Background: Plant immune proteins display complex conformations.
Results: The Prf N-terminal domain forms a homo-dimer, has two binding sites for Pto kinase, and interacts with the Prf leucine-rich repeats domain.
Conclusion: The Prf N-terminal domain coordinates multiple domain interactions to control the activity of the immune complex.
Significance: Additional resolution is supplied to the Prf-Pto complex.
Keywords: Protein Complex, Protein Kinase, Protein Structure, Protein-Protein Interaction, Signal Transduction, N-terminal Domain, NB-LRR, Prf Resistance Protein, Pto Serine/Threonine Kinase, Host-Pathogen Interaction
Abstract
Resistance to Pseudomonas syringae bacteria in tomato (Solanum lycopersicum) is conferred by the Prf recognition complex, composed of the nucleotide-binding leucine-rich repeats protein Prf and the protein kinase Pto. The complex is activated by recognition of the P. syringae effectors AvrPto and AvrPtoB. The N-terminal domain is responsible for Prf homodimerization, which brings two Pto kinases into close proximity and holds them in inactive conformation in the absence of either effector. Negative regulation is lost by effector binding to the catalytic cleft of Pto, leading to disruption of its P+1 loop within the activation segment. This change is translated through Prf to a second Pto molecule in the complex. Here we describe a schematic model of the unique Prf N-terminal domain dimer and its interaction with the effector binding determinant Pto. Using heterologous expression in Nicotiana benthamiana, we define multiple sites of N domain homotypic interaction and infer that it forms a parallel dimer folded centrally to enable contact between the N and C termini. Furthermore, we found independent binding sites for Pto at either end of the N-terminal domain. Using the constitutively active mutant ptoL205D, we identify a potential repression site for Pto in the first ∼100 amino acids of Prf. Finally, we find that the Prf leucine-rich repeats domain also binds the N-terminal region, highlighting a possible mechanism for transfer of the effector binding signal to the NB-LRR regulatory unit (consisting of a central nucleotide binding and C-terminal leucine-rich repeats).
Introduction
Adapted plant pathogens secrete effector proteins into the plant cytoplasm to alter plant cell structure and function to aid the infection (1–3). Effectors can suppress host defenses, but in resistant hosts, they can be perceived by immune recognition complexes, of which the key component is a plant resistance (R)2 protein. Resistance proteins elicit dramatic immune responses such as a local cell death known as the hypersensitive response (4, 5). Resistance proteins are tightly regulated and typically show a high degree of sequence variation, which is likely due to effector diversification. Most R proteins are multidomain proteins consisting of a central nucleotide binding (NB) site and C-terminal leucine-rich repeats (LRR) (NB-LRR proteins) (6, 7). In addition to these conserved domains, numerous additional domains have been described; the most common of these are alternate Toll interleukin 1 receptor (TIR) or coiled-coil (CC) domains (8) immediately N-proximal to the NB-LRR moiety.
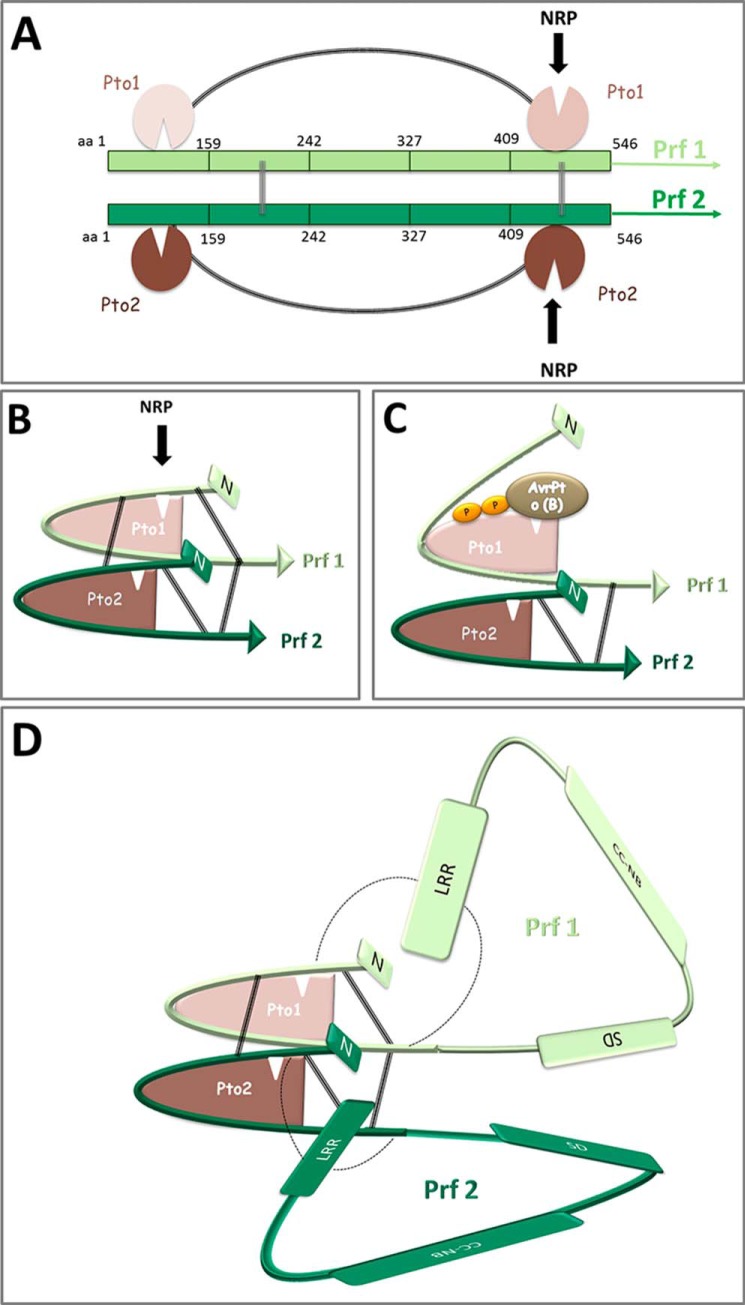
Plant NB-LRR proteins may recognize effectors directly, or indirectly via accessory proteins that are incorporated into recognition complexes (8–10). In the second mechanism, the accessory is described as a molecular bait (or decoy/guard) that provides the recognition capability for the NB-LRR protein (8, 11, 12). The bait is generally a virulence target of the effector, or a mimic of one, and is modified by the effector leading to activation of the NB-LRR moiety. Exactly how effector binding is translated to the NB-LRR regulatory unit is unknown for most effector-R protein interactions. One well studied example for indirect recognition is the Pto-Prf protein complex of tomato. Studies on this complex have revealed important insights into the modification of accessory proteins by effector molecules (13). The Pto kinase is an accessory of the NB-LRR protein Prf, and interacts directly with the unrelated Pseudomonas syringae pv. tomato (Pst) effector proteins AvrPto and AvrPtoB (14–18). The effector binding sites on Pto overlap each other and the catalytic cleft of the enzyme, making important contacts with surface-exposed residues within the kinase P+1 loop and proximal residues known collectively as the negative regulatory patch (NRP). Pto kinase activity is compromised by the effectors. The Prf protein dimerizes, allowing the proximity of at least two Pto kinases. Binding of AvrPto or AvrPtoB to complexed Pto disrupts the P+1 loop. De-repression of the P+1 loop activates a second helper Pto molecule in the Prf complex, which trans-phosphorylates the first, leading to full activation of the complex. Thus, the oligomeric arrangement of the complex constitutes a trap for the effector (19). Pto trans-phosphorylation requires an intact NB motif within Prf, suggesting mutual co-regulation of kinase and NB moieties (20). Interestingly, the NRP is required to maintain Prf in the inactive configuration prior to effector perception, suggesting that there is molecular competition between the effectors and Prf for this binding surface. However, effector binding does not change the Pto-Prf interaction, and the two kinases are kept in close proximity (13, 21). This is mediated by the unique Prf N-terminal domain (previously abbreviated as N-term, here named N, residues 1–546), which is the interaction site for Pto (21) and the contact surface for Prf oligomerization (22). Further, N expressed as an isolated domain can reassemble with the remainder of the Prf molecular comprising the solanaceous domain-coiled-coil-NB-LRR (SCNL) domains (see Fig. 1A), and the hybrid molecule is functional (21).
