Abstract
Telomeres are nucleoprotein structures that protect chromosome ends from degradation and recombination. Cancers often have critically shortened telomeres, contributing to genomic instability. Many of these tumors activate telomerase to stabilize telomeric ends and achieve a capacity for unlimited replication. Telomere shortening has been reported in in situ and invasive carcinomas, including breast, and has been associated with disease recurrence after surgical resection. However, previous studies have not evaluated breast cancer subtypes. The objective of this study was to evaluate telomere lengths in different subtypes of breast cancer. Breast carcinomas (n = 103) identified between 2001 and 2010 from patients seen at the Johns Hopkins Hospital were categorized into luminal A (n = 18), luminal B (n = 28), HER-2-positive (n = 20) and triple-negative carcinomas (n = 37) based on tumor characteristics. Telomere lengths were assessed directly at the single cell level by fluorescence in situ hybridization, and patient groups were compared using Fisher’s exact tests. ER-negative status (P = 0.022), PR-negative status (P = 0.008), HER-2-positive status (P = 0.023) and p53-positive status (P = 0.022) were associated with shorter telomere length. A larger proportion of luminal A cancers had normal or long telomere lengths as compared with luminal B cases (P = 0.002), HER-2-positive cases (P = 0.011) or triple-negative cases (P = 0.0003). Luminal B, HER-2-positive and triple-negative cases did not differ significantly. Telomere length was shorter in more aggressive subtypes, such as luminal B, HER-2-positive and triple-negative tumors, suggesting that tumor telomere length may have utility as a prognostic and/or risk marker for breast cancer.
Keywords: breast cancer, HER-2, luminal A, luminal B, subtype, telomere, triple-negative
Telomeres, specialized nucleoprotein structures, function to protect and stabilize the ends of eukaryotic chromosomes by preventing chromosome fusions, exonucleolytic degradation and masking telomere-induced double-strand DNA break damage signals. However, telomeres can become critically shortened, and dysfunctional, by several mechanisms including incomplete replication, wherein they are shortened during each cycle of chromosome replication.1 In normal cells, telomere shortening induces tumor-suppressive checkpoint pathways, such as cellular senescence or apoptosis, which halts cell-cycle progression before telomeres become destabilized.2,3 However, abrogation of these checkpoints leads to continued cell division. Consequently, there is a limit to the number of doublings somatic cells can undergo before triggering successive rounds of chromosome breakage–fusion–bridge cycles, thus driving chromosome amplification and loss of structural rearrangements.4
It is well established that telomere shortening is present in the majority of in situ and invasive carcinomas,5 including breast.6,7 Thus, telomere shortening is an early event in malignant transformation. In addition to tumor initiation, short dysfunctional telomeres may also affect disease progression. Previous studies have shown reduced telomere length in grade III tumors,8 and reduced telomere DNA content, a surrogate for telomere length, correlates with aneuploidy and lymph node metastasis,9 and shorter telomeres were associated with higher stage and histological grade.10 A retrospective study (n = 77) showed that short telomeres were associated with tumor size, nodal involvement, TNM stage and was an independent predictor of 5-year overall survival and 5-year breast cancer-free survival.11 A larger population-based prospective study (n = 530) showed that short telomeres conferred a relative hazard of breast cancer recurrence of 2.88, after adjusting for prognostic factors and adjuvant therapies.12 However, none of these studies took into consideration specific breast cancer subtypes that are now used to help guide treatment decisions.
Four main molecular classes of breast cancer were first identified by gene expression profiling.13 On further validation, these subtypes correlate well with the clinical characterization of ER, PR and HER-2 protein expression status.14,15 These groups are luminal A carcinomas (ER and PR positive, HER-2 negative), luminal B carcinomas (ER or PR positive, HER-2 positive), HER-2 carcinomas (ER and PR negative, HER-2 positive) and triple-negative carcinomas (ER/PR/HER-2 negative). Important for clinical care, these subtypes predict prognosis and therapeutic response.16–18 Luminal A tumors respond well to selective estrogen receptor modulators, such as tamoxifen.19 Luminal B tumors tend to be less sensitive to hormonal therapies and thus carry a worse prognosis than do luminal A tumors. HER-2-positive cancers tend to be high grade, aggressive and carry a poor prognosis; however, they respond well to trastuzumab, an anti-HER-2 monoclonal antibody. Conversely, triple-negative carcinomas are extremely aggressive and currently do not respond to any hormonal or antibody-based targeted therapy.20
The objective of this study was to evaluate telomere lengths in subtypes of breast carcinoma. This was accomplished using a fluorescence in situ hybridization (FISH) assay that allows telomere length assessments in formalin-fixed, paraffin-embedded archival material, while providing single-cell resolution and keeping the tissue architecture intact. We evaluated telomere length in 103 cases of invasive breast cancer and correlated telomere length with established molecular markers.
