Abstract
Purpose.
The pathophysiology of ocular hypertension (OH) leading to primary open-angle glaucoma shares many features with a secondary form of OH caused by treatment with glucocorticoids, but also exhibits distinct differences. In this study, a pharmacogenomics approach was taken to discover candidate genes for this disorder.
Methods.
A genome-wide association study was performed, followed by an independent candidate gene study, using a cohort enrolled from patients treated with off-label intravitreal triamcinolone, and handling change in IOP as a quantitative trait.
Results.
An intergenic quantitative trait locus (QTL) was identified at chromosome 6p21.33 near the 5′ end of HCG22 that attained the accepted statistical threshold for genome-level significance. The HCG22 transcript, encoding a novel mucin protein, was expressed in trabecular meshwork cells, and expression was stimulated by IL-1, and inhibited by triamcinolone acetate and TGF-β. Bioinformatic analysis defined the QTL as an approximately 4 kilobase (kb) linkage disequilibrium block containing 10 common single nucleotide polymorphisms (SNPs). Four of these SNPs were identified in the National Center for Biotechnology Information (NCBI) GTEx eQTL browser as modifiers of HCG22 expression. Most are predicted to disrupt or improve motifs for transcription factor binding, the most relevant being disruption of the glucocorticoid receptor binding motif. A second QTL was identified within the predicted signal peptide of the HCG22 encoded protein that could affect its secretion. Translation, O-glycosylation, and secretion of the predicted HCG22 protein was verified in cultured trabecular meshwork cells.
Conclusions.
Identification of two independent QTLs that could affect expression of the HCG22 mucin gene product via two different mechanisms (transcription or secretion) is highly suggestive of a role in steroid-induced OH.
Keywords: ocular hypertension, glucocorticoid, GWAS, gene
A pharmacogenomics approach was taken to discover candidate genes for steroid-induced ocular hypertension. Two independent quantitative trait loci were identified in association with a novel mucin gene HCG22, which was characterized. The loci could affect expression of the HCG22 mucin gene product via two different mechanisms (transcription or secretion).
Glaucoma is a leading cause of irreversible blindness, presently affecting approximately 70 million people worldwide.1,2 The most common form is primary open-angle glaucoma (POAG), which accounts for approximately 90% of all cases. Ocular hypertension (OH) is the major risk factor for POAG and lowering IOP is the only effective treatment.3 However, many patients remain refractory to existing IOP-lowering interventions and eventually may become blind, underscoring the unmet medical need for novel approaches to control IOP.
Intraocular pressure is a product of the rate of aqueous humor production, resistance to outflow, and episcleral venous pressure.3,4 The aqueous outflow pathways in the angle of the eye are composed of a spongy tissue called the trabecular meshwork (TBM), which leads into Schlemm's canal. Much of the resistance to outflow resides within the TBM, putatively within 7 to 14 μm of the inner wall of Schlemm's canal in a region known as the juxtacanalicular or cribriform region.5–8 Primary open-angle glaucoma is caused by an increase in outflow resistance9,10 due to poorly understood defects in this region.11–14 Ocular hypertension in POAG has been associated with dysregulated intracellular signaling pathways controlled by IL-1/NF-κB,15 TGF-β superfamily,16 and Wnt.17,18
The pathophysiology of OH leading to POAG shares many features with a secondary form of OH caused by treatment with glucocorticoids (GCs).19–25 Glucocorticoids are a class of steroid hormones released in response to stress. As part of the natural feedback mechanism that turns down the inflammatory response, they are useful for treating a wide variety of diseases.26 Complicating this, however, is the considerable interindividual variability and tissue-specific GC sensitivity among individuals, which can cause a variety of systemic side effects.27–31 Treatment with steroids, such as dexamethasone or triamcinolone acetonide (TA), in the eye causes elevated IOP in predisposed individuals. It has been documented that approximately 40% of the normal population develops an IOP increase > 6 mm Hg above baseline following topical administration of GCs, 4 times a day for 4 to 6 weeks.32 When GCs are administered intravitreally, IOP may increase by 30% or more in up to half of patients.33 These individuals are considered to be “steroid responders.” Interestingly, almost all POAG patients are steroid responders25,32,34–40; conversely, steroid-responders who do not have POAG are at much higher risk of developing POAG compared to nonresponders.37–41 A better understanding of genetic and molecular mechanisms underlying individual variation in the response to steroids could shed light on our understanding of steroid-induced OH and OH leading to POAG. That being said, mechanisms of steroid-induced OH and OH leading to POAG also show distinct differences; therefore, studying the genetics of steroid-induced OH specifically could lead to discovery of genes involved in IOP regulation that could not have been revealed any other way.
The genome-wide association study (GWAS) represents an agnostic approach for prioritization of genes that are associated with a disease process, and makes it possible to discover novel genes that could not otherwise be identified. In the current study, we performed a pharmaco-GWAS for this purpose, using a cohort enrolled from patients treated with off-label intravitreal TA (IVTA) for various retinal indications, and followed up with an independent candidate gene study. We identified two independent quantitative trait loci (QTLs) at chromosomal locus 6p21.33 associated with the novel mucin gene HCG22.
Materials and Methods
Ethics Statement
This study was compliant with the Declaration of Helsinki, the Health Insurance Portability and Accountability Act (HIPAA), and the Institutional Review Boards of the University of Miami (Miami, FL, USA), Lee Memorial Health System (Cape Coral, FL, USA) and the University of Southern California (Los Angeles, CA, USA). Informed consent was obtained from all research subjects after explanation of the nature and possible consequences of the study. The Institutional Review Board of the University of Southern California reviewed the protocol for cell isolation from the discarded human corneal rims (which are coded specimens) and designated it as nonhuman subject research.
Study Cohorts
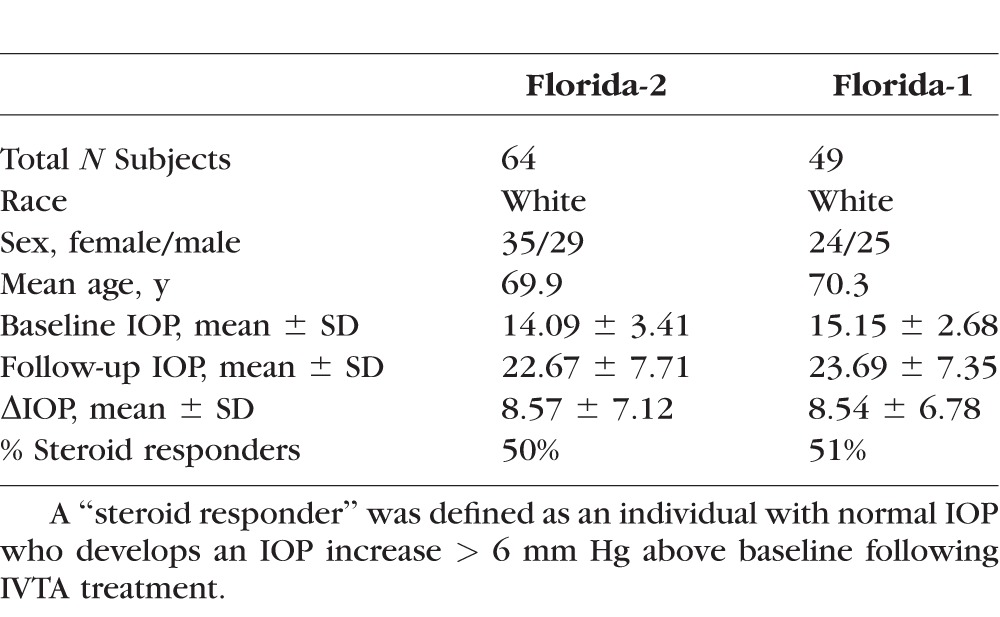
Research subjects for the “Florida-2” cohort served as the discovery group for the current study. Research subjects were enrolled at the Bascom Palmer Eye Institute, University of Miami (Miami, FL, USA; 13 subjects), the Retina Health Center (Fort Myers, FL, USA; 61 subjects), or the California Retina Associates (Santa Barbara, CA, USA; 5 subjects). Most subjects reported their race as “white.” Patients were offered entry into the study if they were to be treated with an injection of IVTA (approximately 4 mg in 0.1 cc) for various retinal diseases or previously had been treated with IVTA. All treatments were similar: approximately IVTA, 4 mg in 0.1 cc. Inclusion criteria were age 18 years or greater, at least two IOP measurements over six months post injection, and the ability to give appropriate informed consent. Exclusion criteria were prior pars plana vitrectomy in the study eye, previous trabeculectomy or glaucoma tube shunt surgery in the study eye, anterior segment neovascularization in the study eye, and uveitis other than cystoid macular edema status post cataract or scleral buckle surgery in the study eye.
The previously described “Florida-1” cohort,42 enrolled at the Bascom Palmer Eye Institute, University of Miami served as the replicate group for the current study. Inclusion/exclusion criteria were similar to the Florida-2 cohort, as described previously.42 Exit criteria described in the published study included subsequent intravitreal injection, any intervention intended to lower IOP, or subsequent intraocular surgery; however, for the current study this was revised to allow subsequent IVTA or interventions intended to lower IOP, to parallel the Florida-2 cohort. Clinical data was updated by retrospective chart review.
Measurement of IOP
Intraocular pressure was recorded in the study eye before treatment with IVTA (baseline IOP). All IOP measurements were performed according to the treating physician's standard practice, typically applanation tonometry or Tono-Pen (Reichert Technologies, Buffalo, NY, USA). On each follow-up visit, IOP was recorded (follow-up IOP) for up to 1 year, or until the study eye met any of the predetermined exit criteria.
