Summary
Ribosome biogenesis can modulate protein synthesis; a process heavily relied upon for cancer cell proliferation. In this study, involvement of large subunit ribosomal proteins (RPLs) in melanoma has been dissected and RPLs categorized based on modulation of cell proliferation and therapeutic targeting potential. Based on these results, two categories of RPLs were identified; the first causing negligible effects on cell viability, p53 expression, and protein translation; while the second category decreased cell viability and inhibited protein synthesis mediated with or without p53 protein stabilization. RPL13 represents the second category, where siRNA-mediated targeting inhibited tumor development through decreased cellular proliferation. Mechanistically, decreased RPL13 levels increased p53 stability mediated by RPL5 and RPL11 binding to and preventing MDM2 from targeting p53 for degradation. The consequence was p53-dependent cell cycle arrest and decreased protein translation. Thus, targeting certain category 2 RPL proteins can inhibit melanoma tumor development mediated through the MDM2-p53 pathway.
Keywords: p53, cell cycle, melanoma, ribosomal proteins, RP-MDM2-p53, protein synthesis
Introduction
Although melanoma accounts for approximately 5% of skin cancer cases, it is the cause of 75% of skin cancer-related deaths (Tarver, 2012). For the past 30 years, the incidence and mortality rates of melanomas continued to rise (Siegel et al., 2012). Effective treatments that would lead to a cure have been lacking but recent progress has been made with the approval of new targeted therapies (Bollag et al., 2012), most recently with ipilimumab (Hodi et al., 2010), a human immunoglobulin specific for CTLA-4, and vemurafenib (Flaherty et al., 2010) which targets mutant V600EBRaf, a mutation present in approximately 50% of melanoma patients. These therapies are only effective for a certain proportion of melanoma patients (Rughani et al., 2013) and most eventually relapse with drug resistant recurrent disease (Yauch and Settleman, 2012). With these limitations, the search continues for more effective treatment options.
The ribosome is an essential component of all cells that is necessary for protein synthesis. The eukaryotic ribosome contains approximately 79 ribosomal proteins in addition to four ribosomal RNAs (rRNAs), and is organized into an 80S structure consisting of the 60S large and 40S small subunits (Ben-Shem et al., 2011). Oncogenes and tumor suppressors can directly regulate ribosome production and protein translation (Ruggero and Pandolfi, 2003). Furthermore, the rate of protein synthesis is directly proportional to the rate of cellular proliferation and growth (Baxter and Stanners, 1978) (Rudra and Warner, 2004) (Thomas, 2000). Thus, cancer cells rely heavily on protein synthesis for survival.
Ribosome biogenesis is the process by which ribosomes are synthesized. It constitutes a highly complex process involving assembling rRNA and ribosomal proteins, requiring substantial energy (Warner et al., 2001). A novel pathway termed the ribosomal protein-MDM2-p53 (RP-MDM2-p53) pathway has begun to be elucidated that serves as a stress response to the disruption of ribosome biogenesis (Deisenroth and Zhang, 2010). Upon any stress to ribosome biogenesis, this pathway is activated, and certain ribosomal proteins, such as RPL5 (Dai and Lu, 2004), RPL11 (Zhang et al., 2003), RPL23 (Dai et al., 2004), RPL26 (Zhang et al., 2010) and RPS7 (Zhu et al., 2009) bind to MDM2 and inhibit ubiquitination of p53. This causes stabilization and accumulation of p53 in cells, resulting in cell cycle arrest, apoptosis, or senescence (Vogelstein et al., 2000). In fact, depletion of certain ribosomal proteins in mice produced a variety of phenotypes that could be restored by the deletion of p53 (McGowan et al., 2008; Terzian and Box, 2013).
The greater reliance of cancer cells on protein synthesis and the presence of mechanisms to activate a tumor suppressor during the surveillance of ribosome biogenesis provide a foundation from which to work in order to develop potential therapeutic approaches to target melanoma. Ribosomal proteins make up, along with ribosomal RNA, the large and small subunits of the ribosome. Once thought to be exclusively involved in protein synthesis, ribosomal proteins are being identified in extra-ribosomal functions, such as in RP-MDM2-p53 pathway regulation (Warner and McIntosh, 2009), mRNA translation regulation (Kondrashov et al., 2011; Mazumder et al., 2003), and modulation of regulatory proteins (Wan et al., 2007). Since ribosomal proteins are likely linked to p53 through the RP-MDM2-p53 pathway and to translation as part of the ribosome, they may offer a unique therapeutic potential in a diverse array of tumor types.
In this study, the involvement of RPLs has been comprehensively dissected for the first time and categories defined based on role in melanoma cells, as well as efficacy for therapeutically targeting these proteins has been evaluated. RPLs can be grouped into two categories based on cellular responses upon siRNA knockdown. Category 1 contains RPLs that do not participate in the RP-MDM2-p53 pathway. Targeting these RPLs caused little change in cell viability, no change in p53 expression, and little inhibition of protein synthesis. Category 2 contains RPLs that participate in the RP-MDM2-p53 pathway. Targeting these RPLs, comprising three-fourths of RPLs, decreased cell viability and caused an inhibition of protein synthesis. These RPLs can be subcategorized based on ability (category 2A) or inability (category 2B) of the various proteins to increase p53 stability. RPL13 was selected as a candidate to represent the 2A category to determine the therapeutic potential of targeting this class of proteins. siRNA mediated knockdown of RPL13 significantly decreased tumor growth mediated through decreased cell proliferation. Targeting the 2A category of RPLs caused increased p53 stability. RPL13 silencing demonstrated that the effects were mediated by the RP-MDM2-p53 pathway and by decreasing protein synthesis. Thus, targeting of category 2A RPL proteins has been shown to be effective for inhibiting cancer development, which is mediated by activating the RP-MDM2-p53 pathway and decreasing protein synthesis.
Results
Targeting ribosomal proteins in the large subunit decreased melanoma cell viability
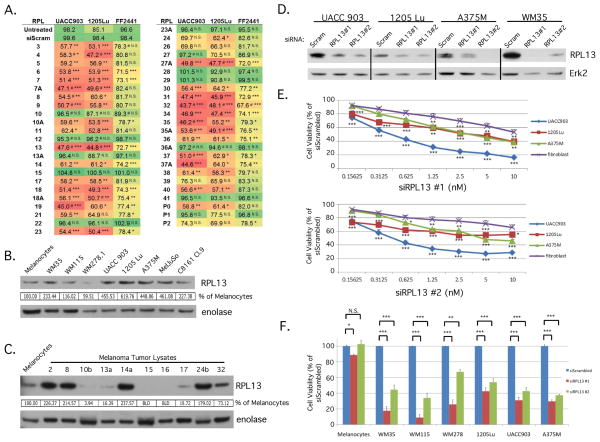
To dissect the role of large subunit ribosomal proteins (RPLs) for the first time in melanoma and evaluate the therapeutic potential of targeting these proteins, a siRNA screen was undertaken to observe the effects of inhibiting each individual member of the RPL gene family. Knocking down RPLs in two metastatic melanoma cell lines UACC 903 and 1205 Lu, led to decreased melanoma cell viability for 74% and 79% of the RPL genes, respectively (Fig. 1A). Targeting these same genes in the normal foreskin fibroblast FF2441 cell line produced a less inhibitory response than in the melanoma cell lines, with an average difference of 22% and a maximum difference of 35% between fibroblast and melanoma cell viability. A subset of RPLs were knocked down in melanocytes and demonstrate that, as with fibroblasts, there is a less inhibitory response (Fig. S1). This difference was not due to variability in protein knockdown between cell lines as all three cell lines had similar knockdown efficiencies for each respective siRNA (Figs. S2). RPL7 protein levels were decreased from 70–95%, RPL11 from 35–65%, and RPL31 from 74–80%.
Figure 1. siRNA targeting of ribosomal proteins in the large subunit decreased melanoma cell viability.