Materials and methods
Case Selection
All of the 103 cases evaluated in this study were incident breast carcinomas that were surgically resected at the Johns Hopkins Hospital from 2001 to 2010. At the time of resection, specimens were freshly sectioned, fixed overnight in 10% neutral buffered formalin and uniformly processed. Women treated with neoadjuvant chemotherapy were not included in this study. Clinical characteristics, such as age at diagnosis, weight, ethnicity, parity status and menopausal status were obtained from patient’s medical records. Pathological characteristics, such as Elston grade and TNM stage, were obtained from patient’s pathology records. This study was approved by the Institutional Review Board of the Johns Hopkins School of Medicine.
Tissue Microarray Construction
For the majority of cases (n = 72), tissue microarrays were constructed as described previously.21,22 In brief, each tissue microarray consisted of 99 tissue cores, each 1.4mm in diameter. These cores were arranged in 9 rows and 11 columns. Column 6 consisted of various unrelated control tissues, leaving 90 cores on the array for breast carcinoma samples. For each carcinoma case, 5 areas were identified on the hematoxylin and eosin slides, punched from the corresponding donor blocks and placed on the array. Among the five samples of each case, we attempted to include the normal tissue and carcinoma in situ in one sample if possible, leaving four to five cores of invasive ductal carcinoma per case. Any case that displayed a processing artifact was excluded. In addition, as not to exhaust the tissue, small-size (<1 cm) cases were excluded. For an additional 31 cases, whole sections from surgical specimen blocks were used.
Immunohistochemistry
All immunohistochemistry was performed as described previously.21,22 The slides were reviewed by two pathologists (APS and PA) to confirm the interpretation of results. Immunohistochemistry for ER (monoclonal, 1:1 dilution, Ventana) and PR (monoclonal, 1:60 dilution, Dako) were performed on all cases as part of a routine panel. Cases demonstrating weak, moderate or strong nuclear labeling for ER or PR in >1% of cells were considered ER positive or PR positive, respectively. Immunohistochemistry for HER-2 was performed on all cases as part of a routine panel using the Dako Herceptest kit and following the manufacturer’s guidelines. Cases were scored using established criteria as 0 or 1 + (negative), 2 + (equivocal) and 3 + (positive). FISH analysis for HER-2 amplification was performed on all cases with 2 + (equivocal) immunohistochemistry results using the Path Vysion kit (Vysis-Abbott Molecular). Cases with either a 3 + (strong positive) immunohistochemistry score or a HER-2 FISH amplification ratio >4 were considered HER-2-positive. Cases with low-level amplification (ratios 2.2–4.0) were excluded from this study because of their uncertain clinical significance. To determine the basal subtype, CK 5/6 (monoclonal, Dako) and EGFR (monoclonal, 1:50 dilution, Zymed) immunohistochemistry was performed on whole sections from a subset of cases (29 of 37), which were negative for ER, PR and HER-2. For CK 5/6, cases were scored on the basis of the percentage of positive cells: 1 + (1–25%), 2 + (26–50%), 3 + (51–75%) and 4 + (76–100%). Cases with membranous or cytoplasmic labeling in >25% of neoplastic cells were considered positive. For EGFR, cases were scored on the basis of the percentage of positive cells: 1 + (1–25%), 2 + (26–50%), 3 + (51–75%) and 4 + (76–100%). Any strong membranous staining for EGFR was considered positive, generally labeling 10–50% of neoplastic cells. p53 (monoclonal, Ventana) and Ki-67 (monoclonal, Ventana) immunohistochemistry were also performed and only nuclear labeling was scored. For p53, labeling of >30% of nuclei was considered aberrant overexpression, and labeling of <30% of nuclei was considered negative for aberrant overexpression; 30% labeling of nuclei was considered equivocal. For Ki-67, the labeling of ≥20% of nuclei was considered high and the labeling of <20% of nuclei was considered low.
