Abstract
Species recognition in lichen-forming fungi has been a challenge because of unsettled species concepts, few taxonomically relevant traits, and limitations of traditionally used morphological and chemical characters for identifying closely related species. Here we analyze species diversity in the cosmopolitan genus Protoparmelia s.l. The ~25 described species in this group occur across diverse habitats from the boreal -arctic/alpine to the tropics, but their relationship to each other remains unexplored. In this study, we inferred the phylogeny of 18 species currently assigned to this genus based on 160 specimens and six markers: mtSSU, nuLSU, ITS, RPB1, MCM7, and TSR1. We assessed the circumscription of species-level lineages in Protoparmelia s. str. using two coalescent-based species delimitation methods – BP&P and spedeSTEM. Our results suggest the presence of a tropical and an extra-tropical lineage, and eleven previously unrecognized distinct species-level lineages in Protoparmelia s. str. Several cryptic lineages were discovered as compared to phenotype-based species delimitation. Many of the putative species are supported by geographic evidence.
Introduction
Lichens are symbiotic organisms consisting of a fungal partner (mycobiont), one or more photosynthetic partners (photobionts; [1]), and diverse bacterial communities [2]. Lichens contribute to ecosystem functioning by nutrient recycling [3], weathering rocks, preventing soil erosion, and acting as pioneer species in barren areas. They inhabit diverse ecosystems from the arctic to the tropics and commonly form an integral part of terrestrial biodiversity [4]. Lichens are preferred model systems for ecological, evolutionary, phylogeographic and population genetic studies of symbiotic associations on the account of their wide, often cosmopolitan, distribution, intriguing eco-physiological interdependence and co-evolutionary and adaptive strategies [5]. Almost one fifth of all known fungi and half of all ascomycetes are lichenized, consisting of approximately 28,000 species worldwide [6,7]. However, studies suggest that the estimate of existing lichen diversity might represent only 50–60% of the real diversity [8,9], as current species recognition in lichen-forming fungi appears to vastly underestimate the true number of species. According to Galloway [8], the number of known taxa in different genera has increased from 20% (Parmelia sensu stricto, [10]) to 86% in the New World Oropogon [11]. Recent molecular studies have demonstrated the presence of many distinct lineages subsumed under a single species name (e.g., [12–15]). In the basidiolichen fungus Dictyonema glabratum a single taxon was found to be composed of at least 126 species [9], thus showing a tremendous amount of unexplored diversity in lichen-forming fungi.
Species recognition in lichen-forming fungi has been a challenge because of i) the few taxonomically relevant characters (reviewed by [16,17]), ii) unsettled species concepts [18,19], iii) and unexplored regions containing high levels of diversity, especially in the tropics [20,21]. Morphological and chemical characters that have commonly been used to circumscribe species may not be useful for identifying closely-related species and often fail to accurately characterize species-level diversity [2,19,22,23]. Accurate species delimitation may be obscured by cryptic speciation [24,25], incongruence between morphology and molecular data [26,27], or incongruence between gene trees and species trees [28]. Moreover, morphological and chemical variations may constitute morpho- or chemotypes of the same species with no molecular differentiation, thus blurring our understanding of species boundaries [29,30]. The implementation of molecular techniques and availability of markers for amplifying phylogenetically informative loci have provided great insights into otherwise unrecognized species complexes. Improved species recognition has important implications for understanding diversity, ecological and biogeographical patterns, factors promoting diversification, and for devising better conservation policies [31].
Different studies have utilized varied combinations of the available techniques for unraveling hidden diversity. For example, Harrington and Near [32] used STEM [33] to explore the independent evolutionary lineages within snubnose darters (Etheostoma simoterum species complex). Giarla et al. [15] used two coalescent-based approaches (BP&P and spedeSTEM) for delimiting species in Andean mouse opossums (Thylamys spp) using three nuclear loci and found three additional lineages than previously recognized. Leavitt et al. [34,35] used Bayesian population clustering, genealogical concordance, Bayesian species delimitation, and a DNA barcode approach to support the presence of five previously unrecognized species in the lichen-forming fungus Rhizoplaca melanophthalma species-complex (Lecanoraceae). Parnmen et al. [36] used a 4-locus phylogenetic approach, combined with GMYC [37,38] and STEM [33] and found at least 12 species in the Cladia aggregata complex. Mounting evidence continues to support the perspective that traditional phenotype-based species boundaries fail to adequately characterized species-level diversity in many lichen-forming fungi (reviewed in [22]).
We implemented a molecular approach for species recognition in the cosmopolitan lichen-forming genus Protoparmelia s. str., combining phylogenetic trees and coalescent-based species delimitation methods. The phylogenetic relationships of the heterogeneous genus Protoparmelia have been a matter of debate. Morphological and anatomical characters of this genus show similarity to both Lecanoraceae and Parmeliaceae. Protoparmelia was initially placed in Lecanoraceae because it includes crustose lichens, with one-celled hyaline ascospores and Lecanora-type ascus [39,40]. Later, secondary metabolite profiles showing the presence of lobaric acid brought into question its placement in Lecanoraceae [41]. Studies on the ascoma ontogeny [42,43] further showed the presence of a typical character of Parmeliaceae in Protoparmelia, i.e. cupular exciple, a cup-shaped structure below the hymenium [44]. DNA sequence-based studies suggested Protoparmelia to be the sister-group to Parmeliaceae [45–47]. Tropical species of Protoparmelia with multispored asci were previously placed in the genus Maronina [48]. The authors indicated a close relationship of Protoparmelia and Maronina on the basis of similar ascus types, and suggested the former to be the multi-spore derivative of Protoparmelia. Subsequently, Papong et al. [49] proposed the inclusion of Maronina in Protoparmelia based on molecular data. However, the tropical clade differs from other species in Protoparmelia in being predominantly corticolous, having alectoronic acid as a major compound, and containing many isidiate or sorediate species, whereas most species in the traditional circumscription of Protoparmelia are saxicolous and occur in boreal-arctic/alpine and temperate regions.
Protoparmelia s.l. offers an interesting study system for a variety of reasons. This genus is morphologically and chemically heterogeneous [43,50], and in a previous study [47], we showed that Protoparmelia s.l. is polyphyletic. In addition, the relationships of most taxa to each other remain largely unexplored. Members of this genus inhabit ecologically diverse habitats, such as boreal-arctic/alpine, temperate, Mediterranean, subtropical, and tropical regions and also vary greatly in their distribution range with some species being cosmopolitan (e.g., P. badia, P. memnonia), whereas other, mainly tropical species being locally restricted (e.g., P. orientalis, P. multifera). Furthermore, congeners occur on various substrates, with some species growing on bark or decorticated wood, and others on rocks. Protoparmelia species exhibit varied life styles. For example, some species are lichenicolous and parasitize other lichen-forming fungi during early parts of their life cycle [50]. Sexual reproduction is common in some species (P. badia and P. orientalis), whereas others propagate mainly via asexual propagules (P. isidiata, P. corallifera and P. capitata) with or without any sexual reproduction.
The heterogeneity of characters makes Protoparmelia s.l. [51] an interesting candidate for testing species delimitation scenarios using multi-locus DNA sequence data. Protoparmelia s. str. [47] although being a small genus, is sister to the largest family of lichen-forming fungi, i.e., Parmeliaceae [45–47], consisting of approximately 2,800 species distributed in 80 genera [52,53]. Resolving relationships of Protoparmelia s. str. may contribute to understanding character evolution in an important clade of lichen-forming fungi. The aims of the current study are two-fold: 1) exploring the phylogenetic relationships of Protoparmelia s.l. species by constructing a multi-locus phylogeny, and 2) assessing the circumscription of lineages in Protoparmelia s. str. based on multi-locus species-tree inference and coalescent approaches.
Materials and Methods
Taxon sampling
This study includes a total of 160 samples of Protoparmelia s.l. from 18 currently described species. About 70% of the total described species were included in this study. Additionally, three unidentified species, most likely new to science, were also included in the study. We selected 73 taxa from reportedly close relatives of Protoparmelia s.l. [45,47], namely Parmeliaceae (40 taxa), Lecanoraceae (4 taxa), Gypsoplacaceae ([54]; 2 taxa), Miriquidica group (12 taxa), and Ramboldia (10 taxa) to infer the relationship of Protoparmelia s.l. with other taxa within related groups within Lecanorales. Cladoniaceae (5 taxa) were selected as outgroup. Details of the study material and GenBank accession numbers are given in S1 Table.
DNA amplification and sequencing
Genomic DNA was extracted from lichen thalli using the CTAB method [55]. PCR amplification was performed using general, previously published primers for RPB1, TSR1, MCM7, nuLSU, mtSSU and ITS (Table 1). For some species of Protoparmelia s.l. and Miriquidica group specific primers were designed (Table 1). PCR reactions were carried out in a volume of 25 μl. Each reaction mix contained 2.5 μl buffer, 0.13 μl (0.65 U) Ex Taq polymerase, 1.0 μl dNTP mix (2.5 mM each), 1.0 μl each (10 mM) of the primer set, ca. 20 ng of template, and 16 μl H2O. Reactions were performed with the following cycling conditions: initial denaturation at 95°C for 4 min, followed by 35 cycles of 95°C for 30 s, 50°C for 40 s, 72°C for 1 min, and final elongation at 72°C for 5 min. PCR products were checked for amplification on 1% agarose gels. Bands of expected size were extracted using the peqGOLD Gel Extraction Kit. All PCR products were labeled with the Big Dye Terminator v3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, CA, USA) and cycle sequenced as follows: (1) 1 min 96°C, (2) 26 cycles of 20 s 96°C, 5 s 50°C, and (3) 2 min 60°C. Products were purified using the Big Dye XTerminator Purification Kit (Life Technologies) and then detected on ABI PRISM 3730 DNA Analyzer (Applied Biosystems).