FIGURE 1.
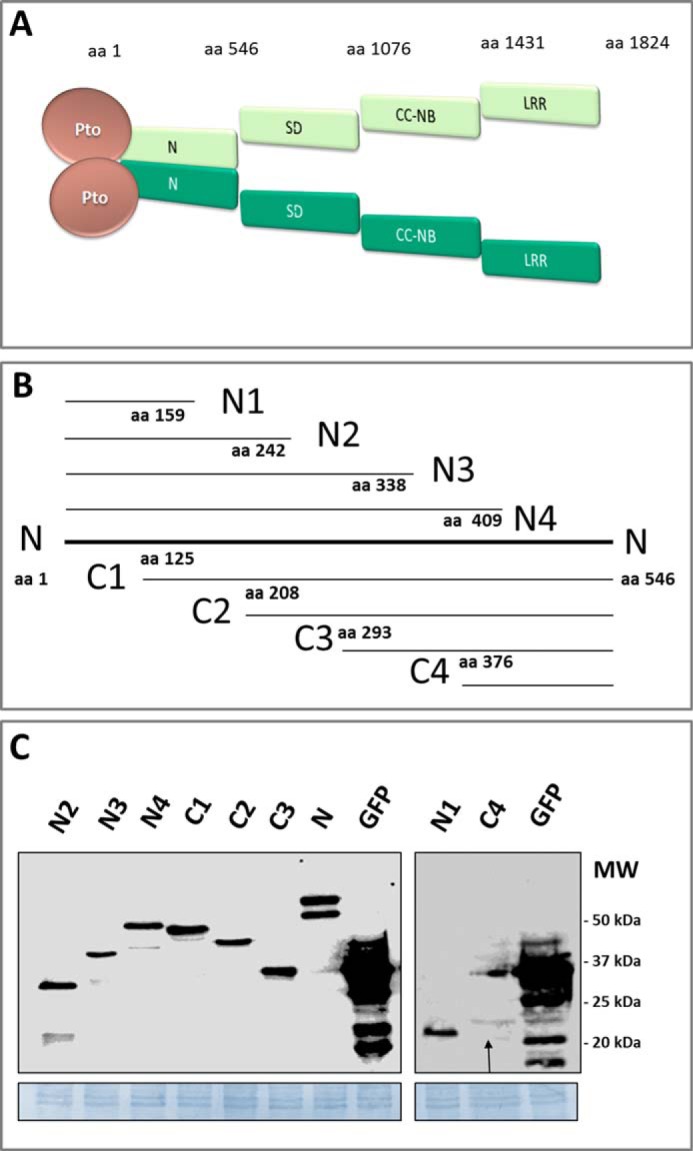
The Pto-Prf protein complex and generation of N-terminal deletion constructs. A, schematic structure of the Pto-Prf protein complex illustrating the Prf domains and homodimerization mediated by the N domain (21, 22). NBS, nucleotide binding site. B, the Prf N domain deletion proteins. N1, N2, N3, and N4 carry the N terminus of N with N1 being the shortest fragment. C1, C2, C3, and C4 carry the C terminus of the N domain with C4 being the shortest fragment. The aa coordinate of each fragment is indicated as a number at the terminus of each fragment. C, immunoblot detection of the N fragments in N. benthamiana protein extracts and comparison of their expression levels to GFP using an anti-HA antibody. N. benthamiana plants were transformed transiently with a construct expressing each gene from the 35S promoter, and leaf tissue was harvested 2 days after infiltration. For detection of N1 and C4, their expression levels were enhanced by the proteasome inhibitor MG132. The arrow points to the C4 band. MW: molecular weight.
The Prf N domain coordinates the activity of the receptor complex via multiple molecular interactions. However, our understanding of how this occurs is limited, and in particular, we lack an understanding of how effector perception by Pto is transferred to the NB motif of Prf. To gain further insight into these questions, we generated deletion fragments of N to determine which regions are important for homotypic interactions and to delineate binding sites for Pto kinase. We found multiple molecular interactions that give a schematic structure of the N-Pto complex and show how the kinase is positioned for effector interaction. Finally, we detect a novel N-LRR interaction, which suggests how the effector signal can be transferred to the NB-LRR unit of the complex.
EXPERIMENTAL PROCEDURES
Generation of Constructs
All constructs were cloned into the binary vectors pT70 (a pTFS-40 derivative containing the 35S promoter) or pT60 (pTFS-40 derivative containing the genomic Pto promoter) carrying 3′-terminal sequences encoding 5×Myc or 3×HA-1×FLAG (3×HAF) epitope tags and transformed into Agrobacterium tumefaciens GV3101 pMp90 (23, 24). Primers used are given in Table 1. Agroinfiltration for transient protein expression and trypan blue staining were performed as described (21).
TABLE 1.
