Background: Sortilin 1 mediates lysosome-dependent apoB100 degradation in hepatocytes.
Results: Blocking insulin/PI3K/AKT signaling caused posttranslational hepatic sortilin 1 degradation.
Conclusion: Insulin resistance is an underlying cause of decreased hepatic Sort1 in diabetes.
Significance: This study suggests a new mechanism underlying altered hepatic apoB100 metabolism in type 2 diabetes.
Keywords: Apolipoprotein, Cholesterol, Diabetes, Lipid Metabolism, Triglyceride, Cholesterol, Lipid Metabolism
Abstract
Insulin promotes hepatic apolipoprotein B100 (apoB100) degradation, whereas insulin resistance is a major cause of hepatic apoB100/triglyceride overproduction in type 2 diabetes. The cellular trafficking receptor sortilin 1 (Sort1) was recently identified to transport apoB100 to the lysosome for degradation in the liver and thus regulate plasma cholesterol and triglyceride levels. Genetic variation of SORT1 was strongly associated with cardiovascular disease risk in humans. The major goal of this study is to investigate the effect and molecular mechanism of insulin regulation of Sort1. Results showed that insulin induced Sort1 protein, but not mRNA, in AML12 cells. Treatment of PI3K or AKT inhibitors decreased Sort1 protein, whereas expression of constitutively active AKT induced Sort1 protein in AML12 cells. Consistently, hepatic Sort1 was down-regulated in diabetic mice, which was partially restored after the administration of the insulin sensitizer metformin. LC-MS/MS analysis further revealed that serine phosphorylation of Sort1 protein was required for insulin induction of Sort1 in a casein kinase 2-dependent manner and that inhibition of PI3K signaling or prevention of Sort1 phosphorylation accelerated proteasome-dependent Sort1 degradation. Administration of a PI3K inhibitor to mice decreased hepatic Sort1 protein and increased plasma cholesterol and triglyceride levels. Adenovirus-mediated overexpression of Sort1 in the liver prevented PI3K inhibitor-induced Sort1 down-regulation and decreased plasma triglyceride but had no effect on plasma cholesterol in mice. This study identified Sort1 as a novel target of insulin signaling and suggests that Sort1 may play a role in altered hepatic apoB100 metabolism in insulin-resistant conditions.
Introduction
Diabetic dyslipidemia significantly increases the risk of cardiovascular disease, which is the leading cause of morbidity and mortality in type 2 diabetes (1). Hepatic insulin resistance is a characteristic feature of type 2 diabetes and a key underlying cause of hepatic triglyceride (TG),2 rich very low density lipoprotein (VLDL) overproduction, dyslipidemia, and an atherogenic lipoprotein profile (2, 3). Apolipoprotein B100 (apoB100) is the principal apolipoprotein component of VLDL and plays a key role in promoting hepatic VLDL assembly and secretion. It is well known that insulin, via activation of the PI3K pathway, promotes postendoplasmic reticulum presecretory apoB100 degradation and thus inhibits hepatic VLDL production by limiting the apoB100 availability for VLDL assembly (4). Under physiological conditions, it is thought that insulin inhibition of hepatic apoB100 production is necessary to allow rapid plasma clearance of gut-derived TG-rich chylomicrons and thus prevent postprandial hyperlipidemia. However, hepatic insulin resistance is considered a major cause of reduced apoB100 degradation and increased apoB100 secretion in type 2 diabetes. Existing evidence suggests that insulin regulation of apoB100 degradation requires the endosome/lysosome system; however, the downstream factors involved are still not well defined (4).
Recent studies revealed that hepatic sortilin 1 (Sort1), a transmembrane multiligand sorting receptor, is a novel regulator of lipid metabolism. Genetic variations of SORT1 showed strong and reproducible association with plasma LDL cholesterol (LDL-C), TG, and cardiovascular disease risk in large human populations (5–9). Sort1 mainly localizes in the trans-Golgi network (TGN) and facilitates the transport of various target proteins for lysosome targeting (10). A minor fraction of Sort1 localized to the plasma membrane is involved in receptor-mediated endocytosis. Studies in both mice and cell models showed that liver Sort1 directed intracellular apoB100 for lysosomal degradation (5, 11). In addition, plasma membrane-bound Sort1 can also mediate circulating apoB100 endocytosis and lysosome targeting (6, 11). Liver-specific Sort1 overexpression attenuated hepatic apoB100 secretion and decreased plasma TG and LDL-C in various diabetic and hyperlipidemic mouse models, and conversely, liver-specific Sort1 knockdown resulted in hyperlipidemia in these models (5, 12–14).
In this study, we investigated the effect and mechanism of insulin regulation of hepatic Sort1. Our results revealed that hepatic Sort1 was a novel target of the insulin/PI3K/AKT signaling cascade. Furthermore, hepatic Sort1 was highly sensitive to insulin resistance-induced posttranslational degradation, which may play a role in altered hepatic apoB100 metabolism in insulin-resistant conditions.
EXPERIMENTAL PROCEDURES
Reagents
Antibodies against Sort1 and TGN46 were purchased from Abcam (Cambridge, MA). Antibodies against AKT, phospho-AKT (Ser-473), histone 3, HA, and the PI3K inhibitor wortmannin were purchased from Cell Signaling Technology (Danvers, MA). Anti-FLAG M2 antibody, actin antibody, metformin, cycloheximide, AKT1/2 inhibitor VIII, streptozotocin (STZ), tetrabromocinnamic acid (TBCA), chloroquine, and bafilomycin A1 were purchased from Sigma. PX866 was purchased from Cayman Chemical (Ann Arbor, MI). Anti-Lamp1 antibody was purchased from the Developmental Studies Hybridoma Bank (University of Iowa). Alexa Fluor 488 IgG and Alexa Fluor 594 IgG were purchased from Life Technologies, Inc.
Animals
Male wild type C57BL/6J mice and ob/ob mice at 10 weeks old were purchased from the Jackson Laboratory (Bar Harbor, ME). All mice were maintained on a standard chow diet and water ad libitum for 2 weeks before experiments began. The Western diet (TD.88137, Harlan Teklad) contains 21% milk fat (w/w) and 0.2% cholesterol. Metformin was mixed with either chow diet or Western diet at 0.25% (w/w). Insulin-deficient C57BL/6 mice were generated via intraperitoneal injection with 7.5 mg/kg STZ once daily for 5 consecutive days. Control mice were injected with vehicle only (sodium citrate buffer, pH 4.5). Hyperglycemia was confirmed at 1 week after the last injection. Liver tissues were collected at 2 weeks after the last injection. PX866 was dissolved in sterile 1× PBS with 5% EtOH and administered through intraperitoneal injection at 8 mg/kg in a 100-μl volume. Further details regarding individual experiments are given in the figure legends. All study protocols were approved by the Institutional Animal Care and Use Committee.
Cell Culture, Transfection, and Treatment
The mouse hepatocyte cell line AML12 cells were a kind gift from Dr. Yanqiao Zhang (Northeast Ohio Medical University, Rootstown, OH). Cells were maintained in DMEM supplemented with 10% FBS and 1× insulin-transferrin-selenium solution (Life Technologies). Cells were cultured in DMEM without any supplements overnight before insulin treatment. Sort1 mutant plasmids were generated with a PCR-based QuikChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). Transient transfection was performed with Lipofectamine 2000 reagent (Life Technologies).
Immunofluorescent Staining
AML12 cells were plated in chamber slides and transfected or treated as indicated in the figure legends. Cells were fixed with 4% paraformaldehyde, permeabilized, and blocked in 10% donkey serum with 0.3 m glycine and 0.1% Tween 20, followed by incubation with primary antibodies at 4 °C overnight. Cells were then washed in 1× PBS and incubated with Alexa Fluor-conjugated secondary antibodies. After washing with 1× PBS, cells were mounted and imaged with a Leica DM 5500 confocal microscope.