Table 1. Primers used in this study.
| Taxa | Locus | Primer name | Sequence | Reference |
|---|---|---|---|---|
| Protoparmelia | RPB1 | fRPB1cR | CNGGCDATNTCRTTRTCCATRTA | [87] |
| RPB1 | gRPB1Af | GADTGTCCDGGDCATTTTGG | [88] | |
| RPB1 | RPB1PPsp FOR | GTGCTTTGCTTCAGCAGTGCTC | [47] | |
| RPB1 | RPB1PPsp REV | AGCGACGAACATTGCCGTTCGCAC | [47] | |
| RPB1 | PPRPB1 FOR | GATGCGGTYTGGCGGCTTTGCAAGCC | This study | |
| RPB1 | PPRPB1 REV | GGCTTGCAAAGCCGCCARACCGCATC | This study | |
| TSR1 | *120040PP_TSR1_FOR | CAGTGTTTTGCCCAGAGAAAGGCTTTCAAG | This study | |
| TSR1 | *120082PP_TSR1_FOR | TAACGTCCTTGCGAAAGAACGATTAGCGAG | This study | |
| MCM7 | MCM7 709 (f) | ACIMGIGTITCVGAYGTHAARCC | [89] | |
| MCM7 | MCM7-1348 | GAYTTDGCIACICCIGGRTCWCCCAT | [89] | |
| MCM7 | PPspecMCM7 FOR | GAICGDTGIGGITRIGARRTITTIC | [47] | |
| MCM7 | PPspecMCM7 REV | GIIARRTAITCRTACATGKIRCC | [47] | |
| MCM7 | PPMCM7FOR | CTATCGACACGAGCATCCAAG | This study | |
| MCM7 | PPMCM7REV | CATGTGACCGRAATGCTTGTATTTC | This study | |
| nuLSU | LR6 (r) | LR6: CGCCAGTTCTGCTTACC | [90,91] | |
| nuLSU | AL1R (f) | GGGTCCGAGTTGTAATTTGT | [90,91] | |
| nuLSU | LR5: | TCCTGAGGGAAACTTCG | [91] | |
| nuLSU | L3 | CCGTGTTTCAAGACGGG | [91] | |
| nuLSU | LROR | ACCCGCTGAACTTAAGC | [91] | |
| nuLSU | LSUPPspFOR2 | GAAACCCCTTCGACGAGTCGAG | [47] | |
| nuLSU | LSUPPspREV1 | AGATGGTTCGATTAGTCTTTCG | [47] | |
| ITS | ITS1-F | CTTGGTCATTTAGAGGAAGTAA | [92] | |
| ITS | ITS2 | GCTGCGTTCTTCATCGATGC | [93] | |
| ITS | ITS3 | GCATCGATGAAGAACGCAGC | [93] | |
| ITS | ITS4 | TCCTCCGCTTATTGATATGC | [93] | |
| ITS | PPITSFFOR1A | GAAGGATCATTATCGAGAGAGG | This study | |
| ITS | PPITSFREV1A | CTTTCAAAGCGGGAGAAATTTACTAC | This Study | |
| ITS | PPITSFFOR1Anested | GATCATTATCGAGAGAGGGGCTTC | This Study | |
| ITS | PPITSFREV1Anested | GGAGAAATTTACTACGCTTAAAG | This Study | |
| mtSSU | mrSSU1 | AGCAGTGAGGGATATTGGTC | [94] | |
| mtSSU | MSU7: | GTCGAGTTACAGACTACAATCC | [95] | |
| mtSSU | mrSSU2 | CTGACGTTGAAGGACGAAGG | [94] | |
| mtSSU | mrSSU2R | CCTTCGTCCTTCAACGTCAG | [94] | |
| mtSSU | mrSSU3R | ATGTGGCACGTCTATAGCCC | [94] | |
| mtSSU | MSU1 | GATGATGGCTCTGATTGAAC | [95] | |
| Miriquidica | RPB1 | RPB1MIRI FOR | CTACAGATGATATCAAGCTCATG | [47] |
| RPB1 | RPB1MIRI REV | CATGAGCTTGATATCATCTGTAG | [47] | |
| RPB1 | RPB1MIRIint FOR | CATGACGAAAATCAAGAAACTGCTG | This study | |
| RPB1 | RPB1MIRIint REV | CATGCCGTCGCCTATCTCCTTAGTC | Thus study | |
| RPB1 | RPB1MIRIFOR1new | TAGCACAACAATCCGGCATTCAAG | This study | |
| RPB1 | RPB1MIRIREV1new | TCATTGCTGAGTCCCATGAGCTTG | This study | |
| RPB1 | RPB1MIRIREV2new | GCACGAATAATGTCCCCAAGCTTG | This study | |
| TSR1 | MIRI_TSR1_FOR | CAACGTTCTGGCTAGAGAGCGTCTGGCAAG | This study | |
| TSR1 | *MIRI_40_82_TSR1_REV | CADAGYTGMAGHGYTTGAACCARTTSAC | This study | |
| TSR1 | *MIRI_82_TSR1_REV | CAKAGYTGCAGMGCTTTGAACCAGTTGAC | This study | |
| TSR1 | TSRMIRIFOR1 | TGAGCTGCATCCAAAYGTWCTKGC | This study | |
| TSR1 | TSRMIRIINTREV | TAGCGRTYGAATTTGTGGACGTTG | This study | |
| TSR1 | TSRMIRIREV1 | AACATGTAGCGRAYIGTSACGAG | This study | |
| TSR1 | GS1_22TSR1_FOR | GAKCCCATGARCCAGAAGAWTG | This study | |
| TSR1 | GS1_22TSR1_REV | GAAGAACATGTASCGGACSGTCAC | This study | |
| MCM7 | MCM7MIRI FOR | CAATTTACTCCAATGACTGAATGTC | [47] | |
| MCM7 | MCM7MIRI REV | CATGCCGTCGCCTATCTCCTTAGTC | [47] | |
| nuLSU | NULSUMIRIINT FOR | CTCGGACCGAGGATCGCGCTTC | This study | |
| nuLSU | NULSUMIRIINT REV | GAAGCGCGATCCTCGGTCCGAG | This study | |
| nuLSU | NULSUMIRIFOR1 | CAGAGACCGATAGCGCACAAGTAGAG | This study | |
| nuLSU | NULSUMIRIREV1 | GAGCCTCCACCAGAGTTTCCTCTG | This study | |
| ITS | ITSfMIRI FOR | TATCGAGTGGAGGGGCTTCGCTC | [47] | |
| ITS | ITSfMIRI REV | TAACGTTTAGGCGGTTGTTGGC | [47] | |
| ITS | ITSFMIRIFOR1 | GAATTCAGTGAATCATCGAATCTTTG | This study | |
| ITS | ITSFMIRIREV1 | AGAGTGTAATGACGCTCGAACAGG | This study | |
| Ramboldia | RPB1 | RPB1RAMBINTFOR | GTCTGCCATAATTGYGGCAAGATC | This study |
| RPB1 | RPB1RAMBINTREV | GAYATTTCCACAACCRCCATGATC | This study | |
| RPB1 | RPB1RAMFORgroup1 | GTYTGCCATAATTGCGGCAAGATC | This study | |
| RPB1 | RPB1RAMREVgroup2 | ATGTGRCGAAARATRTTKAGSGCC | This study |
Taxon specific primers were designed for some Protoparmelia, Ramboldia and Miriquidica species.
For each locus, consensus sequences were assembled separately and aligned using MAFFT [56] as implemented in Geneious v5.4 [57], followed by manual editing. Gaps were treated as missing data and ambiguously aligned nucleotides were excluded.
Phylogenetic analyses
Model selection
Model selection was performed to find the best-fitting model for each data set. We used the Corrected Akaike Information Criterion (AICc) [58] as implemented in jModelTest v2.1.1 [59].
Congruence among loci
To test the level of congruence among loci, we used the Congruence Among Distance Matrices test (CADM, [60]), as implemented in the package ape in R. The null hypothesis assumes that all tested phylogenetic trees are completely incongruent. Incongruence here refers to phylogenetic trees with different topologies among loci, which suggests completely distinct evolutionary histories. The level of congruence ranges from 0 to 1. In addition, maximum likelihood (ML) analyses were performed individually on each locus with RAxML-HPC BlackBox v8.1.11 [61] on the Cipres Science gateway [62] using the default GTR + G model with 1,000 bootstrap (BS) replicates. Conflicts were considered significant if individuals grouped in a clade with ≥ 70% BS support in one data set, but in a different clade with high support in another locus.
Phylogeny of Protoparmelia s.l.
Since no supported conflicts were observed in single locus trees and CADM analysis rejected the hypothesis of incongruence among loci, data sets were concatenated (see Results). The maximum likelihood search was performed on the concatenated 6-locus data set including all the relatives of Protoparmelia s.l. with RAxML-HPC BlackBox v8.1.11 [61] on the Cipres Scientific gateway [62]. Only those taxa for which the sequence information was available for at least three loci were included in the concatenated data set. The default GTR + G model was used as the substitution model and data was partitioned according to the different genes. RPB1, TSR1 and MCM7 data were also partitioned by codon position.
Bayesian inference was performed using the best fitting model as inferred by jModelTest, for the single as well as concatenated data sets as implemented in MrBayes v3.2.1 [63,64] on the Cipres Scientific gateway [62]. Two parallel MCMCMC runs were performed each using four chains and 5,000,000 generations, sampling trees every 100th generation. A 50% majority rule consensus tree was generated from the combined sampled trees of both runs after discarding the first 25% as burn-in (12,500 trees, likelihoods below stationary level).
