Abstract
Background
Enterovirus 71 (EV71) is one of the major etiological pathogens of hand, foot and mouth disease (HFMD) and can cause severe cerebral and pulmonary complications and even fatality. MicroRNAs (miRNAs), a class of small non-coding RNA molecules, play an important role in post-transcriptional regulation of gene expression and thereby influencing various physiological and pathological processes. Increasing evidence suggests that miRNAs act as key effector molecules in the complicated pathogen-host interactions. However, the roles of miRNAs in EV71 infection and pathogenesis are not well understood.
Methods
To identify special miRNAs involved in EV71 infection, a microarray assay was performed to study the expression pattern of miRNAs in EV71-infected human rhabdomyosarcoma cells (RD cells) and uninfected RD cells. We further predicted the putative target genes for the dysregulated miRNAs using the online bioinformatic algorithms (TargetScan, miRanda and PicTar) and carried out functional annotation including GO enrichment and KEGG pathway analysis for miRNA predicted targets. Then, the results of microarray were further confirmed by quantitative RT-PCR.
Results
Totally, 45 differentially expressed miRNAs ware identified by microarray, among which 36 miRNAs were up-regulated and 9 were down-regulated. 7166 predicted target genes for the dysregulated miRNAs were revealed by using TargetScan in conjunction with miRanda and PicTar. The GO annotation suggested that predicted targets of miRNAs were enriched into the category of signal transduction, regulation of transcription, metabolic process, protein phosphorylation, apoptotic process and immune response. KEGG pathway analysis suggested that these predicted target genes were involved in many important pathways, mainly including endocytosis and focal adhesion, MAPK signaling pathway, hypertrophic cardiomyopathy, melanogenesis and ErbB signaling pathway. The expression levels of 8 most differentially up-regulated miRNAs and 3 most differentially down-regulated miRNAs were confirmed by qRT-PCR. The expressions of hsa-miR-4530, hsa-miR-4492, hsa-miR-6125, hsa-miR-494-3p, hsa-miR-638, hsa-miR-6743-5p, hsa-miR-4459 and hsa-miR-4443 detected by qRT-PCR were consistent with the microarray data.
Conclusion
These results might extend our understanding to the regulatory mechanism of miRNAs underlying the pathogenesis of EV71 infection, thus strengthening the preventative and therapeutic strategies of HFMD caused by EV71.
Keywords: Enterovirus 71; Hand, foot and mouth disease; MicroRNA; Microarray
Background
Enterovirus 71 (EV71) is a single-positive-stranded RNA virus belonging to the Enterovirus genus of the Picornaviridae family, and has become one of the most important neurotropic enteroviruses after the near eradication of poliovirus [1,2]. It is the major causative pathogen of hand, foot and mouth disease (HFMD) which is a common exanthematous and febrile disease in children. But for some patients especially in children under 5 years old, EV71 can cause severe complications, including myocarditis, aseptic meningitis, encephalitis, acute flaccid paralysis, pulmonary oedema or haemorrhage, and even fatality. The underlying mechanisms of serious cerebral and pulmonary complications and even death are not very clear. And up to now, no specific vaccines, effective antiviral agents or therapies against EV71 infection are available [3]. Therefore, the needs of better understanding the mechanism of EV71 pathogenesis are warranted.
Increasing evidence suggests that microRNAs (miRNAs), an abundant class of endogenous small noncoding RNA molecules (~22 nucleotides), modulate gene expression at the posttranscriptional level [4]. miRNAs are processed from the primary transcripts (pri-miRNA) which are subsequently cleaved by Drosha into pre-miRNA of approximately 70 nucleotides and exported to the cytoplasm [5]. In the cytoplasm, cleavage of pre-miRNA by Dicer proteins yields a mature miRNA of approximately 22 nucleotides [6]. Then, the mature miRNAs bind to 3′ untranslated regions (3′ UTR) of target mRNAs, which is able to recruit target mRNA and initiate mRNA degradation or translation inhibition [7]. Therefore, a large number of diverse biological processes, such as cell proliferation, cell cycle, apoptosis, immune response, differentiation, metabolic processes and cellular response to stress are regulated [4,8-10].
It has been demonstrated that miRNAs act as key effector molecules in the complicated interaction network between virus and host [11,12]. On one hand, miRNAs encoded by human cells could directly affect the virus replication. On the other hand, virus could change cell normal functions by influencing cellular gene expressions through miRNAs. To identify cellular miRNAs involved in the host response to EV71 infection and to elucidate the pathogenesis mechanism of EV71 virus, miRNAs profiles in the EV71-infected cells were examined in this study. The differences in expression levels were confirmed by quantitative real-time RT-PCR (qRT-PCR) assay. The computational methods were further performed to identify potential targets of dysregulated miRNAs, With gene ontology (GO) and KEGG biological pathway analysis, several pathways were enriched. These results may enhance our understanding on the prevention and treatment of hand-foot-and-mouth disease caused by EV71.
Results
Virus infection and observation of cytopathic effect
RD cells which are frequently used to isolate EV71 from clinical specimens are highly susceptible to EV71. After 48 h of infection, EV71 induced a severe CPE in RD cells (Figure 1).
Figure 1.
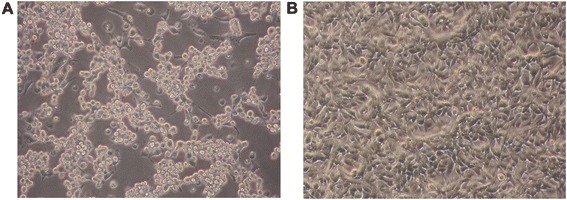
The morphological changes of EV71-induced cytopathic effect in RD cells were observed under a light microscope at 20× magnification at 48 h p.i.. RD cells were infected with EV71 at a m.o.i. of 0.1. (A) EV71-infected RD cells exhibited severe CPE appearance in response to virus replication at 48 h p.i..; (B) Uninfected RD cells showed normal morphology.
