Background: Nuclear actin dynamics may function during basic cellular processes such as adhesion, differentiation, cell shape, and motility.
Results: Integrin signaling induces nuclear actin polymerization via the LINC complex in response to fibronectin stimulation or cell spreading.
Conclusion: Nuclear actin reorganization occurs during cell spreading and fibronectin stimulation.
Significance: This may have important implications for understanding mechanotransduction and nuclear plasticity for matrix-directed differentiation.
Keywords: Actin, Fibronectin, Formin, Mechanotransduction, Nuclear Envelope, Nucleus
Abstract
We recently discovered signal-regulated nuclear actin network assembly. However, in contrast to cytoplasmic actin regulation, polymeric nuclear actin structures and functions remain only poorly understood. Here we describe a novel molecular tool to visualize real-time nuclear actin dynamics by targeting the Actin-Chromobody-TagGFP to the nucleus, thus establishing a nuclear Actin-Chromobody. Interestingly, we observe nuclear actin polymerization into dynamic filaments upon cell spreading and fibronectin stimulation, both of which appear to be triggered by integrin signaling. Furthermore, we show that nucleoskeletal proteins such as the LINC (linker of nucleoskeleton and cytoskeleton) complex and components of the nuclear lamina couple cell spreading or integrin activation by fibronectin to nuclear actin polymerization. Spreading-induced nuclear actin polymerization results in serum response factor (SRF)-mediated transcription through nuclear retention of myocardin-related transcription factor A (MRTF-A). Our results reveal a signaling pathway, which links integrin activation by extracellular matrix interaction to nuclear actin polymerization through the LINC complex, and therefore suggest a role for nuclear actin polymerization in the context of cellular adhesion and mechanosensing.
Introduction
Although actin is one of the most abundant proteins in the cytosol of mammalian cells, its presence in the nuclear compartment is comparably low, which complicates the investigation of its nuclear functions. In addition, actin dynamics in the cytosol are clearly determined by the spatiotemporal control of factors that orchestrate assembly and disassembly of actin filaments, whereas the regulation of nuclear actin is far more complex due to the fact that most proteins including actin regulators are subjected to nucleocytoplasmic shuttling in a highly dynamic manner (1, 2). Actin itself is imported and exported by specific mechanisms involving importin 9 and exportin 6 (3, 4). Thus, modulation of nuclear actin likely depends and is critically influenced by the signaling context as well as shuttling dynamics of its regulating factors. It is therefore not surprising that the formation of nuclear F-actin has been controversially discussed up to the point that nuclear actin was considered to be entirely or mainly monomeric in its nature (5–9). Recently, we and others have demonstrated the existence of nuclear F-actin structures in somatic cell nuclei using actin probes, which are specifically targeted to the nucleus (10, 11). This enabled us to directly visualize the formation of a highly dynamic endogenous actin network that rapidly formed upon serum stimulation in intact mammalian cell nuclei (10, 12). Thus, cells possess the ability to assemble long nuclear actin filaments into organized network-like structures within intact nuclear compartments in response to extracellular ligands. However, whether other and more general cellular processes would involve nuclear actin polymerization remains to be elucidated (13). Here we show that cells can form nuclear F-actin upon adhesion and spreading, which depends on functional integrin signaling and components of the LINC2 (linker of nucleoskeleton and cytoskeleton) complex. Nuclear actin polymerization can additionally be triggered by direct integrin activation using fibronectin and appears to be mediated by mDia formins. Our data uncover a cell spreading-induced nuclear actin response that involves the nucleoskeleton and components of the nuclear lamina.
EXPERIMENTAL PROCEDURES
Reagents, Antibodies, and Plasmids
Cell culture reagents were obtained from Invitrogen. Antibodies were obtained from Santa Cruz Biotechnology (MRTF-A, lamin A/C, emerin), Abcam (HDAC2), Sigma (β-actin, FLAG), and Cell Signaling (α-tubulin).
Adding the SV40 large T antigen nuclear localization signal (NLS) (PPKKKRKV) to the Actin-Chromobody-TagGFP (ChromoTek) using standard molecular cloning techniques generated the nuclear actin probe nAC (nuclear Actin-Chromobody. Nucleocytoplasmic shuttling constructs (AC-NLS-NES and LifeAct-GFP-NLS-NES) were obtained by adding the nuclear export signal (NES) of HIV-1 Rev (LPPLERLTL) to the pre-existing NLS.
Cell Lines, Transfections, and Treatments
NIH3T3 and NIH3T3-nAC cells were maintained in DMEM supplemented with 10% (v/v) serum (FCS) at 37 °C in a 5% CO2 atmosphere. NIH3T3 cells stably expressing nAC where generated by lentiviral transduction. The eGFP in pWPXL-eGFP was replaced by nAC using BamHI and SpeI. Viruses were produced in HEK293T cells by simultaneous transfection of pWPXL-nAC as well as the packaging vector psPAX2 and the envelope plasmid pMD2.G. Supernatant containing lentiviral particles was harvested 48 h after transfection. NIH3T3 cells were transduced with viral particles in the presence of 10 μg ml−1 Polybrene (Sigma).
