Background: 3,4-Dihydroxy-2-butanone-4-phosphate synthase (DHBPS) is essential for many pathogens and is absent in humans.
Results: We have characterized DHBPS from Vibrio cholerae in the presence of substrate d-ribulose 5-phosphate (Ru5P) and inhibitor 4-phospho-d-erythronohydroxamic acid (4PEH).
Conclusion: 4PEH inhibits DHBPS competitively and interacts with enzyme similarly to the substrate.
Significance: 4PEH can be used as a lead molecule for designing novel antibiotics.
Keywords: Crystal Structure, Enzyme Inhibitor, Enzyme Kinetics, Enzyme Mechanism, Riboflavin, DHBPS, ribB, Ribulose 5-Phosphate
Abstract
The riboflavin biosynthesis pathway has been shown to be essential in many pathogens and is absent in humans. Therefore, enzymes involved in riboflavin synthesis are considered as potential antibacterial drug targets. The enzyme 3,4-dihydroxy-2-butanone-4-phosphate synthase (DHBPS) catalyzes one of the two committed steps in the riboflavin pathway and converts d-ribulose 5-phosphate (Ru5P) to l-3,4-dihydroxy-2-butanone 4-phosphate and formate. Moreover, DHBPS is shown to be indispensable for Mycobacterium, Salmonella, and Helicobacter species. Despite the essentiality of this enzyme in bacteria, no inhibitor has been identified hitherto. Here, we describe kinetic and crystal structure characterization of DHBPS from Vibrio cholerae (vDHBPS) with a competitive inhibitor 4-phospho-d-erythronohydroxamic acid (4PEH) at 1.86-Å resolution. In addition, we also report the structural characterization of vDHBPS in its apo form and in complex with its substrate and substrate plus metal ions at 1.96-, 1.59-, and 2.04-Å resolution, respectively. Comparison of these crystal structures suggests that 4PEH inhibits the catalytic activity of DHBPS as it is unable to form a proposed intermediate that is crucial for DHBPS activity. Furthermore, vDHBPS structures complexed with substrate and metal ions reveal that, unlike Candida albicans, binding of substrate to vDHBPS induces a conformational change from an open to closed conformation. Interestingly, the position of second metal ion, which is different from that of Methanococcus jannaschii, strongly supports an active role in the catalytic mechanism. Thus, the kinetic and structural characterization of vDHBPS reveals the molecular mechanism of inhibition shown by 4PEH and that it can be explored further for designing novel antibiotics.
Introduction
Riboflavin (vitamin B2) is a universal precursor for the synthesis of flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD). FMN and FAD are essential cofactors for many proteins that are involved in cellular and physiological processes including carbohydrate and amino acid metabolism (1). Humans and animals obtain riboflavin from their diet, whereas it is biosynthesized in most bacteria as they are devoid of any riboflavin transporters (2–4). Furthermore, it has been shown that the riboflavin-deficient mutants of Escherichia coli and Salmonella sp. require high amounts of exogenous riboflavin for their growth (5). As a consequence, it is speculated that the enzymes involved in bacterial riboflavin biosynthesis are potential antibacterial drug targets because this pathway is essential for bacteria and absent in humans (2–4, 6–9).
In bacteria, biosynthesis of riboflavin is carried out by a total of six enzymes from two branches of the pathway starting with guanosine 5′-triphosphate (GTP) and d-ribulose 5-phosphate (Ru5P)5 as substrates. From one branch, GTP is converted into 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione by three enzymes namely GTP cyclohydrolase II, bifunctional pyrimidine deaminase/reductase, and a yet unknown pyrimidine phosphatase (10–16). In another branch, 3,4-dihydroxy-2-butanone-4-phosphate synthase catalyzes the conversion of Ru5P to 3,4-dihydroxy-2-butanone 4-phosphate (DHBP) and formate (17, 18). The condensation (19, 20) of 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione and DHBP by 6,7-dimethyl-8-ribityllumazine synthase catalyzes the formation of 6,7-dimethyl-8-ribityllumazine. Riboflavin synthase catalyzes the formation of riboflavin and 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione using two molecules of 6,7-dimethyl-8-ribityllumazine by a dismutation reaction (21–23). In the final step, bifunctional riboflavin kinase/FAD synthase converts riboflavin to FMN and FAD, respectively (1, 24–26).
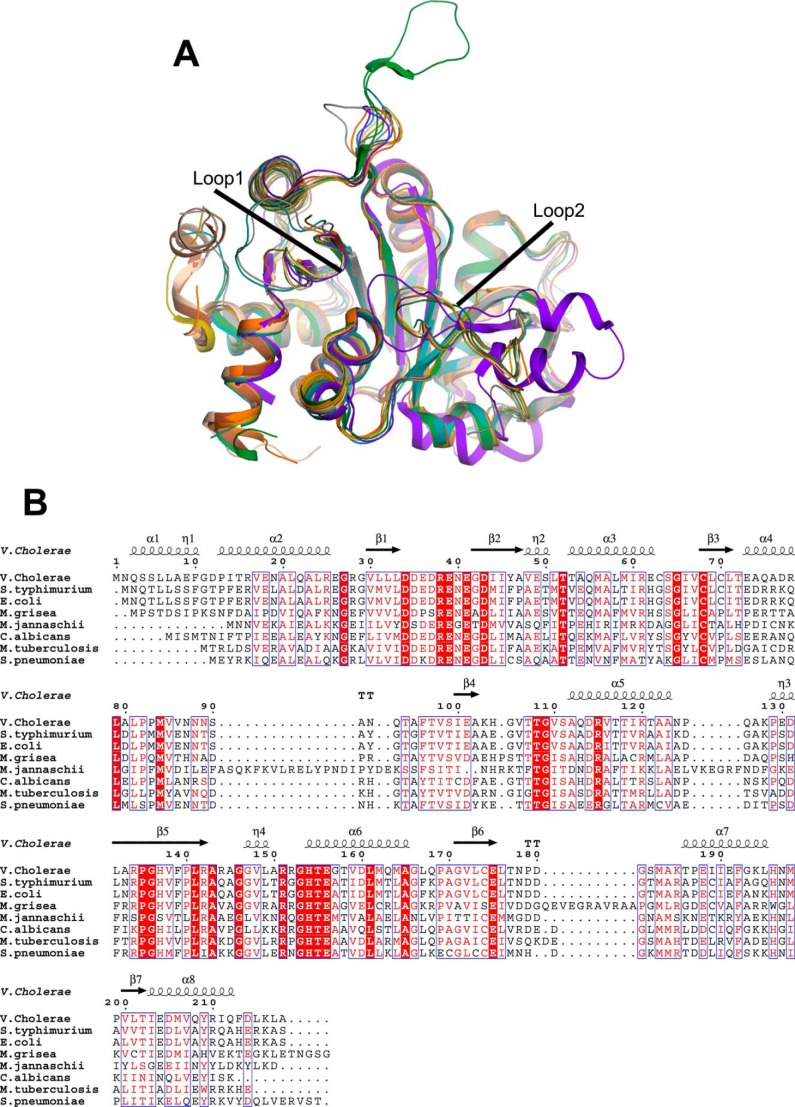
The gene ribB encodes for the enzyme l-3,4-dihydroxy-2-butanone-4-phosphate synthase (DHBPS; EC 4.1.99.12) in bacteria. The proposed mechanism for the conversion of Ru5P to DHBP by DHBPS involves several steps including enolization, protonation, dehydration, skeletal rearrangement, and release of formate in the presence of metal ions (17, 18) (see Fig. 1A). In most of Gram-negative bacteria, DHBPS exists as monofunctional form, whereas in Gram-positive bacteria including Mycobacterium tuberculosis DHBPS co-exists with GTP cyclohydrolase II and thus is found in a bifunctional form (9, 16, 27–29). Crystal structures of DHBPS from E. coli (30, 31), Magnaporthe grisea (32), Methanococcus jannaschii (33), Candida albicans (34), Salmonella typhimurium (35), M. tuberculosis (28, 36), and Streptococcus pneumoniae (27) have been reported. All these structures reveal that DHBPS is a homodimer where each monomer forms an α + β fold and that its active site is located at two topologically equivalent positions at the dimeric interface of each monomer (27, 28, 30–36). The DHBPS structure complexed with substrate and metal ions indicates that an acidic active site loop (Loop1) and another loop (Loop2) undergo a conformational change upon substrate and/or metal binding (32, 33, 35). Furthermore, these structures also reveal the amino acids involved in the proposed reaction mechanism of DHBPS (27, 28, 32–36). Nevertheless, certain differences observed in the existing structures prevent us from predicting the molecular mechanism of DHBPS completely. For example, DHBPS from C. albicans (34) does not show any conformational change of Loop1 upon substrate binding, whereas other structures show a conformational change of Loop1 upon substrate/metal binding (32, 33, 35). Furthermore, the two metal ions that are required for the catalytic activity of DHBPS are positioned differently in M. jannaschii compared with M. grisea and S. typhimurium (33–35). Importantly, despite DHBPS being shown to be essential for many pathogens including M. tuberculosis (7), Helicobacter pylori (37), and Salmonella (38) and its requirement in disease models like typhoid fever and enteritis (39), no inhibitors have been identified until now for this enzyme. Here, we demonstrate for the first time that 4-phospho-d-erythronohydroxamic acid (4PEH), an analog of substrate Ru5P (Fig. 1B), inhibits DHBPS competitively. Furthermore, to understand the molecular mechanism of inhibition shown by 4PEH, we have characterized the structure of DHBPS from Vibrio cholerae (vDHBPS) in its apo form and in complex with phosphate, d-ribulose 5-phosphate, d-ribulose 5-phosphate plus zinc ions, 4PEH, and 4PEH plus zinc ions. vDHBPS shares a sequence identity of about 26–60% with other DHBPS sequences whose structures have been reported thus far (Table 1).
