Background: Barley yellow dwarf virus, a positive strand RNA virus, contains a 3′-translational enhancer element (BTE).
Results: Ribosomes bind the 5′-UTR only in the presence of the 3′-element.
Conclusion: The BTE is the site of initial ribosome recruitment.
Significance: Ribosome recruitment to the 3′-BTE provides a feedback loop for the switch between translation and transcription of the viral mRNA.
Keywords: Plant Virus, Protein Synthesis, Protein-Nucleic Acid Interaction, RNA Structure, RNA Virus
Abstract
Barley yellow dwarf virus mRNA, which lacks both cap and poly(A) tail, has a translation element (3′-BTE) in its 3′-UTR essential for efficient translation initiation at the 5′-proximal AUG. This mechanism requires eukaryotic initiation factor 4G (eIF4G), subunit of heterodimer eIF4F (plant eIF4F lacks eIF4A), and 3′-BTE-5′-UTR interaction. Using fluorescence anisotropy, SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) analysis, and toeprinting, we found that (i) 40S subunits bind to BTE (Kd = 350 ± 30 nm), (ii) the helicase complex eIF4F-eIF4A-eIF4B-ATP increases 40S subunit binding (Kd = 120 ± 10 nm) to the conserved stem-loop I of the 3′-BTE by exposing more unpaired bases, and (iii) long distance base pairing transfers this complex to the 5′-end of the mRNA, where translation initiates. Although 3′-5′ interactions have been recognized as important in mRNA translation, barley yellow dwarf virus employs a novel mechanism utilizing the 3′-UTR as the primary site of ribosome recruitment.
Introduction
All viruses are parasites of the translational machinery (1). When a positive strand RNA virus first enters a cell to initiate infection, a minuscule number of invading viral genomic RNAs must compete with the overwhelming number of host mRNAs to gain a foothold in the translational machinery. Before RNA replication can commence, viral RNA must be translated in order to synthesize the viral replicase. To outcompete host mRNAs for the ribosomes and to evade host translational control mechanisms, positive strand RNA viruses have evolved a plethora of non-canonical translation initiation mechanisms (2, 3); these include the powerful internal ribosome entry sites (IRESs)2 of picornaviruses (4, 5), hepatitis C virus (6, 7), and dicistroviruses (8) as well as 3′-cap-independent translation elements (3′-CITEs) in plant viruses (9).
Numerous plant and animal viral mRNAs do not possess a 5′-cap structure (m7G(5′)ppp(5′)N) in the mRNA and translate efficiently using alternative cap-independent pathways. Many viruses harbor sequences and a complex secondary structure within their untranslated regions (UTRs) that allow them to bypass cellular translation control steps to produce viral proteins. The 5′-terminal regions of animal and some plant viral mRNAs contain an IRES that recruits the 40S subunit to promote cap-independent translation (10–12). These IRESs interact with a subset of initiation factors that vary depending on the IRES to recruit the 40S subunit in a cap-independent manner (6, 13–15). The IRES of hepatitis C virus binds directly to the 40S ribosomal subunit and eIF3 but does not require any eIF4 factors (7, 16). Even more extreme, dicistroviruses can assemble 80S ribosomes without any eIFs (17, 18). Plant potyviruses contain a 5′ covalently linked protein (VPg) and an adjacent IRES, both of which interact with eIFs and recruit translational machinery (19–21).
Instead of containing IRESs, uncapped RNAs of many plant viruses contain a 3′-cap-independent translation element (3′-CITE) in their 3′-UTR that confers efficient translation initiation. Unlike with IRESs, ribosome scanning from the 5′-end of the mRNA is required for translation of mRNAs relying on a 3′-CITE (9, 22–24). These include RNAs of plus-strand RNA viruses in the Tombusviridae family and the Luteovirus and Umbravirus genera, all of which lack both a 5′-cap and a 3′-poly(A) tail (23–25). The 3′-CITEs fall into about seven apparently unrelated structural classes. 3′-CITEs in five of these classes bind to and require a component of eIF4F (24, 26–28). It should be noted that plant eIF4F is a heterodimer of eIF4G and eIF4E. Although eIF4A is present in plants, it is not part of the eIF4F complex. In addition, plants contain a homolog of eIF4F termed eIF(iso)4F, consisting of eIF(iso)4E and eIF(iso)4G. The tRNA-shaped 3′-CITE of turnip crinkle virus binds directly to ribosomal 60S subunits and 80S ribosomes, but its interactions with initiation factors are unknown (29). The factor requirements of the most recently discovered 3′-CITE (from the Xinjiang isolate of Cucurbit aphid-borne yellows virus) are also unknown (30).
One of the well characterized CITEs is the barley yellow dwarf virus (BYDV)-like cap-independent translation element (3′-BTE) found in the Luteovirus, Dianthovirus, and Necrovirus genera. 3′-BTEs contain a 17-nucleotide (nt) conserved sequence (CS) GGAUCCUGGGAAACAGG that forms stem-loop I (3′-SL-I) due to base pairing of the underlined sequences (Fig. 1). In Fig. 1, the bases in italic type are the CS, and those in the green box are complementary to a sequence near the 3′-end of 18S rRNA (Fig. 1B) (31). Translation initiation factor eIF4G binds the 3′-BTE with high affinity (24, 32) and protects SL-I from modification by SHAPE reagents (33). The loop of stem-loop III (3′-SL-III) of the 3′-BTE forms a long distance RNA-RNA “kissing” stem-loop base-pairing interaction with a loop in the 5′-end of the mRNA (25, 34) (Fig. 1C). Binding of the 3′-BTE to eIF4G and to the 5′-UTR are required for efficient cap-independent translation. Finally, the 3′-BTE does not appear to be an IRES, because this translation initiation requires ribosome scanning from the 5′-end of the mRNA (22, 23). To accommodate these observations, two models have been proposed. Either eIF4F (via its eIF4G subunit) binds the 3′-BTE and is delivered to the 5′-UTR, via long distance base pairing, where it recruits the 40S ribosomal subunit (in the 43 S preinitiation complex), or eIF4F recruits the 40S subunit to the 3′-BTE. The BTE then base-pairs to the 5′-UTR to deliver the 43 S preinitiation complex to the 5′-end (23). In addition, the involvement of any other initiation factors in BYDV translation initiation remains unknown (24). The results described here address these questions.
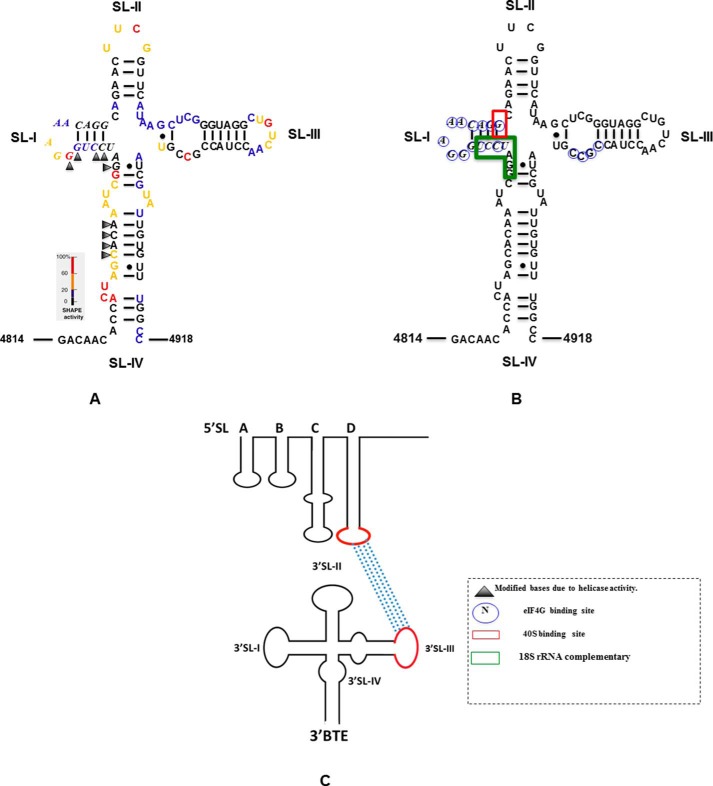
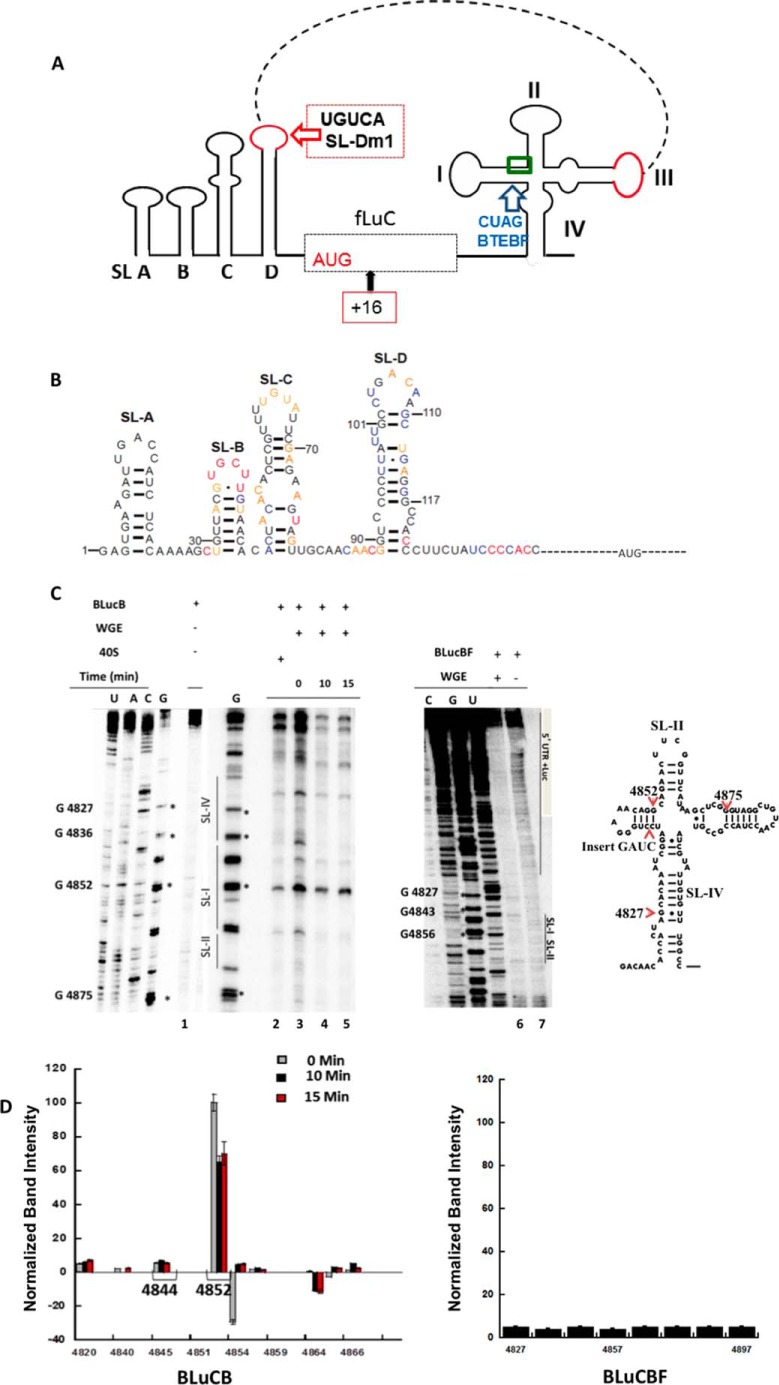
FIGURE 1.