Transfection of plasmids was performed in 6-well plates or μ-slides (ibidi) with Lipofectamine LTX with Plus reagent (Invitrogen). siRNA was transfected with RNAiMAX reagent (Invitrogen). The following sequences were used: 5′-CCGTGCTCCTGGGGCTGGG-3′ (murine emerin); 5′-CTGGACTTCCAGAAGAACATT-3′ (murine lamin A/C); 5′-GACCTTAAAGGTGGAAATAAA-3′ (murine Sun1); and 5′-CAGGATTGGAATGGTGGATTA-3′ (murine Sun2).
When indicated, cells were treated with 80 μg ml−1 soluble fibronectin (derived from bovine plasma, Sigma). In Fig. 4C, NIH3T3-nAC cells were preincubated with antibodies (5 μg/ml) for 1 h prior to fibronectin (FN) stimulation. The integrin inhibitory antibody (4B4, Beckman Coulter) was compared with nonspecific IgG (Santa Cruz Biotechnology). To observe nuclear actin network assembly upon serum stimulation (see Fig. 1C), cells were starved for 16 h in serum-free medium before stimulation with 20% serum (FCS).
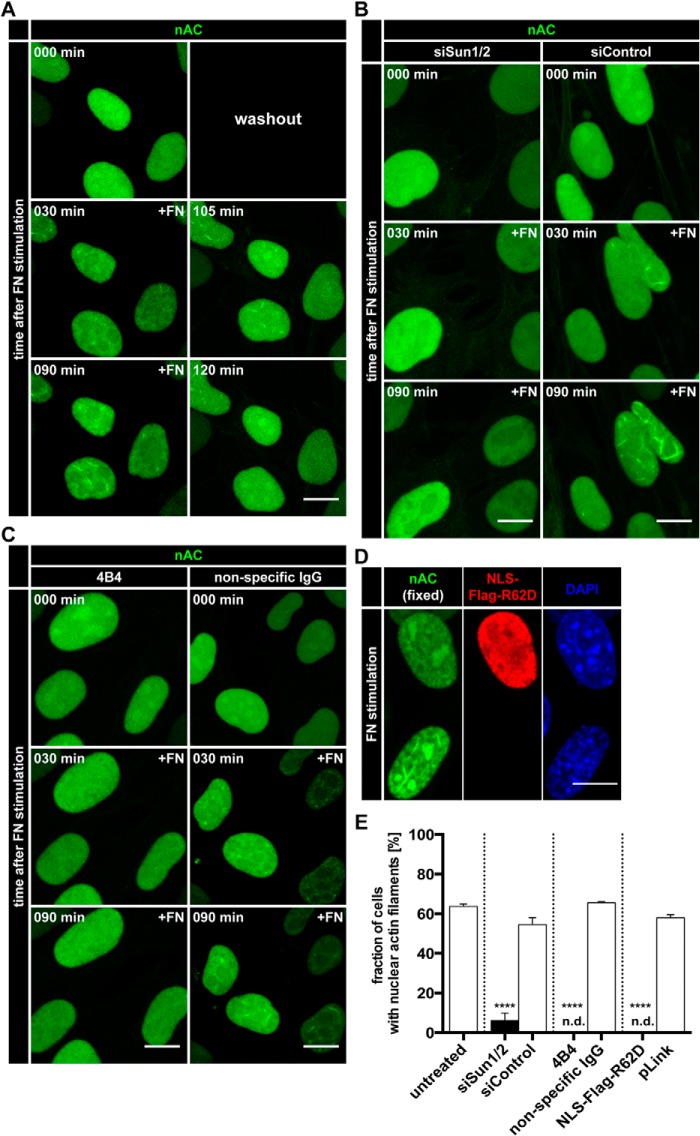
FIGURE 4.
Fibronectin triggers nuclear F-actin formation via Sun proteins and integrin signaling. A, NIH3T3-nAC cells were stimulated with soluble FN and imaged over time using an LSM700. After 90 min, FN-containing medium was replaced with normal medium. Individual frames reveal nAC-probed endogenous actin dynamics at the indicated time points. Maximum intensity projections are shown. Scale bar, 10 μm. See also supplemental Movie S3. B, NIH3T3-nAC cells were treated with the indicated siRNAs prior to stimulation with soluble FN. Individual frames reveal endogenous actin dynamics at the indicated time points. Maximum intensity projections are shown. Scale bar, 10 μm. See also supplemental Movie S4. C, NIH3T3-nAC cells were preincubated with an integrin-inhibitory antibody (4B4) or nonspecific IgG prior to stimulation with FN. Individual frames reveal endogenous actin detected by nAC at the indicated time points. Maximum intensity projections are shown. Scale bar, 10 μm. See also supplemental Movie S5. D, NIH3T3-nAC cells were transfected with NLS-Flag-R62D. Cells were stimulated with soluble FN for 90 min, fixed, and stained (anti-FLAG, red). Nuclei were labeled using DAPI (blue). Scale bar, 10 μm. E, quantifications of nuclear F-actin formation in NIH3T3-nAC cells. Cells were treated as indicated (see also panels A–D), and quantifications were performed 90 min after FN stimulation. n.d., not detected; error bars represent S.E.; ****, p < 0.001. Data were collected from three independent experiments; treated cells were compared with their respective control using a two-sided, unpaired Student's t test. siSun1/2, siRNA targeting Sun1/Sun2. siControl, non-targeting siRNA.