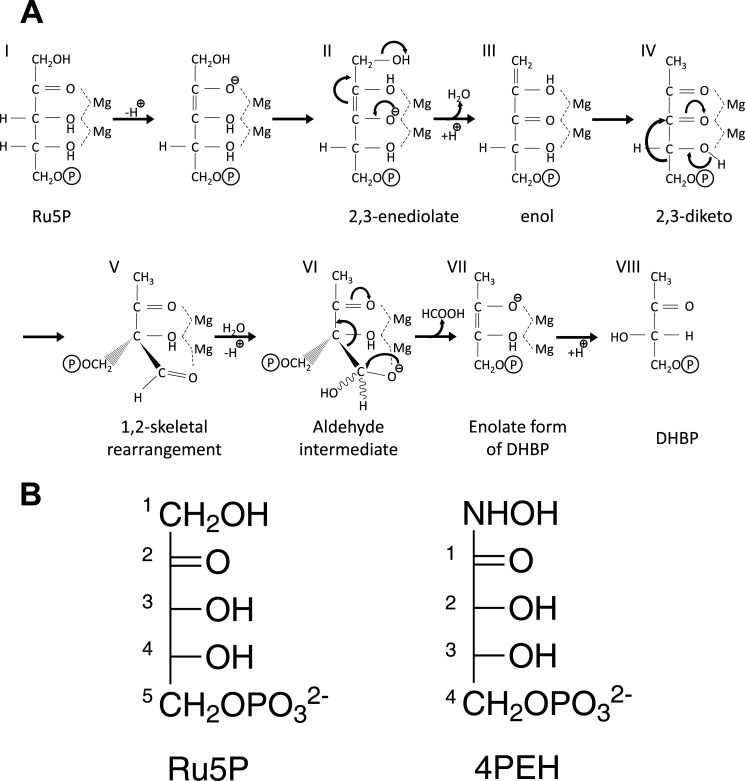
FIGURE 1.
Reaction catalyzed by DHBPS. A, proposed mechanism for the conversion of Ru5P to DHBP by DHBPS (18). B, chemical structures of substrate Ru5P and inhibitor 4PEH.
TABLE 1.
Comparison of amino acid sequence similarity of vDHBPS with other homologs
| Protein Data Bank code | Species | No. of residues | Sequence |
r.m.s.d.a | |
|---|---|---|---|---|---|
| Identity | Similarity | ||||
| % | |||||
| 4P8E | V. cholerae (from this work) | 218 | 100 | 100 | 0.00 (218)b |
| 3LS6 | S. typhimurium | 216 | 61 | 71 | 0.63 (205) |
| 1G57 | E. coli | 212 | 60 | 72 | 0.58 (204) |
| 3MIO | M. tuberculosis | 196 | 43 | 63 | 0.93 (181) |
| 1TKU | C. albicans | 197 | 43 | 64 | 1.04 (187) |
| 4FFJ | S. pneumoniae | 199 | 42 | 66 | 1.04 (189) |
| 1K4I | M. grisea | 214 | 38 | 55 | 1.08 (200) |
| 1PVY | M. jannaschii | 227 | 26 | 47 | 1.30 (187) |
a Root mean square deviation.
b Number of Cα atoms used for superposition.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of ribB from V. cholerae
A pair of oligonucleotides representing forward (5′-ATC GAC AGA CAT ATG AAT CAG TCC TCC TTG-3′) and reverse (5′-ACT AAG TAC CTC GAG TTA AGC CAG TTT TAG ATC-3′) sequences along with NdeI and XhoI restriction sites (underlined) were synthesized (Integrated DNA Technologies) to amplify the ribB gene (vribB) from genomic DNA of V. cholerae E1 Tor (strain N16961) using a standard polymerase chain reaction (PCR) protocol. The amplified PCR product was purified by agarose gel electrophoresis and subsequently digested with restriction enzymes NdeI and XhoI (New England Biolabs) for 12 h at 37 °C. The digested PCR product was ligated into pET-28c vector (Novagen), which was digested with the same set of restriction enzymes. The integration of vribB gene with the vector was verified by DNA sequencing. In addition, the mutants E39A, E41A, R61A, S64A, M84A, N89A, V98A, H154A, and F139A of vribB were created by site-directed mutagenesis by using an overlap extension PCR method (40) with suitable primers. The amplified mutants were cloned into pET-28c vector similarly to cloning of wild type and verified by DNA sequencing. The resultant plasmids for wild type and mutants were transformed into E. coli BL21(DE3).
For the expression of vribB (vDHBPS), a single colony of E. coli BL21(DE3) strain carrying pET28c-vribB was inoculated in 20 ml of Luria-Bertani medium supplemented with 30 μg/ml kanamycin and grown overnight at 37 °C in a rotary shaking incubator. An aliquot (1%) of the overnight-grown seed culture was inoculated in fresh Luria-Bertani medium supplemented with 30 μg/ml kanamycin and allowed to grow further until the absorbance (A) at 600 nm reached a value of 0.6–0.8. vribB expression was induced by adding 0.5 mm isopropyl β-d-1-thiogalactopyranoside, and the cells were allowed to grow further for 3 h. The cells were harvested by centrifuging the culture at 5,000 rpm for 15 min at 4 °C. The cell pellet was resuspended in 40 ml of lysis buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 1 mm DTT, and 1 mg/ml lysozyme) along with 0.2 ml of a mixture of protease inhibitors (Sigma). The resuspended cells were incubated on ice for 30 min and disrupted by sonication for 30 min on ice (30-s pulse on/off with 20% amplitude) (Sonics). The soluble and insoluble cell fractions were separated by centrifuging the cell lysate at 12,000 × g for 30 min at 4 °C. The supernatant was loaded onto a nickel-nitrilotriacetic acid column (Qiagen, Germany) that was pre-equilibrated with equilibration buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, and 1 mm DTT). The column was washed with 25 column volumes of wash buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 1 mm DTT, and 20 mm imidazole) to get rid of any nonspecifically bound proteins. Finally, the bound vDHBPS protein was eluted with elution buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 1 mm DTT, and 250 mm imidazole) and subsequently dialyzed overnight against 25 mm Tris-HCl, pH 8.0 at 25 °C. The dialyzed vDHBPS was further purified by gel filtration chromatography using a Sephacryl S-200 column (GE Healthcare) equilibrated with 25 mm Tris-HCl, pH 8.0. The purified vDHBPS was concentrated up to 20 mg/ml as measured by the Bradford method (41) using an Amicon concentrator with a 10-kDa-molecular mass cutoff membrane (Millipore). The mutant proteins E39A, E41A, R61A, S64A, M84A, N89A, and V98A were expressed and purified similarly to wild-type vDHBPS. Because the protein expression and stability were very poor for F139A and H154A mutants, their respective cells were induced with 1 mm isopropyl β-d-1-thiogalactopyranoside and further allowed to grow overnight at 20 °C. Both F139A and H154A proteins were prepared similarly to wild-type vDHBPS except that they were maintained in 50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 1 mm DTT, and 10% glycerol buffer throughout the process. The purity of the protein at each stage was checked by 15% SDS-PAGE (42).
Size Exclusion Chromatography Studies
The oligomeric status of wild-type and mutants of vDHBPS was determined by gel filtration chromatography using a prepacked Superdex 200 analytical column (GE Healthcare) installed on a BioLogic Duo Flow system (Bio-Rad). For each purified protein, an approximately 4–6 mg/ml concentration in a 1-ml volume was injected into the column. The flow rate was maintained at 0.3 ml/min. To establish a molecular mass and elution volume relationship, known molecular mass proteins were run on the same column. The proteins used as standards were ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), and ribonuclease A (13.7 kDa) (GE Healthcare).
Kinetic Studies of vDHBPS
The vDHBPS enzyme activity was measured by a colorimetric method as reported earlier (43). Briefly, in a 125-μl reaction, 2.5 μg of vDHBPS was mixed with 50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, and a variable concentration of d-ribulose 5-phosphate and incubated at 37 °C for 15 min. The enzyme reaction was quenched, and color was developed by addition of 100 μl of saturated creatine solution (30 mg/ml) followed by 50 μl of α-naphthol (35 mg/ml in 1.0 n NaOH). The color was allowed to develop for 30 min at room temperature before measuring the absorbance at 525 nm using an ELISA plate reader (BioTek PowerWave XS plate reader). The absorbance values were converted into nanomoles of DHBP using a standard plot that was established by mixing 0–55 nmol of 2,3-butanedione in a 125-μl reaction volume and incubating the samples in creatine and naphthol for the color development (43). The Km and Vmax for Ru5P were calculated from a Michaelis-Menten saturation curve by non-linear curve fitting using Origin 8.5. Similarly, the inhibitory effect of 4PEH on vDHBPS activity was measured using the same colorimetric assay except the protein concentration was kept at 20 μg and the reaction was carried out for 30 min. The inhibitor constant (Ki) was determined using a constant inhibitor concentration and varying the substrate concentration from 10 μm to 2 mm.
Crystallization of vDHBPS
The vDHBPS at a concentration of 20 mg/ml in 25 mm Tris-HCl, pH 8.0 was used for crystallization experiments using the sitting drop vapor diffusion method in a 96-well plate (MRC plates, Molecular Dimensions, UK). Initial crystallization screens were carried out using PEG/Ion Screen (Hampton Research) with each drop containing 1 μl of protein mixed with 1 μl of precipitant and equilibrated against 60 μl of reservoir solution followed by incubation at 20 °C. Crystals appeared within 3–4 days in a number of conditions. The crystallization conditions obtained from the initial screen were optimized in 24-well sitting drop crystallization plates (Combi Clover Jr., Emerald BioSystems) at 20 °C. Drops containing 2 μl of protein and 2 μl of reservoir solutions were equilibrated against 200 μl of reservoir solution. The apo form of vDHBPS (E-Apo) crystals were grown in 0.2 m Na2HPO4, pH 9.1 and 20% PEG 3350, and phosphate-bound (E-PO4) crystals were grown in 0.2 m NaH2PO4, pH 4.5 and 20% PEG 3350. The Ru5P and 4PEH complex crystals were obtained by mixing vDHBPS with 10 mm Ru5P (E-Ru5P) and 4PEH (E-4PEH) separately and incubated for 30 min at 20 °C. The crystals were grown in 0.2 m Na2HPO4, pH 9.1 and 20% PEG 3350. Finally, the E-Ru5P-Zn2+ and E-4PEH-Zn2+ complex crystals were obtained by soaking the E-Ru5P and E-4PEH crystals with 100 mm zinc chloride (ZnCl2) for 12 h at 20 °C.
Data Collection and Processing
The x-ray diffraction data set for the E-PO4 complex crystal was collected at 100 K using synchrotron radiation at BM14 beamline (European Synchrotron Radiation Facility, Grenoble, France) equipped with a MAR Mosaic charge-coupled device detector. The diffraction data sets for all other crystals (E-Apo, E-Ru5P, E-Ru5P-Zn2+, E-4PEH, and E-4PEH-Zn2+) were collected using a MAR345 image plate detector (MAR Research, Germany) mounted on a MicroMax-007HF (Rigaku) rotating anode x-ray generator operated at 40 kV and 30 mA. The data sets were collected at 100 K using an Oxford Cryostream. Before diffraction, each crystal was soaked for 10 s in a cryoprotectant solution consisting of 30% glycerol mixed with their corresponding mother liquor (44). The diffraction images of all the data sets were indexed, integrated, and scaled using the XDS suite of programs (45). The scaled intensities were converted into structure factors using CTRUNCATE (46) as implemented in CCP4 (Collaborative Computational Project Number 4) (47).