Phylogeny of Protoparmelia s. str.
Maximum likelihood analysis was performed individually on each locus of Protoparmelia s. str. (excluding Lecanoraceae, Parmeliaceae, Miriquidica group and Ramboldia clades), with RAxML-HPC BlackBox v8.1.11 [61] on the Cipres Science gateway [62], using the default GTR + G model, with 1,000 BS replicates. Gypsoplacaceae was used as outgroup. Only taxa for which sequence information was available for at least three loci were included in the concatenated data set. The default GTR + G model was used as the substitution model and the data was partitioned according to the different genes. For RPB1, TSR1 and MCM7 data were also partitioned by codon position. Since no supported conflicts were observed in single locus trees and CADM analysis rejected the hypothesis of incongruence among loci, data sets were concatenated. Maximum likelihood search was then performed on the concatenated 6-locus data set using RAxML-HPC BlackBox v8.1.11 [61] on the Cipres Scientific gateway v3.3 [62].
We performed jModelTest for each locus on the reduced data set to select the best locus-specific models of evolution.
Bayesian inference was performed using the best fitting model as suggested by jModelTest, for the single and concatenated data sets separately as implemented in MrBayes v3.2.1 [63,64] on the Cipres Scientific gateway [62]. Two parallel MCMCMC runs were performed each using four chains and 5,000,000 generations, sampling trees every 100th generation. A 50% majority rule consensus tree was generated from the combined sampled trees of both runs after discarding the first 25% as burn-in (12,500 trees).
*BEAST as implemented in BEAST v2.1 [65] was used to estimate the species tree for BP&P [66]. We used a Birth-Death process and gamma-distributed population sizes for the species tree prior and a pairwise linear population size model with a constant root. *BEAST incorporates the coalescent process and the uncertainty associated with gene trees and nucleotide substitution model parameters and estimates the species tree directly from the sequence data. For each locus, the closest model to the best-suggested model from jModelTest under the AICc criterion was selected as the best substitution model for *BEAST. Two independent Markov chain Monte Carlo (MCMC) analyses were performed for a total of 100,000,000 generations, sampling every 5,000 steps. Default values were used for the remaining priors. Convergence of the runs to the same posterior distribution and the adequacy of sampling (using the Effective Sample Size [ESS] diagnostic) were assessed with Tracer v1.4 [67]. After removing the first 20% of the samples as burn-in, all runs were combined to generate posterior probabilities of nodes from the sampled trees using TreeAnnotator v1.7.4 [68]. The species tree produced by *BEAST was subsequently used for inferring speciation probabilities by BP&P [66].
Species delimitation in Protoparmelia s. str.
For testing the species boundaries in Protoparmelia s. str. [47], currently accepted taxa were taken as putative species (12 described species). In addition well-supported (BS ≥ 70%, PP ≥ 0.94) monophyletic clades from ML and Bayesian phylogenies were taken as putative species, resulting in a 25-species scenario (Figs 1 and 2).
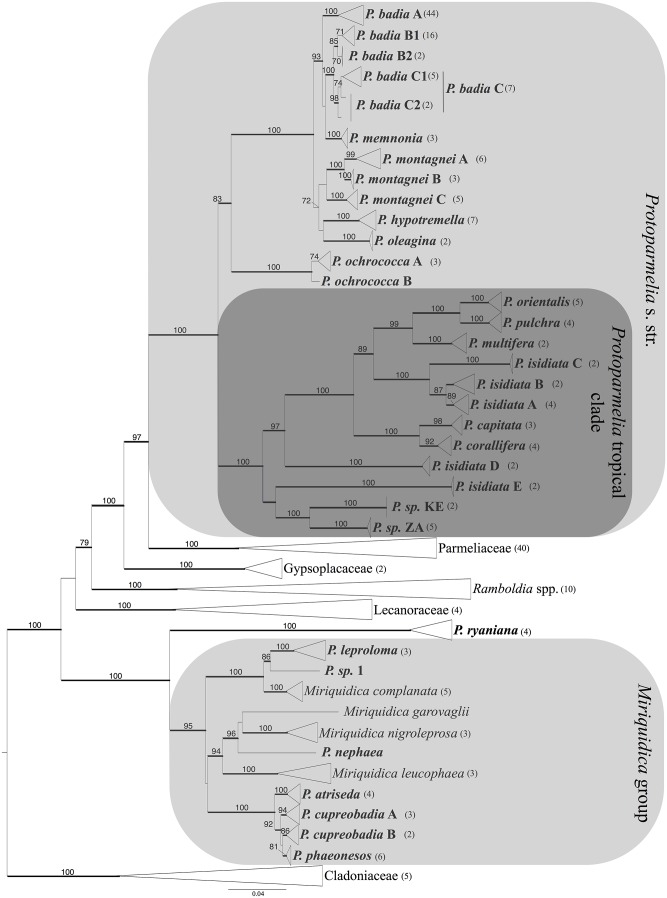
Fig 1. Phylogeny of Protoparmelia sensu lato and its allies based on a concatenated 6-locus data set including ITS, nuLSU, mtSSU, MCM7, TSR1 and RPB1 sequences.
This is a maximum likelihood tree. Numbers above branches indicate ML BS ≥ 70%. Branches in bold indicate Bayesian posterior probabilities (PP) ≥ 0.94. Terminal clades were collapsed for clarity of presentation. The length of the triangle corresponds to branch lengths. Numbers in parentheses indicate number of specimens included in collapsed clade. Identity of each specimen in a clade is given in Supporting information S1 Table. Protoparmelia s.l. species are in bold.
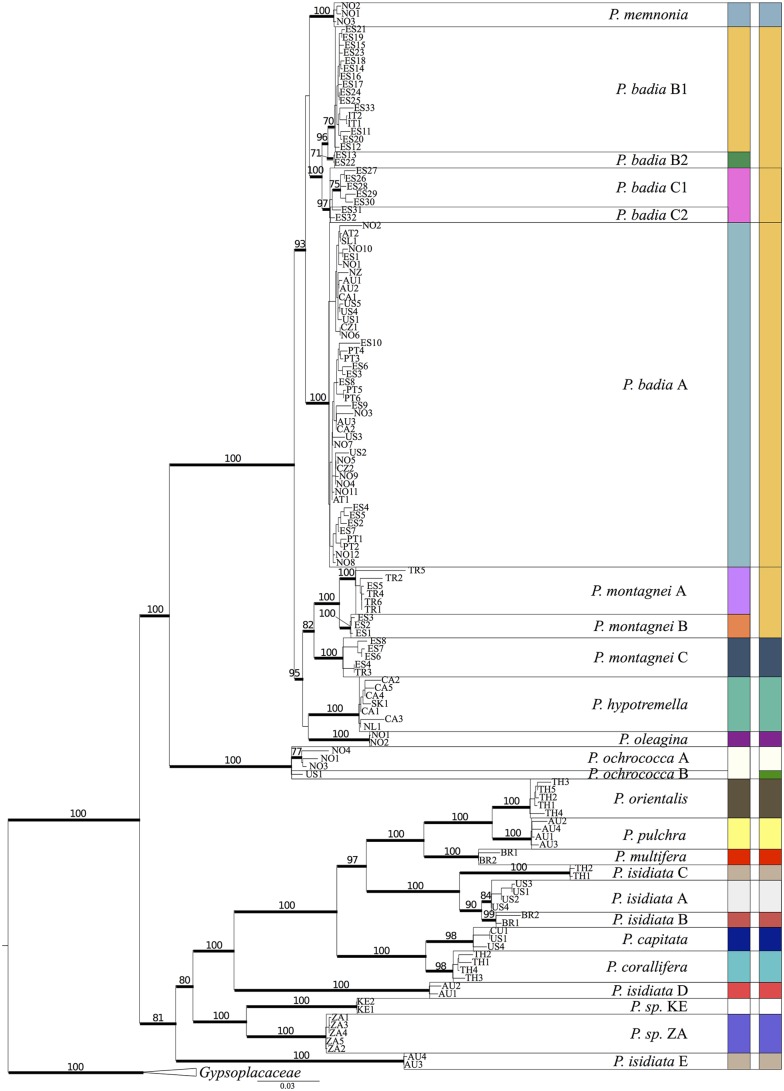
Fig 2. Phylogeny of Protoparmelia s. str. based on six concatenated loci.
Numbers above branches indicate ML BS ≥ 70%. Branches in bold indicate Bayesian posterior probabilities (PP) ≥ 0.94. Specimen indicators include country codes (see Supporting information S1 Table). Taxon names refer to putative species supported by ML BS ≥ 70% or Bayesian Inference (PP ≥ 0.94), and tested for speciation probabilities using BP&P and spedeSTEM. Colored boxes indicate species supported by BP&P (left) and spedeSTEM (right), respectively.
The marginal posterior probability of 25-species scenario suggested by molecular data was estimated using the program BP&P v3 [66]. BP&P utilizes reversible-jump Bayesian Markov chain Monte Carlo (MCMC) algorithms for analyzing phylogenetic data from multiple loci to generate speciation probabilities of assigned species. It takes into account uncertainties due to unknown gene trees and ancestral coalescent processes. This method accommodates the species phylogeny as well as incomplete lineage sorting due to ancestral polymorphism. Species tree from *BEAST was used to infer the speciation probabilities by BP&P. BP&P v3 incorporates nearest-neighbor interchange (NNI) algorithm allowing changes in the species tree topology, eliminating the need for a fixed user-specified guide tree [66]. BP&P gives the posterior probability of each delimited species and the posterior probability for the number of delimited species. A gamma prior G (1, 10), with mean 1/10 = 0.1 (one difference per 10 bp) was used on the population size parameters (s). The age of the root in the species tree (τ0) was assigned the gamma prior G (2, 2000) which means 0.1% of sequence divergence, while the other divergence time parameters were assigned the Dirichlet prior [66]. Each analysis was run twice to confirm consistency between runs.
