385 of the Kex2 protein found in yeast; (B) the Subtilisin Ser_221; (C) the Proteinase K Ser_224. This conserved Serine located in the catalytic site is bound to an inhibitor in the Kex2 structure. The JPD display of all the parameters for the catalytic Serine in all three structures is shown. In addition, in the lower part of each panel (A–C), the details of the interatomic contacts that each Serine makes with surrounding amino acids adds more information concerning how the difference in specificity is achieved.
In order to get more biological content out of the JPD presentation, a case study of several proteins is presented. First, the two highly evolutionarily conserved regions of the Taq pol I protein are presented (Figure 1). Based on the data from experiments in which many residue substitutions were performed for this particular region, we questioned why only a single residue from the region could not be mutated without changing the wild-type activity. By quick inspection of the parameters shown by JPD, one can see that conservation is very low for a number of residues (dark boxes in the Conservation row); however, only the residue under investigation has, in addition to low relative entropy, a high value for internal contacts energy (the red box in the Contacts Energy row). Consequently, the JPD display is able to quickly spot the reasons and/or predict why a certain sequence position is more crucial in terms of biological activity than others.
We have also studied two different enzymes bound to the same inhibitor. In this case, JPD can be of great use since it can show the parameters of both chains according to the structural alignment (with gaps introduced to satisfy optimal structural alignment). A most useful option that JPD offers in structural alignment mode is one which permits the user to see the information on interface residues (IFRs). In Figure 2 the JPD pattern shows only the residues at the interface area of the two aligned chains. It is necessary to say that the IFRs (underlined by the red wavy line) for each of the chains shown are calculated for the complex of the E (enzyme) chain with the I (inhibitor) chain for the respective PDB files. After the user selects to view only the IFR region, JPD continues to preserve the alignment mode, so that the user can inspect the spatial position of these important ‘interfacing’ residues (of course in comparison with the homologous and aligned structures). This clearly offers a rapid insight into the mechanism of the specificity for certain substrates/inhibitors. In this specific case, we see two different enzymes binding to the same inhibitor. Both enzymes demonstrate a similar pattern of IFRs, yet variations between the two ensembles of IFRs should suggest the key elements responsible for the successful binding of the inhibitor to both enzyme chains.
Finally, we used JPD to identify the most important differences among three representative enzymes of a conserved family of eukaryotic serine proteases (focusing on their respective active site residues). These proteins perform similar, yet different enough functions that different substrates/inhibitors react with them. Key to understanding the specificity with which these enzymes interact with their substrates is probably complete understanding of all the interactions, and the parameters that describe these interactions, for all the residues involved. In this case we show (Figure 3) all the parameters that JPD can display for the catalytic Serine in all three structures.
CONCLUDING REMARKS
JavaProtein Dossier is described here as a new interactive tool for browsing a newly assembled, structure-related database, JPD_DB. This database is, as far as we are aware, the largest collection of its kind. No other web server provides such a volume of data integrated into a graphic environment and with the same level of interactivity as JPD. Our objective with JPD is to facilitate information extraction and knowledge growth. The powerful Select tool is one of the key features available to the user in JPD. This tool is able to group amino acids into subsets which have a definite relationship to biological function. The applications discussed illustrate how powerful JPD can be when used in research and teaching.
FUTURE DEVELOPMENTS
JPD is able to accept users' local files as input to calculate most of the parameters to be displayed. However, depending on the size of the protein, this procedure may take up to 5 min on our SGI Origin 3400 server with eight R14000 processors (the most CPU-intensive procedure being aligning the sequences homologous to a query sequence). Consequently, if we keep this service enabled, our server can become overloaded with users' requests for processing their local files. Since our SGI server is currently being used for both development and production, we have decided to disable the local file processing option on the JPD web page for a while. As an alternative, users may employ the stand-alone Gold Sting modules (such as Contacts, Scorpion and Formiga) to calculate some of the parameters for their local PDB files. We plan to offer full local file processing capabilities in JPD in the next release of Sting: Diamond Sting. By that time, we hope to have obtained a separate server for this purpose.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to express special thanks to Phil Bourne and Wolfgang Bluhm from RCSB/PDB in San Diego, Jose Valverde from CNB in Madrid, Spain, Akinori Sarai from Kyushu Fukuoka University in Japan and Oscar Grau from LaPlata University in Argentina for their collaboration in testing and maintaining Gold Sting at the respective Gold Sting mirror sites. Special thanks are expressed to Barry Honig for his continuous interest in this project. We are thankful to Jorge Hernandez, Sandra Krauchenko and Andrea Guelfi for testing JPD. We are also thankful to the LNCC-CENAPAD for making available to us their High Performance Computing Resources.