Structure Determination and Refinement
The structure of vDHBPS in the apo form was solved by a molecular replacement method using PHASER (48) with one subunit of E. coli DHBPS as the search model (Protein Data Bank code 1G57). The Matthews' coefficient (VM = 2.1) (49) and solvent content (42%) as calculated by CCP4 (47) indicated two molecules per asymmetric unit. The PHASER with default parameters gave a single solution with two DHBPS molecules in the asymmetric unit. The initial model was refined as a rigid body followed by restraint refinement using REFMAC5 (50). To calculate the free R factor (51), 5% of total reflection data were excluded from the refinement for each structure. The vDHBPS model was further refined using PHENIX (52) and built using the program COOT (53). During the initial stages of refinement, the model was subjected to simulated annealing refinement (54) to minimize the model bias. Moreover, composite omit maps (55) were calculated intermittently during refinements and used to improve the model. The refinement and model building were carried out iteratively until the vDHBPS model was completely built and R and free R factors were converged. The solvent molecules were added in the difference Fourier electron density map at above the 3.0 σ level and formed at least one hydrogen bond with the protein or other solvent molecule. For the E-PO4, E-Ru5P, E-Ru5P-Zn2+, E-4PEH, and E-4PEH-Zn2+ complexes, the structures were determined either by the molecular replacement method or by the difference Fourier method using the vDHBPS apo form as the model. Similar to apo-vDHBPS, all the complex crystal structures were refined using the PHENIX suite of programs except E-Ru5P and E-Ru5P-Zn2+, which were refined with REFMAC5. Well defined electron densities for bound phosphate, Ru5P, and 4PEH were seen in their respective difference Fourier map at the 3.0 σ level, and the same were included in the model and further refined. The zinc ion was added in the E-Ru5P-Zn2+ and E-4PEH-Zn2+ models if the difference Fourier map showed electron density at above the 5.0 σ level. The Ramachandran plot statistics were calculated using PROCHECK (56), and superposition of the molecules was carried out by LSQMAN (57). The program PyMOL (58) was used to visualize and analyze the structures and for making figures.
RESULTS
Characterization of vDHBPS with Substrate and Inhibitor
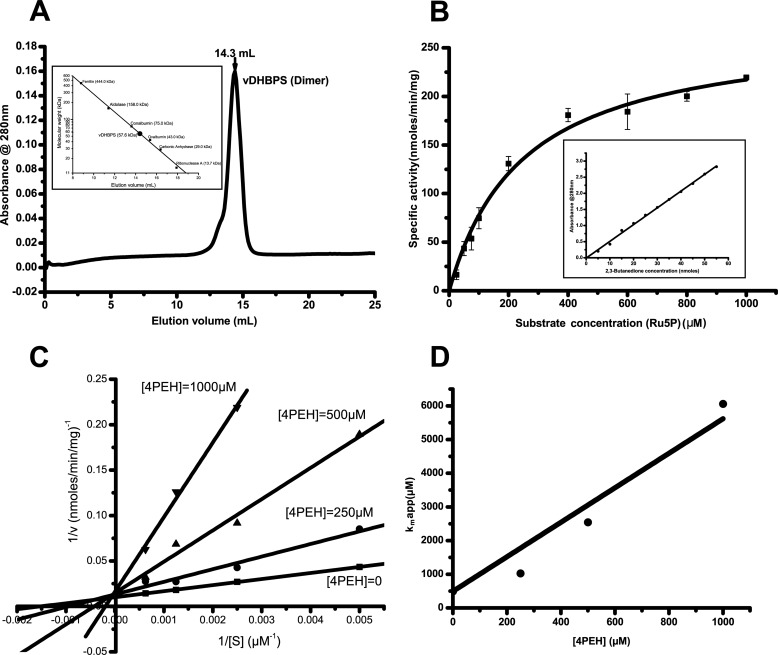
The vribB gene encoding vDHBPS was PCR-amplified, cloned, expressed as a His-tagged protein, and purified to homogeneity. The size exclusion chromatography studies suggest that vDHBPS exists as a dimer in solution (Fig. 2A). Furthermore, the kinetic characterization of vDHBPS with substrate Ru5P shows a typical Michaelis-Menten saturation behavior with Km and Vmax values corresponding to 250 ± 30 μm and 270 ± 10 nmol/min/mg, respectively (Fig. 2B). The compound 4PEH is an analog of substrate in which the first hydroxymethyl group of Ru5P is replaced with a hydroxylamine group (Fig. 1B), and therefore it is anticipated to inhibit the catalytic activity of DHBPS. To determine the efficacy of such inhibition, if any, we measured the catalytic activity of vDHBPS in the presence of different concentrations of 4PEH. The Lineweaver-Burk plot of measured values strongly suggests that 4PEH inhibits vDHBPS competitively (Fig. 2C). Furthermore, a plot of the apparent Km values derived from the Lineweaver-Burk plot against inhibitor concentration reveals a Ki value of 100 ± 10 μm (Fig. 2D).
FIGURE 2.
Oligomeric and kinetic characterization of vDHBPS. A, size exclusion chromatography elution profile of vDHBPS with a Superdex 200 column. The inset shows the elution profile of standard molecular masses from the same column. B, Michaelis-Menten plot for vDHBPS with various substrate (Ru5P) concentrations. Error bars represent S.D. The inset shows the standard plot generated with known concentration of 2,3-butanedione and used for converting absorbance to DHBP. C, Lineweaver-Burk plot for different concentrations of substrate (Ru5P) at each inhibitor (4PEH) concentration. D, the calculated apparent Km is plotted against inhibitor (4PEH) concentration to estimate Ki.
Quality of vDHBPS Model
To investigate the structural changes that take place during catalysis and inhibition in a systematic manner, we initiated structure analysis of vDHBPS in various forms. Accordingly, we determined the crystal structures of vDHBPS in apo form (E-Apo) and in complex with phosphate (E-PO4), substrate (E-Ru5P), inhibitor (E-4PEH), substrate plus metal ions (E-Ru5P-Zn2+), and inhibitor plus metal ions (E-4PEH-Zn2+). All vDHBPS crystals were crystallized in orthorhombic space group P212121 with unit cell parameters in the range of a = 52–56 Å, b = 73–78 Å, and c = 95–99 Å, and x-ray diffraction data sets were collected in the resolution range of 1.59–2.04 Å. The data collection statistics for all the crystals are shown in Table 2. The vDHBPS structure was solved by the molecular replacement method using E. coli DHBPS (30, 31) as the search model. The vDHBPS monomer consists of 218 amino acid residues and exists as a dimer both in solution and in the crystal structure, thus consisting of a total of 436 amino acid residues. In addition, at the N terminus of vDHBPS, an extra 20 amino acid residues (MGSSHHHHHHSSENLYFQGH) were introduced from the cloning vector. The electron density for all the structures was generally good and allowed us to build the vDHBPS model without any ambiguity. However, the electron density for the N- and C-terminal (1–4 and 216–218, respectively) residues was poor and thus not included in the model for most of the crystal structures. Furthermore, in the apo (E-Apo) and phosphate (E-PO4) complexes, residues from 83 to 97 were not traceable in the electron density map due to their known flexibility and were not included in the model. The refinement and statistical parameters for all crystal structures are within the allowed limits and are shown in Table 3. The coordinates and structure factors for all the crystal structures are deposited in the Protein Data Bank and corresponding Protein Data Bank codes are given in Table 3.
TABLE 2.
vDHBPS data collection and processing details
| Particulars | E-Apo | E-PO4 | E-Ru5P | E-Ru5P-Zn2+ | E-4PEH | E-4PEH-Zn2+ |
|---|---|---|---|---|---|---|
| Wavelength (Å) | 1.5418 | 0.9780 | 1.5418 | 1.5418 | 1.5418 | 1.5418 |
| No. crystals used | 1 | 1 | 1 | 1 | 1 | 1 |
| Resolution range (Å) | 20.0–1.96 | 20.0–1.59 | 20.0–2.04 | 20.0–2.04 | 20.0–1.86 | 20.0–1.86 |
| Last resolution shell (Å) | 2.08-1.96 | 1.68-1.59 | 2.16-2.04 | 2.16-2.04 | 1.97-1.86 | 1.97-1.86 |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 |
| Unit cell parameters (Å) | a = 54.270, b = 78.050, c = 97.943 | a = 56.554, b = 73.597, c = 95.560 | a = 54.408, b = 74.086, c = 99.331 | a = 52.079, b = 77.090, c = 97.531 | a = 52.674, b = 72.654, c = 100.085 | a = 52.079, b = 76.805, c = 97.634 |
| Total no. of reflections | 237,158 | 286,972 | 181,735 | 184,298 | 251,374 | 274,912 |
| Unique reflections | 30,192 | 52,849 | 26,194 | 25,330 | 32,615 | 33,367 |
| Mosaicity (°) | 0.5 | 0.1 | 0.3 | 0.6 | 0.3 | 0.2 |
| Redundancy | 7.8 (6.8)a | 5.4 (5.0) | 6.9 (6.6) | 7.3 (6.1) | 7.7 (7.6) | 8.2 (7.8) |
| Overall I/σ(I) | 14.7 (2.6) | 32.1 (7.4) | 19.2 (3.7) | 23.1 (6.4) | 15.7 (5.5) | 25.3 (6.8) |
| Completeness (%) | 98.5 (93.5) | 96.6 (93.5) | 99.4 (97.8) | 98.3 (92.7) | 99.3 (97.7) | 99.6 (98.2) |
| Rmerge (%)b | 10.9 (77.0) | 3.6 (19.7) | 8.5 (53.5) | 6.5 (25.8) | 10.5 (36.1) | 7.1 (31.4) |
a Values for the last resolution shell are in parentheses.
b Rmerge = ΣhklΣi|Ii(hkl) − I(hkl)|/ΣhklΣiIi(hkl) where I(hkl) is the intensity of reflection hkl.