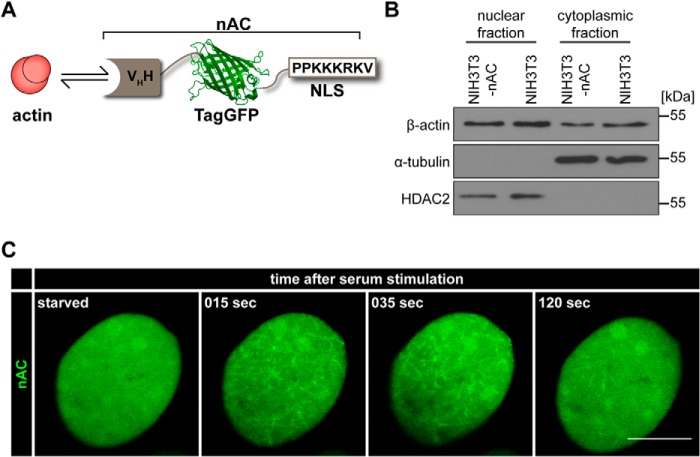
FIGURE 1.
A nAC monitors actin dynamics in somatic cell nuclei. A, graphic of the nAC. VHH, variable domain of heavy chain antibody (antigen-recognizing domain binding to actin); TagGFP, green fluorescent protein. B, nuclear and cytoplasmic fractions of NIH3T3 and NIH3T3-nAC cells were immunoblotted for endogenous actin, α-tubulin (cytoplasmic loading control), and HDAC2 (nuclear loading control). C, NIH3T3-nAC cells were starved in serum-free medium for 16 h and monitored before as well as during serum (FCS) stimulation. In resting or starved cells, nuclear actin was diffusely distributed as visualized by nAC (left panel). Individual time frames reveal nAC-probed endogenous F-actin assembly and distribution at the indicated time points as assessed using an LSM700. Scale bar, 10 μm. See also supplemental Movie S1.
Cell Spreading Assay
NIH3T3 and NIH3T3-nAC cells were detached using a trypsin/EDTA solution (Invitrogen). Cells were centrifuged, and the cell pellet was resuspended in complete medium (DMEM + 10% (v/v) serum (FCS)) before plating onto glass coverslips or fibronectin- (40 μg ml−1) and gelatin- (0.1% in double-distilled H2O) coated μ-slides.
Cell spreading was analyzed by live cell imaging or in fixed samples. Spreading cells are characterized by a more rounded shape, whereas cells with an overall flat profile were considered as control cells. In any case, quantifications were performed from three independent experiments by counting at least 50 nuclei per experiment. Cells that displayed a diffusely distributed nAC signal (i.e. see Fig. 1C, left panel) were quantified as negative, whereas cells displaying nuclear actin filaments of any type or size were quantified as positive.
MRTF-A/SRF Luciferase Reporter Assay
MRTF-A/SRF activity was assayed as described previously (14) using p3DA.luc and pRL-TK reporter plasmids. After transfection, cells were cultured for 24 h, subsequently used in a cell spreading assay, and lysed at the indicated time points.
Immunofluorescence and Image Analysis
Cells grown on coverslips were fixed in 3.7% formaldehyde and permeabilized with 0.3% Triton X-100 before incubation with desired antibodies. Secondary antibodies labeled with Alexa Fluor 488 and Alexa Fluor 555 were from Invitrogen. Slides were mounted with fluorescent mounting medium (Dako). Images were collected with an LSM700 confocal microscope (Zeiss) equipped with a 63×, 1.4 NA oil objective. A 10× objective was used in Fig. 5D. Image processing was performed with Fiji (National Institutes of Health).
