Background: Yeast Pah1 phosphatidate phosphatase required for triacylglycerol synthesis is subject to proteasome-mediated degradation.
Results: Pah1 is degraded by the 20S proteasome in a ubiquitin-independent manner that is governed by its phosphorylation state.
Conclusion: 20S proteasomal degradation of Pah1 is regulated by phosphorylation and dephosphorylation.
Significance: Pah1 function in lipid metabolism is regulated by the 20S proteasome.
Keywords: Diacylglycerol, Lipid, Phosphatase, Phosphatidate, Proteasome, Triacylglycerol, Yeast, Phosphatidate Phosphatase
Abstract
Saccharomyces cerevisiae Pah1 phosphatidate phosphatase, which catalyzes the conversion of phosphatidate to diacylglycerol for triacylglycerol synthesis and simultaneously controls phosphatidate levels for phospholipid synthesis, is subject to the proteasome-mediated degradation in the stationary phase of growth. In this study, we examined the mechanism for its degradation using purified Pah1 and isolated proteasomes. Pah1 expressed in S. cerevisiae or Escherichia coli was not degraded by the 26S proteasome, but by its catalytic 20S core particle, indicating that its degradation is ubiquitin-independent. The degradation of Pah1 by the 20S proteasome was dependent on time and proteasome concentration at the pH optimum of 7.0. The 20S proteasomal degradation was conserved for human lipin 1 phosphatidate phosphatase. The degradation analysis using Pah1 truncations and its fusion with GFP indicated that proteolysis initiates at the N- and C-terminal unfolded regions. The folded region of Pah1, in particular the haloacid dehalogenase-like domain containing the DIDGT catalytic sequence, was resistant to the proteasomal degradation. The structural change of Pah1, as reflected by electrophoretic mobility shift, occurs through its phosphorylation by Pho85-Pho80, and the phosphorylation sites are located within its N- and C-terminal unfolded regions. Phosphorylation of Pah1 by Pho85-Pho80 inhibited its degradation, extending its half-life by ∼2-fold. The dephosphorylation of endogenously phosphorylated Pah1 by the Nem1-Spo7 protein phosphatase, which is highly specific for the sites phosphorylated by Pho85-Pho80, stimulated the 20S proteasomal degradation and reduced its half-life by 2.6-fold. These results indicate that the proteolysis of Pah1 by the 20S proteasome is controlled by its phosphorylation state.
Introduction
Pah1 PAP2 (1), the Saccharomyces cerevisiae ortholog of the mammalian lipin 1, 2, and 3 PAP enzymes (2, 3), catalyzes the Mg2+-dependent dephosphorylation of PA to form DAG and Pi (4) (Fig. 1). It has emerged as one of the most highly regulated enzymes that controls lipid synthesis in yeast (5–7). This may be explained by the fact that PA is a common substrate that is partitioned to DAG and CDP-DAG, which are used for the synthesis of the neutral lipid TAG and membrane phospholipids, respectively (5–7) (Fig. 1A). In yeast supplemented with choline or ethanolamine, the DAG produced by PAP activity may also be used to synthesize the phospholipids phosphatidylcholine or phosphatidylethanolamine, respectively, via the Kennedy pathway (5, 6). In contrast to PAP, the Cds1 (8)/Tam41 (9) CDP-DAG synthases, which catalyze the synthesis of CDP-DAG from PA (10) (Fig. 1A), are not known to be highly regulated lipid biosynthetic enzymes (6).
FIGURE 1.
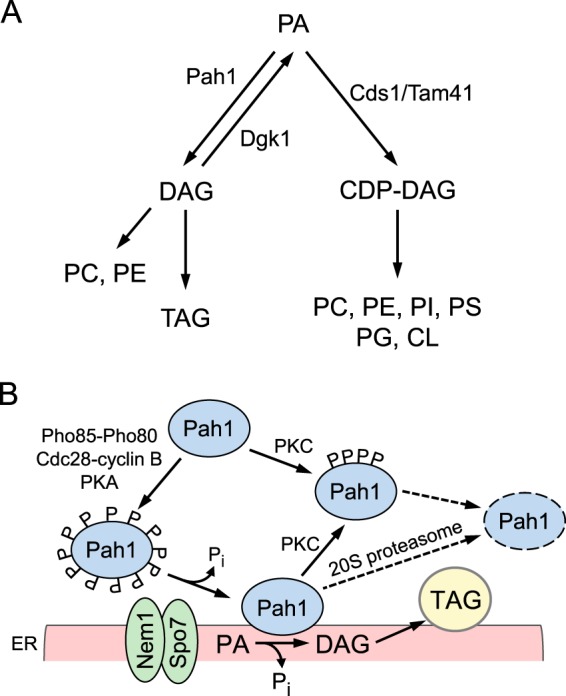
Lipid synthesis pathways in S. cerevisiae and a model for the regulation of Pah1 by phosphorylation, dephosphorylation, and 20S proteasomal degradation. A, the pathways shown in the figure include the relevant steps discussed in this work. More comprehensive pathways for lipid synthesis may be found (5, 6). Pah1 PAP is found at a branch point where PA is partitioned to DAG and CDP-DAG for the synthesis of TAG or membrane phospholipids, respectively. B, Pah1 in the cytosol is phosphorylated by multiple protein kinases (e.g. Pho85-Pho80, Cdc28-cyclin B, protein kinase A, and protein kinase C). The phosphorylated enzyme (indicated by the letter P) translocates to the ER membrane through its dephosphorylation by the Nem1-Spo7 phosphatase complex. Dephosphorylated Pah1 associated with the membrane catalyzes the conversion of PA to DAG, which is acylated to form TAG. Dephosphorylated Pah1 or protein kinase C-phosphorylated Pah1 that is not phosphorylated at the target sites for Pho85-Pho80/Cdc28-cyclin B is degraded by the 20S proteasome (indicated by the dashed line arrows and ellipse). PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine, PG, phosphatidylglycerol; CL, cardiolipin; PKA, protein kinase A; PKC, protein kinase C.
Roles of PAP for lipid synthesis in S. cerevisiae are manifested during cell growth. In the exponential phase, PAP activity is relatively low, and PA is primarily partitioned to CDP-DAG for the synthesis of membrane phospholipids (11–13). As the cells progress into the stationary phase, PAP activity is increased, and PA is primarily converted to DAG for the synthesis of TAG (11–13). In addition, the different levels of PAP activity play an important role in the PA-mediated control of Opi1, a transcriptional repressor that attenuates the expression of several phospholipid synthesis genes by binding to Ino2 of the Ino2-Ino4 transcriptional activator complex (5, 6, 14–16). The affinity of PA to the Opi1 repressor at the nuclear/ER membrane prevents its nuclear translocation and thereby inhibits its repressor function (15). Thus, the elevated PA content caused by lower PAP activity effects the tethering of Opi1 to the nuclear/ER membrane and the derepression of gene expression, whereas reduced PA content caused by higher PAP activity effects the Opi1 nuclear translocation and the repression of gene expression (17–19).
The importance of PAP activity for regulating lipid synthesis in yeast is epitomized by distinct phenotypes of the cells lacking the enzyme, many of which are intimately related to the increased level of PA and the decreased levels of DAG and TAG (1, 12, 17, 19, 20). In particular, elevated PA content causes the derepression of phospholipid synthesis genes (e.g. INO1 and OPI3) and the aberrant expansion of the nuclear/ER membrane, whereas reduced DAG and TAG contents cause the susceptibility to fatty acid-induced toxicity and defects in lipid droplet formation (1, 12, 17, 19, 20). Some of these phenotypes require expression of Dgk1 DAG kinase (12, 20, 21), the enzyme that converts DAG back to PA (Fig. 1A). The impact of PAP on overall cell physiology is further shown by the fact that PAP mutant cells are unable to grow on non-fermentable carbon sources (e.g. respiratory deficiency) (1, 22) or at elevated temperatures (1, 17, 22) and exhibit defects in cell wall integrity (23, 24) and vacuole fusion (e.g. as related to protein trafficking) (25). With respect to mammalian cell physiology, defects in lipin PAP enzymes result in metabolic disorders that include lipodystrophy, insulin resistance, peripheral neuropathy, rhabdomyolysis, and inflammation (2, 26–37).
Studies with Pah1 PAP have significantly advanced the understanding of its mode of action and regulation (7, 38). PAP activity, which is dependent on Mg2+, is directed by the conserved DXDX(T/V) catalytic motif within a HAD-like domain and by the conserved glycine residue within the NLIP domain (1, 19). Pah1 is regulated by phosphorylation and dephosphorylation for its subcellular localization, catalytic activity, and abundance (17, 18, 39–45). With more than 30 phosphorylation sites (18, 46–51), it is one of the most heavily phosphorylated proteins in S. cerevisiae and has been shown to be a target for multiple protein kinases in vitro (49, 52). Pah1 as a bona fide substrate has been confirmed for Pho85-Pho80 (40), Cdc28-cyclin B (39), protein kinase A (41), and protein kinase C (42). The physiological relevance of Pah1 phosphorylation by these protein kinases has been shown by analysis of cells expressing phosphorylation-deficient mutant forms of the enzyme (18, 39–43). Phosphorylation of Pah1 by Pho85-Pho80, Cdc28-cyclin B, and protein kinase A sequesters the enzyme in the cytosol apart from the membrane, where its substrate PA is present (39–41, 43) (Fig. 1B). Pah1 is dephosphorylated by the Nem1-Spo7 protein phosphatase complex located at the nuclear/ER membrane (17, 53). It is this dephosphorylation that activates Pah1 on the membrane surface (17, 18, 39–41, 43, 44, 53) (Fig. 1B). In this process, the acidic tail of Pah1 plays a role for interaction with Nem1-Spo7 (44), and its N-terminal amphipathic helix mediates association with the membrane surface (43). Phosphorylation of Pah1 by Pho85-Pho80 or protein kinase A inhibits PAP activity (40, 41), whereas its dephosphorylation by Nem1-Spo7 stimulates the enzyme activity (54). Pah1 is also regulated for PAP activity by membrane-associated and cytosolic factors. Negatively charged phospholipids (e.g. CDP-DAG and phosphatidylinositol) stimulate PAP activity by a mechanism that increases the affinity of Pah1 for PA (55), whereas the positively charged sphingoid bases (e.g. sphinganine and phytosphingosine) inhibit the enzyme activity by a mechanism that excludes PA from the enzyme binding site (56). ATP and CTP, nucleotides that serve as precursors in membrane phospholipid synthesis (5, 6), inhibit PAP activity by a complex mechanism that affects catalytic efficiency and by chelation of Mg2+ (57).