TABLE 3.
vDHBPS refinement details
| Parameters | E-Apo | E-PO4 | E-Ru5P | E-Ru5P-Zn2+ | E-4PEH | E-4PEH-Zn2+ |
|---|---|---|---|---|---|---|
| Resolution range (Å) | 20.0–1.96 | 20.0–1.59 | 20.0–2.04 | 20.0–2.04 | 20.0–1.86 | 20.0–1.86 |
| Rcryst (%)a | 18.10 | 16.88 | 17.34 | 17.56 | 16.28 | 14.43 |
| Rfree (%)b | 22.50 | 19.70 | 22.36 | 23.22 | 20.69 | 19.09 |
| r.m.s.d.c | ||||||
| Bond length (Å) | 0.007 | 0.006 | 0.009 | 0.006 | 0.007 | 0.007 |
| Bond angle (°) | 1.036 | 1.089 | 1.310 | 1.141 | 1.141 | 1.052 |
| Ramachandran plot, residues in | ||||||
| Most favored region (%) | 97.75 | 98.01 | 97.35 | 98.14 | 97.20 | 97.72 |
| Additionally allowed region (%) | 2.25 | 1.99 | 2.65 | 1.86 | 2.80 | 2.28 |
| No. of residues (out of 436) | 406 | 403 | 423 | 431 + 2d | 430 | 432 + 2d |
| No. of solvent molecules | 332 | 425 | 284 | 277 | 344 | 416 |
| Wilson B-factor (Å2) | 21.40 | 11.92 | 24.43 | 21.00 | 14.62 | 13.89 |
| Average B for protein atoms (Å2) | 23.80 | 13.97 | 29.43 | 22.35 | 13.40 | 14.50 |
| Average B for Loop1 (for A and B, Å2) | 24.54 | 18.66 | 30.17 | 25.35 | 13.13 | 15.35 |
| Average B for Loop2 (for A and B, Å2) | 36.80 | 25.35 | 42.68 | 28.12 | 20.39 | 21.06 |
| Average B for Solvents (Å2) | 30.43 | 23.76 | 36.49 | 26.84 | 19.92 | 23.30 |
| Ligands (in active site) | PO4 (2) | Ru5P (2) | Ru5P (2) | 4PEH (2) | 4PEH (2) | |
| Average B for ligands (Å2) | 19.87 | 43.06 | 25.41 | 20.59 | 12.59 | |
| Average occupancy for ligands | 0.64 | 1.00 | 1.00 | 1.00 | 1.00 | |
| No. of Zn2+ (in active site) | 10 (4) | 11 (4) | ||||
| Average B for Zn2+ (in active site) (Å2) | 24.63 (21.85) | 19.34 (12.65) | ||||
| Average occupancy for Zn2+ (in active site) | 0.75 (1.00) | 0.70 (0.91) | ||||
| Protein Data Bank code | 4P8J | 4P6D | 4P77 | 4P8E | 4P6C | 4P6P |
a Rcryst = Σhkl‖Fobs| − |Fcalc‖/Σ|Fobs|.
b Rfree is the cross-validation R-factor computed for the test set of 5% of unique reflections.
c Root mean square deviation.
d From His tag.
Overall Structure of vDHBPS
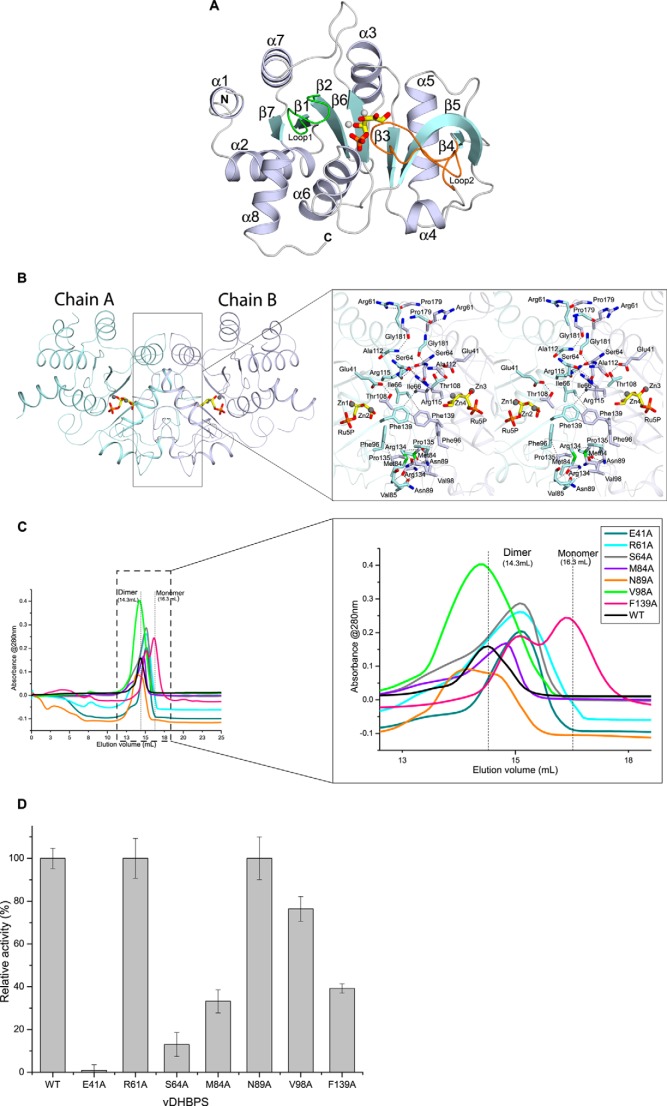
The overall three-dimensional structure of vDHBPS shows an α + β fold (Fig. 3A) similar to other reported DHBPS structures (Table 1) (28, 30–36). Each monomer consists of a central β-sheet formed by seven β-strands, which are oriented in a parallel and antiparallel manner. The central β-sheet is surrounded by eight α-helices, and both are connected through several loops. The asymmetric unit consists of two molecules related by 2-fold symmetry (Fig. 3B) that can superimpose with each other with a root mean square deviation of about 0.25–0.50 Å, indicating that the two monomers are nearly identical to each other in the respective vDHBPS structures. The two molecules interact with each other mainly through hydrogen bond and hydrophobic interactions. The residues involved in hydrogen bonds are Glu-41, Arg-61, Ser-64, Gly-65, Asn-89, Thr-108, Ala-112, Arg-115, Arg-134, Pro-179, and Gly-181 from one monomer and Thr-108, Pro-179, Arg-115, Ser-64, Arg-134, Glu-41, Gly-181, Ser-64, Asn-89, Arg-61, and Ala-112 of the other monomer, respectively (Fig. 3b). Similarly, the hydrophobic interactions are found between Ile-66, Met-84, Val-85, Phe-96, Val-98, Pro-135, Phe-139, and Phe-139 from one monomer and Phe-139, Met-84, Val-98, Pro-135, Val-85, Phe-96, Ile-66, and Phe-139 of the other monomer, respectively (Fig. 3B). The PISA server (59) predicts that the two molecules of vDHBPS in the asymmetric unit form a stable homodimer (Fig. 3B) with total buried surface area of 2,645 Å2 (16% of the surface area of the complex).
FIGURE 3.
Structural characterization of vDHBPS. A, the overall structure of vDHBPS. Secondary structures are shown in white-blue for α-helix, cyan for β-strands, and white for loops. Ru5P and metal ions are represented as stick and sphere models, respectively. Loop1 and Loop2 are shown in green and orange, respectively. B, the association of vDHBPS as a dimer. Each monomer is shown in different colors. The Ru5P and metal ions are shown at the corresponding active site of each monomer. The closer view shows the amino acid residues involved in dimerization, and their interactions are shown in dashed lines. C, size exclusion chromatography elution profile of different mutants of vDHBPS from a Superdex 200 column. D, the relative catalytic activity of different mutants of vDHBPS with respect to wild-type (WT) vDHBPS. Error bars represent S.D.
The structural characterization of monofunctional DHBPS reported till now reveals that it exists as a dimer both in the crystal structure and in solution, and dimerization is required for its catalytic activity as its active site is located at the dimeric interface and contributed by residues from both monomers (28, 30–35). Moreover, the two monomers of DHBPS interact with each other mainly through hydrophobic interactions and with some hydrogen bond interactions (31).
To identify the amino acid residues that contribute to the dimer formation of vDHBPS in solution and thus its catalytic activity, we examined all DHBPS crystal structures reported till now. The analysis of amino acid sequences and structures of various monofunctional DHBPSs (28, 30–36) revealed that the residues and their interactions involved in dimer formation are more or less similar to those of vDHBPS. Therefore, we chose three nearly conserved hydrophilic residues (Glu-41, Arg-61, and Ser-64), three conserved hydrophobic residues (Met-84, Val-98, and Phe-139), and one (Asn-89) non-conserved residue that are involved in dimer formation of the vDHBPS structure and mutated these to alanine individually (E41A, R61A, S64A, M84A, V98A, and F139A) by site-directed mutagenesis. Each mutant was cloned, expressed, and purified as was done for wild-type protein (see “Experimental Procedures”). To assess the oligomerization, each mutant protein was passed through a Superdex 200 column individually. The gel filtration profile of each mutant along with wild type is shown in Fig. 3C. The gel filtration profile of all seven mutant proteins can be classified into three categories: (i) mostly dimeric form (N89A and V98A), (ii) in between expected monomeric and dimeric form (E41A, R61A, S64A, and M84A), and (iii) mostly monomeric form (F139A). Although most of the mutant proteins showed a shift in their elution profile (compared with wild-type vDHBPS), indicating that their dimerization was disturbed, they did not elute absolutely as a monomeric peak. Therefore, to check whether the mutation had any effect on its catalytic activity, we measured the DHBPS activity (with 20 μg of protein and 400 μm Ru5P) for all seven mutant proteins (Fig. 3D). The relative catalytic activity of hydrophobic mutants with respect to wild-type vDHBPS was 30, 40, and 80% for M84A, F139A, and V98A, respectively. Analysis of the crystal structure of vDHBPS indicated that both Met-84 and Phe-139 are involved in hydrophobic interactions with the same residues in another monomer, and therefore mutation of these residues may disturb their dimerization and thus the catalytic activity. Consistent with this, F139A mutant mostly eluted as a monomer peak, whereas a significant amount also exists in a monomer-dimer equilibrium (Fig. 3C). In addition, the M84A mutant eluted as a monomer-dimer equilibrium and showed deficient activity. However, the V98A mutant mostly exists in dimeric form and showed a catalytic activity of about 80% compared with wild type. The analysis of sequences revealed that Val-98 is conserved throughout the species; however, its interaction is not conserved in all the structures. Similarly, the relative catalytic activity of hydrophilic mutants with respect to wild-type vDHBPS was 0, 15, and 100% for E41A, S64A, and R61A, respectively. Analysis of Glu-41 and Ser-64 residues in vDHBPS structures revealed that apart from their involvement in dimerization they are also involved in substrate binding. For example, Glu-41 interacts with Thr-108, which in turn interacts with the substrate Ru5P. Moreover, Glu-41 also interacts with Ru5P through a solvent molecule in the same monomer. Therefore, mutation of Glu-41 to alanine probably disrupts this, and thus catalytic activity is lost. Similarly, although the carbonyl oxygen of Ser-64 interacts with Arg-115 of the other monomer, its hydroxyl group interacts with Glu-175 of the same monomer that in turn interacts with substrate Ru5P. Therefore, the S64A mutant may interfere with this interaction, resulting in a significant loss of catalytic activity. Surprisingly, the R61A mutant showed 100% activity, indicating that mutation of Arg-61 to Ala does not affect its catalytic efficiency. Although analysis of sequences showed that Arg-61 is nearly conserved, its interaction is not conserved in other species, and moreover, Arg-61 is located far from the active site. As anticipated, the N89A mutant did not show any effect on its catalytic activity, remaining equal to that of wild-type vDHBPS.
