Background: The formation of Hsp60 complexes is poorly understood.
Results: The biogenesis of Hsp60 complexes depends on mitochondrial (mt) Hsp70 and Hsp10.
Conclusion: MtHsp70 interacts with Hsp10 to promote Hsp60 biogenesis.
Significance: Coupling to partner proteins like Hsp10 modifies the functional specificity of mtHsp70.
Keywords: 70-kDa Heat Shock Protein (Hsp70), Mitochondria, Molecular Chaperone, Protein Assembly, Protein Folding, Hsp10, Hsp60, Protein Biogenesis
Abstract
Mitochondrial Hsp70 (mtHsp70) mediates essential functions for mitochondrial biogenesis, like import and folding of proteins. In these processes, the chaperone cooperates with cochaperones, the presequence translocase, and other chaperone systems. The chaperonin Hsp60, together with its cofactor Hsp10, catalyzes folding of a subset of mtHsp70 client proteins. Hsp60 forms heptameric ring structures that provide a cavity for protein folding. How the Hsp60 rings are assembled is poorly understood. In a comprehensive interaction study, we found that mtHsp70 associates with Hsp60 and Hsp10. Surprisingly, mtHsp70 interacts with Hsp10 independently of Hsp60. The mtHsp70-Hsp10 complex binds to the unassembled Hsp60 precursor to promote its assembly into mature Hsp60 complexes. We conclude that coupling to Hsp10 recruits mtHsp70 to mediate the biogenesis of the heptameric Hsp60 rings.
Introduction
Heat shock proteins of 70 kDa (Hsp70) fulfill several essential functions in prokaryotic and eukaryotic cells, like protein folding and transport. Hsp70 proteins prevent the aggregation of misfolded proteins and facilitate the removal of protein aggregates. To perform these various tasks, the chaperones bind transiently to hydrophobic patches exposed in non-native conformations of their client proteins (1–3).
Mitochondrial Hsp70 (mtHsp70)2 plays a central role in mitochondrial biogenesis (2, 4). In baker's yeast (Saccharomyces cerevisiae), the major mtHsp70 is encoded by SSC1 and is essential for yeast survival (5). MtHsp70 dynamically binds to the presequence translocase (TIM23 complex) of the mitochondrial inner membrane to drive the import of precursor proteins into the matrix by an ATP-dependent cycle of precursor binding and release (6–12). The chaperone is the core component of the presequence translocase-associated motor. Tim44 forms the docking site for the chaperone at the TIM23 complex (13–16). The J domain-containing protein Pam18/Tim14, together with its partner protein Pam16/Tim16 and the nucleotide exchange factor Mge1, regulates the activity of mtHsp70 (17–24). In addition, the chaperone associates with the J protein Mdj1 and Mge1 to promote the folding of nucleus-encoded and mitochondrially encoded proteins in the matrix (25–31). Recent data identified further interactors of the chaperone. Zinc finger motif protein of 17 kDa (Zim17, also termed mtHsp70 escort protein 1 (Hep1)) supports the folding and function of the chaperone (32–37). Furthermore, mtHsp70 interacts with Mss51 and Cox4 to promote the biogenesis of the cytochrome c oxidase (complex IV of the respiratory chain) (38, 39). MtHsp70 cooperates with other chaperone systems to maintain protein homeostasis. It functions together with Hsp78 in protein disaggregation and proteolytic removal of misfolded proteins (40, 41). MtHsp70 also cooperates with the mitochondrial chaperonin system, consisting of Hsp60 and Hsp10, to promote the folding of a subset of client proteins (42–44).
Mitochondrial Hsp60 exists in single and double ring conformations, with one ring being composed of seven subunits (45–48). Detailed structural and mechanistic insights have been obtained for the bacterial counterpart GroEL and its Hsp10 homolog GroES (1, 3). The ring structure of the chaperonin provides a central cavity for folding of the enclosed client protein. The activity of the Hsp60 rings is driven by ATP-dependent conformational changes of the Hsp60 monomers. The heptameric Hsp10 ring forms the lid of the cavity and regulates the ATP-dependent reaction cycle of Hsp60 (47, 49, 50). Although Hsp60 is essential for cell survival (51), the assembly of the ring structures is poorly understood. MtHsp70 promotes the import of the Hsp60 precursor into the mitochondrial matrix (43). The subsequent formation of the Hsp60 ring structures depends on a pre-existing Hsp60 oligomer (44, 52, 53). Whether other factors support the formation of Hsp60 complexes is not known.
Despite its central role in mitochondrial biogenesis, comprehensive studies of the interaction partners of mtHsp70 are missing up to now. Here we performed affinity purification of His-tagged mtHsp70 and analyzed its binding partners by SILAC-based mass spectrometry. We found that mtHsp70 interacts with Hsp60 and Hsp10. Surprisingly, an mtHsp70-Hsp10 complex forms independently of Hsp60. We found that assembly of the Hsp60 precursor into the mature complexes is impaired in mutants of mtHsp70 and Hsp10. The unassembled Hsp60 precursor binds to both mtHsp70 and Hsp10. Therefore, we propose that coupling to Hsp10 enables mtHsp70 to promote the formation of the mature Hsp60 ring structures.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Conditions
The yeast wild-type strains YPH499, YPH499 arg4Δ, and JK9–3d (the wild type for hsp10ts and hsp60ts); the wild-type strain for ssc1–42; and the mutant strains mtHsp70His, ssc1–42, hsp10ts, and hsp60ts have been described before (39, 44, 54). For SILAC-based mass spectrometric analysis of mtHsp70His purification, a kanMX4 cassette was integrated into the ARG4 locus by homologous recombination in the yeast strain expressing mtHsp70His. For biochemical studies, yeast cells were grown to logarithmic growth phase at 23 °C or 30 °C on YPG medium (1% (w/v) yeast extract, 2% (w/v) bacto peptone, and 3% (v/v) glycerol). For cycloheximide treatment, yeast cells were grown at 30 °C in the presence of 50 μg/ml cycloheximide for 2 h. For in vivo heat shock, hsp60ts, hsp10ts, and wild-type strain cells were shifted to 37 °C for 2 h (44).
Isolation of Mitochondria and in Vitro Protein Import Assays
Mitochondria were isolated by differential centrifugation following a published procedure (39). Mitochondria were stored in SE buffer (10 mm MOPS/KOH (pH 7.2), 1 mm EDTA, and 250 mm sucrose) in a protein concentration of 10 mg/ml at −80 °C until use. To generate 35S-labeled precursor proteins, we used cell-free in vitro translation on the basis of rabbit reticulocyte lysate in the presence of 35S-labeled methionine (Promega). Standard import reactions were performed following established assays (39, 55). In brief, 35S-labeled precursors (5–10% of the reaction volume) were incubated with isolated mitochondria at 25 °C in import buffer (3% (w/v) BSA, 250 mm sucrose, 5 mm methionine, 80 mm KCl, 5 mm MgCl2, 10 mm MOPS/KOH (pH 7.2), and 2 mm KH2PO4). Energy was added to the reaction mixture in the form of 2 mm ATP and 2 mm NADH (final concentration). The import reaction was stopped by addition of an AVO mixture (8 μm antimycin A, 1 μm valinomycin, and 20 μm oligomycin, final concentrations) to dissipate the membrane potential. In the indicated control samples, the AVO mixture was added prior to import. For SDS-PAGE analysis, non-imported precursor proteins were removed by incubation with 50 μg/ml proteinase K for 15 min on ice. Subsequently, mitochondria were reisolated, washed with SE buffer, and lysed with SDS-PAGE loading buffer. When the import reaction was studied by blue native electrophoresis, mitochondria were washed with SE buffer and solubilized under non-denaturing conditions using the non-ionic detergent digitonin to 1% (w/v) final concentration in lysis buffer (20 mm Tris-HCl (pH 7.4), 50 mm NaCl, 10% (v/v) glycerol, and 0.1 mm EDTA).