MiRNA microarray analysis
To identify miRNAs involved in EV71 infection and pathogenic mechanism, microarray analysis was performed using total RNA from EV71 infected RD cells and control cells. According to the miRNA profiling, the expression levels of 45 miRNAs were significantly altered in EV71 infected RD cells compared to uninfected RD cells. Among these, 36 miRNAs were up-regulated and 9 were down-regulated. Group-specific signal intensities were shown in Figure 2, and the information of the 45 miRNAs was listed separately in Table 1 with fold changes and P values.
Figure 2.
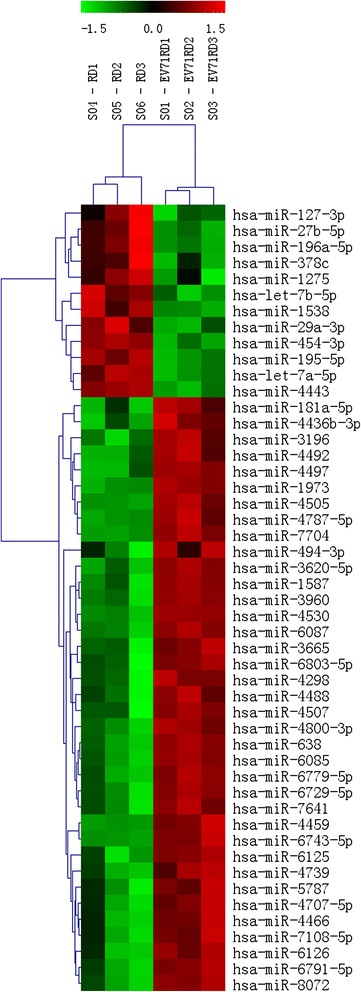
Heat map and unsupervised hierarchical cluster analysis of the differentially expressed miRNAs in EV71-infected cells and control cells. Total RNA from EV71-infected RD cells and uninfected RD cells in triplicate was used for microarray. Differentially expressed miRNAs were chosen with a log2 Ratio ≥ 0.5 and ≤ −0.5 and an adjusted P value < 0.05. Then columns and rows represent samples and particular miRNAs. The samples were correctly grouped together into EV71-infected cells and control cells according to the expression pattern The miRNA clustering tree is shown on the left. The color scale illustrates the relative expression level of miRNAs. Red color represents that the miRNA has higher expression in EV71-infected cells than in control, while green color represents that the miRNA has lower expression in EV71-infected cells than in control. The codes on the legend are log2-transformed values.
Table 1.
Summary of the most significantly differentally expressd miRNA
| miR-name | Fold change | P values | Target sequence (5′ to 3′) |
|---|---|---|---|
| Up-regulated miRNAs | |||
| hsa-miR-4800-3p | 28.83 | 4.19E-03 | CAUCCGUCCGUCUGUCCAC |
| hsa-miR-4530 | 12.00 | 4.64E-03 | CCCAGCAGGACGGGAGCG |
| hsa-miR-4492 | 9.61 | 7.36E-03 | GGGGCUGGGCGCGCGCC |
| hsa-miR-4507 | 8.88 | 3.76E-02 | CUGGGUUGGGCUGGGCUGGG |
| hsa-miR-3620-5p | 8.86 | 1.18E-02 | GUGGGCUGGGCUGGGCUGGGCC |
| hsa-miR-4505 | 8.72 | 6.33E-03 | AGGCUGGGCUGGGACGGA |
| hsa-miR-1587 | 8.54 | 2.03E-02 | UUGGGCUGGGCUGGGUUGGG |
| hsa-miR-7108-5p | 8.19 | 2.11E-02 | GUGUGGCCGGCAGGCGGGUGG |
| hsa-miR-6791-5p | 6.14 | 2.63E-02 | CCCCUGGGGCUGGGCAGGCGGA |
| hsa-miR-6125 | 5.71 | 2.61E-02 | GCGGAAGGCGGAGCGGCGGA |
| hsa-miR-494-3p | 5.49 | 3.58E-02 | UGAAACAUACACGGGAAACCUC |
| hsa-miR-1973 | 5.11 | 4.13E-04 | ACCGUGCAAAGGUAGCAUA |
| hsa-miR-6729-5p | 4.96 | 2.12E-02 | UGGGCGAGGGCGGCUGAGCGGC |
| hsa-miR-638 | 4.61 | 1.17E-02 | AGGGAUCGCGGGCGGGUGGCGGCCU |
| hsa-miR-3960 | 4.15 | 1.55E-02 | GGCGGCGGCGGAGGCGGGGG |
| hsa-miR-6126 | 4.05 | 3.70E-02 | GUGAAGGCCCGGCGGAGA |
| hsa-miR-4787-5p | 4.04 | 1.08E-02 | GCGGGGGUGGCGGCGGCAUCCC |
| hsa-miR-8072 | 4.04 | 2.07E-02 | GGCGGCGGGGAGGUAGGCAG |
| hsa-miR-6085 | 3.90 | 1.49E-02 | AAGGGGCUGGGGGAGCACA |
| hsa-miR-6779-5p | 3.85 | 1.75E-02 | CUGGGAGGGGCUGGGUUUGGC |
| hsa-miR-6087 | 3.49 | 1.02E-02 | UGAGGCGGGGGGGCGAGC |
| hsa-miR-4707-5p | 3.33 | 1.81E-02 | GCCCCGGCGCGGGCGGGUUCUGG |
| hsa-miR-7704 | 3.26 | 4.44E-03 | CGGGGUCGGCGGCGACGUG |
| hsa-miR-3196 | 3.25 | 8.02E-03 | CGGGGCGGCAGGGGCCUC |
| hsa-miR-6743-5p | 3.03 | 6.81E-03 | AAGGGGCAGGGACGGGUGGCCC |
| hsa-miR-4466 | 2.94 | 1.88E-02 | GGGUGCGGGCCGGCGGGG |
| hsa-miR-3665 | 2.77 | 3.44E-02 | AGCAGGUGCGGGGCGGCG |
| hsa-miR-4497 | 2.71 | 1.41E-02 | CUCCGGGACGGCUGGGC |
| hsa-miR-6803-5p | 2.62 | 3.88E-02 | CUGGGGGUGGGGGGCUGGGCGU |
| hsa-miR-5787 | 2.24 | 2.21E-02 | GGGCUGGGGCGCGGGGAGGU |
| hsa-miR-7641 | 2.09 | 2.70E-02 | UUGAUCUCGGAAGCUAAGC |