Interestingly, the bifunctional (DHBPS and GTP cyclohydrolase II) M. tuberculosis RibA2, although shows DHBPS activity, its DHBPS domain exists as monomer in solution (36). The existence of the DHBPS domain of M. tuberculosis RibA2 as a monomer has been mainly attributed to mutation of three hydrophobic residues at the dimeric interface (I66Y, V85Y, and F96Y; vDHBPS numbering). However, the DHBPS domain is observed as a dimer in the crystal structure of M. tuberculosis RibA2 (36). Thus, it is speculated that in the presence of substrate the DHBPS domain may transiently form a dimer in solution; however, the exact mechanism is still unknown (28).
Taken together, we predict that the hydrophobic residues at the dimeric interface play a role in dimer formation, whereas some of the active site residues at the dimeric interface play a role in the catalytic activity of vDHBPS. However, further studies are required to see whether these two properties can be decoupled for vDHBPS.
Conformational Changes among the Structures of vDHBPS
To understand the conformational changes that occurred among different vDHBPS complexes, we analyzed their structures. The superposition of different vDHBPS crystal structures shows root mean square deviations of about 0.24–0.54 Å for 199–214 Cα atoms, indicating that the overall conformations of all vDHBPS structures remain similar (Fig. 4). However, major conformational changes are observed in two loop regions as reported in other species (32, 33, 35). In the crystal structures of E-Apo and E-PO4, Loop1 (residues 34–41) exhibits an ordered open conformation, whereas Loop2 (residues 82–98) is completely disordered (Fig. 4). In the E-Ru5P complex structure, Loop1 adopts a closed conformation and interacts with substrate Ru5P, whereas Loop2 is partially disordered. In E-Ru5P-Zn2+, E-4PEH, and E-4PEH-Zn2+, both Loop1 and Loop2 are observed in a closed and ordered conformation.
FIGURE 4.
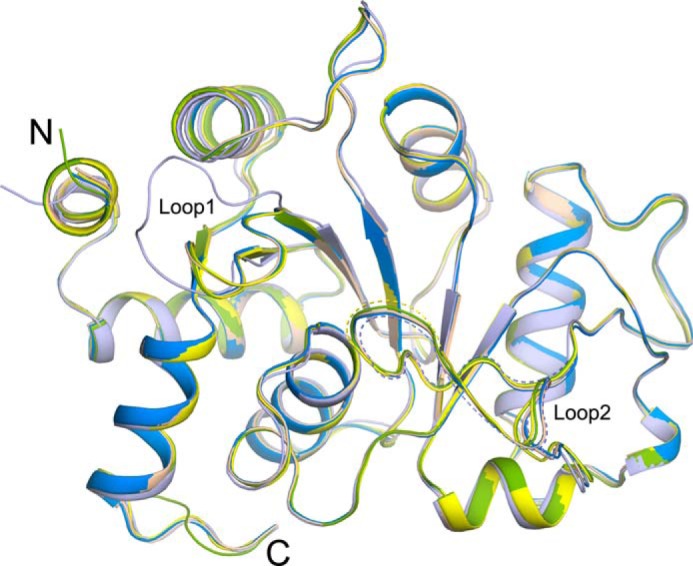
Conformational changes among vDHBPS structures. Superposition of crystal structures of vDHBPS in the apo form (light blue) and in complex with Ru5P (yellow), Ru5P plus zinc ions (wheat), 4PEH (green), and 4PEH plus zinc ions (marine blue) is shown. Dashed lines indicate lack of electron density in that region and not included in the model.
Active Site Architecture of vDHBPS with Ru5P and Metal Ions
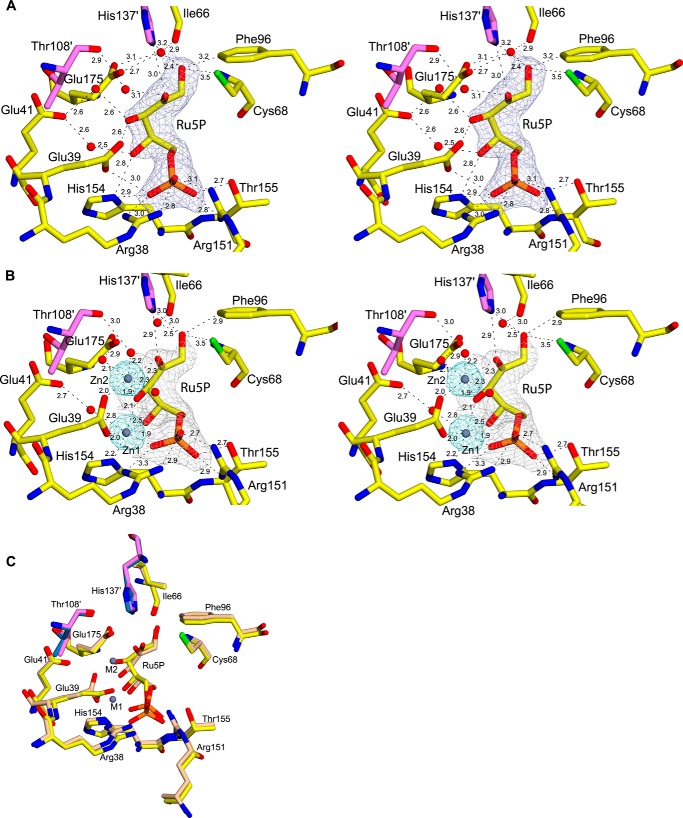
In the E-Ru5P complex, two molecules of substrate (Ru5P) occupy the active site at the dimeric interface of each molecule at topologically equivalent sites and interact with vDHBPS through hydrogen bond and electrostatic interactions (Fig. 5A). Moreover, the observed conformational change of Loop1 upon binding of Ru5P helps in positioning the substrate at the active site (the maximum distance moved is about 12 Å for residue Arg-38). The phosphate moiety of Ru5P interacts with Arg-38, Glu-39, Arg-151, His-154, and Thr-155 through hydrogen bond interactions (Fig. 5A). The first carboxyl group, O1, of Ru5P interacts with SH of Cys-68, and with Nϵ2 of His-137 of the other monomer. Additionally, it interacts with Glu-175 and Ile-66 through a water molecule. The second carbonyl group, O2, of Ru5P interacts with Nϵ2 of His-137 of the other monomer. In addition, it interacts with the carbonyl oxygen of Thr-108 through a water molecule. The carboxyl group O3 of Ru5P interacts with Oϵ2 of Glu-39 in one chain, and in the other chain, it interacts with both Oϵ1 and Oϵ2. Similarly, the carboxyl group O4 of Ru5P interacts with Nδ1 of His-154 and with Oϵ1 of Glu-41 through a water molecule.
FIGURE 5.
Substrate binding site of vDHBPS. A, stereodiagram showing a 2Fo − Fc electron density map contoured at 1.0 σ for Ru5P. Residues interacting with Ru5P are displayed along with their distances. Residues from the other monomer are shown in magenta. B, stereodiagram showing a 2Fo − Fc electron density map for Ru5P and Zn2+ contoured at 1.5 and 5.0 σ, respectively. The distances are shown for the close residues. C, superposition of the active site of E-Ru5P (yellow) and E-Ru5P-Zn2+ (wheat).
Although Ru5P alone binds to vDHBPS, its catalytic activity is observed only in the presence of magnesium ions. To understand the location of metal binding sites in the case of vDHBPS, we attempted to obtain the E-Ru5P complex with magnesium ions by both co-crystallization and soaking experiments. However, in both cases, we were unsuccessful in obtaining the E-Ru5P complex with magnesium ions. In fact, until now, no DHBPS structure has been determined in complex with Ru5P and magnesium ions. It is speculated that both metals are only transiently bound to the enzyme as histidine (His-154) is relatively an unusual ligand for magnesium ions (33). Therefore, to understand the metal ion positions during catalysis, we soaked the E-Ru5P complex crystal with zinc ions and determined its structure (E-Ru5P-Zn2+). Although in the presence of Zn2+ ions vDHBPS does not show any catalytic activity, Zn2+ is predicted to occupy a similar position as magnesium ions (32, 33). This is because both Mg2+ and Zn2+ have the same oxidation state with their last s orbital filled (s2) and have roughly the same ionic radius with 0.74 Å for Zn2+ and 0.72 Å for Mg2+. Moreover, superposition of the E-Ru5P-Zn2+ complex with the Mg2+-bound structure of M. grisea (Protein Data Bank code 1K4I) (32) revealed that both Zn2+ and Mg2+ occupy the same position. Despite such similarity with Mg2+ and the ability to mediate native-like binding, it is predicted that a zinc ion is unable to initiate the reaction as it is not a strong enough Lewis acid (33). The crystal structure of the E-Ru5P-Zn2+ complex clearly shows binding of two zinc ions at each active site, and both Loop1 and Loop2 are observed in a closed and ordered conformation (Fig. 4). The first zinc ion at position M1 is hexacoordinated and interacts with O3 and O4 of Ru5P, one oxygen from phosphate moiety of Ru5P, Nδ1 of His-154, Oϵ2 of Glu-39, and Oϵ1 of Glu-41 through a water molecule (Fig. 5B). The second zinc ion (M2) is coordinated with O2 and O3 of Ru5P, three water molecules, and Oϵ2 of Glu-39 (Fig. 5B). However, the electron densities for O1 and O4 of Ru5P in the E-Ru5P-Zn2+ complex are weak, indicating the flexibility of these atoms in the presence of zinc ions. The superposition of the E-Ru5P and E-Ru5P-Zn2+ structures (Fig. 5C) revealed that Glu-39 undergoes a conformational change upon metal binding and helps in positioning the metals as reported in S. typhimurium DHBPS (35). To understand the contribution of Glu-39 and His-154 in metal binding and thus the catalytic activity of vDHBPS, we created E39A and H154A mutants. Both mutants were cloned, expressed, and purified individually as done for wild-type vDHBPS. However, both mutant proteins did not show any catalytic activity even in the presence of Mg2+ probably due to loss of their interactions with metal ions, indicating their importance in substrate/metal binding. The comparison of E-Ru5P and E-Ru5P-Zn2+ structures also revealed that Ru5P forms a compact structure in the presence of metal ions (Fig. 5C). Earlier reports indicate that the compact conformation of Ru5P in the presence of metal ions may be essential for the easy release of C4 atom from the substrate (33).