Telomere FISH
Telomere lengths were assessed by fluorescence staining for telomeric DNA as described previously.6,23 In brief, deparaffinized slides were hydrated through a graded ethanol series, placed in deionized water, followed by deionized water plus 0.1% Tween-20. Slides were then placed in citrate buffer (catalog no. H-3300; Vector Laboratories), steamed for 14 min (Black and Decker Handy Steamer Plus; Black and Decker), removed and allowed to cool at room temperature for 5 min. Slides were then placed in PBS with Tween (catalog no. P-3563; Sigma Chemical Co.) for 5 min. Slides were thoroughly rinsed with deionized water, followed by 95% ethanol for 5 min and then air dried. In all 25 µl of Cy3-labeled telomere-specific peptide nucleic acid (0.3 µg/ml peptide nucleic acid in 70% formamide, 10 mmol/l Tris, pH 7.5, 0.5% B/M Blocking reagent (catalog no. 1814-320; Boehringer-Mannheim)) was applied to the sample, which was then coverslipped, and denaturation was performed by incubation for 4 min at 83°C. Slides were then transferred to a dark, closed container for hybridization at room temperature for 2 h. Cover-slips were then carefully removed and slides were washed twice in peptide nucleic acid wash solution (70% formamide, 10mmol/l Tris, pH 7.5, 0.1% albumin (from 30% albumin solution, catalog no. A-7284; Sigma Chemical Co.)) for 15 min each, and then rinsed in PBS with Tween and thoroughly rinsed in deionized water. Slides were drained and counterstained with 4′-6-diamidino-2-phenylindole (DAPI) (500 ng/ml in deionized water, Sigma Chemical Co. catalog no. D-8417) for 5 min at room temperature, mounted with Prolong anti-fade mounting medium (catalog no. P-7481; Molecular Probes) and then imaged. The peptide nucleic acid probe complementary to the mammalian telomere repeat sequence was obtained from Applied Bio-systems, and has the sequence (N terminus to C terminus) CCCTAACCCTAACCCTAA with an N-terminal covalently linked Cy3 fluorescent dye. As a positive control for hybridization efficiency, an FITC-labeled peptide nucleic acid probe having the sequence ATTCGTTGGAAACGGGAwith specificity for human centromeric DNA repeats (CENP-B-binding sequence) was also included in the hybridization solution.24
Microscopy and Telomere Length Assessment
Slides were imaged using a Nikon 50i epifluorescence microscope equipped with X-Cite series 120 illuminator (EXFO Photonics Solutions Inc., Ontario, Canada). Fluorescence excitation/emission filters were as follows: Cy3 excitation, 546 nm/10nm BP; emission, 578nm LP (Carl Zeiss Inc.); DAPI excitation, 330 nm; emission, 400nm using an XF02 fluorescence set (Omega Optical, Brattleboro, VT, USA); Alexa Fluor 488 excitation, 475nm; emission, 535nm by a combination of 475RDF40 and 535RDF45 filters (Omega Optical). Gray scale images of representative regions were captured for presentation using Nikon NIS-Elements software and an attached Photometrics CoolsnapEZ digital camera, pseudo-colored and merged. Integration times typically ranged from 500 to 800 ms for Cy3 (telomere) and FITC (centromere) signal capture, and 50–100 ms for the DAPI counterstain. Telomere lengths were qualitatively scored by direct visual assessment of stained slides, comparing telomere signals from tumor cells with telomere signals from benign cells (stromal cells and/or myoepithelial cells) from the same case. In all cases, signals from benign cells were considered 3 +. Telomeres in tumor cells of different cases ranged from short (0 +, 1 +, or 2 +) to normal (3 +) to long (4 +, 5 +).
Statistical Analysis
For all analyses, normal and long telomere groups were combined and compared with the short telomere group. Results were compared using two-sided Fisher’s exact tests. P-values <0.05 were considered to be significant. SAS 9.2 and JMP− statistical packages (SAS Institute, Cary, NC, USA) were used for all analyses.
Results
Clinical and Pathological Characteristics
Table 1 lists the clinical and pathological characteristics of the study population. Of the 103 subjects included in this study, the mean age at diagnosis for all patients was 56 years (range: 30–94 years). Patients were predominantly Caucasian (57%) or African American (31%). Elston grading of tumors showed that 4% were grade I, 23% were grade II and 73% were grade III. According to the established criteria of the American Joint Committee on Cancer (AJCC 2007), 21% were stage I, 59% were stage II, 18% were stage III and 1% were stage IV. Interestingly, the short telomere length group tended to be younger and contained a greater proportion of African-American women when compared with the normal/long group; however, these differences did not reach statistical significance.
