Abstract
Genomic amplification of the gene encoding and phosphorylation of the protein FADD (Fas-associated death domain) is associated with poor clinical outcome in lung cancer and in head and neck cancer. Activating mutations in the guanosine triphosphatase RAS promotes cell proliferation in various cancers. We found that the abundance of phosphorylated FADD correlated with that of mutant KRAS in patient lung cancer tissues. Using immunohistochemistry analysis and in vivo imaging of conditional mouse models of KRASG12D-driven lung cancer, we found that the deletion of the gene encoding FADD suppressed tumor growth, reduced the proliferative index of cells, and decreased the activation of downstream effectors of the RAS–MAPK (mitogen-activated protein kinase) pathway that promote the cell cycle, including retinoblastoma (RB) and cyclin D1. In mouse embryonic fibroblasts, the induction of mitosis upon activation of KRAS required FADD and the phosphorylation of FADD by CK1α (casein kinase 1α). Deleting the gene encoding CK1α in KRAS-mutant mice abrogated the phosphorylation of FADD and suppressed lung cancer development. Phosphorylated FADD was most abundant during the G2/M phase of the cell cycle, and mass spectrometry revealed that phosphorylated FADD interacted with kinases that mediate the G2/M transition, including PLK1 (Polo-like kinase 1), AURKA (Aurora kinase A) and BUB1 (budding uninhibited by benzimidazoles 1). This interaction was decreased in cells treated with a CKI-7, a CK1α inhibitor. Therefore, as the kinase that phosphorylates FADD downstream of RAS, CK1α may be a therapeutic target for KRAS-driven lung cancer.
Introduction
In cancer cells, dysregulated proliferation often occurs through the increased abundance, post-translational modification, or mutation of signaling proteins. RAS is a membrane-associated small guanosine triphosphatase (GTPase), which functions as a signaling mediator of receptor and non-receptor tyrosine kinases, activating cytoplasmic and nuclear effector pathways. Oncogenic activating mutations in RAS are implicated in approximately 30% of all cancers. Mutations in the KRAS gene are common in non-small-cell lung cancer (NSCLC), colorectal cancer, and pancreatic cancer. Approximately 15–25% of patients with lung adenocarcinoma exhibit tumor-associated KRAS mutations (1). Uncontrolled cell proliferation as a result of RAS mutations is attributed to a cascade of signaling kinases known as the mitogen-activated protein kinase (MAPK) pathway. The MAPK pathway includes RAF, mitogen and extracellular signal-regulated kinase (MEK), and extracellular signal-regulated kinase (ERK). Through a series of protein phosphorylation events, the MAPK pathway promotes the activities of several transcription factors, including c-Myc, CREB and c-Fos. This activation leads to the altered transcription of genes that are involved in cell cycle progression. Beyond this, the molecular details of a link between the MAPK cascade and the cell cycle machinery are thus far only partially understood. For example, MEK and ERK – key components of MAPK signaling – have a role in M-phase progression (2), but the precise molecular mechanisms through which they function in this process are not yet clear.
The protein FADD (Fas-associated death domain) is an adaptor protein that is best known for its role in extrinsic apoptosis. However, depending on its subcellular localization (which is regulated by phosphorylation), FADD can have different roles. In the cytoplasm, its main function is to induce apoptosis; whereas in the nucleus, it can have the opposite effect and instead promote cell survival and proliferation (3, 4). Increased abundance of FADD – specifically that of the phosphorylated, nuclear-localized form – correlates with aggressive disease and poor clinical outcome in patients with lung adenocarcinoma or head and neck cancers (5–10). In addition, amplification of the FADD locus on chromosome 11q13.3 is frequently observed in human cancers, resulting in increased FADD abundance, poor prognosis (6–9), and correlation with overabundant CCND1 and CCNB1, genes that encode proteins involved in the regulation of cell cycle progression (5, 11). Human cells lacking FADD exhibit defects in cell cycle progression (5, 12). In mice, loss of Fadd or expression of a dominant-negative Fadd mutant in peripheral T lymphocytes inhibits mitogen-induced T cell proliferation (12–14). FADD is phosphorylated at Ser194 (Ser191 in mouse cells) during the G2/M phase by casein kinase 1 α (CK1α), which directly interacts with FADD in early mitosis (15). Using conditional mouse models, we investigated the upstream signaling events and their significance in mutant Kras-mediated lung oncogenesis.
Results
FADD is required for Kras-driven lung cancer
A transgenic mouse in which the genomic locus for Fadd is insertionally inactivated and complimented by a conditional transgene (Fadd:GFP) (16) was crossed with KrasLSL-G12D mice (17) and subsequently crossed with Rosa26LSL-Luciferase mice (18). The resulting mouse (KrasLSL-G12D; Luc; Fadd−/−) is herein referred to as KFLuc mice. Intranasal instillation of an adenovirus expressing Cre-recombinase (AdCre) enabled the simultaneous activation of oncogenic Kras (to initiate tumorigenesis), expression of luciferase (to measure proliferation), and deletion of the Fadd transgene specifically in lung cells (Fig. 1A). Mice that were heterozygous for genomic Fadd [(KrasLSL-G12D; Luc; Fadd+/−), herein referred to as KLuc mice] were used as controls because they retained Fadd expression despite deletion of the transgene in the lung. Mice that expressed Fadd and wild-type Kras [(Luc; Fadd+/−), herein referred to as Luc mice], as well as mice that expressed wild-type Kras but not Fadd [(Luc; Fadd−/−), herein referred to as FLuc mice], were used as additional controls.
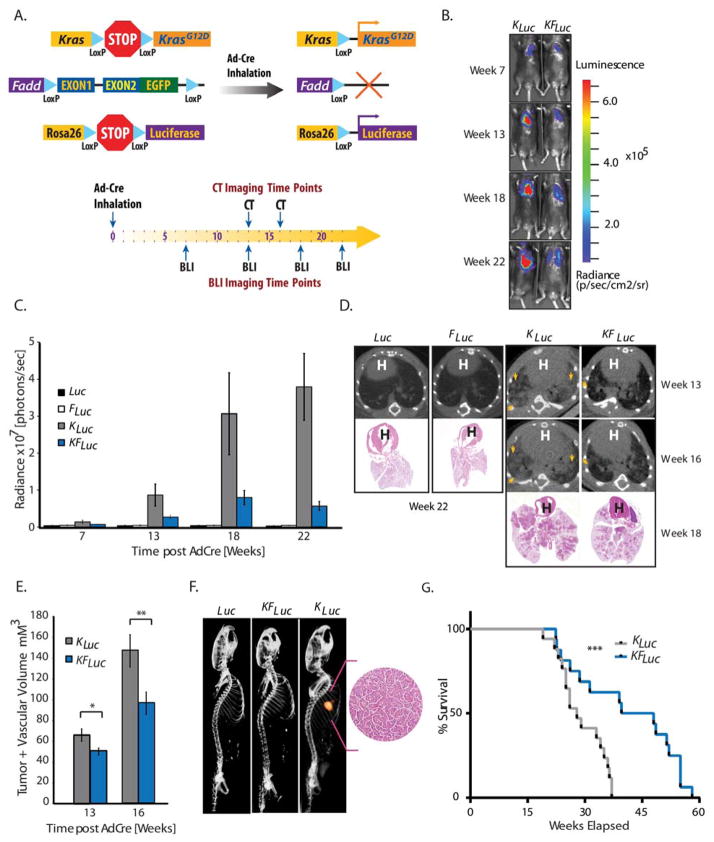
Fig. 1. A requirement for FADD in Kras-driven lung cancer.
(A) Genetic strategy used to activate KrasLSL-G12D, Rosa26LSL-Luciferase and deplete Fadd:GFP expression in a lung-specific manner. (B) Representative bioluminescent images of KLuc, and KFLuc mice at 7 to 22 weeks after intranasal AdCre administration. (C) Average bioluminescence in Luc (n=7), FLuc (n=7), KLuc (n=25), and KFLuc (n=34) mice at specified times. (D) Representative images of CT scans of lungs from Luc, FLuc, KLuc, and KFLuc animals at shown times, with H&E slide corresponding to that animal from week 18 or week 22 as indicated. Arrows, lesions. H, heart. (E) Tumor and vascular volumes in KLuc (n=10) and KFLuc (n=17) mice analyzed by CT at indicated times. Data are means ± SEM. *P=0.009, **P=0.018, unpaired Student’s t-tests. (F) Co-localization of bioluminescence imaging and CT imaging for lung tumors in Luc, KFLuc and KLuc mice. Inset, H&E of lesion in KLuc mouse. (G) Survival plot of KLuc (n=19) and KFLuc (n=15) mice. Median survival time: KFLuc mice, 51.4 weeks; KLuc mice, 34 weeks. ***P=0.005, Wilcoxon log-rank test (95% CI).