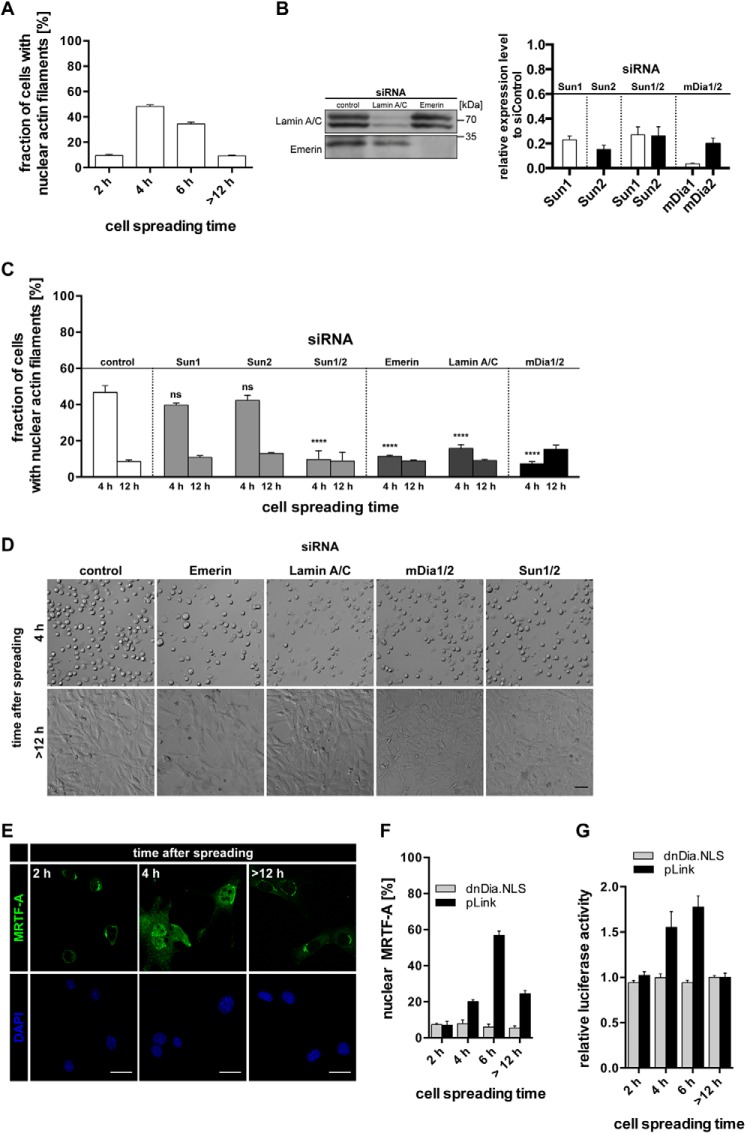
FIGURE 5.
A, quantification of nAC-probed nuclear F-actin formation in NIH3T3 cells at the indicated time points after plating onto glass coverslips. Results are shown as mean ± S.E. from three independently performed experiments. B, knockdown of LINC complex components and mDia1/2 corresponding to panels C and D and Fig. 4, B and E. Left panel, immunoblots for emerin and lamin A/C from siRNA-treated cells as indicated. Right panel, RT-quantitative PCR results relative to control siRNA for Sun1, Sun2, mDia1, and mDia2 from siRNA-treated cells as indicated. C, quantification of nuclear F-actin formation in NIH3T3-nAC cells. Cells were treated with the indicated siRNAs for 48 h prior to plating onto glass coverslips. Error bars represent S.E., ****, p < 0.001; ns, not significant; data were collected from three independent experiments and analyzed using one-way analysis of variance followed by Tukey's honestly significant difference test. D, bright-field images of spreading NIH3T3 cells treated with the indicated siRNAs at 4 h and >12 h after plating. Scale bar, 50 μm. E, representative images of NIH3T3 cells at the indicated time points after plating onto glass coverslips. Cells were stained for endogenous MRTF-A (green). Scale bars, 20 μm. Cells were labeled using DAPI. F, quantification of nuclear MRTF-A during cell spreading in dnDia.NLS- or pLink-transfected cells. Error bars represent S.E. G, relative luciferase activity in NIH3T3 cells at the indicated time points after plating onto glass surfaces. Cells were transfected with luciferase reporter plasmids (pRL-TK and p3DA.luc) and either pLink (control) or dnDia.NLS.
Visualization of nuclear actin filaments using phalloidin was performed as described previously (10). Confocal z-stacks and DAPI stainings were used to confirm the correct localization of the focus plane. Nuclear MRTF-A translocation during cell spreading was quantified by comparing the fraction of cells with a predominant nuclear signal (confirmed by DAPI staining) to cells with a predominant cytoplasmic signal.
Live Cell Imaging
For live cell imaging, cells were grown, transfected, and treated in μ-slides (ibidi). Imaging was performed at 37 °C in a CO2 (5%) humidified chamber using an LSM700 confocal microscope and the ZEN software (Zeiss). Maximum intensity projections were calculated with Fiji.
Immunoblotting and Nuclear Fractionations
Nuclear fractionation was performed as described previously (10).
Statistics
Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software). Data are presented as mean ± S.E. unless stated otherwise. Statistical tests were performed as described in the respective figure legends. Statistical significance is defined as p < 0.05.
RESULTS AND DISCUSSION
We previously generated a nuclear actin probe by fusing LifeAct to an NLS (10). Although this allowed us to reliably detect endogenous nuclear actin polymerization and depolymerization, LifeAct can have some limitations as its expression level needs to be carefully titrated and monitored to prevent any potential stabilization of assembled F-actin structures. We therefore turned to a recently described, antibody-based approach to visualize endogenous proteins utilizing the Chromobody technology (15). We targeted the Actin-Chromobody-TagGFP to the nucleus, generating Actin-Chromobody-TagGFP-NLS, herein termed nAC (Fig. 1A). Transfection of nAC did not affect the abundance of endogenous nuclear actin (Fig. 1B). We validated this approach by inducing nuclear actin network assembly with serum stimulation in living cells (Fig. 1C; supplemental Movie S1). The observed nuclear F-actin structures as well as the temporal resolution were comparable with our previous findings using LifeAct-GFP-NLS (10). Thus, nAC represents a novel and robust tool to visualize and monitor endogenous nuclear actin dynamics in living cells.