We also used spedeSTEM for calculating probabilities of the species scenario. SpedeSTEM [69] is based on the multilocus species-tree method STEM [33]. It assumes all putative species as separate lineages and estimates gene trees in PAUP* [70]. It then calculates the likelihood for alternative species trees in various permutations and combinations of subpopulations by collapsing two or more species into a single lineage using previously estimated gene trees. Species boundaries are then compared using Akaike information criteria and gives probabilities of different species scenarios. We used θ = 0.05 and each analysis included 500 replicates. We tested all 25 possible permutations for clustering within taxonomic species.
Results
DNA sequences
We generated 716 new sequences for this phylogeny, including 142 RPB1, 116 TSR1, 84 MCM7, 150 nuLSU, 127 mtSSU and 107 ITS sequences. The data sets included 310 sequences downloaded from NCBI. A total of 233 taxa were analyzed. The percentage of missing data for each locus was: RPB1- 17.17%, TSR1- 36.48%; MCM7–44.2%, nuLSU- 8.59%, mtSSU—21.45% and ITS- 26.6%.
Model test
Bayesian analysis on the complete data set and the reduced Protoparmelia s. str. data set was performed using the best fitting model for each locus in the concatenated sequence as shown in Table 2.
Table 2. Genetic characteristics of nuclear loci used in this study.
| Full data set | |||
|---|---|---|---|
| Locus | No. of seq | length of alignment | Best model |
| RPB1 | 142 | 696 | 012232+G |
| TSR1 | 196 | 756 | HKY+I+G |
| MCM7 | 131 | 655 | 012212+I+G+F |
| nuLSU | 212 | 1064 | TIM1+I+G |
| mtSSU | 185 | 834 | 012212+I+G+F |
| ITS | 168 | 807 | 012030+I+G |
| Concatenated | 233 | 4812 | NA |
| Protoparmelia s. str. | |||
| Locus | length of alignment | Best model | |
| RPB1 | 114 | 696 | 012232+G+F |
| TSR1 | 98 | 754 | TPM2uf+G |
| MCM7 | 63 | 672 | HKY+G |
| nuLSU | 126 | 972 | TIM1+I+G |
| mtSSU | 93 | 839 | : 012212+I+G+F |
| ITS | 96 | 787 | 011230+I+G+F |
| Concatenated 6 loci | 138 | 4720 | NA |
Genetic characteristics of nuclear loci used in this study, including the total number of sequences per locus, length of the alignment; and best model of evolution selected using the Akaike information criterion as suggested by jModelTest.
For *BEAST, the first available best fitting model for each locus in the concatenated data set, from the models suggested by jModelTest v2.1.1 were the following: RPB1: GTR, TSR1: HKY, MCM7: HKY, nuLSU: GTR, mtSSU: HKY, and ITS: GTR.
Congruence among loci
CADM results showed no significant incongruence among loci, thus allowing concatenation. The null hypothesis of complete incongruence among loci was rejected for both complete (W = 0.75; p<0.0001) and reduced (W = 0.84; p<0.0001) data sets.
Phylogeny of Protoparmelia
Protoparmelia s.l.
Nuclear and mitochondrial gene partitions supported the same overall topology. The concatenated six-locus data set contained 233 specimens. Gene partitions had the following lengths: 696 bp for RPB1, 756 for TSR1, 655 bp for MCM7, 1064 bp for nuLSU rDNA, 834 bp for mtSSU and 807 bp for ITS rDNA. The total length of the concatenated alignment was 4812 bp (dryad doi:10.5061/dryad.0q515). The ML tree for the concatenated data set is presented in Fig 1. Nodes with BS ≥ 70% and Bayesian posterior probability (PP) ≥0.94 were considered as supported.
The 6-locus data set yielded a resolved and well-supported topology of Protoparmelia s.l. (Fig 1). Members of the genus grouped either in Protoparmelia s. str. [47], or with representatives of the genus Miriquidica (“Miriquidica-group” in Fig 1), or as sister to the Miriquidica-group (P. ryaniana). The family Parmeliaceae s. str. formed a well-supported monophyletic group (BS = 100%, PP = 1; Fig 1), which was confirmed to be sister to Protoparmelia s. str. (BS 97% and PP = 1). Within Protoparmelia s. str. we found two distinct clades. One contained species with boreal-arctic/alpine, montane, temperate and Mediterranean distributions (P. badia, P. memnonia, P. hypotremella, P. montagnei, P. oleagina, P. ochrococca), the other contained species with subtropical and tropical distributions (P. capitata, P. corallifera, P. isidiata, P. multifera, P. orientalis, P. pulchra, and two yet undescribed species from Kenya and South Africa, respectively).
Six species of Protoparmelia (P. atriseda, P. cupreobadia, P. leproloma, P. phaeonesos, P. ryaniana and P. sp. 1) including one yet undescribed species formed a monophyletic group together with Miriquidica spp.
Protoparmelia s. str.
The concatenated six-locus data set contained 138 specimens, including two taxa from outgroup Gypsoplacaceae. Gene partitions had the following lengths: 696 bp for RPB1, 754 for TSR1, 672 bp for MCM7, 972 bp for nuLSU rDNA, 839 bp for mtSSU and 787 bp for ITS rDNA. The total length of the concatenated alignment was 4720 bp. Most species as currently circumscribed were monophyletic, except P. isidiata, which formed three independent lineages within the tropical clade (P. isidiata A-C, D and E), and the cosmopolitan species P. badia, which contained multiple supported lineages and formed a species complex with P. memnonia (Fig 2). We found evidence for cryptic species-level diversity in the nominal taxa P. badia, P. montagnei, and P. isidiata (clade P. isidiata A-E). Cryptic diversity corresponded to biogeographic patterns in P. isidiata (clades A-C representing North America, South America and Asia, respectively). Within P. badia, the largest lineage (clade P. badia A) was cosmopolitan, whereas the other supported lineages had a Mediterranean, or Iberian distribution (Fig 2).
Species delimitation in Protoparmelia s. str.
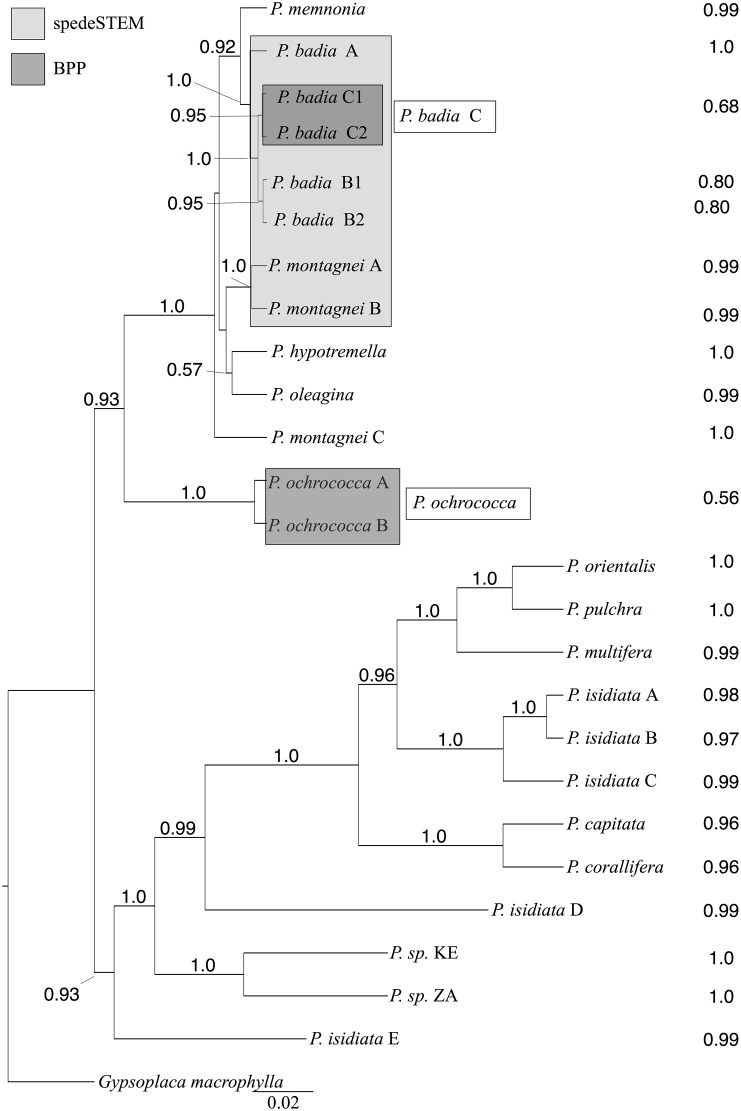
We treated terminal clades supported by ≥ 70% BS and ≥ 0.94 PP (Figs 1 and 2) as putative species for species delimitation analyses. This resulted in a 25-species scenario for Protoparmelia s. str., in contrast to the current 12-species scenario for Protoparmelia s. str., based on morphological and chemical characters. The 25-species scenario in Protoparmelia s. str. was then investigated for species delimitation using BP&P and spedeSTEM. BP&P supported the presence of 23 species with highest probability (PP = 0.41127). Posterior probability of each delimited species is given in Fig 3. Protoparmelia ochrococca A & B, P. badia C1 & C2 were not supported as separate species by BP&P. SpedeSTEM supported 19-species scenario (P. badia A, P. badia B1 & B2, P. badia C1 & C2, P. montagnei A & B collapsed as one species; Fig 3, θ = 0.05, number of runs = 500), using the model that receives the highest support (100% of the model weighting; Table 3). Sixteen putative species (P. memnonia, P. hypotremella, P. oleagina, P. montagnei C, P. orientalis, P. multifera, P. pulchra, P. capitata, P. corallifera, P. sp. KE, P. sp. ZA and the five cryptic isidiate lineages in P. isidiata) were supported as separate lineages by both BP&P and spedeSTEM (Table 4), therefore we suggest these clades to be evolutionary independent. We found conflicting speciation scenarios for P. ochrococca A & B, P. badia A, B1, B2, & C and P. montagnei A & B by the two species delimitation approaches (Fig 2).