Active Site Architecture of vDHBPS with 4PEH and 4PEH-Zn2+
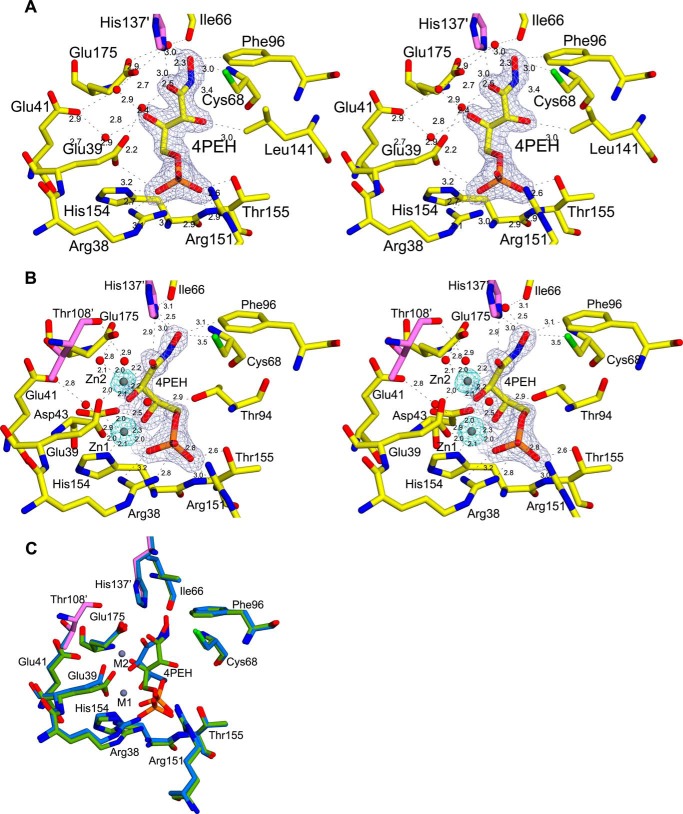
To understand the structural basis for the competitive inhibition shown by 4PEH, we co-crystallized it with vDHBPS to form the E-4PEH complex. In addition, we soaked the crystals of E-4PEH with zinc ions to determine the position of metal ions in the presence of 4PEH (E-4PEH-Zn2+). The overall conformations of these complexes are similar to other vDHBPS structures and show well defined electron density for both 4PEH and zinc ions (Fig. 6, a and b). The hydrogen bond interactions between 4PEH and vDHBPS are similar to the interaction of Ru5P in the E-Ru5P complex. The NHOH oxygen atom of 4PEH interacts with Cys-68, Nϵ2 of His-137, and Glu-175 through a water molecule, whereas O2 does not participate in any interaction except Cδ1 of Leu-141. As observed in the E-Ru5P complex, both Loop1 and Loop2 in the E-4PEH and E-4PEH-Zn2+ complexes are closed and well ordered (Fig. 4). Similar to the E-Ru5P-Zn2+ complex, the binding of zinc ions to E-4PEH revealed that 4PEH also forms a compact conformation (Fig. 6C). Moreover, the two zinc ions in the E-4PEH-Zn2+ complex occupy the same position and exhibit the same coordination as observed in the E-Ru5P-Zn2+ complex. However, unlike E-Ru5P-Zn2+, the E-4PEH-Zn2+ complex shows well defined electron density for both NHOH and O2 oxygen atoms of 4PEH (Fig. 6B).
FIGURE 6.
Inhibitor binding site of vDHBPS. A, stereodiagram showing a 2Fo − Fc electron density map contoured at 1.2 σ for 4PEH. Residues interacting with 4PEH are displayed along with their distances, and residues from the other monomer are shown in magenta. B, stereodiagram showing an electron density map for 4PEH and Zn2+ contoured at 2.0 and 8.0 σ, respectively. The distances are shown for the close residues. C, superposition of the active site of E-4PEH (green) and E-4PEH-Zn2+ (marine blue).
DISCUSSION
Riboflavin biosynthesis pathway is indispensable for many pathogens that lack an efficient riboflavin transporter in their system. Therefore, the enzymes involved in riboflavin biosynthesis are considered as potential drug targets (2–4, 6). One of the rate-limiting enzymes (30) of the riboflavin biosynthesis pathway is DHBPS, which is encoded by the ribB gene in bacteria. In addition, the ribB gene has also been shown to be essential for M. tuberculosis (7), H. pylori (37), and S. typhimurium (39). Despite its essentiality, no inhibitor has been identified till now for the DHBPS enzyme. Here, we characterized vDHBPS with substrate Ru5P and substrate analog inhibitor 4PEH to understand the structural basis for its inhibition.
Earlier, DHBPS structures complexed with Ru5P and non-catalytic metal ions (Zn2+) have been determined to identify the important amino acids that may be involved in the catalysis (30, 36). Comparison of vDHBPS structures with different species (E. coli, M. grisea, C. albicans, M. jannaschii, S. typhimurium, M. tuberculosis, and S. pneumoniae) revealed their identical overall conformations (Table 1 and Fig. 7A) and the conservation of amino acids that are involved in the catalytic activity (Fig. 7B), suggesting a conserved mechanism of catalysis for this enzyme. Moreover, it has been reported that the binding of substrate Ru5P induces conformational changes mainly in two loop regions (Loop1 and Loop2) of DHBPS (33). Loop1 consists of about eight amino acid residues (from 34 to 41) and is conserved in almost all species (Fig. 7B). In general, it has been observed that Loop1 of DHBPS is completely disordered in its apo form, and upon substrate binding, it becomes ordered and is found in a closed conformation, thereby stabilizing the substrate (30–33, 35). This is in contrast with C. albicans DHBPS (34) where there is no change in the conformation of Loop1 both in the presence and absence of the substrate, and it is always found in an ordered and open conformation. This observation is surprising because Loop1 is supposed to interact with substrate Ru5P, thereby stabilizing it during catalysis. Although this observed difference could be species-specific, it is possible that in the case of yeast the substrate Ru5P has been soaked with the DHBPS crystal where this conformational change is probably restricted. In fact, we have shown in S. typhimurium DHBPS (35) that the binding of substrate Ru5P alone is sufficient for the closing of Loop1 into an ordered conformation from a disordered form. Although the movement of Loop1 from an ordered open conformation to a closed conformation upon substrate binding is speculated, no crystal structure showing this effect is yet available. Here, we demonstrate that in both the apo and phosphate-bound forms Loop1 of vDHBPS exists in an ordered open conformation, and binding of substrate Ru5P causes a change from an open to a closed conformation. Therefore, the vDHBPS crystal structure is the first structure per se that confirms the conformational change of Loop1 from an ordered open to a closed conformation in the presence of substrate. However, it is still unclear why in some species Loop1 is disordered and in others it is in an ordered open conformation despite the fact that the amino acid sequence of this loop is almost conserved. Furthermore, in DHBPS, Loop2 (residues from 82–98) has been observed in either partially ordered or disordered conformations in the absence of any metal ions except in the case of M. jannaschii (33) where this loop is significantly large and observed in an ordered conformation. However, binding of metal ions stabilizes Loop2 in the same conformation in almost all species. Although the conformations of Loop1 and Loop2 in the presence of substrate and/or metal ions are variable from species to species, it appears that the closing of both loops is essential for the catalytic activity of DHBPS as these loops in all species are invariably observed in a closed and ordered conformation in the presence of both substrate and metal ions (Fig. 7A). Moreover, although substrate Ru5P shows different conformations in the absence of metal ions from different species (Fig. 8A), its conformation is similar in all species in the presence of metal ions (Fig. 8B). These results strongly suggest that catalytic activity of DHBPS requires the closing of both Loop1 and Loop2 with ordered conformations.
FIGURE 7.
Comparison of vDHBPS with other homologs. A, superposition of crystal structures of DHBPS from V. cholerae (wheat), S. typhimurium (olive green), M. tuberculosis (gray), M. jannaschii (purple), M. grisea (forest green), E. coli (brown), S. pneumoniae (orange), and C. albicans (deep teal). B, multiple sequence alignment of vDHBPS. Sequences of DHBPS from known structures are aligned using Clustal Omega (61). The secondary structures are displayed on the top of the alignment for vDHBPS. Identical residues are shown in white with a red background, whereas similar residues are shown in red. The figure was generated through ESPript (62).
FIGURE 8.
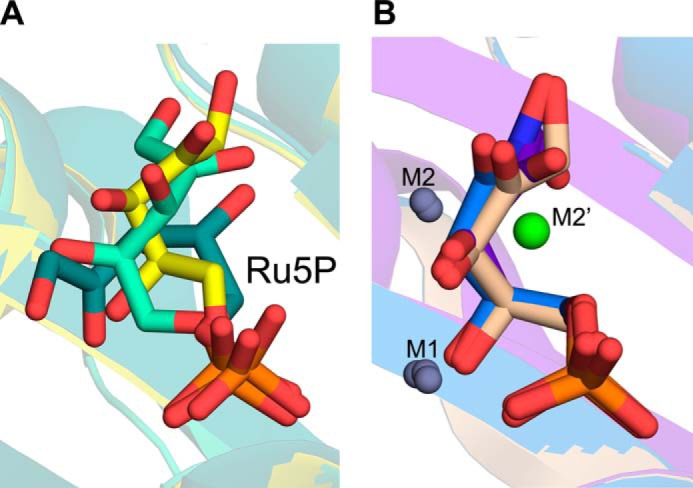
Comparison of conformations of Ru5P. A, superposition of Ru5P from V. cholerae (yellow), S. typhimurium (green-cyan), and C. albicans (deep teal) in the absence of metal ions. B, superposition of Ru5P and 4PEH in the presence of metal ions (M1, M2, and M2′) from E-Ru5P-Zn2+ (wheat), E-4PEH-Zn2+ (marine blue), and M. jannaschii (purple-blue).