Change in IOP (ΔIOP) was defined as the difference between baseline IOP and the maximum IOP measurement for 6 months after each injection. This was used as the quantitative trait (QT) for each research subject. A positive ΔIOP indicated an elevation in IOP following IVTA. If both eyes were treated, the eye with the highest ΔIOP was selected. If a second IVTA injection was given during the 6-month follow up, the last IOP used to determine the ΔIOP was the one the day of the subsequent IVTA injection. If both eyes were treated, the eye with the highest ΔIOP was selected.
For this study, a “steroid responder” was defined, according to Armaly and Becker,32 as an individual with normal IOP (12–22 mm Hg) who has a positive ΔIOP > 6 mm Hg above baseline IOP following injection of IVTA.
DNA Preparation
Peripheral venous blood was obtained from all research subjects. Genomic DNA was extracted from leukocytes using QIAamp DNA Blood Mini Kit from Qiagen, Inc. (Valencia, CA, USA).
Genotyping and Quality Control
DNA samples from the 79 subjects enrolled into the Florida-2 cohort were genotyped using the Illumina HumanOmni2.5-Quad (Omni2.5) BeadChip at the Epigenomics Center Genomics Core facility of the University of Southern California using Illumina GenomeStudio software (v2011.1; Illumina, Inc., San Diego, CA, USA). Genotyping reproducibility was confirmed by including two duplicate samples.
Genome-wide association study quality control (QC) was performed using PLINK (v1.07) software following Illumina protocols and published procedures43,44 at sample and single nucleotide polymorphism (SNP) levels. Of the 79 original subjects, 64 survived the QC procedure with all having a genotyping call rate > 95%. The 15 individuals who failed QC were dropped from further analyses. This included individuals with a substantial amount of African ancestry (n = 5), ambiguous sex (n = 2), unexpected duplicates (n = 2), and individuals with missing IOP phenotype (n = 6) in our genotyped samples.
Of the 2,449,708 SNPs successfully genotyped, a total of 1,545,328 SNPs were included in the final analysis after exclusion from either a low call rate < 90% (65,469), deviation from Hardy-Weinberg Equilibrium with a P value for HWE test < 0.001 (2058), or a minor allele frequency < 0.05 (849,444).
DNA samples from the 52 subjects enrolled into the Florida-1 cohort were previously genotyped using the GeneChip Human Mapping 500K Array Set from Affymetrix in the Genomics Core facility at The Scripps Research Institute (Jupiter, FL, USA).45
Population Stratification
We used PLINK to remove outlier individuals and SNPs in high linkage disequilibrium (LD) or in regions of extended LD. Principal components of genetic ancestry were inferred using EIGENSOFT.46 The genomic control inflation factor47 was calculated and a quantile–quantile (Q–Q) probability plot was generated to visualize the distribution of the test statistics.
Statistical Analysis
Association analysis was conducted using linear regression, treating an inverse-normal transformation of ΔIOP as a continuous variable, with adjustment for age, sex, and principal components of genetic ancestry, and assuming an additive genetic effects model. Genotyped SNPs were analyzed using PLINK software. For a meta-analysis with the Florida-1 and Florida-2 cohorts, a fixed effects model was used, including a test for heterogeneity, as implemented in the meta R package on tests of the overall treatment effect in meta-analysis with normally distributed responses.48
SNP Validation
TaqMan analysis was used to validate genotypes for specific SNPs of interest, or to determine genotypes for SNPs not on the microarrays. Allelic discrimination analysis was performed using SDS 2.3 software and the data were analyzed in TaqMan Genotyper v1.1 (LTI).
Bioinformatics
HaploReg v2 software (update 2013.02.14)49 was used to search for proxy SNPs of the index SNPs identified in this study. ConSite50 was used to identify binding motifs for transcription factor NF-κB. Association of SNPs with gene expression was evaluated using the NCBI GTEx eQTL browser.51 ProtoNet software52 was used to identify proteins with sequence similarity to HCG22.
To assess the likely effect of SNPs on transcription factor response elements, we first identified matches to well-established positional weight matrices (PWM) from Factorbook53 and HOCOMOCO,54 and then determined whether the affected residue is critical within the motif. For consideration, the proposed effect had to be in a response element that scored 80% or better of the maximum PWM score, and the difference of frequency between the reference genome and alternate SNP value had to be greater than 60%. A full mathematical description of the algorithm has been reported previously.55
Cultured Cells
Primary human corneal epithelial cells (purchased from Lifeline Cell Tech, Frederick, MD, USA) were grown in Oculife Complete Medium Kit, provided by the supplier, and used for experiments at passage 4. The immortalized human trabecular meshwork cell line, TM-1, kindly provided by Donna Peters (University of Wisconsin, Madison, WI, USA), was maintained as described.56 Primary human TBM cells were prepared from corneal rims discarded at the time of surgical transplantation of donor corneas, as described.57 Primary TBM cells before passage 8 were used for experiments.
DNA Constructs and Transfections
Total RNA purified from BET1A, a human bronchial epithelial cell line, was used to synthesize cDNA, from which HCG22 cDNA (GenBank AB560771) was PCR-amplified and cloned into the pCR2.1-TOPO vector using the TOPO TA Cloning kit (Invitrogen, Carlsbad, CA, USA), using the oligonucleotide primers 5′-CTA GCA CCA TGT GGA TTC TCT TGA CAT GAT GAG-3′ and 5′-CCA ACA AAG AGC ACA ACA GTG ACT G-3′. The DNA sequence of cloned cDNA was confirmed by DNA sequencing and was found to contain two amino acid substitutions (R to G and E to Q) in the signal peptide region (see Results). EcoR1 and Not I restriction enzyme sites in pcDNA3.1+ vector (Invitrogen) were used to clone and express recombinant HCG22 protein with a His tag at the C-terminus, using forward primer, 5′-GAT CGA ATT CAT GCC CGG CTA CGT CC-3′, and reverse primer, 5′-GCT ACG ATG CGG CCG CTC AGT GGT GGT GGT GGT GGT GCA GAA GGT GAG TGT CAC CAT GTT TA-3′. To express the HCG22-His tag cDNA, TM-1, and HEK293 cells were transfected with 2 μg of the vector plasmid using Lipofectamine LTX reagent (Invitrogen) according to the supplier's protocol. A HEK293 cell line stably expressing HCG22-His was made by culturing the transfected cells in the presence of 800 μg/mL G-418 (a selective agent).
Expression Analysis
The TM-1 cells were grown in 6-well plates at 0.12 millions of cells/well and cultured for 4 days before serum starvation for 7 hours. Cells then were stimulated by adding IL-1β (10 ng/mL), TGF-β2 (10 ng/mL), or TA ((100 ng/mL; Sigma-Aldrich Corp., St. Louis, MO, USA) for 17 hours. Total RNA was purified using TRIZol (Ambion, Grand Island, NY, USA). The cDNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA), and quantitative RT-PCR (qRT-PCR) was performed using iQ SYBR Green Supermix (Bio-Rad) under thermal cycling conditions of 30 seconds at 95°C, 10 seconds at 60°C, and 10 seconds at 72°C. Relative transcript amounts were normalized by GAPDH expression in individual treatment samples. The following PCR primer pairs were used: GCGAAGCCTGGTGGGT and CAGGGACAGGCTGTTTCTCA for MUC21; ACCAACACACCTATGTCAGGC and TCCTGGCACAGTATGGCTG for MUC22; CGAGCGAAGGTACGGAATAGTA and GTCCTCAGTGGTCCTTCCATC for HCG22, which spans exon2-exon3 junction in the coding transcript that is not present in the noncoding HCG22 transcript; GAGTCAACGGATTTGGTCGT and GGCAACAATATCCACTTTACCAGAGT for GAPDH.
O-Glycosylation Test
The TM-1 and HEK293 cells were transiently transfected with HCG22-His vector for 1 day to observe the secretion of HCG22 protein. The HEK293 cells stably transformed with HCG22-His were grown until reaching 80% confluence in a 12-well plate. After washing the wells twice with serum-free Dulbecco's modified Eagle's medium (DMEM), cells were treated with benzyl-2-acetamido-2-deoxy-alpha-D-galactopyranoside (benzyl GalNAc; Sigma-Aldrich Corp.) in serum-free medium for 24 hours at a range of doses. The culture supernatant was collected and subjected to SDS-PAGE and Western blotting. The membrane was probed using anti-His antibody (Abcam, Cambridge, MA, USA) to detect the expressed recombinant protein.
Results
Genome-Wide Association Study
Clinical data on the 64 white subjects from the Florida-2 cohort used for the present analysis are shown in Table 1. A Q–Q plot of SNP association P values (genomic control inflation factor corrected) showed that the observed P values did not differ from their expected values, except at the extreme tail (Supplementary Fig. S1). Thus, this result demonstrates good control of population stratification for our white subjects.
Table 1.
Clinical Data for the Florida-2 (Discovery) Cohort and the Florida-1 (Replication) Cohort
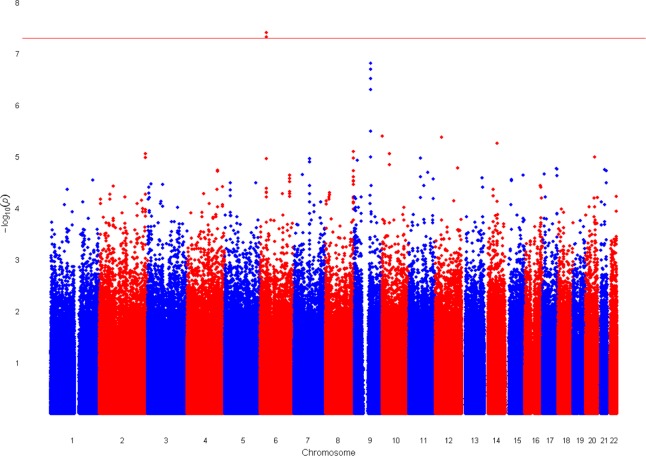
Linear regression analysis was performed to determine association between the quantitative trait and genotype. The results are represented by the Manhattan plot in Figure 1, where the genome-level significance threshold was set at P = 5 × 10−8. Two SNPs, rs2523864 and rs2251830, on chromosome 6 exceeded this threshold and yielded P values of 3.92 × 10−8 and 4.74 × 10−8, respectively. These SNPs are in perfect LD (r2 = 1). A second peak on chromosome 9 reached borderline significance, with the top SNP having a P value of 1.56 × 10−7. This SNP is located in an intragenic region upstream of the TLE4 gene, which encodes a transcription factor. Other regions of the genome with suggestive evidence of association included two SNPs in perfect LD (P = 4.09 × 10−6 for each; r2 = 1) on chromosome 10, located in an intron of the ADAR2 gene (an RNA editing enzyme), and a SNP on chromosome 20, located in an intron of the EYA2 gene (a tyrosine kinase, P = 1.03 × 10−5).