Fig 3. *BEAST species trees for Protoparmelia s. str. as suggested by ML (BS ≥ 70%) or Bayesian (PP ≥ 0.94).
Posterior probabilities at nodes indicate support from the *BEAST analyses. The posterior probability of each delimited species calculated by BP&P are indicated in front of each putative species. Boxes in dark grey indicate clades not supported as separate taxa by BP&P. Protoparmelia badia B1 & B2 were supported as separate species whereas P. badia C1 & C2 were not supported as separate species (referred to as P. badia C) by BP&P. Box in light grey indicates species not supported as separate taxa by spedeSTEM.
Table 3. SpedeSTEM validation results.
| Single AIC calculation | |||||
|---|---|---|---|---|---|
| k | ln | AIC | delta | modelLik | wi |
| 1 | -242465.0193 | 484932.0386 | 187997.2007 | 0.00 | 0.00 |
| 2 | -230753.3543 | 461510.7085 | 164575.8707 | 0.00 | 0.00 |
| 3 | -186257.4103 | 372520.8205 | 75585.98267 | 0.00 | 0.00 |
| 4 | -184345.6627 | 368699.3255 | 71764.48762 | 0.00 | 0.00 |
| 5 | -180035.244 | 360080.4881 | 63145.65023 | 0.00 | 0.00 |
| 6 | -175225.9999 | 350463.9998 | 53529.162 | 0.00 | 0.00 |
| 7 | -164198.5726 | 328411.1453 | 31476.30744 | 0.00 | 0.00 |
| 8 | -160488.1749 | 320992.3498 | 24057.51199 | 0.00 | 0.00 |
| 9 | -160402.6815 | 320823.363 | 23888.52519 | 0.00 | 0.00 |
| 10 | -154132.577 | 308285.1541 | 11350.31624 | 0.00 | 0.00 |
| 11 | -153575.6078 | 307173.2155 | 10238.37768 | 0.00 | 0.00 |
| 12 | -153474.4072 | 306972.8143 | 10037.97648 | 0.00 | 0.00 |
| 13 | -150731.9074 | 301489.8148 | 4554.97696 | 0.00 | 0.00 |
| 14 | -149265.1449 | 298558.2898 | 1623.452 | 0.00 | 0.00 |
| 15 | -149048.2275 | 298126.4551 | 1191.61724 | 0.00 | 0.00 |
| 16 | -148866.247 | 297764.4941 | 829.65624 | 0.00 | 0.00 |
| 17 | -148702.0018 | 297438.0037 | 503.16584 | 0.00 | 0.00 |
| 18 | -148652.4701 | 297340.9402 | 406.10232 | 0.00 | 0.00 |
| 19 | -148448.4189 | 296934.8378 | 0 | 1.00 | 1.00 |
| 20 | -148536.6081 | 297113.2162 | 178.37836 | 0.00 | 0.00 |
| 21 | -148526.4393 | 297094.8785 | 160.04068 | 0.00 | 0.00 |
| 22 | -148522.9071 | 297089.8141 | 154.97628 | 0.00 | 0.00 |
| 23 | -148515.6023 | 297077.2046 | 142.36672 | 0.00 | 0.00 |
| 24 | -148515.525 | 297079.05 | 144.21212 | 0.00 | 0.00 |
| 25 | -148513.4861 | 297076.9721 | 142.13428 | 0.00 | 0.00 |
| Multiple AIC calculation | |||||
| k | ln | AIC | delta | modelLik | wi |
| 1 | -242465.0193 | 484932.0386 | 187997.2007 | 0.00 | 0.00 |
| 2 | -230753.3543 | 461510.7085 | 164575.8707 | 0.00 | 0.00 |
| 3 | -186257.4103 | 372520.8205 | 75585.98267 | 0.00 | 0.00 |
| 4 | -184345.6627 | 368699.3255 | 71764.48762 | 0.00 | 0.00 |
| 5 | -180035.244 | 360080.4881 | 63145.65023 | 0.00 | 0.00 |
| 6 | -175225.9999 | 350463.9998 | 53529.162 | 0.00 | 0.00 |
| 7 | -164198.5726 | 328411.1453 | 31476.30744 | 0.00 | 0.00 |
| 8 | -160488.1749 | 320992.3498 | 24057.51199 | 0.00 | 0.00 |
| 9 | -160402.6815 | 320823.363 | 23888.52519 | 0.00 | 0.00 |
| 10 | -154132.577 | 308285.1541 | 11350.31624 | 0.00 | 0.00 |
| 11 | -153575.6078 | 307173.2155 | 10238.37768 | 0.00 | 0.00 |
| 12 | -153474.4072 | 306972.8143 | 10037.97648 | 0.00 | 0.00 |
| 13 | -150731.9074 | 301489.8148 | 4554.97696 | 0.00 | 0.00 |
| 14 | -149265.1449 | 298558.2898 | 1623.452 | 0.00 | 0.00 |
| 15 | -149048.2275 | 298126.4551 | 1191.61724 | 0.00 | 0.00 |
| 16 | -148866.247 | 297764.4941 | 829.65624 | 0.00 | 0.00 |
| 17 | -148702.0018 | 297438.0037 | 503.16584 | 0.00 | 0.00 |
| 18 | -148652.4701 | 297340.9402 | 406.10232 | 0.00 | 0.00 |
| 19 | -148448.4189 | 296934.8378 | 0 | 1.00 | 1.00 |
| 20 | -148536.6081 | 297113.2162 | 178.37836 | 0.00 | 0.00 |
| 21 | -148526.4393 | 297094.8785 | 160.04068 | 0.00 | 0.00 |
| 22 | -148522.9071 | 297089.8141 | 154.97628 | 0.00 | 0.00 |
| 23 | -148515.6023 | 297077.2046 | 142.36672 | 0.00 | 0.00 |
| 24 | -148515.525 | 297079.05 | 144.21212 | 0.00 | 0.00 |
| 25 | -148513.4861 | 297076.9721 | 142.13428 | 0.00 | 0.00 |
spedeSTEM validation results, using θ = 0.5. The absolute difference between the AICc score for the given model and the best-fitting one is listed under the column labeled ‘‘Di” and the model weighting is listed under the column labeled “wi”.
Table 4. Summary of results of ML, Bayesian and species delimitation analyses (BP&P and spedeSTEM).
| Putative species | BS | BS1 | PP | PP1 | BP&P | spedeSTEM |
|---|---|---|---|---|---|---|
| Protoparmelia badia A | 100 | 1 | 1.0 | - | ||
| Protoparmelia badia B1 | 71 | 85 | 0.61 | 1 | 0.80 | - |
| Protoparmelia badia B2 | 70 | — | 0.80 | - | ||
| Protoparmelia badia C1 | 74 | 98 | 0.97 | 1 | 0.68 | - |
| Protoparmelia badia C2 | 98 | — | - | |||
| Protoparmelia capitata | 98 | 1 | 0.96 | + | ||
| Protoparmelia corallifera | 92 | 1 | 0.96 | + | ||
| Protoparmelia hypotremella | 100 | 1 | 1.0 | + | ||
| Protoparmelia isidiata A | 89 | 0.96 | 0.98 | + | ||
| Protoparmelia isidiata B | 64 | 1 | 0.97 | + | ||
| Protoparmelia isidiata C | 100 | 1 | 0.99 | + | ||
| Protoparmelia isidiata D | 100 | 1 | 0.99 | + | ||
| Protoparmelia isidiata E | 100 | 1 | 0.99 | + | ||
| Protoparmelia memnonia | 100 | 1 | 0.99 | + | ||
| Protoparmelia montagnei A | 99 | 1 | 0.99 | - | ||
| Protoparmelia montagnei B | 100 | 1 | 1.0 | - | ||
| Protoparmelia montagnei C | 100 | 1 | 1.0 | + | ||
| Protoparmelia multifera | 100 | 1 | 1.0 | + | ||
| Protoparmelia ochrococca A | 74 | 0.82 | 1 | 0.56 | + | |
| Protoparmelia ochrococca B | NA | NA | + | |||
| Protoparmelia oleagina | 100 | 1 | 0.99 | + | ||
| Protoparmelia orientalis | 100 | 1 | 1.0 | + | ||
| Protoparmelia pulchra | 100 | 1 | 1.0 | + | ||
| Protoparmelia sp. KE | 100 | 1 | 1.0 | + | ||
| Protoparmelia sp. ZA | 100 | 1 | 1.0 | + |
Clades in Column A represent putative species having ML BS support ≥ 70% or Bayesian PP ≥ 0.94, tested for speciation probabilities using BP&P and spedeSTEM. + represents supported clades;—represents clades not supported. Clades supported by BP&P were considered as separate species. 1 represents support for 22-species scenario (P. badia B1, B2 and P. badia C1, C2, P. ochrococca A, B collapsed), i.e. three instead of five putative species within Protoparmelia badia.