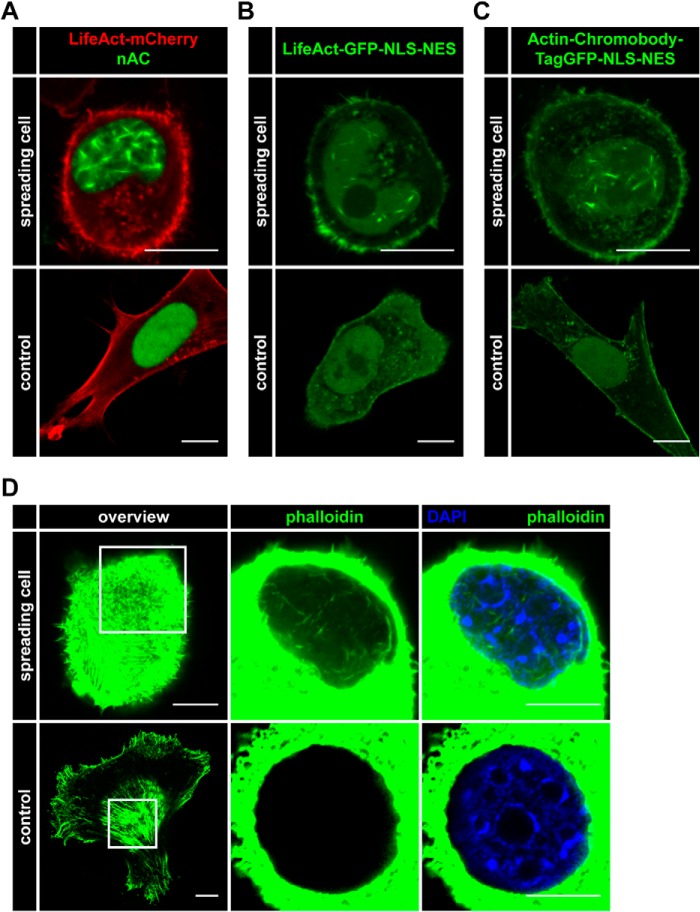
Next we generated stably nAC-expressing NIH3T3 cells to monitor nuclear actin assembly more reliably without transient transfections. While culturing and plating these cell populations, we noticed the formation of distinct nuclear actin filaments during cell spreading (Fig. 2A). These nuclear F-actin structures during spreading were also detectable with a modified nAC probe containing an additional NES (Actin-Chromobody-TagGFP-NLS-NES) to allow for dynamic, nucleocytoplasmic shuttling of the probe (Fig. 2B) or with a comparable LifeAct-GFP-NLS-NES construct (Fig. 2C). Of note, these probes also monitored F-actin structures at the plasma membrane during cell spreading (Fig. 2, B and C). We could confirm the formation of nuclear F-actin during cell spreading using glutaraldehyde fixation and phalloidin staining as the bona fide F-actin marker (Fig. 2D). Taken together, these data reveal the existence of nuclear F-actin formation during cell adhesion and spreading. Of note, spreading-induced nuclear actin filaments appeared to be shorter and thicker than those observed during serum stimulation, indicating that their structure differs significantly from the rapidly and transiently forming network in the serum response.
FIGURE 2.
Nuclear actin polymerization occurs during cell spreading and can be detected by different nuclear actin probes. A–C, NIH3T3 cells were transfected with the indicated (nuclear) actin probes. Spreading cells were assessed for nuclear F-actin formation (green) versus non-spreading (control) cells. LifeAct-mCherry (red) was transfected in A to visualize cytoskeletal actin. Scale bar, 10 μm. D, spreading and non-spreading (control) NIH3T3 cells were assessed for nuclear F-actin formation. Endogenous nuclear F-actin was visualized using phalloidin (Alexa Fluor 488-conjugated, green). Nuclei were labeled using DAPI (blue). Scale bar, 10 μm.