Expression of the PAP-encoding gene PAH1 is regulated by growth phase and nutrient availability (13, 58). Its expression is induced as yeast cells progress into the stationary phase when TAG synthesis is increased at the expense of membrane phospholipids (11), and the growth phase regulation is enhanced by inositol supplementation (13, 58). The PAH1 expression is also induced when exponential phase cells are deprived of the essential mineral zinc. Unlike the growth phase regulation, the zinc-mediated regulation of PAP channels its product DAG to the synthesis of phosphatidylcholine via the CDP-choline branch of the Kennedy pathway (13, 58). Regulation of the PAH1 expression by growth phase and nutrient status involves transcription factors (e.g. Ino2, Ino4, Opi1, Gis1, Rph1, and Zap1) that are known to control several phospholipid synthesis genes (5, 6). These transcription factors exert regulatory effects on PAH1 that are generally opposite to those on the phospholipid synthesis genes (5, 6), suggesting that the opposing regulations coordinate a balanced synthesis of membrane phospholipids and TAG.
The induced expression of PAH1 in the stationary phase and by nutrient status correlates with elevated levels of PAP activity, but the overall abundance of Pah1 is shown to decline (13, 39, 40, 58). Considering that only the membrane-associated Pah1 is physiologically active, the enigmatic regulation seems to ensure that the enzyme level is appropriately controlled on the membrane to preserve PA for phospholipid synthesis and/or prevent the toxic effects of DAG (7, 45). Indeed, the overexpression of hyperactive Pah1 results in lethality (18, 39, 45). We have recently shown that the abundance of Pah1 is controlled in stationary phase cells by proteasomal protease activity (45). By exploring the proteasomal regulation, we show here that Pah1 is degraded by the 20S proteasome in a ubiquitin-independent manner and that the proteasomal degradation is governed by its phosphorylation state.
EXPERIMENTAL PROCEDURES
Reagents
Growth medium supplies were obtained from Difco. Plasmid DNA purification kits and nickel-nitrilotriacetic acid-agarose resin were from Qiagen. PCR primers, protease inhibitors, Triton X-100, MG132, and bovine serum albumin were obtained from Sigma-Aldrich. Clontech was the source of mouse anti-GFP antibodies. IgG-Sepharose, Q-Sepharose, PVDF membrane, mouse anti-His6 antibodies, and the enhanced chemifluorescence Western blotting detection kit were from GE Healthcare. His6-tagged tobacco etch virus protease was purchased from Invitrogen. New England Biolabs was the source of enzymes for DNA manipulations. Protein kinase A catalytic subunit and conventional protein kinase C were from Promega. Bio-Rad was the source of reagents for electrophoresis, Western blotting, protein determination, molecular mass protein standards, and DNA size ladders. Alkaline phosphatase-conjugated goat anti-rabbit antibodies and alkaline phosphatase-conjugated goat anti-mouse antibodies were from Thermo Scientific and Pierce, respectively. Thermo Scientific was the source of S. cerevisiae BY4741 TAP-tagged fusion proteins. All other chemicals were reagent grade.
Strains and Growth Conditions
E. coli strains DH5α and BL21(DE3)pLysS were used for the propagation of plasmids and for the expression of His6-tagged proteins, respectively. Cells were grown at 37 °C in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7.0). For the selection of cells carrying plasmids for the expression of proteins, the growth medium was supplemented with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) (59). The expression of proteins in cells bearing derivatives of plasmid pET-15b was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside. Yeast cells expressing TAP-tagged proteins were grown at 30 °C in YEPD medium (1% yeast extract, 2% peptone, and 2% glucose). Cell numbers in liquid cultures were determined spectrophotometrically at an absorbance of 600 nm.
DNA Manipulations
Isolation of DNA, digestion and ligation of DNA, and PCR amplification of DNA were performed by standard practices (60, 61). The plasmids used in this study are listed in Table 1. Plasmid pGH313, a derivative of pET-15b, directs the isopropyl β-d-1-thiogalactopyranoside-induced expression of His6-tagged Pah1 in E. coli (1). pLH101 and pLH102 were constructed by inserting the GFP-PAH1 and PAH1-GFP coding sequences, respectively, into the XhoI and BamHI sites in pET-15b. pLH103 was constructed by inserting the human SNCA coding sequence, which was amplified by PCR from p426GPD-αSyn (62), into the NdeI and XhoI sites in pET15b. pLH104 was constructed by inserting the GFP coding sequence, which was amplified by PCR from YCplac111-SEC63-GFP (17), into the XhoI and BamHI sites in pET-15b. pGH313(1–821) and pGH313(1–591) were constructed from pGH313 by generating a nonsense mutation at codon 822 and 592, respectively. pGH313(235–862) and pGH313(360–862) were constructed from pGH313 by replacing codons 1–234 and 1–359, respectively, with a start codon. pGH313(360–591) was constructed from pGH313(1–591) by replacing codons 1–359 with a start codon. GFP and PAH1 coding sequences were amplified by PCR from YCplac111-SEC63-GFP (17) and pGH313, respectively. The fusion coding sequences for GFP-PAH1 and PAH1-GFP with XhoI and BamHI sites were generated by overlap extension PCR (63). All of the PAH1 and GFP constructs were confirmed by DNA sequencing. Plasmid transformation of E. coli was performed as described previously (60).
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Source/Reference |
|---|---|---|
| Strain | ||
| E. coli | ||
| DH5α | F− φ80dlacZΔM15Δ (lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rk− mk+) phoA supE44 l−thi-1 gyrA96 relA1 | Ref. 60 |
| BL21(DE3)pLysS | F− ompT hsdSB (rB−mB−) gal dcm (DE3) pLysS | Novagen |
| S. cerevisiae | ||
| BY4741-RPN11-TAP | TAP-tagged Rpn11 expressed in strain BY4741 | Thermo Scientific |
| BY4741-PRE1-TAP | TAP-tagged Pre1 expressed in strain BY4741 | Thermo Scientific |
| BY4741-PAH1-TAP | TAP-tagged Pah1 expressed in strain BY4741 | Thermo Scientific |
| Plasmid | ||
| pGH313 | PAH1 coding sequence inserted (1–862 full length) into pET-15b | Ref. 1 |
| pGH313(1–821) | PAH1 (1–821 truncation) derivative of pGH313 | This study |
| pGH313(1–752) | PAH1 (1–752 truncation) derivative of pGH313 | Ref. 41 |
| pGH313(1–591) | PAH1 (1–591 truncation) derivative of pGH313 | This study |
| pGH313(18–862) | PAH1 (18–862 truncation) derivative of pGH313 | Ref. 41 |
| pGH313(235–862) | PAH1 (235–862 truncation) derivative of pGH313 | This study |
| pGH313(360–862) | PAH1 (360–862 truncation) derivative of pGH313 | This study |
| pGH313(235–752) | PAH1 (235–752 truncation) derivative of pGH313 | Ref. 41 |
| pGH313(360–591) | PAH1 (360–591 truncation) derivative of pGH313 | This study |
| pGH322 | LPIN1α coding sequence inserted into pET-28b (+) | Ref. 76 |
| pGH327 | LPIN1β coding sequence inserted into pET-28b (+) | Ref. 76 |
| pGH321 | LPIN1γ coding sequence inserted into pET-28b (+) | Ref. 76 |
| YCplac111-SEC63-GFP | SEC63-GFP derivative of YCplac111 | Ref. 17 |
| pLH101 | GFP-PAH1 coding sequence inserted into pET-15b | This study |
| pLH102 | PAH1-GFP coding sequence inserted into pET-15b | This study |
| p426GPD-αSyn | SNCA derivative of p426GPD | Ref. 62 |
| pLH103 | SNCA coding sequence inserted into pET-15b | This study |
| pLH104 | GFP coding sequence inserted into pET-15b | This study |
Purification of Enzymes, Proteasomes, and α-Synuclein
All steps were performed at 4 °C. His6-tagged wild type and mutant forms of Pah1, His6-tagged Pho85-Pho80 protein kinase complex, His6-tagged human lipin 1 α, β, and γ isoforms, and His6-tagged human α-synuclein expressed in E. coli were purified by affinity chromatography with nickel-nitrilotriacetic acid-agarose as described by Han et al. (59). For phosphorylation experiments, the affinity-purified Pah1 was further purified by ion-exchange chromatography with Q-Sepharose (54). The endogenously phosphorylated TAP-tagged Pah1 was purified from yeast by IgG-Sepharose affinity chromatography as described by O'Hara et al. (18). Protein A-tagged Nem1-Spo7 protein phosphatase complex (64) was purified from yeast by IgG-Sepharose affinity chromatography (54). The 26 and 20S proteasomes were purified from yeast expressing TAP-tagged Rpn11 and TAP-tagged Pre1, respectively, by IgG-Sepharose affinity chromatography as described previously (65, 66). The protein A tag was removed from the fusion proteins by digestion with His6-tagged tobacco etch virus protease, which was removed from purified protein preparations by nickel-nitrilotriacetic acid-agarose chromatography. SDS-PAGE analysis indicated that the protein and proteasome preparations used in this study were purified. Protein content was estimated by the method of Bradford (67) using bovine serum albumin as a standard.