Discussion
The genus Protoparmelia is more diverse than the traditional taxonomy suggests. This diversity comprises several previously undescribed species, and cryptic lineages within currently accepted species. Most species of Protoparmelia belong to Protoparmelia s. str., consisting of a tropical and an extra-tropical clade. The tropical clade includes several taxa having multispored asci, which were formerly classified in the genus Maronina [48,49,71,72]. All of its members, except the undescribed South African and Keniyan species, are corticolous. Most members of the tropical clade reproduce vegetatively, although a limited number of species propagate predominantly via sexual reproduction. All supported genetic species-level lineages in the tropical clade are congruent with biogeographic origin of the specimens. Evolutionary rates, i.e. rates of base substitutions in an evolutionary lineage over time, appeared to be accelerated in the tropical clade. This phenomenon has also been previously observed in tropical lichens, and attributed to shorter generation times, higher metabolic rates, continuous physiological activity of a poikilohydric organism in a moist environment, and lack of sexuality [73]. The extra-tropical clade contains mostly saxicolous taxa, most of which reproduce sexually. Within this group, while some species show restricted distribution, some other have wide geographic distributions, such as the cosmopolitan P. ‘badia A’ and P. hypotremella which occurs in Europe and North America. Five previously described species and one species putatively new to science group with members of the genus Miriquidica. In contrast to members of Protoparmelia s. str., which produce lobaric or alectoronic acids, these taxa synthesize norstictic acid as major secondary metabolite. Many of these species parasitize other lichens during at least parts of their life cycle [50], a lifestyle not known from members of Protoparmelia s. str. Close affiliations between Miriquidica and Protoparmelia based on shared morphological characteristics have been suggested before [74,75], and a recent molecular study confirmed the close relationship of the P. atriseda-group and Miriquidica [47]. A revision of the genus Miriquidica based on molecular data is currently under way by our colleagues (Timdal, pers. comm.).
Speciation analyses and cryptic diversity
We validated the 25-species scenario for Protoparmelia s. str., which was based on the previously defined species and a few new clades suggested by molecular data (phylogenetic species concept). Based on our sampling, this study largely supported traditionally circumscribed Protoparmelia s. str. species as distinct lineages. However, exceptions included P. isidiata, an asexual tropical species, and P. badia, a sexually reproducing, boreal-arctic/alpine cosmopolitan species. The former was found to be polyphyletic and separated into three distinct lineages, while the later was paraphyletic and formed a species complex with P. memnonia (Figs 1 and 2).
The combined use of species-tree topology and coalescent methods revealed the presence of several cryptic lineages in Protoparmelia s. str. This is in concordance with other studies in which molecular markers in combination with statistical tools revealed many genetically distinct lineages hidden under a single taxon [9,36,76–78]. Studies suggest that cosmopolitan species such as P. badia may reveal high cryptic diversity [79,80], which may or may not correlate to geography. In our study we found that the cosmopolitan P. badia as currently delimited consists of at least four independent evolutionary lineages. Among these newly recognized lineages only P. badia A turns out to be cosmopolitan, inhabiting boreal-arctic/alpine habitats in North America, Europe, New Zealand and Australia. The other lineages of P. badia (P. badia B1, B2 and C) have a more limited distribution, having been collected so far on siliceous substrates in Spain and Italy. Cryptic lineages within P. isidiata (clades A-C) also correspond to broad biogeographic patterns, while lineages identified within P. montagnei co-occur in the Mediterranean region (Fig 2). Thus, geographic evidence supports species delimitation suggested by coalescent-based speciation analyses in most cases. However, current sampling in many lineages is relatively sparse and does not allow conclusions about finer-scale biogeographic patterns, such as endemism. It remains to be seen whether sympatrically-occurring cryptic lineages identified in this study are supported by additional, previously overlooked morphological or chemical characteristics. We have preliminary evidence that the currently recognized P. montagnei chemotypes [81] correspond to the three molecular clades and may thus indeed represent closely related, but separate species.
Conflicts between different methodological approaches to species delimitation are common [13,15,78,82]. In general we follow the approach of adopting the speciation scenario that is supported by both the analyses, in our case 16 species [83]. For some clades, i.e. P. ochrococca A & B, P. badia A, B1, B2 & C, P. montagnei A & B, the most likely speciation scenario given by spedeSTEM deviates from BP&P, and contradicts supported branching patterns in the phylogeny (Figs 2 and 3). For P. badia A, B1, B2 & C, P. montagnei A & B phylogenetic tree and BP&P supported these clades to be evolutionary independent, whereas spedeSTEM suggested them to be a single species. For P. ochrococca A & B phylogenetic tree and spedeSTEM supported these clades to be evolutionary independent, whereas BP&P suggested them to be a single species. Recent studies indicated that spedeSTEM may be less accurate than other species delimitation methods in cases of recent speciation events [84]. For the clades supported by BP&P and not spedeSTEM, we preferred BP&P results as BP&P has been shown to perform well even when putative species were modeled to have diverged from one another only very recently [84]. In addition, BP&P has been shown to outperform other coalescent-based species delimitation approaches especially when using multi-locus DNA sequence data and a modest number of individuals per species [69,83]. Previously the reliability of BP&P has been suggested to be dependent on the accuracy of the user-provided guide tree. However, in the latest version of BP&P the authors addressed this issue and applied the NNI algorithm, which allows flexibility in the species tree. Moreover BP&P is suggested to be conservative in delimiting species, with high probability to be a reliable indicator of evolutionary independence of the lineages [66]. Therefore in case of conflicts we considered BP&P to be more accurate and suggested the lineages supported by BP&P as distinct species.
Our analyses suggest that the sampled specimens of the tropical Protoparmelia s. str. group belong to five distinct species. Two sexually reproducing (apotheciate) species, P. multifera and P. orientalis, traditionally distinguished by having different minor secondary metabolites [49] were supported as different species and were not sister to each other. In fact, the sexually reproducing species P. pulchra was sister to P. orientalis. In addition, we found four distinct asexually reproducing (isidiate) species of Protoparmelia s. str. Two of these species (P. ‘isidiata D’ and P. ‘isidiata E’) occur sympatrically in Australia. Several studies have shown the occurrence of phylogenetically unrelated but morphologically similar lineages thus indicating the presence of high hidden diversity in lichen-forming fungi [25,27,34,85,86].
Conclusions
Our analyses support the presence of 23 distinct lineages in Protoparmelia s. str. in contrast to 12 currently delimited species, revealing much more diversity than currently suggested for this genus. Our study shows that the sister group of the largest family of lichen-forming fungi may harbor a considerable amount of cryptic lineages which can be identified using molecular data. These data highlight the presence of substantial phylogenetic diversity especially in the tropics, and the need for careful re-evaluation of morphological and chemical characters in the group.
Supporting Information
(XLSX)
Acknowledgments
We thank the curators of the herbaria ASCR, BG, CANB, CANL, EA, FR, GZU, HO, LD, MAF, MSC, MSUT, NY, O, OSC, TRH, UPS, UCR and M. Kossowska (Wroclaw, Poland), Pieter P.G. van den Boom (Son, Netherlands), Toby Spribille (Graz), Zdenek Palice (Prague), Kerry Knudsen (Riverside) for sending material used in this study. Maria Gernert (Frankfurt) kindly performed HPLC analyses.
Data Availability
All relevant data are available from Dryad, under the DOI: doi:10.5061/dryad.0q515.
Funding Statement
This study was funded by ‘LOEWE, Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz’ of Hesse’s Ministry of Higher Education, Research, and the Arts. Garima Singh was supported by a fellowship from the German Academic Exchange Service (DAAD). P.K.D., V.J.R. and A.C. thanks the Ministerio de Ciencia e Innovación, Spain for financial support (CGL2011-25003, CGL2013-42498-P).