Table 1.
Clinical and pathological characteristics for all patients and stratified by telomere length
| Characteristics | All subjects (n = 103) | Telomere lengtha | |
|---|---|---|---|
| Short (n = 88) | Normal/long (n = 15) | ||
| Age at diagnosis (years) | |||
| Mean (range) | 56 (30–94) | 55 (30–92) | 61 (34–94) |
| Weight (kg)b | |||
| Mean (range) | 79.1 (44.0–163.5) | 79.9 (46.8–163.5) | 73.8 (44.0–97.7) |
| Ethnicity | |||
| Caucasian | 59 | 46 | 13 |
| African American | 32 | 30 | 2 |
| Other | 12 | 12 | 0 |
| Parity status | |||
| Yes | 82 | 73 | 9 |
| No | 20 | 14 | 6 |
| Missing | 1 | 1 | 0 |
| Menopausal status | |||
| Pre-menopausal | 41 | 35 | 6 |
| Post-menopausal | 60 | 52 | 8 |
| Missing/uncertain | 2 | 1 | 1 |
| Elston grade | |||
| 1 | 4 | 1 | 3 |
| 2 | 24 | 20 | 4 |
| 3 | 75 | 67 | 8 |
| TNM stage | |||
| I | 22 | 22 | 0 |
| II | 61 | 52 | 9 |
| III | 19 | 13 | 6 |
| IV | 1 | 1 | 0 |
There are no statistically significant differences in any of the clinical or pathological characteristics between the telomere length groups.
In all, 18 women are missing weight data.
Telomere-FISH in Breast Tumors
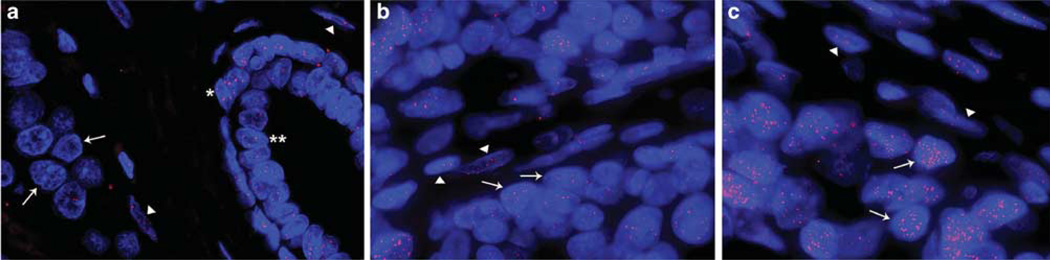
Telomere length was qualitatively scored and grouped into short (n = 88), normal (n = 13) or long (n = 2) categories. Figure 1a shows a representative example of breast cancer with short telomeres as indicated by diminished telomere signals in cancer cells when compared with either stromal cells or myoepithelial cells in an adjacent terminal ductal lobular unit. As shown in Figure 1b, cancer cells from this breast cancer show comparable telomere intensities to that observed in the surrounding benign stroma. Figure 1c, shows an example of a breast tumor with increased telomere signals in cancer cells when compared with the surrounding benign stromal cells. Table 1 shows the clinical and pathological characteristics for the subset of patients with short telomeres (n = 88) and the subset of patients with either normal or long telomeres (n = 15).
Figure 1.
Telomere length analysis by FISH in breast adenocarcinomas. Three representative examples of cases showing short, normal or long telomere lengths in cancer cells are shown. (a) This case shows strikingly diminished telomere signals in tumor cells as compared to the surrounding benign stroma and in an adjacent terminal ductal lobular unit with myoepithelial (*) and luminal (**) cells. (b) This case displays comparable telomere intensities in tumor cells with those observed in the surrounding benign stroma. (c) In this case, cancer cells show extremely bright telomere signals in cancer cells when compared with the surrounding benign stroma. In all the images, the DNA is stained with DAPI (blue) and telomere DNA is stained with the Cy3-labeled telomere-specific peptide nucleic acid probe (red). It is noteworthy that the centromere DNA, stained with the FITC-labeled centromere-specific peptide nucleic acid probe, has been omitted from the image to emphasize the differences in telomere lengths. In all panels, the arrows point to cancer cells and the arrowheads point to benign stromal cells. Original magnification, × 400.