Primers used for generating gfp, Pto mutants, and deletion constructs of Prf for subsequent cloning into pT70 vectors
The SD, NBS, and LRR domains were PCR-amplified from the native Prf sequence; N regions were amplified from a synthetic Prf N-terminal domain sequence. Wild-type Pto, ptoD164N, and ptoL205D sequences were used for site-directed mutagenesis to generate phospho-null and phospho-mimic variants. /5′PHOS/, phosphorylation of 5′ ends used in whole vector amplification for side-directed mutagenesis.
| Gene | Forward primers | Details | Reverse primers | Details |
|---|---|---|---|---|
| gfp | 5′-ctcgagccATGGTGAGCAAGGGCGAG-3′ | XhoI restriction site | 5′-ttctctagaCTTGTACAGCTCGTCCATGCCGAG-3′ | XbaI restriction site -- STOP codon |
| N1 | 5′-gggcgaattgggtaccctcgagccatgGCGAAAGAATGCCGTGAT-3′ | XhoI restriction site | 5′-ttctctagaAATCAGGGCGTTCAGATAAA-3′ | XbaI restriction site -- STOP codon |
| N2 | 5′-gggcgaattgggtaccctcgagccatgGCGAAAGAATGCCGTGAT-3′ | XhoI restriction site | 5′-ttctctagaAATCAGGGCGTTCAGATAAA-3′ | XbaI restriction site -- STOP codon |
| N3 | 5′-gggcgaattgggtaccctcgagccatgGCGAAAGAATGCCGTGAT-3′ | XhoI restriction site | 5′-ttctctagaATCGTAGCTGTAGATGGCATTG-3′ | XbaI restriction site -- STOP codon |
| N4 | 5′-gggcgaattgggtaccctcgagccatgGCGAAAGAATGCCGTGAT-3′ | XhoI restriction site | 5′-ttctctagaGCTCTGAATATAGTTCAGGATA-3′ | XbaI restriction site -- STOP codon |
| C1 | 5′-taccctcgagccatgCCGAGTCGCTCTAGCAG-3′ | XhoI restriction site | 5′-ttctctagaCAGCAGGTTCACCAGAATG-3′ | XbaI restriction site -- STOP codon |
| C2 | 5′-taccctcgagccatgGGTAGCAGCTGCCATATGG-3′ | XhoI restriction site | 5′-ttctctagaCAGCAGGTTCACCAGAATG-3′ | XbaI restriction site -- STOP codon |
| C3 | 5′-taccctcgagccatgGATATTGAAAGCTATC-3′ | XhoI restriction site | 5′-ttctctagaCAGCAGGTTCACCAGAATG-3′ | XbaI restriction site -- STOP codon |
| C4 | 5′ -taccctcgagccatgGTTTGGGAAGAACTGAT-3′ | XhoI restriction site | 5′-ttctctagaCAGCAGGTTCACCAGAATG-3′ | XbaI restriction site -- STOP codon |
| Prf SD | 5′-ctcgagaacaatgATTCCCAAGATGGATGAGATA-3′ | XhoI restriction site | 5′-cgcggatcctaggAGAACGTAAAAGCTCCTCCAA-3′ | BamHI/AvrII restriction site -- STOP codon |
| Prf CC-NB | 5′-taccctcgagccatgATTCCTGCTTGGTATTACACA-3′ | XhoI restriction site | 5′-ttctctagaCTCTTGTTTGGCCTTTTCCAT-3′ | XbaI restriction site -- STOP codon |
| Prf LRR | 5′-taccctcgagccatgAACAAGAGGATTTTCTTCTCCAA-3′ | XhoI restriction site | 5′-ttctctagaagaGTCAAGGGGCTGTTCTTT-3′ | XbaI restriction site -- STOP codon |
| PtoS198A/T199A | /5′-PHOS/CAAACCCATCTTgccgcaGTAGTGAAAGG-3′ | AGC to GCC and ACA to GCA | /5′-PHOS/ATCAAGCTCAGTCCCTTTCTTGGATATTCC-3′ | |
| PtoS198D/T199D | /5′-PHOS/GAGCTTGATCAAACCCATCTTgatgatGTAGTGAAAGG-3′ | GCC to GAT and GCA to GAT | /5′-PHOS/AGTCCCTTTCTTGGATATTCCAAATCAGTAATTTTTG-3′ |
Protein Extraction, Co-immunoprecipitation, and Immunoblotting
For each sample, 300 mg of leaf material was ground in liquid nitrogen and thawed in 2 ml of cold plant protein extraction buffer (150 mm Tris-HCl, pH 7.5, 150 mm sodium chloride, 10 mm EDTA, 10% glycerol, 15 mm DTT, 2% plant protease inhibitor cocktail (Sigma), 2% (w/v) polyvinylpolypyrrolidone). Extracts were centrifuged at 15,000 × g for 10 min at 4 °C. Supernatants were sterile-filtered and centrifuged again at 15,000 × g for 30 min at 4 °C. Antibody-conjugated beads (anti-FLAG M2, Sigma) were incubated in equilibration buffer (Tris-HCl, pH 7.5, 150 mm sodium chloride, 10 mm EDTA, 10% (v/v) glycerol, 1.5% (w/v) BSA) for 1 h at 4 °C and subsequently mixed with the filtered protein extracts for 2 h at 4 °C with slow but constant rotation. Conjugated beads were washed eight times in 1 ml of cold wash buffer (Tris-HCl, pH 7.5, 250 mm sodium chloride, 10 mm EDTA, 10% glycerol, 0.5% plant protease inhibitor) at 4 °C before stripping interacting proteins from the beads by boiling in 50 μl of SDS loading buffer for 5 min. Samples were separated on 8–12% SDS-PAGE gels, blotted onto PVDF membrane, and probed with anti-HA (Roche Applied Science) or anti-Myc (Santa Cruz Biotechnology) primary antibodies, followed by anti-rat IgG-HRP (Sigma) or anti-rabbit IgG-HRP (Sigma) secondary antibodies as appropriate. Labeled proteins were detected by the HRP activity on SuperSignal Femto chemiluminescent substrate (Pierce) using a LAS 4000 luminescence image analyzer (GE Healthcare) for detection. Note that the LAS4000 adjusts appropriate exposure time automatically, and thus protein concentrations between independent blots cannot be compared. All experiments were repeated at least twice, and representative results are shown.
RESULTS
Generation and Expression of Prf N Domain Deletions
The unique Prf N-terminal domain (residues 1–546; hereafter referred to as “N”) is a contact surface for Prf oligomerization and the interaction site for Pto (21, 22). To gain further insight into these interactions, we generated successive genetic deletions from the N and C termini of N (Fig. 1B) and used them to test for protein interactions by co-immunoprecipitation experiments. Unless otherwise indicated, all constructs were expressed in Nicotiana benthamiana leaves under the control of the 35S promoter, using Agrobacterium transient transformation. The smallest fragments N1 and C4 accumulated poorly (Fig. 1C) and were excluded from most subsequent experiments, or tested in separate experiments when absolutely necessary. All deletion proteins lacking the C terminus (N2, N3, and N4) showed N-terminal fragmentation of about 10 kDa (Fig. 1C) as detected previously (21, 22), thus occurring approximately at residue 100.