Secondary structure of the 3′-BTE of BYDV genomic RNA. A, secondary structure of 3′-BTE determined by SHAPE reactivity (35). Bases are color-coded based on the level of modification in the SHAPE reaction, where red color indicates the highest modification. Nucleotides are numbered according to their positions in the viral genome. Bases in boldface italic type comprise the 17-nt CS in all BTEs. Exposed bases due to helicase activity of eIF4F-4A-4B and ATP are designated by triangles and correspond to the 18S rRNA-complementary region (GAUCCU, green box, B). B, circled nucleotides in 3′-BTE are protected from SHAPE reagent by eIF4G binding (33). The green box indicates the 18S rRNA-complementary region (GAUCCU) in the SL-I loop. G (red box) in the SL-I region show a 40S toeprint in a helicase complex-treated BYDV mRNA. C, 3′-UTR BTE interaction with 5′-UTR SL-D loop. The red loops are the SL-III and the 5′-SL-D, which interact through complementary base pairing.
We found that 40S ribosomal subunits bind directly to the 3′-BTE with a low affinity and that this affinity increases significantly in the presence of eIF4F, helicase factors eIF4A and eIF4B, and ATP. We also confirm that 80S ribosome loading to the start codon of the message is possible only in the presence of the 3′-BTE. Furthermore, we observed a ribosome toeprint in the SL-I region of 3′-BTE in the wheat germ in vitro translation system and with purified eIFs. Our results support the model in which the 40S subunit binds the 3′-BTE facilitated by the eIF4 translation factors and in which the long distance base pairing promotes transfer of the 40S complex to the 5′-UTR, where scanning and subsequent translation ensue.
EXPERIMENTAL PROCEDURES
RNA Synthesis and Purification
3′-BTE and mutant 3′-BTE-BF transcripts were generated from synthetic DNA oligonucleotides with a T-7 promoter obtained from Integrated DNA Technologies, Inc. (Coralville, IA). BLucB is a reporter plasmid containing the firefly luciferase gene flanked by the BYDV genomic 5′- and 3′-UTRs. BLucBF is the mutated plasmid containing GATC duplication in the BamHI4837 site. BLucB-SL-Dm1 is a mutated BLucB plasmid where CUGACAA nucleotides of stem-loop D (5′-SL-D) were mutated to CUGUCAA to disrupt the “kissing loop” interaction. Preparation and construction of these plasmids were described previously (23, 31, 35). BYDV 5′-UTR plasmid in a pUC MINUS MCS form was constructed by Blue Heron Biotechnology, Inc. (Bothell, WA) and was also used for in vitro study. BLucB, BLucBF, BLucB-SL-Dm1, or BYDV 5′-UTR templates were linearized using SmaI restriction enzyme (New England Biolabs) and transcribed using the T7 Megscript kit according to the standard protocol (Ambion). Capping of BYDV 5′-UTR mRNAs was done using the T7 mScriptTM (CellScript) kit. All transcripts were purified by the Megaclear kit (Ambion). RNA concentrations were determined using a nanodrop UV-visible spectrometer, and integrity was verified by 8–10% polyacrylamide gel electrophoresis.
Computer-aided Analysis of RNA
The genome sequences of BYDV (NC_004666.1) were obtained from the NCBI GenBankTM database. RNA secondary structures were predicted at 37 °C using MFold (36).
Purification of 80S Ribosomes and 40S and 60S Ribosomal Subunits from Wheat Germ
Ribosomal subunits were isolated from wheat germ according to Goss et al. (37) and Spremulli et al. (38). Briefly, 100 g of raw wheat germ (Bob's Mill) was ground to powder with 100 g of alumina in a cold mortar and resuspended in extraction buffer (20 mm HEPES-KOH, pH 7.6, 100 mm KCl, 1 mm Mg(OAc)2, 2 mm CaCl2, and 5 mm 2-mercaptoethanol) followed by centrifugation for 10 min at 15,000 × g. Subsequent steps involved purification of the supernatant using a (3 × 25-cm) G-25 column and centrifugation of 150 A260 units/ml G-25 fractions for 3 h at 170,000 × g. Further purification was performed as described by Spremulli et al. (38). Ribosomes were salt-washed using a method used for the preparation of yeast ribosomes, as described previously (39, 40). Plant 40S and 60S ribosomal subunits were isolated from purified plant 80S ribosomes by sucrose gradient centrifugation as described previously for the isolation of yeast ribosomal subunits (41). Ribosomes were concentrated using an Amicon Ultra (100k) column (Millipore) for buffer exchange and sample concentration against 20 mm HEPES-KOH (pH 7.6), 5 mm Mg(CH3COO)2, 50 mm NH4Cl, 10% glycerol, and 1 mm dithiothreitol (DTT) and stored at −80 °C at a final concentration of ribosome (∼7 μm) and 20% glycerol.
Protein Expression and Purification
The recombinant proteins eIF4F, -4A, and -4B were expressed and purified as described previously (42, 43). HiTrap Mono-Q ion exchange phosphocellulose and m7GTP-Sepharose columns were used for the purification of eIF4F (43), whereas a Ni2+-nitrilotriacetic acid column and a 5-ml Q-Sepharose column were used for the purification of eIF4A and eIF4B, respectively. Protein purity was analyzed by 10% SDS-PAGE. All protein samples were dialyzed against buffer (20 mm HEPES-KOH (pH 7.6), 150 mm KCl, 1.0 mm MgCl2, and 1.0 mm DTT).
Electrophoretic Mobility Shift Assay (EMSA)
RNA unwinding using helicase (eIF4A-4B-4F) complex was performed as described previously (44). Initiation factors eIF4A, eIF4B, and eIF4F (2.0 μm concentration of each) and 5 mm ATP were incubated with 50 nm 32P-labeled 3′-BTE RNA or 3′-BTEBF RNA for 1 h at 37 °C in buffer containing 20 mm (HEPES)-KOH (pH 7.5), 150 mm KCl, 2 mm dithiothreitol, 5 mm magnesium acetate, 0.1 mm GTP, 20 units of RNAsin (Invitrogen) in a final volume of 10 μl. After a 1-h incubation, 40S ribosomal subunits (final concentration of 500 nm) were added to the helicase reaction mixture and incubated at 37 °C for another 30 min. As a control experiment, mRNAs were incubated with eIF4F and 40S subunits separately under the same experimental conditions. Reactions were stopped by adding loading dye and applied to a native 2% polyacrylamide, 2% agarose gel that had been prerun at 30 mA for 30 min. Electrophoresis was carried out at 50 mA for 2 h at 4 °C. The gel was exposed to a PhosphorImager (Amersham Biosciences) overnight and quantified using ImageQuant software (GE Healthcare).
Solution Structure Mapping
RNA structure probing was done by the selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) method (45, 46). Briefly, RNA (20 pmol) in 6 μl of sterile water was heated at 95 °C for 4 min, quickly cooled on ice, treated with 3 μl of folding buffer (333 mm NaCl, 333 mm HEPES (pH 8.0), 33.3 mm MgCl2), and incubated at 37 °C for 20 min. The RNA solution was treated with benzoyl cyanide (1 μl of 10× BzCN, 400 mm in DMSO) and allowed to react for 2.5 s (equal to five BzCN half-lives). Control reactions contained 1 μl of DMSO. Modified RNAs were used for the primer extension reaction (45).