References
- 1. Richardson DHS (1999) War in the world of lichens: parasitism and symbiosis as exemplified by lichens and lichenicolous fungi. Mycol Res 103: 641–650. [Google Scholar]
- 2. Muggia L, Baloch E, Stabentheiner E, Grube M, Wedin M (2011) Photobiont association and genetic diversity of the optionally lichenized fungus Schizoxylon albescens . FEMS Microbiol Ecol 75: 255–272. 10.1111/j.1574-6941.2010.01002.x [DOI] [PubMed] [Google Scholar]
- 3. Elbert W, Weber B, Burrows S, Steinkamp J, Büdel B, Andreae MO, et al. (2012) Contribution of cryptogamic covers to the global cycles of carbon and nitrogen. Nat Geosci 5: 459–462. [Google Scholar]
- 4. Ahmadjian V (1965) Lichens. Annu Rev Microbiol 19: 1–20. [DOI] [PubMed] [Google Scholar]
- 5. Grube M, Spribille T (2012) Exploring symbiont management in lichens. Mol Ecol 21: 3098–3099. [DOI] [PubMed] [Google Scholar]
- 6. Lutzoni F, Pagel M, Reeb V (2001) Major fungal lineages are derived from lichen symbiotic ancestors. Nature 411: 937–940. [DOI] [PubMed] [Google Scholar]
- 7. Lücking R, Lawrey JD, Sikaroodi M, Gillevet PM, Chaves JL, Sipman HJ, et al. (2009) Do lichens domesticate photobionts like farmers domesticate crops? Evidence from a previously unrecognized lineage of filamentous cyanobacteria. Am J Bot 96: 1409–1418. 10.3732/ajb.0800258 [DOI] [PubMed] [Google Scholar]
- 8. Galloway D (1992) Biodiversity: a lichenological perspective. Biodivers Conserv 1: 312–323. [Google Scholar]
- 9. Lücking R, Dal-Forno M, Sikaroodi M, Gillevet PM, Bungartz F, Moncada B, et al. (2014) A single macrolichen constitutes hundreds of unrecognized species. Proc Natl Acad Sci U S A 111: 11091–11096. 10.1073/pnas.1403517111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hale M (1987) A monograph of the lichen genus Parmelia Acharius sensu stricto (Ascomycotina: Parmeliaceae). Smith. Cont. Bot. Smithson Contrib to Bot 66: 1–55. [Google Scholar]
- 11. Esslinger TL (1989) Systematics of Oropogon (Alectoriaceae) in the New World. Syst Bot Monogr 28: 1–111. [Google Scholar]
- 12. Agarwal I, Bauer AM, Jackman TR, Karanth P (2014) Cryptic species and Miocene diversification of Palaearctic naked-toed geckos (Squamata: Gekkonidae) in the Indian dry zone. Zool Scr 43: 455–471. [Google Scholar]
- 13. Carstens BC, Satler JD (2013) The carnivorous plant described as Sarracenia alata contains two cryptic species. Biol J Linn Soc 109: 737–746. [Google Scholar]
- 14. Carter BE (2012) Species delimitation and cryptic diversity in the moss genus Scleropodium (Brachytheciaceae). Mol Phylogenet Evol 63: 891–903. 10.1016/j.ympev.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 15. Giarla T, Voss R, Jansa S (2014) Hidden diversity in the Andes: comparison of species delimitation methods in montane marsupials. Mol Phylogenet Evol 70: 137–151. 10.1016/j.ympev.2013.09.019 [DOI] [PubMed] [Google Scholar]
- 16. Printzen C (2010) Progress in Botany, Vol. 71 Lüttge UE, Beyschlag W, Büdel B, Francis D, editors Berlin, Heidelberg: Springer Berlin Heidelberg; Available: http://www.springerlink.com/index/10.1007/978-3-642-02167-1. [Google Scholar]
- 17. Crespo A, Divakar PK, Hawksworth DL (2011) Generic concepts in parmelioid lichens, and the phylogenetic value of characters used in their circumscription. Lichenol 43: 511–535. [Google Scholar]
- 18. Crespo A, Pérez-Ortega S (2009) Cryptic species and species pairs in lichens: a discussion on the relationship between molecular phylogenies and morphological characters. An del Jardín Botánico Madrid 66: 71–81. [Google Scholar]
- 19. Grube M, Kroken S (2000) Molecular approaches and the concept of species and species complexes in lichenized fungi. Mycol Res 104: 1284–1294. [Google Scholar]
- 20. Aptroot A, Sipman HJM (2008) Diversity of lichenized fungi in the tropical Andes In: Hong Kong University Press; HK, editor. Biodiversity of tropical microfungi. pp. 220–223. [Google Scholar]
- 21. Sipman HJM, Harris RC (1989) Lichens In: Lieth H. Werger MJA, editor. Tropical rain forest ecosystems. Elsevier Science Publishers, Amsterdam: pp. 303–309. [Google Scholar]
- 22. Lumbsch HT, Leavitt SD (2011) Goodbye morphology? A paradigm shift in the delimitation of species in lichenized fungi. Fungal Divers 50: 59–72. [Google Scholar]
- 23. Baloch E, Grube M (2009) Pronounced genetic diversity in tropical epiphyllous lichen fungi. Mol Ecol 18: 2185–2197. 10.1111/j.1365-294X.2009.04183.x [DOI] [PubMed] [Google Scholar]
- 24. Del-Prado R, Blanco O, Lumbsch HT, Divakar PK, Elix JA, Molina MC, et al. (2013) Molecular phylogeny and historical biogeography of the lichen-forming fungal genus Flavoparmelia (Ascomycota: Parmeliaceae). Taxon 62: 928–939. [Google Scholar]
- 25. Altermann S, Leavitt SD, Goward T (2014) Solve a problem like Letharia? A New look at cryptic species in lichen-forming fungi using bayesian clustering and SNPs from multilocus sequence data. PLoS One 9: e97556 10.1371/journal.pone.0097556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hawksworth DL, Lagreca S (2007) New bottles for old wine: fruit body types, phylogeny, and classification. Mycol Res 111: 999–1000. [DOI] [PubMed] [Google Scholar]
- 27. Spribille T, Klug B, Mayrhofer H (2011) A phylogenetic analysis of the boreal lichen Mycoblastus sanguinarius (Mycoblastaceae, lichenized Ascomycota) reveals cryptic clades correlated with fatty acid profiles. Mol Phylogenet Evol 59: 603–614. 10.1016/j.ympev.2011.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crespo A, Lumbsch HT (2010) Cryptic species in lichen-forming fungi. IMA Fungus 1: 167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Velmala S, Myllys L, Halonen P, Goward T, Ahti T (2009) Molecular data show that Bryoria fremontii and B. tortuosa (Parmeliaceae) are conspecific. Lichenol 41: 231. [Google Scholar]
- 30. Pino-Bodas R, Burgaz AR, Martín MP, Lumbsch HT (2011) Phenotypical plasticity and homoplasy complicate species delimitation in the Cladonia gracilis group (Cladoniaceae, Ascomycota). Org Divers Evol 11: 343–355. [Google Scholar]
- 31. Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, et al. (2007) Cryptic species as a window on diversity and conservation. Trends Ecol Evol 22: 148–155. [DOI] [PubMed] [Google Scholar]
- 32. Harrington RC, Near TJ (2012) Phylogenetic and coalescent strategies of species delimitation in snubnose darters (Percidae: Etheostoma). Syst Biol 61: 63–79. 10.1093/sysbio/syr077 [DOI] [PubMed] [Google Scholar]
- 33. Kubatko LS, Carstens BC, Knowles LL (2009) STEM: species tree estimation using maximum likelihood for gene trees under coalescence. Bioinformatics 25: 971–973. 10.1093/bioinformatics/btp079 [DOI] [PubMed] [Google Scholar]
- 34. Leavitt SD, Johnson L, St Clair LL (2011) Species delimitation and evolution in morphologically and chemically diverse communities of the lichen-forming genus Xanthoparmelia (Parmeliaceae, Ascomycota) in western North America. Am J Bot 98: 175–188. 10.3732/ajb.1000230 [DOI] [PubMed] [Google Scholar]
- 35. Leavitt SD, Lumbsch HT, Stenroos S, St Clair LL (2013) Pleistocene speciation in North American lichenized fungi and the impact of alternative species circumscriptions and rates of molecular evolution on divergence estimates. PLoS One 8: e85240 10.1371/journal.pone.0085240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parnmen S, Rangsiruji A, Mongkolsuk P, Boonpragob K, Nutakki A, Lumbsch HT (2012) Using phylogenetic and coalescent methods to understand the species diversity in the Cladia aggregata complex (Ascomycota, Lecanorales). PLoS One 7: e52245 10.1371/journal.pone.0052245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, et al. (2006) Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol 55: 595–609. [DOI] [PubMed] [Google Scholar]
- 38. Fujisawa T, Barraclough TG (2013) Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: a revised method and evaluation on simulated data sets. Syst Biol 62: 707–724. 10.1093/sysbio/syt033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hafellner J (1984) Studien in Richtung einer naturlicheren Gliederung der Sammelfamilien Lecanoraceae und Lecideaceae. Beihefte zur Nov Hedwigia 79: 241–371. [Google Scholar]
- 40. Eriksson OE, Hawksworth DL (1989) Notes on ascomycete systematics—Nos. 804–888. Syst Ascomycetum 8: 59–86. [Google Scholar]
- 41. Hertel H (1984) Über saxicole, lecideoide Flechten der Subantarctis. Beih. Nov Hedwigia 79: 399–500. [Google Scholar]
- 42. Miyawaki H (1991) Protoparmelia badia in Japan. Hikobia 11: 29–32. [Google Scholar]
- 43. Henssen A (1995) Apothecial structure and development in Protoparmelia badia Parmeliaceae s. lat In: Daniëls FJA, Schulz M, Peine J, editors. Flechten Follmann, contributions to lichenology in honor of Gerhard Follmann. Geobotanical and Phytotaxonomical study group. Botanical Institute. University of Cologne, Cologne. [Google Scholar]
- 44. Henssen A, Jahns HM (1973) Lichens Eine Einführung in die Flechtenkunde. Stuttgart: Thieme-Verlag; pp. 1–467. [Google Scholar]
- 45. Arup U, Ekman S, Grube M, Mattsson J-E, Wedin M (2007) The sister group relation of Parmeliaceae (Lecanorales, Ascomycota). Mycologia 99: 42–49. [DOI] [PubMed] [Google Scholar]
- 46. Crespo A, Lumbsch HT, Mattsson JJ-E, Blanco O, Divakar PK, Articus K, et al. (2007) Testing morphology-based hypotheses of phylogenetic relationships in Parmeliaceae (Ascomycota) using three ribosomal markers and nuclear RPB1 gene. Mol Phylogenet Evol 44: 812–824. [DOI] [PubMed] [Google Scholar]
- 47. Singh G, Divakar PK, Dal Grande F, Otte J, Parnmen S, Wedin M, et al. (2013) The sister-group relationships of the largest family of lichenized fungi, Parmeliaceae (Lecanorales, Ascomycota). Fungal Biol 117: 715–721. 10.1016/j.funbio.2013.08.001 [DOI] [PubMed] [Google Scholar]
- 48. Hafellner J, Rogers RW (1990) Maronina a new genus of lichenized ascomycetes (Lecanorales, Lecanoraceae) with multispored asci. Bibl Lichenol 38: 99–108. [Google Scholar]
- 49. Papong K, Kantvilas G, Lumbsch HT (2011) Morphological and molecular evidence places Maronina into synonymy with Protoparmelia (Ascomycota: Lecanorales). Lichenol 43: 561–567. [Google Scholar]
- 50. Poelt J, Leuckert C (1991) Lichens related to Protoparmeli atriseda (Lichens, Lecanoraceae) in Europe. Nov Hedwigia 52: 39–64. [Google Scholar]
- 51. Ryan BD, Nash TH, Hafellner J (2004) Protoparmelia In: Nash TH, Ryan BD, Diederich P, Gries C, Bungartz F, editors. Lichen flora of the Greater Sonoran desert region. pp. 425–430. [Google Scholar]
- 52. Thell A, Crespo A, Divakar PK, Kärnefelt I, Leavitt SD, Lumbsch HT, et al. (2012) A review of the lichen family Parmeliaceae—history, phylogeny and current taxonomy. Nord J Bot 30: 641–664. [Google Scholar]
- 53. Divakar PK, Crespo A, Wedin M, Leavitt SD, Myllys L, McCune B, et al. (n.d.) Evolution of complex symbiotic relationships in a morphologically derived family of lichen-forming fungi. In press. [DOI] [PubMed] [Google Scholar]
- 54. Timdal E (1990) Gypsoplacaceae and Gypsoplaca, a new family and genus of squamiform lichens. Bibl Lichenol 38: 419–427. [Google Scholar]
- 55. Cubero OF, Crespo A (2002) Isolation of nucleic acids from lichens In: Kranner I, Beckett R, Varma A, editors. Protocols in Lichenology. Berlin: Springer; pp. 381–391. [Google Scholar]
- 56. Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, et al. (2011) Geneious v5.4. Available from http://www.geneious.com/
- 58. Sugiura N (2007) Further analysis of the data by Akaike’s information criterion and the finite corrections. Commun Stat—Theory Methods 7: 13–26. [Google Scholar]
- 59. Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9: 772 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Campbell V, Legendre P, Lapointe F-J (2011) The performance of the Congruence Among Distance Matrices (CADM) test in phylogenetic analysis. BMC Evol Biol 11: 64 10.1186/1471-2148-11-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). pp. 1–8. [Google Scholar]
- 63. Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 64. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 65. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang Z, Rannala B (2014) Unguided species delimitation using DNA sequence data from multiple loci. Mol Biol Evol 31: 3125–3135. 10.1093/molbev/msu279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rambaut A, Suchard MA, Xie D, Drummond AJ (n.d.) Tracer v1.6. 2014. Available: http://beast.bio.ed.ac.uk/Tracer.