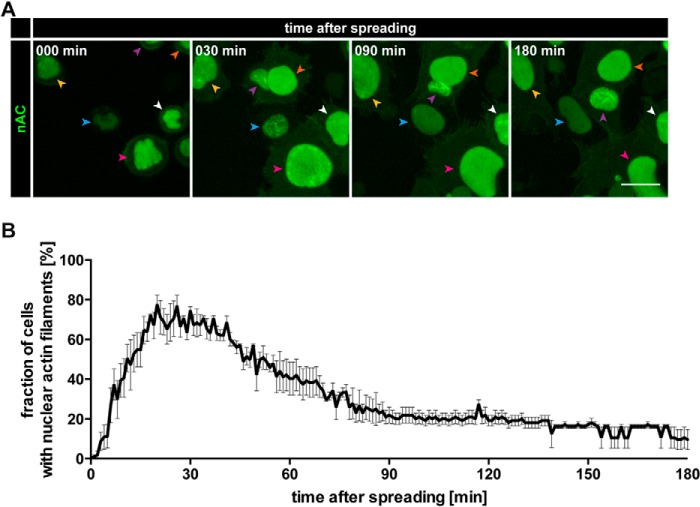
We subsequently analyzed and quantified spreading-induced nuclear actin assembly over time. As shown in Fig. 3A, nuclear F-actin was detectable within 30 min after plating on precoated coverslips and could persist for 2–3 h before disassembly. On average, the maximum response under these conditions occurred at around 25 min after plating before slowly declining over a time period of 3 h or longer (Fig. 3B; supplemental Movie S2). Thus, in contrast to the very rapid and transient F-actin burst during the serum response (Fig. 1C; supplemental Movie S1) (10), spreading-induced nuclear actin assembly is more persistent, and these nuclear actin filaments appear to be shorter and thicker in nature. Of note, these nuclear actin filaments did not colocalize with cofilin (data not shown), which decorates stress-induced nuclear F-actin (16). They were further detectable with LifeAct (Fig. 2, A and B), previously shown not to recognize stress-induced actin-cofilin rods (17). On matrix-coated surfaces, fewer than 80% of all cells displayed detectable nuclear actin filaments during live cell imaging over time, which could indicate that nuclear actin regulation may be dependent on the cell cycle or that it may be influenced by additional, unknown factors.
FIGURE 3.
Temporal characterization of nuclear F-actin formation during cell spreading. A, NIH3T3-nAC cells were plated onto fibronectin- and gelatin-coated surfaces, and their spreading process was monitored over time (see “Experimental Procedures” for details). Individual frames reveal nAC-probed endogenous actin assembly at the indicated time points. Color-coded arrowheads mark the corresponding nuclei of individual cells over time as they move through the visual field. Maximum intensity projections are shown. Scale bar, 10 μm. See also supplemental Movie S2. B, quantification of nuclear F-actin formation during cell spreading as a fraction of cells with nuclear F-actin over a spreading interval of 180 min. Results are shown as mean ± S.E. from three independently performed experiments.
Cell spreading is a mechanosensing process that involves the formation of integrin-based adhesion to the extracellular matrix (18). Thus, we used soluble FN to directly promote and test ligand-induced integrin activation. Interestingly, stimulation of cells with FN readily triggered nuclear F-actin formation within 30 min (Fig. 4A; supplemental Movie S3). This closely resembles the temporal and structural characteristics of spreading-induced nuclear actin assembly as observed in Fig. 3. FN-induced nuclear actin polymerization was reversible after removal of FN-containing medium (Fig. 4A; supplemental Movie S3).
To assess whether the observed nuclear actin filaments were due to de novo actin polymerization, we transfected NIH3T3-nAC cells with the nuclear targeted, non-polymerizable form of actin NLS-Flag-R62D (19, 20). These cells did not form FN-induced nuclear F-actin (Fig. 4, D and E), suggesting that they arise from actin polymerization and not merely from bundling of pre-existing nuclear actin filaments.
To further exclude potential, unspecific effects of fibronectin, we made use of the integrin inhibitory antibody 4B4. Cells preincubated with 4B4 did not display FN-induced nuclear F-actin (Fig. 4C; supplemental Movie S5). Thus, integrin signaling is an integral and necessary component of this pathway.
We then hypothesized that the observed nuclear actin response involves the nucleoskeleton because this structure has recently been shown to be critically involved in mechanosensing and intracellular force transmission via the LINC complex (21–23). The LINC complex provides a physical link between the cytoskeleton and the nuclear envelope (24). This is achieved through the nuclear membrane proteins Sun1 and Sun2, which interact with lamins and other components of the nuclear lamina (21). Therefore, we suppressed Sun1 and Sun2 expression by siRNA and again stimulated cells with FN. Notably, this resulted in a significant loss of FN-promoted nuclear F-actin formation (Fig. 4, B and E; supplemental Movie S4), demonstrating a critical role for the LINC complex in integrin-triggered nuclear actin assembly.
Next we assessed the role of the nucleoskeleton and nuclear lamina during cell spreading-induced nuclear F-actin formation. For this, we used glass coverslips, allowing more convenient analyses and subsequent quantifications. Although as may be expected the overall response of cells displaying nuclear actin assembly during spreading was delayed and decreased as compared with extracellular matrix-coated surfaces (Figs. 3 and 5A), we found that both Sun1 and Sun2 were essentially required for nuclear F-actin formation, but not for silencing of either Sun1 or Sun2 individually (Fig. 5, B and C), which indicates an at least partially redundant function for each Sun protein in this context. Moreover, siRNA against emerin or lamin A/C also resulted in a failure of cells to generate nuclear actin filaments (Fig. 5, B and C), further supporting the notion that the spreading-induced nuclear F-actin response involves a functional nuclear lamina. As may be expected, we found that spreading-induced nuclear actin filaments are sensitive to mDia1/2 silencing (Fig. 5, B and C), indicative of a role for these mDia formins. None of these siRNA treatments prevented cell spreading per se (Fig. 5D).