Recombinant Adenovirus
Adenovirus expressing human full-length Sort1 was a generous gift from Dr. Anders Nykjaer (Aarhus University, Denmark). Adenovirus expressing a C-terminal FLAG-tagged human sortilin 1 was generated using the AdEasy adenoviral vector system (Agilent Technologies). Adenoviral vectors expressing a constitutively active AKT1 or AKT2 were purchased from Vector Biolabs (Philadelphia, PA).
LC-MS/MS Proteomics Analysis
Wild type C57BL/6J mice were injected with 5 × 108 pfu/mouse adenovirus expressing a C-terminal lag-tagged human Sort1. After 7 days, FLAG-Sort1 was immunoprecipitated from the mouse liver lysate with anti-FLAG M2 antibody. The subsequent LC-MS/MS analysis and phosphopeptide verification were performed by MS Bioworks (Ann Arbor, MI).
Measurement of Sort1 Ubiquitination
Cells were transfected with plasmid expressing HA-tagged ubiquitin and infected with Ad-Sort1-FLAG (multiplicity of infection = 1). After 24 h, cells were treated with MG132 and/or wortmannin or TBCA as indicated, followed by immunoprecipitation with anti-FLAG (M2) antibody conjugated to magnetic beads (Sigma). Immunoprecipitated FLAG-Sort1 was detected with anti-FLAG antibody, and ubiquitinated Sort1 was detected with anti-HA antibody.
Lipid Analysis
Cholesterol, triglyceride, and free fatty acids were measured with colorimetric assay kits (Biovision, Milpitas, CA). FPLC analysis of the lipoprotein profile was conducted by the Mouse Metabolic Phenotyping Center at the University of Cincinnati.
Glucose Tolerance Test
Mice were fasted overnight and received a single intraperitoneal injection of glucose at 2 g/kg of body weight. A drop of blood was collected from the tail, and glucose was measured with a glucose monitor.
RNA Isolation and Quantitative Real-time PCR
Real-time PCR were performed with SYBR primers. Amplification of 18 S was used as an internal control. Relative mRNA expression was quantified using the comparative CT (Ct) method and expressed as 2−ΔΔCt.
Immunoblot
Cell or tissue protein samples were prepared in 1× radioimmune precipitation assay buffer containing protease inhibitor mixture followed by brief sonication. Protein concentrations were determined by a BCA assay kit (Life Technologies). Equal amount of protein was used for SDS-PAGE and Western blotting. Densitometry was determined with ImageJ software for Fig. 5G. All experiments were repeated at least three times to confirm reproducibility. A representative blot is shown.
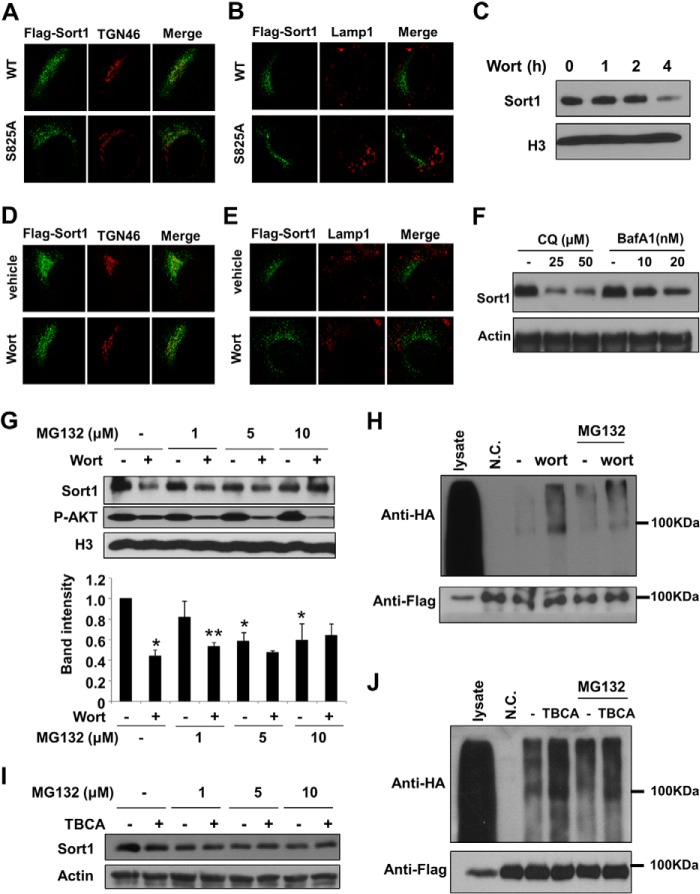
FIGURE 5.
Inhibition of PI3K caused proteasome-dependent Sort1 degradation. A and B, AML12 cells transfected with wild type and S825A mutant human FLAG-Sort1 plasmids. After 24 h, FLAG-Sort1 colocalization with TGN46 or Lamp1 were analyzed by immunofluorescent staining and confocal microscopy as described under “Experimental Procedures.” Anti-FLAG antibody was used to detect WT and mutant FLAG-Sort1. C, AML12 cells were treated with 1 μm wortmannin (wort) for the time indicated, and Sort1 protein was measured by immunoblot. D and E, AML12 cells transfected with wild type human FLAG-Sort1 plasmid. After 24 h, cells were treated with 1 μm wortmannin for 2 h. Sort1 colocalization with TGN46 or Lamp1 were analyzed by immunofluorescent staining and confocal microscopy as described under “Experimental Procedures.” F, AML12 cells were treated with indicated concentration of chloroquine (CQ) or bafilomycin A1 (BafA1) for 8 h. Sort1 protein was determined by immunoblot. G, AML12 cells were pretreated with increasing doses of MG132 for 1 h, followed by 1 μm wortmannin treatment for an additional 4 h. Sort1 protein was measured by immunoblot. Bottom, average Sort1 band intensity of three independent gel blots (normalized to histone H3). Relative intensity was plotted as mean ± S.D. (error bars). *, versus untreated control; **, versus 1 μm MG132 treated sample. H, AML12 cells were infected with Ad-FLAG-Sort1 (multiplicity of infection = 1) and transfected with plasmid expressing HA-tagged ubiquitin. After 24 h, some cells were pretreated with 10 μm MG132 for 1 h, followed by 1 μm wortmannin treatment for an additional 2 h. FLAG-Sort1 was immunoprecipitated with anti-FLAG antibody. Ubiquitinated Sort1 was measured by anti-HA antibody. Immunoprecipitated FLAG-Sort1 was measured by anti-FLAG antibody. Lysate, total cell lysate was used as positive controls for FLAG-Sort1 and HA-ubiquitin. NC, negative control, cells expressing FLAG-Sort1 but not HA-ubiquitin. I, AML12 cells were pretreated with increasing doses of MG132 for 1 h, followed by 15 μm TBCA treatment for an additional 4 h. Sort1 protein was measured by immunoblot. J, AML12 cells were infected with Ad-FLAG-Sort1 (multiplicity of infection = 1) and transfected with plasmid expressing HA-tagged ubiquitin. After 24 h, some cells were pretreated with 10 μm MG132 for 1 h, followed by 15 μm TBCA treatment for an additional 2 h. FLAG-Sort1 was immunoprecipitated with anti-FLAG antibody. Ubiquitinated Sort1 was measured by anti-HA antibody. Immunoprecipitated FLAG-Sort1 was measured by anti-FLAG antibody. Lysate, total cell lysate was used as positive controls for FLAG-Sort1 and HA-ubiquitin. N.C., negative control, cells expressing FLAG-Sort1 but not HA-ubiquitin.
Statistical Analysis
Results were expressed as mean ± S.E. unless noted. Statistical analysis was performed by Student's t test or analysis of variance. A p < 0.05 was considered statistically significant.