SHAPE studies on helicase complex-treated 3′-BTE were done similarly. Folded 3′-BTE (20 pmol) was first incubated with helicase complex prepared according to Ref. 47 at 37 °C for 30 min and was treated with BzCN in DMSO. Control reactions contained folded 3′-BTE and DMSO.
The cDNA primers (a) 5′-AGTTGCTCTCCAGCGGTTC-3′ and (b) 5′-AACGGCGATAACGTGAAG-3′ were used for structure probing as well as toeprinting assays. The primer a is complementary to the luciferase mRNA in all reporter BYDV constructs and was used for 5′ probing. The primer b is complementary to the 3′-UTR region of BYDV mRNA and was used for probing the BTE.
Primer Extension Inhibition (Toeprinting)
Toeprinting assays of 5′-mRNA were performed on the BLucB mRNA, BLucBF mRNA, and BLucB-SL-Dm1 and 5′-UTR BYDV using wheat germ extract (WGE) and using combinations of different eIFs. The assay protocol for toeprinting in WGE was adapted from Ref. 48. Wheat germ extract reactions were prepared according to the Promega wheat germ in vitro translation kit (L-4380) as they were for use in translation assays, except that a complete amino acid mixture was used, and no [35S]Met was used. Each reaction was treated with 5 mm cycloheximide (CHX). RNAs were incubated in the WGE in a total volume of 10 μl for 30 min at 25 °C. Each microliter of the translation reaction was then diluted in primer extension buffer, which contains four parts 1× SSIII FS buffer (Invitrogen), one part 0.1 m DTT, one part 10 mm dNTP mix, and 1 unit/μl RNaseOUT (Invitrogen) (45), and incubated for 2 min at 55 °C. 32P-Labeled primer was then annealed with the RNAs at 37 °C for 2 min, and reverse transcription was done by using SuperScript III reverse transcriptase (Invitrogen) at 48 °C for 20 min. Reverse transcribed cDNAs were purified by phenol/chloroform (25:1) extraction followed by ethanol precipitation. The reactions were separated in an 8% polyacrylamide, 7 m urea sequencing gel. The sequencing ladder and toeprints were visualized by scanning of the dried gel.
Toeprinting reactions for 3′-UTRs were done using WGE or by assembling 40S subunits (16 pmol), mRNAs (6 pmol), and different combinations of eIF4F, eIF4A, and eIF4B (6 pmol each) in 20-μl reaction mixtures. Each reaction was incubated in a binding buffer (20 mm HEPES at pH 7.8, 6 mm MgCl2, 100 mm NaCl) at 37 °C for 30 min. 32P -Labeled primer was then annealed with the RNAs at 37 °C for 2 min, and toeprints were detected using reverse transcription as described (49).
Quantification of Gels
Toeprinting and SHAPE gel images were analyzed by the semiautomated footprinting analysis (SAFA) software (50, 51). The software converts integral band densities of each band into a numerical value. The numerical values were normalized and plotted against sequences (52).
Fluorescence Anisotropy
Fluorescence anisotropy measurements were carried out using a Horiba Spectra ACQ Fluorolog-3 spectrofluorimeter equipped with excitation and emission polarizers. The sample temperature was 25 °C unless otherwise stated. Anisotropy experiments were performed using an L-format detection configuration. Fluorescence anisotropy titrations were employed to study protein-RNA (eIFs-BYDV mRNAs) and ribosome-RNA (40S-BYDV mRNAs) interactions. RNAs were 5′-labeled with fluorescein 5-maleimide using a 5′-end tag nucleic acid labeling system from Vector Laboratories (Burlingame, CA). The fluorescence anisotropy change was monitored when increasing amounts of eIFs or ribosomes were added to 5′-fluorescence-labeled mRNAs in 20 mm HEPES buffer, pH 7.4, 100 mm KCl at 25 °C. The KD was determined by fitting the plot of changes in anisotropy versus ribosome or eIF concentration using the equation, robs = rmin + ((rmax − rmin)/(2[FlRNA]))(b − (b2 − 4[FlRNA][eIFs])0.5), where the robs is the observed anisotropy value for any point in the titration curve, rmin is the minimal anisotropy value in the absence of protein or ribosome, and rmax is final saturated anisotropy value. b = KD + [FlRNA] + [eIFs/ribosome], as described elsewhere (53, 54). Data were fitted using Kaleida Graph (Abelbeck Software).
RESULTS
40S Ribosomal Subunits Bind First to the BYDV mRNA 3′-UTR
Elucidation of the molecular mechanism of BYDV translation initiation requires an understanding of the ribosome binding events. Although it was shown previously that the 3′-BTE interacts specifically with eIF4F with a very high binding affinity (Kd ∼37 nm) (24, 32) and that this binding affinity correlates with translational efficiency (32), the mechanism of ribosome recruitment and the role of associated eIFs during this event remain unclear. As the next step in the establishment of a ribosome recruitment mechanism in BYDV translation, we have quantitatively characterized interactions between the 40S subunit with wild type and mutant forms of the BYDV 3′-BTE and 5′-UTR alone and in the presence of different eIFs.
Previously, we reported the use of fluorescence quenching and anisotropy experiments to study the equilibrium binding between different mRNAs and eIFs (mRNA-eIF interactions) or between mRNAs and ribosomes (32, 55–57). In this study, we use these approaches to determine the equilibrium dissociation constants (Kd values) for the interactions between 40S ribosomal subunits and 3′-BTE, 5′-UTR, and 3′-BTEBF RNAs (3′-BTEBF is a translationally inactive mutant of the BTE, containing a four-base duplication of GAUC in the 17-nt CS (31)) and the effects of different eIFs on 40S-3′-BTE interactions.
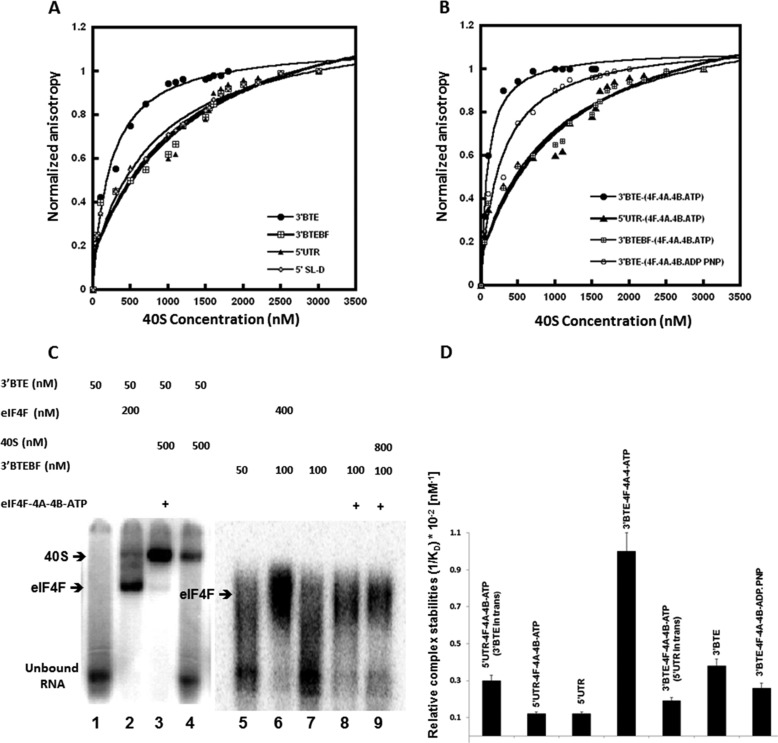
Fluorescence anisotropy measurements of purified 40S ribosomal subunits interacting with fluorescein-labeled 3′-BTE reflected a moderate binding affinity (KD = 350 ± 30 nm) (Fig. 2A). A similar binding affinity was observed for the 3′-BTE with the 80S ribosomes and weaker binding affinity for 60S subunits (KD = 600 ± 20 nm) (data not shown). In contrast, very weak binding affinities to the 40S subunit were observed for the mutant BTEBF (KD = 1200 ± 50 nm) (Fig. 2A), the 5′-UTR of BYDV genomic RNA (KD = 1100 ± 50 nm), or 5′-SL-D of the 5′-UTR (KD = 900 ± 10 nm) (Fig. 2A). These weak binding affinities most likely reflect nonspecific interactions.
FIGURE 2.