We previously demonstrated that nuclear actin assembly critically regulates the SRF coactivator and actin-binding protein MRTF-A (also called MAL or MKL-1) (10). It has been shown that cell adhesion and spreading require transcriptional regulation of MRTF-A (25, 26), which localizes to the nucleus in response to nuclear actin dynamics or F-actin formation and causes the release of monomeric actin from MRTF-A, enabling SRF-mediated transcription (10, 27). We therefore analyzed endogenous MRTF-A distribution during spreading and found that nuclear MRTF-A appearance correlated with the detection of nuclear F-actin (Fig. 5, E and F). Importantly, spreading-induced nuclear MRTF-A localization was blocked after expression of a previously described nuclear dominant negative mDia (dnDia.NLS) (10) (Fig. 5, F and G). This demonstrates that nuclear formin activity is responsible for MRTF-A localization to the nucleus during cell spreading. Nuclear MRTF-A appearance and SRF activity appear to be slightly delayed in comparison with nuclear F-actin detection during spreading, possibly reflecting the notion that MRTF-A/SRF regulation is a consequence of nuclear actin assembly.
Here we identified an adhesion-triggered pathway that promotes the formation of nuclear F-actin during cell spreading (Fig. 6). Interestingly, although the shape of these nuclear filaments differs remarkably from those observed after serum stimulation (10), they appear to be nucleated by the same group of mDia formin regulators. Thus, different pathways may converge at nuclear formin activity to induce linear actin filaments of various length and organization, further suggesting that additional yet unknown actin regulators cooperate. This is consistent with the view that many actin-regulating proteins are detectable in the nucleus (7, 28, 29). We find that spreading-induced nuclear actin assembly can regulate MRTF-A similar to the serum-induced response. However, the spreading response is much slower and more persistent in nature than the very rapid network formation, which occurs within seconds upon serum stimulation (Fig. 1C). Thus, it seems tempting to speculate that additional nuclear functions may be regulated as a consequence of actin polymerization in the nucleus; spreading-mediated nuclear actin dynamics could be involved in changes in chromatin organization (5, 30) or in the control of nuclear shape and positioning such as reported during cell migration (31, 32).
FIGURE 6.
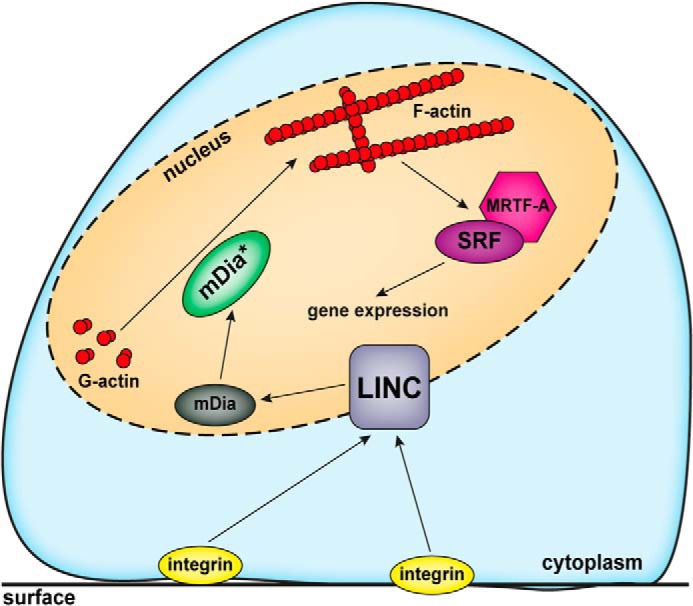
Graphic illustrating the current working model on nuclear F-actin formation induced by mechanotransduction via the LINC complex.
Supplementary Material
Acknowledgments
We thank laboratory members for discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) (GR 2111/7-1).

This article contains supplemental Movies S1–S5.
This article was selected as a Paper of the Week.
- LINC
- linker of nucleoskeleton and cytoskeleton
- MRTF-A
- myocardin-related transcription factor A
- SRF
- serum response factor
- FN
- fibronectin
- nAC
- nuclear Actin-Chromobody
- NLS
- nuclear localization signal
- NES
- nuclear export signal
- TagGFP
- green fluorescent protein.