The N Domain Contains Multiple Interfaces for Homotypic Interactions
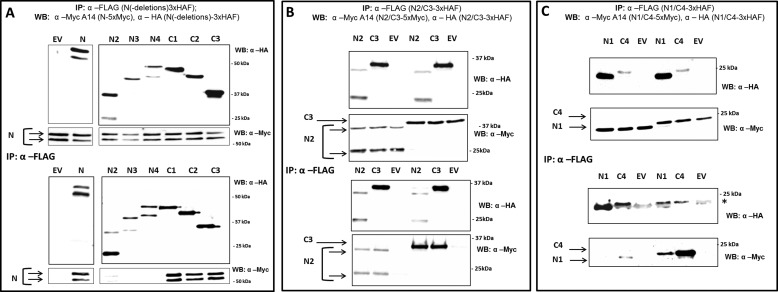
First, we determined which N fragments homodimerize with the full-length N molecule. Only those fragments that carried the C terminus of N (C1, C2, C3) were able to co-precipitate full-length N in the absence of Pto, suggesting that the C terminus contains an important Prf homodimerization site (Fig. 2A). Although N2, N3, and N4 did not interact with full-length N in these tests, it is important to consider that N-N homotypic interactions are competitive in such experiments. Indeed, we found that longer exposure times of the Western blots revealed interactions between N2, N3, and N4 with N (data not shown), suggesting the presence of a homodimerization surface at the N terminus. Importantly, Pto complemented the ability of N2, N3, and N4 to interact with full-length N (Fig. 3B; compare with Fig. 2A). To further investigate N homodimerization, the longest non-overlapping fragments N2 and C3 were co-expressed and their abilities to self-associate or heterodimerize tested. N2 and C3 were each able to self-associate, supporting the results above that showed each terminus in homodimeric associations. Additionally, N2 co-immunoprecipitated C3, and in the opposite pulldown, C3 co-immunoprecipitated N2 (Fig. 2B). Interaction between N2 and C3 demonstrates that non-homologous surfaces in the N- and C-subdomains interact. To narrow this interaction down, potential binding between the shortest terminal deletion fragments, N1 and C4, was tested. Although N1 and C4 are normally unstable when expressed in planta, their accumulation can be enhanced using the proteasome inhibitor MG132 (25). N1 interacted with C4 in both forward and reverse pulldowns (Fig. 2C), suggesting a heterotypic interaction between the N and C termini within the Prf N domain, which may explain at least part of the N2-C3 interaction. On the other hand, and in contrast to N2, the N1 fragment consisting of the 159 N-terminal amino acids of N was unable to homodimerize (Fig. 2C). This defines a further homotypic interaction surface between amino acids (aa) 160–242, in addition to the C4 self-association motif between aa 376–546. Taken together, the Prf N-terminal domain most likely forms a parallel dimer. Additionally, the N dimer apparently folds over crosswise to provide an interaction surface between either end of the molecule.
FIGURE 2.
Homotypic interactions in the Prf N domain. A, the C terminus of N forms a homodimerization site. N. benthamiana plants were transformed transiently with N-5×Myc and a construct encoding each of the N regions fused to a 3×HAF tag, or empty vector (EV). WB, Western blot; IP, immunoprecipitated fraction. B, the C terminus and N terminus of N homo- and heterodimerize. N. benthamiana plants were transformed with N2-5×Myc or C3-5×Myc, and either N2-3×HAF, C3-3×HAF, or EV. C, an N homodimerization site between aa 160 and aa 242. N. benthamiana plants were transformed with N1-5×Myc or C4-5×Myc, and either N1-3×HAF, C4-3×HAF, or EV. N fragments were recovered by anti-FLAG pulldown, and the immunoprecipitates were probed by anti-Myc and anti-HA Western blots after gel electrophoresis. * indicates a cross-reacting band corresponding to the antibody released from the affinity matrix.
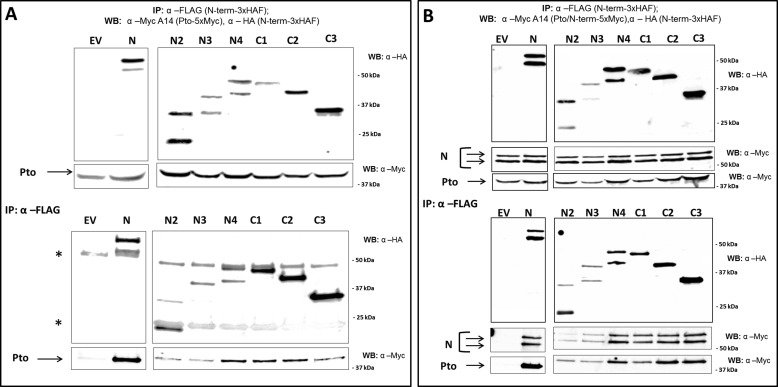
FIGURE 3.
Multiple interactions between N domain fragments and Pto. A, all N deletion fragments bind to Pto. N. benthamiana plants were transformed transiently with Pto-5×Myc, and constructs encoding each of the N deletion fragments were fused to a 3×HAF tag, or EV. WB, Western blot; IP, immunoprecipitated fraction. B, Pto complements the ability of N-terminal fragments to bind full-length N. N. benthamiana plants were transformed transiently with Pto-5×Myc, N-5×Myc, and constructs encoding each of the N deletions fused to a 3×HAF tag, or EV. Leaf tissues were harvested 2 days after infiltration. N and its deletions were recovered by anti-FLAG pulldown, and the immunoprecipitates were probed by anti-Myc and anti-HA Western blots after gel electrophoresis.
The Prf N Domain Contains at Least Two Pto Binding Sites, One of Which Is Affected by the Pto Activation State
The N domain forms the Pto interaction module. To further characterize this important association, the N fragments were assessed for their ability to interact with Pto. All tested regions were able to co-purify Pto (Fig. 3A). N2 and C3 do not share overlapping sequences, which suggests either a very large surface area for Pto interaction or multiple binding sites. We tested the impact of Pto on N-terminal dimerization and found that Pto did not alter the ability of any C-terminal deletion to homodimerize and did not induce N1 homodimerization (data not shown). However, Pto complemented the ability of N2, N3, and N4 to interact with full-length N (Fig. 3B).