| hsa-miR-4298 | 2.00 | 3.56E-02 | CUGGGACAGGAGGAGGAGGCAG |
| hsa-miR-4436b-3p | 1.99 | 1.04E-02 | CAGGGCAGGAAGAAGUGGACAA |
| hsa-miR-4459 | 1.74 | 8.61E-03 | CCAGGAGGCGGAGGAGGUGGAG |
| hsa-miR-4488 | 1.63 | 4.31E-02 | AGGGGGCGGGCUCCGGCG |
| hsa-miR-4739 | 1.45 | 1.30E-02 | AAGGGAGGAGGAGCGGAGGGGCCCU |
| Down-regulated miRNAs | |||
| hsa-let-7a-5p | 0.70 | 3.40E-03 | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-miR-29a-3p | 0.69 | 1.20E-02 | UAGCACCAUCUGAAAUCGGUUA |
| hsa-miR-127-3p | 0.67 | 4.16E-02 | UCGGAUCCGUCUGAGCUUGGCU |
| hsa-miR-196a-5p | 0.59 | 3.35E-02 | UAGGUAGUUUCAUGUUGUUGGG |
| hsa-miR-378c | 0.58 | 4.53E-02 | ACUGGACUUGGAGUCAGAAGAGUGG |
| hsa-let-7b-5p | 0.55 | 8.67E-03 | UGAGGUAGUAGGUUGUGUGGUU |
| hsa-miR-4443 | 0.32 | 1.17E-03 | UUGGAGGCGUGGGUUUU |
| hsa-miR-27b-5p | 0.21 | 4.34E-02 | AGAGCUUAGCUGAUUGGUGAAC |
| hsa-miR-1538 | 0.16 | 2.10E-02 | CGGCCCGGGCUGCUGCUGUUCCU |
Target genes prediction of the differentially expressed miRNAs and functional analysis
The target prediction was performed for the 45 differentially expressed miRNAs to determine the influence of EV71 infection on potential target mRNAs by combining the results from three online free available algorithms. 6950 predicted target genes of 36 up-regulated microRNAs and 216 predicted target genes of 9 down-regulated microRNAs were found respectively.
To further evaluate the biological implications of differential miRNA, we appraised miRNA predicted target genes in gene-term enrichment analysis using Gene Ontology (GO) categories. And KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis also was performed by using the DAVID Functional Annotation Chart tool for the targets identified for differentially expressed miRNAs. GO category is composed of biological process, cellular component, and molecular function. Gene-term enrichment analysis revealed that most predicted target genes were involved in GO category “biological process” dealing with signal transduction, regulation of transcription, cell differentiation, metabolic process, protein phosphorylation, cell cycle, apoptotic process and immune response and so on (Figure 3). Within the GO category “cellular component”, the greatest number of predicted target genes had functions associated with nucleus, cytoplasm and membrane (Figure 3). Regarding the GO category “molecular function”, most of the predicted target genes miRNAs were involved in protein binding, metal ion binding and zinc ion binding (Figure 3). These results were indicated that the targets miRNAs were involved in a wide variety of physiological processes.
Figure 3.
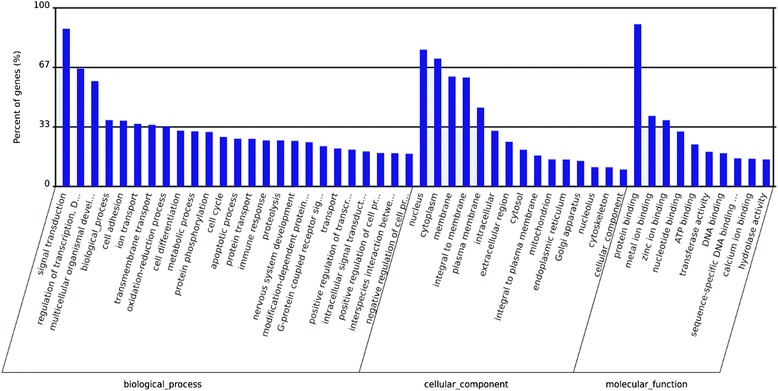
GO categories and distribution for the predicted miRNA targets related with EV71 infeciton. The GO terms were sorted by the number of genes in an ascending order from top to bottom. Biological process is enriched in signaling transduction, regulation of transcription, multicellular organismal development, cell adhesion and ion transport. Cellular components are enriched in nucleus, cytoplasm and membrane. Molecular function is enriched in protein binding, metal ion binding and zinc ion binding.
According to the analysis of enriched KEGG pathways, predicted target genes of differentially expressed miRNAs were related to endocytosis and focal adhesion, MAPK signaling pathway, hypertrophic cardiomyopathy, melanogenesis and ErbB signaling pathway (Table 2). KEGG Pathway analysis illustrated some of the underlying biological processes that may be involved in EV71 infection and might provide useful clues to further research the miRNA targets.
Table 2.