Figure 1.
Manhattan plot of the GWAS for steroid-induced OH. The Manhattan plot depicts the genome-wide P values. The results for the total of 1,545,328 genotyped SNPs that passed the quality control criteria are plotted as −log10(P value) by genomic position. The red line indicates the genome-wide significance level of P = 5 × 10−8.
The statistically significant peak is situated at chromosomal locus 6p21.33 (Fig. 2). This region lies within the 3.6 Mb major histocompatibility complex (MHC) class I locus in the region between the human leukocyte antigen genes HLA-A and HLA-B, approximately 300 kb telomeric of HLA-B. Early studies linked this region to psoriasis, an inflammatory skin disease.58–61 Single nucleotide polymorphisms in this region also have been associated with two inflammatory diseases of the eye. The first is Behçet's disease, a rare immune-mediated systemic vasculitis that often presents with mucous membrane ulceration and ocular involvements that include a secondary form of glaucoma.62 The second is Stevens-Johnson syndrome, a hypersensitivity complex that affects the skin and mucous membranes, including the cornea of the eye.63 Most recently, SNPs in this region have been associated with dilated cardiomyopathy64 and diffuse panbronchiolitis (DPB), a rare, genetically inherited disease of the lungs that primarily affects Asians.65 A complete gene annotation was performed in connection with the DPB study, revealing two novel protein-coding genes PBMUCL1 and PBMUCL2, that mapped to this region.65 Conceptual translation revealed that PBMUCL1 is a new member of the MUC family of mucin genes and has been given the HUGO designation of MUC22. The PBMUCL2 (Genbank: AB560771) gene was found to overlap the gene HCG22, previously associated only with a noncoding transcript (Genbank: AK094433.1). The HUGO Gene Nomenclature Committee (HGCN) subsequently recognized HCG22 as the gene for the coding transcript. The two lead SNPs we identified map to the intergenic region between MUC22 and HCG22 (Fig. 2).
Figure 2.
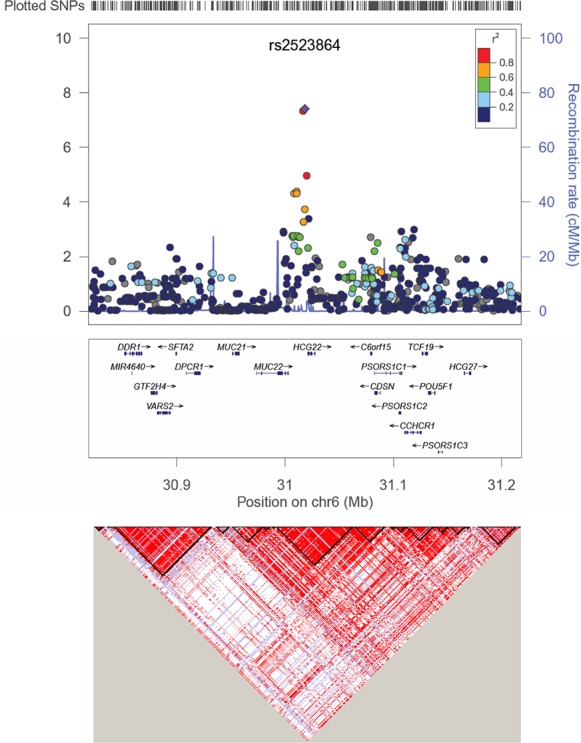
Regional association plot for the lead SNP rs2523864 (marked as purple diamond) on Chr6. All SNPs shown as circles are plotted with their respective P values against their genomic location. The solid diamond indicates the top-ranked SNP rs2523864. The colored box at the right or left corner of each plot indicates the pairwise correlation (r2) between the top SNP and the other SNPs in the region. The gray circles indicate the imputed SNPs from the CEU population of the HapMap. Each plot was created using LocusZoom (available in the public domain at http://csg.sph.umich.edu/locuszoom/) for the top-ranked SNP in each region with a 400-kb region surrounding it. The SNPs are plotted at the top of the figure. The box underneath each plot shows the gene annotations in the region, with the arrow indicated the DNA strand for transcription. The lower LD plot was created using Haploview (available in the public domain at http://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview).
Replication
To confirm the association signal at the chromosome 6p21.33, we used the Florida-1 cohort as an independent replication dataset. Clinical data on the 49 subjects surviving QC analysis are shown in Table 1. Since neither of the peak SNPs for the chromosome 6p21.33 locus was present on the Affymetrix chipset previously used for the Florida-1 cohort, we determined genotypes by Taqman analysis. As noted in the discussion of the Florida-2 cohort, rs2523864 and rs2251830 SNPs are in perfect LD (r2 = 1). Therefore, we only analyzed rs2523864 for replication purposes. Linear regression analysis yielded an effect estimate in a similar direction as in the Florida-2 cohort, with a corresponding P value of 0.18 for association of rs2523864 with IOP. Given the directionally consistent effect of rs2523864, we next carried out a meta-analysis that combined the association results in the Florida-1 and Florida-2 cohorts (n = 113). This analysis significantly strengthened the association of rs2523864 with IOP by one order of magnitude (combined P = 5.34 × 10−9), supporting the notion that this variant influences the response to GCs.
HCG22 Expression Analysis
Glucocorticoids bind the GC receptor (GCR), which acts as a transcription factor to stimulate expression of specific genes. The proximity of the peak SNPs on chromosome 6 to the transcriptional start site for HCG22 suggests that the molecular mechanism for association of these variants with IOP could be through regulation of HCG22 gene expression. If altered transcription of HCG22 is a causal factor in steroid-induced OH, we further reasoned that the gene would be expressed in the TBM. To test this hypothesis, we performed RT-PCR expression analysis with primers specific for the coding transcript of HCG22 using the immortalized TBM cell line, TM-1. For comparison, the adjacent genes for MUC22 and MUC21 also were included in this analysis (Fig. 3). The MUC-type mucins are known to be expressed at high levels in the ocular surface epithelia, but have not been connected previously with the aqueous outflow pathways of the eye. Thus, we compared expression in TM-1 cells to expression in primary HCE cells. The MUC21 mRNA expression was specific for HCE cells, but MUC22 and HCG22 were expressed in HCE and TM-1 cells. This represents a novel finding, since mucins have not previously been associated with cells of the aqueous outflow pathways. These results were confirmed using primary TBM cells (data not shown).
Figure 3.
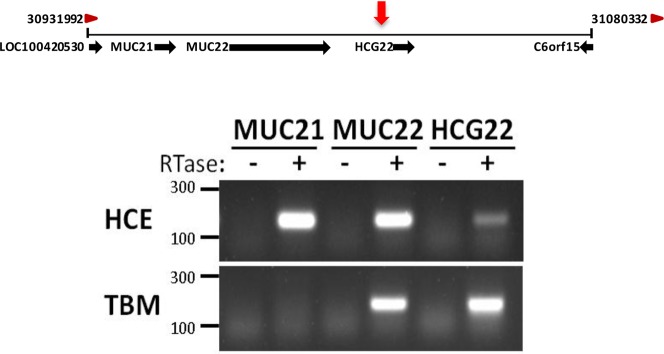
Location and expression of genes surrounding the identified QTL in chromosomal region 6p21.32-33. Top: Schematic of chromosomal region 6p21.32-33 from the NCBI Gene website depicting annotated genes surrounding the identified QTL (red arrow). Bottom: Total RNA was purified from cultured primary HCE and TM-1 cells and used for cDNA synthesis. Reverse transcription-PCR using cDNA was performed using gene-specific primers from MUC21, MUC22, and HCG22, and the products were resolved on a 1.5% agarose gel. Primers for HCG22 were designed to detect only the coding transcript. Similar results were obtained using three primary TBM cell lines (not shown). RTase, reverse transcriptase; HCE, primary corneal epithelial cells obtained from corneal rims.
We next used RT-PCR to quantify HCG22 mRNA expression in TM-1 cells treated with GCs and with IL-1β and TGF-β, agents that are controlled by GCs and associated with OH15,16,65 (Fig. 4). Treatment with IL-1β stimulated HCG22 expression by approximately 4-fold. Treatment with TA inhibited basal and IL-1β–stimulated expression. Transforming growth factor-β inhibited HCG22 expression to approximately 0.3-fold. These results also were confirmed using primary TBM cells (data not shown).
Figure 4.