The analysis of DHBPS crystal structures complexed with metal ions from different species revealed that the positions of metal ions M1 and M2 are nearly conserved (Fig. 9). However, in the case of M. jannaschii (33), the position of the M2 metal ion is slightly different (M2′) as compared with the other structures. Earlier we reported (35) that this difference may be due to the binding of substrate Ru5P because at that time only the M. jannaschii DHBPS structure complexed with substrate and metal ions was available. However, analysis of DHBPS crystal structures complexed with metal ions from V. cholerae, S. typhimurium (35), and M. grisea (32) indicates that the two catalytic metal ions are almost conserved at the M1 and M2 positions (Fig. 9). The only exception is M. jannaschii (33) in which the position of M1 is conserved, whereas M2 is observed at the M2′ position (Fig. 9). In fact, it has been speculated that the first metal ion at M1 is sufficient to initiate the catalytic reaction, and the second ion at the M2′ position may be involved in avoiding unproductive side reactions of highly reactive intermediates (33). In M. jannaschii DHBPS, the interaction of the metal ion at the M1 position is hexacoordinated and similar to the E-Ru5P-Zn2+ complex. However, the metal ion at the M2′ position interacts with DHBPS through somewhat longer bond lengths when compared with the M1 metal ion (33). This observed change in the second metal ion position could be specific to M. jannaschii, or it may be due to the presence of Ca2+ rather than Zn2+. Nevertheless, the proposed mechanism requires (18) both metal ions to participate throughout the catalysis by interacting with the intermediates (Fig. 1A). We speculate that the active participation of metal ions with the intermediates is more appropriate if the two metal ions are positioned at M1 and M2 rather than M1 and M2′. However, more DHBPS structures are required to validate the position of these metal ions, especially in the presence of Mg2+.
FIGURE 9.
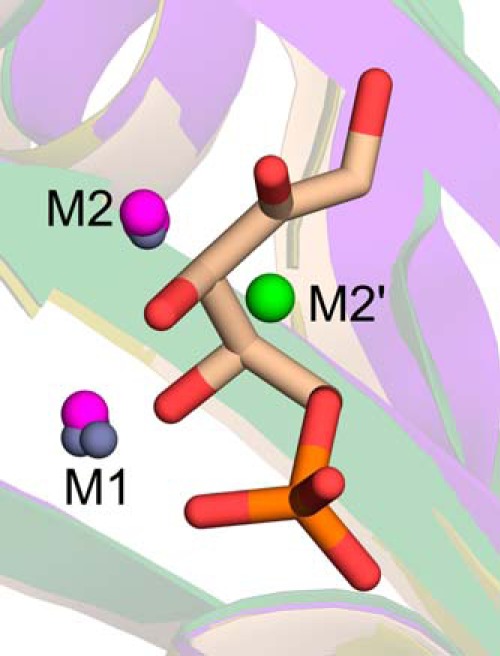
Comparison of position of metal ions in DHBPS. Superposition of two Zn2+ (M1 and M2; gray) from E-Ru5P-Zn2+, one Zn2+ (M1; gray), and one Mg2+ (M2; purple) from S. typhimurium and two Mg2+ (M1 and M2; purple) from M. grisea, one Zn2+ (M1; gray), and one Ca2+ (M2′; green) from M. jannaschii is shown. The Ru5P from E-Ru5P-Zn2+ is shown in sticks for reference. The M1 and M2 positions are observed in all three species, whereas M2′ is observed only in M. jannaschii.
A complex reaction mechanism with several intermediates has been proposed for the catalysis of DHBP formation (Fig. 1A) based on experiments with 13C labeling of substrate (17, 18). Briefly, the reaction starts with the abstraction of the C3 hydrogen atom of Ru5P (I) in concert with the donation of a proton to the formation of 2,3-enediolate (II). The next step involves dehydration of 2,3-enediolate, resulting in enol (III) formation. The enol (III) is converted to 2,3-diketo form (IV) via an acid-base ketonization process. The formation of V by 1,2-skeleton rearrangement of IV is mediated by deprotonation of the C4 hydroxyl group of the substrate. The next step is hydration of V by the Mg2+-associated water molecule to yield an aldehyde intermediate (VI). The retro-aldol reaction of VI with proton donation yields the enolate form of DHBP (VII). Finally, DHBP is generated by protonation of the enolate by a Mg2+-activated water molecule, giving correct stereochemistry (32, 33, 35) for l-DHBP (VII).
Crystal structures of DHBPS from M. grisea complexed with glycerol and metal ions (32) and an active site mutant of DHBPS structure from M. jannaschii complexed with metal ions and substrate Ru5P have revealed the amino acids involved in the catalysis (33). The vDHBPS structure complexed with substrate and metal ions essentially confirms the amino acids involved in the catalysis as reported earlier (32, 33). In the E-Ru5P-Zn2+ complex, Ru5P exists in the 2-keto form along with metal ions (I). The reaction cannot proceed further because it is generally assumed that Zn2+ and Ca2+ ions are not strong enough as Lewis acids, thus freezing the substrate in the 2-keto form (33). Similarly, in the E-4PEH-Zn2+ complex, the 4PEH also exists in the 2-keto form bound with Zn2+ ions. However, in the presence of magnesium ions, which are required for DHBPS catalytic activity, the reaction may not able to form the enol (III) intermediate (Fig. 1) because the hydroxylamine group will not allow the dehydration of 2,3-enediolate, thus inhibiting the catalytic activity of DHBPS. It is also possible that the 2,3-enediolate intermediate is not formed at all with 4PEH as the inhibitor may exist as a stable hydroximate complex with Mg2+, which would therefore not induce dehydration of the NOH group, thus leading to inhibition (Fig. 10) of vDHBPS.
FIGURE 10.
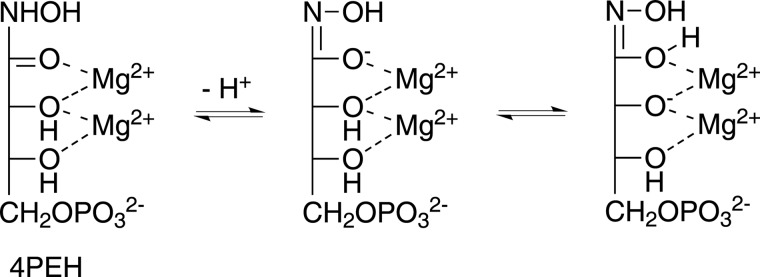
Proposed mechanism of inhibition by 4PEH. Hydroxamic acids can exist in hydroximic acid form or hydroximate following nitrogen deprotonation. The 4PEH may exist as a stable hydroximate complex with Mg2+ that is unable to induce dehydration of the NOH group and thus leads to inhibition.
In summary, for the first time, we have characterized vDHBPS, a potential antibacterial drug target involved in the riboflavin biosynthesis pathway, with a competitive inhibitor, 4PEH. The structural characterization of vDHBPS along with 4PEH revealed that the inhibitor is bound to the active site and offers insights into the mechanism of inhibition. Interestingly, 4PEH has also been shown to be a competitive inhibitor of ribose-5-phosphate isomerase B from M. tuberculosis (60). Taken together, 4PEH or its derivatives could be explored as a potential drug molecule to combat the ever increasing drug-resistant bacterial strains by inhibiting the essential riboflavin biosynthesis pathway.
Acknowledgments
We thank Dr. Hassan Belhrali for help during data collection at European Synchrotron Radiation Facility, Grenoble, France. We also grateful for the help of Dr. Balvinder Singh in improving the manuscript. Dr. Emmanuel Burgos, currently at the Albert Einstein College of Medicine (Bronx, NY) is gratefully acknowledged for the synthesis of 4PEH during his Ph.D. thesis at Orsay (France). We also thank Dr. Saumya Raychaudhuri for providing genomic DNA of V. cholerae.
This work was supported in part by CSIR Network Projects BSC-210 and BSC-104 and by the Department of Biotechnology, Government of India (for synchrotron data collection in France).
- Ru5P
- d-ribulose 5-phosphate
- DHBPS
- 3,4-dihydroxy-2-butanone-4-phosphate synthase
- vDHBPS
- DHBPS from V. cholerae
- 4PEH
- 4-phospho-d-erythronohydroxamic acid
- DHBP
- 3,4-dihydroxy-2-butanone 4-phosphate
- Loop1
- acidic active site loop.