Hormone Receptor Expression Characteristics
Table 2 shows tumor characteristics including ER status, PR status, HER-2, p53 and Ki-67 stratified by telomere length. ER-negative (P = 0.022), PR-negative (P = 0.008) and HER-2-positive (P = 0.023) tumors were significantly more likely to have a greater fraction of short telomeres when compared with the normal/long telomere group. In addition, there was a significant increase in the proportion of p53-positive tumors in the short telomere category (P = 0.022). Shorter telomere length was also more prevalent in tumors with an increased Ki-67 index (≥20% of cells positive), but it did not reach statistical significance.
Table 2.
Association between ER, PR, HER-2, p53 and Ki-67 expression characteristics stratified by telomere length
| Characteristics | Telomere length | P-value | |
|---|---|---|---|
| Short | Normal/long | ||
| ER status | |||
| Positive | 34 | 11 | |
| Negative | 54 | 4 | 0.022 |
| PR status | |||
| Positive | 30 | 11 | |
| Negative | 58 | 4 | 0.008 |
| HER-2 | |||
| Positive | 53 | 4 | |
| Negative | 35 | 11 | 0.023 |
| p53a | |||
| Positive | 40 | 2 | |
| Negative | 46 | 13 | 0.022 |
| Ki-67b | |||
| High (≥20%) | 70 | 8 | |
| Low (<20%) | 17 | 6 | 0.082 |
Two patients with equivocal p53 staining were not included in the analysis.
Two patients are missing Ki-67 data.
Subtype Characterization and Telomere Length Assessment
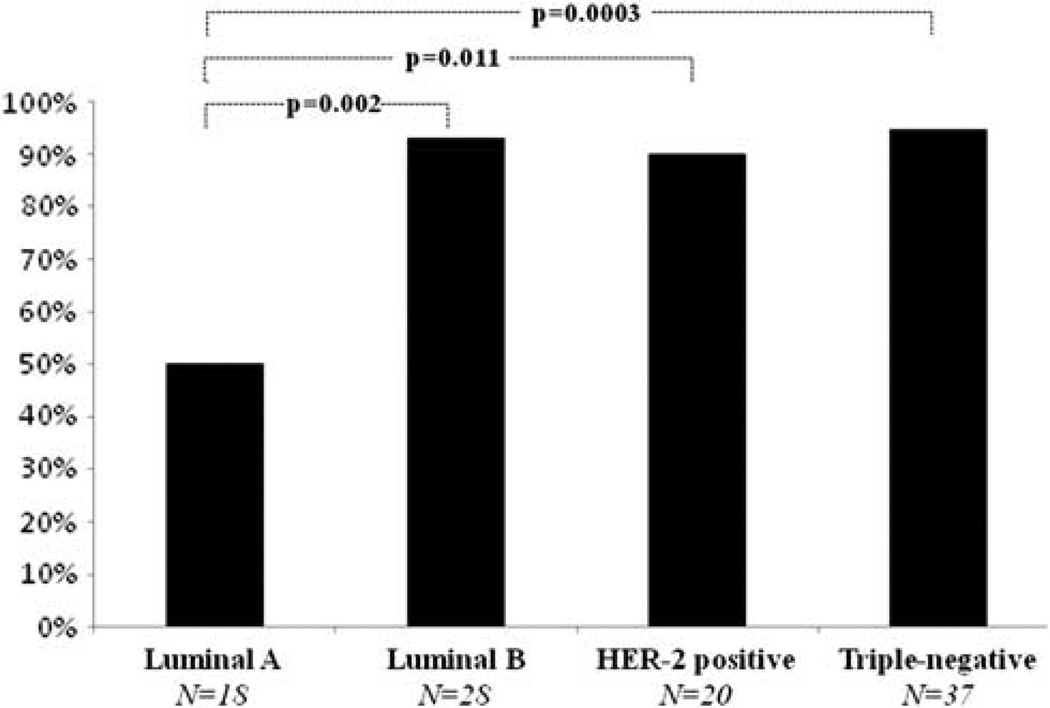
Cases were categorized into one of four groups; luminal A, luminal B, HER-2-positive or triple-negative. In all, 18 cases were ER/PR positive/HER-2 negative, and were considered luminal A cases. There were 28 luminal B cases that were HER-2-positive and ER and/or PR positive. A total of 20 tumors were HER-2-positive and negative for ER and PR, respectively, and were considered HER-2-positive cases. In addition, there were 37 triple-negative carcinoma cases which were negative for ER, PR and HER-2. For 29 of the triple-negative cases, immunohistochemical data on CK 5/6 and EGFR expression were available, allowing assessment of the basal phenotype. In all, 18 of these cases showed staining for CK 5/6 and/or EGFR (basal) and 11 cases were CK 5/6 and EGFR negative (non-basal). Within this triple-negative group, there was no difference in the proportion of cases with short telomeres between the subset of cases determined to be basal (94%) and non-basal (91%) phenotypes. As shown in Figure 2, luminal B cancers (93%), HER-2 cancers (90%) and triple-negative cancers (95%) had an increased proportion of cases with short telomeres when compared with luminal A cancers (50%); these were all statistically significant differences (P = 0.002, P = 0.011 and P = 0.0003, respectively).