Ribosomes bind more tightly to helicase-treated BYDV 3′-UTR compared with the 5′-UTR, BTEBF, or untreated 3′-BTE. A and B, ribosome binding to BYDV-RNA was monitored by changes in 5′-fluorescein-labeled BYDV-RNA fluorescence anisotropy (excitation, 490 nm; emission, 520 nm). A, 40S binding to untreated RNA, 3′-BTE (●), 3′-BTEBF (⊞), 5′-SL-D(♢), and 5′UTR BYDV (▴). KD values are as follows: 3′-BTE (350 ± 30 nm), 3′-BTEBF (1200 ± 50 nm), 5′-SL-D (900 ± 30 nm), and 5′UTR BYDV (1100 ± 50 nm). B, 40S binding to helicase (4F-4B-4A-ATP)-treated RNAs, 3′-BTE (●),3′-BTEBF (⊞), 5′-UTR (▴), and 3′-BTE + 4F-4B-4A-ADPPNP (□). KD values are as follows: 3′-BTE (120 ± 20 nm), 3′-BTEBF (1100 ± 50 nm), 5′-UTR (1100 ± 40 nm), and 3′-BTE + 4F-4B-4A-ADPPNP (350 ± 70 nm). The fluorescein-labeled 3′-BTE, 3′-BTEBF, 5′-SL-D, and 5′-UTR BYDV concentrations were 0.10, 0.5, 0.5, and 0.5 μm, respectively, in titration buffer at 25 °C. The x axis indicates the concentration of ribosomes added. Helicase treatment is described under “Experimental Procedures.” C, EMSA of ribosome binding to helicase-treated BYDV 3′-UTR. 32P-Labeled 3′-BTE or 3′-BTEBF RNAs were incubated in the presence of the indicated proteins or 40S subunits as indicated by a plus sign above each lane. Mobilities of the indicated complexes with labeled RNA are indicated beside the gel. Lane 2 shows the gel shift for eIF4F and RNA. An additional shift occurs when 40S subunits bind (lanes 3 and 4). Helicase treatment increased ribosome-BTE binding (compare lanes 3 and 4). Lanes 5–9 show binding for the non-functional BTEBF, which binds eIF4F but is transnationally inactive (24). D, ribosome-RNA complex stabilities between different BYDV RNAs are compared. Ribosome-BYDV-RNA binding affinity was determined by calculating relative complex stability (1/Kd) from fluorescence anisotropy experiments. Ribosome shows the highest complex stability with helicase-treated 3′-BTE, which decreased when a 3-fold excess of unlabeled 5′-UTR RNAs is present in trans in the reaction. Helicase-treated 5′-UTR shows stronger binding to ribosomes compared with untreated 5′-UTR. This ribosome binding affinity to helicase-treated 5′-UTR increased when unlabeled 3′-BTE RNAs were present in trans in the reaction mixture. To understand whether the presence of unlabeled 5′-UTR affects 40S binding to helicase-treated 3′-BTE, we titrated helicase complex-treated fluorescence-labeled 3′-BTE (50 nm) with 40S in the presence of excess 5′-UTR (150 nm). Similar experiments were done to determine the effect of 3′-BTE in 40S binding to helicase-treated 5′-UTR. Error bars, S.D.
Binding affinity of 40S-3′-BTE or 40S-5′UTR was weaker than expected to account for 3′-BTE-mediated translation. For example, ribosome binding to capped mRNA has a very strong binding affinity (KD ∼2 nm) (58). Because eIFs affect ribosome-binding affinity (58, 59) and eIF4F is required for translation initiation, the effects of eIFs on 40S binding were examined. eIF4F alone did not show a significant effect on 40S binding affinity with 3′-BTE. However, a combination of eIF4A-4B-4F (helicase complex) and ATP enhanced the binding affinity of 40S for 3′-BTE RNA nearly 3-fold (KD = 120 ± 10 nm) (Fig. 2B). In the 17-nt CS, bases 2–7 (GAUCCU) (Fig. 1) are complementary to a tract near the 3′-end of 18S rRNA at the site where the Shine-Dalgarno complementary sequence is located in prokaryotic ribosomes (31). However, the terminal three bases (underlined) are base-paired within the 17-nt CS (Fig. 1A), and some of the complementary bases in 18S rRNA are also embedded in a helix. Thus, perhaps unwinding of RNA by the helicase activity of eIF4A (44) made the CS available to allow base pairing between the GAUCCU and 18S rRNA, enhancing binding of 3′-BTE to the 40S subunit. Further mutagenesis studies will determine the extent to which base sequence and possibly base pairing influences ribosome binding. The specificity of ribosome binding is confirmed in Fig. 2 (B and D), where only helicase complex (eIF4A-4B-4F)-treated 3′-BTE, in the presence of ATP, showed high affinity binding. None of the controls, including a non-hydrolyzable ATP analog (ADPPNP; compare 3′-BTE binding in Fig. 2 (compare A with B and as shown in D), nonfunctional 3′-BTEBF mutant, and the 5′-UTR, showed a similar increase in ribosome binding in the presence of the helicase complex.
Binding selectivity of the 40S with 3′-BTE and 3′-BTEBF in the presence of helicase complex and ATP was verified by EMSAs, in which 32P-labeled 3′-BTE or 3′-BTEBF RNAs, treated with the eIF4A/eIF4B/eIF4F/ATP mixture, were incubated with 40S ribosomes (Fig. 2C). The addition of eIF4F caused a gel shift (lanes 2, 6, 8, and 9), whereas the further addition of 40S ribosomal subunits increased the gel shift (lanes 3 and 4). The EMSA revealed that ribosome binding affinity to the 3′-BTE increased in the presence of the helicase complex (compare lanes 3 and 4), whereas no binding increase was detected for the nonfunctional mutant, 3′-BTEBF (compare lanes 8 and 9), confirming our fluorescence anisotropy measurements.
To further understand the 40S subunit binding, the presence of both 5′- and 3′-elements was examined to test if a complex had higher affinity than either element alone. Fluorescence anisotropy measurements were performed as described in Fig. 2, A and B. Fig. 2D shows the relative stability of the complexes (1/KD). The 40S binding to helicase-treated 3′-BTE was reduced when 150 nm (3-fold excess) 5′-UTR was added in trans prior to titration with ribosomes (Fig. 2D), presumably because the 40S subunit was transferred to the 5′-UTR. This binding to the 5′-UTR did not occur in the absence of 3′-BTE (Fig. 2, B and D). When binding to the 5′-UTR was monitored, there was an increase when a 3-fold excess concentration of 3′-BTE was added prior to titration. Further, we were unable to attain a stable 3′-5′ complex by EMSA even in the presence of eIFs. Taken together, these data suggest a transient interaction.
Structural Probing of 3′-UTR Shows Helicase Treatment Increases RNA Accessibility
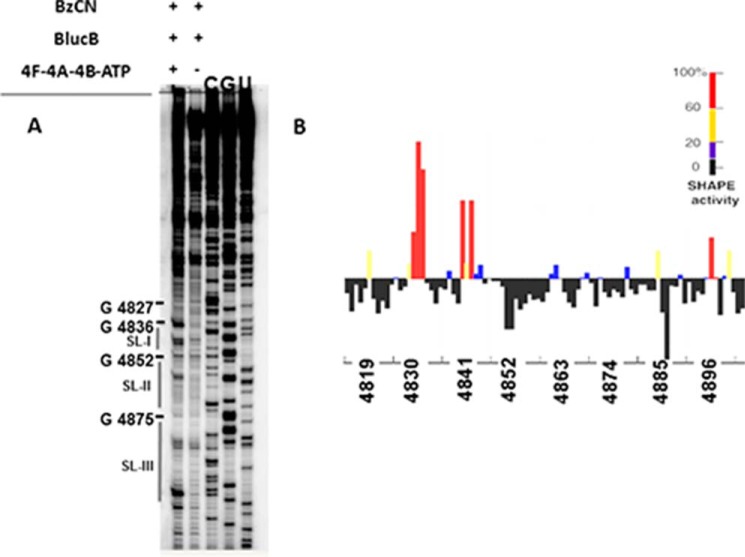
To understand the structural aspect of the requirement of ATP-dependent eIF4A-4B-4F interaction with 3′-BTE for high affinity ribosome binding, 3′-BTE RNA structures were probed using SHAPE in the presence and absence of eIF4A-4B-4F-ATP. The chemical BzCN, which modified flexible and single-stranded nucleotides in a sequence-independent manner (45, 46), was used in SHAPE experiments. Modified residues were mapped by primer extension followed by denaturing gel electrophoresis. SAFA (51) was used to analyze the SHAPE reactivity of each nucleotide, and the results (SHAPE activity higher than 60%) were superimposed onto the known secondary structure of 3′-BTE (Figs. 1 (A and B) and 3).
FIGURE 3.
SHAPE analysis of helicase (4F-4A-4B-ATP) of 3′-BTE. A, denaturing acrylamide gel showing products of primer extension of 3′-BTE after treatment with the benzoyl cyanide. +, helicase; −, no helicase. Sequencing ladders generated with the same primer were run in the gels, and relative coordinates are indicted at the right. B, SHAPE reactivities as a function of nucleotide positions. Normalized and quantified SHAPE reactivities of helicase-treated 3′-BTE are calculated using SAFA and are indicated.
Earlier SHAPE probing in the absence of eIFs and ATP of 3′-BTE (33) (Fig. 1A) revealed the presence of a stem-loop SL-I formed by the 17-nt CS at the distal end of a bulged basal helix, where the first and second guanylate were more exposed than the others (33). The conserved GGAUC of the 17-nt CS, which has 18S rRNA complementarity, showed low SHAPE reactivity except for the first G, which was highly modified by BzCN. Stem-loop SL-III possesses six uninterrupted GC and CG base pairs. It appears that the native fold of 3′-BTE is maintained by the basal helix, which helps to form a more accessible 3′-BTE (33). However, SHAPE studies of helicase complex-treated 3′-BTE (Fig. 1A) showed more modification of nucleotides in the 18S rRNA-complementary region of SL-I as well as nucleotides (nt 4828–4831) in the SL-IV, suggesting that treatment with the helicase complex increased accessibility of the 3′-BTE in regions complementary to the 18S rRNA and possibly explaining the increase in 40S binding affinity.