REFERENCES
- 1. Bacher A. (1991) in Chemistry and Biochemistry of Flavoenzymes (Müller F., ed) Vol. I, pp. 349–370, CRC Press, Boca Raton, FL [Google Scholar]
- 2. Cole S. T., Eiglmeier K., Parkhill J., James K. D., Thomson N. R., Wheeler P. R., Honoré N., Garnier T., Churcher C., Harris D., Mungall K., Basham D., Brown D., Chillingworth T., Connor R., Davies R. M., Devlin K., Duthoy S., Feltwell T., Fraser A., Hamlin N., Holroyd S., Hornsby T., Jagels K., Lacroix C., Maclean J., Moule S., Murphy L., Oliver K., Quail M. A., Rajandream M. A., Rutherford K. M., Rutter S., Seeger K., Simon S., Simmonds M., Skelton J., Squares R., Squares S., Stevens K., Taylor K., Whitehead S., Woodward J. R., Barrell B. G. (2001) Massive gene decay in the leprosy bacillus. Nature 409, 1007–1011 [DOI] [PubMed] [Google Scholar]
- 3. Gerdes S. Y., Scholle M. D., D'Souza M., Bernal A., Baev M. V., Farrell M., Kurnasov O. V., Daugherty M. D., Mseeh F., Polanuyer B. M., Campbell J. W., Anantha S., Shatalin K. Y., Chowdhury S. A., Fonstein M. Y., Osterman A. L. (2002) From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 184, 4555–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sassetti C. M., Boyd D. H., Rubin E. J. (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 [DOI] [PubMed] [Google Scholar]
- 5. Bacher A., Eberhardt S., Richter G. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology (Neidhardt F. C., Ingraham J. L., Low K. B., Magasanik B., Schaechter M., Umbarger H. E., eds), Vol. 1, 2nd Ed., pp. 657–664, ASM Press, Washington, D. C. [Google Scholar]
- 6. Philipp W. J., Poulet S., Eiglmeier K., Pascopella L., Balasubramanian V., Heym B., Bergh S., Bloom B. R., Jacobs W. R., Jr., Cole S. T. (1996) An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc. Natl. Acad. Sci. U.S.A. 93, 3132–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Griffin J. E., Gawronski J. D., Dejesus M. A., Ioerger T. R., Akerley B. J., Sassetti C. M. (2011) High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7, e1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Long Q., Ji L., Wang H., Xie J. (2010) Riboflavin biosynthetic and regulatory factors as potential novel anti-infective drug targets. Chem. Biol. Drug Des. 75, 339–347 [DOI] [PubMed] [Google Scholar]
- 9. Fassbinder F., Kist M., Bereswill S. (2000) Structural and functional analysis of the riboflavin synthesis genes encoding GTP cyclohydrolase II (ribA), DHBP synthase (ribBA), riboflavin synthase (ribC), and riboflavin deaminase/reductase (ribD) from Helicobacter pylori strain P1. FEMS Microbiol. Lett. 191, 191–197 [DOI] [PubMed] [Google Scholar]
- 10. Bacher A., Richter G., Ritz H., Eberhardt S., Fischer M., Krieger C. (1997) Biosynthesis of riboflavin: GTP cyclohydrolase II, deaminase, and reductase. Methods Enzymol. 280, 382–389 [DOI] [PubMed] [Google Scholar]
- 11. Burrows R. B., Brown G. M. (1978) Presence of Escherichia coli of a deaminase and a reductase involved in biosynthesis of riboflavin. J. Bacteriol. 136, 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Foor F., Brown G. M. (1975) Purification and properties of guanosine triphosphate cyclohydrolase II from Escherichia coli. J. Biol. Chem. 250, 3545–3551 [PubMed] [Google Scholar]
- 13. Foor F., Brown G. M. (1980) GTP cyclohydrolase II from Escherichia coli. Methods Enzymol. 66, 303–307 [DOI] [PubMed] [Google Scholar]
- 14. Ren J., Kotaka M., Lockyer M., Lamb H. K., Hawkins A. R., Stammers D. K. (2005) GTP cyclohydrolase II structure and mechanism. J. Biol. Chem. 280, 36912–36919 [DOI] [PubMed] [Google Scholar]
- 15. Richter G., Fischer M., Krieger C., Eberhardt S., Lüttgen H., Gerstenschläger I., Bacher A. (1997) Biosynthesis of riboflavin: characterization of the bifunctional deaminase-reductase of Escherichia coli and Bacillus subtilis. J. Bacteriol. 179, 2022–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Richter G., Ritz H., Katzenmeier G., Volk R., Kohnle A., Lottspeich F., Allendorf D., Bacher A. (1993) Biosynthesis of riboflavin: cloning, sequencing, mapping, and expression of the gene coding for GTP cyclohydrolase II in Escherichia coli. J. Bacteriol. 175, 4045–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Volk R., Bacher A. (1990) Studies on the 4-carbon precursor in the biosynthesis of riboflavin. Purification and properties of l-3,4-dihydroxy-2-butanone-4-phosphate synthase. J. Biol. Chem. 265, 19479–19485 [PubMed] [Google Scholar]
- 18. Volk R., Bacher A. (1991) Biosynthesis of riboflavin. Studies on the mechanism of l-3,4-dihydroxy-2-butanone 4-phosphate synthase. J. Biol. Chem. 266, 20610–20618 [PubMed] [Google Scholar]
- 19. Kis K., Bacher A. (1995) Substrate channeling in the lumazine synthase/riboflavin synthase complex of Bacillus subtilis. J. Biol. Chem. 270, 16788–16795 [DOI] [PubMed] [Google Scholar]
- 20. Kis K., Volk R., Bacher A. (1995) Biosynthesis of riboflavin. Studies on the reaction mechanism of 6,7-dimethyl-8-ribityllumazine synthase. Biochemistry 34, 2883–2892 [DOI] [PubMed] [Google Scholar]
- 21. Plaut G. W. (1960) Studies on the stoichiometry of the enzymic conversion of 6,7-dimethyl-8-ribityllumazine to riboflavin. J. Biol. Chem. 235, PC41–42 [PubMed] [Google Scholar]
- 22. Plaut G. W., Beach R. L., Aogaichi T. (1970) Studies on the mechanism of elimination of protons from the methyl groups of 6,7-dimethyl-8-ribityllumazine by riboflavin synthetase. Biochemistry 9, 771–785 [DOI] [PubMed] [Google Scholar]
- 23. Fischer M., Bacher A. (2008) Biosynthesis of vitamin B2: structure and mechanism of riboflavin synthase. Arch. Biochem. Biophys. 474, 252–265 [DOI] [PubMed] [Google Scholar]
- 24. Wang W., Kim R., Jancarik J., Yokota H., Kim S. H. (2003) Crystal structure of a flavin-binding protein from Thermotoga maritima. Proteins 52, 633–635 [DOI] [PubMed] [Google Scholar]
- 25. Herguedas B., Martínez-Júlvez M., Frago S., Medina M., Hermoso J. A. (2010) Oligomeric state in the crystal structure of modular FAD synthetase provides insights into its sequential catalysis in prokaryotes. J. Mol. Biol. 400, 218–230 [DOI] [PubMed] [Google Scholar]
- 26. Frago S., Martínez-Júlvez M., Serrano A., Medina M. (2008) Structural analysis of FAD synthetase from Corynebacterium ammoniagenes. BMC Microbiol. 8, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li J., Hua Z., Miao L., Jian T., Wei Y., Shasha Z., Shaocheng Z., Zhen G., Hongpeng Z., Ailong H., Deqiang W. (2013) The crystal structure and biochemical properties of DHBPS from Streptococcus pneumoniae, a potential anti-infective target for Gram-positive bacteria. Protein Expr. Purif. 91, 161–168 [DOI] [PubMed] [Google Scholar]
- 28. Singh M., Kumar P., Yadav S., Gautam R., Sharma N., Karthikeyan S. (2013) The crystal structure reveals the molecular mechanism of bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP cyclohydrolase II (Rv1415) from Mycobacterium tuberculosis. Acta Crystallogr. D Biol. Crystallogr. 69, 1633–1644 [DOI] [PubMed] [Google Scholar]
- 29. Richter G., Volk R., Krieger C., Lahm H. W., Röthlisberger U., Bacher A. (1992) Biosynthesis of riboflavin: cloning, sequencing, and expression of the gene coding for 3,4-dihydroxy-2-butanone 4-phosphate synthase of Escherichia coli. J. Bacteriol. 174, 4050–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kelly M. J., Ball L. J., Krieger C., Yu Y., Fischer M., Schiffmann S., Schmieder P., Kühne R., Bermel W., Bacher A., Richter G., Oschkinat H. (2001) The NMR structure of the 47-kDa dimeric enzyme 3,4-dihydroxy-2-butanone-4-phosphate synthase and ligand binding studies reveal the location of the active site. Proc. Natl. Acad. Sci. U.S.A. 98, 13025–13030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liao D. I., Calabrese J. C., Wawrzak Z., Viitanen P. V., Jordan D. B. (2001) Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase of riboflavin biosynthesis. Structure 9, 11–18 [DOI] [PubMed] [Google Scholar]
- 32. Liao D. I., Zheng Y. J., Viitanen P. V., Jordan D. B. (2002) Structural definition of the active site and catalytic mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase. Biochemistry 41, 1795–1806 [DOI] [PubMed] [Google Scholar]
- 33. Steinbacher S., Schiffmann S., Richter G., Huber R., Bacher A., Fischer M. (2003) Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii in complex with divalent metal ions and the substrate ribulose 5-phosphate: implications for the catalytic mechanism. J. Biol. Chem. 278, 42256–42265 [DOI] [PubMed] [Google Scholar]
- 34. Echt S., Bauer S., Steinbacher S., Huber R., Bacher A., Fischer M. (2004) Potential anti-infective targets in pathogenic yeasts: structure and properties of 3,4-dihydroxy-2-butanone 4-phosphate synthase of Candida albicans. J. Mol. Biol. 341, 1085–1096 [DOI] [PubMed] [Google Scholar]
- 35. Kumar P., Singh M., Gautam R., Karthikeyan S. (2010) Potential anti-bacterial drug target: structural characterization of 3,4-dihydroxy-2-butanone-4-phosphate synthase from Salmonella typhimurium LT2. Proteins 78, 3292–3303 [DOI] [PubMed] [Google Scholar]
- 36. Singh M., Kumar P., Karthikeyan S. (2011) Structural basis for pH dependent monomer-dimer transition of 3,4-dihydroxy 2-butanone-4-phosphate synthase domain from Mycobacterium tuberculosis. J. Struct. Biol. 174, 374–384 [DOI] [PubMed] [Google Scholar]
- 37. Salama N. R., Shepherd B., Falkow S. (2004) Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J Bacteriol. 186, 7926–7935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Becker D., Selbach M., Rollenhagen C., Ballmaier M., Meyer T. F., Mann M., Bumann D. (2006) Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440, 303–307 [DOI] [PubMed] [Google Scholar]
- 39. Rollenhagen C., Bumann D. (2006) Salmonella enterica highly expressed genes are disease specific. Infect. Immun. 74, 1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 41. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 42. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 43. Picollelli M. A., Viitanen P. V., Jordan D. B. (2000) Spectrophotometric determination of 3,4-dihydroxy-2-butanone-4-phosphate synthase activity. Anal. Biochem. 287, 347–349 [DOI] [PubMed] [Google Scholar]
- 44. Teng T.-Y. (1990) Mounting of crystals for macromolecular crystallography in a free-standing thin film. J. Appl. Crystallogr. 23, 387–391 [Google Scholar]
- 45. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. French S., Wilson K. (1978) On the treatment of negative intensity observations. Acta Crystallogr. A 34, 517–525 [Google Scholar]
- 47. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kantardjieff K. A., Rupp B. (2003) Matthews coefficient probabilities: improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 12, 1865–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brünger A. T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 52. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 54. Adams P. D., Pannu N. S., Read R. J., Brünger A. T. (1997) Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl. Acad. Sci. U.S.A. 94, 5018–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Adams P. D., Read R. J., Zwart P. H., Hung L.-W. (2008) Iterative-build OMIT maps: map improvement by iterative model building and refinement without model bias. Acta Crystallogr. D Biol. Crystallogr. 64, 515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 57. Kleywegt G. J., Jones T. A. (1995) Where freedom is given, liberties are taken. Structure 3, 535–540 [DOI] [PubMed] [Google Scholar]
- 58. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.2r3pre, Schrödinger LLC, New York [Google Scholar]
- 59. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 60. Roos A. K., Burgos E., Ericsson D. J., Salmon L., Mowbray S. L. (2005) Competitive inhibitors of Mycobacterium tuberculosis ribose-5-phosphate isomerase B reveal new information about the reaction mechanism. J. Biol. Chem. 280, 6416–6422 [DOI] [PubMed] [Google Scholar]
- 61. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robert X., Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]