Phosphorylation and Dephosphorylation of Pah1
Purified unphosphorylated Pah1 was phosphorylated with 100 μm ATP by Pho85-Pho80 (40), protein kinase A (41), or protein kinase C (42) for 2 h at 30 °C as described previously. The purified Pah1 that is endogenously phosphorylated in S. cerevisiae (18) was dephosphorylated by pure Nem1-Spo7 phosphatase complex as described by Su et al. (54).
Proteasome Assays
For the 26S proteasome assay, Pah1 (30 nm) was incubated with the 26S proteasome (2 nm) in a reaction mixture containing 50 mm Tris-HCl (pH 7.0), 1 mm ATP, 1 mm MgCl2, and 2% dimethyl sulfoxide (solvent control for the proteasome inhibitor MG132 (68)). For the 20S proteasome assay, Pah1 (30 nm) was incubated with purified 20S proteasome (2 nm) in a reaction mixture containing 50 mm Tris-HCl (pH 7.0), 0.02% SDS, and 2% dimethyl sulfoxide. Where indicated, 50 μm MG132 (dissolved in 2% dimethyl sulfoxide) was added to the assays to inhibit proteasome activity. The reactions were terminated by the addition of 4× Laemmli's buffer (69), followed by SDS-PAGE and Western blotting analysis.
SDS-PAGE and Western Blotting
SDS-PAGE (69) was performed with 8 or 12.5% slab gels. Western blotting (70, 71) with PVDF membrane was performed by standard procedures using rabbit anti-Pah1 antibodies (39) (1 μg/ml), rabbit anti-lipin 1 antibodies (72) (1:5,000 dilution), mouse anti-His6 antibodies (1:5,000 dilution), or mouse anti-GFP antibodies (1:5,000 dilution). These primary antibodies were detected with secondary antibodies (e.g. goat anti-rabbit or anti-mouse) conjugated with alkaline phosphatase at a dilution of 1:5,000. Immune complexes were detected using the enhanced chemifluorescence Western blotting detection kit. Fluorimaging was used to acquire images from Western blots, and the signal intensity of the image was analyzed using ImageQuant software. Signals were in the linear range of detectability.
Analyses of Data
Statistical analyses were performed with SigmaPlot software. The p values < 0.05 were taken as a significant difference.
RESULTS
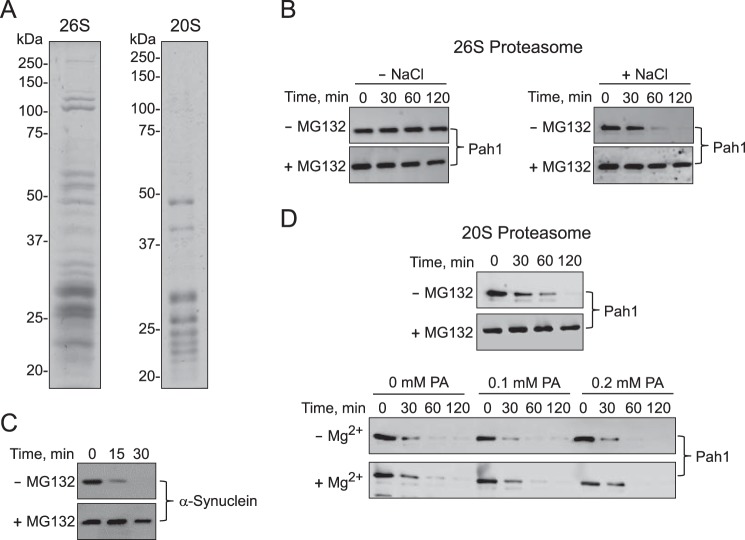
Yeast Pah1 and Human Lipin 1 Are Degraded by the 20S Proteasome but Not by the 26S Proteasome
Our previous work has shown that Pah1 expressed in E. coli, which is not subject to ubiquitination or phosphorylation, is degraded by MG132-inhibitable proteolytic activity in the 100,000 × g pellet fraction of stationary phase cells (45). This result, coupled with the observations that the abundance of Pah1 is stabilized in the stationary phase cells by MG132 or by the lack of Pre1/Pre2 chymotryptic activity of the 20S proteasome, supports the conclusion that Pah1 in stationary phase cells is subject to proteasomal degradation (45). Here, we examined the degradation of purified Pah1 using affinity-purified preparations of 26 and 20S proteasomes (Fig. 2A). The E. coli-expressed Pah1 was resistant to proteolysis by the 26S proteasome (Fig. 2B, left) under assay conditions (e.g. presence of ATP) known to degrade other unmodified proteins (62, 65). As a control, the fidelity of our 26S proteasome preparation was confirmed by its ability to degrade E. coli-expressed α-synuclein (Fig. 2C). The incubation of the 26S proteasome with 0.5 m NaCl dissociates it into the 20S core particle and the 19S regulatory particle (65, 66, 73). When incubated with the salt-treated 26S proteasome, Pah1 was degraded in a time-dependent manner (Fig. 2B, right), suggesting that its degradation occurs by the 20S proteasome. Further analysis with the purified 20S proteasome showed that Pah1 was degraded in a time-dependent manner over a 2-h incubation period (Fig. 2D, top). Next, we questioned whether the degradation was affected by the PAP substrate PA or its cofactor Mg2+ (Fig. 2D, bottom). These experiments were performed in the presence of 2 mm Triton X-100, the non-ionic detergent used to present PA to PAP in uniform Triton X-100/PA-mixed micelles and required for maximum enzyme activity (1, 74, 75). Whereas Triton X-100 stimulated the 20S proteasomal degradation of Pah1 (compare Fig. 2D, top and bottom), neither PA nor Mg2+ ions affected the degradation (Fig. 2D).
FIGURE 2.
Pah1 is degraded by the 20S proteasome, but not by the 26S proteasome. A, the 26S (left) and 20S (right) proteasomes isolated from yeast were separated by SDS-PAGE and stained with Coomassie Brilliant Blue R-250. The electrophoretic patterns of the proteasomal preparations were similar to those described previously (65, 66). The high molecular mass proteins shown in the 26S proteasome preparation are derived from the 19S regulatory particle, and the low molecular mass proteins are derived from the 20S core particle (65, 66). The positions of the molecular mass standards are indicated. B, Pah1 (30 nm) was incubated with the 26S proteasome (2 nm) in a reaction mixture containing 1 mm ATP with or without 50 μm MG132 in the absence or presence of 0.5 m NaCl for the indicated time intervals. C, His6-tagged α-synuclein (400 nm) was incubated with the 26S proteasome (2 nm) and 1 mm ATP with or without 50 μm MG132 for the indicated time intervals. D, top, Pah1 (30 nm) was incubated with the 20S proteasome (2 nm) with or without 50 μm MG132 for the indicated time intervals. D (bottom), the 20S proteasomal degradation of Pah1 was examined with 2 mm Triton X-100 with or without the indicated concentrations of PA in the absence or presence of 1 mm MgCl2. Following the incubations, samples were subjected to SDS-PAGE and Western blotting with anti-Pah1 or anti-His6 antibodies for the analyses of Pah1 or α-synuclein, respectively. The Western blots shown are representative of three independent experiments. The positions of Pah1 and α-synuclein are indicated in the figure.
It is known that SDS facilitates protein entry into the 20S proteasome at low concentrations but denatures and inactivates proteasomal subunits at high concentrations (65, 66, 73). Accordingly, we examined the effects of SDS on the 20S proteasomal degradation of Pah1. SDS was not required for the proteasomal degradation of Pah1, but its addition (0.02–0.09%) to the reaction mixture stimulated (2.5-fold) degradation (Fig. 3A). Accordingly, 0.02% SDS was included in the standard 20S proteasome reaction. The effect of pH on the 20S proteasomal degradation of Pah1 was examined with Tris-maleate-glycine buffer from pH 5.0 to 8.0. Maximum degradation was observed at pH 7.0 (Fig. 3B). Under optimal conditions for SDS and pH, the degradation of Pah1 was dependent on the time of the reaction and the concentration of the 20S proteasome (Fig. 4). The degradation was inhibited by the proteasome inhibitor MG132 (Fig. 4). The 20S proteasomal degradation of Pah1 was not affected by its oxidation with 1 mm hydrogen peroxide, reduction with 1 mm DTT, or denaturation at 100 °C for 5 min (data not shown).
FIGURE 3.