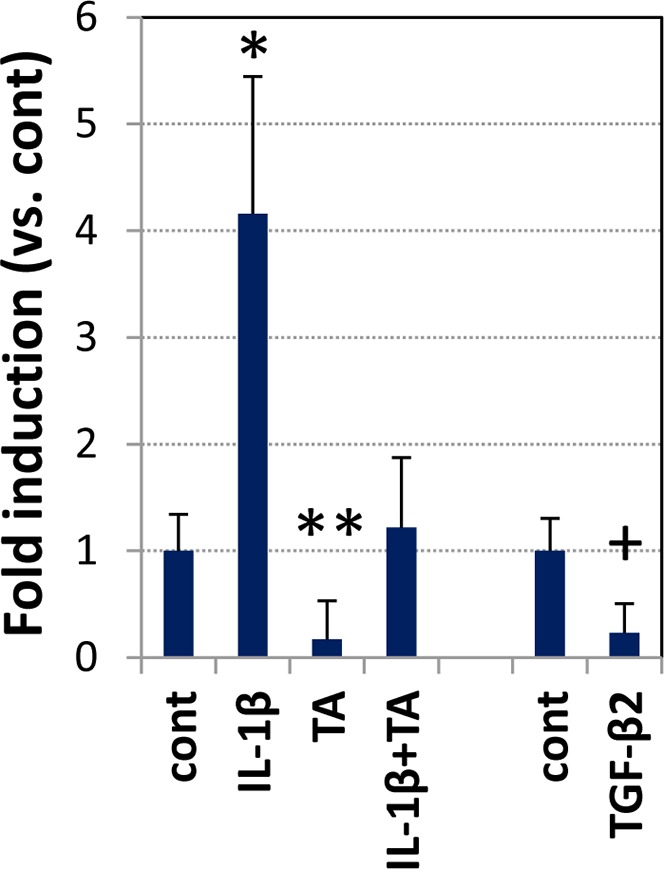
Transcriptional analysis. Trabecular meshwork-1 cells in 12-well plates were treated with IL-1β (10 ng/mL), TGF-β2 (10 ng/mL), and TA (100 ng/mL), individually or in combination, in serum-free media for 17 hours. The HCG22 mRNA levels were measured by qRT-PCR. Relative expression of HCG22 was normalized by the expression of GAPDH in individual samples. Student's t-test was used to identify the statistical significance between treated and untreated samples: *P < 0.01 (n = 5), **P = 0.02 (n = 5), P = 0.07 (n = 4).
To determine any functional impact of the quantitative trait locus on HCG22 expression, we first used bioinformatics tools to search for proxy SNPs of the lead SNP, rs2523864. Using the 1000 Genomes CEU Phase 1 data implemented in HaploReg v2 software,49 we identified 10 SNPs spanning a region of approximately 4 kb that were in high LD with rs2523864 (r2 ≥ 0.8) (Table 2). This LD block is situated upstream of HCG22, located approximately 1.8 kb from the transcriptional start site for all transcripts.
Table 2.
Tag SNPs and Variants in LD (r2 ≥ 0.8) and Functional Associations
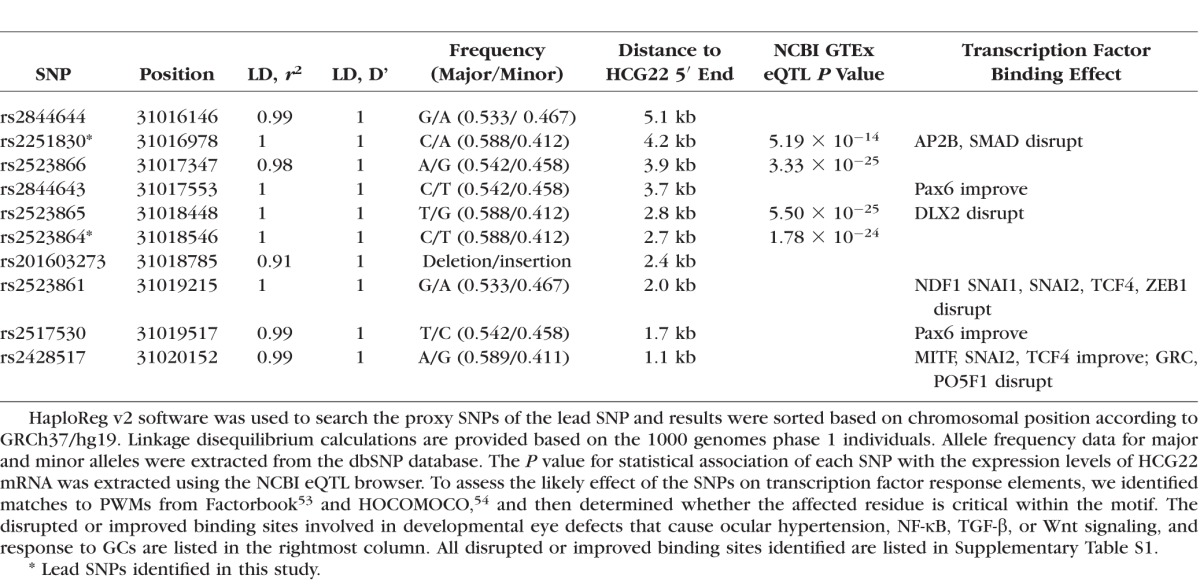
The National Center for Biotechnology Information (NCBI) GTEx eQTL browser is a central resource that archives and displays results of a national research project for determining association between genetic variation and high-throughput molecular-level expression phenotypes, and this information can provide insight into the biological relevance of data from GWA studies.51 We used this resource to determine whether SNPs in the identified LD block were associated with HCG22 expression levels in the lymphoblastoid cell line used by NCBI. This analysis revealed highly significant eQTL P values for four SNPs, two of which are our index SNPs (Table 2).
To assess the likely effect of the SNPs in the LD block on transcription factor response elements, we identified matches to well-established PWMs from Factorbook53 and HOCOMOCO,54 and then determined whether the affected residue is critical within the motif. We found that SNPs in our LD block alter binding motifs for a number of transcription factors (Supplementary Table S1). Those involved in developmental eye defects that cause ocular hypertension, NF-κB, TGF-β or Wnt signaling (pathways associated with OH), and response to GCs are listed in Table 2.
The DLX2 and Pax6 cause Axenfeld-Rieger syndrome, a disorder that manifests as anterior segment dysgenesis and congenital glaucoma.66 The NDF167 and AP268 genes control retinal development. The MITF gene is implicated in microphthalmia and Waardenburg syndrome, a retinal disorder.69 Genes SNAI1 and SNAI2 also are implicated in Waardenburg syndrome.69 Genes TCF4 and ZEB1 are implicated in Fuch's endothelial dystrophy, a disease of the corneal endothelial layer.70,71 Genes TCF4 and ZEB1 also are involved in determining central corneal thickness, which is a modifying genetic factor in OH.72,73
Other altered motifs bind transcription factors are involved in intracellular signaling associated with OH. The SMAD motif mediates TGF-β signaling. The PO5F1 and TCF4 motifs mediate Wnt signaling. Most striking, however, from the perspective of biological relevance, is predicted disruption of the binding motif for the GCR that mediates response to GCs.74,75
Response to IL-1 is mediated by transcription factor NF-κB, and this pathway has been associated with OH in POAG.15 Since no NF-κB or related transcription factor elements appeared in the transcription factor binding site search, we used the web-based bioinformatics tool Consite50 to search the DNA sequence of our approximately 4-kb LD block for transcription factor binding sites that could mediate response to NF-κB. We identified eight canonical NF-κB response elements.
HCG22 Coding Analysis
Previous analysis of the hypothetical protein product of the coding transcript of HCG22 (Genbank: BAJ24155.1; UniProtKB/Swiss-Prot: E2RYF7.1) suggested it as a non-MUC–type mucin.65 Following the N-terminal signal sequence are 15 tandem mucin repeats within a threonine-rich C-terminal region of 160 amino acids. The lack of a transmembrane domain suggests that HCG22 mucin protein is secreted. Glycosylation patterns were predicted by analysis of coding regions of the genes using web-based software, revealing numerous sites for O-linked glycosylation within the tandem repeats. The predicted molecular weight of the HCG22 protein backbone is approximately 26 kDa, but the actual size could be considerably larger due to the addition of sugar chains.
To validate the predictions based on amino acid sequence analysis, HEK293 cells transfected with the HCG22 expression construct were left untreated or treated with Benzyl-GalNAc, which selectively inhibits the elongation and sialylation of mucin-type O-glycan chains.76,77 Conditioned culture media was harvested for Western blotting after 24 hours. Figure 5 shows the results of Western blotting using an antibody to the His tag at the N-terminus, which revealed a band corresponding to 70 to 75 kDa protein. The molecular size of this protein decreased with increasing doses of Benzyl-GalNAc. These results are consistent with HCG22 translation into a protein product that is O-glycosylated and exported outside the cell.
Figure 5.
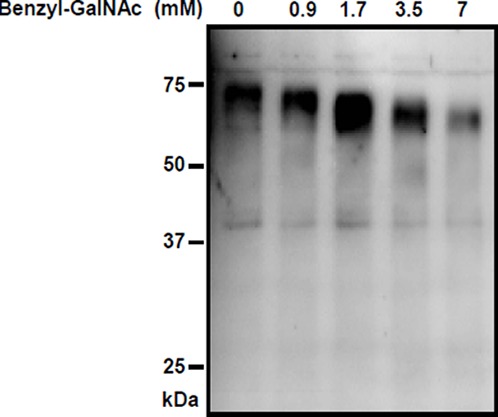
Translation, O-glycosylation and secretion of HCG22 protein. The HEK293 cells expressing HCG22-His tag protein were cultured in the presence of benzyl-GalNAc at indicated doses. After 24 hours, the conditioned media were harvested. Secreted proteins were resolved on a denaturing 10% SDS/PAGE gel, blotted, and probed with anti-His antibody.
Application of the web-based computer program ProtoNet52 reveals that the HCG22 protein product is most closely related to peritrophin-A, a protein found in the insect digestive tract.