Upon AdCre delivery, bioluminescence imaging was performed to monitor tumor growth at indicated time points (Fig. 1, A to C). A time-dependent increase in bioluminescence activity was observed at week 7 in KLuc mice compared with Luc mice (Fig. 1C). In contrast to KLuc mice, we observed minimal or no changes in bioluminescence activity over time in KFLuc or FLuc mice (Fig. 1, B and C). Anatomic visualization of tumor burden in mice was also carried out using micro-computed tomography (μCT). Lesions were identified in KLuc animals, but less so in their KFLuc littermates, while no lung lesions were detectable in mice with wild-type Kras (Luc or FLuc mice) (Fig. 1D). Consistent with these bioluminescence measurements, quantitative analysis of μCT images yielded volumetric measure of the tumor and vascular component (19) that also revealed an increase in the tumor burden at week 13 and 16 in KLuc mice compared with KFLuc mice (Fig. 1E). Using a multimodality reader that allows for simultaneous acquisition of spatially aligned bioluminescence and μCT, we found that the X-ray-dense lesions were also positive for bioluminescence (Fig. 1F). Histological analysis of single lesions in KLuc mice pathologically confirmed a hyper-proliferative phenotype (Fig. 1F, inset). In agreement with these measurements of tumor burden, we observed prolonged survival in KFLuc mice compared to KLuc littermates (Fig. 1G).
Fadd null lung tumors are less proliferative
To confirm our imaging results, we performed hematoxylin and eosin staining analysis of lung tissue from the aforementioned animals. Lung tissue from control Luc or FLuc animals appeared normal with no detectable hyperplasia (Fig. 2A). In contrast, lungs from KLuc mice 18 weeks after AdCre administration exhibited large adenomas (papillary, solid and mixed), large mixed-type adenomas (solid and papillary) within lung parenchyma and atypical adenomatous hyperplasia in the alveoli (Fig. 2A). Histopathology analysis of the H&E stain indicated that tumors from KLuc mice had a greater macrophage and inflammatory infiltrate than did those from KFLuc mice (Fig. 2A)). In contrast, analysis of lung sections from KFLuc mice revealed demonstrably smaller and fewer adenomas (Fig. 2, A and B).
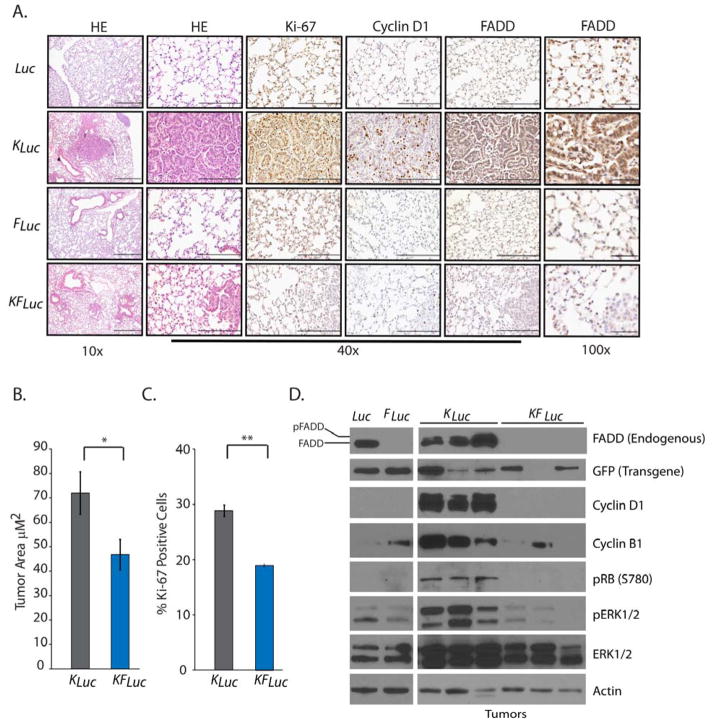
Fig. 2. Fadd null lung tumors are less proliferative.
(A) Representative histology images of lungs removed from mice 18 weeks after AdCre administration. Tissues were stained with hematoxylin and eosin (HE) or antibodies against cyclin D1, Ki-67, and FADD. Scale bars: 10x, 500 μm; 40x, 200 μm; 100x, 50 μm. (B) Average tumor area quantified from H&E-stained lung tissue sections from KLuc (n=6) and KFLuc (n=10) mice. *P=0.04, unpaired Student’s t-test. (C) Average percentage of positive Ki-67 stained cells in lung tissue sections from KLuc and KFLuc mice, 4 fields per 10 mice each. **P=2×10−5, unpaired Student’s t-test. (D) Representative Western blot for endogenous FADD, GFP (Fadd transgene), phosphorylated and total ERK1/2, phosphorylated RB, cyclin D1, cyclin B1 and β-Actin in lung tissue from the indicated mice. Blots are representative of 3 independent experiments.
To assess proliferation, we performed immunohistochemistry for proliferation markers in lung tissue and tumors from these mice. Compared with lung tissue from KFLuc mice, tumors from KLuc mice had significantly increased abundance of the proliferation marker Ki-67 (Fig. 2C), the G1/S phase transition protein cyclin D1 (Fig. 2, A and D), and phosphorylated retinoblastoma (RB) protein (Fig. 2D), which together suggests that Fadd deletion suppresses cell proliferation in Kras mutant cells. We also observed increased FADD abundance in lung tissue from KLuc mice compared with lung tissue from Luc or KFLuc mice (Fig. 2, A and D). High magnification of immunohistochemistry sections revealed that FADD was predominantly nuclear (Fig. 2A). Increased abundance of phosphorylated FADD [evident by the doublet of FADD antibody-reactive protein at 28 kD (5, 20–23)] was observed in tissues from mice with mutant Kras (KLuc mice), whereas only a single band was observed in lung tissue from control Luc mice (Fig. 2D). This finding suggests that mutant KRAS promotes the phosphorylation and nuclear translocation of FADD.
In addition to cyclin D1 and phosphorylated RB, an additional marker of proliferation, cyclin B1, was increased in tumors from KLuc mice compared with controls; however, cyclin B1 abundance was similar to controls in tumors from KFLuc mice (Fig. 2D), suggesting that FADD was required to mediate the induction of cell cycle-associated proteins downstream of mutant KRAS. A loss of Fadd expression resulted in decreased abundance of phosphorylated extracellular signal-regulated kinase 1 and 2 (ERK1/2), a downstream mediator of oncogenic KRAS signaling (Fig. 2D and fig. S1A). FADD is an established adaptor of caspase-dependent apoptosis; however, we did not detect activated (cleaved) caspase 3 in either KLuc or KFLuc lung lesions (fig. S1B), suggesting that the decreased tumor growth in those lacking FADD was not likely the result of apoptotic cell death, consistent with the distinct, non-apoptotic role of nuclear FADD.
On the basis of previous experience with conditional mouse models, we hypothesized that tumors arising in KFLuc animals may be due to persistent FADD abundance because of inefficient Cre recombination of the Fadd transgene. To test this, we analyzed the expression of the Fadd transgene using semi-quantitative PCR of lung tumors isolated from KLuc and KFLuc mice. Although reduced compared to that in control tissue, the Fadd transgene was still present in all samples tested (fig. S2A). Persistent expression the Fadd transgene in KFLuc tumors was also observed by Western blotting and immunohistochemistry (Fig. 2D and fig. S2, B to C). Together, these findings suggest that the development of tumors in the KFLuc mice was due to sustained FADD abundance in KRAS-activated lung cells.
FADD and FADD phosphorylation are required for Kras-driven cell proliferation
Increased abundance of FADD mRNA correlates with poor survival in lung cancer patients (5). We performed transcriptome analysis of published patient data using Oncomine and found that a greater abundance of FADD mRNA was observed in tumors that had mutant KRAS than in those that had wild-type KRAS (fig. S3, A to B). We next investigated whether KRAS signaling affected the phosphorylation of FADD in mouse embryonic fibroblasts (MEFs) derived from the mouse models. FLuc-derived MEFs, which had wild-type Kras, failed to proliferate in culture because of cell cycle arrest, as previously demonstrated (16). Western blotting analysis for FADD in lysates from Luc MEFs revealed a single predominant band at 28 kD, whereas those from KLuc MEFs exhibited a doublet, indicative of the phosphorylated form of FADD (Fig. 3A) (5, 20–23). As expected, we could not detect endogenous FADD in lysates from KFLuc cells. Consistent with our previous finding for a role of FADD in proliferation, we observed increased abundance of cyclin D1 in MEFs from KLuc mice compared to those from KFLuc mice (Fig. 3A). KLuc MEFs exhibited a higher rate of proliferation compared to KFLuc and Luc MEFs (Fig. 3B). This finding was in agreement with previous work, wherein decreased proliferation was observed in Fadd−/− MEFs compared with Fadd+/− MEFs (16, 23, 26). Reconstitution of Fadd expression in KFLuc cells restored cell proliferation (Fig. 3B). To evaluate a requirement for CK1α, the kinase that phosphorylates FADD (14, 27–28), we treated KLuc cells with a CK1α inhibitor, CKI-7., We observed a marked decrease in the proliferative ability of CKI-7-treated KLuc cells, whereas that of CKI-7-treated KFLuc or Luc cells was not substantially affected (Fig. 3C). Because increased abundance of phosphorylated FADD was observed in KLuc cells relative to Luc cells, we hypothesized that MEK and ERK – key effectors of the RAS signaling pathway – may be upstream mediators of FADD phosphorylation. Lonafarnib, which inhibits the farnesylation (and thus cell membrane targeting and function) of Kras (29), as well as the MEK inhibitor PD0325901 (30) inhibited cell proliferation and the phosphorylation of FADD (Fig. 3, C and F). In soft agar colony formation assays, KFLuc MEFs formed 70% fewer colonies than did KLuc MEFs (Fig. 3D). Exogenous Fadd expression in KFLuc cells restored the cells’ capacity to form colonies, confirming a requirement for FADD in proliferation (Fig. 3D). KLuc cells treated with CKI-7, PD0325901 or lonafarnib also reduced colony-forming activity relative to untreated cells (Fig. 3D).