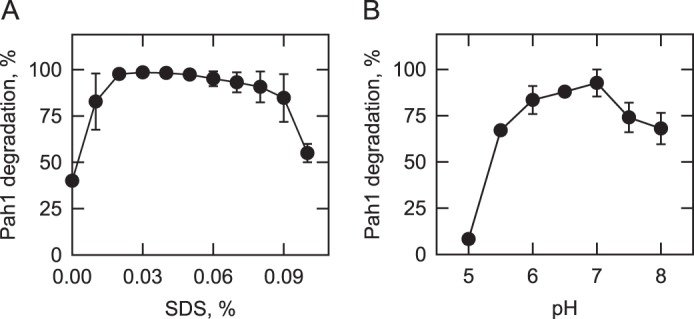
Effects of SDS and pH on the degradation of Pah1 by the 20S proteasome. The proteasomal degradation of Pah1 was examined for 30 min in the presence of the indicated concentrations of SDS (A) and at the indicated pH values with 50 mm Tris-maleate-glycine buffer (B). Following the incubations, samples were subjected to SDS-PAGE and Western blotting with anti-Pah1 antibodies. The relative amounts of Pah1 were quantified using ImageQuant software. The data shown are means ± S.D. (error bars) from triplicate determinations.
FIGURE 4.
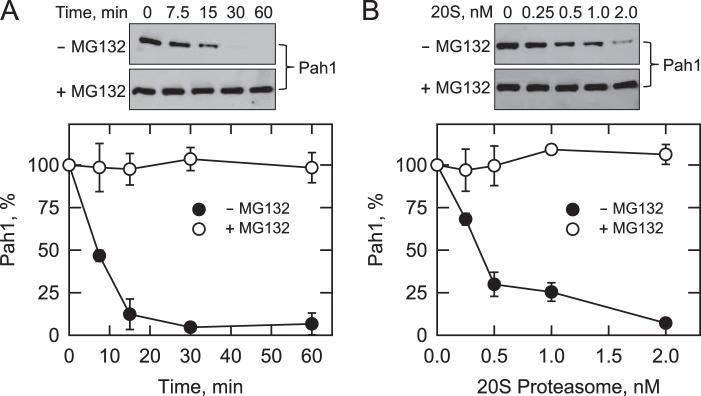
Degradation of Pah1 is dependent on time and the concentration of the 20S proteasome. A, Pah1 (30 nm) was incubated with the 20S proteasome (2 nm) and SDS (0.02%) with or without 50 μm MG132 for the indicated time intervals. B, Pah1 (30 nm) was incubated with the indicated concentrations of the 20S proteasome and SDS (0.02%) for 30 min with or without 50 μm MG132. Following the incubations, samples were subjected to SDS-PAGE and Western blotting with anti-Pah1 antibodies. The relative amounts of Pah1 were quantified using ImageQuant software. The data shown are means ± S.D. (error bars) from triplicate determinations. Representative Western blots of these experiments and the position of Pah1 are shown in the figure.
Because the function of yeast Pah1 is conserved in the mammalian (e.g. mouse and human) lipin PAP enzymes (1, 3, 76) and the biogenesis of the 20S proteasome particle is also conserved (77), we questioned whether E. coli-expressed unmodified human lipin 1 was also subject to degradation by proteasomes. The human lipin 1α isoform was degraded by the 20S proteasome in a time-dependent manner but not degraded by the 26S proteasome (Fig. 5). The other lipin 1 isoforms (β and γ) were similarly degraded by only the 20S proteasome (data not shown).
FIGURE 5.
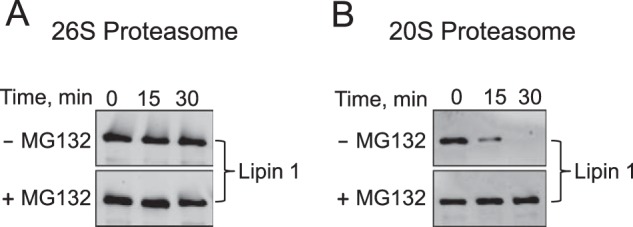
Lipin 1 is degraded by the 20S proteasome but not by the 26S proteasome. A, lipin 1α (30 nm) was incubated with the 26S proteasome (2 nm) in a reaction mixture containing 1 mm ATP with or without 50 μm MG132 for the indicated time intervals. B, lipin 1α (30 nm) was incubated with the 20S proteasome (2 nm) with or without 50 μm MG132 for the indicated time intervals. Following the incubations, samples were subjected to SDS-PAGE and Western blotting with anti-lipin 1 antibodies. The Western blots shown are representative of three independent experiments. The position of lipin 1α is indicated in the figure.
20S Proteasomal Degradation of N- and C-terminal Truncations of Pah1
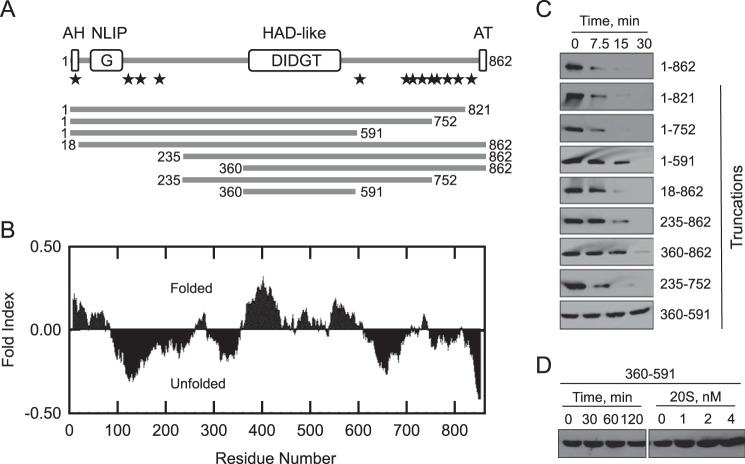
Pah1 contains the evolutionarily conserved NLIP and HAD-like domains (Fig. 6A). Sequence analysis of Pah1 with the FoldIndex algorithm (78) predicts that its folded regions are present at the extreme N terminus, where the amphipathic helix and NLIP domain are found, and in the middle of the protein, where the HAD-like domain is found (Fig. 6B). The program also predicts unfolded regions at both the N and C termini of the protein (Fig. 6B). Because proteins containing intrinsically unstructured regions can be degraded by the 20S proteasome without posttranslational modifications (79, 80), we hypothesized that Pah1 is degraded through its unstructured regions. To explore this hypothesis, a series of N- and C-terminal His6-tagged truncations were expressed and purified from E. coli and were examined for their proteasomal degradation (Fig. 6A). The individual N- or C-terminal truncations containing the unfolded regions were degraded by the 20S proteasome (Fig. 6C). We also examined the proteasomal degradation of Pah1 lacking both the N- and C-terminal regions (Fig. 6C). Truncated Pah1 consisting of residues 235–752 in which the HAD-like domain is flanked by unfolded regions was degraded by the 20S proteasome. The rates of degradation of the various truncations differed, but the reason for this is unclear. Strikingly, the truncated Pah1 consisting of only the HAD-like domain (residues 360–591) was not degraded by the 20S proteasome (Fig. 6C). The resistance of this construct to degradation was observed over a 2-h incubation period and at a 2-fold higher concentration of the 20S proteasome (Fig. 6D).
FIGURE 6.
20S proteasomal degradation of N- and C-terminal truncations of Pah1. A, the domain structure of Pah1 showing the positions of the amphipathic helix (AH); NLIP domain, HAD-like domain containing the DXDX(T/V) catalytic motif; acidic tail (AT); and the positions (denoted by stars) where the enzyme is subject to phosphorylation by Pho85-Pho80 (Ser-110, Ser-114, Ser-168, Ser-602, Thr-723, Ser-744, and Ser-748), Cdc28-cyclin B (Ser-602, Thr-723, and Ser-744), protein kinase A (Ser-10, Ser-677, Ser-773, Ser-774, and Ser-788), and protein kinase C (Ser-677, Ser-769, Ser-773, and Ser-788). Panel A also depicts the truncated forms of Pah1 that were used for 20S proteasomal degradation assays shown in C. B, the Pah1 sequence was analyzed by the FoldIndex algorithm. Regions with positive values or negative values indicate the folded or unfolded forms, respectively (78). C, samples (30 nm) of the full-length and truncated forms of Pah1 were incubated with the 20S proteasome (2 nm) and SDS (0.02%) for the indicated time intervals. D, the truncation of 360–591 (30 nm) was incubated with SDS (0.02%) and 2 nm 20S proteasome for the indicated time intervals or with the indicated concentrations of the 20S proteasome for 120 min. Following the incubations, samples were subjected to SDS-PAGE and Western blotting with anti-His6 antibodies. Portions of the Western blots are shown, which are representative of three independent experiments.