Second Quantitative Trait Locus
The cloned insert of the HCG22 expression construct was sequenced as part of a routine procedure to confirm its identity. Unexpectedly, we identified two nucleotide substitutions in the signal peptide corresponding to two SNPs also present in dbSNP (rs3873352 and rs2523855), each of which predicts an amino acid substitution (NH3-MPRYVPLLLLLLLLRCSERGGG). The substitution due to rs3873352 would cause a change from arginine to glycine at the third amino acid. This represents a change from a positive charge and hydropathy index of −4.5, to a neutral charge and hydropathy index of 0.4. Such a change is likely to affect secretion. The second substitution due to rs2523855 would result in a glutamic acid to glutamine substitution at 18th amino acid, a conservative change that is less likely to have functional consequences. Importantly, the two SNPs are not in LD with each other, or the two SNPs upstream of HCG22 identified in our GWAS analysis (rs2523864 and rs2251830). Given these newly discovered variants are amino acid substitutions that could potentially affect HCG22 function, we carried out a candidate gene analysis to determine whether they were associated with IOP in the Florida-2 cohort. This revealed that the SNP leading to a nonconservative change, rs3873352, also was significantly associated with IOP (P = 2.81 × 10−4), whereas the second SNP, rs2523855, was not significantly associated.
Discussion
The pathophysiology of OH caused by treatment with steroids shares many features with the much more common form of OH leading to POAG, but also has some distinct differences.19–25 Thus, steroid-induced OH provides a valuable model that could help shed light on our understanding of POAG, but also could provide novel insight into the regulation of IOP. Applying a pharmaco-GWAS and a candidate gene approach, we identified two independent QTLs associated with HCG22 at chromosomal locus 6p21.33. Results of follow-up studies indicate that one of these QTLs may affect transcription of HCG22, and one may affect secretion of HCG22 protein in the aqueous outflow pathways. Thus, the first QTL could affect levels of the noncoding and coding transcripts derived from HCG22, while both QTLs could alter levels of the encoded protein. The identification of two independent QTLs that are associated with IOP, possibly through different molecular mechanisms (i.e., gene transcription and protein secretion), is highly suggestive that HCG22 has an important role in steroid-induced OH.
Genetic effects in pharmacologic response can be quite large in comparison with those involved in most complex diseases, and may involve fewer genes.78 In fact, several recent pharmacogenomic case-control studies of approximately 100 cases have detected polymorphisms of genome-wide significance.78 Our 2-stage GWAS comprised two cohorts, analyzed as discovery and replication groups, then combined by meta-analysis for a total of 113 subjects. Analysis of the discovery cohort dataset resulted in a P value of genome-wide significance and the overall P value increased in magnitude (P = 5.34 × 10−9) when the cohorts were combined. However, these findings warrant further investigation, since GWA studies are inherently hypothesis-generating exercises, and function is the ultimate test of significance. We note that the genetic associations we identified are supported by initial functional follow up studies reported here, which serve as a starting point for more detailed characterization of HCG22 and its role in the development of OH in response to GCs.
The method used in this study to assess the quantitative trait was conservative, and this could partially explain why the Florida-2 results did not replicate in the Florida-1 cohort. The ΔIOP was calculated as the maximum difference between baseline and follow-up IOPs and the timing of the follow-up visits may not have corresponded with the highest IOP attained. Moreover, high responders are treated early with IOP-lowering medications, meaning these patients would never demonstrate their full potential. Our discovery and replication sets were obtained in two different physician practices, where steroid response may have been handled with different levels of aggressiveness. Each of these factors could lead to an underestimate of the ΔIOP in the Florida-1 cohort. Adding to this, the Florida-1 cohort was smaller than the Florida-2 cohort. However, the identified variants had directionally consistent effects in both cohorts and the combined meta-analysis increased the strength of the association by one order of magnitude, supporting the notion that variants at the HCG22 locus influence the response to GCs.
The two QTLs identified in this study are located at chromosomal locus 6p21.33 within the 3.6 Mb MHC class I locus, between the genes for HLA-A and HLA-B, a little more than 300 Kb telomeric of HLA-B. Interestingly, studies published more than 35 years ago, identified association between HLA-A, HLA-B, and POAG79–81 and a study comparing cellular responsiveness to GCs and HLA classification for 25 POAG patients and 25 individuals who respond to GCs with IOP > 31 mm Hg found an association with HLA-B12.82,83
The first QTL identified in this study mapped to an LD block of approximately 4.0 kb, which is situated in an intergenic area approximately 1.1 kb upstream of the transcriptional start site of the HCG22 gene. This suggested that the QTL might affect the regulation of HCG22 transcription by GCs. We found that TBM cells express HCG22, and expression is stimulated by IL-1 and inhibited by TGF-β. We also found that GCs inhibit constitutive and IL-1–stimulated expression of HCG22. Elevated IL-1/NF-κB and TGF-β signaling have been associated with OH leading to POAG, and GCs act in part by modifying IL-1 and TGF-β signaling. Thus, these results are consistent with a role in the disease process.
Gene expression phenotypes are heritable84 and can be mapped as eQTLs.85,86 The basis of eQTL analysis is that SNPs could have a tangible effect on gene regulation by altering the coding or promoter sequences of genes, their splicing junctions, or other regulatory elements that determine the final expression level of a gene or genes. Thus, SNPs suspected of affecting gene expression can be tested with associative statistics. Using NCBI's GTEx eQTL browser to search genome-wide datasets, we identified four SNPs within our identified LD block that affect unstimulated HCG22 mRNA levels. The datasets were obtained using a lymphoblastoid cell line, however GC response in lymphocytes has been shown to predict GC response in other organs,87>–89 including the eye.81–83,90 Thus, these eQTLs are likely to be relevant to the aqueous outflow pathways. Additional experiments will be needed to confirm this hypothesis.
Bioinformatics analysis was used to predict disruption/improvement of transcription factor binding by each of the SNPs in the LD block. Several of the factors that bind to the motifs identified are expressed in the eye and are involved in eye development and developmental disorders and, thus, are likely to be relevant to eye physiology. Several others are involved in signaling pathways altered in OH. However, most appealing with regards to potential mechanism was the GCR binding motif disruption that was identified. Functional analysis linking the DNA in our LD block to a transcriptional reporter gene can be used in future studies to determine how this and other candidate SNPs specifically affect GC-induced transcription in TBM cells. Since the SNPs in the identified LD block are perfectly correlated genetically, it is possible that more than one SNP contributes to the phenotype associated with this QTL.
The GCR gene NR3C1 generates two transcription factor isoforms: GCRα and GCRβ.91 Isoform GCRβ is a truncated form of GCRα that lacks the ligand-binding domain. It has a dominant-negative effect on GCRα-induced transactivation of promoters activated by GCs. However, GCRβ also has intrinsic, GCRα-independent transcriptional activity, regulating mRNA expression of a large number of genes negatively or positively.74,75 The GCRβ/GCRα ratio is reduced in a variety of diseases of GC sensitivity, including asthma, rheumatoid arthritis, and ulcerative colitis.35,36,92 The GCRβ/GCRα ratio also is reduced in glaucomatous TBM cells, and it was shown recently that this contributes to their GC sensitivity.39,93 It is conjectured that the relative reduction of GCRβ in relation to GCRα might be caused by elevated proinflammatory cytokines.94 This notion also is relevant to OH in POAG, which is associated with elevated IL-1α/NF-κB signaling.15
An important outcome of these studies is the implication that the HCG22 mucin protein may be a causative factor in steroid-induced OH. Previous analysis of the predicted amino acid sequence of the HCG22 coding transcript revealed features consistent with a small, secreted, O-glycosylated mucin-like protein.65 Here, we showed for the first time to our knowledge that this transcript can be translated, O-glycosylated, and exported outside the cell as an approximately 70 to 75 kDa mucin protein. Our application of the web-based computer program ProtoNet reveals that HCG22 is most closely related to peritrophin-A, a mucin protein found in the peritrophic matrix of the insect digestive tract. Comprised of a network of glycoproteins, proteoglycans, and chitin fibrils, the peritrophic matrix functions as a molecular sieve to partially digested protein and carbohydrate, as a barrier to ingested pathogens, as a scaffold for proteases and glycosidases, and as a sink for toxic substances.95 As noted above, we found that GCs inhibit constitutive and IL-1-stimulated expression of HCG22. Thus, it is tempting to speculate that disrupted binding of the GCR in individuals with the minor genotype may prevent the GC-mediated inhibition of HCG22 expression and contribute to GC-induced OH. One idea is that too much HCG22 protein is detrimental to the trabecular meshwork cell under conditions of GC treatment or physiological stress. Further studies will be needed to test this hypothesis.
Sequencing of the cDNA insert of the expression construct used for these studies revealed two SNPs within the putative signal peptide of HCG22, one that would cause a nonconservative amino acid change, and one that would cause a conservative change. Signal peptides are surprisingly complex and single amino acid substitutions can have major effects on their function in secretion.96 Statistical analysis revealed a significant association between the nonconservative SNP and steroid response. Future studies will be needed to quantify the SNP effects on secretion efficiency and/or intracellular distribution of HCG22 in cultured TBM cells.
While this manuscript was under review, sequences of three new HCG22 noncoding transcripts were submitted to Genbank (HG499131.1, HG499132.1, and HG499135.1). It has been reported that only approximately 20% of transcription progress across the human genome is associated with protein-coding genes.97 Because of recent advances in RNA sequencing and computational methods, long noncoding RNAs are emerging as key regulators of diverse cellular processes.98–100 The relative involvement in steroid-induced ocular hypertension of the HCG22 coding mRNA and the HCG22 noncoding mRNAs remains to be investigated in future studies.
Only a few other published studies have used genetics to prioritize genes associated with steroid-induced OH. Myocilin (MYOC), the first gene identified for hereditary forms of POAG,101 had already been defined as a highly induced protein in TBM cell cultures treated with GCs.102,103 However, subsequent analysis revealed no evidence for a link between steroid-induced OH and POAG-causing mutations.104,105 A candidate gene study using a cohort from trials of topically administered GCs following photorefractive keratectomy found a significant correlation between N363S heterozygosity and steroid-induced OH in a small subgroup treated with prednisolone acetate.106 However, using the Florida-1 cohort described here, we found no statistically significant associations of response to IVTA with six candidate polymorphisms in NR3C1 previously associated with systemic GC sensitivity, including N363S.42 A similar study from another group on response to IVTA also found no significance.107 Thus, this result remains to be replicated by other researchers. To our knowledge, none of the currently ongoing GWA studies for POAG, OH, or related disorders has identified this locus. This suggests the possibility that it is specific for steroid-induced OH.