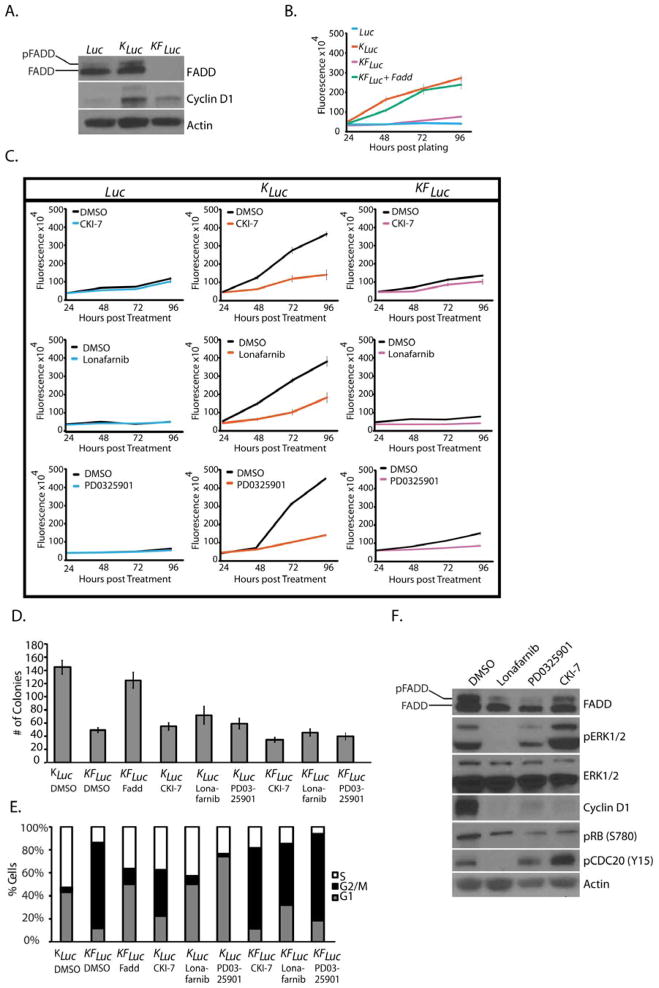
Fig. 3. FADD and FADD phosphorylation are required for Kras-driven cell proliferation.
(A) Western blotting for FADD, Cyclin D1 and β-Actin in lysates from Luc, KLuc and KFLuc MEFs. (B and C) Alamar blue proliferation assay in cultures of Luc, KLuc and KFLuc MEFs that were either (B) untreated or (C) treated with DMSO, 250 μM CKI-7, 10 μM lonafarnib, or 200 nM PD0325901. (D) Agar colony formation of KLuc, KFLuc, and FADD-reconstituted KFLuc MEF cells treated as indicated for 2 weeks, concentrations as in (C). (E) Fluorescence-activated cell sorting (FACS) analysis of cell cycle distribution in KLuc and KFLuc MEFs 24 hours after the indicated treatment, concentrations as in (C). (F) Western blotting for the indicated proteins in KLuc MEFs treated for 6 hours as in (C). Blots (A and F) and FACS data (E) are representative of 3 independent experiments. Data are means ± SEM from 3 independent experiments.
Examination of the cell cycle distribution revealed that compared to KLuc cells, a greater fraction of KFLuc cells were observed in the G2/M phase (Fig. 3E), and reconstitution of Fadd restored normal cell cycle distribution (Fig. 3E). Treating KLuc cells with CKI-7 caused the distribution of cells to shift toward G2/M similar to untreated KFLuc (Fadd-deficient) cells, whereas treating KLuc cells with PD0325901 or lonafarnib caused the distribution to shift toward G1 (Fig. 3E). CKI-7-treated KLuc cells showed a marked decrease in the abundance of cyclin D1 and phosphorylated RB and an increase in that of phosphorylated CDC2 (cell division cycle 2) (Fig. 3F), together suggesting an arrest of cells at G2/M. The amount of phosphorylated FADD was also decreased upon treatment with CKI-7 (Fig. 3F). Treatment with lonafarnib or PD0325901 also resulted in decreased amount of cyclin D1, phosphorylated RB, and phosphorylated FADD (Fig. 3F). Together, these data suggest that, upon constutive KRAS activation, FADD is phosphorylated in a MEK- and CK1α-dependent manner and that this signaling pathway is required for progression of cells through G2/M.
FADD interacts with key mediators of the G2/M transition
To study the phosphorylation of FADD as cells transit G2/M, H1975 human lung cancer cells were synchronized by a double thymidine block. Phosphorylation of FADD was prominent eight hours after cells were released from this treatment, which coincided with a peak in the abundance of cyclin B1 and AURKA (Aurora kinase A), but was preceded by a an increase in PLK1 (polo-like kinase 1) abundance (Fig. 4A), events which mark the transition of cells from G2 into M phase (31–32).
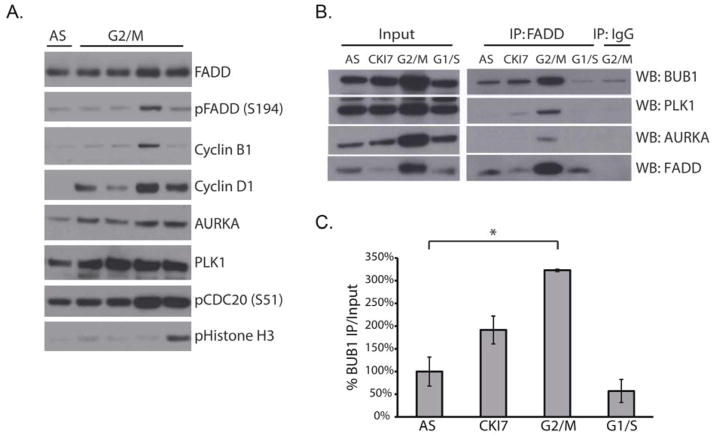
Fig. 4. FADD interacts with key mediators of G2/M transition.
(A) Western blotting for the indicated proteins in H1975 cells after double thymidine block and release (G2/M). Cells were harvested at 6, 7, 8, and 9 hours after synchronization. AS, asynchronous cells. (B and C) Western blotting after FADD immunoprecipitation from A549 cells (B) and quantification of the BUB1-FADD interaction (C). Cells were asynchronous (AS) or treated with CKI-7, nocodozole (G2/M), or hydroxyurea (G1/S). BUB1 abundance was normalized to that in asynchronous cells. Data are means ± SEM from 3 independent experiments,*P = 0.005, unpaired Student’s t-test.
We next conducted an interactome study using Halo-tagged FADD in human embryonic kidney (HEK) 293T cells. Mass spectrometry analysis of co-precipitating polypeptides (Data file S1) revealed the interaction of FADD with CK1α as well as several proteins involved in the cell cycle, including PLK1, CDC20 and Aurora kinase B (AURKB) (Table 1). To evaluate whether the interaction of FADD with these proteins was specific for G2/M, A549 lung cancer cells were treated with hydroxyurea to induce a G1/S arrest, or with nocodozole to induce a G2/M arrest. Immunopreciptiation analysis of these cells revealed an interaction of FADD with PLK1 predominantly in G2/M (Fig. 4B). Because PLK1, AURKB and CDC20 interact with BUB1 (budding uninhibited by benzimidazoles 1, a regulator of the spindle assembly checkpoint) during G2/M (35–37), we investigated whether BUB1 also interacted with FADD. Indeed, a greater amount of BUB1 and AURKA interacted with FADD in G2/M-arrested than in G1/S-arrested or asynchronous cells. Inhibition of CK1α using CKI-7 decreased the interaction of FADD with AURKA andPLK1 (Fig. 4, B and C). Functional annotation analysis of the interactome using DAVID (fig. S4, A to B, 33–34) also identified other FADD-interacting nodes, including proteins involved in cell death, a pathway in which FADD was originally studied in (20).
Table 1. FADD interacts with proteins involved in the G2/M transition.