20S Proteasomal Degradation of GFP-Pah1 and Pah1-GFP Fusion Proteins
To further explore the 20S proteasomal degradation of Pah1, we analyzed the protein fused at the N or C terminus with GFP, a highly structured protein that is not subject to degradation by the 20S proteasome (81). Because both fusion proteins were tagged with the His6 epitope at the N terminus, the full-length protein and the degradation products of the reactions were analyzed with anti-His6 and anti-GFP antibodies. The abundance of full-length His6-GFP-Pah1 was reduced in a time-dependent manner along with the production of 70- and 40-kDa proteolytic fragments (Fig. 7A). This degradation pattern was observed in the Western blotting analysis by both the anti-His6 and the anti-GFP antibodies. As mentioned above, Pah1 is predicted to have unfolded regions on both sides of the HAD-like domain (Fig. 6B). The production of the 70-kDa proteolytic fragment indicated that the degradation occurred at the C-terminal region of the fusion protein and stopped at the HAD-like domain. The production of the 40-kDa proteolytic fragment indicated that the degradation of the full-length fusion protein or 70-kDa proteolytic fragment occurred at the N-terminal side of the HAD-like domain and stopped at the NLIP domain. The detection of the 27-kDa proteolytic fragment only by the anti-GFP antibodies indicated that the His6 tag and the N-terminal region of Pah1 are also susceptible to degradation by the proteasome. In a reciprocal analysis, the His6-Pah1-GFP fusion protein was also degraded by the 20S proteasome to produce 51- and 38-kDa proteolytic fragments (Fig. 7B), indicating that degradation of the fusion protein occurred at the C-terminal side of the HAD-like domain. The poor detection of the His6-tagged proteolytic fragments by anti-His6 antibodies indicated that they were too small in size to be detected in our electrophoretic conditions. Overall, these results substantiated the conclusion that the degradation of Pah1 by the 20S proteasome occurs from both the N- and C-terminal unfolded regions of the protein.
FIGURE 7.
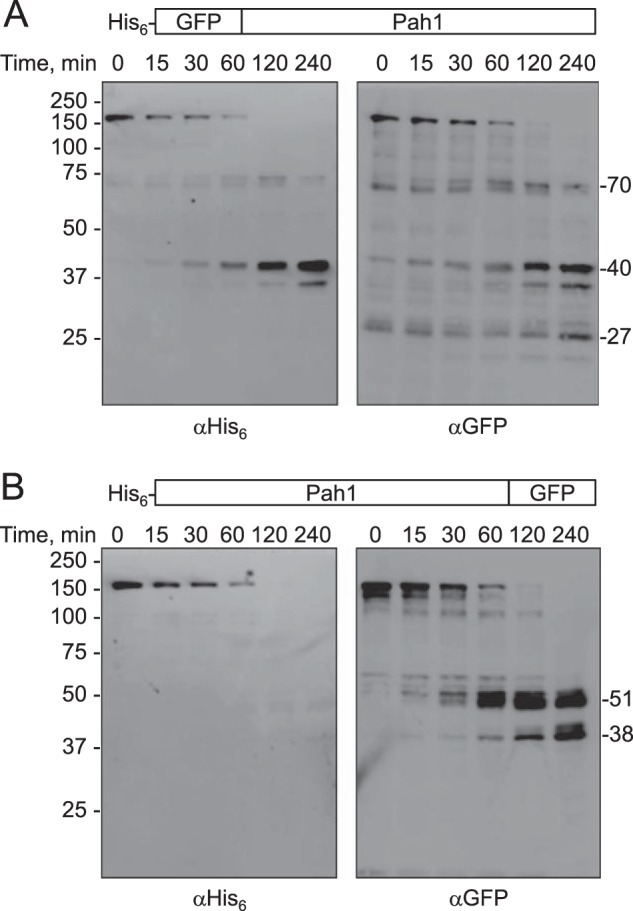
20S proteasomal degradation of GFP-Pah1 and Pah1-GFP fusion proteins. GFP-Pah1 (A) or Pah1-GFP (B) fusion proteins (30 nm each) were incubated with the 20S proteasome (2 nm) and SDS (0.02%) for the indicated time intervals. Following the incubations, samples were subjected to SDS-PAGE and Western blotting with anti-His6 antibodies or anti-GFP antibodies where indicated. The Western blots shown are representative of three independent experiments.
20S Proteasomal Degradation of Pah1 Is Regulated by Its Phosphorylation and Dephosphorylation
Studies with yeast carrying Pah1 with alanine mutations for phosphorylation sites indicate that its abundance in vivo is stabilized through its phosphorylation by Pho85-Pho80 (40), Cdc28-cyclin B (39), or protein kinase A (41). In Pah1, the target phosphorylation sites of the protein kinases are located in the regions that are predicted to be unfolded (Fig. 6A). In addition, Pah1 phosphorylated by Pho85-Pho80 and Cdc28-cyclin B exhibits a shift in electrophoretic mobility, reflecting a structural change by phosphorylation (39, 40, 54). Accordingly, we questioned whether phosphorylation has any effect on the degradation of Pah1 by the 20S proteasome (Fig. 8). Pah1 phosphorylated by Pho85-Pho80, which causes the shift in electrophoretic mobility (Fig. 8, top), was partially resistant to proteasomal degradation and showed a half-life (11 min) nearly 2-fold longer than that of the unphosphorylated enzyme (6 min). The effect of Cdc28-cyclin B on Pah1 degradation was not examined because its three phosphorylation sites are among the seven sites phosphorylated by Pho85-Pho80 (39, 40). Phosphorylation by protein kinase A had a small effect on the reduction of Pah1 degradation, extending its half-life from 6 min (unphosphorylated) to 7.5 min (phosphorylated). The effect of protein kinase A was not observed when Pah1 was phosphorylated by Pho85-Pho80. In contrast to the aforementioned protein kinases, protein kinase C has the effect of decreasing Pah1 abundance when it is not phosphorylated by Pho85-Pho80 (42). Consistent with this finding, the phosphorylation of Pah1 by protein kinase C had a small stimulatory effect on its 20S proteasomal degradation, reducing the half-life of the protein from 6 min (unphosphorylated) to 5 min (phosphorylated).
FIGURE 8.
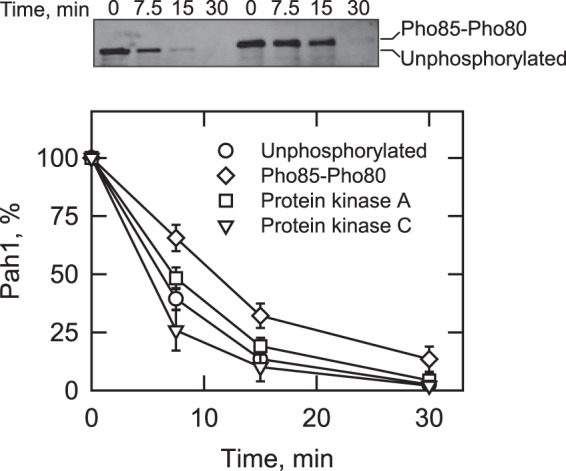
Phosphorylation by Pho85-Pho80, protein kinase A, or protein kinase C regulates the 20S proteasomal degradation of Pah1. Pah1 (30 nm) was phosphorylated with Pho85-Pho80, protein kinase A, or protein kinase C. The unphosphorylated and phosphorylated forms of Pah1 were incubated with the 20S proteasome (2 nm) and SDS (0.02%) for the indicated time intervals. Following the incubations, samples were subjected to SDS-PAGE and Western blotting with anti-Pah1 antibodies. The relative amounts of Pah1 were quantified using ImageQuant software. The data shown are means ± S.D. (error bars) from triplicate determinations. The top panel shows the effect of Pho85-Pho80-mediated phosphorylation on the electrophoretic mobility of Pah1; the positions of the phosphorylated and unphosphorylated forms are indicated. The phosphorylations with protein kinase A (41) or protein kinase C (42) do not affect the electrophoretic mobility of Pah1 (not shown).
The abundance and phosphorylation state of Pah1 are also regulated through its dephosphorylation by Nem1-Spo7 protein phosphatase activity (18, 39–42, 54). Here, we examined the effect of Nem1-Spo7 on the proteasomal degradation of Pah1. The S. cerevisiae-expressed preparation of Pah1, which is heterogeneously phosphorylated in vivo (18, 54), was dephosphorylated by the Nem1-Spo7 protein phosphatase and then examined for its degradation by the 26 and 20S proteasomes. Neither the phosphorylated nor the dephosphorylated form of Pah1 was degraded by the 26S proteasome (Fig. 9A). However, both forms of Pah1 were degraded by the 20S proteasome (Fig. 9B). Moreover, the dephosphorylation of Pah1 stimulated its degradation by the 20S proteasome; the half-life of the dephosphorylated enzyme (12 min) was 2.6-fold shorter than that of the endogenously phosphorylated enzyme (32 min) (Fig. 9B).
FIGURE 9.

Phosphorylated and dephosphorylated forms of Pah1 are not degraded by the 26S proteasome, but dephosphorylation of Pah1 by the Nem1-Spo7 phosphatase complex stimulates degradation by the 20S proteasome. Pah1, which is endogenously phosphorylated in S. cerevisiae (18), was dephosphorylated by the Nem1-Spo7 protein phosphatase. The phosphorylated (left) and dephosphorylated (right) forms of Pah1 (30 nm each) were incubated with 2 nm 26S proteasome plus 1 mm ATP (A) or the 20S proteasome (2 nm) plus SDS (0.02%) (B) with or without 50 μm MG132 for the indicated time intervals. Following the incubations, samples were subjected to SDS-PAGE and Western blotting with anti-Pah1 antibodies. The Western blots shown are representative of three independent experiments. The position of Pah1 is indicated in the figure. B, bottom, the relative amounts of Pah1 in B from incubations without MG132 were quantified using ImageQuant software. The data shown are means ± S.D. (error bars) from triplicate determinations.