RESULTS
Hepatic Insulin/PI3K/AKT Signaling Regulates Sort1 in Vitro and in Vivo
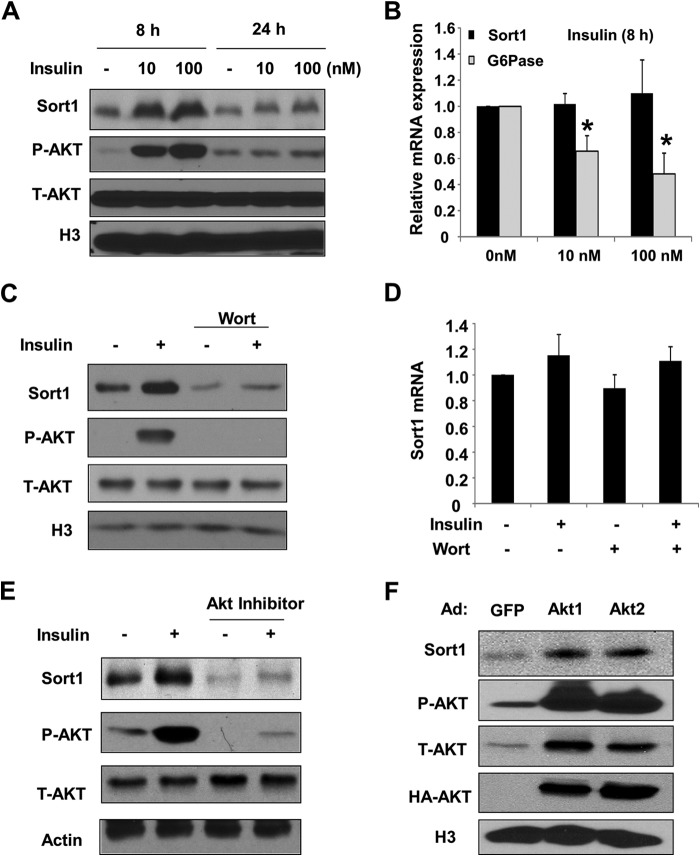
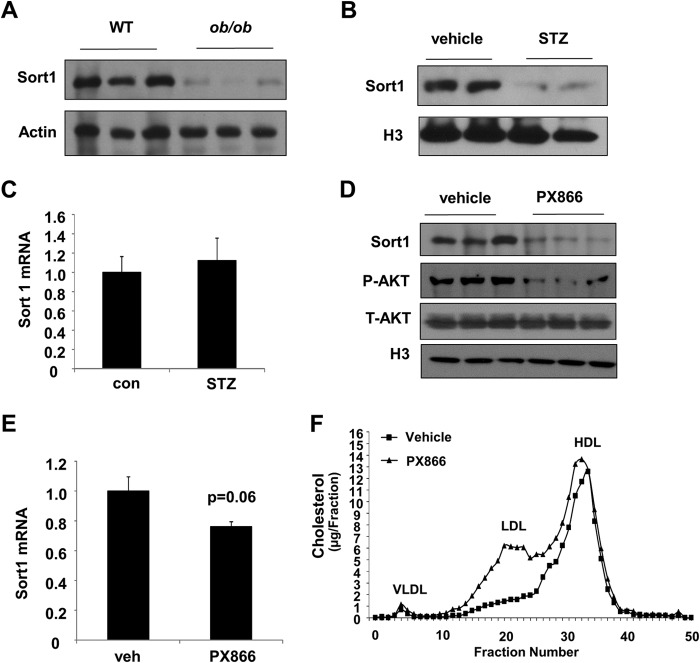
We first found that treating AML12 cells with insulin resulted in a dose-dependent increase in Sort1 protein, but not mRNA, which closely correlated with cellular AKT activation (Fig. 1, A and B). The mRNA of glucose-6-phosphatase was measured as a positive control for insulin treatment (Fig. 1B). Treatment of AML12 cells with 100 nm insulin in time course showed that maximal induction of Sort1 was observed at 6–8 h but not at 4 h or earlier (not shown). Treating cells with the PI3K inhibitor, wortmannin, or an AKT1/2 inhibitor not only blocked insulin induction of Sort1 protein but also significantly decreased basal Sort1 protein without altering the Sort1 mRNA (Fig. 1, C–E). In addition, adenovirus-mediated expression of constitutively active AKT1 or AKT2 induced Sort1 protein in AML12 cells (Fig. 1F). Consistent with these in vitro observations, obese mice with hepatic steatosis and insulin resistance showed markedly decreased hepatic Sort1 protein (Fig. 2A). Importantly, STZ-treated mice also showed decreased hepatic Sort1 protein but not mRNA (Fig. 2, B and C), suggesting that decreased hepatic insulin signaling independent of hepatic steatosis was sufficient to reduce hepatic Sort1 levels.
FIGURE 1.
Insulin/PI3K/AKT signaling cascade induces Sort1 protein in AML12 cells. A, AML12 cells were serum-starved for 16 h, followed by insulin treatment as indicated. Sort1, phospho-AKT (Ser-473) (P-AKT), total AKT (T-AKT), and histone 3 (H3) were determined by immunoblot. B, AML12 cells were treated with insulin for 8 h. Sort1 and glucose-6-phosphatase (G6Pase) mRNA expressions were measured by real-time PCR. Results were expressed as the mean ± S.D. (error bars) of three replicates. C and D, AML12 cells were serum-starved for 16 h. Cells were pretreated with 1 μm wortmannin (wort), followed by 100 nm insulin treatment for 8 h. Sort1 protein or mRNA was determined by immunoblot or real-time PCR, respectively. Real-time PCR results were expressed as mean ± S.D. of three replicates. E, AML12 cells were serum-starved for 16 h and pretreated with 10 μm AKT1/2 inhibitor for 1 h, followed by 100 nm insulin treatment for 8 h. Sort1 protein was determined by immunoblot. F, AML12 cells were infected with Ad-GFP or adenovirus expressing HA-tagged constitutively active AKT1 or AKT2 at a multiplicity of infection of ∼1 in serum-free and insulin-free medium for 16 h. Sort1 protein was measured by immunoblot.
FIGURE 2.
Inhibition of PI3K signaling reduced Sort1 protein in mouse livers. A, hepatic Sort1 protein in male 12-week-old WT and ob/ob mice. B and C, hepatic Sort1 protein (B) and mRNA (C) in vehicle- and STZ-treated mice. Real-time PCR results were expressed as mean ± S.E. (error bars) n = 4. D and E, male 12-week-old C57BL6J mice were intraperitoneally injected with PX866 (8 mg/kg) or vehicle (n = 4). After 8 h, liver Sort1 protein (D) and liver Sort1 mRNA (E) were measured. F, male 12-week-old C57BL6J mice were intraperitoneally injected with PX866 (8 mg/kg) or vehicle once daily for 3 days. After the last injection, mice were then fasted for 8 h, and pooled plasma (n = 4) was used for FPLC cholesterol analysis. P-AKT, phospho-AKT; T-AKT, total AKT.