The past decade has seen the emergence of a first generation of effective pharmaceuticals for retinal diseases. Among these, GC drugs have shown much promise. For example, off-label use of TA, delivered by intravitreal injection (i.e., IVTA), has been used as primary or adjunctive treatment for a variety of retinal diseases, including choroidal neovascularization,108 macular edema secondary to diabetic retinopathy,109 and retinal vein occlusion.110 However, when GCs are administered intravitreally, IOP may increase by 30% or more in up to half of patients,33 usually within 3 months.111 The IOP elevation may be prolonged33,112 and intractable,113,114 requiring surgical intervention.115 Identification of SNPs associated with response to steroids in the eye ultimately may provide a valuable predictive tool that could make GC drugs safer, by identifying patients who are steroid responders before treatment. Expansion of the current datasets and additional independent replication will be needed to determine whether the tag SNPs reported here might be used for this purpose.
Supplementary Material
Acknowledgments
The authors thank William Smiddy, MD (University of Miami, Miami, FL, USA), for patient referral to the Florida-1 cohort, Sabrina Gerzenstein (University of Miami, Miami, FL, USA) for technical assistance in preparation and banking of the Florida-1 cohort DNA samples, Donna Peters, MD (University of Wisconsin, Madison, WI, USA), for the gift of the TM-1 cell line, and Pragna Patel, MD (University of Southern California, Los Angeles, CA, USA), for helpful discussions on the genetic approaches.
Supported by National Institutes of Health (NIH; Bethesda, MD, USA) Grants R01-EY009828 (MEF), R01-EY022651 (XG), R01- CA136924 (GAC), UL1-RR031986 (pilot project subaward, MEF), P30-EY014801 (University of Miami), and P30-CA014089 (NIH/National Cancer Institute, USC); Department of Defense Grants W81XWH-09-1-0675 and W81XWH-13-1-0048 (University of Miami); a Schaffer Innovative Glaucoma Research Grant from the Glaucoma Research Foundation (available in the public domain at http://www.glaucoma.org; MEF); a Senior Scientific Investigator Award from Research to Prevent Blindness (available in the public domain at http://www.rpbusa.org/rpb; MEF); an unrestricted grant from Research to Prevent Blindness (available in the public domain at http://www.rpbusa.org/rpb, University of Miami); an Investigator Award from Prevent Blindness America, Chicago, Illinois, United States (available in the public domain at http://www.preventblindness.org/; SGS); an Investigator Award from Prevent Blindness Florida, Tampa, Florida, United States (available in the public domain at http://florida.preventblindness.org/; SGS); the Retina Health Center, Fort Myers, Florida, United States (available in the public domain at http://www.retinahealthcenter.com/; to support research staff involved in sample and data acquisition, AME); IC Labs, Fort Myers, Florida, United States (costs of the Illumina chip screening in the USC Epigenomics Center Core Facility, AME); Walter G. Ross Distinguished Chair in Ophthalmic Research at the University of Miami (MEF); and the Keck School of Medicine of USC (start-up funding for MEF).
Disclosure: S. Jeong, None; N. Patel, None; C.K. Edlund, None; J. Hartiala, None; D.J. Hazelett, None; T. Itakura, None; P.-C. Wu, None; R.L. Avery, IC Labs (F, I, S), P; J.L. Davis, None; H.W. Flynn, IC Labs (F, I, S), Retina Health Center (F, I, S), P; G. Lalwani, None; C.A. Puliafito, None; H. Wafapoor, None; M. Hijikata, None; N. Keicho, None; X. Gao, None; P. Argüeso, None; H. Allayee, None; G.A. Coetzee, None; M.T. Pletcher, None; D.V. Conti, None; S.G. Schwartz, None; A.M. Eaton, IC Labs (F, I, S), Retina Health Center (F, I, S), P; M.E. Fini, None
References
- 1.Congdon N, O'Colmain B, Klaver CC, et al.; Eye Diseases Prevalence Research Group. Causes and prevalence of visual impairment among adults in the United States. Arch Ophthalmol. 2004; 122: 477–485. [DOI] [PubMed] [Google Scholar]
- 2.Quigley HA,, Broman AT.The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006; 90: 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fautsch MP,, Johnson DH.Aqueous humor outflow: what do we know? Where will it lead us? Invest Ophthalmol Vis Sci. 2006; 47: 4181–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowston JG,, Weinreb RN.Glaucoma medication and aqueous humor dynamics. Curr Opin Ophthalmol. 2005; 16: 94–100. [DOI] [PubMed] [Google Scholar]
- 5.Acott TS,, Kelley MJ.Extracellular matrix in the trabecular meshwork. Exp Eye Res. 2008; 86: 543–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ethier CR.The inner wall of Schlemm's canal. Exp Eye Res. 2002; 74: 161–172. [DOI] [PubMed] [Google Scholar]
- 7.Johnson M.What controls aqueous humour outflow resistance? Exp Eye Res. 2006; 82: 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maepea O,, Bill A.Pressures in the juxtacanalicular tissue and Schlemm's canal in monkeys. Exp Eye Res. 1992; 54: 879–883. [DOI] [PubMed] [Google Scholar]
- 9.Epstein DL,, Allingham RR,, Schuman JS.Chandler and Grant's Glaucoma. Baltimore MD: Williams and Wilkins;1996. [Google Scholar]
- 10.Grant WM.Experimental aqueous perfusion in enucleated human eyes. Arch Ophthalmol. 1963; 69: 783–801. [DOI] [PubMed] [Google Scholar]
- 11.Alvarado J,, Murphy C,, Polansky J,, Juster R.Age-related changes in trabecular meshwork cellularity. Invest Ophthalmol Vis Sci. 1981; 21: 714–727. [PubMed] [Google Scholar]
- 12.Alvarado JA,, Wood I,, Polansky JR.Human trabecular cells. II. Growth pattern and ultrastructural characteristics. Invest Ophthalmol Vis Sci. 1982; 23: 464–478. [PubMed] [Google Scholar]
- 13.Alvarado J,, Murphy C,, Juster R.Trabecular meshwork cellularity in primary open-angle glaucoma and nonglaucomatous normals. Ophthalmology. 1984; 91: 564–579. [DOI] [PubMed] [Google Scholar]
- 14.Alvarado JA,, Alvarado RG,, Yeh RF, et al. A new insight into the cellular regulation of aqueous outflow: how trabecular meshwork endothelial cells drive a mechanism that regulates the permeability of Schlemm's canal endothelial cells. Br J Ophthalmol. 2005; 89: 1500–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang N,, Chintala SK,, Fini ME,, Schuman JS.Activation of a tissue-specific stress response in the aqueous outflow pathway of the eye defines the glaucoma disease phenotype. Nat Med. 2001; 7: 304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wordinger RJ,, Sharma T,, Clark AF.The role of TGF-beta2 and bone morphogenetic proteins in the trabecular meshwork and glaucoma. J Ocul Pharmacol Ther. 2014; 30: 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang WH,, McNatt LG,, Pang IH, et al. Increased expression of the WNT antagonist sFRP-1 in glaucoma elevates intraocular pressure. J Clin Invest. 2008; 118: 1056–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao W,, Millar JC,, Wang WH,, et al. Existence of the canonical Wnt signaling pathway in the human trabecular meshwork. Invest Ophthalmol Vis Sci. 2012; 53: 7043–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson DH,, Bradley JM,, Acott TS.The effect of dexamethasone on glycosaminoglycans of human trabecular meshwork in perfusion organ culture. Invest Ophthalmol Vis Sci. 1990; 31: 2568–2571. [PubMed] [Google Scholar]
- 20.Partridge CA,, Weinstein BI,, Southren AL,, Gerritsen ME.Dexamethasone induces specific proteins in human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 1989; 30: 1843–1847. [PubMed] [Google Scholar]
- 21.Rohen JW,, Linner E,, Witmer R.Electron microscopic studies on the trabecular meshwork in two cases of corticosteroid-glaucoma. Exp Eye Res. 1973; 17: 19–31. [DOI] [PubMed] [Google Scholar]
- 22.Steely HT, Browder SL, Julian MB, Miggans ST, Wilson KL. . The effects of dexamethasone on fibronectin expression in cultured human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 1992; 33: 2242–2250. [PubMed] [Google Scholar]
- 23.van Rossum EF,, Lamberts SW.Polymorphisms in the glucocorticoid receptor gene and their associations with metabolic parameters and body composition. Recent Prog Horm Res. 2004; 59: 333–357. [DOI] [PubMed] [Google Scholar]
- 24.Yun AJ,, Murphy CG,, Polansky JR,, Newsome DA,, Alvarado JA.Proteins secreted by human trabecular cells. Glucocorticoid and other effects. Invest Ophthalmol Vis Sci. 1989; 30: 2012–2022. [PubMed] [Google Scholar]
- 25.Zhou L,, Li Y,, Yue BY.Glucocorticoid effects on extracellular matrix proteins and integrins in bovine trabecular meshwork cells in relation to glaucoma. Int J Mol Med. 1998; 1: 339–346. [PubMed] [Google Scholar]
- 26.Rhen T,, Cidlowski JA.Antiinflammatory action of glucocorticoids--new mechanisms for old drugs. N Engl J Med. 2005; 353: 1711–1723. [DOI] [PubMed] [Google Scholar]
- 27.Berry A,, Matthews L,, Jangani M, et al. Interferon-inducible factor 16 is a novel modulator of glucocorticoid action. FASEB J. 2010; 24: 1700–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chriguer RS,, Elias LL,, da Silva IM, Jr,, et al. Glucocorticoid sensitivity in young healthy individuals: in vitro and in vivo studies. J Clin Endocrinol Metab. 2005; 90: 5978–5984. [DOI] [PubMed] [Google Scholar]