DISCUSSION
Proteasomal degradation is a novel mechanism by which Pah1 abundance in yeast is controlled in the stationary phase of growth (45). In this work, we characterized the proteolytic degradation of Pah1 using purified preparations of enzyme and proteasomes. The E. coli-expressed (i.e. not subject to posttranslational modifications) and yeast-expressed (i.e. subject to endogenous posttranslational modifications) forms of Pah1 were degraded by the 20S proteasome, but not by the 26S proteasome. Although a function of the 20S proteasome is to process damaged and oxidized proteins (82, 83), the rate of Pah1 degradation was not affected by boiling or by treatments with hydrogen peroxide or DTT. Our previous work indicated that increased levels of PA, which are brought about by catalytic site mutations that reduce PAP activity or by the overexpression of Dgk1 DAG kinase activity, stabilize Pah1 abundance (45). However, the in vitro analysis performed here indicates that PA has no significant effect on the 20S proteasomal degradation of Pah1, suggesting that PA levels affect the stability of Pah1 through an indirect regulatory mechanism. The 20S proteasomal degradation was clearly greatest when Pah1 was not phosphorylated, either by a lack of phosphorylation (e.g. E. coli-expressed enzyme) or by dephosphorylation of inherently phosphorylated (yeast-expressed) enzyme with the Nem1-Spo7 protein phosphatase. In contrast, the 20S proteasomal degradation was significantly attenuated when Pah1 was endogenously phosphorylated or phosphorylated in vitro by Pho85-Pho80 and by protein kinase A. The half-lives of the unphosphorylated and dephosphorylated forms of Pah1 shown in Figs. 8 and 9 were not the same. This may be explained if the dephosphorylation of the yeast-expressed Pah1 was not complete. Overall, these findings indicate that the phosphorylation state of Pah1 governs its degradation by the 20S proteasome and define more clearly why phosphorylation/dephosphorylation affects Pah1 abundance in vivo.
Unfolded proteins are targeted for proteasomal degradation (84–86). Generally, the posttranslational modification of ubiquitination provides a signal and facilitates unfolding for degradation by the 26S proteasome (84–86). However, proteins may also be subjected to proteasomal degradation through a ubiquitin-independent mechanism that is mediated by unfolded regions of the protein (79, 87). Pah1 is identified as an unstable protein based on its primary structure (88), and the FoldIndex algorithm predicts that Pah1 is a highly unstructured protein with the exception of the conserved NLIP and HAD-like domains. Consistent with this prediction, Pah1 was shown to be degraded from the unfolded regions at both the N and C termini of the protein, and the HAD-like domain was resistant to degradation. In addition to being targeted for proteasomal degradation, the unfolded regions of a protein are generally susceptible to posttranslational modifications that may induce a structural change (89). In the case of Pah1, it is the unfolded regions where the phosphorylation target sites of Pho85-Pho80 (40), Cdc28-cyclin B (39), protein kinase A (41), and protein kinase C (42) are located (Fig. 6A). That Pho85-Pho80 and protein kinase A caused Pah1 to be less susceptible to 20S proteasomal degradation indicates that phosphorylation induces a change to a more ordered state. Conversely, the increased degradation of Pah1 through its dephosphorylation by Nem1-Spo7 indicates a change in structure to a more disordered state.
Protein kinase A (41) and protein kinase C (42) share three sites of phosphorylation (e.g. Ser-677, Ser-773, and Ser-788) at the C terminus of Pah1 (Fig. 6A). However, the phosphorylations by these kinases had opposing effects on the 20S proteasomal degradation of the enzyme; protein kinase A had an inhibitory effect, whereas protein kinase C had a stimulatory effect. These results indicate that the unique protein kinase A sites (e.g. Ser-10 and Ser-774) may play a role in protecting Pah1 from degradation, and the unique protein kinase C site (e.g. Ser-769) may play a role in stimulating enzyme degradation. In vivo, the S10A mutation destabilizes Pah1 abundance (41), whereas alanine mutations of the protein kinase C sites stabilize abundance, but only when Pah1 is not already phosphorylated by Pho85-Pho80 (42).
We posit that the 20S proteasomal degradation of Pah1 is physiologically relevant. In glucose-grown yeast cells, the 26S proteasome is primarily localized to the nucleus (90). However, when the cells are depleted for glucose and progress into the stationary phase, the decrease in cellular pH (to ∼5.5) results in the dissociation of the 26S proteasome into the 20S core and 19S regulatory particles, promoting the formation of cytoplasmic proteasome storage granules (90–93). The acidic environment in the stationary phase also favors the Nem1-Spo7-mediated dephosphorylation of Pah1 (54), and during this phase of growth, phosphorylations by cyclin-dependent protein kinases (e.g. Pho85-Pho80 and Cdc28-cyclin B) and protein kinase A are expected to be less prevalent (94, 95). Although the pH optimum for the 20S proteasomal degradation of Pah1 was 7.0, about 70% of the maximum activity remains at pH 5.5. Thus, the non-phosphorylated state of Pah1, coupled with the increased levels of the 20S proteasome, are conducive to the degradation of Pah1 in stationary phase cells.
Like Pah1 (45), its mammalian ortholog lipin 1 is stabilized in cells treated with the proteasome inhibitor MG132 (96). However, it is unclear whether lipin 1 is subject to ubiquitin-dependent or ubiquitin-independent mechanisms of proteasomal degradation. Because the NLIP and HAD-like domains and PAP function of Pah1 and lipin 1 are conserved (7, 97), we examined whether the E. coli-expressed unmodified lipin 1 could be degraded by proteasomes. Similar to Pah1, human lipin 1 (α, β, or γ isoform) was degraded by the 20S proteasome but not by the 26S proteasome. Thus, lipin 1 is also subject to ubiquitin-independent proteasomal degradation. The HAD-like domain of lipin 1 is located at the C terminus (2), and the FoldIndex algorithm predicts that the central region between the NLIP and HAD-like domains to be unfolded (43%). Akin to Pah1, lipin 1 is a highly phosphorylated PAP enzyme (98), and most of the phosphorylation sites are positioned within the unfolded region of the protein. Studies to examine the role of phosphorylation/dephosphorylation on the proteasomal degradation of lipin 1 are warranted.
There is no clear indication that Pah1 is degraded by the ubiquitin-dependent 26S proteasomal degradation pathway. Pah1 is not among the >1,000 ubiquitinated proteins identified in exponential phase cells through proteomic analysis (99, 100), and our attempts to unequivocally establish the ubiquitination of Pah1 from stationary phase cells have thus far been unsuccessful. Considering that Pah1 is partially stabilized in the stationary phase of some mutants defective in the ubiquitination pathway (45), we do not exclude the possibility that Pah1 is also regulated by a ubiquitin-dependent process.
As discussed previously (45), Pah1 is not the only lipid metabolic enzymes in yeast that is subject to proteasomal degradation. The abundance of Ole1 Δ-9 fatty acid desaturase (101) and Hmg2 hydroxymethylglutaryl-CoA reductase (102) is known to be regulated by proteasomal degradation. In addition, studies profiling the proteomics of ubiquitination in yeast (99) have identified lipid biosynthetic enzymes (e.g. Gpt2p, Slc1, and Cho1) and proteins (e.g. Opi1 and Scs2) that control lipid synthesis. Interestingly, these putative proteasome targets are intimately related to the metabolism of PA, the Pah1 PAP substrate. Gpt2 and Slc1 are acyltransferase enzymes that catalyze the conversion of glycerol 3-phosphate to PA, whereas the Cho1 phosphatidylserine synthase utilizes the PA-derived CDP-DAG to produce phosphatidylserine, which is converted to the major membrane phospholipids phosphatidylethanolamine and phosphatidylcholine (5, 6). Opi1 is a transcriptional repressor controlling phospholipid synthesis, and its function is controlled through its localization governed by interaction with PA and Scs2 (6, 103). Thus, proteasomal degradation is emerging as an additional mechanism by which lipid metabolism is regulated in yeast.
Acknowledgments
We thank Symeon Siniossoglou for anti-lipin 1 antibodies and the plasmid YCplac111-SEC63-GFP. Susan Lindquist is acknowledged for the plasmid p426GPD-αSyn. We also thank Karin Römisch for helpful discussions about ubiquitin-independent degradation by the 20S proteasome.
This work was supported, in whole or in part, by National Institutes of Health Grants GM028140 and GM050679.
- PAP
- phosphatidate phosphatase
- PA
- phosphatidate
- DAG
- diacylglycerol
- TAG
- triacylglycerol
- ER
- endoplasmic reticulum
- HAD
- haloacid dehalogenase
- TAP
- tandem affinity purification.