Figure 2.
Proportion of cases with short telomeres among different subtypes of breast cancer. P-values were determined using two-sided Fisher’s exact tests.
Discussion
The first principal conclusion emerging from this study is the observation that telomere shortening is associated with other established breast cancer prognostic factors. It is well established that breast cancer patients with tumors that are ER and/or PR negative have an increased risk of mortality.25–27 We observed an increased proportion of tumors in the short telomere group that were negative for ER and PR. Although temporality cannot be determined, these results suggest that telomere shortening may contribute to the selection of cells capable of growing in the absence of hormone receptors. Another molecular marker associated with poor breast cancer prognosis is the presence of mutations in the p53 gene, predominantly missense mutations leading to conformational alterations of the protein and accumulation in the tumor cell nuclei.28–30 We observed an increased proportion of tumors in the short telomere group that were p53 positive, suggesting that less-aggressive tumors are characterized by normal length telomeres and no mutations in the p53 gene. Conversely, short, dysfunctional telomeres may provide a strong selective pressure for abrogation of the p53 pathway. Normally, critical telomere shortening leads to p53-dependent tumor-suppressive cellular responses, such as cellular senescence and apoptosis.2 However, if this checkpoint is abrogated and the telomeres are partially stabilized by upregulation of telomerase, then further proliferation occurs and genomic instability may accumulate.31,32
The second principal conclusion emerging from this study is that telomere shortening occurs in the vast majority of luminal B, HER-2 and triple-negative tumors, but in a smaller fraction of luminal A type tumors. A recent large patient cohort demonstrated that luminal A tumors are associated with a lower risk of local or regional recurrence when compared with other molecular subtypes.18 It has long been recognized that breast adenocarcinomas are characterized by genomic instability. The molecular mechanisms leading to genomic instability are not fully elucidated; however, one potential initiator of genomic instability is telomere dysfunction due to critical telomere shortening.4,33 Thus, telomere shortening may be reflective of the degree of underlying genomic instability, a feature shared by higher-grade HER-2 and triple-negative carcinomas.34 Another possibility is that telomere shortening follows tumor progression as evidenced by the association with higher stage and histological grade.8,10,11 It is noteworthy that recent data have demonstrated that not only can short telomeres lead to telomere dysfunction but abnormally long telomeres may also do so.35,36 In this context, we previously demonstrated the presence of alternative lengthening of telomeres, a recombination-based mechanism that lengthens telomeres, in three cases of breast carcinoma.21 Interestingly, all three cases were also HER-2-positive, suggesting a possible common underlying mechanism. The significance of long telomeres in cancers lacking evidence of the alternative lengthening of telomeres phenotype is currently not clear and would need to be addressed in future studies.
This study is the first assessment of telomere lengths in breast cancer subtypes. In addition, we analyzed telomere lengths directly, at the single cell level, within breast cancer tissues using a FISH assay. Previous studies have measured telomeres using bulk assays such as Southern blot and terminal restriction fragment analysis,8 quantitative PCR10 or with a chemiluminescent-based slot blot assay that measures telomere DNA content, a proxy of telomere length.9,11,12 These types of measurements do not allow for single cell resolution. Study limitations include the absence of small tumors (<1 cm), thus selecting for higher stage and histological grade cases, and the limited demographic and lifestyle information, as this was obtained retrospectively. In addition, the lack of complete data on follow-up time, because of the fact that part, but not all, of the patients’ treatment occurred at our Institution, prevented us from determining an association between telomere length and outcome.