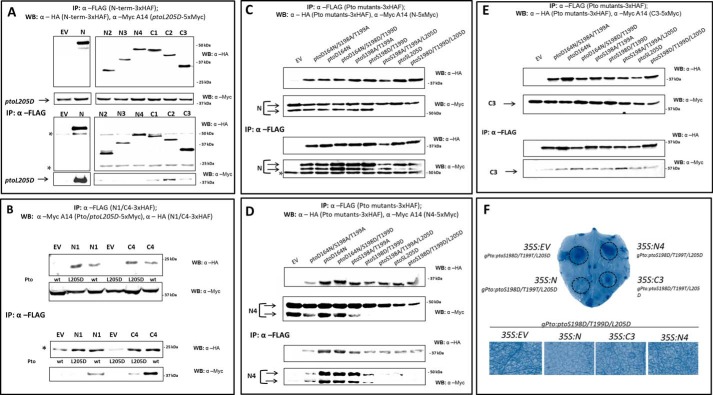
Signaling by the Pto-Prf complex is dependent on two key events during effector-mediated activation: first, disruption of the NRP by effector binding, and second, Pto trans-phosphorylation, which in turn requires an intact NB domain within Prf (19, 23). Pto is phosphorylated on Ser-198 and Thr-199, which can be mimicked by the ptoS198D/T199D double mutant (19). Disruption of the P+1 loop is mimicked by the L205D mutation, which causes a constitutive gain-of-function (CGF) phenotype when the mutants are expressed in N. benthamiana (15). The L205D mutant lacks kinase activity but is phosphorylated in trans by the N. benthamiana Pto homologue 1 (NbPth1) within a NbPrf protein complex (19). We previously speculated that the Pto NRP patch is mirrored by a complementary region in an unknown binding protein to provide inhibitory regulation of Pto signaling (15). As Prf is a constitutive partner of Pto, this sequence likely resides within the N-terminal domain. To locate this region, we co-expressed the N-terminal domain deletion constructs together with ptoL205D and tested the proteins for interaction. Interestingly, N-terminal proteolysis of N was largely absent when ptoL205D was co-expressed (Fig. 4A), implying that the cleavage site may no longer be accessible perhaps due to a change in conformation. The fragments N1, N2, N3, and N4, which commonly lack the C-terminal residues 410–546 but contain the amino acids 1–159, were unable to interact with ptoL205D (Fig. 4, A and B). This suggests that the P+1 loop mutation affects the interaction with Prf residues 1–159, which may interact directly with the Pto catalytic cleft to repress signaling (15). Importantly, C1, C2, and C3 (and even the unstable C4 protein) were all able to bind both Pto and ptoL205D. This defines a second Pto binding site between residues 410–546, which is not affected by P+1 loop mutation. To extend these observations, we tested whether the N4 interaction is affected by phosphorylation status. For this, phospho-mimic (S198D/T199D) and phospho-null (S198A/T199A) Pto mutations were combined with or without the kinase inactivation mutation D164N (15) or the CGF mutant L205D, and the ability of the composite mutants to bind N, N4, and C3 was tested. Whereas all mutants bound to the full-length N domain and the N-terminal deletion C3, no mutant containing the L205D mutation (ptoL205D, ptoS198A/T199A/L205D, ptoS198D/T199D/L205D) was able to co-precipitate N4 (Fig. 4, C–E). Again, N-terminal processing of the 10-kDa fragment was not detected when Pto molecules containing the L205D mutation were paired with Prf N fragments. Interestingly, the phospho-mimic mutant ptoS198D/T198D showed an impaired interaction with N4, but this was not seen with the triple mutant containing the D164N kinase knock-out mutation or with the phospho-null mutant ptoS198A/T198A. This suggests that the kinase activity of ptoS198D/T198D can contribute to the diminished interaction with N4. Overall, the data are suggestive of an interaction between the Pto catalytic cleft mediated particularly by Pto Leu-205 and the first 159 amino acid residues of Prf.
FIGURE 4.
The Prf N domain encodes two binding sites for Pto. A and B, Prf N molecules lacking the C-terminal residues from 409 to 546 do not bind ptoL205D. N. benthamiana plants were transformed transiently with Pto-5×Myc (A) or ptoL205D-5×Myc (A and B) and one of the N regions as indicated fused to a 3×HAF tag, or EV. WB, Western blot; IP, immunoprecipitated fraction. * indicates a cross-reacting band corresponding to the antibody released from the affinity matrix. C, key Pto mutations do not change the interaction with Prf N. N. benthamiana plants were transformed transiently with N-5×Myc and one of the Pto mutants as indicated fused to a 3×HAF tag, or EV. Tissues were processed as above. Note the absence of the cleaved N form in the presence of the Pto L205D mutation. D, Pto molecules containing the L205D mutation do not interact with Prf N4. N. benthamiana plants were transformed transiently with N4-5×Myc and one of the Pto mutants as indicated fused to a 3×HAF tag, or EV. Tissues were processed as above. Note again the absence of the cleaved N form in the presence of the Pto L205D mutation. E, Pto molecules containing the L205D mutation retain the interaction with Prf C3. N. benthamiana plants were transformed transiently with C3-5×Myc and one of the Pto mutants as indicated fused to a 3×HAF tag, or EV. Tissues were processed as above. F, the C terminus of the N domain inhibits CGF signaling by ptoL205D. N. benthamiana plants were transiently transformed with gPto:ptoL205D/S198D/T199D, and either EV, full-length N, or the deletion constructs C3 or N4. All N constructs were expressed from the 35S promoter. Hypersensitive response symptoms from ptoL205D/S198D/T199D expression were visible 5 days after infiltration, and cell death was visualized by staining with trypan blue.
The overexpressed N domain can outcompete NbPrf for Pto binding, causing accumulation of a non-functional Pto-N complex. Overexpression of 35S:N compromised the cell death induced by the most active form of Pto, ptoS198D/T199D/L205D, after weak expression of this mutant from its native promoter (gPto:ptoS198D/T199D/L205D). Interestingly, co-expression of N4, which lacks the C-terminal 136 aa of full-length N (Fig. 1), was unable to inhibit this hypersensitive response (Fig. 4F). In contrast, the N-terminal deletion C3 (aa 293–546, Fig. 1), completely inhibited the hypersensitive response (Fig. 4F). These results are consistent with identification of a Pto binding site within residues 410–546, whereas the additional interaction in the N terminus site is disrupted by the activation state of Pto and cannot inhibit activated Pto molecules. Taken together, the change of conformation during effector activation probably affects the regulatory interaction between Pto and Prf, rather than either molecule alone.