Affinity Purification of mtHsp70His
Mitochondria from wild-type and mtHsp70His cells were isolated and lysed with lysis buffer containing 1% (w/v) digitonin and 10 mm imidazole at a final protein concentration of 1 mg/ml. Lysis was performed for 15 min on ice. When indicated, solubilization was performed in the presence of 10 mm ADP or ATP in buffer A (20 mm Tris-HCl (pH 7.4), 50 mm KCl, 10 mm MgCl2, and 10% (v/v) glycerol) containing 1% (w/v) digitonin. In all cases, unsoluble material was removed by centrifugation. The mitochondrial lysate was incubated with Ni2+-NTA-agarose (Qiagen) under constant rotation for 1 h at 4 °C. Unbound proteins were discarded, and the Ni2+-NTA-agarose beads were washed intensively with lysis buffer containing 0.1% (w/v) digitonin and 40 mm imidazole. To elute bound proteins, the Ni2+-NTA-agarose beads were incubated with lysis buffer containing 250 mm imidazole and 0.1% (w/v) digitonin.
For characterization of the binding of precursor proteins to mtHsp70His, a 5-fold standard import reaction with the 35S-labeled precursor was performed before affinity purification via Ni2+-NTA-agarose. The affinity purification followed the procedure described above. In the case of detection of protein complexes by blue native electrophoresis, bound proteins were eluted by addition of buffer A containing 20 mm ATP.
SILAC-based Affinity Purification
YPH499 arg4Δ (wild-type) and mtHsp70His arg4Δ cells were grown in synthetic medium (0.67% (w/v) bacto-yeast nitrogen base, amino acid mix, 3% (v/v) glycerol, and 0.2% (w/v) glucose) at 30 °C to an early logarithmic growth phase. The medium was supplemented with either 15N213C6-lysine or 15N413C6-arginine (Euriso-Top) for wild-type or 14N212C6-lysine and 14N412C6-arginine for mtHsp70His arg4Δ cells (39). Mitochondria were isolated following the standard procedure (39). Lysis and purification via Ni2+-NTA-agarose were performed as described above. Subsequently, the elution fractions were pooled and subjected to mass spectrometric analysis.
Mass Spectrometry and Data Analysis
Mass spectrometric analyses of affinity-purified mtHsp70His complexes were performed as described previously (56) with minor modifications. Protein complexes (n = 3) were precipitated with ice-cold acetone and resuspended in urea buffer (8 m urea in 50 mm NH4HCO3). Cysteine residues were reduced with 5 mm Tris-2-carboxyethylphosphine (30 min at 37 °C) and subsequently alkylated with 50 mm iodoacetamide (30 min at room temperature in the dark). 50 mm NH4HCO3 was added to reach a final urea concentration of 2 m. Proteins were then subjected to proteolysis with trypsin overnight at 37 °C. Peptides were dried, reconstituted in 0.1% (v/v) trichloroacetic acid, and subjected to LC/MS analysis on an LTQ-Orbitrap XL (Thermo Scientific, Bremen, Germany) directly coupled to an UltiMateTM 3000 RSLCnano system (Thermo Scientific). Peptide separation was performed using a C18 reverse-phase nano LC column (50 cm × 75 μm; Acclaim PepMap; particle size, 2 μm; pore size, 100 Å; Thermo Scientific). For peptide elution, a 130-min linear LC gradient ranging from 1.5–21% (v/v) acetonitrile and 2.5–35% (v/v) methanol in 4% (v/v) dimethyl sulfoxide/0.1% (v/v) formic acid was applied. The flow rate was 250 nl/min.
Full scan MS spectra in the range of m/z 370–1700 were acquired in the Orbitrap at a resolution of 60,000 at m/z 400. Simultaneously with completion of the full scan, up to five of the most intense peptide ions (z ≥ +2) were fragmented further by low-energy collision-induced dissociation in the linear ion trap. A dynamic exclusion time of 45 s was applied to prevent repeated fragmentation of previously selected precursor ions.
For protein identification and SILAC-based relative protein quantification, MS data were analyzed with MaxQuant (version 1.4.1.2) and its search algorithm Andromeda (57, 58). MS/MS data were searched against the Saccharomyces Genome Database (version 02/03/2011) (59) using the following parameters: mass tolerances of 4.5 ppm for precursor and 0.5 Da for fragment ions, tryptic specificity, maximum of two missed cleavages, acetylation of protein N termini and oxidation of methionine as variable modifications, carbamidomethylation of cysteine residues as fixed modification, and Arg-10 and Lys-8 as heavy labels. A false discovery rate of <0.01 on the peptide and protein level was applied. Proteins were identified on the basis of at least one unique peptide with at least seven amino acids. For the calculation of mtHsp70His/WT protein ratios, only unique peptides were taken into account, and the minimum number of ratio counts (i.e. SILAC peptide pairs) was one. Data were visualized by plotting the mean log10 mtHsp70His/WT ratios across all three replicates against the p value (determined using one-sided Student's t test) for each protein. Potential mtHsp70His -interacting proteins were required to have a sequence coverage of >4%, a ratio mtHsp70/WT of >5, and a p value of <0.05.
Coimmunoprecipitation of Imported Precursor Proteins
Protein A-Sepharose (GE Healthcare) was coated with anti-Hsp10 antibodies or its corresponding preimmune serum by covalent coupling with dimethylpimelidate. To detect association to imported precursor proteins, a 5-fold import reaction was performed following the procedure described above. After washing, the mitochondria were lysed with lysis buffer containing 1% (w/v) digitonin at a final protein concentration of 1 mg/ml. After removal of unsoluble material, the mitochondrial lysate was incubated with the indicated beads for 1 h under constant rotation at 4 °C. Unbound material was removed, and beads were washed intensively with lysis buffer containing 0.1% (w/v) digitonin. Finally, bound proteins were eluted at low pH using 0.1 m glycine (pH 2.5). Eluted samples were immediately neutralized with Tris base and analyzed by SDS-PAGE.
In the case of two-step purification, the 35S-labeled Hsp60 precursor was imported into mtHsp70His mitochondria. Subsequently, mitochondria were lysed and proteins were purified via Ni2+-NTA-agarose as described above. The elution sample in lysis buffer containing 250 mm imidazole was incubated with protein A-Sepharose coated with anti-Hsp10 or the corresponding preimmune serum. Coimmunoprecipitation was performed as described above.
Binding Assay to the Mge1 Affinity Matrix
For the preparation of the Mge1 affinity matrix, His-tagged Mge1 lacking the presequence was recombinantly expressed and purified as described previously (39). Purified Mge1His was coupled to Ni2+-NTA-agarose. Subsequently, affinity beads were washed several times with lysis buffer containing 0.1% (w/v) digitonin and 10 mm imidazole. Mitochondria were lysed in 1% (w/v) digitonin in lysis buffer at a final protein concentration of 1 mg/ml. After removal of unsoluble material by centrifugation, the mitochondrial lysate was incubated with the Mge1 affinity matrix for 1 h under constant rotation at 4 °C. Subsequently, unbound material was removed, and the beads were washed intensively with lysis buffer containing 0.1% (w/v) digitonin and 40 mm imidazole. To elute bound proteins, the affinity matrix was incubated with 250 mm imidazole and 0.1% (w/v) digitonin in lysis buffer.
Protein Aggregation Assay
To study protein aggregation in wild-type and mutant mitochondria, isolated mitochondria were solubilized in lysis buffer containing 1% (w/v) digitonin. After incubation on ice for 15 min, protein aggregates were collected by centrifugation for 10 min at 4 °C and 18,000 × g. Proteins of the supernatant were precipitated with trichloroacetic acid. All pellet fractions were denatured by solubilization with SDS-PAGE loading buffer and subjected to SDS-PAGE and Western blotting.