The top cell cycle- related proteins that were pulled down with Halo-tagged FADD in HEK 293T cells. The cut off that determined FADD-interacting proteins was a spectral count (SpC) of 5 or greater and a control SpC of 0. NSAF is the normalized spectral abundance factor.
| Identified Proteins | Gene Symbol | Molecular Weight (kD) | FADD peptide SpC | Control peptide SpC | FADD NSAF | Control NSAF |
|---|---|---|---|---|---|---|
|
| ||||||
| FADD | FADD | 23 | 555 | 0 | 0.104876887 | 0 |
| CK1α | CSNK1A1 | 39 | 20 | 0 | 0.002228846 | 0 |
| Polo-Like Kinase 1 | PLK1 | 68 | 5 | 0 | 0.000319577 | 0 |
| Aurora Kinase B | AURKB | 39 | 6 | 0 | 0.000668654 | 0 |
| Cell Division Cycle 20 | CDC20 | 55 | 17 | 0 | 0.001343386 | 0 |
| Centrosomal Protein 170 | CEP170 | 175 | 11 | 0 | 0.000273193 | 0 |
| Structural Maintence of Chromosomes 1A | SMC1A | 143 | 13 | 0 | 0.000395114 | 0 |
| Non-SMC condensing II complex, subunit D3 | NCAPD3 | 169 | 19 | 0 | 0.000166099 | 0 |
| Non-SMC condensing II complex, subunit G2 | NCAPG2 | 131 | 18 | 0 | 0.000597195 | 0 |
| Non-SMC condensing II complex, subunit D2 | NCAPD2 | 157 | 6 | 0 | 0.000166099 | 0 |
| Dynactin 2 | DCTN2 | 44 | 5 | 0 | 0.000493892 | 0 |
| Cell Division Cycle 73 | CDC73 | 61 | 7 | 0 | 0.0004875 | 0 |
| Fanconi anemia complementation group D2 | FANCD2 | 165 | 15 | 0 | 0.000392733 | 0 |
| Fanconi anemia complementation group I | FANCI | 149 | 36 | 0 | 0.001050101 | 0 |
| Cytoskeleton associated protein 5 | CKAP5 | 226 | 7 | 0 | 0.000134618 | 0 |
CK1α is required in Kras-driven lung cancer
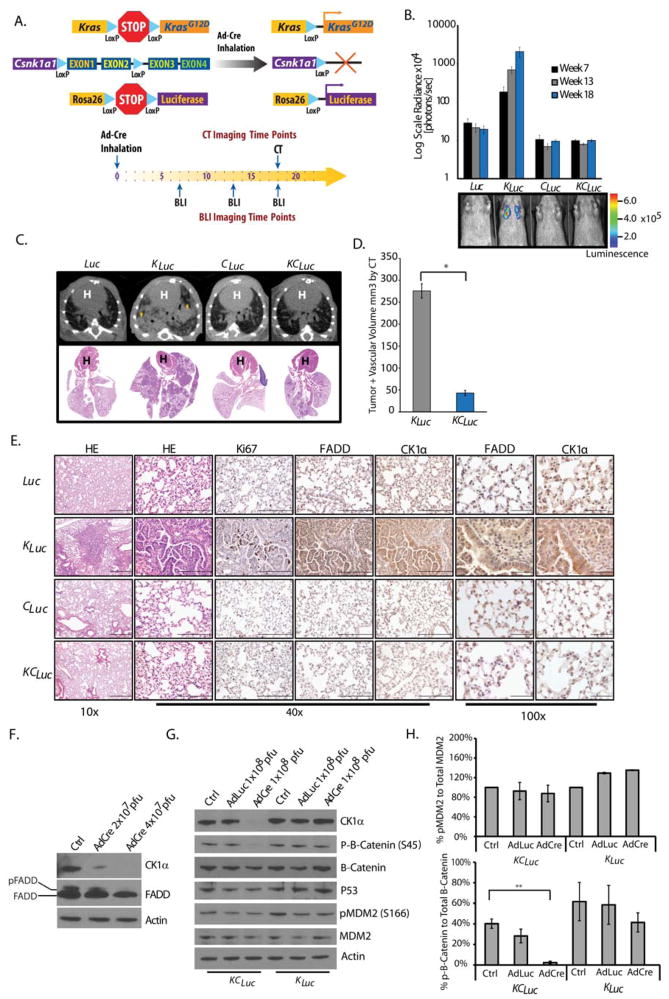
To investigate whether CK1α is required for Kras-driven lung oncogenesis, a mouse model was generated wherein the genomic locus of the gene encoding CK1α could be conditionally deleted in the presence of mutant Kras expression (Csnk1afl/fl mice) (38). KrasLSL-G12D; Rosa26LSL-Luciferase mice (17, 18) were crossed with Csnk1afl/fl to generate KCLuc mice (KrasLSL-G12D; Luc; Csnk1afl/fl). Intranasal inhalation of an adenovirus for expression of AdCre initiated Kras-mediated tumorigenesis in the presence (KLuc) or absence of Csnk1a1 expression (KCLuc, Fig. 5A). Littermates with wild-type Kras in the presence (Luc) or absence of Csnk1a1 (CLuc) were used as controls (Fig. 5A). Bioluminescence imaging revealed a robust increase in tumor burden in KLuc mice compared to Luc mice (Fig. 5B), as shown above (Fig. 1, B and C), whereas no increase in bioluminescence was observed over time in KCLuc and CLuc mice, confirmed by μCT (Fig. 5, C and D), suggesting that Csnk1a1 knockout suppressed tumorigenesis.
Fig. 5. CK1α mediates Kras-driven lung cancer.
(A) Genetic strategy used to activate KrasLSL-G12D, Rosa26LSL-Luciferase and delete Csnk1a expression in a lung specific manner. Bioluminescence and/or CT imaging was performed at the indicated weeks after administration of Cre recombinase. (B) Bioluminescence of Luc (n=8), CLuc (n=5), KLuc (n=38), and KCLuc (n=25) mice at the specified times. Data are means ± SEM. Pseudocolor scale is radiance as photons/second/cm2/steradian (p/sec/cm2/sr). (C) Representative images of CT scans of lungs from Luc, KLuc, CLuc, and KCLuc mice, with corresponding H&E slide. Arrows, lesions. H, heart. (D) Average tumor and vascular volumes of KLuc (n=8) and KCLuc (n=7) mice analyzed by CT 18 weeks after AdCre administration. Data are means ± SEM; *P=7.47×10−6, unpaired Student’s t-test. (E) Representative histology for H&E or indicated proteins in lungs from mice 18 weeks after AdCre administration. Scale bars: 10x, 500 μm; 40x, 200 μm; 100x, 50 μm. (F) Representative Western blot for CK1α, FADD and β-actin, in lysates from KCLuc MEFs treated with various concentrations of AdCre. Pfu, plaque-forming units (G and H) Representative Western blot (G) and quantification of phosphorylated fractions (H) for the indicated proteins in lysates of KLuc and KCLuc MEFs treated with AdCre or AdLuc. Data are means ± SEM from three independent experiments. **P =0.003, unpaired Student’s t-test.
Histological analysis of lung sections from the KCLuc mice at week 18 demonstrated minimal hyperplasia and a rare adenoma compared to KLuc mice (Fig. 5E and fig. S5A). Lesions present in KCLuc mice still showed some positive staining for CK1α, suggesting that Cre-mediated recombination of Csnk1a1 was incomplete (fig. S5A). Both Luc and CLuc mice showed healthy lung tissue as expected. Compared with tumors from KCLuc, CLuc and Luc mice, KLuc lung tumors had increased abundance of FADD, which was also often nuclear localized (Fig. 5E).
We confirmed that ablation of Csnk1a1 resulted in decreased phosphorylation of FADD by isolating and treating MEFs from KCLuc mice with increasing titers of AdCre (Fig. 5F). Because CK1α is also known to play a role in p53 signaling by phosphorylating MDM2 (mouse double minute 2) and MDMX (mouse double minute X), as well as in Wnt signaling by phosphorylating β-catenin (39–41), we next asked whether the ablation of Csnk1a1 in our KCLuc MEFs stimulated these signaling pathways. The abundance of p53 and that of phosphorylated MDM2 were unchanged in KCLuc MEFs compared to KLuc MEFs (Fig. 5, G and H). Similarly, the amount of β-catenin was unchanged although phosphorylated β-Catenin (Ser45) levels were lower (Fig. 5H).
Discussion
The constitutive activation of KRAS, through either dysregulated epidermal growth factor receptor (EGFR) signaling or mutations in KRAS, leads to enhanced proliferation through the RAS-MEK-ERK signaling axis. Mutually exclusive to this, genomic amplification and/or overexpression of EGFR are also commonly found in patients with lung cancer (1). Erlotinib and gefitinib, small molecule kinase inhibitors targeting the EGFR, have shown significant efficacy but in a limited set of patients. In addition, development of drug resistance due to mutations of EGFR and compensatory signaling through alternate receptor tyrosine kinases has limited the broad applicability of these therapies. Recent progress in targeting KRASG12C is encouraging (42), but KRAS inhibitors are currently not clinically available. Efforts to target downstream effectors of KRAS, including MEK, are in development however, preliminary evidence indicates that these may not be effective as monotherapies (43) due to compensatory signaling through the PI3K/AKT pathway. The EGFR-KRAS-MEK-ERK signaling axis represents a major mitogenic pathway in cancer, yet the molecular details of signaling events downstream of RAS and MEK that lead to cell cycle proliferation are not well understood. This may explain the paucity of molecular targets for abrogating EGFR and KRAS mitogenic signaling.
The studies presented here demonstrate that phosphorylation of FADD by CK1α represents a key signaling event in KRAS-mediated mitogenic signaling. Using genetically engineered conditional mouse models, we found that the ablation of Fadd or Csnk1a1 resulted in decreased proliferation and oncogenic potential of mutant Kras cells in the lung. Expression of mutant Kras resulted in an increase in FADD protein, as well as its phosphorylated form in tumor tissue and in MEFs derived from these animals. Inhibition of KRAS, MEK [also reported previously (44)] or CK1α decreased the phosphorylation of FADD, which further affirmed our finding that phosphorylation of FADD represents an important signaling event downstream of the KRAS-MEK-ERK pathway.