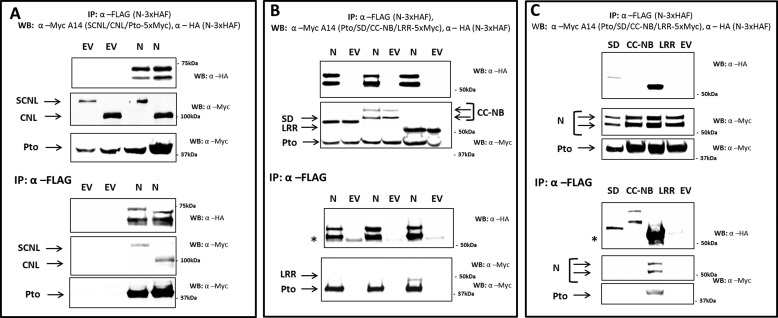
The LRR Domain of Prf Binds the N-terminal Domain
Prf requires both N and SCNL domains for its function (21). Their interaction restores the functional Prf molecule when the two components are expressed separately (21) (Fig. 5A). Reconstitution of subdomains by separate expression has also been shown extensively for the potato R protein Rx (26). Moreover, the LRR domain has been implicated in several systems as controlling R protein activation (8, 12, 27–29). To explore this further, constructs encoding the CC-NB-LRR (CNL, SCNL domain lacking SD) SD (solanaceous domain), CC-NB, and LRR domains of Prf were generated, and their abilities to interact with N were tested. The CNL and the isolated LRR domain of Prf, but not the isolated SD or CC-NB domains, were able to co-precipitate full-length N, and vice versa (Fig. 5). Thus, changes in the Pto-N interaction during effector binding may be translated to Prf via the LRR domain. Co-expression of Pto or its L205D mutant did not change the N-LRR interaction (data not shown). Thus, either Pto and ptoL205D do not affect this interaction, or they may facilitate a change in the N-LRR interaction from cis to trans or vice versa.
FIGURE 5.
Prf N interacts with the LRR domain. A, N. benthamiana plants were transiently transformed with constructs encoding N-5×HAF or EV, and SCNL or CNL (SCNL without SD) fused to a 5×Myc tag. WB, Western blot; IP, immunoprecipitated fraction. B, N. benthamiana plants were transiently transformed with constructs encoding N-5×Myc or EV, and one of the SD, CC-NB, or LRR domains fused to a 3×HAF tag. C, reverse co-immunoprecipitation of B. N. benthamiana plants were transiently transformed with N fused to a 3×HAF tag or EV, and co-transformed with one of the SD, 5×Myc, CC-NB5×Myc, or LRR-5×Myc. All plants were co-transformed with Pto-5×Myc. Transformed leaves were harvested 2 days after infiltration. Proteins tagged with 3×HAF were immunoprecipitated with anti-FLAG, and immunoprecipitates were analyzed after Western blotting with anti-HA and anti-Myc antibodies after gel electrophoresis. * indicates a cross-reacting band corresponding to the antibody released from the affinity matrix.
DISCUSSION
We describe here a proposed scheme for the structure of the Prf N domain. Using deletion analysis, we detected a number of sites of homotypic interaction that underlie the ability of the domain to homodimerize. In addition, we found an unexpected ability of the extreme N and C termini of the molecule to interact, which suggests a folded structure. Consistent with this, we found binding sites for Pto at both the N and C termini. Moreover, the N-terminal Pto binding site was abrogated by a catalytic cleft mutation in Pto that causes the CGF phenotype, whereas the C-terminal site was unaffected by this mutation. The data give insight into how the activity of Pto is controlled prior to effector interaction. Finally, we found a novel interaction between the N and LRR domains, which may underlie regulatory interaction between the Pto-N and NB switches of the protein complex. Studies on the structure and function of the Pto-Prf protein complex are limited by the current constraints of the research system. Heterologous expression of the proteins in N. benthamiana is a powerful tool to understand the mode of action of Pto-Prf and other R proteins. However, this does not rule out possible discrepancies in the structure of the native Pto-Prf complex and the schematic structure generated here. We interpret our data as N forming a parallel homodimer (Fig. 6, A and B). The existence of an antiparallel relationship cannot be excluded. Our data are suggestive of two major homo-interaction areas: the C4 fragment comprising the C-terminal residues 376–546, and a second region by inference based on the ability of N2 but not N1 to dimerize, placing the interaction site between residues 160 and 242. We cannot exclude that the region between 242 and 374 also self-associates. The deletion analysis performed here was limited by the fact that each construct represents a deletion of a deletion, and very often the encoded proteins failed to accumulate. This restricted our ability to make even smaller constructs to test some of our hypotheses directly, particularly with respect to internal deletion constructs of the N domain. In addition to this basic structure, we propose that the domain folds across the long axis, allowing the N and C termini to come into proximity, which is supported by the ability of the smallest terminal fragments to interact with each other. The likely outcome of this interaction is to position two Pto molecules together in an inhibited state, ready for effector perception and transphosphorylation (Fig. 6).
FIGURE 6.
Structure of the dimeric Pto-Prf protein complex. A, schematic illustrating the parallel N homodimer and its interaction with two Pto molecules. Homotypic intermolecular interaction sites on each N-terminal domain are located between aa 159–242 and 376–546. Two independent Pto binding sites on either end of the Prf N domain are located between aa 1–159 and aa 410–546. Each Pto molecule is depicted twice to indicate the two different binding sites on N that interact with different parts of the Pto molecules. Black curves indicate the interaction between and the N and C termini of N. Numbers represent aa positions counting from the N terminus. B, three-dimensional representation of the schematic in A. C, effector-mediated changes within the Prf N-Pto complex. Binding of AvrPto or AvrPtoB to the Pto NRP leads to a loss of interaction with aa 1–159 of the Prf1 N domain. This is translated through Prf Pto2 for trans-phosphorylation of Pto1 and full complex activation (reviewed in Ntoukakis et al. (13)). D, three-dimensional representation of the Pto-Prf complex. The LRR domain of Prf interacts with N, bringing the LRR domain into proximity to the Pto-N activation hub. The N-LRR interaction is shown in cis. Prf, light and dark green; Pto, light and dark red.
Self-association of plant NB-LRR proteins has been described in a number of examples. This includes proteins of both the CC-NB-LRR class, such as Prf, RPS5, and MLA10, and the Toll interleukin 1 receptor, including tobacco N, flax L6, and the heterologous pair RPS4/RRS1. On the other hand, multimerization is absent for CC-NB-LRRs such as Mi and Rx (30). Not enough is understood about these models to generalize roles for dimerization (19, 22, 31). In animal systems, the NB domain is the oligomerizing structure, although unrelated N-terminal motifs such as the CARD domain can also mediate self-interaction (36, 37). Oligomerization of the Pto-Prf complex occurs at least in part to bring two kinase molecules into proximity, possibly for Pto transphosphorylation. Identification of binding sites for Pto within the N domain provides further insight into this important level of regulation.