REFERENCES
- 1. Han G.-S., Wu W.-I., Carman G. M. (2006) The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281, 9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Péterfy M., Phan J., Xu P., Reue K. (2001) Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27, 121–124 [DOI] [PubMed] [Google Scholar]
- 3. Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., Reue K. (2007) Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282, 3450–3457 [DOI] [PubMed] [Google Scholar]
- 4. Smith S. W., Weiss S. B., Kennedy E. P. (1957) The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem. 228, 915–922 [PubMed] [Google Scholar]
- 5. Carman G. M., Han G.-S. (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80, 859–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henry S. A., Kohlwein S. D., Carman G. M. (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pascual F., Carman G. M. (2013) Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim. Biophys. Acta 1831, 514–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shen H., Heacock P. N., Clancey C. J., Dowhan W. (1996) The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. J. Biol. Chem. 271, 789–795 [DOI] [PubMed] [Google Scholar]
- 9. Tamura Y., Harada Y., Nishikawa S., Yamano K., Kamiya M., Shiota T., Kuroda T., Kuge O., Sesaki H., Imai K., Tomii K., Endo T. (2013) Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metab. 17, 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carter J. R., Kennedy E. P. (1966) Enzymatic synthesis of cytidine diphosphate diglyceride. J. Lipid Res. 7, 678–683 [PubMed] [Google Scholar]
- 11. Taylor F. R., Parks L. W. (1979) Triacylglycerol metabolism in Saccharomyces cerevisiae relation to phospholipid synthesis. Biochim. Biophys. Acta 575, 204–214 [DOI] [PubMed] [Google Scholar]
- 12. Fakas S., Qiu Y., Dixon J. L., Han G.-S., Ruggles K. V., Garbarino J., Sturley S. L., Carman G. M. (2011) Phosphatidate phosphatase activity plays a key role in protection against fatty acid-induced toxicity in yeast. J. Biol. Chem. 286, 29074–29085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pascual F., Soto-Cardalda A., Carman G. M. (2013) PAH1-encoded phosphatidate phosphatase plays a role in the growth phase- and inositol-mediated regulation of lipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 288, 35781–35792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wagner C., Dietz M., Wittmann J., Albrecht A., Schüller H. J. (2001) The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol. Microbiol. 41, 155–166 [DOI] [PubMed] [Google Scholar]
- 15. Loewen C. J. R., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., Levine T. P. (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304, 1644–1647 [DOI] [PubMed] [Google Scholar]
- 16. Carman G. M., Henry S. A. (2007) Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282, 37293–37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., Siniossoglou S. (2005) The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24, 1931–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Hara L., Han G.-S., Peak-Chew S., Grimsey N., Carman G. M., Siniossoglou S. (2006) Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281, 34537–34548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han G.-S., Siniossoglou S., Carman G. M. (2007) The cellular functions of the yeast lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 282, 37026–37035 [DOI] [PubMed] [Google Scholar]
- 20. Adeyo O., Horn P. J., Lee S., Binns D. D., Chandrahas A., Chapman K. D., Goodman J. M. (2011) The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han G.-S., O'Hara L., Carman G. M., Siniossoglou S. (2008) An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J. Biol. Chem. 283, 20433–20442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Irie K., Takase M., Araki H., Oshima Y. (1993) A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein. Mol. Gen. Genet. 236, 283–288 [DOI] [PubMed] [Google Scholar]
- 23. Lussier M., White A. M., Sheraton J., di Paolo T., Treadwell J., Southard S. B., Horenstein C. I., Chen-Weiner J., Ram A. F., Kapteyn J. C., Roemer T. W., Vo D. H., Bondoc D. C., Hall J., Zhong W. W., Sdicu A. M., Davies J., Klis F. M., Robbins P. W., Bussey H. (1997) Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147, 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ruiz C., Cid V. J., Lussier M., Molina M., Nombela C. (1999) A large-scale sonication assay for cell wall mutant analysis in yeast. Yeast 15, 1001–1008 [DOI] [PubMed] [Google Scholar]
- 25. Sasser T., Qiu Q. S., Karunakaran S., Padolina M., Reyes A., Flood B., Smith S., Gonzales C., Fratti R. A. (2012) The yeast lipin 1 orthologue Pah1p regulates vacuole homeostasis and membrane fusion. J. Biol. Chem. 287, 2221–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Phan J., Reue K. (2005) Lipin, a lipodystrophy and obesity gene. Cell Metab. 1, 73–83 [DOI] [PubMed] [Google Scholar]
- 27. Lindegaard B., Larsen L. F., Hansen A. B., Gerstoft J., Pedersen B. K., Reue K. (2007) Adipose tissue lipin expression levels distinguish HIV patients with and without lipodystrophy. Int. J. Obes. 31, 449–456 [DOI] [PubMed] [Google Scholar]
- 28. Nadra K., de Preux Charles A.-S., Médard J.-J., Hendriks W. T., Han G.-S., Grès S., Carman G. M., Saulnier-Blache J.-S., Verheijen M. H. G., Chrast R. (2008) Phosphatidic acid mediates demyelination in Lpin1 mutant mice. Genes Dev. 22, 1647–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zeharia A., Shaag A., Houtkooper R. H., Hindi T., de Lonlay P., Erez G., Hubert L., Saada A., de Keyzer Y., Eshel G., Vaz F. M., Pines O., Elpeleg O. (2008) Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am. J. Hum. Genet. 83, 489–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reue K., Brindley D. N. (2008) Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J. Lipid Res. 49, 2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reue K., Dwyer J. R. (2009) Lipin proteins and metabolic homeostasis. J. Lipid Res. 50, S109–S114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Donkor J., Zhang P., Wong S., O'Loughlin L., Dewald J., Kok B. P., Brindley D. N., Reue K. (2009) A conserved serine residue is required for the phosphatidate phosphatase activity but not transcriptional coactivator functions of lipin-1 and lipin-2. J. Biol. Chem. 284, 29968–29978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim H. B., Kumar A., Wang L., Liu G. H., Keller S. R., Lawrence J. C., Jr., Finck B. N., Harris T. E. (2010) Lipin 1 represses NFATc4 transcriptional activity in adipocytes to inhibit secretion of inflammatory factors. Mol. Cell. Biol. 30, 3126–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grkovich A., Dennis E. A. (2009) Phosphatidic acid phosphohydrolase in the regulation of inflammatory signaling. Adv. Enzyme Regul. 49, 114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mul J. D., Nadra K., Jagalur N. B., Nijman I. J., Toonen P. W., Médard J. J., Grès S., de Bruin A., Han G.-S., Brouwers J. F., Carman G. M., Saulnier-Blache J. S., Meijer D., Chrast R., Cuppen E. (2011) A hypomorphic mutation in Lpin1 induces progressively improving neuropathy and lipodystrophy in the rat. J. Biol. Chem. 286, 26781–26793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nadra K., Médard J. J., Mul J. D., Han G.-S., Grès S., Pende M., Metzger D., Chambon P., Cuppen E., Saulnier-Blache J. S., Carman G. M., Desvergne B., Chrast R. (2012) Cell autonomous lipin 1 function is essential for development and maintenance of white and brown adipose tissue. Mol. Cell. Biol. 32, 4794–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Michot C., Mamoune A., Vamecq J., Viou M. T., Hsieh L.-S., Testet E., Lainé J., Hubert L., Dessein A. F., Fontaine M., Ottolenghi C., Fouillen L., Nadra K., Blanc E., Bastin J., Candon S., Pende M., Munnich A., Smahi A., Djouadi F., Carman G. M., Romero N., de Keyzer Y., de Lonlay P. (2013) Combination of lipid metabolism alterations and their sensitivity to inflammatory cytokines in human lipin-1-deficient myoblasts. Biochim. Biophys. Acta 1832, 2103–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Siniossoglou S. (2013) Phospholipid metabolism and nuclear function: roles of the lipin family of phosphatidic acid phosphatases. Biochim. Biophys. Acta 1831, 575–581 [DOI] [PubMed] [Google Scholar]
- 39. Choi H.-S., Su W.-M., Morgan J. M., Han G.-S., Xu Z., Karanasios E., Siniossoglou S., Carman G. M. (2011) Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of Ser602, Thr723, and Ser744 as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem. 286, 1486–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choi H.-S., Su W.-M., Han G.-S., Plote D., Xu Z., Carman G. M. (2012) Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J. Biol. Chem. 287, 11290–11301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Su W.-M., Han G.-S., Casciano J., Carman G. M. (2012) Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjunction with the Pho85p-Pho80p and Cdc28p-cyclin B kinases to regulate lipid synthesis in yeast. J. Biol. Chem. 287, 33364–33376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Su W. M., Han G. S., Carman G. M. (2014) Cross-talk phosphorylations by protein kinase C and Pho85p-Pho80p protein kinase regulate Pah1p phosphatidate phosphatase abundance in Saccharomyces cerevisiae. J. Biol. Chem. 289, 18818–18830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Karanasios E., Han G.-S., Xu Z., Carman G. M., Siniossoglou S. (2010) A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. U.S.A. 107, 17539–17544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karanasios E., Barbosa A. D., Sembongi H., Mari M., Han G.-S., Reggiori F., Carman G. M., Siniossoglou S. (2013) Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol. Biol. Cell 24, 2124–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pascual F., Hsieh L.-S., Soto-Cardalda A., Carman G. M. (2014) Yeast Pah1p phosphatidate phosphatase is regulated by proteasome-mediated degradation. J. Biol. Chem. 289, 9811–9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chi A., Huttenhower C., Geer L. Y., Coon J. J., Syka J. E., Bai D. L., Shabanowitz J., Burke D. J., Troyanskaya O. G., Hunt D. F. (2007) Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 104, 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li X., Gerber S. A., Rudner A. D., Beausoleil S. A., Haas W., Villén J., Elias J. E., Gygi S. P. (2007) Large-scale phosphorylation analysis of α-factor-arrested Saccharomyces cerevisiae. J. Proteome. Res. 6, 1190–1197 [DOI] [PubMed] [Google Scholar]