KEGG pathway analysis for the predicted miRNA targets related with EV71 infeciton
| Pathway Id | Pathway description | S gene number | TS gene number | B gene number | TB gene number | P value of Fisher’s exact test |
|---|---|---|---|---|---|---|
| 4144 | Endocytosis | 158 | 3220 | 181 | 4524 | 0.000000 |
| 4510 | Focal adhesion | 163 | 3220 | 193 | 4524 | 0.000008 |
| 4010 | MAPK signaling pathway | 209 | 3220 | 257 | 4524 | 0.000084 |
| 5410 | Hypertrophic cardiomyopathy (HCM) | 74 | 3220 | 84 | 4524 | 0.000168 |
| 4916 | Melanogenesis | 86 | 3220 | 100 | 4524 | 0.000344 |
| 4012 | ErbB signaling pathway | 73 | 3220 | 84 | 4524 | 0.000490 |
| 5214 | Glioma | 57 | 3220 | 64 | 4524 | 0.000509 |
| 5210 | Colorectal cancer | 72 | 3220 | 83 | 4524 | 0.000605 |
| 4512 | ECM-receptor interaction | 70 | 3220 | 81 | 4524 | 0.000917 |
| 4142 | Lysosome | 95 | 3220 | 113 | 4524 | 0.000946 |
| 5220 | Chronic myeloid leukemia | 63 | 3220 | 73 | 4524 | 0.001796 |
| 5223 | Non-small cell lung cancer | 47 | 3220 | 53 | 4524 | 0.002023 |
| 4910 | Insulin signaling pathway | 109 | 3220 | 133 | 4524 | 0.002627 |
| 5212 | Pancreatic cancer | 61 | 3220 | 71 | 4524 | 0.002697 |
| 4912 | GnRH signaling pathway | 78 | 3220 | 93 | 4524 | 0.003094 |
| 4722 | Neurotrophin signaling pathway | 101 | 3220 | 123 | 4524 | 0.003313 |
| 4130 | SNARE interactions in vesicular transport | 32 | 3220 | 35 | 4524 | 0.003616 |
| 4520 | Adherens junction | 62 | 3220 | 73 | 4524 | 0.004499 |
| 4514 | Cell adhesion molecules (CAMs) | 105 | 3220 | 129 | 4524 | 0.004825 |
| 5414 | Dilated cardiomyopathy | 72 | 3220 | 86 | 4524 | 0.004880 |
MiRNA expression validation by qRT-PCR
In order to validate the microarray results, the expression levels of 8 most differentially up-regulated miRNAs and 3 most differentially down-regulated miRNAs were examined by qRT-PCR, using the same RNA samples that were used for the microarrays. As shown in Figure 4, the expressions of hsa-miR-4530, hsa-miR-4492, hsa-miR-6125, hsa-miR-494-3p, hsa-miR-638, hsa-miR-6743-5p, hsa-miR-4459 and hsa-miR-4443 detected by qRT-PCR were consistent with the microarray data with significance (P < 0.05). The expression levels of hsa-miR-4505 and hsa-miR-29a-3p were not proven to be significant. The expression patterns of hsa-miR-27b-5p was not in line with the microarray results.
Figure 4.
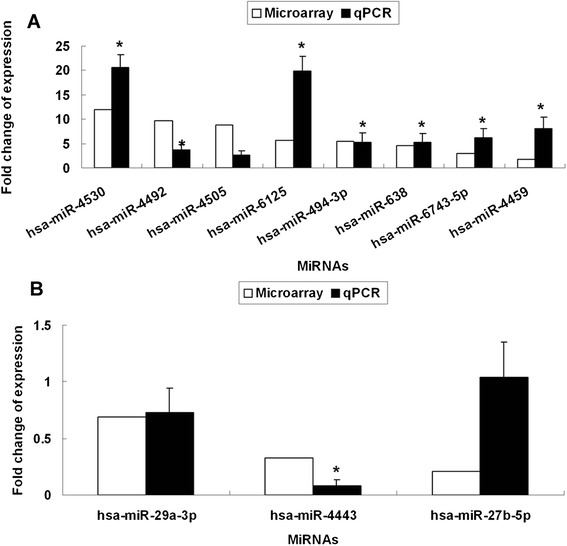
Validation of the differential expression of 11 miRNAs identified in the microarray by qRT-PCR. U6 was used as a reference gene. The expression levels of miRNAs in uninfected RD cells were set as 1. The expression levels of most miRNAs detected by qRT-PCR were correlated with those detected by microarray. (A) 8 up-regulated miRNAs (hsa-miR-4530, hsa-miR-4492, hsa-miR-4505, hsa-miR-6125, hsa-miR-494-3p, hsa-miR-638, hsa-miR-6743-5p, hsa-miR-4459). (B) 3 down-regulated miRNAs (hsa-miR-29a-3p, hsa-miR-4443, hsa-miR-27b-5p). *P < 0.05.
Discussion
Recently, the roles of miRNAs in pathogen-host interactions have been paid more attention than before. Not only the miRNAs encoded by virus but also cellular miRNAs have been proven to participate in the interactions. Through remodelling cellular miRNAs expression, viruses influence cellular microenvironment and metabolism, which are likely to play a very important role in the viral pathogenesis. To date, a few studies have already been done to explore the effects of miRNAs in EV71 infection, inferring that miRNAs may be involved in the pathogenesis of EV71 infection. By using deep sequencing, Cui [13] revealed that certain miRNAs might be essential in the EV71 infection. The research results of Li et al. suggested that miRNA-548 regulates host antiviral response via direct targeting of IFN-λ1 [14]. Using a human colorectal adenocarcinoma cell line (HT29) based EV71 infection model, Lui et al. showed that knockdown of DGCR8, an essential cofactor for microRNAs biogenesis, resulted in a reduction of EV71 replication [15]. Zhang [16] has demonstrated that miR-27a may have antiviral activity against EV71 by inhibiting EGFR. Zheng et al. provned that hsa-miR-296-5p suppressed EV71 replication by targeting the viral genome [17]. And Wen et al. reported that miR-23b inhibited EV71 replication by down-regulating EV71 VPl protein [18]. Recently, Bian et al. [19] compared the miRNA profiles between interferon (IFN)-α or IFN-γ treated RD cells and EV71-infected RD cells. miR-124 and miR-491-3p were regulated in opposite manners by the IFNs and EV71. Wnt signaling cascade, platelet-derived growth factor receptor (PDGFR)/PDGF, phosphatidylinositol 3-kinase(PI3K) and Jun N-terminal kinase (JNK)/mitogen-activated protein kinase (MAPK) were predicted in both EV71 infection and IFN treatment. These results indicated that cellular miRNAs participated in the virus life cycle as critical host factors by targeting the downstream genes or signaling pathways. The identified miRNAs and signal transduction patyway would be helpful to understand the interaction between the virus and host.