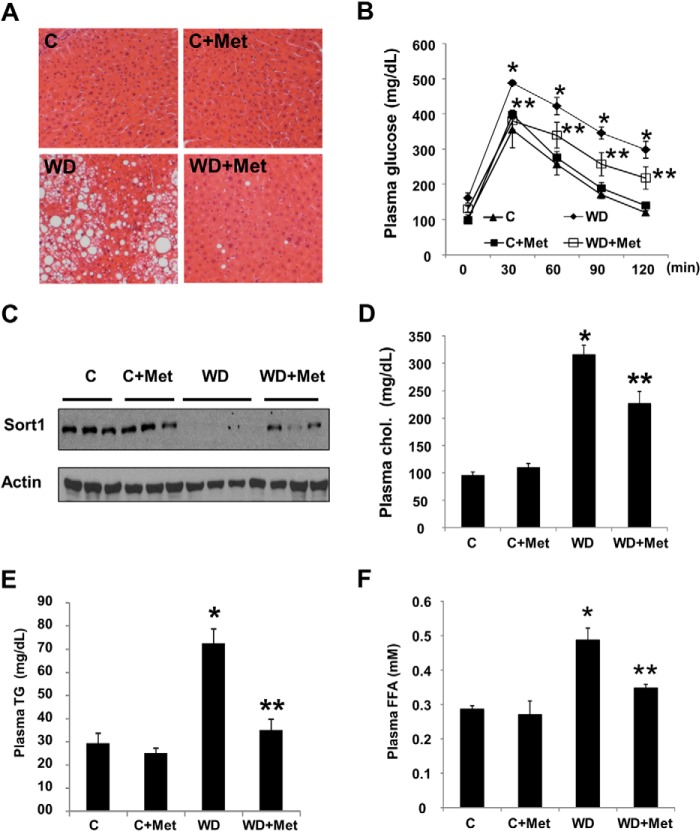
To obtain further in vivo evidence to prove that decreased PI3K signaling causes hepatic Sort1 down-regulation, we injected mice with a PI3K inhibitor, PX866, a wortmannin analogue with higher potency and lower cytotoxicity (15). A single injection of PX866 rapidly decreased liver AKT phosphorylation and liver Sort1 protein at 8 h postinjection without causing a significant reduction in the Sort1 mRNA levels (Fig. 2, D and E). PX866 injection once daily for 3 days yielded a similar reduction in liver Sort1 protein in mice (data not shown). Consistent with a previous report that wortmannin significantly increased hepatic apoB100/VLDL secretion and caused hyperlipidemia in mice (16), PX866 injection for 3 days caused significantly elevated plasma TG and cholesterol levels (see Fig. 6). FPLC analysis showed that PX866 treatment primarily raised plasma LDL-C with little effect on plasma HDL-cholesterol (Fig. 2F). These results suggested that blocking hepatic insulin/PI3K signaling caused rapid and marked down-regulation of Sort1, which inversely correlated with plasma lipid levels. To further test if improving insulin sensitivity in insulin-resistant mice could increase hepatic Sort1 levels, we administered the insulin sensitizer metformin to mice because a recent study showed that metformin significantly improved hepatic insulin sensitivity in obese mice via AMP-activated protein kinase-dependent inhibition of hepatic lipogenesis (17). Consistently, metformin decreased hepatic steatosis (Fig. 3A) and improved whole body insulin sensitivity, as indicated by a glucose tolerance test (Fig. 3B). Metformin did not induce hepatic Sort1 in insulin-sensitive chow-fed mice but partially restored hepatic Sort1 levels in Western diet-fed mice (Fig. 3C). In addition, metformin treatment at 0.5 and 1 mm for 8 or 18 h did not affect Sort1 protein in AML12 cells (not shown), suggesting that metformin probably increased hepatic Sort1 protein in mice indirectly by improving hepatic insulin sensitivity. Metformin also decreased plasma cholesterol, TG, and free fatty acid levels in Western diet-fed mice (Fig. 3, D–F), which may be at least partly due to improved hepatic insulin sensitivity and steatosis.
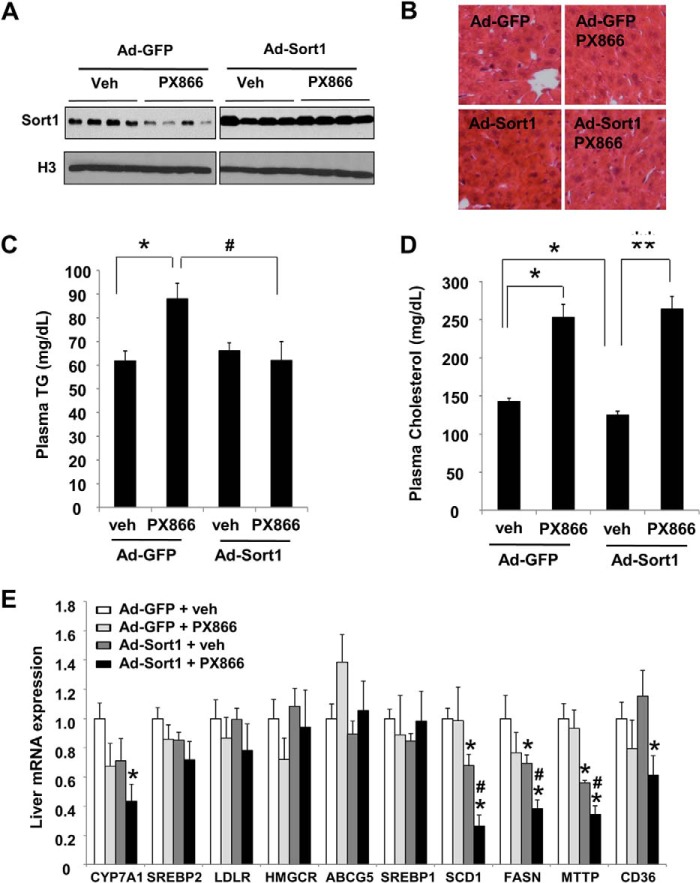
FIGURE 6.
Hepatic Sort1 overexpression prevented the triglyceride-raising effect of PX866 in wild type mice. Male 12-week-old C57BL6J mice were intravenously injected with Ad-GFP or Ad-Sort1 at 5 × 108 pfu/mouse. After 5 days, mice received either vehicle (veh) or 8 mg/kg PX866 intraperitoneal injection once daily for an additional 3 days. Mice were sacrificed after an 8-h fast. A, liver Sort1 protein was measured by immunoblot. B, H&E staining of liver sections. C and D, plasma triglyceride (C) and cholesterol (D) were measured with assay kits. E, liver mRNA expression was measured by real-time PCR. Results in C–E are plotted as mean ± S.E. (error bars) (n = 4). p ≤ 0.05 is considered as statistical significance; *, versus Ad-GFP + vehicle group; **, versus Ad-Sort1 + vehicle group; #, versus Ad-GFP + PX866 group.
FIGURE 3.
Metformin improved insulin sensitivity and partially restored hepatic Sort1 levels in Western diet-fed mice. Male C57BL6J mice at 12 weeks of age were fed a chow diet (C) or Western diet (WD) with or without 0.25% (w/w) metformin (Met) for 8 weeks. A, H&E staining of liver section. B, a glucose tolerance test was performed in overnight-fasted mice. C, the liver Sort1 protein was measured by immunoblot. D–F, plasma cholesterol, triglyceride, and free fatty acid (FFA) were measured with assay kits. All results are plotted as mean ± S.E. (error bars) (n = 4). p ≤ 0.05 is considered as statistical significance; *, versus chow-fed group; **, versus Western diet-fed group.