Miscellaneous
In general, the signals detected by the antibodies used in this study were confirmed intensively using wild-type and the corresponding mutant mitochondria. The antibodies have been described previously (39, 44, 60). For immunodetection, proteins were transferred by semidry Western blotting to a PVDF membrane (Millipore). After immunodecoration with the indicated antibodies, the signals were detected using the chemiluminescence kit (ECL, GE Healthcare). 35S-labeled proteins and protein complexes were detected by digital autoradiography (Storm imaging system, GE Healthcare). Analysis and quantification of protein signals were performed with ImageQuant 5.2 software (GE Healthcare).
RESULTS AND DISCUSSION
MtHsp70 Interacts with Hsp60 and Hsp10
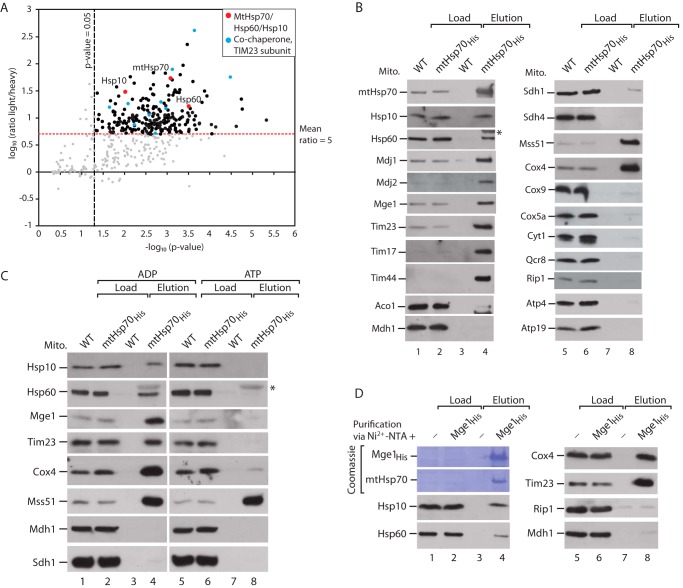
To identify novel binding partners of the major mitochondrial Hsp70, we used a yeast strain expressing a His-tagged mtHsp70 (mtHsp70His). We have demonstrated previously that the His-tagged mtHsp70 is functional and interacts with known cochaperones (39). Isolated mitochondria were lysed with the non-ionic detergent digitonin and subjected to affinity purification via Ni2+-NTA chromatography. The elution fractions from wild-type and mtHsp70His mitochondria were mixed and analyzed by stable isotope labeling by amino acids in cell culture (SILAC)-based mass spectrometry (61) (Fig. 1A, supplemental Table S1). We used the SILAC ratios to determine proteins that were specifically enriched in the mtHsp70His-bound fraction (Fig. 1A, supplemental Table S1). A large number of different proteins were copurified along with mtHsp70His, comprising potential interaction partners and substrate proteins of the chaperone. Known partner proteins, like subunits of the TIM23 complex and several cochaperones, were enriched in the mtHsp70His purification (Fig. 1A, supplemental Table S1). Surprisingly, Hsp10 and Hsp60 were found among the strongly enriched proteins. We analyzed the affinity purification by Western blotting and immunodetection using antibodies against various mitochondrial proteins (Fig. 1B). We could demonstrate the copurification of Hsp60 and Hsp10 along with mtHsp70His, indicating that a fraction of Hsp60 and Hsp10 interacts with mtHsp70 (Fig. 1B). Known mtHsp70 interactors, like the cochaperones Mge1, Mdj1, and Mdj2 and components of the TIM23 complex, bound to mtHsp70His with high efficiency. We also confirmed the binding of mtHsp70 to Cox4 and Mss51 (Fig. 1B) (38, 39). MtHsp70 binds to several interaction partners, like Tim44, Cox4, Zim17, and Mge1, in an ATP-sensitive manner (13–15, 27, 31, 35, 39, 62). Therefore, we performed the affinity purification via mtHsp70His in the presence of ATP or ADP. The association of both Hsp60 and Hsp10 to mtHsp70His is abolished in the presence of ATP (Fig. 1C, lanes 4 and 8). As reported, binding of Mss51 to Hsp70His is not affected upon addition of ATP, whereas other control proteins, like Tim23, Cox4, and Mge1, lose their contact with mtHsp70 (Fig. 1C, lanes 4 and 8) (38, 39). Next we analyzed whether binding of the cochaperone Mge1 to mtHsp70 interferes with the association of Hsp60 and Hsp10. To this end, we recombinantly expressed tagged Mge1 (Mge1His) and coupled the purified protein to Ni2+-NTA-agarose (39). Subsequently, the Mge1-affinity matrix was incubated with lysed mitochondria. MtHsp70 bound in stoichiometric amounts to the Mge1-coated column, as detected by Coomassie staining (Fig. 1D, lane 4). Hsp60 and Hsp10, as well as control proteins, were copurified along with the Mge1 affinity matrix, indicating that the chaperonin system binds to Mge1-bound mtHsp70 (Fig. 1D, lanes 4 and 8). Therefore, we conclude that Hsp10 and Hsp60 specifically bind to mature mtHsp70 in an ATP-sensitive manner.
FIGURE 1.
MtHsp70 interacts with the mitochondrial chaperonin system. A, SILAC-based quantitative mass spectrometric analysis of mtHsp70His affinity purifications. The mean log10 light/heavy ratio and −log10 p values of three independent experiments were determined and plotted against each other. The thresholds for the light/heavy ratio >5 and p > 0.05 are indicated. Red dots, mitochondrial chaperones; blue dots, cochaperones of mtHsp70 and TIM23 subunits; black dots, other proteins; gray dots, unspecific proteins (see supplemental Table S1). B, WT and mtHsp70His mitochondria (Mito) were lysed with digitonin and subjected to affinity purification via Ni2+-NTA-agarose. Bound proteins were eluted with imidazole and analyzed by SDS-PAGE, Western blotting, and immunodetection with the indicated antisera. Load, 0.5%; elution, 100%. Aco1, aconitase 1; Mdh1, mitochondrial malate dehydrogenase 1; Sdh, succinate dehydrogenase; Cox, cytochrome c oxidase; Atp, subunits of the F1FO-ATP synthase. The asterisk marks cross-reactivity of anti-Hsp60 antibody with mtHsp70. C, WT and mtHsp70His mitochondria were lysed in digitonin in the presence of ADP or ATP and subjected to affinity purification via Ni2+-NTA-agarose. Bound proteins were eluted with imidazole and analyzed by SDS-PAGE, Western blotting, and immunodetection with the indicated antisera. Load, 0.5%; elution, 100%. The asterisk marks cross-reactivity of anti-Hsp60 antibody with mtHsp70. D, WT mitochondria were lysed in digitonin and incubated with empty or Mge1His-coated Ni2+-NTA-agarose. Bound complexes were eluted with imidazole, and samples were analyzed by SDS-PAGE, Western blotting, and immunodetection with the indicated antisera. Load, 0.5%; elution, 100%.
MtHsp70 and the chaperonins capture substrate proteins during protein folding (31, 43, 63). Along this line, it has been reported that mtHsp70 binds to the Hsp60 precursor to promote its import (43). We asked whether the association between the chaperones persists upon prolonged incubation and would, therefore, reflect a stable binding to the mature Hsp60 and Hsp10 rather than transient binding to premature forms. To this end, yeast cells expressing mtHsp70His were incubated with cycloheximide, which blocks de novo synthesis of nucleus-encoded proteins (64). After incubation with cycloheximide, mitochondria were isolated, lysed with digitonin, and subjected to affinity purification via Ni2+-NTA-agarose (Fig. 2). Incubation with cycloheximide did not affect the association of Hsp60 and Hsp10 with mtHsp70 (Fig. 2, lanes 6 and 7). Similarly, the stable interactions with Mge1 and the presequence translocase were not affected (Fig. 2, lanes 6 and 7). We conclude that the association of mtHsp70 to Hsp60 and Hsp10 does not depend on the import of newly synthesized proteins. Therefore, these mitochondrial chaperones interact stably with each other.