(A) A screen containing three independent siRNA targeting each RPL was transfected into 1205 Lu, UACC 903, or FF2441 cells. Cell viability was measured by a MTS assay and values were averaged for each RPL. The heatmap colors visually depict effect of respective RPL knockdown on cell viability. Green = least effect on cell viability. Red = largest effect on cell viability. # indicates instances where 2 of 3 siRNA displayed the growth effect instead of 3 of 3. * p<0.05, ** p<0.01, ***p<0.001. (B) Western blot comparing RPL13 protein levels between FOM103 melanocytes and melanoma cell lines. Protein band intensity was quantified using ImageJ software and RPL13 values normalized to the alpha enolase control and expressed as a percentage of FOM103 control. (C) Western blot comparing RPL13 protein levels between FOM103 melanocytes and melanoma tumor lysates. Blot intensities were quantitated using ImageJ software, normalized to alpha enolase loading control and expressed as a percentage of FOM103 control. BLD = below level of detection. (D) siRNA to RPL13 or scrambled control were transfected into UACC 903, 1205 Lu, A375M, and WM35 cells to demonstrate the ability of each siRNA to knockdown RPL13 protein expression. Erk2 was used as a control for equal protein loading. (E,F) siRNA to RPL13 or scrambled control were reverse transfected into fibroblast and melanoma cell lines at varying concentrations (E) or at 10 nM (F) and cell viability measured by MTS to compare differences in extent of cell viability decrease. Fibroblasts represent the average of four different fibroblast cell lines; MRC5, FF2441, and two primary neonatal fibroblast cell lines (Neonatal Fibroblasts, and FibroNeoSkin). * p<0.05, ** p<0.01, ***p<0.001. (N≥3) Bars; average, ±SEM.
Since knockdown was similar in melanoma and normal fibroblast cells, shown in Fig. 1A, the RPL genes were grouped into two categories: category 1 included all of the RPLs that have no effect on cell viability when knocked down and category 2 included all RPLs that decrease viability when targeted. Category 1 RPLs were not simply instances where knockdown was inefficient as the siRNA were able to knockdown protein levels, which is illustrated for RPL28 and RPL29 (Fig. S3). Since effectiveness of siRNA for protein knockdown has not been determined for the other RPLs in this category, it cannot be conclusively determined whether they belong in this category or that the siRNA targeting each respective mRNA were ineffective. A representative protein of category 2, ribosomal protein L13 (RPL13), was selected to dissect the process leading to growth inhibition for this group of proteins. RPL13 was expressed in melanocytes, and to a greater extent in most melanoma cell lines tested, with advanced stage melanoma cell lines having four to six times greater RPL13 protein expression (Fig. 1B), and 40% of melanoma tumors taken directly from patients, having increased RPL13 protein compared to normal human melanocytes (Fig. 1C), suggesting it was important for advanced melanoma cell survival. Interestingly, not all ribosomal proteins were expressed equally in the tumors (Fig. S4), however the significance remains to be determined.
RPL13 protein levels can be decreased upon transfection with two independent siRNA targeting different regions of the RPL13 mRNA (Fig. 1D). UACC 903, 1205 Lu, and A375M melanoma cells were inhibited to 20–50% greater levels at siRNA concentrations ranging from 1.25–10 nM than normal fibroblast cells (Fig. 1E). Targeting RPL13 in normal human melanocytes did not decrease viability, while viability of melanoma cell lines from the radial (WM35) vertical (WM115 and WM278) and metastatic (UACC 903, 1205 Lu, and A375M) stages of growth were inhibited by 40 to 90% (Fig. 1F). Viability in UACC 903 was also tested using an SRB assay and a DNA assay to verify MTS results (data not shown). These effects were not constrained to melanoma cells as cell lines from cancers of the prostate, brain, lung, cervix, connective tissue, and bone also demonstrated decreased cell viability upon RPL13 silencing ranging from 36% to 71% (Fig. S5).
siRNA mediated inhibition of RPL13 decreased tumor growth by reducing the proliferative potential of tumor cells
Reducing RPL13 protein levels in melanoma cells (Fig. 1D) decreased the viability of three cultured melanoma cell lines derived from tumors of advanced stage patients (Fig. 1E). To determine whether similar effects occurred on tumor growth, UACC 903 and 1205 Lu cells transfected with siRPL13 were subcutaneously injected into the left and right flanks of nude mice three days after transfection and tumor sizes were measured on alternate days following an established published approach (Sharma et al., 2005). Tumor size was inhibited by up to 90% for UACC 903 cells after 28 days (Fig. 2A) and up to 60% by 24 days for 1205 Lu cells (Fig. 2B) following siRNA mediated inhibition of RPL13, which is the first time the effect of RPL13 on tumor development has been examined. To identify the process regulated by targeting RPL13, temporally and spatially matched UACC 903 tumors were harvested 13 days after injection and rates of apoptosis, angiogenesis, and cell proliferation compared. Apoptosis, measured by TUNEL staining of tumor sections, showed no significant differences compared to controls (Fig. 2C). Similarly, angiogenesis, quantified by CD31 staining, showed no significant difference between control cells and those in which RPL13 protein levels were knocked down (Fig. 2D). In contrast, there was a significant decrease in cellular proliferation observed by Ki-67 staining (Fig. 2E), suggesting that decreased tumor growth following siRNA mediated RPL13 knockdown was being modulated by changes in the proliferative potential of the tumor cells.
Figure 2. RPL13 protein knockdown decreased tumor growth by reducing cellular proliferation.
(A,B) Transfection of RPL13 siRNA into UACC 903 (A) or 1205 Lu (B) led to a statistically significant decrease in tumor growth. Cells transfected with siRNA or buffer control were incubated for 3 days before s.c. injection into nude mice. Tumor volumes were measured every other day (N=6) Points; mean, ± SEM. (C, D, E) Tissue sections from size and time matched tumors were imaged and quantified after staining with antibodies to Ki-67, CD31, or TUNEL (N=10) Bars; mean, ±SEM (statistics, One-way ANOVA followed by Tukey’s test for multiple comparisons). *p<0.05, ** p<0.01, ***p<0.001.
RPL13 protein knockdown induced p53-dependent G0/G1 and G2/M arrest
Targeting ribosomal proteins such as RPS9 and RPL37 has been reported to lead to p53 activation through the RP-MDM2-p53 pathway (Lindstrom and Nister, 2010; Llanos and Serrano, 2010). To determine whether p53 activation is a larger phenomenon occurring across all category 2 ribosomal proteins, a subset of RPLs from category 2 was selected for further study, which included RPL5, RPL7, RPL11, RPL13, RPL18, RPL31, and RPL34, whose targeting decreased viability when knocked down using siRNA (Fig. 1A). RPL5 and RPL11 were chosen because they have a unique function by directly binding to MDM2 in the RP-MDM2-p53 pathway (Deisenroth and Zhang, 2010; Sun et al., 2010; Zhang et al., 2003). Proteins representing the first category, RPL28 and RPL29, which do not significantly alter viability when targeted using siRNA served as controls. When the selected RPL protein levels were knocked down using siRNA, a unique pattern of p53 protein expression was observed (Fig. 3A). None of the RPLs from category 1 significantly increased p53 protein levels. However, category 2 could be subdivided into category 2A, which dramatically increased p53 protein levels, or category 2B, that included RPL5 and RPL11, which did not increase p53 protein levels even though targeting decreased viability. p21, a downstream target of p53, followed the same pattern (Fig. 3A). The fibroblast cell line FF2441 and all melanoma cell lines tested showed an increase in p53 and p21 protein levels upon knockdown of RPL13, a category 2A protein (Fig. 3B). To determine whether p53 protein levels were increased due to stabilization of p53 in category 2A proteins, cells were treated with cycloheximide (CHX) to inhibit protein synthesis. In control lysates, p53 protein levels decreased dramatically within 30 minutes of CHX treatment; however, in cells transfected with siRPL13, p53 protein levels were stabilized for two to four fold longer periods of time (Fig. 3C). p53 can function as a tumor suppressor regulating cell cycle arrest (Vogelstein et al., 2000). To examine whether cell cycle arrest was affected by p53 stabilization, propidium iodide was used to stain DNA in cells to measure changes in the various cell cycle phases. Targeting RPL13 decreased the percentage of cells progressing through the S phase of the cell cycle with an accumulation of cells in G0/G1 and G2/M (Fig. 3D and S6). When p53 protein levels were knocked down using siRNA in combination with RPL13 siRNA, a partial restoration of cells in S phase was observed, suggesting that p53 partially accounts for the effects observed when targeting category 2A RPL proteins affecting cell proliferation. Interestingly, knockdown of RPL13 in melanoma cell lines decreased CDK2 phosphorylation at Thr160, a requirement for CDK2 activation (Gu et al., 1992), and multiple cyclin protein levels involved in the cell cycle consistent with the observation of cell cycle arrest, however this was not detected in the normal fibroblast FF2441 cell line (Fig. S7).