Previous findings demonstrate a role for FADD, specifically its phosphorylated form, in the proliferation of T cells (12, 14, 16, 22). This complements our findings that FADD and its phosphorylation are required for Kras-mediated cell proliferation. Additionally, in FADD null Jurkat cells (which arrest in G2/M), reconstitution of either a FADD phospho-mimetic (FADD-Asp) or a non-phosphorylatable form (FADD-Ala) fails to complement the FADD null phenotype (5). (23). A study by Kim et al. showed that reduced amounts of the AK2/DUSP26 phosphatase protein complex that dephosphorylates FADD correlates with increased phosphorylation of FADD and cell proliferation in MEFs, tumor cells and human cancer tissues; in MEFs, proliferation and FADD phosphorylation is reversed by ectopic expression of AK2 (23). These reports further support a role for phosphorylated FADD in mitogenic signaling.
Controlled phosphorylation/dephosphorylation reactions of complexes such as the cyclin B/cyclin-dependent kinase 1 (CDK1) are central to the control of mitosis (45). Data here and elsewhere showing that the phosphorylation of FADD in a cell cycle-dependent manner (5, 15), the G2/M arrest of cells lacking FADD (5), and the G2/M arrest of cells in which the phosphorylation of FADD is inhibited are all consistent with a role for phosphorylated FADD in promoting cell cycle progression downstream of KRAS activation. Maximal phosphorylation of FADD before that of histone H3 (Ser10) (46) and the interaction of FADD with PLK1, AURKA, CDC20 and BUB1 (all mediators of the G2/M transition) further support its role in promoting the G2/M transition. The interaction of FADD with PLK1 and AURKA has been demonstrated previously (31), but our findings show that these occur in a G2/M-specific manner and in the context of mitogenic signaling by mutant KRAS.
Previous studies showed that the conditional deletion of Csnk1a1 in the intestine results in Wnt pathway activation, as does concurrent deletion of p53, which contributes to the development of invasive carcinomas (38). Our analysis of MEFs derived from KCLuc or CLuc mice did not reveal dysregulated Wnt signaling. This discrepancy could be explained by the cellular context (intestinal tissue compared to lung tissue or MEFs), because the Wnt pathway has been primarily associated with colon cancer and lung metastasis (24, 49).
In summary, using a murine model of lung cancer we demonstrate a requirement for FADD in Kras-mediated tumorigenesis, and demonstrate that phosphorylation of FADD by CK1α is required for mitogenic activity of the KRAS-MEK-ERK signaling pathway. Upon phosphorylation, FADD is translocated to the nucleus (5–6, 16, 22, 26), wherein it is required for G2/M progression through its interaction with PLK1, AURKA, CDC20 and BUB1 (Fig. 6). Our finding of increased in FADD mRNA expression in patients with mutant KRAS emphasizes the significance of the signaling pathway in KRAS-mediated oncogenesis. We propose that CK1α may be an attractive downstream target for inhibition of oncogenic signaling in EGFR and mutant KRAS-driven tumors.
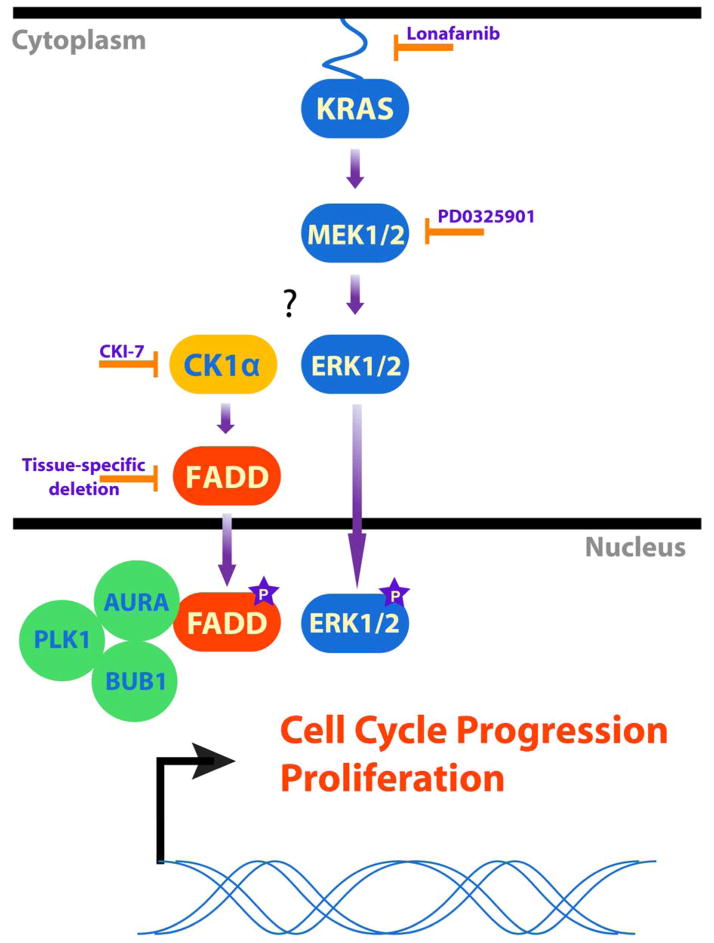
Fig. 6. Model for the role of FADD in KRAS-mediated cell proliferation.
We propose that FADD and CK1α are necessary in mediating mitogenic KRAS signaling in cancer cells. Inhibiting KRAS, MEK or CK1α, or deleting Csnk1a1 or Fadd decreased the abundance of phosphorylated FADD and decreased cell proliferation. We propose cells lacking FADD and CK1α fail to progress through G2/M because the interaction of FADD with key G2/M transition proteins like AURKA, PLK1 or BUB1 is lost.
Materials and Methods
Mice
All animal protocols were approved by the University of Michigan University Committee on Use and Care of Animals (UCUCA). All mouse work was performed in accordance with the University of Michigan protocol P00004781. Animals were housed in specific pathogen-free facilities of the University of Michigan Biomedical Science Research Building. KrasLSL-G12D mice (17) from the National Cancer Institute repository were intercrossed with Rosa26LSL-Luciferase (Jackson Laboratory, stock# 005125) and Fadd−/−; Fadd:GFP (16) mice to create KrasG12D; Luc; Fadd−/− or Fadd+/− animals (KFLuc or KLuc, respectively). Combinations of mutant littermates were used as controls: Luc; Fadd+/− or Luc; Fadd−/− (Luc or FLuc). A Kaplan-Meier survival curve reflects all animals that needed to be euthanized due to humane endpoints (such as labored breathing), or died during the time of experiments. Csnk1alfl/fl mice (38) (provided by Y. Ben-Neriah and E. Pikarsky) were also crossed with KrasLSL-G12D mice and Rosa26LSL-Luciferase mice to create KrasLSL-G12D; Luc; Csnk1a1fl/fl or Csnk1a1+/+ animals (KCLuc or KLuc respectively) and combinations of littermates were used as controls Luc; Csnk1a+/+ or Luc; Csnk1afl/fl (Luc or CLuc).
Genotyping and PCR
Mice were genotyped using tail DNA. For the genotyping of mutant KrasLSL-G12D, wild-type Kras and Cre-recombined KrasLSL-G12D, three primers were used: 5′-GTCTTTCCCCAGCACAGTGC, 5′-CTCTTGCCTACGCCACCAGCT and 5′-AGCTAGCCACCATGGCTTGAGTAAGTCTGCA. For Rosa26LSL-Luciferase the primers used were 5′-CGTGATCTGCAACTCCAGTC and 5′-GGAGCGGGAGAAATGGATATG. For the Fadd wild type allele the following primers were used: 5′-TGCGCCGACACGATCTACTG and 5′-TGTCAGGGTGTTTCTGAGGA. For the Fadd knockout neo cassette, the primers were: 5′-CGCTCGGTGTTCGAGGCCACACGC and 5′-ACTGTAGTGCCCAGCAGAGACCAGC. For the Fadd transgene (Fadd:GFP), primers were 5′-GTTGTCTTCGAAGTGCTCAGGC and 5′-GAACTTGTGGCCGTTTACGTC. For the genotyping of wild-type and Csnk1a1fl/fl mice, the following primers were used 5′-TCCACAGTTAACCGTAATCGT and 5′-AACTGCAAATGAAAGCCCTG. The Fadd:GFP primers were used for semi-quantitative PCR to determine Cre recombination efficiency. 10 ng of template were used and the PCR was run for 25 cycles.
Induction of lung cancer
Lung tumors were initiated by intranasal inhalation of 3×107 plaque-forming units of Adenovirus-Cre Recombinase (AdCre) in 6 to 8 week-old mice (48).
Bioluminescent imaging
Mice were imaged at 7, 13, 18 and 22 weeks after intranasal inhalation of AdCre. Imaging was performed on an IVIS Spectrum from Perkin Elmer. Mice were injected with D-Luciferin (150 mg/kg, Promega) solution in PBS and anesthetized with 1 to 2% isoflurane/air while imaged. Serial images were acquired at 30 s to 2 min intervals for up to 20 min after injection to capture the peak luminescence. Regions of interest were drawn around each lung and a highest photon emission value for each image was used for analysis.