REFERENCES
- 1. Gama-Carvalho M., Carmo-Fonseca M. (2001) The rules and roles of nucleocytoplasmic shuttling proteins. FEBS Lett. 498, 157–163 [DOI] [PubMed] [Google Scholar]
- 2. Floch A. G., Palancade B., Doye V. (2014) Fifty years of nuclear pores and nucleocytoplasmic transport studies: multiple tools revealing complex rules. Methods Cell Biol. 122, 1–40 [DOI] [PubMed] [Google Scholar]
- 3. Dopie J., Skarp K. P., Rajakylä E. K., Tanhuanpää K., Vartiainen M. K. (2012) Active maintenance of nuclear actin by importin 9 supports transcription. Proc. Natl. Acad. Sci. U.S.A. 109, E544–E552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stüven T., Hartmann E., Görlich D. (2003) Exportin 6: a novel nuclear export receptor that is specific for profilin·actin complexes. EMBO J. 22, 5928–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grosse R., Vartiainen M. K. (2013) To be or not to be assembled: progressing into nuclear actin filaments. Nat. Rev. Mol. Cell Biol. 14, 693–697 [DOI] [PubMed] [Google Scholar]
- 6. Treisman R. (2013) Shedding light on nuclear actin dynamics and function. Trends Biochem. Sci. 38, 376–377 [DOI] [PubMed] [Google Scholar]
- 7. de Lanerolle P., Serebryannyy L. (2011) Nuclear actin and myosins: life without filaments. Nat. Cell Biol. 13, 1282–1288 [DOI] [PubMed] [Google Scholar]
- 8. Visa N., Percipalle P. (2010) Nuclear functions of actin. Cold Spring Harb. Perspect. Biol. 2, a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skarp K. P., Vartiainen M. K. (2010) Actin on DNA: an ancient and dynamic relationship. Cytoskeleton 67, 487–495 [DOI] [PubMed] [Google Scholar]
- 10. Baarlink C., Wang H., Grosse R. (2013) Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 340, 864–867 [DOI] [PubMed] [Google Scholar]
- 11. Belin B. J., Cimini B. A., Blackburn E. H., Mullins R. D. (2013) Visualization of actin filaments and monomers in somatic cell nuclei. Mol. Biol. Cell 24, 982–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baarlink C., Grosse R. (2014) Formin' actin in the nucleus. Nucleus 5, 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hendzel M. J. (2014) The F-act's of nuclear actin. Curr. Opin. Cell Biol. 28, 84–89 [DOI] [PubMed] [Google Scholar]
- 14. Brandt D. T., Baarlink C., Kitzing T. M., Kremmer E., Ivaska J., Nollau P., Grosse R. (2009) SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of β1-integrin. Nat. Cell Biol. 11, 557–568 [DOI] [PubMed] [Google Scholar]
- 15. Rocchetti A., Hawes C., Kriechbaumer V. (2014) Fluorescent labelling of the actin cytoskeleton in plants using a cameloid antibody. Plant Methods 10, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nishida E., Iida K., Yonezawa N., Koyasu S., Yahara I., Sakai H. (1987) Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc. Natl. Acad. Sci. U.S.A. 84, 5262–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Munsie L. N., Caron N., Desmond C. R., Truant R. (2009) Lifeact cannot visualize some forms of stress-induced twisted F-actin. Nat. Methods 6, 317. [DOI] [PubMed] [Google Scholar]
- 18. Humphrey J. D., Dufresne E. R., Schwartz M. A. (2014) Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 15, 802–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kabsch W., Mannherz H. G., Suck D., Pai E. F., Holmes K. C. (1990) Atomic structure of the actin:DNase I complex. Nature 347, 37–44 [DOI] [PubMed] [Google Scholar]
- 20. Posern G., Sotiropoulos A., Treisman R. (2002) Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol. Biol. Cell 13, 4167–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lombardi M. L., Jaalouk D. E., Shanahan C. M., Burke B., Roux K. J., Lammerding J. (2011) The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 286, 26743–26753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simon D. N., Wilson K. L. (2011) The nucleoskeleton as a genome-associated dynamic 'network of networks'. Nat. Rev. Mol. Cell Biol. 12, 695–708 [DOI] [PubMed] [Google Scholar]
- 23. Kaminski A., Fedorchak G. R., Lammerding J. (2014) The cellular mastermind(?)-mechanotransduction and the nucleus. Prog. Mol. Biol. Transl. Sci. 126, 157–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lombardi M. L., Lammerding J. (2011) Keeping the LINC: the importance of nucleocytoskeletal coupling in intracellular force transmission and cellular function. Biochem. Soc. Trans. 39, 1729–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Medjkane S., Perez-Sanchez C., Gaggioli C., Sahai E., Treisman R. (2009) Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat. Cell Biol. 11, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olson E. N., Nordheim A. (2010) Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol 11, 353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vartiainen M. K., Guettler S., Larijani B., Treisman R. (2007) Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316, 1749–1752 [DOI] [PubMed] [Google Scholar]
- 28. Weston L., Coutts A. S., La Thangue N. B. (2012) Actin nucleators in the nucleus: an emerging theme. J. Cell Sci. 125, 3519–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vartiainen M. K. (2008) Nuclear actin dynamics: from form to function. FEBS Lett. 582, 2033–2040 [DOI] [PubMed] [Google Scholar]
- 30. Kapoor P., Shen X. (2014) Mechanisms of nuclear actin in chromatin-remodeling complexes. Trends Cell Biol. 24, 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fedorchak G. R., Kaminski A., Lammerding J. (2014) Cellular mechanosensing: getting to the nucleus of it all. Prog. Biophys. Mol. Biol. 115, 76–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gundersen G. G., Worman H. J. (2013) Nuclear positioning. Cell 152, 1376–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.