In our present study, a comprehensive miRNA profile was performed in EV71-infected RD cells through microarray assay to identify cellular miRNAs and downstream signaling pathway involved in the host response to EV71 infection. 36 up-regulated and 9 down-regulated miRNAs were found. By using qRT-PCR, the expression level of 11 miRNAs were identified. Among them, the expression patterns of 7 up-regulated (hsa-miR-4530, hsa-miR-4492, hsa-miR-6125, hsa-miR-494-3p, hsa-miR-638, hsa-miR-6743-5p, hsa-miR-4459) and 1 down-regulated miRNA (hsa-miR-4443) were consistent with the microarray data.
Among the 8 miRNAs, some miRNAs have been reported in other research field. Hsa-miR-4530 was validated as a miRNA marker that differentiated pancreato-biliary cancers from other clinical conditions including healthy controls, non-malignant abnormalities, and other types of cancers [20]. Hsa-miR-4459 could decrease the expression of its targets, CDC20B and ATG13, and thus altered stemness via cell cycle and autophagy in human embryonic stem cells [21]. Hsa-miR-494-3p, a well-known miRNA, plays different roles in different malignancies and was found to be implicated in multiple cell processes including cell proliferation, apoptosis, and invasion. For example, miR-494-3p was found to be act as a cancer gene that could promote glioma cell proliferation through the down-expression of PTEN, a tumor-suppressor gene [22,23]. While miR-494 acts as an antioncogene in gastric carcinoma by targeting c-myc in gastric cancer [24,25]. Furthermore, hsa-miR-638 has previously been reported to be associated with virus infection. Up-expression of hsa-miR-638 was identified by microarray technology in chikungunya virus infection in human cells [26]. Kumar et al. [27] reported hsa-miR-638 decreased the levels of HBV transcripts or HBV gene products. Liu et al. [28] demonstrated inhibition of hsa-miR- 638 slightly increased HCV entry during in vitro acute HCV infection. But all of the 8 miRNAs including hsa-miR-4530, hsa-miR-4492, hsa-miR-6125, hsa-miR-494-3p, hsa-miR-638, hsa-miR-6743-5p, hsa-miR-4459 and hsa-miR-4443 were found to be related with EV71 infection for the first time. This may provide new clues for EV71 infection research.
According to the result of GO analysis, the predicted target genes were involved in signal transduction, regulation of transcription, cell differentiation, metabolic process, protein phosphorylation, cell cycle, apoptotic process and immune response et al.. These biological processes are reported to be crucial in the interplay between host and virus. As for biological pathways, endocytosis and focal adhesion, MAPK signaling pathway and ErbB signaling pathway et al. were among the enriched pathways of the predicted target genes.
Mitogen-activated protein kinase (MAPK) are a family of protein kinases responsible for phosphorylating serine and threonine in many proteins [29]. It is widely conserved in eukaryotes and involved in many cellular functions such as inflammation, cell proliferation, differentiation, movement and death [30-32]. Three major MAPK signaling pathways have been identified, including extracellular regulated kinases (ERK1/2), JNK(JNK1/2) and p38 MAPK (p38 α/β/γ/δ). It has been reported that JNK1/2 and p38 MAPK signal pathways plays an important role in response to infection and replication of human immunodeficiency virus type 1, encephalomyocarditis virus, coxsackievirus B3, hepatitis C virus, herpes simplex virus 1, and the severe acute respiratory syndrome coronavirus [33-36]. The diverse effects of activates signaling cascades include induction of apoptosis in infected cells and enhancement of viral replication. MEK1 is crucial in the ERK signaling cascade and is required to promote EV71 replication [37]. EV71 infection induces the up-regulated gene expressions of MAPK signaling pathway such as ERK, JNK and PI3K/AKT, which may be associated with the secretions of inflammatory cytokines and host cell apoptosis, which might be implicated in CNS inflammation and disorders such as encephalitis or meningitis [38]. Similarly, Tung et al. [39] domenstrated that cellular ERK levels have been shown to be the major causal factor for upregulation of cyclooxygenase-2 (COX-2) expression induced by EV71 infection which may participate in EV71-induced CNS damage. COX-2 and its metabolite, prostaglandin E2 (PGE2), which are considered a tumor biomarker [40], are up-regulated by EV71 infection via the activated c-Src/PDGFR/PI3K/Akt/p42/p44 MAPK/AP1 and NF-κB pathways in rat brain astrocytes [41]. These evidence together with our study supports the notion that MAPK signalling cascade as key moleculars in EV71 replication cycle and pathogenesis.
As well known, the miRNA targets predicition merely by the means of bioinformatics is not enough. MiRNA targets should be verified by further experiments, such as miRNA transfection or knockdown and luciferase assay. Therefore, our next task is to carry out these experiments to confirm the miRNA targets which were predicted in this present study. If the function of differentially expressed miRNAs and their corresponding predicted target genes have been identified, it will improve the protection and treatment strategies of EV71 infection.