FIGURE 2.
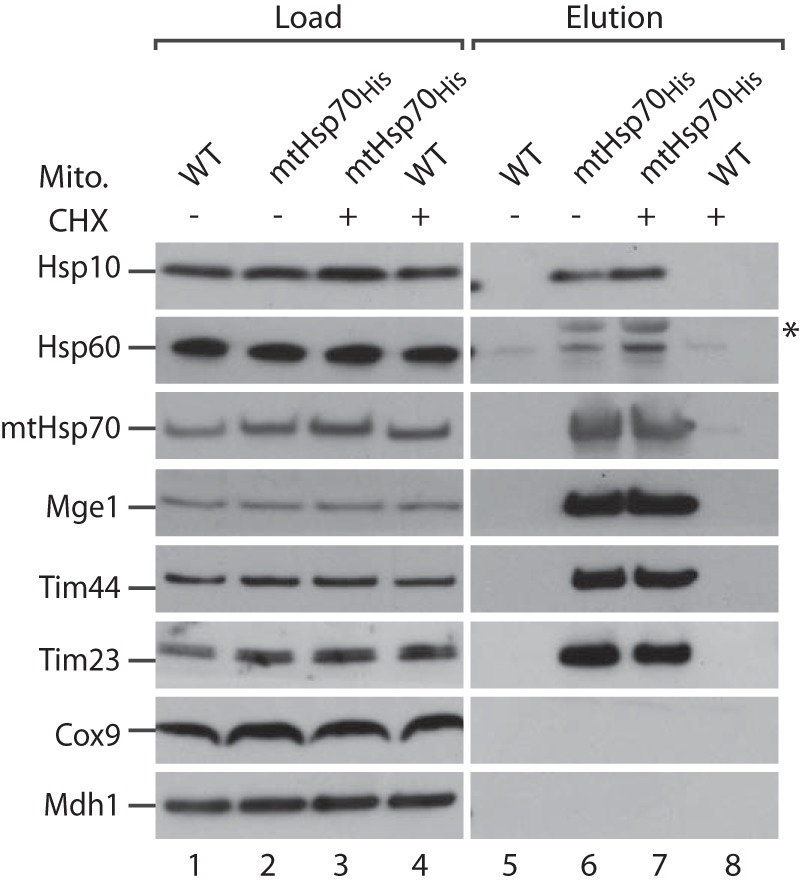
MtHsp70 interacts stably with mitochondrial Hsp60 and Hsp10. WT and mtHsp70His yeast cells were grown in the presence or absence of cycloheximide (CHX) as described under “Experimental Procedures.” Mitochondria (Mito) were isolated and subjected to affinity purification via Ni2+-NTA-agarose. Bound proteins were eluted with imidazole and analyzed by SDS-PAGE, Western blotting, and immunodetection with the indicated antisera. Load, 0.5%; elution, 100%. The asterisk marks cross-reactivity of anti-Hsp60 antibody with mtHsp70.
MtHsp70 Interacts with Hsp10 Independently of Hsp60
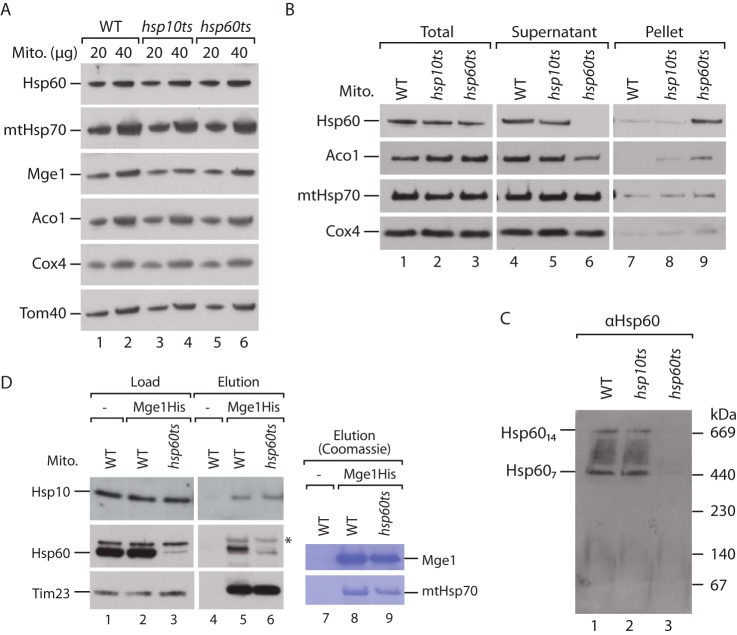
We have shown that mtHsp70 interacts with Hsp60 and Hsp10. The association of both chaperone systems may facilitate substrate channeling, as suggested for the bacterial homologs (65). However, Hsp10 interacts stably with Hsp60 in the presence, but not in the absence, of ATP or ADP (47, 49, 66, 67). The observed binding of Hsp10 to mtHsp70 occurred without the addition of nucleotides and was lost in the presence of ATP (Fig. 1, B and C). Therefore, the mode of interaction of Hsp10 with Hsp60 and with mtHsp70 differs. We wondered whether Hsp10 associates with mtHsp70 independently of Hsp60. To address this question, we employed mitochondria isolated from hsp60ts cells, which were shifted to non-permissive growth conditions. In these hsp60ts mitochondria, the steady-state levels of Hsp60 are unchanged, but the vast majority of Hsp60 aggregates (Figs. 3, A and B) (44). Consequently, Hsp60 ring complexes cannot be detected in hsp60ts mitochondria by blue native gel electrophoresis (Fig. 3C, lane 3). We analyzed whether the interaction of endogenous Hsp10 with mtHsp70 was affected in the mutant mitochondria by utilizing an Mge1His affinity matrix. The association of mtHsp70 with the Mge1His affinity matrix was largely unchanged in hsp60ts mitochondria (Fig. 3D, lanes 8 and 9). Surprisingly, copurification of Hsp10 to an Mge1His affinity matrix in hsp60ts mitochondria remained unaltered (Fig. 3D, lanes 5 and 6). In contrast, binding of Hsp60 to mtHsp70 was strongly diminished (Fig. 3D, lanes 5 and 6). Therefore, Hsp10 interacts with Mge1-bound mtHsp70 independently of Hsp60.
FIGURE 3.
MtHsp70 interacts with Hsp10 independently of Hsp60. A, the indicated amounts of WT, hsp10ts, and hsp60ts mitochondria (Mito) were analyzed by SDS-PAGE, Western blotting, and immunodetection with the indicated antisera. B, wild-type, hsp10ts, and hsp60ts mitochondria were lysed with detergent, and protein aggregates were separated by centrifugation. Total, pellet, and supernatant fractions were analyzed by SDS-PAGE, Western blotting, and immunodetection with the indicated antisera. C, WT, hsp10ts, and hsp60ts mitochondria were lysed with digitonin, and mitochondrial chaperonin complexes were studied by blue native electrophoresis, Western blotting, and immunodecoration with Hsp60-specific antiserum. D, WT and hsp60ts mitochondria were subjected to affinity purification via empty or Mge1His-coated Ni2+ NTA-agarose. Proteins were analyzed by SDS-PAGE, Coomassie staining, or Western blotting and immunodetection with the indicated antisera. Load, 0.5%; elution, 100%. The asterisk marks cross-reactivity of anti-Hsp60 antibody with mtHsp70.