This work was supported in part by the following grants: FAPESP 01/08895-0, FINEP 1945/01 and CNPq 521093/2001-5 (NV).
REFERENCES
- 1.Berman H.M., Bourne,P.E. and Westbrook,J. (2004) The PDB: a case study in management of community data. Curr. Proteomics, 1, 49–57. [Google Scholar]
- 2.Tufte E.R. (2001) The Visual Display of Quantitative Information. Graphics Press, Cheshire, CT. [Google Scholar]
- 3.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laskowski R.A., Hutchinson,E.G., Michie,A.D., Wallace,A.C., Jones,M.L. and Thornton,J.M. (1997) PDBsum: a web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci., 22, 488–490. [DOI] [PubMed] [Google Scholar]
- 5.Hogue C.W.V. (1997) Cn3D: a new generation of three-dimensional molecular structure viewer. Trends Biochem. Sci., 22, 314–316. [DOI] [PubMed] [Google Scholar]
- 6.Neshich G., Togawa,R., Mancini,A.L., Kuser,P.R., Yamagishi,M.E.B., Pappas,G., Jr, Torres,W.V., Campos,T.F., Ferreira,L.L., Luna,F.M., Oliveira,A.G., Miura,R.T., Inoue,M.K., Horita,L.G., de Souza,D.F., Dominiquini,F., Álvaro,A., Lima,C.S., Ogawa,F.O., Gomes,B.G., Palandrani,J.C.F., dos Santos,G.F., de Freitas,E.M., Mattiuz,A.R., Costa,I.C., de Almeida,C.L., Souza,S., Baudet,C. and Higa,R.H. (2003) STING Millennium: a web based suite of programs for comprehensive and simultaneous analysis of protein structure and sequence. Nucleic Acids Res., 31, 3386–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shindyalov I. and Bourne,P. (1998) Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng., 11, 739–747. [DOI] [PubMed] [Google Scholar]
- 8.Sander C. and Schneider,R. (1991) Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins, 9, 56–68. [DOI] [PubMed] [Google Scholar]
- 9.Schneider R., de Daruvar,A. and Sander,C. (1997) The HSSP database of protein structure—sequence alignments. Nucleic Acids Res., 25, 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider R. and Sander,C. (1996) The HSSP database of protein structure—sequence alignments. Nucleic Acids Res., 24, 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bucher P. and Bairoch,A. (1994) A generalized profile syntax for biomolecular sequences motifs and its function in automatic sequence interpretation. In Altman,R., Brutlag,D., Karp,P., Lathrop,R. and Searls,D. (eds), ISMB-94; Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp. 53–61. [PubMed] [Google Scholar]
- 12.Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- 13.Tsodikov O.V., Record,M.T.,Jr and Sergeev,Y.V. (2002) Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem., 23, 600–609. [DOI] [PubMed] [Google Scholar]
- 14.Sridharan S., Nicholls,A. and Honig,B. (1992) A new vertex algorithm to calculate solvent accessible surface areas. Biophys. J., 61, A174. [Google Scholar]
- 15.Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 16.Frishman D. and Argos,P. (1995) Knowledge-based protein secondary structure assignment. Proteins, 23, 566–579. [DOI] [PubMed] [Google Scholar]
- 17.Radzicka A. and Wolfenden,R. (1988) Comparing the polarities of the amino acids: side-chain distribution coefficients between the vapor phase, cyclohexane, 1-octanol, and neutral aqueous solution. Biochemistry, 27, 1664–1670. [Google Scholar]
- 18.Gattiker A., Gasteiger,E. and Bairoch,A. (2002) ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformatics, 1, 107–108. [PubMed] [Google Scholar]
- 19.Eisenhaber F., Lijnzaad,P., Argos,P., Sander,C. and Scharf,M. (1995) The double cubic lattice method: efficient approaches to numerical integration of surface area and volume and to dot surface contouring of molecular assemblies. J. Comput. Chem., 16, 273–284. [Google Scholar]
- 20.Pupko T., Bell,R.E., Mayrose,I., Glaser,F. and Ben-Tal,N. (2002) Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics, 18 (Suppl. 1), S71–S77. [DOI] [PubMed] [Google Scholar]
- 21.Patel P.H. and Loeb,L.A. (2000) DNA polymerase active site is highly mutable: evolutionary consequences. Proc. Natl Acad. Sci. USA, 97, 5095–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rockwell N.C. and Thorner,J.W. (2000) The kindest cuts of all: crystal structures of Kex2 and furin reveal secrets of precursor processing. Trends Biochem. Sci., 29, 80–87. [DOI] [PubMed] [Google Scholar]