Cycloheximide Stalls Ribosomes near the Start Codon
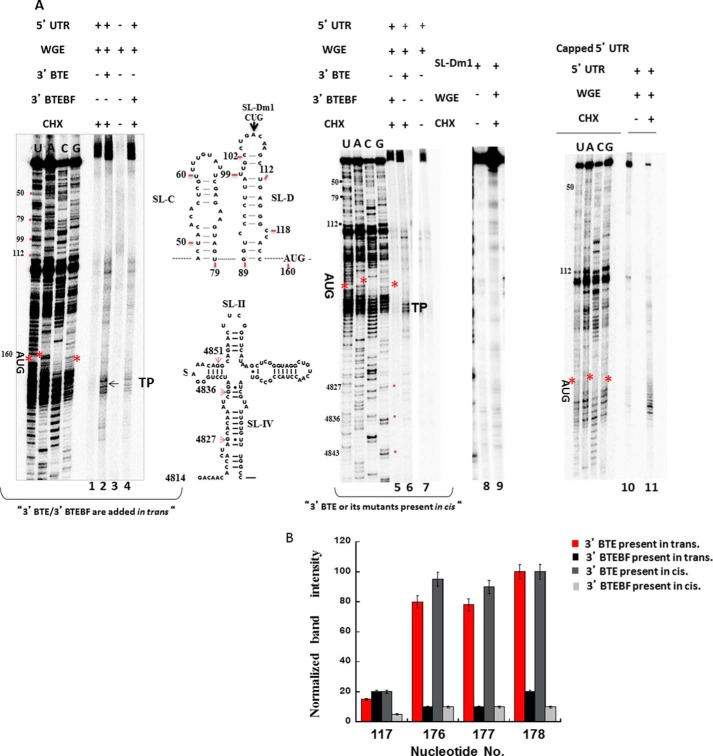
Strong secondary structures or AUG codons in the 5′-UTR of BYDV RNA can greatly reduce translation initiation (15, 22, 23), suggesting that 3′-BTE-mediated translation requires scanning from the 5′-end. To further investigate the role of the 5′-UTR in translation initiation, we performed toeprinting analysis of translation reactions in WGE. Initially, we analyzed a 5′-capped version of the BYDV 5′-UTR linked to a luciferase reporter gene (B-Luc) to authenticate the functionality of the system. When capped B-Luc RNA was incubated in WGE treated with CHX, which stalls 80S initiation complexes ∼16–18 nt downstream of the P-site codon by inhibiting elongation, toeprints were observed at the expected location (Fig. 4, lane 11). We next characterized ribosome recruitment on uncapped B-Luc mRNA (mRNA lacking the 3′-BTE). When incubated in CHX-treated WGE, no significant toeprints were observed (Fig. 4A, lane 1). However, when 3′-BTE was either added in trans (lane 2) or present in the reporter mRNA (lane 6), strong toeprints were observed ∼16–18 nt downstream of the start codon in the uncapped message. A much weaker toeprint was observed at residue 117 prior to the AUG codon, which may be the result of secondary structure. This toeprint was observed even for the nonfunctional BTEBF mutant (Fig. 4, A (lane 4) and B). Observation of several bands in toeprint analysis (∼+16 to +18) has been reported by others (26, 48) and is seen in cell extract (48).
FIGURE 4.
80S complex formation on 5′-UTR BYDV mRNAs depends on the presence of 3′-BTE. A, denaturing PAGE showing the products of primer extension generated by reverse transcription of the uncapped BYDV 5′-UTR upstream of the Luc coding region. 5′-UTR-containing RNAs were incubated in WGE containing CHX for 20 min at 25 °C. The AUG codon where ribosome stalling occurs is indicated by asterisks. Different conditions are indicated at the top of the gels. Toeprints of the uncapped 5′-UTR in the presence of 3′-BTE (lanes 2 and 6) are shown. Mutant 3′-BTEBF (lanes 4 and 5) and mutant SL-Dm1 in which kissing loop base pairing between 5′-SL-D and 3′-BTE is disrupted (lane 9) do not show corresponding toeprints. Nucleotides are numbered according to BYDV genomic sequence. Lane 11 shows toeprints of capped BLucB. Nucleotides 1–159 correspond to the 5′-UTR, whereas the 3′-BTE starts from nt 4814. Primers are described under “Experimental Procedures.” B, graph of quantified, normalized, and background-corrected bands from A. Numbers on the x axis indicate 5′-UTR residues; 117 is the weak toeprint, and 176–178 are the +16–18 residues, which show much stronger toeprints. 5′-UTR + 3′-BTE in trans (red), as expected, is similar to 5′UTR + 3′-BTE in cis (gray) and negative control 5′-UTR + 3′-BTEBF in trans (black) and 5′UTR + 3′-BTEBF in cis (light gray) are similar, neither of which show significant toeprints. Error bars, S.D.
As a negative control, we investigated the effect of the nonfunctional BTEBF on ribosome recruitment to the 5′-end of the reporter mRNA in WGE. No corresponding toeprints were observed when mutant 3′-BTEBF was present in cis on BLucBF mRNA (Fig. 4A, lane 5) or when BTEBF was added to the WGE system in trans (Fig. 4A, lane 4), although a week toeprint was observed at residue 117, as noted above. Normalized band intensities are shown in Fig. 4B. The weak toeprint mentioned above is at residue 117, whereas residues 176–178 are the +16–18 nucleotides past the AUG. Furthermore, primer extension inhibition of mutant BLucB-SL-Dm1 containing a mutation in 5′-SL-D that prevents the kissing loop interaction between the 5′-UTR and 3′-BTE gave no prominent toeprint at the ∼+16–18 or any other 5′-UTR nucleotide position (Fig. 4A, lane 9), indicating the requirement for RNA-RNA interaction in addition to the 3′-BTE, consistent with earlier reports of translational inhibition when the 5′-3′ interaction was disrupted (22, 23). Our toeprinting assay data confirm that the 3′-BTE is necessary and essential to allow for 5′-ribosomal entrance and delivery to the start codon.
Next, we wanted to differentiate between the two possible mechanisms for the ribosome recruitment to the 5′-UTR. One mechanism is the ribosome binding to the 3′-BTE directly, followed by delivery to the 5′-end via long distance base pairing, or, alternatively, it could bind directly at the 5′-UTR only in the presence of eIF4F (or eIF4G) bound to the BTE. We observed toeprints in the SL-I region of the 3′-BTE when the in vitro translation reaction was quenched in a time frame of 0–15 min in the BLucB mRNA construct, indicating loading of translation machinery to the 3′-end (Figs. 5 (A and C, lanes 3–5) and Fig. 1B show the 40S binding site). Additionally, no toeprints were observed in the SL-I region when the assay was done in WGE with mutant BLucBF (Fig. 5C, lane 6). Normalized band intensities are shown in Fig. 5D. A weak toeprint was observed at residue 4844 in SL-I that is not seen when purified 40S subunits and eIFs are used (Fig. 6, lanes 2 and 3). Taken together, these data indicate that the ribosome binds to the SL-I region of the 3′-BTE both in the presence and absence of the 5′-UTR. However, binding to the 5′-UTR only occurs when the 3′-BTE is present, and the 5′-UTR is not required for 3′-BTE binding, suggesting that the ribosomes bind to the 3′-BTE and are subsequently delivered to the 5′-end of the message, requiring the RNA-RNA kissing loop interaction for successful transfer.
FIGURE 5.
Map of BLucB and 40S interaction with 3′- and 5′-UTR. A, map of BLucB mRNA is shown with indications of mutations in the SL-I and SL-D loop, the 80S-loading site (+16) (red box), and the 40S binding site in SL-I region (green box). B, secondary structure of BYDV 5′-UTR. Bases are color-coded according to the level of modification in the SHAPE reaction; red indicates the highest modification. Nucleotides are numbered according to their positions in the viral genome. C, BYDV mRNA (BlucB) was incubated in translationally active WGE, and the reaction was quenched in a 0–15 min time scale (lanes 3–5). BLucB and BLucBF controls are shown in lanes 1 and 6, respectively. Lane 2 shows a weak 40S toeprint in the 3′-SL-I region of BLucB mRNA when 6 pmol of mRNA was incubated with 16 pmol of 40 S. Lanes 3–5 show a distinct toeprint at 4852. Weaker stops are seen at 4818 and 4844 (D). BLucBF does not show a toeprint. D, graph of quantified, normalized, and background-corrected toeprints of BLucB and BLucBF. Normalized toeprint band intensity (BLucB) of the zero time WGE reaction (gray) is stronger than the 10 min (black) and 15 min (red) reaction band intensities. Error bars, S.E.