MtHsp70 Promotes the Formation of Hsp60 Complexes
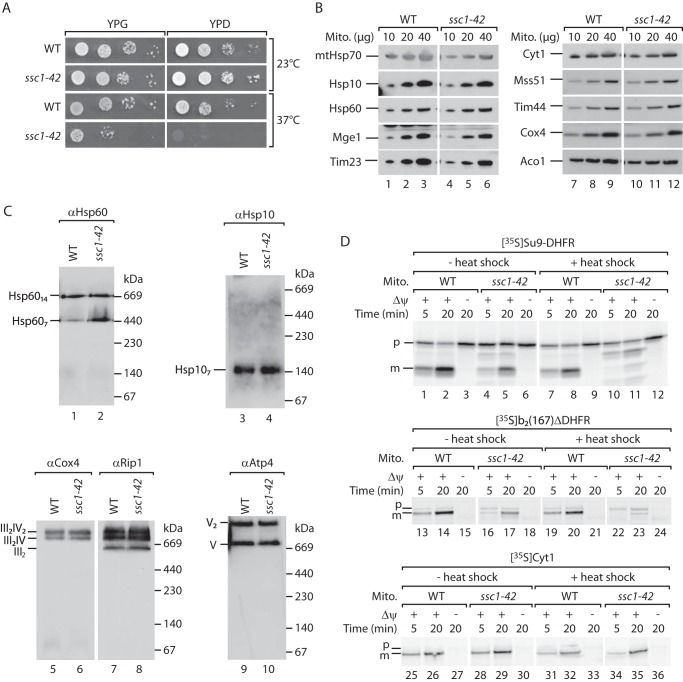
We speculated that the mtHsp70-Hsp10 interaction is involved in the biogenesis of Hsp60. So far, it has been reported that mtHsp70 is required for the import of Hsp60, but a potential role of the chaperone in the formation of Hsp60 complexes has not been addressed (43). To distinguish between the role of mtHsp70 in import or folding and assembly of the Hsp60 precursors, we used a temperature-sensitive mutant of mtHsp70, ssc1–42 (54). The ssc1–42 mutant mtHsp70 harbors six point mutations that are distributed randomly in the ATPase, substrate-binding, and C-terminal domains of the chaperone (I101T, I114N, I485L, N560D, V588A, and K626R). The mutant ssc1–42 grows normally at the permissive temperature but reveals a strong growth defect at 37 °C on non-fermentable and fermentable carbon sources (Fig. 4A). We grew the cells at the permissive temperature and isolated mitochondria for biochemical characterization. Steady-state levels of mtHsp70 were mildly reduced in the mutant, whereas the levels of other mitochondrial proteins, including Hsp10 and Hsp60, were similar to wild-type mitochondria (Fig. 4B). We analyzed the levels of Hsp60 and Hsp10 by blue native electrophoresis. A previous study revealed that the single (Hsp607) and double (Hsp6014) ring complexes of Hsp60 can be detected on a blue native gel (60). Here we show that the heptameric Hsp10 oligomer can also be resolved on blue native gels (Fig. 4C). The levels of the Hsp60 and Hsp10 complexes as well as of respiratory chain complexes were unaffected in ssc1–42 mutant mitochondria (Fig. 4C). To functionally characterize the mutant mtHsp70, we tested whether the protein import capacity is affected in ssc1–42 mitochondria. Model precursor proteins were labeled with [35S]methionine in a cell-free translation system and incubated with isolated mitochondria under import conditions. Subsequently, non-imported precursor proteins were proteolytically removed. We selected two precursors for our import studies that are transported, via mtHsp70, into the mitochondrial matrix: the precursor subunit 9 of the F1FO-ATP synthase fused to DHFR (pSu9-DHFR) and the precursor of a cytochrome b2 variant lacking the transmembrane segment fused to DHFR (b2Δ167-DHFR). The import of both precursor proteins was moderately affected in the mutant mitochondria under permissive conditions (Fig. 4D, lanes 4, 5, 16, and 17). However, upon in vitro heat shock, the transport of both model substrates into the mutant mitochondria was largely compromised (Fig. 4D, lanes 10, 11, 22, and 23). In contrast, import of the 35S-labeled precursor of cytochrome c1 (Cyt1) was not affected by in vitro heat shock (Fig. 4D, lanes 34 and 35). The precursor of cytochrome c1 is sorted into the inner membrane in an mtHsp70-independent manner. We conclude that the import capability of the mutated mtHsp70 can be specifically inactivated in ssc1–42 mitochondria by in vitro heat shock.
FIGURE 4.
The temperature-sensitive mutant of mtHsp70, ssc1–42, can be inactivated by in vitro heat shock. A, WT and ssc1–42 cells were serially diluted and spotted on glucose (YPD)- or glycerol (YPG)-containing medium. Cells grew at the indicated temperatures. B, the indicated amounts of WT and ssc1–42 mitochondria (Mito) were analyzed by SDS-PAGE, Western blotting, and immunodetection with the indicated antisera. C, WT and ssc1–42 mitochondria were lysed with digitonin, and mitochondrial protein complexes were studied by blue native electrophoresis, Western blotting, and immunodetection with the indicated antisera. Cox, cytochrome c oxidase; Rip1, Rieske iron-sulfur protein; Atp, subunits of the F1FO-ATP synthase. D, 35S-labeled precursors of subunit 9 of the F1Fo-ATP-synthase fused to DHFR (Su9-DHFR, top panel), cytochrome b2(167)Δ fused to DHFR (center panel), and cytochrome c1 (Cyt1, bottom panel) were imported into WT and ssc1–42 mitochondria at the indicated time points. Where indicated, isolated mitochondria were subjected to in vitro heat shock prior to import. In control reactions, the membrane potential (Δψ) was dissipated prior to import. Proteins were detected by SDS-PAGE and autoradiography. p, precursor protein; m, mature protein.
Next, we established an assay to study the biogenesis of 35S-labeled Hsp60 in mitochondria. The import of the 35S-labeled Hsp60 precursor occurs rapidly and is strictly dependent on the membrane potential (Fig. 5A, bottom panel). We employed blue native electrophoresis to study the assembly of the Hsp60 precursor into the mature complexes. Indeed, we observed an efficient integration of the 35S-labeled Hsp60 precursor into two high molecular weight complexes (Fig. 5A, top panel), which correspond in size to the single (Hsp607) and double (Hsp6014) rings of Hsp60 (Fig. 4C). Import and assembly studies at different temperatures revealed that formation of the mature Hsp60 complexes occurs in a delayed manner compared with the import of Hsp60 into the mitochondrial matrix (Fig. 5A). Interestingly, a low molecular weight form of 150 kDa can be detected at a lower temperature on blue native gels (Fig. 5A). Because the 150-kDa band remained stable upon solubilization with SDS, it most likely represents monomeric Hsp60 (Fig. 5B). Depending on the isoelectric point and capability to bind the Coomassie dye, the running behavior of water-soluble proteins on blue native gels can differ from molecular weight markers (68). This could explain the difference between the size of the potential monomeric Hsp60 of 60 kDa and its running behavior on the blue native gel above the 140 kDa mass marker. The blue native system allows following the formation of Hsp60 complexes from the unassembled Hsp60 precursor.
FIGURE 5.
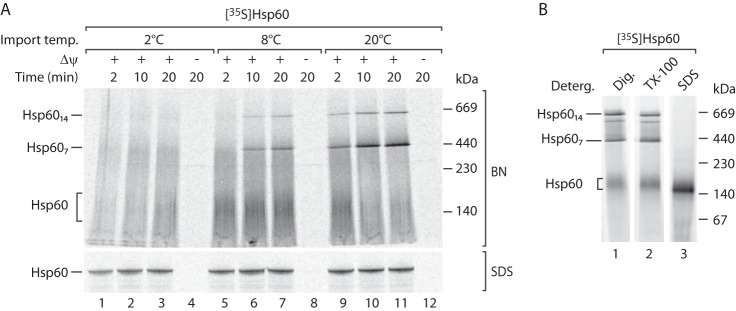
Assay for the assembly of Hsp60 complexes. A, 35S-labeled Hsp60 was imported into isolated WT mitochondria at the indicated temperatures and time points. Proteins were analyzed either by SDS-PAGE (SDS) or by blue native (BN) electrophoresis and autoradiography. B, 35S-labeled Hsp60 was imported into isolated WT mitochondria. Mitochondria were lysed with the indicated detergents (Deterg.) and subjected to blue native electrophoresis. Protein complexes were detected by autoradiography. Dig., digitonin; TX-100, Triton X-100.