Micro-computed tomography
μCT imaging was performed at 13 and 16 or 18 weeks after intranasal administration of AdCre using a Siemens Inveon System with the following parameters: 80 kVp, 500 μA, 400 ms exposure, 360 projections over 360 degrees, and 49.2 mm field of view (56 μm voxel size). Quantitative analysis was performed on automatically segmented lung volumes as the sum of lung, tumor, and vascular tissues. Following image calibration to Hounsfield units (HU) using air and a water phantom, segmentation of the lungs was accomplished using a connected threshold algorithm developed in-house (Matlab) with a threshold of −200 HU. This volume was subtracted from the total chest volume to approximate the tumor plus vascular volume. An assumption with this analysis is that vasculature should be similar between all subjects, with changes in this volume indicative of tumor volume changes (19).
Combined bioluminescence and CT imaging
CT and bioluminescence imaging was performed to demonstrate co-localization of luminescence with CT-determined lung lesions at 22 weeks after administration of AdCre using an IVIS Spectrum CT from Perkin-Elmer. Mice were injected and anesthetized as above for bioluminescent imaging, and imaged at 1 min intervals. The following parameters for CT imaging were used: 50 kVp, 1 mA, 720 projections over 360 degrees, and 12 cm field of view (150 μm voxel size).
Histology
Animals were euthanized and the lung was perfused with PBS through the heart. The lung was removed and fixed in formalin for 24 hours. Samples were embedded in paraffin and sliced into 5 μm sections. Samples were stained with hematoxylin and Eosin Y (H&E). Briefly, samples were hydrated in a sequence of xylene, 100% EtOH, 95% EtOH then water. Samples were then stained in Gill Hematoxylin and washed in tap water. Samples were immersed in 95% EtOH and then stained with acidic Eosin Y. All antibodies were used at a dilution of 1:200, and biotinylated-rabbit or biotinylated-goat secondary antibodies were used in conjunction with the Vector systems Vectastain ABC and DAB (3, 3′-diaminobenzidine) HRP system for visualization and Alexa488-conjugated chicken IgG was used for immunofluorescence.
Antibodies and Reagents
Rabbit polyclonal antibodies to GFP, phosphorylated ERK1/2, phosphorylated RB, ERK1/2, cyclin D1, phosphorylated FADD at Ser194, AURKA, phosphorylated β-Catenin, β-Catenin, phosphorylated MDM2, and PLK1 were purchased from Cell Signaling Technology. The antibody for human FADD was obtained from BD Biosciences. Antibodies against cyclin B1, CK1α, MDM2 and p53 were purchased from Santa Cruz Biotechnology. The antibody against mouse FADD used for western blotting was initially purchased from Epitomics (cat# 3523-1), and then from Abcam (cat# ab124812). Antibodies for BUB1, β-Actin, mouse FADD (cat# ab24533) and GFP were from Abcam. Ki-67 antibody was obtained from Vector Labs. CKI-7 was from Sigma. PD0325901 was purchased from Selleck, and Lonafarnib was bought from Cayman Chemical. Alamar Blue was purchased from Invitrogen. Luciferin was obtained from Promega. Adenovirus-Cre was bought from the University of Michigan Vector core.
Tissue Isolation and Western blotting
Animals were euthanized with CO2. Tumors were harvested and snap frozen in liquid nitrogen. Tumor samples were manually homogenized in RIPA buffer (Invitrogen) with protease inhibitor (Complete-Roche), PhosSTOP (Roche), and PMSF (Sigma). Lysates were centrifuged at 4°C for 30 min at 16,000xg and the supernatant collected for analysis. Cells from culture dishes were collected in cold 1x PBS and centrifuged at 1,800 × g for 5 min at 4°C. The cell pellet was washed with cold PBS and then lysed with the same buffer as above. Cells in lysis buffer were incubated in ice for 30 min. The lysates were then cleared by centrifugation at 16,000 × g for 15 min. The collected supernatants were estimated for protein content by Lowry assay (Biorad). Lysates with equal amounts of protein were resolved by 12% SDS/PAGE, and protein abundance was detected by Western blot analysis using the appropriate primary and secondary antibodies. Specific signals were visualized by using ECL Clarity western detection system by Biorad.
Cell culture and MEF isolation
A549 (lung epithelial carcinoma), NCI H1975 (lung epithelial adenocarcinoma) and HEK293T cells were purchased from the American Type Culture Collection (ATCC). A549 and H1975 cell lines were cultured in RPMI-1640 (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin (pen/strep), and glutamine (Invitrogen). HEK293T cells were cultured in DMEM (Invitrogen) and supplemented with 10% FBS, pen/strep, and glutamine. Mouse embryonic fibroblasts (MEFs) were isolated from E13-14 embryos from one pregnant mouse. Briefly, embryos were placed in ice cold PBS, and the placenta and visceral tissue was removed. Embryos were then minced in trypsin and incubated at 37°C. The minced embryos were then further disrupted by vigorous pipetting, plated with DMEM supplemented with 10% FBS, pen/strep, glutamine, and non-essential amino acids (Invitrogen), and cultured at 37°C with 5% CO2. MEFs were subjected to AdCre 1×108 pfu and were FACs sorted for GFP negative cells 24 to 48 hours later.
Analysis of cell proliferation, soft agar colony formation, and cell cycle
To assess cell proliferation, 3×103 MEFs were plated per well in a 96-well dish in DMEM supplemented with 10% FBS, pen/strep, glutamine and 1% non-essential amino acids. 5 hours after plating, 250 μM CKI-7, 10 μM Lonafarnib or 200 nM PD0325901 were added to designated wells. 24 hours after adding CKI-7 Lonafarnib or PD0325901, 10 μl Alamar blue was added to the first set of wells. Fluorescence was measured an hour later on a Perkin Elmer Envision plate reader. This was repeated at 48, 72 and 96 hours after addition of inhibitors. For the soft agar assay, 2×104 MEFs were plated in 0.3% agarose DMEM complete, with 20% FBS, on top of 0.7% agar supplemented with DMEM. Media was added to the cells twice a week for 2 weeks. 0.005% crystal violet was added to the cells and colonies were counted 24 hours later using a dissecting microscope and ImageJ. For cell cycle analysis, MEFs were plated in supplemented DMEM as described above. 24 hours later 250 uM CKI-7, 10uM Lonafarnib or 200nM PD0325901 were added. 24 hours after adding the inhibitors, cells were trypsinized and resuspended with PBS. Cells were then fixed with 100% ice cold ethanol for 20 min then centrifuged and resuspended in a propidium iodide and RNAse Type I-A solution (final concentration 50 μg/ml and 100 μg/ml, respectively, in PBS). Cells were incubated at room temperature for 20 min then over night at 4°C in the dark. Cells were analyzed as described above using a FACSAria III machine and with the FlowJo software.
Synchronization with thymidine and Coimmunoprecipitation
To detect the cell cycle-dependent endogenous association between FADD and other cell cycle-related proteins, NCI H1975 cells were synchronized using a double thymidine block. Cells were incubated with 4 mM thymidine (Sigma) for 16 hours, released into fresh media for 9 hours, followed by 16 hours thymidine incubation then released into fresh media. Cells were harvested at the indicated time points. Synchronized cells were harvested in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 1% NP40) supplemented with protease and phosphatase inhibitors. Protein concentration was determined by Biorad Protein Assay (Bio-Rad). Immunoprecipitation was carried out by incubating cell lysates with antibodies against human FADD or normal mouse IgG for 2 hours at 4°C. Antibody-protein complexes were captured with protein A or G-sepharose (GE Healthcare) by incubating 50 μl of beads per 200 μg of protein at 4°C for 2 to 3 hours. The resulting pellet was collected by centrifugation at 2,000 g for 2 min, washed three times with lysis buffer, boiled in 2X NuPage LDS sample buffer (Invitrogen) and resolved by SDS-PAGE. Western blots were carried out as outlined above.
Mass Spectrometry
12 million HEK 293T cells in 15 cm dishes were transfected with 30 μg pFN21A-FADD-HaloTag and Ctrl-HaloTag using Fugene HD. 24 hours post transfection, cells were scraped into DPBS and cell pellets were frozen at −70°C for at least 20 min. Previously frozen cell pellets were lysed in Promega Mammalian Lysis Buffer supplemented with protease inhibitor cocktail (G65A). The cleared lysate was bound to the pre-equilibrated HaloLink resin for 15 min at room temperature. Resin-bound proteins were eluted with Promega SDS Elution Buffer (G651A). Samples were analyzed by LC-MS/MS analysis according to manufacturer’s guidelines (Promega TM342 Section 5A).
Statistics
Differences between groups were assessed by an unpaired Student’s t-test. Results were declared statistically significant at the two-tailed 5% comparison-wise significance level (P<0.05). All error bars represent the standard error of the mean (SEM).
Supplementary Material
Figure S1: Fadd null lesions have lower abundance of phosphorylated ERK1/2 but no detectable difference for an apoptotic marker. (A) Representative images of lung tissue from Luc, KLuc and KFLuc stained with an antibody against phosphorylated ERK1/2 (pERK1/2) or Hematoxylin and Eosin Y (H&E). Scale bar, 200 μm. (B) Representative images of lung tissue from KLuc and KFLuc animals stained with an antibody against cleaved-caspase 3 at 40x magnification. Scale bar, 200 μm.