Figure 3. Effect of silencing RPL13 induced a p53-dependent cell cycle arrest.
(A) UACC 903 cells were transfected with siRNA targeting selected RPLs or scrambled control followed by western blotting. Erk2 served as the control for equal protein loading. (B) FF2441 fibroblast and UACC 903, 1205 Lu, or WM35 melanoma cell lines were transfected with RPL13 or scrambled control siRNA to measure increases in p53 and p21 levels by western blotting. Erk2 was used as a control for equal protein loading. (C) UACC 903 and 1205 Lu cells transfected with RPL13 or scrambled control siRNA were treated with cycloheximide (CHX) to stop protein synthesis. Western blotting from 0–2 hours after CHX treatment showed stability of p53. The siScramble 0–2hr blots were intentionally overexposed to enable comparison to the other blots. Enolase served as the control for equal protein loading. (D) UACC 903 or 1205 Lu cells were transfected with RPL13 siRNA alone or in combination with p53 siRNA and after 3 days incubation, cells were stained with propidium iodide, run on a BD FACSCalibur and results analyzed by ModFit LT (N≥3).
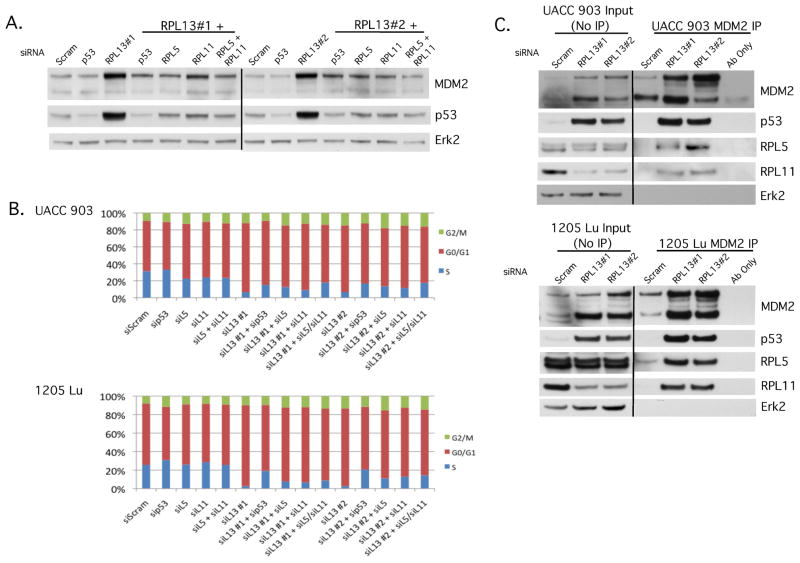
RPL5 and RPL11 mediated the p53 stability increase as well as cell cycle effects observed following RPL13 silencing
p53 serves as a monitor of ribosome biogenesis through the RP-MDM2-p53 pathway, which utilizes the extraribosomal effects of RPL5, RPL11, and possibly other RPs to bind to MDM2 to inhibit the ubiquitination of p53 (Deisenroth and Zhang, 2010; Sun et al., 2010; Zhang et al., 2003). To determine whether RPL5 and RPL11 were necessary for p53 stabilization upon RPL13 knockdown, RPL5 and RPL11 were siRNA targeted in combination with RPL13 siRNA to observe the effects on p53 protein levels. When RPL13 was silenced, p53 protein levels increased, but combining it with the silencing of RPL5 or RPL11 prevented increases in p53 protein levels (Fig. 4A), suggesting that both RPL5 and RPL11 are a necessary link for regulating p53 stabilization after RPL13 knockdown. To test whether p53-dependent cell cycle arrest is dependent upon RPL5 and RPL11, RPL5 or RPL11 was knocked down using siRNA in combination with RPL13 siRNA. This led to a partial restoration of the percentage of cells in S phase as seen when p53 and RPL13 were knocked down in combination (Fig. 4B).
Figure 4. RPL5 and RPL11 mediated p53 and cell cycle effects following RPL13 silencing.
(A) UACC 903 cells were transfected with RPL13 siRNA in the presence or absence of p53, RPL5, or RPL11 siRNA. Western blotting of lysates shows changes in MDM2 and p53 protein levels. Erk2 served as a control for equal protein loading. (B) UACC 903 and 1205 Lu cells were transfected with RPL13 siRNA alone or in combination with p53, RPL5, or RPL11 siRNA and after 3 days, cells were stained with propidium iodide, run on a BD FACSCalibur and results analyzed by ModFit LT (N≥3). (C) Lysates of UACC 903 and 1205 Lu cells transfected with RPL13 or scrambled siRNA control were coimmunoprecipitated using an MDM2 primary Ab, followed by western blotting, to examine the extent of p53, RPL5, and RPL11 binding to MDM2. Erk2 was used as a loading control for equal protein loading.
Since ribosome biogenesis disruption can cause certain ribosomal proteins to bind to MDM2 and inhibit its ability to ubiquitinate p53, MDM2 was co-immunoprecipitated and probed for the presence of RPL5 and RPL11 to determine whether RPL13 knockdown caused a similar effect. Lysates from cells transfected with RPL13 siRNA showed no change in RPL5 protein levels and a decrease in RPL11 protein levels compared to the control; however, co-immunoprecipitation of MDM2 in these lysates pulled down significantly more RPL5 and RPL11 protein when RPL13 is knocked down compared to the control (Fig. 4C). p53 was also able to be pulled down, which was consistent with the known interactions between MDM2 and p53 (Vogelstein et al., 2000).
Consistent with the observation that p53 knockdown only partially restores the percentage of cells in S phase, we also observed that cell viability could not be significantly restored when p53 was knocked down in combination with RPL13 knockdown (Fig. S8). Thus, while the RP-MDM2-p53 pathway may explain some of the effects seen after RPL knockdown, it cannot explain the observations completely. This interesting finding suggests that other factors also exist to mediate the effects that we have subsequently identified.