Materials and methods
Cell culture, virus infection and observation of cytopathic effect
Human rhabdomyosarcoma cells (RD cells; ATCC CCL-136, Manassas, VA) were grown in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Logan, UT) supplemented with 10% fetal bovine serum (FBS; Gibco, USA) plus 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C with 5% CO2. When the cells had grown to 90% confluence, cells were infected with EV71 (Human enterovirus 71 strain 87–2008 isolated and identified from Shaanxi Province, China, in 2010, Genbank accession no. HM003207) at a m.o.i. of 0.1 and maintained in medium containing 2% fetal calf serum following 1 h adsorption at 37°C. After infection, the morphological changes of virus-induced cytopathic effect were observed and photographed under microscopy.
Total RNA extraction and quality control
EV71-infected RD Cells and uninfected RD cells in triplicate were harvested at 48 h postinfection and resuspended in 1 ml of TRIzol reagent (life technology), strongly shaken and stored at −20°C. Total RNA, including small RNA, was extracted using Total RNA Kit (LC Sciences, USA) according to the manufacturer’s protocol. After purification, the concentration of RNA samples was determined by measuring the absorbance at 260 and 280 nm with the NanoDrop 1000. And the integrity was assayed by Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Only cases with RNA Integrity Numbers (RIN) ≥7–10 were further used.
MicroRNA microarray assay
MiRNA microarray, covering all miRNAs in the miRBase database (v.20.0) (www.mirbase.org), was conducted by LC Sciences (Houston, TX, USA) to study the expression profiling. 5 ug of RNA from each sample was size fractionated by using an YM-100 Microcon centrifugal filter (Millipore, Bedford, MA, USA). The small RNAs(<300 nt) were 3′-extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for later fluorescent dye staining. Hybridization was performed overnight on a μParaflo microfluidic chip using a micro-circulation pump (Atactic Technologies) [42]. On the microfluidic chip, each detection probe consisted of a chemically modified nucleotide coding segment complementary to target microRNA from miRBase and a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. The detection probes were made by in situ synthesis using PGR (photogenerated reagent) chemistry. The hybridization melting temperatures were balanced by chemical modifications of the detection probes. The hybridization buffer was 100 μL 6 × SSPE buffer (0.90 M NaCl, 60 mM Na2HPO4, 6 mM EDTA, pH 6.8) containing 25% formamide at 34°C. After RNA hybridization, tag-conjugating Cy3 dye were circulated through the microfluidic chip for dye staining. Fluorescence images were collected using a laser scanner (GenePix 4000B, Molecular Device) and digitize analysis was performed using Array-Pro image analysis software (Media Cybernetics, Bethesda, MD). Data were analyzed by first subtracting the background and then normalizing the signals using a LOWESS filter (Locally-weighted Regression) [43]. The differentially expressed miRNAs were defined using the ratio of detected signals log2-fold changes [log2(infected/control)] and the Student’s t-test was used to calculate P values. Those with a log2 ratio >0.5 or ≤ −0.5 and P values <0.05 were considered as differentially expressed miRNAs. The cluster analysis based on the relative expression levels of miRNAs was also carried out.
Target gene prediction, GO enrichment and KEGG pathway analysis
Prediction of putative miRNA targets was performed by using the online softwares, TargetScan (http://www.targetscan.org/) in conjunction with miRanda (http://www.microrna.org/microrna/home.do) and PicTar (http://pictar.mdc-berlin.de/). The enriched GO (http://www.geneontology.org/) terms and KEGG pathways database (http://www.genome.jp/kegg/) for the predicted target genes of the differentially expressed miRNAs were identified by using DAVID (The Database for Annotation, Visualization and Integrated Discovery; http://david.abcc.ncifcrf.gov/) gene annotation tool [44]. Fisher’s two-side exact test and Chi-square test were used to classify the GO categories and KEGG pathway categories, and the false discovery rate (FDR) was also calculated to correct the P values. We chose only GOs and the enriched pathways that had a P value of <0.05 and a FDR of < 0.05. The result may reveal the functions, metabolic pathways or signal transduction pathways significantly associated to predicted targets.
Validation of microarray data with qPCR analysis
Validations of differentially expressed miRNAs were carried out by qPCR with SYBR green [45]. 8 of the most significantly up-regulated miRNAs (hsa-miR-4530, hsa-miR-4492, hsa-miR-4505, hsa-miR-6125, hsa-miR-494-3p, hsa-miR-638, hsa-miR-6743-5p and hsa-miR-4459) and 3 of the most significantly down-regulated microRNAs (hsa-miR-29a-3p, hsa-miR-4443, hsa-miR-27b-5p) were selected as representatives for confirmation. Total RNA, which was used for microarray, was polyadenylated and reverse-transcribed with a poly(T) adapter into cDNAs following the manufacturer’s directions with All in OneTM miRNA qRT-PCR detection kit (GeneCopoeia). For real-time PCR, the miRNA-specific forward primer and the Universal Adaptor PCR Primer as the reverse primer were used. All of primers were purchased from GeneCopoeia (Rockville, Md, USA) and the miRNA-specific forward primers are listed in Table 3. Real-time PCR was runing on Stratagene mx3005p (Agilent Technologies, USA) in triplicate for each sample. The relative amount of each miRNA was normalized against U6 by the 2-ΔΔCt method.
Table 3.