Our results imply binding sites for Pto on the N and C termini of the Prf N domain. One of these lies C-terminal of residue 409, which also contains a strong homo-interaction site. Thus, the Pto binding site might be on the obverse side of the homo-interaction site, or as an alternative, formed in the cleft between each monomer. We also identified a second interaction site within the residues 1–159. Importantly, deletion fragments containing only the N-terminal site were unable to interact with a CGF mutant of the Pto kinase that is mutated in the P+1 loop substrate binding domain. This is an intriguing observation because it suggests that the N-terminal region of Prf carries the proposed complementary surface patch to the Pto NRP that normally acts to repress Prf signaling (15, 20). Furthermore, as the NRP region overlaps the effector binding sites on Pto (15, 17), it is likely that this region of Prf also contacts each effector, and indeed may also play a role in inhibition of kinase activity. It will be intriguing to test these ideas in future experiments.
The existence of two Pto interaction surfaces may explain why loss of the Pto-Prf interaction after effector binding has never been observed, despite the fact that conformational changes are clearly involved in receptor complex activation (15, 21). Unlike the C-terminal binding site, the N-terminal site is not part of a homo-interaction surface, implying that it is more mobile. This is supported by the observation that N-terminal proteolysis of N is largely absent when ptoL205D is co-expressed. Thus, the cleavage site is likely surface-exposed before P+1 loop disruption, perhaps by distortion of the polypeptide chain. It seems likely that effector binding also releases the N-terminal interaction for subsequent Pto transphosphorylation, but this remains to be tested. In this scenario, the stable C-terminal interaction surface of Prf N would allow close proximity and transphosphorylation of Pto molecules even after disruption of the N-terminal interaction (Fig. 5C). Further evidence that Pto binding at the N-terminal site induces a conformational shift is provided by the observation that Pto complemented the ability of deletion mutants lacking the C terminus to interact with full-length N. The phospho-mimic ptoS198D/T199D mutant retained some ability to interact with the N-terminal Prf region, supporting a previous conclusion that Pto trans-phosphorylation alone is not sufficient for activation of the protein complex and requires P+1 loop disruption (19). Despite this, when compared with the kinase inactive form ptoD164N/S198D/T199D or the phospho-null form ptoS198A/T199A, ptoS198D/T199D repeatedly showed reduced interaction with the N-terminal binding site. This suggests that Pto kinase activity plays a further role beyond the known transphosphorylation sites, and we note that phosphorylation of Prf is also a possibility.
Further support for our structure is provided by the observation that both full-length N and the C3 subdomain lacking the N-terminal interaction site can suppress the autoactivity of ptoL205D. We interpret this result in terms of ptoL205D forming an inactive complex with the N molecules, and hence becoming unavailable to interact with NbPrf. In contrast, the N4 molecule lacking the C-terminal interaction site did not inhibit ptoL205D because it is unable to bind to it.
We found a novel interaction between the Prf LRR and N domains. This may explain functional restoration of the Prf molecule after co-expression of the isolated N and SCNL domains. Full-length N bound to the SCNL, CNL, and LRR domains, but not the SD or CC-NB domains. The interactions did not require Pto, and we were unable to map the interaction site further using N deletion molecules, as all of them interacted with the LRR domain (data not shown), suggesting two independent binding sites for LRR (Fig. 6). Whether the N-LRR interaction occurs in cis or trans within the dimeric full-length Prf complex is so far undetermined. The inability of SCNL to bind Prf (21) does not give insight into this question, as a mature Prf complex would most likely homodimerize through its N domain in preference to binding the unstable SCNL molecule. However, deletion of either the N termini or the C termini of Prf N led to loss of AvrPto- or AvrPtoB-dependent activation, in the presence of both Pto and SCNL (data not shown). It is thus possible that the N-LRR interaction changes from cis to trans or vice versa during complex activation. The LRR domain is often the effector binding determinant in direct recognition by NB-LRR proteins (31–33), but this is not the case for those NB-LRRs that recognize effectors indirectly. Mediating certain intra- or intermolecular interactions might be a general feature of LRR domains from NB-LRR proteins. Importantly, placing the LRR domain close to the Pto-N regulatory hub could provide a potential link between the two molecular switches of this intriguing immune complex (Fig. 6D). As such, differential affinity of LRR to N may activate the NB site of the complex as found for other NB-LRRs (26, 31, 34, 35). Because of multiple interaction sites between N with Pto and LRR, we are prevented from testing this hypothesis for Pto and Prf by the current constraints of the research system.
Acknowledgment
We thank Dr. Vardis Ntoukakis for insightful discussions.
This work was supported by Grants FT0992129 and DP110103322 from the Australian Research Council (to J. P. R.). This work was also supported by a Tuition Fee Exemption Sponsorship from the Australian National University (to I. M. L. S.).
- R
- resistance
- NB
- nucleotide binding
- LRR
- leucine-rich repeats
- CC
- coiled-coiled
- SD
- solanaceous domain
- NRP
- negative regulatory patch
- N
- Prf N-terminal domain
- SCNL
- SD-CC-NB-LRR
- CNL
- CC-NB-LRR
- CGF
- constitutive gain-of-function
- aa
- amino acids
- EV
- empty vector.