- 29.Donn R,, Berry A,, Stevens A, et al. Use of gene expression profiling to identify a novel glucocorticoid sensitivity determining gene, BMPRII. FASEB J. 2007; 21: 402–414. [DOI] [PubMed] [Google Scholar]
- 30.Huizenga NA,, Koper JW,, de Lange P,, et al. Interperson variability but intraperson stability of baseline plasma cortisol concentrations, and its relation to feedback sensitivity of the hypothalamo-pituitary-adrenal axis to a low dose of dexamethasone in elderly individuals. J Clin Endocrinol Metab. 1998; 83: 47–54. [DOI] [PubMed] [Google Scholar]
- 31.Stevens A,, Ray DW,, Zeggini E, et al. Glucocorticoid sensitivity is determined by a specific glucocorticoid receptor haplotype. J Clin Endocrinol Metab. 2004; 89: 892–897. [DOI] [PubMed] [Google Scholar]
- 32.Armaly MF,, Becker B.Intraocular pressure response to topical corticosteroids. Fed Proc. 1965; 24: 1274–1278. [PubMed] [Google Scholar]
- 33.Rhee DJ,, Peck RE,, Belmont J, et al. Intraocular pressure alterations following intravitreal triamcinolone acetonide. Br J Ophthalmol. 2006; 90: 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker B.Intraocular pressure response to topical corticosteroids. Invest Ophthalmol. 1965; 4: 198–205. [PubMed] [Google Scholar]
- 35.Hamid QA,, Wenzel SE,, Hauk PJ, et al. Increased glucocorticoid receptor beta in airway cells of glucocorticoid-insensitive asthma. Am J Respir Crit Care Med. 1999; 159: 1600–1604. [DOI] [PubMed] [Google Scholar]
- 36.Honda M,, Orii F,, Ayabe T,, et al. Expression of glucocorticoid receptor beta in lymphocytes of patients with glucocorticoid-resistant ulcerative colitis. Gastroenterology. 2000; 118: 859–866. [DOI] [PubMed] [Google Scholar]
- 37.Lewis JM,, Priddy T,, Judd J, et al. Intraocular pressure response to topical dexamethasone as a predictor for the development of primary open-angle glaucoma. Am J Ophthalmol. 1988; 106: 607–612. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y,, Song L,, Li B.The expression of glucocorticoid receptor beta messenger RNA in peripheral white blood cells of hormone-resistant nephrotic syndrome patients [in Chinese]. Zhonghua Nei Ke Za Zhi. 2001; 40: 725–728. [PubMed] [Google Scholar]
- 39.Zhang X,, Clark AF,, Yorio T.Regulation of glucocorticoid responsiveness in glaucomatous trabecular meshwork cells by glucocorticoid receptor-beta. Invest Ophthalmol Vis Sci. 2005; 46: 4607–4616. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H,, Zhao GH,, Zhang Q, et al. Relationship between glucocorticoid receptors in the peripheral blood lymphocytes and trabecular meshwork and glucocorticoid induced glaucoma [in Chinese]. Zhonghua Yan Ke Za Zhi. 2006; 42: 431–434. [PubMed] [Google Scholar]
- 41.Kitazawa Y,, Horie T.The prognosis of corticosteroid-responsive individuals. Arch Ophthalmol. 1981; 99: 819–823. [DOI] [PubMed] [Google Scholar]
- 42.Gerzenstein SM,, Pletcher MT,, Cervino AC, et al. Glucocorticoid receptor polymorphisms and intraocular pressure response to intravitreal triamcinolone acetonide. Ophthalmic Genet. 2008; 29: 166–170. [DOI] [PubMed] [Google Scholar]
- 43.Laurie CC,, Doheny KF,, Mirel DB,, et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010; 34: 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner S,, Armstrong LL,, Bradford Y, et al. Quality control procedures for genome-wide association studies. Curr Protoc Hum Genet 2011; Chapter 1:Unit1.19. [DOI] [PMC free article] [PubMed]
- 45.Patel N,, Itakura T,, Gonzalez JM, Jr,, Schwartz SG,, Fini ME.GPR158, an orphan member of G protein-coupled receptor Family C: glucocorticoid-stimulated expression and novel nuclear role. PLoS One. 2013; 8: e57843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patterson N,, Price AL,, Reich D.Population structure and eigenanalysis. PLoS Genet. 2006; 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Devlin B,, Roeder K.Genomic control for association studies. Biometrics. 1999; 55: 997–1004. [DOI] [PubMed] [Google Scholar]
- 48.Hartung J,, Knapp G.On tests of the overall treatment effect in meta-analysis with normally distributed responses. Stat Med. 2001; 20: 1771–1782. [DOI] [PubMed] [Google Scholar]
- 49.Ward LD,, Kellis M.HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012; 40: D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandelin A,, Wasserman WW,Lenhard BConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004; 32: W249–W252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Consortium GT.The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013; 45: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rappoport N,, Karsenty S,, Stern A,, Linial N,, Linial M.ProtoNet 6.0: organizing 10 million protein sequences in a compact hierarchical family tree. Nucleic Acids Res. 2012; 40: D313–D320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J,, Zhuang J,, Iyer S, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012; 22: 1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kulakovskiy IV,, Medvedeva YA,, Schaefer U,, et al. HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res. 2013; 41: D195–D202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hazelett DJ,, Rhie SK,, Gaddis M, et al. Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet. 2014; 10: e1004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Filla MS,, Liu X,, Nguyen TD,, et al. In vitro localization of TIGR/MYOC in trabecular meshwork extracellular matrix and binding to fibronectin. Invest Ophthalmol Vis Sci. 2002; 43: 151–161. [PubMed] [Google Scholar]
- 57.Diskin S,, Kumar J,, Cao Z, et al. Detection of differentially expressed glycogenes in trabecular meshwork of eyes with primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2006; 47: 1491–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan X,, Yang S,, Huang W,, et al. Fine mapping of the psoriasis susceptibility locus PSORS1 supports HLA-C as the susceptibility gene in the Han Chinese population. PLoS Genet. 2008; 4: e1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tiala I,, Wakkinen J,, Suomela S, et al. The PSORS1 locus gene CCHCR1 affects keratinocyte proliferation in transgenic mice. Hum Mol Genet. 2008; 17: 1043–1051. [DOI] [PubMed] [Google Scholar]
- 60.Nair RP,, Stuart PE,, Nistor I,, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006; 78: 827–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Genetic Analysis of Psoriasis Consortium and the Wellcome Trust Case Control Consortium 2; Strange A Capon F, Spencer CC et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010; 42: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hou S,, Yang Z,, Du L,, et al. Identification of a susceptibility locus in STAT4 for Behcet's disease in Han Chinese in a genome-wide association study. Arthritis Rheum. 2012; 64: 4104–4113. [DOI] [PubMed] [Google Scholar]
- 63.Ueta M,, Tokunaga K,, Sotozono C, et al. HLA-A*0206 with TLR3 polymorphisms exerts more than additive effects in Stevens-Johnson syndrome with severe ocular surface complications. PLoS One. 2012; 7: e43650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meder B,, Ruhle F,, Weis T,, et al. A genome-wide association study identifies 6p21 as novel risk locus for dilated cardiomyopathy. Eur Heart J. 2014; 35: 1069–1077. [DOI] [PubMed] [Google Scholar]
- 65.Hijikata M,, Matsushita I,, Tanaka G, et al. Molecular cloning of two novel mucin-like genes in the disease-susceptibility locus for diffuse panbronchiolitis. Hum Genet. 2011; 129: 117–128. [DOI] [PubMed] [Google Scholar]
- 66.Amendt BA.The Molecular Mechanisms of Axenfeld-Rieger Syndrome. New York, NY: Lands Bioscience;2005. [Google Scholar]
- 67.Conte I,, Marco-Ferreres R,, Beccari L, et al. Proper differentiation of photoreceptors and amacrine cells depends on a regulatory loop between NeuroD and Six6. Development. 2010; 137: 2307–2317. [DOI] [PubMed] [Google Scholar]
- 68.Bassett EA,, Pontoriero GF,, Feng W,, et al. Conditional deletion of activating protein 2alpha (AP-2alpha) in the developing retina demonstrates non-cell-autonomous roles for AP-2alpha in optic cup development. Mol Cell Biol. 2007; 27: 7497–7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Potterf SB,, Furumura M,, Dunn KJ,, Arnheiter H,, Pavan WJ.Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum Genet. 2000; 107: 1–6. [DOI] [PubMed] [Google Scholar]
- 70.Lechner J,, Dash DP,, Muszynska D, et al. Mutational spectrum of the ZEB1 gene in corneal dystrophies supports a genotype-phenotype correlation. Invest Ophthalmol Vis Sci. 2013; 54: 3215–3223. [DOI] [PubMed] [Google Scholar]