Phosphorylation of Sort1 Mediates Insulin Regulation of Sort1
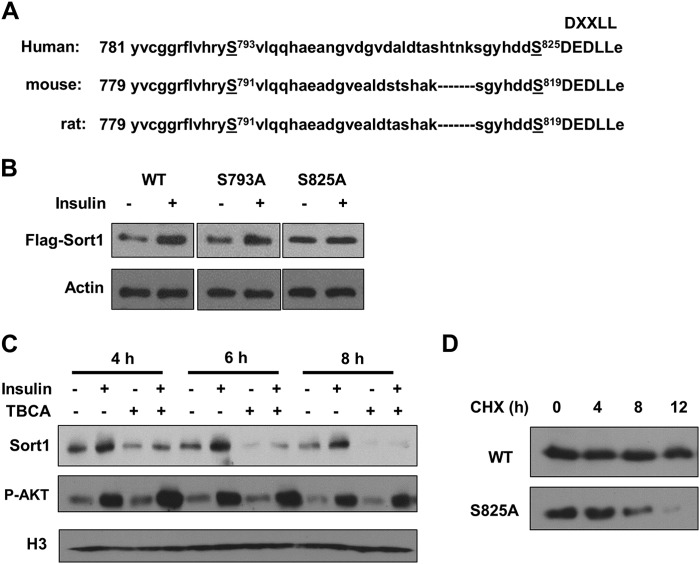
After establishing a regulatory relationship between insulin signaling and Sort1 protein in both in vitro and in vivo models, we next investigated the molecular mechanisms underlying the insulin/PI3K/AKT signaling regulation of Sort1. We first tested the hypothesis that insulin may regulate Sort1 localization and/or protein stability via protein phosphorylation. LC-MS/MS analyses showed that Sort1 was phosphorylated at serine residues Ser-793 and Ser-825 on the cytoplasmic tail (Table 1 and Fig. 4A). No verified phosphorylation sites were identified on the luminal/extracellular domain of the Sort1 protein. To test whether phosphorylation at these two residues is involved in insulin regulation of Sort1, we first expressed FLAG-tagged wild type or S793A or S825A phosphomutant forms of Sort1 in AML12 cells. As shown in Fig. 4B, insulin treatment induced ectopically expressed wild type and S793A mutant Sort1 but not S825A mutant Sort1, which provided the initial evidence that phosphorylation of Ser-825, but not Ser-793, was required for insulin induction of Sort1 (Fig. 4B).
TABLE 1.
LC-MS/MS identification of phosphorylation sites on human Sort1 purified from mouse liver
| Site | Peptide sequence | Localization probability | MI score | Δppm | A score | Start | Stop |
|---|---|---|---|---|---|---|---|
| Ser-825 | HDDS*DEDLL | 100% | 38.04 | 0.39 | 1000 | 822 | 830 |
| Ser-793 | YS*VLQQHAEANGVDGVDA | 100% | 71.67 | −0.64 | 33.26 | 792 | 818 |
FIGURE 4.
Phosphorylation of Ser-825 is required for insulin to induce Sort1 in AML12 cells. A, sequence illustrates phosphorylated serine residues Ser-793 and Ser-825 (S) identified in human Sort1 protein by LC-MS/MS and the conserved DXXLL motifs among species. B, AML12 cells were transfected with wild type and mutant human FLAG-Sort1 plasmids, as indicated. Cells were serum-starved for 16 h and treated with 100 nm insulin for 8 h. Ectopically expressed Sort1 was detected with anti-FLAG antibody in immunoblot. C, serum-starved (16 h) AML12 cells were pretreated with 15 μm TBCA for 1 h followed by 100 nm insulin treatment as indicated. Sort1 protein was measured by immunoblot. D, AML12 cells transfected with wild type and mutant human FLAG-Sort1 plasmids were treated with 100 μg/ml cycloheximide (CHX). FLAG-Sort1 protein was measured by immunoblot. P-AKT, phospho-AKT.
Despite the fact that constitutively active AKT directly induced Sort1 protein (Fig. 1F), Ser-825 is not within an RXRXX(S/T)-like consensus AKT phosphorylation sequence and thus is probably not directly phosphorylated by AKT (Fig. 4A). Instead, previous studies have established that Ser-825 is a consensus casein kinase 2 (CK2) target and was directly phosphorylated by CK2 in an in vitro kinase assay (18). Interestingly, previous studies have also demonstrated that CK2 is an insulin-sensitive kinase, and insulin induced CK2 activity in liver cells and rat livers, whereas CK2 activity decreased in diabetic rat livers (19, 20). We therefore asked whether CK2 was involved in insulin regulation of Sort1. Results in Fig. 4C showed that a CK2 inhibitor TBCA largely prevented insulin induction of Sort1 in AML12 cells (Fig. 4C). Furthermore, TBCA treatment alone resembled the effects of wortmannin and AKT inhibitor and strongly decreased basal Sort1 protein (Fig. 4C). These results not only suggested that CK2 was required for insulin induction of Sort1 but also indicated that blocking CK2 phosphorylation of Ser-825 may promote Sort1 protein degradation. Indeed, when cells were treated with cycloheximide to block protein synthesis, phosphorylation-defective S825A mutant Sort1 showed a faster degradation rate than wild type Sort1 (Fig. 4D). Unfortunately, we could not directly test the effect of insulin on Ser-825 phosphorylation because of the lack of a commercially available phospho-specific antibody. However, existing knowledge from previous studies (18–20) and new results presented here suggested that CK2-dependent phosphorylation of Ser-825 was probably involved in the insulin regulation of Sort1 protein stability.
PI3K Inhibition Results in Proteasome-dependent Sort1 Degradation
As illustrated in Fig. 4A, Ser-825 is adjacent to the N-terminal of the DXXLL dileucine motif, which is highly conserved among a large number of trafficking proteins (21). Previous studies have established that the DXXLL motif mediates the highly specific interaction with an adaptor protein called Golgi-localized, γ-ear-containing Arf-binding protein (GGA), and the DXXLL-GGA interaction was required for clathrin-coated vesicle formation and subsequent transport from TGN to endosomes/lysosomes (18, 22, 23). Structural and functional studies further found that phosphorylation of Ser-825 by CK2 significantly increased the binding affinity of the DXXLL motif with GGAs (24), whereas abolishing serine phosphorylation decreased the DXXLL-GGA interaction and impaired receptor lysosome transport efficiency (25). Based on these previous findings, we next asked whether impaired insulin/PI3K signaling alters Sort1 cellular trafficking, an event that could predispose Sort1 protein to degradation. We first tested whether the phosphorylation-defective S825A mutant Sort1 showed altered cellular localization patterns and found that both wild type and S825A mutant Sort1 co-localized primarily with the Golgi marker TGN46 (Fig. 5A) with minimal co-localization with lysosome marker Lamp1 (Fig. 5B). These results are consistent with previous findings of a primary TGN localization of Sort1 in many cell types (9, 11) and further suggest that preventing Ser-825 phosphorylation did not cause a major shift of Sort1 cellular localization. We next asked whether blocking PI3K by wortmannin alters Sort1 localization before wortmannin-induced Sort1 degradation. In AML12 cells, wortmannin treatment resulted in a marked Sort1 down-regulation at 4 h but not at the 2 h time point (Fig. 5C). However, wortmannin treatment for 2 h did not significantly alter Sort1 cellular localization, which further suggested that the majority of the Sort1 protein still resided in the TGN, but not lysosomes, shortly before wortmannin-induced protein degradation (Fig. 5, D and E). In addition, treatment with the lysosome inhibitor chloroquine or bafilomycin A1 for 8 h did not increase but rather markedly decreased Sort1 protein in AML12 cells (Fig. 5F). Despite the fact that Sort1 primarily traffics between TGN and lysosomes, these results suggested that the involvement of lysosomes in mediating such rapid and marked Sort1 degradation seemed unlikely and prompted us to further test the involvement of proteasome-mediated Sort1 degradation. Indeed, proteasome inhibition by MG132 completely prevented wortmannin-induced Sort1 degradation (Fig. 5G). Similarly, MG132 also prevented the CK2 inhibitor TBCA-induced Sort1 degradation (Fig. 5I). It is noted that MG132 treatment alone caused some reduction in basal Sort1 protein levels (Fig. 5, G and I), but the underlying mechanism was not fully clear. We then tested the effect of wortmannin or TBCA treatment on Sort1 protein ubiquitination. Results showed that wortmannin treatment (Fig. 5H) or TBCA treatment (Fig. 5J) for 2 h significantly increased the amount of ubiquitinated Sort1, which provides additional evidence to suggest that the proteasome-dependent pathway may be involved in Sort1 protein degradation.