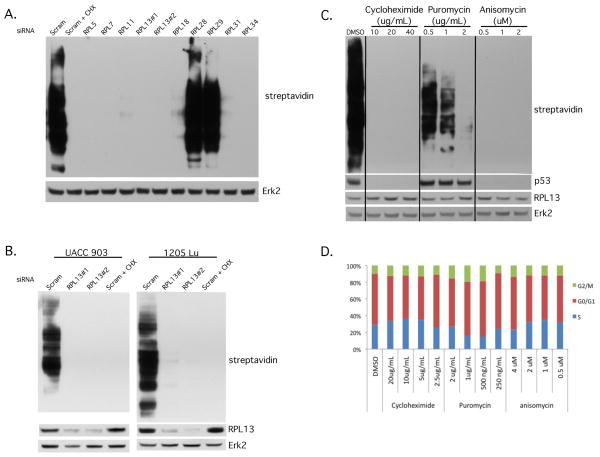
siRNA mediated RPL13 knockdown caused protein synthesis inhibition, which is unique from the p53 stabilization role
Disruption of ribosomal protein function could affect ribosome-related functions, i.e. protein synthesis (Dresios et al., 2000). Using the methionine analog azidohomoalanine (AHA) to measure the extent of new protein synthesis (Dieterich et al., 2006), every targeted RPL that decreased viability in vitro was found to inhibit protein synthesis, while those that had little to no effect on viability did not completely inhibit protein synthesis (Figs. 5A, 5B). Significantly, with the exception of RPL5 and RPL11, protein synthesis correlated with p53 stabilization. This did not occur with RPL5 and RPL11 because of the direct involvement of these proteins in the stabilization of p53. To determine whether increased p53 stabilization was modulated by protein synthesis inhibition, three different protein synthesis inhibitors that act on the 60S subunit, cycloheximide (Ennis and Lubin, 1964), anisomycin (Grollman, 1967), and puromycin (Nathans, 1964), were tested and effect on p53 examined. Complete inhibition of protein synthesis with cycloheximide and anisomycin treated cells decreased p53 protein levels (Fig. 5C). Concentrations of puromycin tested did not completely inhibit protein synthesis therefore p53 protein levels were detectable. Importantly however, no protein synthesis inhibitor was able to increase p53 protein levels as was observed with category 2A RPLs. Cell cycle arrest is likely not caused by the inhibition of protein synthesis either because treatment with protein synthesis inhibitors did not cause cell cycle arrest seen with RPL13 silencing (Fig. 5D). The slightly decreased percentage of cells in the S phase in puromycin-treated cells was accompanied by an accumulation of cells in G2/M, whereas upon RPL13 knockdown the accumulation was observed in G0/G1 and G2/M cell populations. Thus, we have found that inhibition of protein synthesis from RPL knockdown is an additional effect alongside the activation of the RP-MDM2-p53 pathway rather than a consequence of p53 stabilization. Likewise, protein synthesis inhibition is not the cause of the activation of the RP-MDM2-p53 pathway. This may explain why effects of RPL13 silencing cannot be restored completely by co-knockdown of RPL13 with p53, RPL5, or RPL11.
Figure 5. siRNA mediated RPL13 knockdown caused protein synthesis inhibition, and effects seen were not due to an effect on general protein synthesis inhibition.
(A, B) Cells transfected with RPL siRNA were subjected to methionine starvation then azidohomoalanine (AHA) incubation before collecting lysates. Biotin was attached to the AHA in newly synthesized proteins before performing western blotting and probing membranes with streptavidin-HRP to identify the extent of new protein synthesis in UACC 903 (A) or the effect of siRPL13 inhibition in UACC 903 and 1205 Lu (B). Erk2 served as the control for equal protein loading. (C) UACC 903 cells were treated with varying concentrations of the protein synthesis inhibitors cycloheximide, puromycin, and anisomycin for 4 hours before subjecting cells to methionine starvation then AHA incubation prior to collecting lysates. Biotin was attached to the AHA in newly synthesized proteins before performing western blotting and probing the membrane with streptavidin-HRP to identify the extent of new protein synthesis. Erk2 was used as a control for equal protein loading. (D) UACC 903 cells were treated with varying concentrations of cycloheximide, puromycin or anisomycin for 24 hours before staining cells with propidium iodide, running on a BD FACSCalibur and analyzing results using ModFit LT (N=3).
Discussion
In this study, for the first time, all large subunit ribosomal proteins were studied to elucidate RPL protein function in melanoma cells to identify potential therapeutic targets. Results suggest that RPLs can be grouped into two categories. Category 1 consists of RPLs that do not participate in the RP-MDM2-p53 pathway having little effect on cell proliferation or on inhibition of protein synthesis. Category 2 contains RPLs that participate in the RP-MDM2-p53 pathway and causes protein synthesis inhibition, affecting cell proliferation (Fig. 6). Category 2 can be further delineated based on differences in p53 stabilization into category 2A, which induce the RP-MDM2-p53 pathway upon knockdown, and category 2B, which enables the pathway. Results described in this report suggest that category 2A proteins, which induce the RP-MDM2-p53 pathway and reduce protein production, may be the most promising therapeutic targets in melanoma and other cancer cell types. This is because targeting them increased p53 protein expression and disrupted protein synthesis. Because of the importance of proliferation for cancer, these cells require greater protein synthesis (Baxter and Stanners, 1978; Stumpf and Ruggero, 2011). Therefore, protein synthesis can directly contribute to tumor formation (Ruggero, 2009) and would be a reasonable cancer cell target. Furthermore, this effect is not limited to melanoma as many other cancer types demonstrated decreased viability upon RPL13 knockdown. Therefore, simultaneously targeting a normal process essential to a cancer cell and increasing expression of a tumor suppressor may provide an effective means to halt tumor growth.
Figure 6. Large subunit ribosomal protein categories based on functional roles. RPL function in melanoma cells can be grouped into two categories.
Category 1 contains all RPLs that, when targeted, have little to no effect on cell viability and protein synthesis inhibition. These are also the proteins that do not participate in the RP-MDM2-p53 pathway. Category 2 contains all RPLs that, when targeted, decrease cell viability and inhibit protein synthesis. These are proteins that participate in the RP-MDM2-p53 pathway. Based on differences in effect on p53 stabilization, category 2 RPLs can be further grouped into 2A, those that induce the RP-MDM2-p53 pathway, and 2B, those that enable the RP-MDM2-p53 pathway. Most, if not all, RPLs of the second category could be grouped into either those that induce or enable the RP-MDM2-p53 pathway but only a subset was studied in detail, so the remaining cannot be conclusively categorized without further study. Highlighted in blue, green, and red are validated components of categorys 1, 2A, and 2B, respectively, from this study.
Targeting a ribosomal protein that activated the RP-MDM2-p53 pathway caused cell cycle arrest. Cell cycle arrest could be attributed in part to p53 stabilization because decreasing p53 protein expression partially alleviated arrest and enabled cells to progress through the S phase. In addition, RPL5 and RPL11 were also necessary mediators of this effect. This is in line with evidence linking other disruptions of ribosome biogenesis and cell cycle arrest (Deisenroth and Zhang, 2010). Variability in the recovery of the S phase may be because siRNA targeting p53 more completely knocked down p53 expression than siRPL5 and siRPL11, or that the knockdown of p53, RPL5, or RPL11 has additional consequences by itself. RPL13 knockdown is not solely dependent on p53 status since knockdown in p53-null cells were still able to decrease viability (data not shown). This suggests that the disruption of protein synthesis is an important aspect of targeting ribosomal proteins which p53 is not directly affecting.
Because of the role of category 2B RPLs such as RPL5 and RPL11 as enablers of the RP-MDM2-p53 pathway, this report suggests that the proteins would not be ideal targets for wildtype p53 melanoma tumors since targeting these proteins would prevent p53 accumulation. It is not yet clear how many RPLs would act as activators or enablers of the RP-MDM2-p53 pathway since only a subset was tested. However, the studies of others infer that RPL23 (Dai et al., 2004), RPL26 (Zhang et al., 2010), and RPL37 (Daftuar et al., 2013) as well as small ribosomal subunit proteins RPS3 (Yadavilli et al., 2009), RPS7 (Zhu et al., 2009), RPS15 (Daftuar et al., 2013), RPS20 (Daftuar et al., 2013), RPS27 (Xiong et al., 2011) and RPS27A (Sun et al., 2011) may also be included as enablers and thus would also not be good therapeutic targets. While it would be ideal to be able to both disrupt protein synthesis and activate a tumor suppressor, a therapeutic strategy targeting ribosomal proteins would not rely predominantly on p53 stabilization and therefore the therapeutic approach of targeting category 2A RPLs would not be constrained to wildtype p53 melanomas. Since there are p53-independent mechanisms of cell cycle arrest following disruption of ribosome biogenesis (Donati et al., 2012) and because ribosomal protein knockdown disrupts protein synthesis, a greater majority of melanomas should be able to be targeted using this therapeutic strategy.