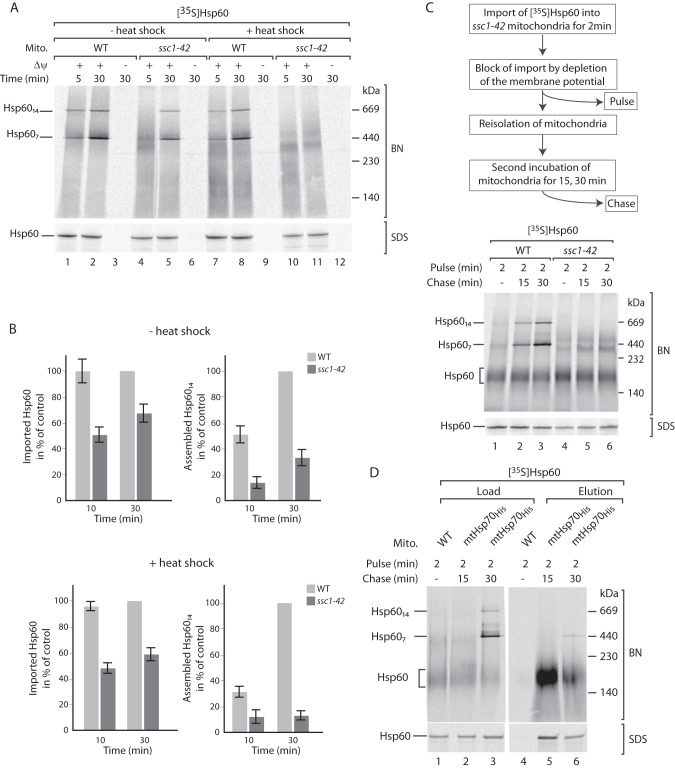
Having established an assay to separately analyze the import and assembly of the 35S-labeled Hsp60 precursor, we asked which step of Hsp60 biogenesis is affected in ssc1–42 mutant mitochondria. The 35S-labeled Hsp60 precursor is imported into untreated and in vitro heat shock-treated ssc1–42 mitochondria, although with reduced efficiency (Fig. 6, A and B). This observation supports the role of mtHsp70 in the import of the Hsp60 precursor (42). Strikingly, the assembly of 35S-labeled Hsp60 is more severely impaired in both untreated and heat-treated ssc1–42 mitochondria (Fig. 6, A and B), indicating an additional role of mtHsp70 in the formation of Hsp60 complexes. To test this notion, 35S-labeled Hsp60 was imported into wild-type and mutant mitochondria for a short time period (Fig. 6C, Pulse). Subsequently, the membrane potential was depleted to prevent further import of 35S-labeled Hsp60 precursors. Mitochondria were reisolated and incubated under import conditions to allow the assembly of the imported 35S-labeled Hsp60 (Fig. 6C, Chase). Although 35S-labeled Hsp60 was efficiently integrated into the mature complexes in wild-type mitochondria, its assembly was blocked in ssc1–42 mutant mitochondria (Fig. 6C). To substantiate a role of mHsp70 in the formation of Hsp60 complexes, we analyzed which oligomeric state of Hsp60 binds to mtHsp70. 35S-labeled Hsp60 was imported into mtHsp70His mitochondria for a short time period (pulse), followed by dissipation of the membrane potential and further incubation (chase). After the import reaction, mitochondria were lysed with digitonin and subjected to affinity purification via Ni2+-NTA agarose. Bound proteins were eluted from mtHsp70His by incubation with ATP. After a short import period, mtHsp70His binds to unassembled 35S-labeled Hsp60 (Fig. 6D, lane 5), whereas, upon chase incubation, a heptameric ring of 35S-labeled Hsp60 associates with mtHsp70His (Fig. 6D, lane 6). We conclude that mtHsp70 binds to two different assembly stages of Hsp60, revealing an important role of mtHsp70 in the formation of the Hsp60 complexes.
FIGURE 6.
MtHsp70 promotes the assembly of Hsp60 complexes. A, 35S-labeled Hsp60 was imported into WT and ssc1–42 mitochondria at the indicated time points. Where indicated, isolated mitochondria (Mito) were subjected to in vitro heat shock, or the membrane potential (Δψ) was dissipated prior to import. Proteins were analyzed by SDS-PAGE (SDS) or blue native (BN) electrophoresis and autoradiography. B, quantification of the import and assembly (Hsp6014) experiments in A. The longest import time point in WT mitochondria was set to 100% (control). Mean ± S.E. of four independent experiments is depicted. C, 35S-labeled Hsp60 was imported into WT and ssc1–42 mitochondria for a short time (Pulse), followed by further incubation after block of the import (Chase). Proteins were analyzed by SDS-PAGE or blue native electrophoresis and autoradiography. D, 35S-labeled Hsp60 was imported into WT and mtHsp70His mitochondria for a short time (Pulse), followed by further incubation after block of the import (Chase). Mitochondria were lysed with digitonin, and proteins were purified via Ni2+-NTA-agarose. Bound proteins were eluted with ATP and analyzed by SDS-PAGE or blue native electrophoresis and autoradiography.
MtHsp70 Cooperates with Hsp10 to Promote the Formation of the Hsp60 Ring Structures
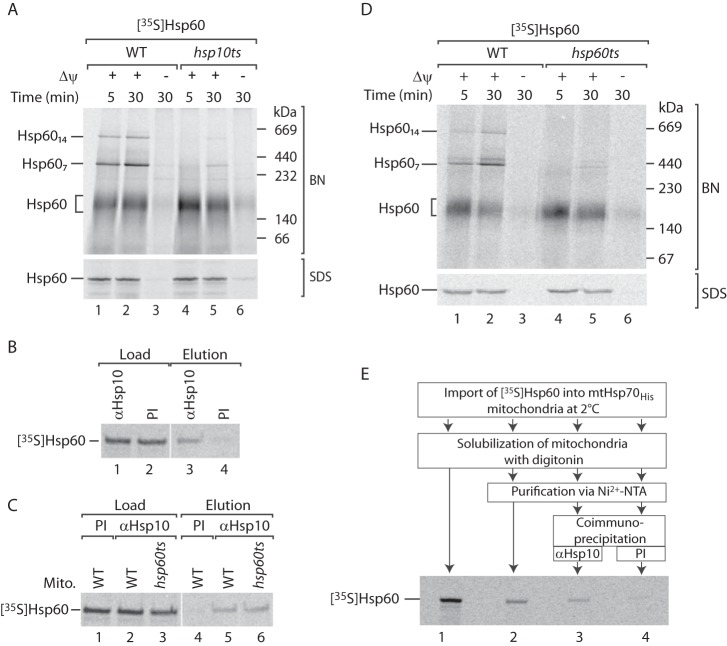
We wondered whether Hsp10 functions in the assembly of Hsp60 complexes as well. To this end, we imported 35S-labeled Hsp60 into hsp10ts mitochondria (44). Indeed, similar to ssc1–42 mitochondria, the assembly of Hsp60 into the high molecular weight complexes is compromised in hsp10ts mitochondria, whereas the overall import efficiency is not altered (Fig. 7A). Steady-state levels of Hsp60 complexes in hsp10ts mitochondria are unaffected (Fig. 3C), pointing to a direct role of Hsp10 in the biogenesis of Hsp60. To substantiate a role of Hsp10 in the biogenesis of Hsp60 complexes, we wondered whether Hsp10 is capable to bind to the unassembled Hsp60 precursor. Therefore, we imported 35S-labeled Hsp60 at 2 °C to prevent formation of the mature Hsp60 complexes (Fig. 5A). Under these conditions, the 35S-labeled Hsp60 precursor is coimmunoprecipitated by antisera against Hsp10, indicating that Hsp10 interacts with an unassembled form of Hsp60 (Fig. 7B, lanes 3 and 4). Supporting this view, coimmunoprecipitation of the 35S-labeled Hsp60 precursor by Hsp10-specific antiserum is not affected in hsp60ts mitochondria (Fig. 7C, lanes 5 and 6). In contrast, the imported 35S-labeled Hsp60 precursor does not assemble into the mature Hsp60 complexes in hsp60ts mitochondria, likely because of aggregation and malfunction of the mutated endogenous Hsp60 (Figs. 7D and 3, B and C) (44, 52). We conclude that Hsp10 promotes early steps in the formation of Hsp60 complexes. This observation is supported by the increased protease accessibility of imported Hsp60 in the Hsp10 temperature-sensitive strain (hsp10ts) (44). We asked whether mtHsp70 and Hsp10 cooperate in the biogenesis of Hsp60. To address this question, we imported 35S-labeled Hsp60 at 2 °C into mtHsp70His mitochondria to prevent the formation of mature Hsp60 complexes. Subsequently, the imported Hsp60 was copurified along with mtHsp70His via Ni2+-NTA-agarose (Fig. 7E, lane 2). Bound proteins were eluted and subjected to coimmunoprecipitation with Hsp10-specific antibodies or their corresponding preimmune serum. Following this approach, we found that a portion of the mtHsp70-bound Hsp60 was associated with Hsp10 (Fig. 7E, lane 3). We conclude that a fraction of Hsp10 interacts with mtHsp70 to promote the biogenesis of Hsp60.