Figure S2: Fadd null lesions still express Fadd transgene. (A) Semi-quantitative PCR Fadd transgene (Fadd:GFP) in lung tumor samples from individual KLuc or KFLuc mice compared to a lung and liver sample from KFLuc mice. Controls were water (H2O) and tail DNA from a non-transgenic mouse (FVB-NJ). (B) Representative Western blot of tumor samples from individual KLuc and KFLuc mice probed for FADD, GFP (Fadd transgene) and β-actin. (C) Tumor tissue stained with DAPI and an antibody against GFP. Scale bar, 500 μm.
Figure S3: Increased FADD mRNA correlates with KRAS mutation status in lung cancer patients. (A and B) Box and whiskers graph of FADD expression, separated on the basis of KRAS status, in patient data from (A) Ding et al., 2008 (22) or (B) Okayama et al 2012 (23), analyzed with Oncomine. Patients with unknown KRAS status (“No Value” group): n = 34 (A), 20 (B); patients with mutant KRAS: n = 12 (A), 20 (B); patients with wild-type KRAS: n = 29 (A), 206 (B). *P=0.009;**P=0.007, unpaired Student’s t-test.
Figure S4: FADD interacts with proteins involved in the cell cycle (A) Analysis of mass spectrometry data (data file S1) revealing top cellular processes to which FADD contributes in HEK 293T cells. Data are ranked by P values as analyzed from DAVID gene ontology (GO) enrichment. (B) Interactome of cell cycle proteins from mass spectrometry experiment prepared using STRING. Lines represent confidence of interaction.
Figure S5: Immunohistochemistry of Csnk1a1 null lesions reveal residual CK1α protein. Representative images of lung histology from week 18 post AdCre administration, stained with (H&E) and antibodies against Ki-67, FADD, and CK1α. Scale bars: 10x, 500 μm; 40x, 200 μm; 100x, 50 μm.
Data file S1: Mass spectrometry data. Raw data from the mass spectrometry analysis of the Halo-tagged FADD peptide in HEK 293T cells. Analysis is presented in Table 1 and fig. S4. Excel file is provided online in the additional supplementary materials.
Acknowledgments
We would like to thank Kathleen Cho and for the Rosa26LSL-Luciferase mice, and Marina Pasca di Magliano for the KrasLSL-G12D mice. We would like to thank the Lautenberg Center for Immunology at the Hebrew University-Hadassah Medical School and especially Yinon Ben-Neriah and Eli Pikarsky for their generous gift of the Csnk1a1fl/fl mice. We are thankful to the MIL and FACs cores at the University of Michigan for assisting with all of the microscopy and cell cycle data. Also thanks to Mike Lafferty, James Stevenson, Paul Joseph, Katrin Vetter and Julie Blossom for their technical assistance.
Funding: This work was supported by NIH grants R01CA129623 and P50CA093990.
Footnotes
Author contributions: B.M.B., B.D.R., A.R., and S.G. planned and wrote the paper; B.M.B., K.A.S., B.A.H., J.L.B., D.L.D, K.A.H. and S.G. planned and performed experiments; R.M.P. performed pathological analysis of mouse lung cancer histology; C.J.C., D.G.B., and J.Z. provided materials and Fadd−/−; Fadd:GFP mice, expertise and discussions over the course of the experiments and during the preparation of the manuscript.
Competing interests: The authors declare no competing interests.
Data and materials availability: The raw mass spectrometry data (Data file S1) is also available at [URL and accession #TBD].
References and Notes
- 1.Herbst RS, Heymach JV, Lippman SM. Lung Cancer. N Engl Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu X, Yan S, Zhou T, Terada Y, Erikson RL. The MAP kinase pathway is required for entry into mitosis and cell survival. Oncogene. 2004;23:763–776. doi: 10.1038/sj.onc.1207188. [DOI] [PubMed] [Google Scholar]
- 3.Tourneur L, Chiocchia G. FADD: a regulator of life and death. Trends Immunol. 2010;31:260–269. doi: 10.1016/j.it.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Gómez-Angelats M, Cidlowski JA. Molecular evidence for the nuclear localization of FADD. Cell Death Differ. 2003;10:791–797. doi: 10.1038/sj.cdd.4401237. [DOI] [PubMed] [Google Scholar]
- 5.Chen G, Bhojani MS, Heaford AC, Chang DC, Laxman B, Thomas DG, Griffin LB, Yu J, Coppola JM, Giordano TJ, Lin L, Adams D, Orringer MB, Ross BD, Beer DG, Rehemtulla A. Phosphorylated FADD induces NF-κB, perturbs cell cycle, and is associated with poor outcome in lung adenocarcinomas. PNAS. 2005;102:12507–12512. doi: 10.1073/pnas.0500397102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhojani MS, Chen G, Ross BD, Beer DG, Rehemtulla A. Nuclear localized phosphorylated FADD induces cell proliferation and is associated with aggressive lung cancer. Cell Cycle. 2005;4:1478–1481. doi: 10.4161/cc.4.11.2188. [DOI] [PubMed] [Google Scholar]
- 7.Gibcus JH, Menkema L, Mastik MF, Hermsen MA, de Bock GH, van Velthuysen ML, Takes RP, Kok K, Alvarez Marcos CA, van der Laan BF, van den Brekel MW, Langendijk JA, Kluin PM, van der Wal JE, Schuuring E. Amplicon mapping and expression profiling identify the Fas-associated death domain gene as a new driver in the 11q13.3 amplicon in laryngeal/pharyngeal cancer. Clin Cancer Res. 2007;13:6257–6266. doi: 10.1158/1078-0432.CCR-07-1247. [DOI] [PubMed] [Google Scholar]
- 8.Prapinjumrune C, Morita K, Kuribayashi Y, Hanabata Y, Qi Shi Q, Nakajima Y, Inazawa J, Omura K. DNA amplification and expression of FADD in oral squamous cell carcinoma. J Oral Pathol Med. 2010;39:525–532. doi: 10.1111/j.1600-0714.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- 9.Drakos E, Leventaki V, Atsaves V, Schlette EJ, Lin P, Vega F, Miranda RN, Claret FX, Medeiros LJ, Rassidakis GZ. Expression of serine 194-phosphorylated Fas-associated death domain protein correlates with proliferation in B-cell non-Hodgkin lymphomas. Hum Pathol. 2011;42:1117–1124. doi: 10.1016/j.humpath.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrijvers ML, Pattje WJ, Slagter-Menkema L, Mastik MF, Gibcus JH, Langendijk JA, van der Wal JE, van der Laan BF, Schuuring E. FADD expression as a prognosticator in early-stage glottic squamous cell carcinoma of the larynx treated primarily with radiotherapy. Int J Radiation Oncol Biol Phys. 2012;83:1220–1226. doi: 10.1016/j.ijrobp.2011.09.060. [DOI] [PubMed] [Google Scholar]
- 11.Rasamny JJ, Allak A, Krook KA, Jo VY, Policarpio-Nicolas ML, Sumner HM, Moskaluk CA, Frierson HF, Jr, Jameson MJ. Cyclin D1 and FADD as biomarkers in head and neck squamous cell carcinoma. Otolaryngology Head Neck Surg. 2012;146:923–931. doi: 10.1177/0194599811435052. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Kabra NH, Cado D, Kang C, Winoto A. FADD-deficient T cells exhibit a disaccord in regulation of the cell cycle machinery. J Bio Chem. 2001;32:29815–29818. doi: 10.1074/jbc.M103838200. [DOI] [PubMed] [Google Scholar]
- 13.Hueber AO, Zornig M, Bernard AM, Chautan M, Evan G. A dominant negative Fas-associated death domain protein mutant inhibits proliferation and leads to impaired calcium mobilization in both T-cells and fibroblasts. J Bio Chem. 2000;275:10453–10462. doi: 10.1074/jbc.275.14.10453. [DOI] [PubMed] [Google Scholar]
- 14.Newton K, Kurts C, Harris AW, Strasser A. Effects of a dominant interfering mutant of FADD on signal transduction in activated T cells. Current Biology. 2001;11:273–276. doi: 10.1016/s0960-9822(01)00067-7. [DOI] [PubMed] [Google Scholar]
- 15.Alappat EC, Feig C, Boyerinas B, Volkland J, Samuels M, Murmann AE, Thronburn A, Kidd VJ, Slaughter CA, Osborn SL, Winoto A, Tang WJ, Peter ME. Phosphorylation of FADD at serine 194 by CKIα regulates its nonapoptotic activities. Molecular Cell. 2005;19:321–332. doi: 10.1016/j.molcel.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Rosenberg S, Wang H, Imtiyaz HZ, Hou YJ, Zhang J. Conditional Fas-associated death domain protein (FADD): GFP knockout mice reveal FADD is dispensable in thymic development but essential in peripheral T cell homeostasis. J Immunol. 2005;175:3033–3044. doi: 10.4049/jimmunol.175.5.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Safran M, Kim WY, Kung AL, Horner JW, DePinho RA, Kaelin WG. Mouse Reporter Strain for Noninvasive Bioluminescent Imaging of Cells that Have Undergone Cre-Mediated Recombination. Mol Imaging. 2003;2:297–302. doi: 10.1162/15353500200303154. [DOI] [PubMed] [Google Scholar]