Ribosome biogenesis and auxiliary processes associated with the ribosome utilize a lot of energy (Warner, 1999), which necessitates that any disruption of these processes be met with strategies to avoid wasting energy. Activation of p53 is one response to arrest affected cells but others exist. Knockdown of ribosomal proteins may have an effect on the ribosome itself, either during ribosome biogenesis or during protein production, which would ultimately affect protein synthesis. Results reported here suggest that targeting the majority of RPLs affect protein synthesis, which could be detrimental to highly proliferating cancer cells compared to normal ones. One caveat, however, is that since cancer stem cells may lie dormant (Schillert et al., 2013), treatment may not be as effective at targeting these actively proliferating cells. It could also be affecting proteins having short half-lives by disrupting levels disproportionately. This report uniquely suggests that not all RPL targets affected cell viability or protein synthesis when knocked down. One possibility to explain this observation is that these ribosomal proteins are not essential for the assembly, structure, or function of the ribosome as is the case in yeast for RPL26 and RPL29 (Babiano et al., 2012; DeLabre et al., 2002). As such, they may not cause aberrant ribosome biogenesis when disrupted. It is interesting, however, that RPL29 has been shown to inhibit pancreatic cancer cell proliferation (Li et al., 2012), but it has been deemed a non-essential ribosomal protein in yeast (DeLabre et al., 2002), and appears to be non-essential in melanoma as well. This suggests that different cell types may have varying dependencies on certain ribosomal proteins.
Stresses on ribosome biogenesis include inhibition of rRNA synthesis or processing (Act D, 5FU (Sun et al., 2007), mycophenolic acid (Sun et al., 2008)) and silencing of nucleolar or ribosomal proteins (Bee et al., 2011; Lindstrom and Nister, 2010; Llanos and Serrano, 2010; Wu et al., 2011). Targeting ribosome biogenesis through disruption of RNA polymerase I can be a viable option to therapeutically and selectively target cancer cells (Drygin et al., 2011). Likewise, silencing of certain ribosomal proteins can affect cancer growth (Bee et al., 2011; Lindstrom and Nister, 2010; Wu et al., 2011), and targeting the translation apparatus could also be a therapeutic approach (Malina et al., 2011). Since we have shown that many ribosomal proteins are linked to p53 through the RP-MDM2-p53 pathway and to protein translation as part of ribosome function, they may serve as unique therapeutic targets for a diverse array of cancer types. We must, however, keep in mind that haploinsufficiency of certain RPs in zebrafish elevated cancer incidence (Amsterdam et al., 2004) and that Diamond-Blackfan anemia is related to defects in ribosome biogenesis (Choesmel et al., 2007), therefore any therapeutic strategy must strive to avoid off-target effects.
Although the majority of RPLs caused a similar effect on p53 protein levels and protein synthesis when targeted, it is likely that there are other yet unknown effects as well, including ones unique to particular ribosomal proteins. For instance, RPL38 has been implicated in the translation of a specific subset of Hox mRNA suggesting that ribosomal proteins may have more specific functions than simply being one component of the large or small ribosomal subunits (Kondrashov et al., 2011). These, and other, extra-ribosomal roles of RPs are important to consider if a therapeutic strategy is developed because there may be unintended consequences of knocking down an RP that has more functions than the typical ribosomal activities. Furthermore, disruption of one RP may affect the function of another RP in turn, especially if they cannot come together to form intact subunits after disruption, so this potential impact on the cell would need to be taken into consideration. In addition, it is important to note that prokaryotic and eukaryotic ribosomes serve the same function, however eukaryotic ribosomes contain many more proteins and rRNA segments. The purpose of these additional entities is yet to be conclusively established. Research on ribosomes suggests that its protein components may serve unique functions (Gilbert, 2011), which occurs with RPL19, a protein overexpressed in prostate cancer (Bee et al., 2011), and RPL6, a protein overexpressed in gastric cancer (Wu et al., 2011), having unique roles and may be novel therapeutic targets. Furthermore, multiple mouse models of RP deficiencies have attributed particular phenotypic abnormalities to suppression of protein synthesis and extra-ribosomal functions, many of which could be reduced following p53 deletion (Terzian and Box, 2013). In this study, only large subunit ribosomal proteins were examined, however, small subunit proteins, ribosome assembly factors, and nucleolar proteins might also be important (Pestov et al., 2001). These are promising therapeutic targets in cancer cells that not only can activate the RP-MDM2-p53 pathway and affect protein synthesis, but may also regulate unique functions relied upon by cancer cells.
In summary, this report shows that targeting ribosomal proteins in melanoma and other cancers can be a promising therapeutic option for cancer treatment. These data uniquely suggest that targeting most large subunit ribosomal proteins not only increased p53 stability leading to cell cycle arrest, an effect mediated by RPL5 and RPL11, but also inhibited protein synthesis. This led to decreased cell viability in culture and significantly decreased tumor growth in animals. In contrast, targeting these genes in melanocytes or fibroblasts had a marginal effect on cell viability. Therefore, increasing expression of a tumor suppressor while targeting a normal process more essential to cancer than normal cells may provide an effective means to inhibit tumor growth.
Materials and Methods
Cell lines
UACC 903, 1205 Lu, FF2441, A375M, MRC5, and two primary neonatal fibroblast cell lines (Neonatal Fibroblasts, and FibroNeoSkin) cell lines were maintained in DMEM (Thermo) supplemented with 10% fetal bovine serum (FBS) (Thermo HyClone, Logan, UT, USA) and 1% glutaMAX (Life Technologies, Carlsbad, California, USA). WM35, WM115, and WM278 cell lines were grown in media #2 (MCDB media supplemented with heat-inactivated FBS, glutaMAX, and insulin) (Satyamoorthy et al., 1997). FOM103 melanocytes were grown in DermaLife media (Lifeline Cell Technology, Walkersville, MD, USA) containing LifeFactors DermaLife M (Lifeline Cell Technology). All cell lines were maintained at 37°C with 5% CO2 in a humidified incubator. All melanoma cell lines used are p53 wildtype (Ji et al., 2012; Khlgatian et al., 2002) as few human melanomas contain mutant p53.
siRNA Transfections
Silencer Select siRNA (Life Technologies) were used in preliminary screening (Table S1). Duplexed Stealth siRNA (Life Technologies) were used in all subsequent transfection experiments.
siRNA sequences:
Scrambled – AAUUCUCCGAACGUGUCACGUGAGA
RPL13 #1 – AGGAAGAGAAGAAUUUCAAAGCCUU
RPL13 #2 – CACUGAGGAAGAGAAGAAUUUCAAA
p53 – UCCACACGCAAAUUUCCUUCCACUC
RPL5 – GAAGACGACGAGAGGGUAAAACUGA
RPL7 – UGAUUGCUCGAUCUCUUGGUAAAUA
RPL11 – CAUGCGGGAACUUCGCAUCCGCAAA
RPL15 – UAGCUUACCAAUCUUAAGGCUAAUA
RPL18 – GAGGCUGUUGGUCAAGUUAUACAGG
RPL28 #2– UUCCUGAUCAAGAGGAAUAAGCAGA
RPL28 #3– CUACAGCACUGAGCCCAAUAACUUG
RPL29 #1– AAACCCCGAUCACAAAGAUACGAAU
RPL29 #2– UUGGCAAAGCGCAUGUUCCUCAGGA
RPL29 #3– CCUGUGCUAUUUGUACAAAUAAACC
RPL31 – CGGGCACUCAAAGAGAUUCGGAAAU
RPL34 – CUGGUAAUAGAAUUGUUUACCUUUA
MTS assays were carried out by reverse-transfecting 5,000 cells/well with RNAiMAX (Life Technologies) and 10 nM siRNA in 96-well plates according to manufacturer’s recommendations. For FOM103 melanocytes, 20,000 cells/well and 40 nM siRNA were used. After 48 hours, plates were replaced with serum-free media and incubated for 72 hours before performing an MTS assay (Promega, Madison, Wisconsin, USA).
Transfections to collect lysates were carried out using the Amaxa Nucleofector (Lonza, Basel Switzerland) using Solution R/program K-17(Stahl et al., 2004). 1 × 106 cells were nucleofected with 100 pmoles of respective siRNA followed by incubation for 72 hours at 37°C with 5% CO2 in a humidified incubator.