- 71.Kuot A,, Hewitt AW,, Griggs K,, et al. Association of TCF4 and CLU polymorphisms with Fuchs' endothelial dystrophy and implication of CLU and TGFBI proteins in the disease process. Eur J Hum Genet. 2012; 20: 632–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao X,, Gauderman WJ,, Liu Y, et al. A genome-wide association study of central corneal thickness in Latinos. Invest Ophthalmol Vis Sci. 2013; 54: 2435–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Igo RP Jr,, Kopplin LJ,, Joseph P,, et al. Differing roles for TCF4 and COL8A2 in central corneal thickness and fuchs endothelial corneal dystrophy. PLoS One. 2012; 7: e46742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kino T,, Manoli I,, Kelkar S, et al. Glucocorticoid receptor (GR) beta has intrinsic, GRalpha-independent transcriptional activity. Biochem Biophys Res Commun. 2009; 381: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kino T,, Su YA,, Chrousos GP.Human glucocorticoid receptor isoform beta: recent understanding of its potential implications in physiology and pathophysiology. Cell Mol Life Sci. 2009; 66: 3435–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ulloa F,, Franci C,, Real FX.GalNAc-alpha-O-benzyl inhibits sialylation of de Novo synthesized apical but not basolateral sialoglycoproteins and blocks lysosomal enzyme processing in a post-trans-Golgi network compartment. J Biol Chem. 2000; 275: 18785–18793. [DOI] [PubMed] [Google Scholar]
- 77.Huang J,, Byrd JC,, Yoon WH,, Kim YS.Effect of benzyl-alpha-GalNAc, an inhibitor of mucin glycosylation, on cancer-associated antigens in human colon cancer cells. Oncol Res. 1992; 4: 507–515. [PubMed] [Google Scholar]
- 78.Daly AK.Genome-wide association studies in pharmacogenomics. Nat Rev Genet. 2010; 11: 241–246. [DOI] [PubMed] [Google Scholar]
- 79.Aviner Z,, Henley WL,, Fotino M,, Leopold IH.Histocompatibility (HL-A) antigens and primary open-angle glaucoma. Tissue Antigens. 1976; 7: 193–200. [DOI] [PubMed] [Google Scholar]
- 80.Bigger JF,, Palmberg PF,, Becker B.Increased cellular sensitivity to glucocorticoids in primary open-angle glaucoma. Invest Ophthalmol. 1972; 11: 832–837. [PubMed] [Google Scholar]
- 81.Bigger JF,, Palmberg PF,, Zink HA.In vitro corticosteroid: correlation response with primary open-angle glaucoma and ocular corticosteroid sensitivity. Am J Ophthalmol. 1975; 79: 92–97. [DOI] [PubMed] [Google Scholar]
- 82.Becker B,, Shin DH,, Palmberg PF,, Waltman SR.HLA antigens and corticosteroid response. Science. 1976; 194: 1427–1428. [DOI] [PubMed] [Google Scholar]
- 83.Becker B,, Palmberg F,, Shin DH.Glucocorticoid responsiveness associated with HLA-B12. Invest Ophthalmol Vis Sci. 1977; 16: 61–63. [PubMed] [Google Scholar]
- 84.Yan H,, Yuan W,, Velculescu VE,, Vogelstein B,, Kinzler KW.Allelic variation in human gene expression. Science. 2002; 297: 1143. [DOI] [PubMed] [Google Scholar]
- 85.Emilsson V,, Thorleifsson G,, Zhang B, et al. Genetics of gene expression and its effect on disease. Nature. 2008; 452: 423–428. [DOI] [PubMed] [Google Scholar]
- 86.Cheung VG,, Spielman RS.Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat Rev Genet. 2009; 10: 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hagg PM,, Hurskainen T,, Palatsi R,, Ilves M,, Oikarinen A.Increased expression of glucocorticoid receptor beta in lymphocytes of patients with severe atopic dermatitis unresponsive to topical corticosteroid. Br J Dermatol. 2010; 162: 318–324. [DOI] [PubMed] [Google Scholar]
- 88.Hakonarson H,, Bjornsdottir US,, Halapi E, et al. Profiling of genes expressed in peripheral blood mononuclear cells predicts glucocorticoid sensitivity in asthma patients. Proc Natl Acad Sci U S A. 2005; 102: 14789–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xie YC,, Jiao YJ,, Xu XH,, Wang H,, Yin J.The expression and clinical implications of glucocorticoid receptor isoforms in peripheral blood mononuclear cells in myasthenia gravis [in Chinese]. Zhonghua Nei Ke Za Zhi. 2007; 46: 56–59. [PubMed] [Google Scholar]
- 90.Palmberg P,, Becker B.Lymphocyte sensitivity to glucocorticoids. Am J Ophthalmol. 1977; 83: 599–601. [DOI] [PubMed] [Google Scholar]
- 91.Oakley RH,, Sar M,, Cidlowski JA.The human glucocorticoid receptor beta isoform. Expression biochemical properties, and putative function. J Biol Chem. 1996; 271: 9550–9559. [DOI] [PubMed] [Google Scholar]
- 92.Derijk RH,, Schaaf MJ,, Turner G,, et al. A human glucocorticoid receptor gene variant that increases the stability of the glucocorticoid receptor beta-isoform mRNA is associated with rheumatoid arthritis. J Rheumatol. 2001; 28: 2383–2388. [PubMed] [Google Scholar]
- 93.Jain A,, Wordinger RJ,, Yorio T,, Clark AF.Role of the alternatively spliced glucocorticoid receptor isoform GRbeta in steroid responsiveness and glaucoma. J Ocul Pharmacol Ther. 2014; 30: 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lewis-Tuffin LJ,, Jewell CM,, Bienstock RJ,, Collins JB,, Cidlowski JA.Human glucocorticoid receptor beta binds RU-486 and is transcriptionally active. Mol Cell Biol. 2007; 27: 2266–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wijffels G,, Eisemann C,, Riding G, et al. A novel family of chitin-binding proteins from insect type 2 peritrophic matrix. cDNA sequences, chitin binding activity, and cellular localization. J Biol Chem. 2001; 276: 15527–15536. [DOI] [PubMed] [Google Scholar]
- 96.Hegde RS,, Bernstein HD.The surprising complexity of signal sequences. Trends Biochem Sci. 2006; 31: 563–571. [DOI] [PubMed] [Google Scholar]
- 97.Kaur P,, Karolina DS,, Sepramaniam S,, Armugam A,, Jeyaseelan K.Expression profiling of RNA transcripts during neuronal maturation and ischemic injury. PLoS One. 2014; 9: e103525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cao J.The functional role of long non-coding RNAs and epigenetics. Biol Proced Online. 2014; 16: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Clark BS,, Blackshaw S.Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front Genet. 2014; 5: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Antoniou D,, Stergiopoulos A,, Politis PK.Recent advances in the involvement of long non-coding RNAs in neural stem cell biology and brain pathophysiology. Front Physiol. 2014; 5: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fingert JH,, Stone EM,, Sheffield VC,, Alward WL.Myocilin glaucoma. Surv Ophthalmol. 2002; 47: 547–561. [DOI] [PubMed] [Google Scholar]
- 102.Nguyen TD,, Chen P,, Huang WD, et al. Gene structure and properties of TIGR, an olfactomedin-related glycoprotein cloned from glucocorticoid-induced trabecular meshwork cells. J Biol Chem. 1998; 273: 6341–6350. [DOI] [PubMed] [Google Scholar]
- 103.Polansky JR,, Fauss DJ,, Chen P,, et al. Cellular pharmacology and molecular biology of the trabecular meshwork inducible glucocorticoid response gene product. Ophthalmologica. 1997; 211: 126–139. [DOI] [PubMed] [Google Scholar]
- 104.Fingert JH,, Clark AF,, Craig JE, et al. Evaluation of the myocilin (MYOC) glaucoma gene in monkey and human steroid-induced ocular hypertension. Invest Ophthalmol Vis Sci. 2001; 42: 145–152. [PubMed] [Google Scholar]
- 105.Kim BS,, Savinova OV,, Reedy MV,, et al. Targeted disruption of the myocilin gene (Myoc) suggests that human glaucoma-causing mutations are gain of function. Mol Cell Biol. 2001; 21: 7707–7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Szabo V,, Borgulya G,, Filkorn T, et al. The variant N363S of glucocorticoid receptor in steroid-induced ocular hypertension in Hungarian patients treated with photorefractive keratectomy. Mol Vis. 2007; 13: 659–666. [PMC free article] [PubMed] [Google Scholar]
- 107.Fingert JH,, Alward WL,, Wang K,, Yorio T,, Clark AF.Assessment of SNPs associated with the human glucocorticoid receptor in primary open-angle glaucoma and steroid responders. Mol Vis. 2010; 16: 596–601. [PMC free article] [PubMed] [Google Scholar]
- 108.Spaide RF,, Sorenson J,, Maranan L.Photodynamic therapy with verteporfin combined with intravitreal injection of triamcinolone acetonide for choroidal neovascularization. Ophthalmology. 2005; 112: 301–304. [DOI] [PubMed] [Google Scholar]
- 109.Martidis A,, Duker JS,, Greenberg PB, et al. Intravitreal triamcinolone for refractory diabetic macular edema. Ophthalmology. 2002; 109: 920–927. [DOI] [PubMed] [Google Scholar]
- 110.Ip MS,, Gottlieb JL,, Kahana A,, et al. Intravitreal triamcinolone for the treatment of macular edema associated with central retinal vein occlusion. Arch Ophthalmol. 2004; 122: 1131–1136. [DOI] [PubMed] [Google Scholar]
- 111.Smithen LM,, Ober MD,, Maranan L,, Spaide RF.Intravitreal triamcinolone acetonide and intraocular pressure. Am J Ophthalmol. 2004; 138: 740–743. [DOI] [PubMed] [Google Scholar]
- 112.Quiram PA,, Gonzales CR,, Schwartz SD.Severe steroid-induced glaucoma following intravitreal injection of triamcinolone acetonide. Am J Ophthalmol. 2006; 141: 580–582. [DOI] [PubMed] [Google Scholar]
- 113.Kaushik S,, Gupta V,, Gupta A,, Dogra MR,, Singh R.Intractable glaucoma following intravitreal triamcinolone in central retinal vein occlusion. Am J Ophthalmol. 2004; 137: 758–760. [DOI] [PubMed] [Google Scholar]
- 114.Kiddee W,, Trope GE,, Sheng L, et al. Intraocular pressure monitoring post intravitreal steroids: a systematic review. Surv Ophthalmol. 2013; 58: 291–310. [DOI] [PubMed] [Google Scholar]
- 115.Viola F,, Morescalchi F,, Staurenghi G.Argon laser trabeculoplasty for intractable glaucoma following intravitreal triamcinolone. Arch Ophthalmol. 2006; 124: 133–134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.