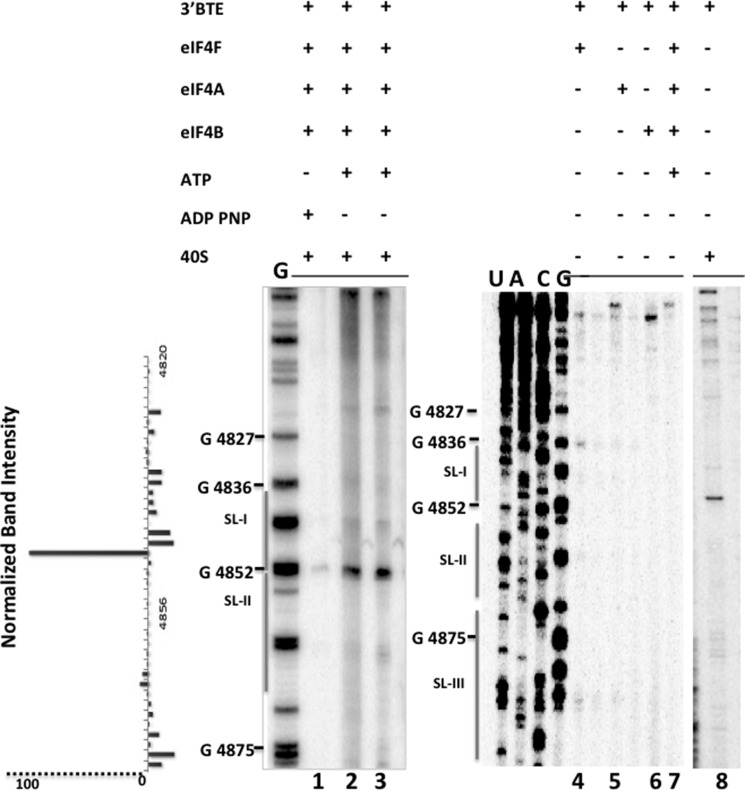
FIGURE 6.
eIFs 4A/4B/4F and ATP recruit 40S ribosomes to the 3′-BTE SL-I region. In the presence of the indicated combinations of factors and ATP, strong 40S toeprints are observed in the SL-I loop of 3′-BTE at position 4852 (lanes 2 and 3) (Fig. 1B). The presence of factors, 40S, and non-hydrolyzable ATP (lane 1) is indicated by a plus sign above the lanes. Reactions were done by assembling 6 pmol of mRNA, 16 pmol of 40S, and 6 pmol of various proteins in a final volume of 20 μl, and 4 μl of each reaction was loaded for gel electrophoresis. A weak toeprint for 40S is shown in Lane 8. Lanes 4–7 show that none of the eIFs alone or in combination produced a toeprint. Similarly, no toeprint was observed in the absence of ATP (lane 1). Normalized band intensities are shown on the left.
eIF4A, 4B, 4F, and ATP Are Necessary for Efficient Ribosome Binding to the 3′-end of the Message
To determine the initiation factor requirements for ribosome binding, toeprinting studies were performed using the purified components of the wheat germ translation system. Toeprints were observed in the SL-I region when ribosomes were incubated with BLucB in the presence of eIF4F, eIF4A, eIF4B, and ATP (Fig. 6, lanes 2 and 3), and a weak toeprint was observed when only 40S subunits were present in the reaction (Figs. 5C (lane 2) and 6 (lane 8)). Very weak or no toeprinting was observed when ribosomes were incubated in the presence of eIF4F-4A-4B and a non-hydrolyzable form of ATP (Fig. 6, lane 1). Efficient stops in the presence of eIF4F/4B/4A were not identified at the SL-I region under similar reaction conditions when individual factors in the absence of ribosomes were used for toeprinting experiments (Fig. 6, lanes 4–7).
Overall, we found good agreement between the binding of ribosomes observed during primer extension inhibition assays and fluorescence binding assays. Efficient binding of 40S subunits to the 3′-BTE was achieved only in the presence of helicase complex and ATP, as shown with both assays.
DISCUSSION
In many well characterized mammalian viruses, 5′-UTRs have IRESs (17), which recruit the ribosome either by directly interacting with 40S subunits (18, 60) or with the help of various initiation factors (2, 6). Here we provide direct evidence of a completely different ribosome recruitment pathway to an mRNA. In this case, initiation factors and the mRNA structure (3′-BTE) facilitate recruitment of the 40S subunit to the 3′-UTR, from which it is delivered to the 5′-end by kissing loop base pairing. It was shown previously that the 3′-BTE facilitates translation by directly interacting with eIF4F and base-pairing to the 5′-end of the message (24). Here we show that the 40S subunit is also recruited to the 3′-BTE.
Using fluorescence anisotropy and gel mobility shift-based binding studies, we determined that 3′-BTE, which contains a sequence complementary to 18S rRNA (GAUCCU) shows moderate binding affinity with purified 40S ribosomes and that mutation of this sequence to GAUCGAUCU (mutant BTEBF) weakens this binding affinity. Earlier studies showed that this mutation dramatically reduced translation in vitro and in cells (23, 31, 61). The 5′-UTR also showed a low affinity binding to 40S ribosomal subunits in fluorescence anisotropy and gel mobility shift assays. These binding affinities appear to be nonspecific based on both the low affinity and inability to toeprint the 40S subunit on the RNAs. Binding enhancement of 40S ribosomes and 3′-BTE was observed in the presence of eIF4F-4A-4B-ATP. This led us to propose that the helicase activity of this complex disrupts the secondary structure of RNA, exposing the BTE sequence complementary to that of 18S rRNA and also that of rRNA because in both RNAs some of the complementary bases are in stem-loops, making the complementary sequences accessible for possible base pairing between the BTE and 18S rRNA. Further mutagenesis experiments will be necessary to test the extent of base pairing.
For mammalian eIF4G, the “ratcheting” motion of swiveling eIF4A between two anti-cooperative binding sites not only propels the helicase motion but also recruits eIF3 for association with eIF4G. Although this may occur in WGE, it is not the case for 40S subunit binding with the purified factors because no eIF3 was added.
We found that 80S ribosome stalling occurs close to the AUG start codon in cycloheximide-treated wheat germ translation system only when the 3′-BTE is present. These studies suggest two possible mechanisms of ribosome loading to the 5′-end of the message. One possible mechanistic pathway is that the ribosome interacts with 3′-BTE first and subsequently is transferred to the 5′-end of the message. The second possible mechanism is that the 3′-BTE transfers eIF4F to the 5′-end to which the 40S subunit is recruited, as predicted for Tombusviridae genera (26). In both models, eIF4F is required for 40S recruitment, and long distance base pairing between the 3′-BTE and 5′-UTR is required for delivery of host components, either initiation factors or initiation factors and the 40S subunit. The data presented here support the first mechanism, because toeprinting assays using purified factors and 40S ribosomal subunits showed toeprints (made by 40S subunit binding) in the 3′-BTE SL-I (18S rRNA-complementary) region. Furthermore, we were unable to show significant direct ribosome binding to the 5′-end of the message, upstream of the start codon, with either toeprints or fluorescence binding assays. Strong toeprints close to the start codon for CHX stalled ribosomes were observed only in the presence of 3′-BTE and when the kissing loop between 3′-UTR and 5′-UTR was not disrupted.
We further considered a model where ribosomes formed an RNP particle with both 5′-UTR and 3′-BTE. Although such an interaction must occur for ribosomal transfer or interaction with the AUG, we were unable to obtain a stable complex as judged by EMSA even in the presence of factors, and when 40S subunits bound to fluorescently labeled 3′-BTE were treated with 5′-UTR, the anisotropy decreased, presumably because the 40S subunits were transferred to the unlabeled 5′-UTR. Taken together, we conclude that the 5′-3′ interaction is a transient interaction and the 40S subunits bind initially to the 3′-BTE.
Our data lead us to propose a general model of BTE-mediated cap-independent translation for the ribosome recruitment pathway and delivery of the translation machinery to the 5′-end of the BYDV message. Our model (Fig. 7) suggests the following: (i) eIF4F and possibly eIFiso4Fs are recruited directly to the folded 3′-BTE; (ii) helicase complex eIF4F-4A-4B-ATP improves 40S binding affinity with 3′-SL-I by exposing more accessible sites of the 3′-BTE; (iii) 40S ribosomes bind to the 3′-BTE; and then (iv) via long distance RNA-RNA interaction between 5′-SL-D and 3′-SL-III, the translation machinery transfers to the 5′-end of the message to start scanning.
FIGURE 7.