- 19.Haines BB, Bettano KA, Chenard M, Sevilla RS, Ware C, Angagaw MH, Winkelmann CT, Tong C, Reilly RF, Sur C, Zhang W. Micro–Computed Tomography Method to Analyze Lung Tumors in Genetically Engineered Mouse Models. Neoplasia. 2009;11:39–47. doi: 10.1593/neo.81030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Winoto A. A mouse Fas-associated protein with homology to the Mort1/FADD protein is essential for Fas-induced apoptosis. Mol Cell Biol. 1996;16:2756–2763. doi: 10.1128/mcb.16.6.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rochat-Steiner V, Becker K, Micheau O, Schneider P, Burns K, Tschopp J. FIST/HIPK3: a Fas/FADD-serine/threonine kinase that induces FADD phosphorylation and inhibits fas-mediated Jun NH(2)-terminal kinase activation. J Exp Med. 2000;192:1165–1174. doi: 10.1084/jem.192.8.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osborn SL, Sohn SJ, Winoto A. Constitutive phosphorylation mutation in Fas-associated death domain (FADD) results in early cell cycle defects. J Bio Chem. 2007;28:22786–22792. doi: 10.1074/jbc.M703163200. [DOI] [PubMed] [Google Scholar]
- 23.Kim H, Lee H, Oh Y, Choi S, Hong S, Kim H, Lee S, Choi J, Hwang J, Kim K, Kim H, Zhang J, Youn H, Noh D, Jung Y. The DUSP26 phosphatase activator adenylate kinase 2 regulates FADD phosphorylation and cell growth. Nat Commun. 2014;5:3351. doi: 10.1038/ncomms4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba II, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;7216:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S, Sakamoto H, Kumamoto K, Takenoshita S, Gotoh N, Mizuno H, Sarai A, Kawano S, Yamaguchi R, Miyano S, Yokota J. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;1:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 26.Cheng W, Wang L, Zhang R, Du P, Yang B, Zhuang H, Tang B, Yao C, Yu M, Wang Y, Zhang J, Yin W, Li J, Zheng W, Lu M, Hua Z. Regulation of PKC inactivation by FADD. J Bio Chem. 2012;287:26126–26135. doi: 10.1074/jbc.M112.342170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schinske KA, Nayti S, Khan AP, Williams TM, Johnson TD, Ross BD, Tomas RP, Rehemtulla A. A novel kinase inhibitor of FADD phosphorylation chemosensitizes through the inhibition of NF-κB. Mol Cancer Ther. 2011;10:1807–1817. doi: 10.1158/1535-7163.MCT-11-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan AP, Schinske KA, Nyati S, Bhojani MS, Ross BD, Rehemtulla A. High-throughput molecular imaging for the identification of FADD kinase inhibitors. J Biomol Screen. 2010;15:1063–1070. doi: 10.1177/1087057110380570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun S, Zhou Z, Wang R, Fu H, Khuri FR. The Farnesyltransferase inhibitor Lonafarnib induces growth arrest or apoptosis of human lung cancer cells without down regulation of Akt. Cancer Bio & Therapy. 2013;3:1092–1098. doi: 10.4161/cbt.3.11.1176. [DOI] [PubMed] [Google Scholar]
- 30.Akinleye A, Furqan M, Mukhi N, Ravella P, Liu D. MEK and the inhibitors: from bench to beside. J Hematol Oncol. 2013;6:27. doi: 10.1186/1756-8722-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang M, Lee S, Kang NS, Kim E. Cooperative phosphorylation of FADD by Aur-A and Plk1 in response to taxol triggers both apoptotic and necrotic cell death. Cancer Res. 2011;71:7207–7215. doi: 10.1158/0008-5472.CAN-11-0760. [DOI] [PubMed] [Google Scholar]
- 32.Anger M, Kues WA, Klima J, Mielenz M, Kubelka M, Motlik J, Esner M, Dvorak P, Carnwath JW, Niemann H. Cell cycle dependent expression of Plk1 in synchronized porcine fetal fibroblasts. Mol Rep Dev. 2003;65:245–253. doi: 10.1002/mrd.10289. [DOI] [PubMed] [Google Scholar]
- 33.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi W, Tang Z, Yu H. Phosphorylation – and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol Biol Cell. 2007;17:3705–3716. doi: 10.1091/mbc.E06-03-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang Z, Shu H, Oncel D, Chen S, Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol Cell. 2004;16:387–397. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 37.Ricke RM, Jeganathan KB, van Deursen JM. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J Cell Biol. 2011;193:1049–1064. doi: 10.1083/jcb.201012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elyada E, Pribluda A, Goldstein RE, Morgenstern Y, Brachya G, Cojocaru G, Snir-Alkalay I, Burstain I, Haffner-Krausz R, Jung S, Wiener Z, Alitalo K, Oren M, Pikarsky E, Ben-Neriah Y. CKIα ablation highlights a critical role for p53 in invasiveness control. Nature Letters. 2011;470:409–413. doi: 10.1038/nature09673. [DOI] [PubMed] [Google Scholar]
- 39.Huart AA, MacLaine NJ, Meek DW, Hupp TR. CK1alpha plays a central role in mediating MDM2 control of p53 and E2F-1 protein stability. J Biol Chem. 2009;284:32384–32394. doi: 10.1074/jbc.M109.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L, Li C, Pan Y, Chen J. Regulation of p53-MDMX interaction by Casein Kinase 1 Alpha. Mol Cell Bio. 2005;15:6509–6520. doi: 10.1128/MCB.25.15.6509-6520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Del Valle-Pérez B, Arqués O, Vinyoles M, de Herreros AG, Duñach M. Coordinated action of CK1 isoforms in canonical Wnt signaling. Mol Cell Biol. 2011;31:2877–2888. doi: 10.1128/MCB.01466-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adjei AA. Blocking oncogenic Ras signaling for cancer therapy. J Nat Cancer Institute. 2001;93:1062–1074. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 43.Riely GJ, Kris MG, Rosenbaum D, Marks J, Li A, Chitale DA, Nafa K, Riedel ER, Hsu M, Pao W, Miller VA, Ladanyi M. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;18:5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng XW, Chandra J, Loegering D, Van Becelaere K, Kottke TJ, Gore SD, Karp JE, Sebolt-Leopold J, Kaufmann SH. Central role of Fas-associated death domain protein in apoptosis induction by mitogen-activated protein kinase kinase inhibitor CI-1040 (PD184352) in acute lymphocytic leukemia cells in vitro. J Bio Chem. 2003;278:47326–47339. doi: 10.1074/jbc.M304793200. [DOI] [PubMed] [Google Scholar]
- 45.Fung TK, Poon RYC. A roller coaster ride with the mitotic cyclins. Sem Cell Dev Biol. 2007;16:335–342. doi: 10.1016/j.semcdb.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 46.Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 47.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nature Reviews Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 48.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral antiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Fadd null lesions have lower abundance of phosphorylated ERK1/2 but no detectable difference for an apoptotic marker. (A) Representative images of lung tissue from Luc, KLuc and KFLuc stained with an antibody against phosphorylated ERK1/2 (pERK1/2) or Hematoxylin and Eosin Y (H&E). Scale bar, 200 μm. (B) Representative images of lung tissue from KLuc and KFLuc animals stained with an antibody against cleaved-caspase 3 at 40x magnification. Scale bar, 200 μm.
Figure S2: Fadd null lesions still express Fadd transgene. (A) Semi-quantitative PCR Fadd transgene (Fadd:GFP) in lung tumor samples from individual KLuc or KFLuc mice compared to a lung and liver sample from KFLuc mice. Controls were water (H2O) and tail DNA from a non-transgenic mouse (FVB-NJ). (B) Representative Western blot of tumor samples from individual KLuc and KFLuc mice probed for FADD, GFP (Fadd transgene) and β-actin. (C) Tumor tissue stained with DAPI and an antibody against GFP. Scale bar, 500 μm.
Figure S3: Increased FADD mRNA correlates with KRAS mutation status in lung cancer patients. (A and B) Box and whiskers graph of FADD expression, separated on the basis of KRAS status, in patient data from (A) Ding et al., 2008 (22) or (B) Okayama et al 2012 (23), analyzed with Oncomine. Patients with unknown KRAS status (“No Value” group): n = 34 (A), 20 (B); patients with mutant KRAS: n = 12 (A), 20 (B); patients with wild-type KRAS: n = 29 (A), 206 (B). *P=0.009;**P=0.007, unpaired Student’s t-test.
Figure S4: FADD interacts with proteins involved in the cell cycle (A) Analysis of mass spectrometry data (data file S1) revealing top cellular processes to which FADD contributes in HEK 293T cells. Data are ranked by P values as analyzed from DAVID gene ontology (GO) enrichment. (B) Interactome of cell cycle proteins from mass spectrometry experiment prepared using STRING. Lines represent confidence of interaction.
Figure S5: Immunohistochemistry of Csnk1a1 null lesions reveal residual CK1α protein. Representative images of lung histology from week 18 post AdCre administration, stained with (H&E) and antibodies against Ki-67, FADD, and CK1α. Scale bars: 10x, 500 μm; 40x, 200 μm; 100x, 50 μm.
Data file S1: Mass spectrometry data. Raw data from the mass spectrometry analysis of the Halo-tagged FADD peptide in HEK 293T cells. Analysis is presented in Table 1 and fig. S4. Excel file is provided online in the additional supplementary materials.