Preventing Hepatic Sort1 Down-regulation Attenuated Hypertriglyceridemia in PX866-treated Mice
Previous studies showed that liver-specific Sort1 overexpression decreased plasma cholesterol and TG levels in various genetic and diet-induced hyperlipidemic mouse models (5, 11, 12). Last, we asked how preventing hepatic Sort1 down-regulation by adenovirus-mediated Sort1 overexpression could affect plasma lipid levels in the PX866-induced hyperlipidemic mouse model. As shown in Fig. 6A, Ad-Sort1 injection increased hepatic Sort1 protein ∼2.5-fold over baseline levels and overcame PX866-induced Sort1 down-regulation. Unlike pathological hepatic insulin resistance associated with obesity and diabetes, pharmacological inhibition of PI3K signaling did not cause hepatic steatosis in mice (Fig. 6B). Interestingly, Sort1 overexpression completely prevented PX866-induced plasma TG elevation (Fig. 6C). This is consistent with previous findings that blocking PI3K signaling by wortmannin increased hepatic TG secretion in mice (16) and that hepatic Sort1 overexpression inhibited hepatic TG secretion in mice (5, 12). However, Sort1 overexpression only decreased plasma cholesterol in vehicle-treated mice and did not prevent PX866-induced plasma cholesterol elevation (Fig. 6D), suggesting that redundant mechanisms mediated the cholesterol-raising effect of PX866. To obtain further molecular insights into the TG-lowering effect of Sort1 in PX866-treated mice, we measured the mRNA expression of genes involved in hepatic lipid metabolism (Fig. 6E). Among all genes measured, we found that the mRNA levels of stearoyl-CoA desaturase 1, fatty acid synthase, CD36, and microsomal triglyceride transfer protein were markedly decreased in PX866/Ad-Sort1-treated mice, which was consistent with lower plasma TG levels in this group of mice. We also found a strong reduction of the cholesterol 7α-hydroxylase mRNA in the Ad-Sort1/PX866-treated group, suggesting that decreased hepatic cholesterol catabolism may offset the cholesterol-lowering effect of Sort1 overexpression in this group of mice. The mRNA of sterol regulatory element-binding proteins 1 and 2, LDL receptor, HMG-CoA reductase, and ATP-binding cassette subfamily G member 5 were not significantly different between groups.
DISCUSSION
This study suggests that hepatic Sort1 acts as a novel molecular link between insulin resistance and altered hepatic apoB100 metabolism in hepatocytes. Hepatic apoB100/VLDL overproduction is considered as the primary defect in the pathogenesis of hyperlipidemia in type 2 diabetes. Although the inhibitory effect of insulin on hepatic apoB100 production has been well established (4, 26), the downstream mediators have not been well defined. Recent studies have shown that hepatic Sort1 lowered plasma cholesterol and TG levels by inhibiting hepatic apoB100/VLDL production in mice (5, 11, 27). New results presented here demonstrated that hepatic Sort1 was highly sensitive to insulin resistance-induced degradation. Of note, pharmacological inhibition of PI3K signaling and STZ-induced insulin deficiency in lean mice caused hepatic Sort1 down-regulation in the absence of hepatic fat accumulation, which is another causal factor for VLDL overproduction in type 2 diabetes. This is consistent with previous findings that insulin-dependent hepatic apoB100 degradation depends on the PI3K signaling and that treatment with the PI3K inhibitor wortmannin significantly increases hepatic VLDL production in mice (16).
The major goal of this study was to delineate the molecular mechanisms by which insulin regulates Sort1 in the liver. To this end, we have demonstrated that insulin signaling regulated Sort1 via PI3K/AKT-dependent posttranslational mechanisms. This study presented important evidence to support that phosphorylation of Ser-825 was required for insulin induction of Sort1 and that inhibition of each component in the insulin/PI3K/AKT/CK2 axis or abolishing Ser-825 phosphorylation was sufficient to promote Sort1 protein degradation. Thus, in addition to the previous findings that CK2-dependent Ser-825 phosphorylation critically regulated receptor cellular trafficking function (18, 22–25), this study further showed that Ser-825 phosphorylation regulated Sort1 protein stability. We recognize that our mechanistic study was somewhat limited by the lack of a commercially available Ser-825 phospho-specific Sort1 antibody. However, this novel signaling mechanism of Sort1 regulation is highly consistent with and thus supported by previous findings on the regulation of Sort1 trafficking and function (18–20, 22–25). Furthermore, this study suggests that blocking PI3K signaling probably increased Sort1 E3-ligase recognition and subsequent ubiquitination and proteasome-mediated degradation. Previous studies suggested that preventing Ser-825 phosphorylation may decrease Sort1 lysosome targeting (24, 25). However, our data were not sufficient to conclude whether increased Sort1 degradation was secondary to decreased Sort1 exit from the TGN because both WT and S825A mutant Sort1 showed a primary TGN localization. Two other studies have shown that lysosomes may also mediate Sort1 protein turnover (28, 29). It is known that Sort1 protein that reached the lysosome compartment is rapidly and efficiently recycled back to the TGN via the interaction with a retromer complex for reuse, and these two studies showed that decreasing Sort1 transport from the lysosome back to the TGN by preventing Sort1 palmitoylation increased Sort1 degradation in the lysosomes (28, 29). In our study, the primary TGN localization of Sort1 shortly before wortmannin-induced rapid degradation excluded a major role of the lysosome-mediated degradation pathway. Of note, both chloroquine and bafilomycin A1 decreased cellular Sort1 protein levels, suggesting that normal endosome/lysosome function was actually required for Sort1 trafficking between TGN and lysosomes. In addition, this evidence collectively supports the possibility that impaired Sort1 trafficking may be closely linked to Sort1 protein stability.
In this study, we further tested how preventing hepatic Sort1 down-regulation by adenovirus-mediated gene expression could affect plasma lipids in a PX866-induced hyperlipidemic mouse model. We found that Sort1 overexpression in the liver completely prevented PX866-induced plasma TG elevation (Fig. 6C). In addition to the known effect of Sort1 on hepatic TG production, it was interesting to find decreased expression of several hepatic lipogenic genes in the Ad-Sort1/PX866-treated group. Although the mechanism underlying these changes is not clear and is beyond the scope of the current study, we speculate that reduced lipogenic gene expression could be secondary to the Sort1 effect on hepatic apoB100 metabolism because several previous studies have consistently shown that inhibition of hepatic apoB100 by antisense oligonucleotide results in decreased hepatic lipogenic gene expressions, which explains the absence of hepatic steatosis despite significantly reduced hepatic TG secretion (30–32). Furthermore, PX866 treatment could also contribute to further decreased gene expression because insulin/PI3K is known to positively regulate lipogenic gene expressions (33–35).
In contrast to the strong TG-lowering effect, Sort1 overexpression did not prevent the PX866-induced hypercholesterolemia (Fig. 6D). One possibility for such a discrepancy compared with a few previous studies may be the non-pathological hyperlipidemic mouse model used in this study (5, 11, 13). In addition, we speculate that the cholesterol-lowering effect of Sort1 overexpression could be masked in this mouse model due to other pleiotropic effects of PX866 on cholesterol metabolic pathways that could not be regulated by Sort1. Of note, hepatic cholesterol 7α-hydroxylase was markedly decreased in Ad-Sort1/PX866-treated mice. In addition, it was previously shown that blocking PI3K signaling by insulin receptor knockout caused not only hepatic TG overproduction but also impaired hepatic LDL-C uptake (36, 37). It was thus possible that PX866 could also impair hepatic LDL uptake in our experimental setting. In contrast, hepatic VLDL overproduction was probably the primary defect underlying pathophysiological diabetic hyperlipidemia because hepatic cholesterol catabolism and LDL-C uptake did not decrease in obese and diabetic mice (33, 36). Therefore, due to the different model used in the study, our plasma cholesterol data do not necessarily contradict a number of previous reports showing that hepatic Sort1 overexpression significantly lowered plasma cholesterol in obese and hyperlipidemic mice (5, 11–14).