Primers used for qRT-PCR
| Primer name | Forward primer sequence (5′–3′) | Product size (bp) |
|---|---|---|
| Up-regulated miRNAs | ||
| hsa-miR-4530 | CAGGACGGGAGCGAAAA | 66 |
| hsa-miR-4492 | CTGGGCGCGCGCCAAA | 66 |
| hsa-miR-4505 | CTGGGCTGGGACGGAAAA | 68 |
| hsa-miR-6125 | GCGGAGCGGCGGAAAA | 67 |
| hsa-miR-494-3p | GAAACATACACGGGAAACCTCAA | 74 |
| hsa-miR-638 | GGGCGGGTGGCGGCCTAAA | 69 |
| hsa-miR-6743-5p | AGGGACGGGTGGCCCA | 69 |
| hsa-miR-4459 | GCGGAGGAGGTGGAGAAA | 68 |
| Down-regulated miRNAs | ||
| hsa-miR-29a-3p | CACCATCTGAAATCGGTTAA | 72 |
| hsa-miR-4443 | TGGAGGCGTGGGTTTTAAA | 69 |
| hsa-miR-27b-5p | AGAGCTTAGCTGATTGGTGAACAA | 75 |
Statistical analysis
All statistical analysis was performed using the SPSS 17.0 statistical software (SPSS Inc., Chicago, IL, USA). The difference between two groups was determined by a two-tailed Student’s t test, with P values of <0.05 considered to be statistically significant.
Footnotes
Meng Xun and Chao-Feng Ma contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MX designed the study, and performed qRT-PCR. C-FM provided the constructs, and evaluated the results. Cell culture and virus infection were conducted by Q-LD. The manuscript was written by MX. J-RX and Y-HJ critically evaluated the results and revised the manuscript. All authors have read and approved the final version of the manuscript.
Authors’ information
Meng Xun and Chao-Feng Ma are joint first authors.
Contributor Information
Meng Xun, Email: xunmeng@mail.xjtu.edu.cn.
Chao-Feng Ma, Email: mark7447@tom.com.
Quan-Li Du, Email: 4270714@qq.com.
Yan-Hong Ji, Email: jiyanhong@mail.xjtu.edu.cn.
Ji-Ru Xu, Email: xujiru@mail.xjtu.edu.cn.
References
- 1.Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis. 2010;10:778–90. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- 2.Yi L, Lu J, Kung HF, He ML. The virology and developments toward control of human enterovirus 71. Crit Rev Microbiol. 2011;37:313–27. doi: 10.3109/1040841X.2011.580723. [DOI] [PubMed] [Google Scholar]
- 3.Zhu FC, Liang ZL, Li XL, Ge HM, Meng FY, Mao QY, et al. Immunogenicity and safety of an enterovirus 71 vaccine in healthy Chinese children and infants: a randomised, double-blind, placebo-controlled phase 2 clinical trial. Lancet. 2013;381:1037–45. doi: 10.1016/S0140-6736(12)61764-4. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 7.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 8.Palumbo S, Miracco C, Pirtoli L, Comincini S. Emerging roles of microRNA in modulating cell-death processes in malignant glioma. J Cell Physiol. 2014;229:277–86. doi: 10.1002/jcp.24446. [DOI] [PubMed] [Google Scholar]
- 9.Qin W, Shi Y, Zhao B, Yao C, Jin L, Ma J, et al. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One. 2010;5:e9429. doi: 10.1371/journal.pone.0009429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witwer KW, Sisk JM, Gama L, Clements JE. MicroRNA regulation of IFN-beta protein expression: rapid and sensitive modulation of the innate immune response. J Immunol. 2010;184:2369–76. doi: 10.4049/jimmunol.0902712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurzynska-Kokorniak A, Jackowiak P, Figlerowicz M, Figlerowicz M. Human- and virus-encoded microRNAs as potential targets of antiviral therapy. Mini Rev Med Chem. 2009;9:927–37. doi: 10.2174/138955709788681573. [DOI] [PubMed] [Google Scholar]
- 12.Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123–41. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui L, Guo X, Qi Y, Qi X, Ge Y, Shi Z, et al. Identification of microRNAs involved in the host response to enterovirus 71 infection by a deep sequencing approach. J Biomed Biotechnol. 2010;2010:425939. doi: 10.1155/2010/425939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Xie J, Xu X, Wang J, Ao F, Wan Y, et al. MicroRNA-548 down-regulates host antiviral response via direct targeting of IFN-lambda1. Protein Cell. 2013;4:130–41. doi: 10.1007/s13238-012-2081-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lui YL, Tan TL, Woo WH, Timms P, Hafner LM, Tan KH, et al. Enterovirus71 (EV71) utilise host microRNAs to mediate host immune system enhancing survival during infection. PLoS One. 2014;9:e102997. doi: 10.1371/journal.pone.0102997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Chen X, Shi Y, Zhou B, Du C, Liu Y, et al. miR-27a suppresses EV71 replication by directly targeting EGFR. Virus Genes. 2014;49:373–382. doi: 10.1007/s11262-014-1114-4. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Z, Ke X, Wang M, He S, Li Q, Zheng C, et al. Human microRNA hsa-miR-296-5p suppresses enterovirus 71 replication by targeting the viral genome. J Virol. 2013;87:5645–56. doi: 10.1128/JVI.02655-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen BP, Dai HJ, Yang YH, Zhuang Y, Sheng R. MicroRNA-23b inhibits enterovirus 71 replication through downregulation of EV71 VPl protein. Intervirology. 2013;56:195–200. doi: 10.1159/000348504. [DOI] [PubMed] [Google Scholar]
- 19.Bian L, Wang Y, Liu Q, Xia J, Long JE. Prediction of signaling pathways involved in enterovirus 71 infection by algorithm analysis based on miRNA profiles and their target genes. Arch Virol. 2015;160:173–82. doi: 10.1007/s00705-014-2249-2. [DOI] [PubMed] [Google Scholar]