- 68.Rambaut A, Drummond AJ (2009) TreeAnnotator. Available: http://beast.bio.ed.ac.uk/TreeAnnotator.
- 69. Ence DD, Carstens BC (2011) SpedeSTEM: a rapid and accurate method for species delimitation. Mol Ecol Resour 11: 473–480. 10.1111/j.1755-0998.2010.02947.x [DOI] [PubMed] [Google Scholar]
- 70.Swafford DL (2003) PAUP*: Phylogenetic analysis using parsimony (and other methods) 4.0 Beta. Available: http://www.sinauer.com/paup-phylogenetic-analysis-using-parsimony-and-other-methods-4-0-beta.html.
- 71. Kantvilas G, Elix JA (2007) The genus Ramboldia (Lecanoraceae): a new species, key and notes. Lichenol 39: 135. [Google Scholar]
- 72. Kantvilas G, Papong K, Lumbsch HT (2010) Further observations on the genus Maronina, with descriptions of two new taxa from Thailand. Lichenol 42: 557–561. [Google Scholar]
- 73. Lumbsch HT, Hipp AL, Divakar PK, Blanco O, Crespo A (2008) Accelerated evolutionary rates in tropical and oceanic parmelioid lichens (Ascomycota). BMC Evol Biol 8: 257 10.1186/1471-2148-8-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rambold G (1989) A Monograph of the saxicolous lecideoid lichens of Australia. Bibl Lichenol 34. [Google Scholar]
- 75. Hertel H, Rambold G (1987) Miriquidica genus novum Lecanoracearum (Ascomycetes lichenisati). Mitteilungen der Bot Staatssammlung München 23: 377–392. [Google Scholar]
- 76. Dal Grande F, Alors D, Divakar PK, Bálint M, Crespo A, Imke Schmitt (2014) Insights into intrathalline genetic diversity of the cosmopolitan lichen symbiotic green alga Trebouxia decolorans Ahmadjian using microsatellite markers. Mol Phylogenet Evol 72: 54–60. 10.1016/j.ympev.2013.12.010 [DOI] [PubMed] [Google Scholar]
- 77. Welton LJ, Siler CD, Oaks JR, Diesmos AC, Brown RM (2013) Multilocus phylogeny and Bayesian estimates of species boundaries reveal hidden evolutionary relationships and cryptic diversity in Southeast Asian monitor lizards. Mol Ecol 22: 3495–3510. 10.1111/mec.12324 [DOI] [PubMed] [Google Scholar]
- 78. Domingos FMCB, Bosque RJ, Cassimiro J, Colli GR, Rodrigues MT, Santos MG, et al. (2014) Out of the deep: cryptic speciation in a Neotropical gecko (Squamata, Phyllodactylidae) revealed by species delimitation methods. Mol Phylogenet Evol 80: 113–124. 10.1016/j.ympev.2014.07.022 [DOI] [PubMed] [Google Scholar]
- 79. Murtagh GJ, Dyer PS, Furneaux PA, Crittenden PD (2002) Molecular and physiological diversity in the bipolar lichen-forming fungus Xanthoria elegans . Mycol Res 106: 1277–1286. [Google Scholar]
- 80. Myllys L, Stenroos S, Thell A, Ahti T (2003) Phylogeny of bipolar Cladonia arbuscula and Cladonia mitis (Lecanorales, Euascomycetes). Mol Phylogenet Evol 27: 58–69. [DOI] [PubMed] [Google Scholar]
- 81. Barber M, Giralt M, Elix A, Gomez-bolea A, Llimona X (2006) A taxonomic study of Protoparmelia montagnei (syn. P. psarophana) centered in the Eastern Iberian Peninsula. Mycotaxon 97: 299–320. [Google Scholar]
- 82. Satler JD, Carstens BC, Hedin M (2013) Multilocus species delimitation in a complex of morphologically conserved trapdoor spiders (mygalomorphae, antrodiaetidae, Aliatypus). Syst Biol 62: 805–823. 10.1093/sysbio/syt041 [DOI] [PubMed] [Google Scholar]
- 83. Carstens BC, Pelletier TA, Reid NM, Satler JD (2013) How to fail at species delimitation. Mol Ecol 22: 4369–4383. 10.1111/mec.12413 [DOI] [PubMed] [Google Scholar]
- 84. Camargo A, Morando M, Avila LJ, Sites JW (2012) Species delimitation with ABC and other coalescent-based methods: a test of accuracy with simulations and an empirical example with lizards of the Liolaemus darwinii complex (Squamata: Liolaemidae). Evolution 66: 2834–2849. 10.1111/j.1558-5646.2012.01640.x [DOI] [PubMed] [Google Scholar]
- 85. Molina MC, Del-Prado R, Divakar PK, Sánchez-Mata D, Crespo A (2011) Another example of cryptic diversity in lichen-forming fungi: the new species Parmelia mayi (Ascomycota: Parmeliaceae). Org Divers Evol 11: 331–342. [Google Scholar]
- 86. Argüello A, Del-Prado R, Cubas P, Crespo A (2007) Parmelina quercina (Parmeliaceae, Lecanorales) includes four phylogenetically supported morphospecies. Biol J Linn Soc 91: 455–467. [Google Scholar]
- 87. Matheny PB, Liu YJ, Ammirati JF, Hall BD (2002) Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am J Bot 89: 688–698. 10.3732/ajb.89.4.688 [DOI] [PubMed] [Google Scholar]
- 88. Stiller JW, Hall BD (1997) The origin of red algae: Implications for plastid evolution. Proc Natl Acad Sci 94: 4520–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Schmitt I, Crespo A, Divakar PK, Fankhauser JD, Herman-Sackett E, Kalb K, et al. (2009) New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 23: 35–40. 10.3767/003158509X470602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Döring H, Clerc P, Grube M, Wedin M (2000) Mycobiont-specific PCR primers for the amplification of nuclear ITS and LSU rDNA from lichenized ascomycetes. Lichenol 32: 200–204. [Google Scholar]
- 91. Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2: 113–118. [DOI] [PubMed] [Google Scholar]
- 93. White TJ, Bruns TD, Lee SB, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis M, Gelfand H, Sninsky J, White T, editors. PCR Protocols: a guide to methods and applications. Academic Press, NY, USA: pp. 315–322. [Google Scholar]
- 94. Zoller S, Lutzoni F, Scheidegger C (1999) Genetic variation within and among populations of the threatened lichen Lobaria pulmonaria in Switzerland and implications for its conservation. Mol Ecol 8: 2049–2059. [DOI] [PubMed] [Google Scholar]
- 95. Zhou S, Stanosz GR (2001) Primers for amplification of mt SSU rDNA, and a phylogenetic study of Botryosphaeria and associated anamorphic fungi. Mycol Res 105: 1033–1044. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are available from Dryad, under the DOI: doi:10.5061/dryad.0q515.