- 48. Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., Shah K., Shokat K. M., Morgan D. O. (2003) Targets of the cyclin-dependent kinase Cdk1. Nature 425, 859–864 [DOI] [PubMed] [Google Scholar]
- 49. Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., McCartney R. R., Schmidt M. C., Rachidi N., Lee S. J., Mah A. S., Meng L., Stark M. J., Stern D. F., De Virgilio C., Tyers M., Andrews B., Gerstein M., Schweitzer B., Predki P. F., Snyder M. (2005) Global analysis of protein phosphorylation in yeast. Nature 438, 679–684 [DOI] [PubMed] [Google Scholar]
- 50. Dephoure N., Howson R. W., Blethrow J. D., Shokat K. M., O'Shea E. K. (2005) Combining chemical genetics and proteomics to identify protein kinase substrates. Proc. Natl. Acad. Sci. U.S.A. 102, 17940–17945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mah A. S., Elia A. E., Devgan G., Ptacek J., Schutkowski M., Snyder M., Yaffe M. B., Deshaies R. J. (2005) Substrate specificity analysis of protein kinase complex Dbf2-Mob1 by peptide library and proteome array screening. BMC Biochem. 6, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu Z., Su W.-M., Carman G. M. (2012) Fluorescence spectroscopy measures yeast PAH1-encoded phosphatidate phosphatase interaction with liposome membranes. J. Lipid Res. 53, 522–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Siniossoglou S., Santos-Rosa H., Rappsilber J., Mann M., Hurt E. (1998) A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17, 6449–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Su W.-M., Han G.-S., Carman G. M. (2014) Yeast Nem1-Spo7 protein phosphatase activity on Pah1 phosphatidate phosphatase is specific for the Pho85-Pho80 protein kinase phosphorylation sites. J. Biol. Chem. 289, 34699–34708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu W.-I., Carman G. M. (1996) Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by phospholipids. Biochemistry 35, 3790–3796 [DOI] [PubMed] [Google Scholar]
- 56. Wu W.-I., Lin Y.-P., Wang E., Merrill A. H., Jr., Carman G. M. (1993) Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by sphingoid bases. J. Biol. Chem. 268, 13830–13837 [PubMed] [Google Scholar]
- 57. Wu W.-I., Carman G. M. (1994) Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by nucleotides. J. Biol. Chem. 269, 29495–29501 [PubMed] [Google Scholar]
- 58. Soto-Cardalda A., Fakas S., Pascual F., Choi H. S., Carman G. M. (2012) Phosphatidate phosphatase plays role in zinc-mediated regulation of phospholipid synthesis in yeast. J. Biol. Chem. 287, 968–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Han G.-S., Sreenivas A., Choi M. G., Chang Y. F., Martin S. S., Baldwin E. P., Carman G. M. (2005) Expression of human CTP synthetase in Saccharomyces cerevisiae reveals phosphorylation by protein kinase A. J. Biol. Chem. 280, 38328–38336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 61. Innis M. A., Gelfand D. H. (1990) in PCR Protocols: A Guide to Methods and Applications (Innis M. A., Gelfand D. H., Sninsky J. J., White T. J., eds) pp. 3–12, Academic Press, Inc., San Diego [Google Scholar]
- 62. Outeiro T. F., Lindquist S. (2003) Yeast cells provide insight into α-synuclein biology and pathobiology. Science 302, 1772–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 64. Siniossoglou S., Hurt E. C., Pelham H. R. (2000) Psr1p/Psr2p, two plasma membrane phosphatases with an essential DXDX(T/V) motif required for sodium stress response in yeast. J. Biol. Chem. 275, 19352–19360 [DOI] [PubMed] [Google Scholar]
- 65. Leggett D. S., Glickman M. H., Finley D. (2005) Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 301, 57–70 [DOI] [PubMed] [Google Scholar]
- 66. Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., Finley D. (2002) Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 67. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 68. Lee D. H., Goldberg A. L. (1996) Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 271, 27280–27284 [DOI] [PubMed] [Google Scholar]
- 69. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 70. Burnette W. (1981) Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112, 195–203 [DOI] [PubMed] [Google Scholar]
- 71. Haid A., Suissa M. (1983) Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 96, 192–205 [DOI] [PubMed] [Google Scholar]
- 72. Grimsey N., Han G.-S., O'Hara L., Rochford J. J., Carman G. M., Siniossoglou S. (2008) Temporal and spatial regulation of the phosphatidate phosphatases lipin 1 and 2. J. Biol. Chem. 283, 29166–29174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tanaka K., Ii K., Ichihara A., Waxman L., Goldberg A. L. (1986) A high molecular weight protease in the cytosol of rat liver. I. Purification, enzymological properties, and tissue distribution. J. Biol. Chem. 261, 15197–15203 [PubMed] [Google Scholar]
- 74. Lin Y.-P., Carman G. M. (1989) Purification and characterization of phosphatidate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 264, 8641–8645 [PubMed] [Google Scholar]
- 75. Lin Y.-P., Carman G. M. (1990) Kinetic analysis of yeast phosphatidate phosphatase toward Triton X-100/phosphatidate mixed micelles. J. Biol. Chem. 265, 166–170 [PubMed] [Google Scholar]
- 76. Han G.-S., Carman G. M. (2010) Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J. Biol. Chem. 285, 14628–14638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen P., Hochstrasser M. (1995) Biogenesis, structure and function of the yeast 20S proteasome. EMBO J. 14, 2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Prilusky J., Felder C. E., Zeev-Ben-Mordehai T., Rydberg E. H., Man O., Beckmann J. S., Silman I., Sussman J. L. (2005) FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21, 3435–3438 [DOI] [PubMed] [Google Scholar]
- 79. Asher G., Reuven N., Shaul Y. (2006) 20S proteasomes and protein degradation “by default”. BioEssays 28, 844–849 [DOI] [PubMed] [Google Scholar]
- 80. Tar K., Dange T., Yang C., Yao Y., Bulteau A. L., Salcedo E. F., Braigen S., Bouillaud F., Finley D., Schmidt M. (2014) Proteasomes associated with the Blm10 activator protein antagonize mitochondrial fission through degradation of the fission protein Dnm1. J. Biol. Chem. 289, 12145–12156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liu C. W., Corboy M. J., DeMartino G. N., Thomas P. J. (2003) Endoproteolytic activity of the proteasome. Science 299, 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Davies K. J. (2001) Degradation of oxidized proteins by the 20S proteasome. Biochimie 83, 301–310 [DOI] [PubMed] [Google Scholar]
- 83. Pickering A. M., Davies K. J. (2012) Degradation of damaged proteins: the main function of the 20S proteasome. Prog. Mol. Biol. Transl. Sci. 109, 227–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 85. Hochstrasser M. (1996) Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30, 405–439 [DOI] [PubMed] [Google Scholar]
- 86. Hochstrasser M. (2009) Introduction to intracellular protein degradation. Chem. Rev. 109, 1479–1480 [DOI] [PubMed] [Google Scholar]
- 87. Asher G., Shaul Y. (2005) p53 proteasomal degradation: poly-ubiquitination is not the whole story. Cell Cycle 4, 1015–1018 [DOI] [PubMed] [Google Scholar]
- 88. Dinkel H., Van Roey K., Michael S., Davey N. E., Weatheritt R. J., Born D., Speck T., Krüger D., Grebnev G., Kuban M., Strumillo M., Uyar B., Budd A., Altenberg B., Seiler M., Chemes L. B., Glavina J., Sánchez I. E., Diella F., Gibson T. J. (2014) The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 42, D259–D266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Iakoucheva L. M., Radivojac P., Brown C. J., O'Connor T. R., Sikes J. G., Obradovic Z., Dunker A. K. (2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32, 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Laporte D., Salin B., Daignan-Fornier B., Sagot I. (2008) Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 181, 737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Orij R., Urbanus M. L., Vizeacoumar F. J., Giaever G., Boone C., Nislow C., Brul S., Smits G. J. (2012) Genome-wide analysis of intracellular pH reveals quantitative control of cell division rate by pHc in Saccharomyces cerevisiae. Genome Biol. 13, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Laporte D., Lebaudy A., Sahin A., Pinson B., Ceschin J., Daignan-Fornier B., Sagot I. (2011) Metabolic status rather than cell cycle signals control quiescence entry and exit. J. Cell Biol. 192, 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Peters L. Z., Hazan R., Breker M., Schuldiner M., Ben-Aroya S. (2013) Formation and dissociation of proteasome storage granules are regulated by cytosolic pH. J. Cell Biol. 201, 663–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. De Virgilio C. (2012) The essence of yeast quiescence. FEMS Microbiol. Rev. 36, 306–339 [DOI] [PubMed] [Google Scholar]
- 95. Gray J. V., Petsko G. A., Johnston G. C., Ringe D., Singer R. A., Werner-Washburne M. (2004) “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68, 187–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhang P., Verity M. A., Reue K. (2014) Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab. 20, 267–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Csaki L. S., Dwyer J. R., Fong L. G., Tontonoz P., Young S. G., Reue K. (2013) Lipins, lipinopathies, and the modulation of cellular lipid storage and signaling. Prog. Lipid Res. 52, 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., Kumar A., Lawrence J. C., Jr. (2007) Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 282, 277–286 [DOI] [PubMed] [Google Scholar]
- 99. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 100. Lee W. C., Lee M., Jung J. W., Kim K. P., Kim D. (2008) SCUD: Saccharomyces cerevisiae ubiquitination database. BMC Genomics 9, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Braun S., Matuschewski K., Rape M., Thoms S., Jentsch S. (2002) Role of the ubiquitin-selective CDC48(UFD1/NPL4) chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 21, 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hampton R. Y., Rine J. (1994) Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J. Cell Biol. 125, 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Chirala S. S., Zhong Q., Huang W., al-Feel W. (1994) Analysis of FAS3/ACC regulatory region of Saccharomyces cerevisiae: identification of a functional UASINO and sequences responsible for fatty acid mediated repression. Nucleic Acids Res. 22, 412–418 [DOI] [PMC free article] [PubMed] [Google Scholar]