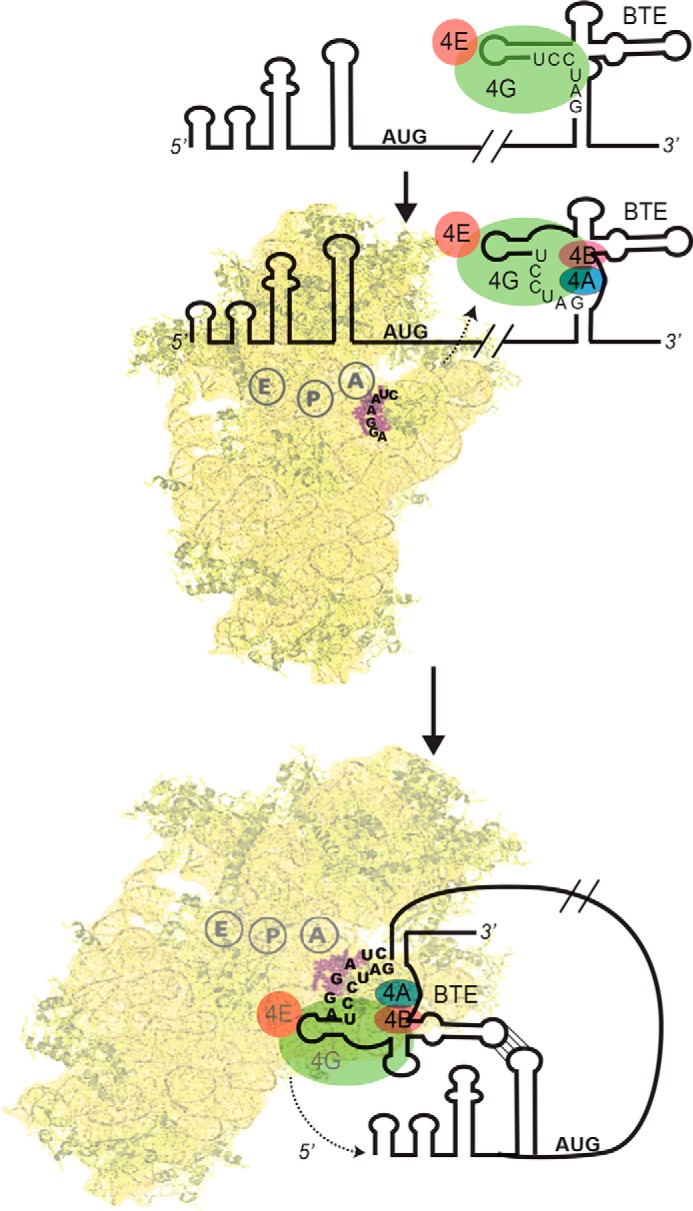
Model for recruitment of the 40 S subunit to the 5′-UTR via 3′-BTE. A, eIF4G binds the SL-I region of the 3′-BTE. eIF4E remains bound to eIF4G but is not required for BTE binding. B, the six-base sequence (GAUCCU) complementary to 18S rRNA is mostly base-paired. In the presence of ATP, eIF4A, eIF4B, and eIF4F bind the complex and facilitate unwinding of the helices that contain GAUCCU sequence. C, the 43 S preinitiation complex then binds the BTE, possibly via base pairing of the AGGAUC sequence in the 18S rRNA to GAUCCU in the BTE. Subsequently (or simultaneously), RNA-RNA long distance base pairing between the 3′-BTE and the 5′-UTR delivers the translation machinery to the 5′-terminus of the message, from which it scans to the start codon.
Other plant viruses utilize various other RNA structures to bind eIF4F and ultimately deliver the ribosome to the 5′-end of the genome (26). A tRNA-shaped 3′-CITE of TCV (TSS) was shown to bind directly to the 60S subunit and 80S ribosome, but not the 40S subunit, which leaves the question of how and where the 40S subunit is recruited (29). A TSS-like domain was also found in the 3′-UTR of pea enation mosaic virus RNA2 (62), adjacent to another 3′-CITE that binds eIF4E (63). This TSS was reported to bind the 40S as well as the 60S ribosomal subunits and the 80S ribosome (62). Binding affinity of the PEMV RNA2 TSS to the 40S subunit (Kd = 360 nm) was similar to BTE binding to the 40S subunit that we observed in the absence of initiation factors (Kd = 400 nm). However, the roles of translation factors in the function of either of the TSS elements have not been determined.
Our model (Fig. 7) bears an interesting resemblance to interaction of hepatitis C virus RNA with the ribosome to facilitate cap-independent translation but with different binding sites and factor requirements. Of particular relevance is that the 3′-UTR of HCV also binds to the 40S subunit. This is not essential for but greatly enhances activity of the IRES at the 5′-end (64). Thus, as in BTE-containing mRNAs, the 5′- and 3′-UTRs can interact simultaneously with the 40S subunit, similar to the cyclization proposed for capped and poly(A) tail mRNA, where the cap and poly(A) tail act synergistically to promote efficient translation. For the HCV IRES, it is proposed that this interaction is for ribosome recycling. In the case of the BTE, the 3′ interaction is much more important, because the 3′-BTE is the only 40S subunit-recruiting domain in BTE-dependent translation, in contrast to HCV, in which the IRES in the 5′-UTR is the primary binding site of the 40S subunit.
The interaction of the 3′-UTR with the 5′-UTR to control translation of BTE-containing RNAs may indicate that viruses use this interaction in a mechanism to switch viral RNA from translation to replication. A newly translated replicase would be expected to bind the extreme 3′-terminus (not needed for BTE- or IRES-driven translation), and proceed in the 5′ direction on its template as the template is still undergoing translation. Upon reaching the element in the 3′-UTR required for any BTE translation, the replicase would disrupt this structure and shut off BTE translation initiation at the 5′-end. This, in turn, would free the coding region of the viral RNA of ribosomes, making it available for full-length negative strand synthesis by the replicase. As described previously (25, 65), this potentiality provides an elegant feedback loop to assure a productive balance between replication and translation. Our data provide evidence supporting this model. Additionally our model could be applied to other viruses containing BTE-like elements or other 3′-CITEs.
Acknowledgments
We thank Dr. K. Treder, Dr. R. Bachu, and Dr. E. Gustillo for insightful comments.
This work was supported, in whole or in part, by National Institutes of Health Grant 2R01 GM067104 (to W. A. M.). This work was also supported by National Science Foundation Grant MCB1157632 (to D. J. G.) and United States Department of Agriculture National Institute of Food and Agriculture Grant 2011-67012-30715 (to J. J. K.).
- IRES
- internal ribosome entry site
- eIF
- eukaryotic initiation factor
- CITE
- cap-independent translation element
- BYDV
- barley yellow dwarf virus
- nt
- nucleotide(s)
- CS
- conserved sequence
- SL-I
- -III, and -D, stem-loop I, III, and D, respectively
- SHAPE
- selective 2′-hydroxyl acylation analyzed by primer extension
- BzCN
- benzoyl cyanide
- CHX
- cycloheximide
- ADPPNP
- 5′-adenylyl-β,γ-imidodiphosphate
- SAFA
- semiautomated footprinting analysis
- BTE
- barley 3′-translational enhancer element.
REFERENCES
- 1. Flint S. J., Enquist L. W., Racaniello V. R., Skalka A. M. (eds) (2009) Principles of Virology, 3rd Ed., pp. 156–170, ASM Press, Washington, D. C. [Google Scholar]
- 2. Reineke L. C., Lloyd R. E. (2011) Animal virus schemes for translation dominance. Curr. Opin. Virol. 1, 363–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Firth A. E., Brierley I. (2012) Non-canonical translation in RNA viruses. J. Gen. Virol. 93, 1385–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sweeney T. R., Abaeva I. S., Pestova T. V., Hellen C. U. (2014) The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO. J. 33, 76–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fernández N., García-Sacristán A., Ramajo J., Briones C., Martínez-Salas E. (2011) Structural analysis provides insights into the modular organization of picornavirus IRES. Virology 409, 251–261 [DOI] [PubMed] [Google Scholar]
- 6. Fraser C. S., Doudna J. A. (2007) Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 5, 29–38 [DOI] [PubMed] [Google Scholar]
- 7. Hashem Y., des Georges A., Dhote V., Langlois R., Liao H. Y., Grassucci R. A., Pestova T. V., Hellen C. U., Frank J. (2013) Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 503, 539–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costantino D. A., Pfingsten J. S., Rambo R. P., Kieft J. S. (2008) tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat. Struct. Mol. Biol. 15, 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simon A. E., Miller W. A. (2013) 3′ cap-independent translation enhancers of plant viruses. Annu. Rev. Microbiol. 67, 21–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pelletier J., Sonenberg N. (1988) Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325 [DOI] [PubMed] [Google Scholar]
- 11. Hellen C. U., Sarnow P. (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15, 1593–1612 [DOI] [PubMed] [Google Scholar]
- 12. Jackson R. J., Hellen C. U., Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Otto G. A., Puglisi J. D. (2004) The pathway of HCV IRES-mediated translation initiation. Cell 119, 369–380 [DOI] [PubMed] [Google Scholar]
- 14. Kieft J. S., Zhou K., Jubin R., Murray M. G., Lau J. Y., Doudna J. A. (1999) The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 292, 513–529 [DOI] [PubMed] [Google Scholar]
- 15. Pestova T. V., Hellen C. U., Shatsky I. N. (1996) Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell Biol. 16, 6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pestova T. V., Shatsky I. N., Fletcher S. P., Jackson R. J., Hellen C. U. (1998) A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12, 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pfingsten J. S., Costantino D. A., Kieft J. S. (2006) Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science 314, 1450–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hertz M. I., Thompson S. R. (2011) Mechanism of translation initiation by Dicistroviridae IGR IRESs. Virology 411, 355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khan M. A., Miyoshi H., Gallie D. R., Goss D. J. (2008) Potyvirus genome-linked protein, VPg, directly affects wheat germ in vitro translation: interactions with translation initiation factors eIF4F and eIFiso4F. J. Biol. Chem. 283, 1340–1349 [DOI] [PubMed] [Google Scholar]
- 20. Gallie D. R. (2001) Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J. Virol. 75, 12141–12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khan M. A., Goss D. J. (2012) Poly(A)-binding protein increases the binding affinity and kinetic rates of interaction of viral protein linked to genome with translation initiation factors eIFiso4F and eIFiso4F.4B complex. Biochemistry 51, 1388–1395 [DOI] [PubMed] [Google Scholar]
- 22. Guo L., Allen E. M., Miller W. A. (2001) Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell 7, 1103–1109 [DOI] [PubMed] [Google Scholar]
- 23. Rakotondrafara A. M., Polacek C., Harris E., Miller W. A. (2006) Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′-cap-independent translation element. RNA 12, 1893–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Treder K., Kneller E. L., Allen E. M., Wang Z., Browning K. S., Miller W. A. (2008) The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA 14, 134–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller W. A., White K. A. (2006) Long-distance RNA-RNA interactions in plant virus gene expression and replication. Annu. Rev. Phytopathol. 44, 447–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nicholson B. L., Wu B., Chevtchenko I., White K. A. (2010) Tombusvirus recruitment of host translational machinery via the 3′ UTR. RNA 16, 1402–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gazo B. M., Murphy P., Gatchel J. R., Browning K. S. (2004) A novel interaction of Cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J. Biol. Chem. 279, 13584–13592 [DOI] [PubMed] [Google Scholar]