In conclusion, we have demonstrated that telomere lengths vary among different subtypes of breast cancer in a manner consistent with their aggressiveness. Prospective studies are required to further evaluate the usefulness of telomere length as a prognostic and/or predictive marker of disease progression and treatment response within the various subtypes and determine temporality.
Acknowledgements
This work was supported by a National Institutes of Health (NIH) Postdoctoral Training Fellowship (T32 CA067751) in Pathobiology of Cancer (to CMH), a Department of Defense Breast Cancer Research Program (W81XWH-09-1-0650) Postdoctoral Fellowship (to CMH) and by the National Cancer Institute with a SPORE in Breast Cancer Grant (P50 CA88843) to The Johns Hopkins Hospital.
Footnotes
Disclosure/conflict of interest
The authors declare no conflict of interest.
References
- 1.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 2.Vaziri H. Critical telomere shortening regulated by the ataxia-telangiectasia gene acts as a DNA damage signal leading to activation of p53 protein and limited lifespan of human diploid fibroblasts. A review. Biochemistry (Mosc) 1997;62:1306–1310. [PubMed] [Google Scholar]
- 3.Wright WE, Shay JW. Cellular senescence as a tumor-protection mechanism: the essential role of counting. Curr Opin Genet Dev. 2001;11:98–103. doi: 10.1016/s0959-437x(00)00163-5. [DOI] [PubMed] [Google Scholar]
- 4.Feldser DM, Hackett JA, Greider CW. Telomere dysfunction and the initiation of genome instability. Nat Rev Cancer. 2003;3:623–627. doi: 10.1038/nrc1142. [DOI] [PubMed] [Google Scholar]
- 5.Meeker AK, Hicks JL, Iacobuzio-Donahue CA, et al. Telomere length abnormalities occur early in the initiation of epithelial carcinogenesis. Clin Cancer Res. 2004;10:3317–3326. doi: 10.1158/1078-0432.CCR-0984-03. [DOI] [PubMed] [Google Scholar]
- 6.Meeker AK, Hicks JL, Gabrielson E, et al. Telomere shortening occurs in subsets of normal breast epithelium as well as in situ and invasive carcinoma. Am J Pathol. 2004;164:925–935. doi: 10.1016/S0002-9440(10)63180-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin K, de Solorzano CO, Knowles D, et al. In situ analyses of genome instability in breast cancer. Nat Genet. 2004;36:984–988. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- 8.Odagiri E, Kanada N, Jibiki K, et al. Reduction of telomeric length and c-erbB-2 gene amplification in human breast cancer, fibroadenoma, and gynecomastia. Relationship to histologic grade and clinical parameters. Cancer. 1994;73:2978–2984. doi: 10.1002/1097-0142(19940615)73:12<2978::aid-cncr2820731215>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 9.Griffith JK, Bryant JE, Fordyce CA, et al. Reduced telomere DNA content is correlated with genomic instability and metastasis in invasive human breast carcinoma. Breast Cancer Res Treat. 1999;54:59–64. doi: 10.1023/a:1006128228761. [DOI] [PubMed] [Google Scholar]
- 10.Radpour R, Barekati Z, Haghighi MM, et al. Correlation of telomere length shortening with promoter methylation profile of p16/Rb and p53/p21 pathways in breast cancer. Mod Pathol. 2010;23:763–772. doi: 10.1038/modpathol.2009.195. [DOI] [PubMed] [Google Scholar]
- 11.Fordyce CA, Heaphy CM, Bisoffi M, et al. Telomere content correlates with stage and prognosis in breast cancer. Breast Cancer Res Treat. 2006;99:193–202. doi: 10.1007/s10549-006-9204-1. [DOI] [PubMed] [Google Scholar]
- 12.Heaphy CM, Baumgartner KB, Bisoffi M, et al. Telomere DNA content predicts breast cancer-free survival interval. Clin Cancer Res. 2007;13:7037–7043. doi: 10.1158/1078-0432.CCR-07-0432. [DOI] [PubMed] [Google Scholar]
- 13.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 14.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 16.Sotiriou C, Neo SY, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voduc KD, Cheang MC, Tyldesley S, et al. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol. 2010;28:1684–1691. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- 19.Anderson WF, Chatterjee N, Ershler WB, et al. Estrogen receptor breast cancer phenotypes in the Surveillance, Epidemiology, and End Results database. Breast Cancer Res Treat. 2002;76:27–36. doi: 10.1023/a:1020299707510. [DOI] [PubMed] [Google Scholar]