In summary, this study revealed a novel posttranslational regulation of hepatic Sort1 by insulin/PI3K/AKT signaling and thus provided new mechanistic insights into the altered hepatic lipid metabolism under insulin-resistant conditions. Sort1 is a novel sorting receptor implicated in hepatic and plasma lipid metabolism in humans. However, despite being a multifunctional receptor, the roles of liver Sort1 beyond the regulation of hepatic lipid secretion are still incompletely understood. Future studies that aim at better defining both the function and the regulation of Sort1 in metabolism are necessary to better define the role of Sort1 in the regulation of lipid homeostasis.
Acknowledgment
We thank the Mouse Metabolic Phenotyping Center at the University of Cincinnati (supported by National Institutes of Health Grant U24 DK059630) for the FPLC analysis of the plasma lipoprotein profile.
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grant 1R01DK102487-01 (to T. L.), NIH, NCRR, Grant 5P20RR021940-07, and NIH, NIGMS, Grant 8 P20 GM103549-07. This work was also supported by an American Diabetes Association Junior Faculty Award (to T. L.).
- TG
- triglyceride
- Sort1
- sortilin 1
- apoB100
- apolipoprotein B100
- TGN
- trans-Golgi network
- CK2
- casein kinase 2
- LDL-C
- LDL-cholesterol
- STZ
- streptozotocin
- TBCA
- tetrabromocinnamic acid
- GGA
- Golgi-localized, γ-ear-containing Arf-binding protein.
REFERENCES
- 1. Reaven G. M. (2002) Multiple CHD risk factors in type 2 diabetes: beyond hyperglycaemia. Diabetes Obes. Metab. 4, S13–S18 [DOI] [PubMed] [Google Scholar]
- 2. Cohen J. C., Horton J. D., Hobbs H. H. (2011) Human fatty liver disease: old questions and new insights. Science 332, 1519–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choi S. H., Ginsberg H. N. (2011) Increased very low density lipoprotein (VLDL) secretion, hepatic steatosis, and insulin resistance. Trends Endocrinol. Metab. 22, 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fisher E. A. (2012) The degradation of apolipoprotein B100: multiple opportunities to regulate VLDL triglyceride production by different proteolytic pathways. Biochim. Biophys. Acta 1821, 778–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Musunuru K., Strong A., Frank-Kamenetsky M., Lee N. E., Ahfeldt T., Sachs K. V., Li X., Li H., Kuperwasser N., Ruda V. M., Pirruccello J. P., Muchmore B., Prokunina-Olsson L., Hall J. L., Schadt E. E., Morales C. R., Lund-Katz S., Phillips M. C., Wong J., Cantley W., Racie T., Ejebe K. G., Orho-Melander M., Melander O., Koteliansky V., Fitzgerald K., Krauss R. M., Cowan C. A., Kathiresan S., Rader D. J. (2010) From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 466, 714–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Linsel-Nitschke P., Heeren J., Aherrahrou Z., Bruse P., Gieger C., Illig T., Prokisch H., Heim K., Doering A., Peters A., Meitinger T., Wichmann H. E., Hinney A., Reinehr T., Roth C., Ortlepp J. R., Soufi M., Sattler A. M., Schaefer J., Stark K., Hengstenberg C., Schaefer A., Schreiber S., Kronenberg F., Samani N. J., Schunkert H., Erdmann J. (2010) Genetic variation at chromosome 1p13.3 affects sortilin mRNA expression, cellular LDL-uptake and serum LDL levels which translates to the risk of coronary artery disease. Atherosclerosis 208, 183–189 [DOI] [PubMed] [Google Scholar]
- 7. Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., Rieder M. J., Cooper G. M., Roos C., Voight B. F., Havulinna A. S., Wahlstrand B., Hedner T., Corella D., Tai E. S., Ordovas J. M., Berglund G., Vartiainen E., Jousilahti P., Hedblad B., Taskinen M. R., Newton-Cheh C., Salomaa V., Peltonen L., Groop L., Altshuler D. M., Orho-Melander M. (2008) Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40, 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Myocardial Infarction Genetics Consortium, Kathiresan S., Voight B. F., Purcell S., Musunuru K., Ardissino D., Mannucci P. M., Anand S., Engert J. C., Samani N. J., Schunkert H., Erdmann J., Reilly M. P., Rader D. J., Morgan T., Spertus J. A., Stoll M., Girelli D., McKeown P. P., Patterson C. C., Siscovick D. S., O'Donnell C. J., Elosua R., Peltonen L., Salomaa V., Schwartz S. M., Melander O., Altshuler D., Ardissino D., Merlini P. A., Berzuini C., Bernardinelli L., Peyvandi F., Tubaro M., Celli P., Ferrario M., Fetiveau R., Marziliano N., Casari G., Galli M., Ribichini F., Rossi M., Bernardi F., Zonzin P., Piazza A., Mannucci P. M., Schwartz S. M., Siscovick D. S., Yee J., Friedlander Y., Elosua R., Marrugat J., Lucas G., Subirana I., Sala J., Ramos R., Kathiresan S., Meigs J. B., Williams G., Nathan D. M., MacRae C. A., O'Donnell C. J., Salomaa V., Havulinna A. S., Peltonen L., Melander O., Berglund G., Voight B. F., Kathiresan S., Hirschhorn J. N., Asselta R., Duga S., Spreafico M., Musunuru K., Daly M. J., Purcell S., Voight B. F., Purcell S., Nemesh J., Korn J. M., McCarroll S. A., Schwartz S. M., Yee J., Kathiresan S., Lucas G., Subirana I., Elosua R., Surti A., Guiducci C., Gianniny L., Mirel D., Parkin M., Burtt N., Gabriel S. B., Samani N. J., Thompson J. R., Braund P. S., Wright B. J., Balmforth A. J., Ball S. G., Hall A., Wellcome Trust Case Control, C., Schunkert H., Erdmann J., Linsel-Nitschke P., Lieb W., Ziegler A., Konig I., Hengstenberg C., Fischer M., Stark K., Grosshennig A., Preuss M., Wichmann H. E., Schreiber S., Schunkert H., Samani N. J., Erdmann J., Ouwehand W., Hengstenberg C., Deloukas P., Scholz M., Cambien F., Reilly M. P., Li M., Chen Z., Wilensky R., Matthai W., Qasim A., Hakonarson H. H., Devaney J., Burnett M. S., Pichard A. D., Kent K. M., Satler L., Lindsay J. M., Waksman R., Knouff C. W., Waterworth D. M., Walker M. C., Mooser V., Epstein S. E., Rader D. J., Scheffold T., Berger K., Stoll M., Huge A., Girelli D., Martinelli N., Olivieri O., Corrocher R., Morgan T., Spertus J. A., McKeown P., Patterson C. C., Schunkert H., Erdmann E., Linsel-Nitschke P., Lieb W., Ziegler A., Konig I. R., Hengstenberg C., Fischer M., Stark K., Grosshennig A., Preuss M., Wichmann H. E., Schreiber S., Holm H., Thorleifsson G., Thorsteinsdottir U., Stefansson K., Engert J. C., Do R., Xie C., Anand S., Kathiresan S., Ardissino D., Mannucci P. M., Siscovick D., O'Donnell C. J., Samani N. J., Melander O., Elosua R., Peltonen L., Salomaa V., Schwartz S. M., Altshuler D. (2009) Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41, 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kjolby M., Andersen O. M., Breiderhoff T., Fjorback A. W., Pedersen K. M., Madsen P., Jansen P., Heeren J., Willnow T. E., Nykjaer A. (2010) Sort1, encoded by the cardiovascular risk locus 1p13.3, is a regulator of hepatic lipoprotein export. Cell Metab. 12, 213–223 [DOI] [PubMed] [Google Scholar]