- 20.Kojima M, Sudo H, Kawauchi J, Takizawa S, Kondou S, Nobumasa H, et al. MicroRNA Markers for the Diagnosis of Pancreatic and Biliary-Tract Cancers. PLoS One. 2015;10:e118220. doi: 10.1371/journal.pone.0118220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu W, Han L, Su L, Zhao J, Zhang Y, Zhang S, Zhao B, Miao J. A 3′UTR-Associated RNA, FLJ11812 Maintains Stemness of Human Embryonic Stem Cells by Targeting miR-4459. Stem Cells Dev 2014. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 22.Kwak SY, Yang JS, Kim BY, Bae IH, Han YH. Ionizing radiation-inducible miR-494 promotes glioma cell invasion through EGFR stabilization by targeting p190B rhoGAP. Biochim Biophys Acta. 1843;2014:508–16. doi: 10.1016/j.bbamcr.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Lai L, Chen Q, Song Y, Xu S, Ma F, et al. MicroRNA-494 is required for the accumulation and functions of tumor-expanded myeloid-derived suppressor cells via targeting of PTEN. J Immunol. 2012;188:5500–10. doi: 10.4049/jimmunol.1103505. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Li Z, Kong X, Xie D, Jia Z, Jiang W, et al. Down-regulation of microRNA-494 via loss of SMAD4 increases FOXM1 and beta-catenin signaling in pancreatic ductal adenocarcinoma cells. Gastroenterology. 2014;147:485–97. doi: 10.1053/j.gastro.2014.04.048. [DOI] [PubMed] [Google Scholar]
- 25.Shen PF, Chen XQ, Liao YC, Chen N, Zhou Q, Wei Q, et al. MicroRNA-494-3p targets CXCR4 to suppress the proliferation, invasion, and migration of prostate cancer. Prostate. 2014;74:756–67. doi: 10.1002/pros.22795. [DOI] [PubMed] [Google Scholar]
- 26.Saxena T, Tandon B, Sharma S, Chameettachal S, Ray P, Ray AR, et al. Combined miRNA and mRNA signature identifies key molecular players and pathways involved in chikungunya virus infection in human cells. PLoS One. 2013;8:e79886. doi: 10.1371/journal.pone.0079886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar M, Sharma Y, Bandi S, Gupta S. Endogenous antiviral microRNAs determine permissiveness for hepatitis B virus replication in cultured human fetal and adult hepatocytes. J Med Virol 2015. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 28.Liu X, Wang T, Wakita T, Yang W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology. 2010;398:57–67. doi: 10.1016/j.virol.2009.11.036. [DOI] [PubMed] [Google Scholar]
- 29.Olavarria VH, Valdivia S, Salas B, Villalba M, Sandoval R, Oliva H, et al. ISA virus regulates the generation of reactive oxygen species and p47phox expression in a p38 MAPK-dependent manner in Salmo salar. Mol Immunol. 2015;63:227–234. doi: 10.1016/j.molimm.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 30.Levy DE, Marie IJ, Durbin JE. Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol. 2011;1:476–86. doi: 10.1016/j.coviro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kyriakis JM, Avruch J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol Rev. 2012;92:689–737. doi: 10.1152/physrev.00028.2011. [DOI] [PubMed] [Google Scholar]
- 32.Guma M, Stepniak D, Shaked H, Spehlmann ME, Shenouda S, Cheroutre H, et al. Constitutive intestinal NF-kappaB does not trigger destructive inflammation unless accompanied by MAPK activation. J Exp Med. 2011;208:1889–900. doi: 10.1084/jem.20110242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong J, Shen XH, Chen C, Qiu H, Yang RG. Down-regulation of HIV-1 infection by inhibition of the MAPK signaling pathway. Virol Sin. 2011;26:114–22. doi: 10.1007/s12250-011-3184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillis PA, Okagaki LH, Rice SA. Herpes simplex virus type 1 ICP27 induces p38 mitogen-activated protein kinase signaling and apoptosis in HeLa cells. J Virol. 2009;83:1767–77. doi: 10.1128/JVI.01944-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pleschka S, Wolff T, Ehrhardt C, Hobom G, Planz O, Rapp UR, et al. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat Cell Biol. 2001;3:301–5. doi: 10.1038/35060098. [DOI] [PubMed] [Google Scholar]
- 36.Spaziani A, Alisi A, Sanna D, Balsano C. Role of p38 MAPK and RNA-dependent protein kinase (PKR) in hepatitis C virus core-dependent nuclear delocalization of cyclin B1. J Biol Chem. 2006;281:10983–9. doi: 10.1074/jbc.M512536200. [DOI] [PubMed] [Google Scholar]
- 37.Wang B, Zhang H, Zhu M, Luo Z, Peng Y. MEK1-ERKs signal cascade is required for the replication of Enterovirus 71 (EV71) Antiviral Res. 2012;93:110–7. doi: 10.1016/j.antiviral.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Peng H, Shi M, Zhang L, Li Y, Sun J, Zhang L, et al. Activation of JNK1/2 and p38 MAPK signaling pathways promotes enterovirus 71 infection in immature dendritic cells. BMC Microbiol. 2014;14:147. doi: 10.1186/1471-2180-14-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tung WH, Hsieh HL, Yang CM. Enterovirus 71 induces COX-2 expression via MAPKs, NF-kappaB, and AP-1 in SK-N-SH cells: Role of PGE(2) in viral replication. Cell Signal. 2010;22:234–46. doi: 10.1016/j.cellsig.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 40.Cao C, Gao R, Zhang M, Amelio AL, Fallahi M, Chen Z, et al. Role of LKB1-CRTC1 on glycosylated COX-2 and response to COX-2 inhibition in lung cancer. J Natl Cancer Inst. 2015;107:358. doi: 10.1093/jnci/dju358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tung WH, Lee IT, Hsieh HL, Yang CM. EV71 induces COX-2 expression via c-Src/PDGFR/PI3K/Akt/p42/p44 MAPK/AP-1 and NF-kappaB in rat brain astrocytes. J Cell Physiol. 2010;224:376–86. doi: 10.1002/jcp.22133. [DOI] [PubMed] [Google Scholar]
- 42.Gao X, Gulari E, Zhou X. In situ synthesis of oligonucleotide microarrays. Biopolymers. 2004;73:579–96. doi: 10.1002/bip.20005. [DOI] [PubMed] [Google Scholar]
- 43.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 44.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 45.Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–25. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]