Western Blotting
Cell lysates were harvested three days after transfection by washing plates with PBS followed by addition of RIPA lysis buffer containing Halt Protease & Phosphatase Inhibitor Cocktail (Thermo Scientific, Rockford, IL, USA). Lysates were centrifuged (10,000 ×g) for 10 minutes at 4°C to remove cell debris. Protein concentration was determined using the bicinchoninic acid assay kit (Thermo). 25 μg of respective lysates were loaded onto 4–12% Bis-Tris NuPAGE gels (Life Technologies) and run in an XCell SureLock Mini-Cell gel apparatus (Life Technologies). Following electrophoresis, protein was transferred to polyvinylidene difluoride membranes (PVDF) (Pall Corporation, Port Washington, NY, USA). Blots were probed according to company’s specifications and exposed using ECL Western Blotting Substrate (Thermo Scientific) or Supersignal West Femto Chemiluminescent Substrate (Thermo Scientific). Primary antibodies used: RPL13 (sc-133960), RPL29 (sc-103166), α enolase (sc-7455), p53 (sc-6243), p21 (sc-756), Erk2 (sc-1647), MDM2 (SMP14) (sc-965), and MDM2 (D-12) (sc-5304) from Santa Cruz Biotechnology (Dallas, TX, USA), RPL7 (ab72550), RPL28 (ab112609), and RPL31 (ab103991) from Abcam (Cambridge, UK), RPL11 (37-3000) from Invitrogen, RPL5 from Dr. Mushui Dai, and cyclin E1 (#4129), cyclin E2 (#4132), cyclin B1 (#4138), cyclin H (#2927), cyclin A2 (#4656), CDK2 (#2546), and pCDK2 (T160) (#2561) from Cell Signaling (Danvers, MA). Secondary antibodies goat anti-rabbit IgG-HRP (sc-2004), goat anti-mouse IgG-HRP (sc-2005), and donkey anti-goat IgG-HRP (sc-2020) were purchased from Santa Cruz Biotechnology.
Tumor Lysate Studies
Melanoma tumors were collected from human patients according to protocols approved by he Penn State Human Subjects Protection Office as described previously (Stahl et al., 2004). Pulverized tumor samples were lysed with RIPA lysis buffer containing Halt Protease & Phosphatase Inhibitor Cocktail and subsequently sonicated. Tumor lysates were centrifuged (10,000 xg) for 10 minutes at 4°C to remove cell debris.
Co-Immunoprecipitation
Lysates were co-immunoprecipitated using the Co-Immunoprecipitation Kit (26149, Thermo Scientific). MDM2 (SMP14) primary Ab (sc-965, Santa Cruz) cross-linking to beads and co-IP of lysates were performed according to manufacturer’s protocol. Briefly, 5 μg of the MDM2 (SMP14) antibody was incubated with 20 μL AminoLink Plus coupling resin for covalent coupling to immobilize antibody. The antibody-coupled resin was washed, and then incubated with protein lysate at 4°C overnight. After washing the resin, antigen complexes were eluted using elution buffer. Samples were analyzed by western blotting as described above. A negative control antibody only lane was run to ensure antibody was properly crosslinked so that no heavy or light chains interfered during western blotting.
Flow cytometry for cell cycle analysis
Three days after transfection, 1 × 106 asynchronously proliferating cells per treatment were collected. Cell pellets were washed twice with serum-free DMEM before resuspending in propidium iodide staining solution (0.1 mg/mL propidium iodide, 0.02 mg/mL Ribonuclease A, 1 mg/mL sodium citrate, and 0.3% Triton-X-100 in 1X PBS). Cells were run on a BD FACSCalibur (BD Biosciences, San Jose, California, USA) and results analyzed by ModFit LT (Verity Software House, Topsham, Maine).
Protein Synthesis Analysis
After transfection and incubation at 37°C with 5% CO2 in a humidified incubator for three days, cells were starved of methionine by washing plates with PBS then incubating with methionine-free DMEM for 1 hour. 25 μM L-azidohomoalanine (AHA, Life Technologies), a methionine analog, was added to the media for 4–6 hours so that all new protein synthesis will incorporate AHA. As a negative control, 80 μg/mL cycloheximide was added immediately before the addition of AHA to stop protein synthesis. Cell lysates were then collected using RIPA buffer. The Click-iT Protein Reaction Buffer Kit (Life Technologies) and Biotin Alkyne (Life Technologies) were used to attach biotin to AHA via a click reaction according to manufacturer’s protocol. Biotin-attached lysates were run on 4–12% Bis-Tris gels, transferred to PVDF membranes, and probed with streptavidin-HRP (R&D Systems, Minneapolis, MN, USA) to visualize the level of protein synthesis.
Animal studies
200 pmol siRNA to RPL13 or Scrambled control was reverse transfected into 2 × 106 UACC 903 and 1205 Lu cell lines per plate using RNAiMAX. Sterile, nuclease free DEPC H2O (EMD Chemicals, Gibbstown, NJ, USA) was used as the Buffer control. 72 hours after transfection, 1 × 106 cells in 0.2 mL of DMEM media containing 10% FBS were subcutaneously injected into the left and right flanks of 4–6 week old athymic female nude mice. Tumors were measured every other day beginning at day 6 using calipers to measure length, width, and depth.
For size and time matched tumors, 7.5 times as many siRPL13-treated cells were injected into nude mice than siScramble and buffer-treated nude mice in order to obtain similar tumor sizes by two weeks post injection. 13 days after injection, tumors were collected for fixation.
Tissue staining
Formalin-fixed, paraffin-embedded tumor sections from size and time matched tumors were used to measure apoptosis, cell proliferation, and vessel density. Apoptosis was visualized using the In Situ Cell Death Detection Kit TMR Red (Roche, Indianapolis, IN, USA) (Stahl et al., 2003). Cell proliferation rates were measured by Ki-67 staining using Ki-67 mouse anti-human primary Ab (BD Pharmingen) with a biotinylated goat anti-mouse IgG secondary Ab (BD Pharmingen) (Stahl et al., 2003). Vessel density quantification was performed using the rat anti-mouse CD31 primary Ab (BD Pharmingen) and biotinylated goat anti-rat secondary Ab (BD Pharmingen) on zinc-fixed, paraffin-embedded tumor sections (Stahl et al., 2003). Density was calculated on the IP Lab imaging software program by taking the proportion of area of tumor occupied by the vessels divided by the total tumor area. In all analyses, 6 different tumors from each treatment group with a minimum of three fields per tumor were quantitated.
Supplementary Material
Fig. S1: RPL protein knockdown had a negligible effect on melanocyte cell viability. siRNA to various RPLs or scrambled control were transfected into normal melanocyte FOM103 cells to measure cell viability decrease after RPL silencing via MTS assay. **p<0.01, *** p<0.001 (N=4) Bars; mean, ±SEM (statistics, One-way ANOVA followed by Tukey’s test for multiple comparisons).
Fig. S2: Knockdown of RPL protein levels is comparable between cell lines. siRNA to various RPLs were transfected into UACC 903, 1205 Lu, and FF2441 cell lines. Blot intensities were quantitated using ImageJ software, normalized to Erk2 protein loading control, and expressed as a percentage of siScrambled control.
Fig. S3: siRNA targeting of RPL28 and RPL29 decreased protein levels. Transfection of siRNA targeting RPL28 and RPL29 into UACC 903 or 1205 Lu led to knockdown of protein levels as measured by western blotting. Blots were quantitated by ImageJ, normalized to the Erk2 protein loading control, and expressed as a percentage of the siScrambled blot intensity.
Fig. S4: Ribosomal protein expression levels in melanoma tumors from patients. Western blot from Figure 1C comparing protein levels of various ribosomal proteins between FOM103 melanocytes and melanoma tumor lysates. Blot intensities were quantitated using ImageJ software, normalized to alpha enolase protein loading control and expressed as a percentage of FOM103 control. BLD = below level of detection.
Fig. S5: RPL13 protein knockdown decreased the viability of cells from multiple cancer types. siRNA to RPL13 or scrambled control were reverse transfected into prostate (PC-3), brain (LN-229), lung adenocarcinoma (A549), cervix (HeLa), connective tissue (HT-1080), bone (U-2 OS), or lung non-small cell (H1299) cells to measure cell viability decrease after RPL13 silencing via MTS assay. *** p<0.001 (N=4) Bars; mean, ±SEM (statistics, One-way ANOVA followed by Tukey’s test for multiple comparisons).