FIGURE 7.
MtHsp70 and Hsp10 cooperate in the assembly of Hsp60. A, 35S-labeled Hsp60 was imported into WT and hsp10ts mitochondria at the indicated time points. Proteins were analyzed by SDS-PAGE (SDS) or blue native (BN) electrophoresis and autoradiography. Where indicated, the membrane potential (Δψ) was dissipated prior to import. B, 35S-labeled Hsp60 was imported into WT mitochondria at 2 °C, followed by coimmunoprecipitation with antibodies against Hsp10 (αHsp10) or preimmune serum (PI). Load, 1%; elution, 100%. C, 35S-labeled Hsp60 was imported into WT and hsp60ts mitochondria (Mito) at 2 °C, followed by coimmunoprecipitation with antibodies against Hsp10 or preimmune serum. Load, 1%; elution, 100%. D, 35S-labeled Hsp60 was imported into WT and hsp60ts mitochondria at the indicated time points. After import, the samples were analyzed by SDS-PAGE or blue native electrophoresis and autoradiography. E, 35S-labeled Hsp60 was imported into WT and mtHsp70His mitochondria at 2 °C, followed by affinity purification via Ni2+-NTA-agarose. Bound proteins were subjected to coimmunoprecipitation with antibodies against Hsp10 or preimmune serum. Load, 0.5%; elution Ni2+-NTA-agarose, 0.5%; elution coimmunoprecipitation, 100%.
CONCLUSIONS
MtHsp70 mediates multiple functions in mitochondrial protein homeostasis (2, 4). To carry out its different tasks mtHsp70 interacts with various interaction partners. Recent studies have indicated more specialized functions of mtHsp70 in the biogenesis of cytochrome c oxidase. Here mtHsp70 interacts with Mss51 and Cox4, which do not belong to the classical cochaperones of Hsp70 (38, 39). These observations indicate that further interaction partners of the chaperone may exist. Indeed, we found a robust interaction of mtHsp70 with Hsp60 and Hsp10. In bacteria, an association of the Hsp70 protein DnaK with the Hsp60 homolog GroEL has been reported (65). Similar to bacteria, the interaction between the two different types of chaperones may facilitate the transfer of substrate proteins. Strikingly, the interaction of Hsp10 with mtHsp70 occurs independently of Hsp60. MtHsp70 and Hsp10 cooperate to facilitate the integration of the Hsp60 precursor into the mature Hsp60 complexes. Two populations of mtHsp70 mediate Hsp60 biogenesis. First, mtHsp70 drives the import of Hsp60 at the presequence translocase. Second, mtHsp70 and Hsp10 promote the maturation of Hsp60 complexes. We propose that coupling to dedicated partner proteins like Hsp10, Tim44, Cox4, or Mss51 enables mtHsp70 to carry out a plethora of specific functions in mitochondrial biogenesis.
Supplementary Material
Acknowledgments
We thank Drs. Nikolaus Pfanner and Martin van der Laan for discussion and Nicole Zufall and Bettina Knapp for technical assistance. The work included in this study has also been performed in partial fulfillment of the requirements for the doctoral thesis of L. B. at the University of Freiburg.
This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 746 and the Excellence Initiative of the German Federal and State Governments (EXC 294 BIOSS).

This article contains supplemental Table S1.
- mtHsp70
- mitochondrial Hsp70
- SILAC
- stable isotope labeling by amino acids in cell culture
- Ni2+-NTA
- nickel-nitrilotriacetic acid
- DHFR
- dihydrofolate reductase.
REFERENCES
- 1. Bukau B., Weissman J., Horwich A. (2006) Molecular chaperones and protein quality control. Cell 125, 443–451 [DOI] [PubMed] [Google Scholar]
- 2. Kampinga H. H., Craig E. A. (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 4. Voos W. (2013) Chaperone-protease networks in mitochondrial protein homeostasis. Biochim. Biophys. Acta 1833, 388–399 [DOI] [PubMed] [Google Scholar]
- 5. Craig E. A., Kramer J., Kosic-Smithers J. (1987) SSC1, a member of the 70-kDa heat shock protein multigene family of Saccharomyces cerevisiae, is essential for growth. Proc. Natl. Acad. Sci. U.S.A. 84, 4156–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dolezal P., Likic V., Tachezy J., Lithgow T. (2006) Evolution of the molecular machines for protein import into mitochondria. Science 313, 314–318 [DOI] [PubMed] [Google Scholar]
- 7. Neupert W., Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 8. Baker M. J., Frazier A. E., Gulbis J. M., Ryan M. T. (2007) Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 17, 456–464 [DOI] [PubMed] [Google Scholar]
- 9. Endo T., Yamano K. (2009) Multiple pathways for mitochondrial protein traffic. Biol. Chem. 390, 723–730 [DOI] [PubMed] [Google Scholar]
- 10. Becker T., Böttinger L., Pfanner N. (2012) Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem. Sci. 37, 85–91 [DOI] [PubMed] [Google Scholar]
- 11. Sokol A. M., Sztolsztener M. E., Wasilewski M., Heinz E., Chacinska A. (2014) Mitochondrial protein translocases for survival and wellbeing. FEBS Lett. 588, 2484–2495 [DOI] [PubMed] [Google Scholar]
- 12. Schulz C., Schendzielorz A., Rehling P. (2015) Unlocking the presequence import pathway. Trends Cell Biol. 10.1016/j.tcb.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 13. Schneider H.-C., Berthold J., Bauer M. F., Dietmeier K., Guiard B., Brunner M., Neupert W. (1994) Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature 371, 768–774 [DOI] [PubMed] [Google Scholar]
- 14. Rassow J., Maarse A. C., Krainer E., Kübrich M., Müller H., Meijer M., Craig E. A., Pfanner N. (1994) Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J. Cell Biol. 127, 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Q., D'Silva P., Walter W., Marszalek J., Craig E. A. (2003) Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science 300, 139–141 [DOI] [PubMed] [Google Scholar]
- 16. Slutsky-Leiderman O., Marom M., Iosefson O., Levy R., Maoz S., Azem A. (2007) The interplay between components of the mitochondrial protein translocation motor studied using purified components. J. Biol. Chem. 282, 33935–33942 [DOI] [PubMed] [Google Scholar]