- 20.Schneider BP, Winer EP, Foulkes WD, et al. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14:8010–8018. doi: 10.1158/1078-0432.CCR-08-1208. [DOI] [PubMed] [Google Scholar]
- 21.Subhawong AP, Heaphy CM, Argani P, et al. The alternative lengthening of telomeres phenotype in breast carcinoma is associated with HER-2 overexpression. Mod Pathol. 2009;22:1423–1431. doi: 10.1038/modpathol.2009.125. [DOI] [PubMed] [Google Scholar]
- 22.Subhawong AP, Subhawong T, Nassar H, et al. Most basal-like breast carcinomas demonstrate the same Rb-/p16+ immunophenotype as the HPV-related poorly differentiated squamous cell carcinomas which they resemble morphologically. Am J Surg Pathol. 2009;33:163–175. doi: 10.1097/PAS.0b013e31817f9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meeker AK, Gage WR, Hicks JL, et al. Telomere length assessment in human archival tissues: combined telomere fluorescence in situ hybridization and immunostaining. Am J Pathol. 2002;160:1259–1268. doi: 10.1016/S0002-9440(10)62553-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, Hong YK, Ontiveros SD, et al. Single base discrimination of CENP-B repeats on mouse and human chromosomes with PNA-FISH. Mamm Genome. 1999;10:13–18. doi: 10.1007/s003359900934. [DOI] [PubMed] [Google Scholar]
- 25.Parl FF, Schmidt BP, Dupont WD, et al. Prognostic significance of estrogen receptor status in breast cancer in relation to tumor stage, axillary node metastasis, and histopathologic grading. Cancer. 1984;54:2237–2242. doi: 10.1002/1097-0142(19841115)54:10<2237::aid-cncr2820541029>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 26.Anderson WF, Chu KC, Chatterjee N, et al. Tumor variants by hormone receptor expression in white patients with node-negative breast cancer from the surveillance, epidemiology, and end results database. J Clin Oncol. 2001;19:18–27. doi: 10.1200/JCO.2001.19.1.18. [DOI] [PubMed] [Google Scholar]
- 27.Dunnwald LK, Rossing MA, Li CI. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res. 2007;9:R6. doi: 10.1186/bcr1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuda H. Gene and chromosomal alterations in sporadic breast cancer: correlation with histopathological features and implications for genesis and progression. Breast Cancer. 2009;16:186–201. doi: 10.1007/s12282-009-0124-x. [DOI] [PubMed] [Google Scholar]
- 29.Soussi T, Legros Y, Lubin R, et al. Multifactorial analysis of p53 alteration in human cancer: a review. Int J Cancer. 1994;57:1–9. doi: 10.1002/ijc.2910570102. [DOI] [PubMed] [Google Scholar]
- 30.Sjogren S, Inganas M, Norberg T, et al. The p53 gene in breast cancer: prognostic value of complementary DNA sequencing versus immunohistochemistry. J Natl Cancer Inst. 1996;88:173–182. doi: 10.1093/jnci/88.3-4.173. [DOI] [PubMed] [Google Scholar]
- 31.Romanov SR, Kozakiewicz BK, Holst CR, et al. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- 32.Holst CR, Nuovo GJ, Esteller M, et al. Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res. 2003;63:1596–1601. [PubMed] [Google Scholar]
- 33.Artandi SE, DePinho RA. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr Opin Genet Dev. 2000;10:39–46. doi: 10.1016/s0959-437x(99)00047-7. [DOI] [PubMed] [Google Scholar]
- 34.Hu X, Stern HM, Ge L, et al. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res. 2009;7:511–522. doi: 10.1158/1541-7786.MCR-08-0107. [DOI] [PubMed] [Google Scholar]
- 35.Reddel RR. Alternative lengthening of telomeres, telomerase, and cancer. Cancer Lett. 2003;194:155–162. doi: 10.1016/s0304-3835(02)00702-4. [DOI] [PubMed] [Google Scholar]
- 36.Stewart SA. Telomere maintenance and tumorigenesis: an ‘ALT’ernative road. Curr Mol Med. 2005;5:253–257. doi: 10.2174/1566524053586653. [DOI] [PubMed] [Google Scholar]