- 10. Hermey G. (2009) The Vps10p-domain receptor family. Cell Mol. Life Sci. 66, 2677–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Strong A., Ding Q., Edmondson A. C., Millar J. S., Sachs K. V., Li X., Kumaravel A., Wang M. Y., Ai D., Guo L., Alexander E. T., Nguyen D., Lund-Katz S., Phillips M. C., Morales C. R., Tall A. R., Kathiresan S., Fisher E. A., Musunuru K., Rader D. J. (2012) Hepatic sortilin regulates both apolipoprotein B secretion and LDL catabolism. J. Clin. Invest. 122, 2807–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ai D., Baez J. M., Jiang H., Conlon D. M., Hernandez-Ono A., Frank-Kamenetsky M., Milstein S., Fitzgerald K., Murphy A. J., Woo C. W., Strong A., Ginsberg H. N., Tabas I., Rader D. J., Tall A. R. (2012) Activation of ER stress and mTORC1 suppresses hepatic sortilin-1 levels in obese mice. J. Clin. Invest. 122, 1677–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bi L., Chiang J. Y., Ding W. X., Dunn W., Roberts B., Li T. (2013) Saturated fatty acids activate ERK signaling to down-regulate hepatic sortilin 1 in obese and diabetic mice. J. Lipid Res. 54, 2754–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li J., Bi L., Hulke M., Li T. (2014) Fish oil and fenofibrate prevented phosphorylation-dependent hepatic sortilin 1 degradation in Western diet-fed mice. J. Biol. Chem. 289, 22437–22449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ihle N. T., Williams R., Chow S., Chew W., Berggren M. I., Paine-Murrieta G., Minion D. J., Halter R. J., Wipf P., Abraham R., Kirkpatrick L., Powis G. (2004) Molecular pharmacology and antitumor activity of PX-866, a novel inhibitor of phosphoinositide-3-kinase signaling. Mol. Cancer Ther. 3, 763–772 [PubMed] [Google Scholar]
- 16. Chirieac D. V., Davidson N. O., Sparks C. E., Sparks J. D. (2006) PI3-kinase activity modulates apo B available for hepatic VLDL production in apobec-1−/− mice. Am. J. Physiol. Gastrointest. Liver Physiol. 291, G382–G388 [DOI] [PubMed] [Google Scholar]
- 17. Fullerton M. D., Galic S., Marcinko K., Sikkema S., Pulinilkunnil T., Chen Z. P., O'Neill H. M., Ford R. J., Palanivel R., O'Brien M., Hardie D. G., Macaulay S. L., Schertzer J. D., Dyck J. R., van Denderen B. J., Kemp B. E., Steinberg G. R. (2013) Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat. Med. 19, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nielsen M. S., Madsen P., Christensen E. I., Nykjaer A., Gliemann J., Kasper D., Pohlmann R., Petersen C. M. (2001) The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 20, 2180–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sommercorn J., Mulligan J. A., Lozeman F. J., Krebs E. G. (1987) Activation of casein kinase II in response to insulin and to epidermal growth factor. Proc. Natl. Acad. Sci. U.S.A. 84, 8834–8838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martos C., Plana M., Guasch M. D., Itarte E. (1985) Effect of starvation, diabetes and insulin on the casein kinase 2 from rat liver cytosol. Biochem. J. 225, 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bonifacino J. S., Traub L. M. (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447 [DOI] [PubMed] [Google Scholar]
- 22. Puertollano R., Aguilar R. C., Gorshkova I., Crouch R. J., Bonifacino J. S. (2001) Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science 292, 1712–1716 [DOI] [PubMed] [Google Scholar]
- 23. Zhu Y., Doray B., Poussu A., Lehto V. P., Kornfeld S. (2001) Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science 292, 1716–1718 [DOI] [PubMed] [Google Scholar]
- 24. Kato Y., Misra S., Puertollano R., Hurley J. H., Bonifacino J. S. (2002) Phosphoregulation of sorting signal-VHS domain interactions by a direct electrostatic mechanism. Nat. Struct. Biol. 9, 532–536 [DOI] [PubMed] [Google Scholar]
- 25. Johnson K. F., Kornfeld S. (1992) The cytoplasmic tail of the mannose 6-phosphate/insulin-like growth factor-II receptor has two signals for lysosomal enzyme sorting in the Golgi. J. Cell Biol. 119, 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vergès B. (2010) Abnormal hepatic apolipoprotein B metabolism in type 2 diabetes. Atherosclerosis 211, 353–360 [DOI] [PubMed] [Google Scholar]
- 27. Chamberlain J. M., O'Dell C., Sparks C. E., Sparks J. D. (2013) Insulin suppression of apolipoprotein B in McArdle RH7777 cells involves increased sortilin 1 interaction and lysosomal targeting. Biochem. Biophys. Res. Commun. 430, 66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dumaresq-Doiron K., Jules F., Lefrancois S. (2013) Sortilin turnover is mediated by ubiquitination. Biochem. Biophys. Res. Commun. 433, 90–95 [DOI] [PubMed] [Google Scholar]
- 29. McCormick P. J., Dumaresq-Doiron K., Pluviose A. S., Pichette V., Tosato G., Lefrancois S. (2008) Palmitoylation controls recycling in lysosomal sorting and trafficking. Traffic 9, 1984–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mullick A. E., Fu W., Graham M. J., Lee R. G., Witchell D., Bell T. A., Whipple C. P., Crooke R. M. (2011) Antisense oligonucleotide reduction of apoB-ameliorated atherosclerosis in LDL receptor-deficient mice. J. Lipid Res. 52, 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crooke R. M., Graham M. J., Lemonidis K. M., Whipple C. P., Koo S., Perera R. J. (2005) An apolipoprotein B antisense oligonucleotide lowers LDL cholesterol in hyperlipidemic mice without causing hepatic steatosis. J. Lipid Res. 46, 872–884 [DOI] [PubMed] [Google Scholar]
- 32. Lee R. G., Fu W., Graham M. J., Mullick A. E., Sipe D., Gattis D., Bell T. A., Booten S., Crooke R. M. (2013) Comparison of the pharmacological profiles of murine antisense oligonucleotides targeting apolipoprotein B and microsomal triglyceride transfer protein. J. Lipid Res. 54, 602–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li T., Francl J. M., Boehme S., Ochoa A., Zhang Y., Klaassen C. D., Erickson S. K., Chiang J. Y. (2012) Glucose and insulin induction of bile acid synthesis: mechanisms and implication in diabetes and obesity. J. Biol. Chem. 287, 1861–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Horton J. D., Goldstein J. L., Brown M. S. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Owen J. L., Zhang Y., Bae S. H., Farooqi M. S., Liang G., Hammer R. E., Goldstein J. L., Brown M. S. (2012) Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc. Natl. Acad. Sci. U.S.A. 109, 16184–16189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ai D., Chen C., Han S., Ganda A., Murphy A. J., Haeusler R., Thorp E., Accili D., Horton J. D., Tall A. R. (2012) Regulation of hepatic LDL receptors by mTORC1 and PCSK9 in mice. J. Clin. Invest. 122, 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Biddinger S. B., Hernandez-Ono A., Rask-Madsen C., Haas J. T., Alemán J. O., Suzuki R., Scapa E. F., Agarwal C., Carey M. C., Stephanopoulos G., Cohen D. E., King G. L., Ginsberg H. N., Kahn C. R. (2008) Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metab. 7, 125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]