- 28. Truniger V., Nieto C., González-Ibeas D., Aranda M. (2008) Mechanism of plant eIF4E-mediated resistance against a Carmovirus (Tombusviridae): cap-independent translation of a viral RNA controlled in cis by an (a)virulence determinant. Plant J. 56, 716–727 [DOI] [PubMed] [Google Scholar]
- 29. Stupina V. A., Meskauskas A., McCormack J. C., Yingling Y. G., Shapiro B. A., Dinman J. D., Simon A. E. (2008) The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA 14, 2379–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miras M., Sempere R. N., Kraft J. J., Miller W. A., Aranda M. A., Truniger V. (2014) Interfamilial recombination between viruses led to acquisition of a novel translation-enhancing RNA element that allows resistance breaking. New Phytol. 202, 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang S., Browning K. S., Miller W. A. (1997) A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 16, 4107–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Banerjee B., Goss D. J. (2014) Eukaryotic initiation factor (eIF) 4F binding to barley yellow dwarf virus (BYDV) 3′-untranslated region correlates with translation efficiency. J. Biol. Chem. 289, 4286–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kraft J. J., Treder K., Peterson M. S., Miller W. A. (2013) Cation-dependent folding of 3′ cap-independent translation elements facilitates interaction of a 17-nucleotide conserved sequence with eIF4G. Nucleic Acids Res. 41, 3398–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kneller E. L., Rakotondrafara A. M., Miller W. A. (2006) Cap-independent translation of plant viral RNAs. Virus Res. 119, 63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Z., Kraft J. J., Hui A. Y., Miller W. A. (2010) Structural plasticity of Barley yellow dwarf virus-like cap-independent translation elements in four genera of plant viral RNAs. Virology 402, 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goss D. J., Rounds D. J. (1988) A kinetic light-scattering study of the binding of wheat germ protein synthesis initiation factor 3 to 40S ribosomal subunits and 80S ribosomes. Biochemistry 27, 3610–3613 [DOI] [PubMed] [Google Scholar]
- 38. Spremulli L. L., Walthall B. J., Lax S. R., Ravel J. M. (1977) Purification and properties of a Met-tRNAf binding factor from wheat germ. Arch. Biochem. Biophys. 178, 565–575 [DOI] [PubMed] [Google Scholar]
- 39. McCormack J. C., Yuan X., Yingling Y. G., Kasprzak W., Zamora R. E., Shapiro B. A., Simon A. E. (2008) Structural domains within the 3′ untranslated region of Turnip crinkle virus. J. Virol. 82, 8706–8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Acker M. G., Kolitz S. E., Mitchell S. F., Nanda J. S., Lorsch J. R. (2007) Reconstitution of yeast translation initiation. Methods Enzymol. 430, 111–145 [DOI] [PubMed] [Google Scholar]
- 41. Stupina V. A., Yuan X., Meskauskas A., Dinman J. D., Simon A. E. (2011) Ribosome binding to a 5′ translational enhancer is altered in the presence of the 3′ untranslated region in cap-independent translation of turnip crinkle virus. J. Virol. 85, 4638–4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Heerden A., Browning K. S. (1994) Expression in Escherichia coli of the two subunits of the isozyme form of wheat germ protein synthesis initiation factor 4F: purification of the subunits and formation of an enzymatically active complex. J. Biol. Chem. 269, 17454–17457 [PubMed] [Google Scholar]
- 43. Mayberry L. K., Dennis M. D., Leah Allen M., Ruud Nitka K., Murphy P. A., Campbell L., Browning K. S. (2007) Expression and purification of recombinant wheat translation initiation factors eIF1, eIF1A, eIF4A, eIF4B, eIF4F, eIF(iso)4F, and eIF5. Methods Enzymol. 430, 397–408 [DOI] [PubMed] [Google Scholar]
- 44. Rozen F., Edery I., Meerovitch K., Dever T. E., Merrick W. C., Sonenberg N. (1990) Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell Biol. 10, 1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilkinson K. A., Merino E. J., Weeks K. M. (2006) Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1, 1610–1616 [DOI] [PubMed] [Google Scholar]
- 46. Merino E. J., Wilkinson K. A., Coughlan J. L., Weeks K. M. (2005) RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127, 4223–4231 [DOI] [PubMed] [Google Scholar]
- 47. Jaramillo M., Dever T. E., Merrick W. C., Sonenberg N. (1991) RNA unwinding in translation: assembly of helicase complex intermediates comprising eukaryotic initiation factors eIF-4F and eIF-4B. Mol. Cell Biol. 11, 5992–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sachs M. S., Wang Z., Gaba A., Fang P., Belk J., Ganesan R., Amrani N., Jacobson A. (2002) Toeprint analysis of the positioning of translation apparatus components at initiation and termination codons of fungal mRNAs. Methods 26, 105–114 [DOI] [PubMed] [Google Scholar]
- 49. Yu Y., Marintchev A., Kolupaeva V. G., Unbehaun A., Veryasova T., Lai S. C., Hong P., Wagner G., Hellen C. U., Pestova T. V. (2009) Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 37, 5167–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Das R., Laederach A., Pearlman S. M., Herschlag D., Altman R. B. (2005) SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11, 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Laederach A., Das R., Vicens Q., Pearlman S. M., Brenowitz M., Herschlag D., Altman R. B. (2008) Semiautomated and rapid quantification of nucleic acid footprinting and structure mapping experiments. Nat. Protoc. 3, 1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Filbin M. E., Vollmar B. S., Shi D., Gonen T., Kieft J. S. (2013) HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat Struct. Mol. Biol. 20, 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Luo Y., Goss D. J. (2001) Homeostasis in mRNA initiation: wheat germ poly(A)-binding protein lowers the activation energy barrier to initiation complex formation. J. Biol. Chem. 276, 43083–43086 [DOI] [PubMed] [Google Scholar]
- 54. Khan M. A., Goss D. J. (2005) Translation initiation factor (eIF) 4B affects the rates of binding of the mRNA m7G cap analogue to wheat germ eIFiso4F and eIFiso4F.PABP. Biochemistry 44, 4510–4516 [DOI] [PubMed] [Google Scholar]
- 55. Firpo M. A., Connelly M. B., Goss D. J., Dahlberg A. E. (1996) Mutations at two invariant nucleotides in the 3′-minor domain of Escherichia coli 16 S rRNA affecting translational initiation and initiation factor 3 function. J. Biol. Chem. 271, 4693–4698 [DOI] [PubMed] [Google Scholar]
- 56. Ray S., Yumak H., Domashevskiy A., Khan M. A., Gallie D. R., Goss D. J. (2006) Tobacco etch virus mRNA preferentially binds wheat germ eukaryotic initiation factor (eIF) 4G rather than eIFiso4G. J. Biol. Chem. 281, 35826–35834 [DOI] [PubMed] [Google Scholar]
- 57. Yumak H., Khan M. A., Goss D. J. (2010) Poly(A) tail affects equilibrium and thermodynamic behavior of tobacco etch virus mRNA with translation initiation factors eIF4F, eIF4B and PABP. Biochim. Biophys. Acta 1799, 653–658 [DOI] [PubMed] [Google Scholar]
- 58. Maag D., Fekete C. A., Gryczynski Z., Lorsch J. R. (2005) A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell 17, 265–275 [DOI] [PubMed] [Google Scholar]
- 59. Pestova T. V., Kolupaeva V. G., Lomakin I. B., Pilipenko E. V., Shatsky I. N., Agol V. I., Hellen C. U. (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 98, 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Spahn C. M., Kieft J. S., Grassucci R. A., Penczek P. A., Zhou K., Doudna J. A., Frank J. (2001) Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291, 1959–1962 [DOI] [PubMed] [Google Scholar]
- 61. Guo L., Allen E., Miller W. A. (2000) Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA 6, 1808–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gao F., Kasprzak W., Stupina V. A., Shapiro B. A., Simon A. E. (2012) A ribosome-binding, 3′ translational enhancer has a T-shaped structure and engages in a long-distance RNA-RNA interaction. J. Virol. 86, 9828–9842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang Z., Parisien M., Scheets K., Miller W. A. (2011) The cap-binding translation initiation factor, eIF4E, binds a pseudoknot in a viral cap-independent translation element. Structure 19, 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bai Y., Zhou K., Doudna J. A. (2013) Hepatitis C virus 3′UTR regulates viral translation through direct interactions with the host translation machinery. Nucleic Acids Res. 41, 7861–7874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Barry J. K., Miller W. A. (2002) A −1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl. Acad. Sci. U.S.A. 99, 11133–11138 [DOI] [PMC free article] [PubMed] [Google Scholar]