Fig. S6: Silencing RPL13 induced a p53-dependent cell cycle arrest. Data from Figure 3D presented as bar graphs to more clearly represent differences between groups (N≥3). *p<0.05, ** p<0.01, ***p<0.001.
Fig. S7: siRNA mediated RPL13 knockdown altered expression levels of cell cycle proteins. (A and B) Transfection of siRNA targeting RPL13 or scrambled control into melanoma cells (UACC 903, 1205 Lu, and WM35) or normal fibroblast cells (FF2441). Western blotting shows results of RPL13 protein knockdown on the levels of cell cycle-related proteins. Erk2 served as a protein loading control.
Fig. S8: Combining p53 with RPL13 protein knockdown did not completely restore cell viability. siRNA targeting RPL13, p53, RPL13 and p53, or scrambled control were transfected into UACC 903, 1205 Lu, or A375M melanoma cells. Change in cell viability was measured after five days via a MTS assay. *p<0.05, ** p<0.01, ***p<0.001. (N=4) Bars; mean, ±SEM (statistics, One-way ANOVA followed by Tukey’s test for multiple comparisons).
Table S1: Silencer Select siRNA sequences and target information used for screening RPL genes that would affect melanoma cell viability.
Significance.
Little is known regarding involvement of large subunit ribosomal proteins (RPLs) in melanoma development or the potential to target these proteins for therapeutic purposes. To unravel the involvement of these proteins in melanoma, a screen of all RPLs was undertaken, showing that not all RPLs affect melanoma cell survival in a similar manner. Certain RPL proteins represented by RPL13 can be targeted to stabilize p53 in melanoma cells to induce p53-dependent cell cycle arrest, and decrease protein translation. Thus, for the first time in melanoma RPLs are shown to a potential therapeutic target capable of inhibiting tumor development.
Acknowledgments
Financial Support: This work was supported by The Foreman Foundation for Melanoma Research, Melanoma Research Foundation and NIH grants CA-127892, CA-136667, and CA-138634.
The authors thank Arati Sharma for technical assistance.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS biology. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiano R, Gamalinda M, Woolford JL, Jr, De La Cruz J. Saccharomyces cerevisiae ribosomal protein L26 is not essential for ribosome assembly and function. Molecular and cellular biology. 2012;32:3228–41. doi: 10.1128/MCB.00539-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter GC, Stanners CP. The effect of protein degradation on cellular growth characteristics. Journal of cellular physiology. 1978;96:139–45. doi: 10.1002/jcp.1040960202. [DOI] [PubMed] [Google Scholar]
- Bee A, Brewer D, Beesley C, Dodson A, Forootan S, Dickinson T, Gerard P, Lane B, Yao S, Cooper CS, et al. siRNA knockdown of ribosomal protein gene RPL19 abrogates the aggressive phenotype of human prostate cancer. PloS one. 2011;6:e22672. doi: 10.1371/journal.pone.0022672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau De Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science (New York, NY) 2011;334:1524–9. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, Hirth P. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nature reviews. 2012 doi: 10.1038/nrd3847. Drug discovery. [DOI] [PubMed] [Google Scholar]
- Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, Leblanc T, Tchernia G, Da Costa L, Gleizes PE. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109:1275–83. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daftuar L, Zhu Y, Jacq X, Prives C. Ribosomal Proteins RPL37, RPS15 and RPS20 Regulate the Mdm2-p53-MdmX Network. PloS one. 2013;8:e68667. doi: 10.1371/journal.pone.0068667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. The Journal of biological chemistry. 2004;279:44475–82. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Molecular and cellular biology. 2004;24:7654–68. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–60. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- Delabre ML, Kessl J, Karamanou S, Trumpower BL. RPL29 codes for a non-essential protein of the 60S ribosomal subunit in Saccharomyces cerevisiae and exhibits synthetic lethality with mutations in genes for proteins required for subunit coupling. Biochimica et biophysica acta. 2002;1574:255–61. doi: 10.1016/s0167-4781(01)00372-4. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9482–7. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G, Montanaro L, Derenzini M. Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer research. 2012;72:1602–7. doi: 10.1158/0008-5472.CAN-11-3992. [DOI] [PubMed] [Google Scholar]
- Dresios J, Derkatch IL, Liebman SW, Synetos D. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry. 2000;39:7236–44. doi: 10.1021/bi9925266. [DOI] [PubMed] [Google Scholar]
- Drygin D, Lin A, Bliesath J, Ho CB, O’brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A, et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer research. 2011;71:1418–30. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- Ennis HL, Lubin M. CYCLOHEXIMIDE: ASPECTS OF INHIBITION OF PROTEIN SYNTHESIS IN MAMMALIAN CELLS. Science (New York, NY) 1964;146:1474–6. doi: 10.1126/science.146.3650.1474. [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, Mcarthur GA, Sosman JA, O’dwyer PJ, Lee RJ, Grippo JF, Nolop K, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WV. Functional specialization of ribosomes? Trends in biochemical sciences. 2011;36:127–32. doi: 10.1016/j.tibs.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman AP. Inhibitors of protein biosynthesis. II. Mode of action of anisomycin. The Journal of biological chemistry. 1967;242:3226–33. [PubMed] [Google Scholar]
- Gu Y, Rosenblatt J, Morgan DO. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. The EMBO journal. 1992;11:3995–4005. doi: 10.1002/j.1460-2075.1992.tb05493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi FS, O’day SJ, Mcdermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. The New England journal of medicine. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Njauw CN, Taylor M, Neel V, Flaherty KT, Tsao H. p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibition. The Journal of investigative dermatology. 2012;132:356–64. doi: 10.1038/jid.2011.313. [DOI] [PubMed] [Google Scholar]
- Khlgatian MK, Hadshiew IM, Asawanonda P, Yaar M, Eller MS, Fujita M, Norris DA, Gilchrest BA. Tyrosinase gene expression is regulated by p53. The Journal of investigative dermatology. 2002;118:126–32. doi: 10.1046/j.0022-202x.2001.01667.x. [DOI] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–97. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Ge M, Yin Y, Luo M, Chen D. Silencing expression of ribosomal protein L26 and L29 by RNA interfering inhibits proliferation of human pancreatic cancer PANC-1 cells. Molecular and cellular biochemistry. 2012;370:127–39. doi: 10.1007/s11010-012-1404-x. [DOI] [PubMed] [Google Scholar]
- Lindstrom MS, Nister M. Silencing of ribosomal protein S9 elicits a multitude of cellular responses inhibiting the growth of cancer cells subsequent to p53 activation. PloS one. 2010;5:e9578. doi: 10.1371/journal.pone.0009578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llanos S, Serrano M. Depletion of ribosomal protein L37 occurs in response to DNA damage and activates p53 through the L11/MDM2 pathway. Cell cycle. 2010;9:4005–12. doi: 10.4161/cc.9.19.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malina A, Cencic R, Pelletier J. Targeting translation dependence in cancer. Oncotarget. 2011;2:76–88. doi: 10.18632/oncotarget.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V, Maitra RK, Dicorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–98. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- Mcgowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, De Angelis MH, Myers RM, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nature genetics. 2008;40:963–70. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans D. PUROMYCIN INHIBITION OF PROTEIN SYNTHESIS: INCORPORATION OF PUROMYCIN INTO PEPTIDE CHAINS. Proceedings of the National Academy of Sciences of the United States of America. 1964;51:585–92. doi: 10.1073/pnas.51.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Molecular and cellular biology. 2001;21:4246–55. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Warner JR. What better measure than ribosome synthesis? Genes & development. 2004;18:2431–6. doi: 10.1101/gad.1256704. [DOI] [PubMed] [Google Scholar]