REFERENCES
- 1. Mudgett M. B. (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56, 509–531 [DOI] [PubMed] [Google Scholar]
- 2. Panstruga R., Dodds P. N. (2009) Terrific protein traffic: the mystery of effector protein delivery by filamentous plant pathogens. Science 324, 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Staskawicz B. J. (2001) Genetics of plant-pathogen interactions specifying plant disease resistance. Plant Physiol. 125, 73–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Doorn W. G., Woltering E. J. (2005) Many ways to exit? Cell death categories in plants. Trends Plant Sci. 10, 117–122 [DOI] [PubMed] [Google Scholar]
- 5. Meyers B. C., Dickerman A. W., Michelmore R. W., Sivaramakrishnan S., Sobral B. W., Young N. D. (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20, 317–332 [DOI] [PubMed] [Google Scholar]
- 6. Deslandes L., Rivas S. (2012) Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 17, 644–655 [DOI] [PubMed] [Google Scholar]
- 7. Chisholm S. T., Coaker G., Day B., Staskawicz B. J. (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814 [DOI] [PubMed] [Google Scholar]
- 8. Collier S. M., Moffett P. (2009) NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 14, 521–529 [DOI] [PubMed] [Google Scholar]
- 9. Dodds P. N., Rathjen J. P. (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548 [DOI] [PubMed] [Google Scholar]
- 10. Takken F. L., Goverse A. (2012) How to build a pathogen detector: structural basis of NB-LRR function. Curr. Opin. Plant Biol. 15, 375–384 [DOI] [PubMed] [Google Scholar]
- 11. Dangl J. L., Jones J. D. (2001) Plant pathogens and integrated defence responses to infection. Nature 411, 826–833 [DOI] [PubMed] [Google Scholar]
- 12. van der Hoorn R. A., Kamoun S. (2008) From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ntoukakis V., Saur I. M., Conlan B., Rathjen J. P. (2014) The changing of the guard: the Pto/Prf receptor complex of tomato and pathogen recognition. Curr. Opin. Plant Biol. 20, 69–74 [DOI] [PubMed] [Google Scholar]
- 14. Kim Y. J., Lin N. C., Martin G. B. (2002) Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109, 589–598 [DOI] [PubMed] [Google Scholar]
- 15. Wu A.-J., Andriotis V. M. E., Durrant M. C., Rathjen J. P. (2004) A patch of surface-exposed residues mediates negative regulation of immune signaling by tomato Pto kinase. Plant Cell 16, 2809–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tang X., Frederick R. D., Zhou J., Halterman D. A., Jia Y., Martin G. B. (1996) Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274, 2060–2063 [DOI] [PubMed] [Google Scholar]
- 17. Dong J., Xiao F., Fan F., Gu L., Cang H., Martin G. B., Chai J. (2009) Crystal structure of the complex between Pseudomonas effector AvrPtoB and the tomato Pto kinase reveals both a shared and a unique interface compared with AvrPto-Pto. Plant Cell 21, 1846–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xing W., Zou Y., Liu Q., Liu J., Luo X., Huang Q., Chen S., Zhu L., Bi R., Hao Q., Wu J. W., Zhou J. M., Chai J. (2007) The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature 449, 243–247 [DOI] [PubMed] [Google Scholar]
- 19. Ntoukakis V., Balmuth A. L., Mucyn T. S., Gutierrez J. R., Jones A. M., Rathjen J. P. (2013) The tomato Prf complex is a molecular trap for bacterial effectors based on Pto transphosphorylation. PLoS Pathog. 9, e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mucyn T. S., Wu A. J., Balmuth A. L., Arasteh J. M., Rathjen J. P. (2009) Regulation of tomato Prf by Pto-like protein kinases. Mol. Plant Microbe Interact. 22, 391–401 [DOI] [PubMed] [Google Scholar]
- 21. Mucyn T. S., Clemente A., Andriotis V. M., Balmuth A. L., Oldroyd G. E., Staskawicz B. J., Rathjen J. P. (2006) The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 18, 2792–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gutierrez J. R., Balmuth A. L., Ntoukakis V., Mucyn T. S., Gimenez-Ibanez S., Jones A. M., Rathjen J. P. (2010) Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 61, 507–518 [DOI] [PubMed] [Google Scholar]
- 23. Rathjen J. P., Chang J. H., Staskawicz B. J., Michelmore R. W. (1999) Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J. 18, 3232–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Balmuth A., Rathjen J. P. (2007) Genetic and molecular requirements for function of the Pto/Prf effector recognition complex in tomato and Nicotiana benthamiana. Plant J. 51, 978–990 [DOI] [PubMed] [Google Scholar]
- 25. Ntoukakis V., Mucyn T. S., Gimenez-Ibanez S., Chapman H. C., Gutierrez J. R., Balmuth A. L., Jones A. M., Rathjen J. P. (2009) Host inhibition of a bacterial virulence effector triggers immunity to infection. Science 324, 784–787 [DOI] [PubMed] [Google Scholar]
- 26. Moffett P., Farnham G., Peart J., Baulcombe D. C. (2002) Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21, 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Warren R. F., Henk A., Mowery P., Holub E., Innes R. W. (1998) A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10, 1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Banerjee D., Zhang X., Bent A. F. (2001) The leucine-rich repeat domain can determine effective interaction between RPS2 and other host factors in Arabidopsis RPS2-mediated disease resistance. Genetics 158, 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Du X., Miao M., Ma X., Liu Y., Kuhl J. C., Martin G. B., Xiao F. (2012) Plant programmed cell death caused by an autoactive form of Prf is suppressed by co-expression of the Prf LRR domain. Mol. Plant 5, 1058–1067 [DOI] [PubMed] [Google Scholar]
- 30. Hao W., Collier S. M., Moffett P., Chai J. (2013) Structural basis for the interaction between the potato virus X resistance protein (Rx) and its co-factor Ran GTPase-activating protein 2 (RanGAP2). J. Biol. Chem. 288, 35868–35876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bernoux M., Ve T., Williams S., Warren C., Hatters D., Valkov E., Zhang X., Ellis J. G., Kobe B., Dodds P. N. (2011) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9, 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Césari S., Kanzaki H., Fujiwara T., Bernoux M., Chalvon V., Kawano Y., Shimamoto K., Dodds P., Terauchi R., Kroj T. (2014) The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 33, 1941–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cesari S., Thilliez G., Ribot C., Chalvon V., Michel C., Jauneau A., Rivas S., Alaux L., Kanzaki H., Okuyama Y., Morel J. B., Fournier E., Tharreau D., Terauchi R., Kroj T. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Ooijen G., Mayr G., Albrecht M., Cornelissen B. J., Takken F. L. (2008) Transcomplementation, but not physical association of the CC-NB-ARC and LRR domains of tomato R protein Mi-1.2 is altered by mutations in the ARC2 subdomain. Mol. Plant 1, 401–410 [DOI] [PubMed] [Google Scholar]
- 35. Slootweg E. J., Spiridon L. N., Roosien J., Butterbach P., Pomp R., Westerhof L., Wilbers R., Bakker E., Bakker J., Petrescu A. J., Smant G., Goverse A. (2013) Structural determinants at the interface of the ARC2 and leucine-rich repeat domains control the activation of the plant immune receptors Rx1 and Gpa2. Plant Physiol. 162, 1510–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Linhoff M. W., Harton J. A., Cressman D. E., Martin B. K., Ting J. P. (2001) Two distinct domains within CIITA mediate self-association: involvement of the GTP-binding and leucine-rich repeat domains. Mol. Cell. Biol. 21, 3001–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Inohara N., Chamaillard M., McDonald C., Nuñez G. (2005) NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 74, 355–383 [DOI] [PubMed] [Google Scholar]