- Ruggero D. The role of Myc-induced protein synthesis in cancer. Cancer research. 2009;69:8839–43. doi: 10.1158/0008-5472.CAN-09-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nature reviews. Cancer. 2003;3:179–92. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- Rughani MG, Gupta A, Middleton MR. New treatment approaches in melanoma: current research and clinical prospects. Therapeutic advances in medical oncology. 2013;5:73–80. doi: 10.1177/1758834012463260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyamoorthy K, Dejesus E, Linnenbach AJ, Kraj B, Kornreich DL, Rendle S, Elder DE, Herlyn M. Melanoma cell lines from different stages of progression and their biological and molecular analyses. Melanoma research. 1997;7(Suppl 2):S35–42. [PubMed] [Google Scholar]
- Schillert A, Trumpp A, Sprick MR. Label retaining cells in cancer--the dormant root of evil? Cancer letters. 2013;341:73–9. doi: 10.1016/j.canlet.2013.04.019. [DOI] [PubMed] [Google Scholar]
- Sharma A, Trivedi NR, Zimmerman MA, Tuveson DA, Smith CD, Robertson GP. Mutant V599EB-Raf regulates growth and vascular development of malignant melanoma tumors. Cancer research. 2005;65:2412–21. doi: 10.1158/0008-5472.CAN-04-2423. [DOI] [PubMed] [Google Scholar]
- Siegel R, Desantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al. Cancer treatment and survivorship statistics, 2012. CA: a cancer journal for clinicians. 2012;62:220–41. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- Stahl JM, Cheung M, Sharma A, Trivedi NR, Shanmugam S, Robertson GP. Loss of PTEN promotes tumor development in malignant melanoma. Cancer research. 2003;63:2881–90. [PubMed] [Google Scholar]
- Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, Kester M, Sandirasegarane L, Robertson GP. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer research. 2004;64:7002–10. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- Stumpf CR, Ruggero D. The cancerous translation apparatus. Current opinion in genetics & development. 2011;21:474–83. doi: 10.1016/j.gde.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, Dai MS, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. The Journal of biological chemistry. 2007;282:8052–9. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- Sun XX, Dai MS, Lu H. Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. The Journal of biological chemistry. 2008;283:12387–92. doi: 10.1074/jbc.M801387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, Devine T, Challagundla KB, Dai MS. Interplay between ribosomal protein S27a and MDM2 protein in p53 activation in response to ribosomal stress. The Journal of biological chemistry. 2011;286:22730–41. doi: 10.1074/jbc.M111.223651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, Wang YG, Xirodimas DP, Dai MS. Perturbation of 60 S ribosomal biogenesis results in ribosomal protein L5- and L11-dependent p53 activation. The Journal of biological chemistry. 2010;285:25812–21. doi: 10.1074/jbc.M109.098442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarver T. Cancer Facts & Figures 2012. American Cancer Society (ACS) Journal of Consumer Health on the Internet. 2012;16:366–367. [Google Scholar]
- Terzian T, Box N. Genetics of ribosomal proteins: “curiouser and curiouser”. PLoS genetics. 2013;9:e1003300. doi: 10.1371/journal.pgen.1003300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. An encore for ribosome biogenesis in the control of cell proliferation. Nature cell biology. 2000;2:E71–2. doi: 10.1038/35010581. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, Hegde V, Lam LT, Staudt LM, Levens D, et al. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131:927–39. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends in biochemical sciences. 1999;24:437–40. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Warner JR, Mcintosh KB. How common are extraribosomal functions of ribosomal proteins? Molecular cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, Vilardell J, Sohn JH. Economics of ribosome biosynthesis. Cold Spring Harbor symposia on quantitative biology. 2001;66:567–74. doi: 10.1101/sqb.2001.66.567. [DOI] [PubMed] [Google Scholar]
- Wu Q, Gou Y, Wang Q, Jin H, Cui L, Zhang Y, He L, Wang J, Nie Y, Shi Y, et al. Downregulation of RPL6 by siRNA inhibits proliferation and cell cycle progression of human gastric cancer cell lines. PloS one. 2011;6:e26401. doi: 10.1371/journal.pone.0026401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Zhao Y, He H, Sun Y. Ribosomal protein S27-like and S27 interplay with p53-MDM2 axis as a target, a substrate and a regulator. Oncogene. 2011;30:1798–811. doi: 10.1038/onc.2010.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadavilli S, Mayo LD, Higgins M, Lain S, Hegde V, Deutsch WA. Ribosomal protein S3: A multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA repair. 2009;8:1215–24. doi: 10.1016/j.dnarep.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch RL, Settleman J. Recent advances in pathway-targeted cancer drug therapies emerging from cancer genome analysis. Current opinion in genetics & development. 2012;22:45–9. doi: 10.1016/j.gde.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Yuan Y, Zhang W, Guan W, Wu Z, Jin C, Chen H, Zhang L, Yang X, et al. Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic acids research. 2010;38:6544–54. doi: 10.1093/nar/gkq536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Molecular and cellular biology. 2003;23:8902–12. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Molecular cell. 2009;35:316–26. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: RPL protein knockdown had a negligible effect on melanocyte cell viability. siRNA to various RPLs or scrambled control were transfected into normal melanocyte FOM103 cells to measure cell viability decrease after RPL silencing via MTS assay. **p<0.01, *** p<0.001 (N=4) Bars; mean, ±SEM (statistics, One-way ANOVA followed by Tukey’s test for multiple comparisons).
Fig. S2: Knockdown of RPL protein levels is comparable between cell lines. siRNA to various RPLs were transfected into UACC 903, 1205 Lu, and FF2441 cell lines. Blot intensities were quantitated using ImageJ software, normalized to Erk2 protein loading control, and expressed as a percentage of siScrambled control.
Fig. S3: siRNA targeting of RPL28 and RPL29 decreased protein levels. Transfection of siRNA targeting RPL28 and RPL29 into UACC 903 or 1205 Lu led to knockdown of protein levels as measured by western blotting. Blots were quantitated by ImageJ, normalized to the Erk2 protein loading control, and expressed as a percentage of the siScrambled blot intensity.
Fig. S4: Ribosomal protein expression levels in melanoma tumors from patients. Western blot from Figure 1C comparing protein levels of various ribosomal proteins between FOM103 melanocytes and melanoma tumor lysates. Blot intensities were quantitated using ImageJ software, normalized to alpha enolase protein loading control and expressed as a percentage of FOM103 control. BLD = below level of detection.
Fig. S5: RPL13 protein knockdown decreased the viability of cells from multiple cancer types. siRNA to RPL13 or scrambled control were reverse transfected into prostate (PC-3), brain (LN-229), lung adenocarcinoma (A549), cervix (HeLa), connective tissue (HT-1080), bone (U-2 OS), or lung non-small cell (H1299) cells to measure cell viability decrease after RPL13 silencing via MTS assay. *** p<0.001 (N=4) Bars; mean, ±SEM (statistics, One-way ANOVA followed by Tukey’s test for multiple comparisons).
Fig. S6: Silencing RPL13 induced a p53-dependent cell cycle arrest. Data from Figure 3D presented as bar graphs to more clearly represent differences between groups (N≥3). *p<0.05, ** p<0.01, ***p<0.001.
Fig. S7: siRNA mediated RPL13 knockdown altered expression levels of cell cycle proteins. (A and B) Transfection of siRNA targeting RPL13 or scrambled control into melanoma cells (UACC 903, 1205 Lu, and WM35) or normal fibroblast cells (FF2441). Western blotting shows results of RPL13 protein knockdown on the levels of cell cycle-related proteins. Erk2 served as a protein loading control.
Fig. S8: Combining p53 with RPL13 protein knockdown did not completely restore cell viability. siRNA targeting RPL13, p53, RPL13 and p53, or scrambled control were transfected into UACC 903, 1205 Lu, or A375M melanoma cells. Change in cell viability was measured after five days via a MTS assay. *p<0.05, ** p<0.01, ***p<0.001. (N=4) Bars; mean, ±SEM (statistics, One-way ANOVA followed by Tukey’s test for multiple comparisons).
Table S1: Silencer Select siRNA sequences and target information used for screening RPL genes that would affect melanoma cell viability.