- 17. Westermann B., Prip-Buus C., Neupert W., Schwarz E. (1995) The role of the GrpE homologue, Mge1p, in mediating protein import and protein folding in mitochondria. EMBO J. 14, 3452–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Laloraya S., Dekker P. J., Voos W., Craig E. A., Pfanner N. (1995) Mitochondrial GrpE modulates the function of matrix Hsp70 in translocation and maturation of preproteins. Mol. Cell Biol. 15, 7098–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. D'Silva P. D., Schilke B., Walter W., Andrew A., Craig E. A. (2003) J protein cochaperones of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl. Acad. Sci. 100, 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Truscott K. N., Voos W., Frazier A. E., Lind M., Li Y., Geissler A., Dudek J., Müller H., Sickmann A., Meyer H. E., Meisinger C., Guiard B., Rehling P., Pfanner N. (2003) A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 163, 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mokranjac D., Sichting M., Neupert W., Hell K. (2003) Tim14, a key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 22, 4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frazier A. E., Dudek J., Guiard B., Voos W., Li Y., Lind M., Meisinger C., Geissler A., Sickmann A., Meyer H. E., Bilanchone V., Cumsky M. G., Truscott K. N., Pfanner N., Rehling P. (2004) Pam16 has an essential role in the mitochondrial protein import motor. Nat. Struct. Mol. Biol. 11, 226–233 [DOI] [PubMed] [Google Scholar]
- 23. Kozany C., Mokranjac D., Sichting M., Neupert W., Hell K. (2004) The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat. Struct. Mol. Biol. 11, 234–241 [DOI] [PubMed] [Google Scholar]
- 24. Li Y., Dudek J., Guiard B., Pfanner N., Rehling P., Voos W. (2004) The presequence translocase-associated protein import motor of mitochondria. Pam16 functions in an antagonistic manner to Pam18. J. Biol. Chem. 279, 38047–38054 [DOI] [PubMed] [Google Scholar]
- 25. Nakai M., Kato Y., Ikeda E., Toh-e A., Endo T. (1994) Yge1p, a eukaryotic Grp-E homolog, is localized in the mitochondrial matrix and interacts with mitochondrial Hsp70. Biochem. Biophys. Res. Comm. 200, 435–442 [DOI] [PubMed] [Google Scholar]
- 26. Laloraya S., Gambill B. D., Craig E. A. (1994) A role for a eukaryotic GrpE-related protein, Mge1p, in protein translocation. Proc. Natl. Acad. Sci. 91, 6481–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Voos W., Gambill B. D., Laloraya S., Ang D., Craig E. A., Pfanner N. (1994) Mitochondrial GrpE is present in a complex with hsp70 and preproteins in transit across membranes. Mol. Cell Biol. 14, 6627–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rowley N., Prip-Buus C., Westermann B., Brown C., Schwarz E., Barrell B., Neupert W. (1994) Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell 77, 249–259 [DOI] [PubMed] [Google Scholar]
- 29. Herrmann J. M., Stuart R. A., Craig E. A., Neupert W. (1994) Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J. Cell Biol. 127, 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Westermann B., Gaume B., Herrmann J. M., Neupert W., Schwarz E. (1996) Role of the mitochondrial DnaJ homolog Mdj1p as a chaperone for mitochondrially synthesized and imported proteins. Mol. Cell Biol. 16, 7063–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Horst M., Oppliger W., Rospert S., Schönfeld H. J., Schatz G., Azem A. (1997) Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 16, 1842–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burri L., Vascotto K., Fredersdorf S., Tiedt R., Hall M. N., Lithgow T. (2004) Zim17, a novel zinc finger protein essential for protein transport into mitochondria. J. Biol. Chem. 279, 50243–50249 [DOI] [PubMed] [Google Scholar]
- 33. Yamamoto H., Momose T., Yatsukawa Y., Ohshima C., Ishikawa D., Sato T., Tamura Y., Ohwa Y., Endo T. (2005) Identification of a novel member of yeast mitochondrial Hsp70-associated motor and chaperone proteins that facilitates protein translocation across the inner membrane. FEBS Lett. 579, 507–511 [DOI] [PubMed] [Google Scholar]
- 34. Sanjuán Szklarz L. K., Guiard B., Rissler M., Wiedemann N., Kozjak V., van der Laan M., Lohaus C., Marcus K., Meyer H. E., Chacinska A., Pfanner N., Meisinger C. (2005) Inactivation of the mitochondrial heat shock protein Zim17 leads to aggregation of matrix Hsp70s followed by pleiotropic effects on morphology and protein biogenesis. J. Mol. Biol. 351, 206–218 [DOI] [PubMed] [Google Scholar]
- 35. Sichting M., Mokranjac D., Azem A., Neupert W., Hell K. (2005) Maintenance of structure and function of mitochondrial Hsp70 chaperones requires the chaperone Hep1. EMBO J. 24, 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blamowska M., Neupert W., Hell K. (2012) Biogenesis of the mitochondrial Hsp70 chaperone. J. Cell Biol. 199, 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lewrenz I., Rietzschel N., Guiard B., Lill R., van der Laan M., Voos W. (2013) The functional interaction of mitochondrial Hsp70s with the escort protein Zim17 is critical for Fe/S biogenesis and substrate interaction at the inner membrane preprotein translocase. J. Biol. Chem. 288, 30931–30943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fontanesi F., Soto I. C., Horn D., Barrientos A. (2010) Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol. Cell Biol. 30, 245–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Böttinger L., Guiard B., Oeljeklaus S., Kulawiak B., Zufall N., Wiedemann N., Warscheid B., van der Laan M., Becker T. (2013) A complex of Cox4 and mitochondrial Hsp70 plays an important role in the assembly of the cytochrome c oxidase. Mol. Biol. Cell 24, 2609–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmitt M., Neupert W., Langer T. (1995) Hsp78, a Clp homologue within mitochondria, can substitute for chaperone functions of mt-Hsp70. EMBO J. 14, 3434–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moczko M., Schönfisch B., Voos W., Pfanner N., Rassow J. (1995) The mitochondrial ClpB homolog Hsp78 cooperates with matrix Hsp70 in maintenance of mitochondrial function. J. Mol. Biol. 254, 538–543 [DOI] [PubMed] [Google Scholar]
- 42. Ostermann J., Horwich A. L., Neupert W., Hartl F. U. (1989) Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature 341, 125–130 [DOI] [PubMed] [Google Scholar]
- 43. Manning-Krieg U. C., Scherer P. E., Schatz G. (1991) Sequential action of mitochondrial chaperones in protein import into the matrix. EMBO J. 10, 3273–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dubaquié Y., Looser R., Fünfschilling U., Jenö P., Rospert S. (1998) Identification of in vivo substrates of the yeast mitochondrial chaperonins reveals overlapping but non-identical requirement for hsp60 and hsp10. EMBO J. 17, 5868–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hutchinson E. G., Tichelaar W., Hofhaus G., Weiss H., Leonard K. R. (1989) Identification and electron microscopic analysis of a chaperonin oligomer from Neurospora crassa mitochondria. EMBO J. 8, 1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Viitanen P. V., Lorimer G. H., Seetharam R., Gupta R. S., Oppenheim J., Thomas J. O., Cowan N. J. (1992) Mammalian mitochondrial chaperonin 60 functions as a single toroidal ring. J. Biol. Chem. 267, 695–698 [PubMed] [Google Scholar]
- 47. Nielsen K. L., Cowan N. J. (1998) A single ring is sufficient for productive chaperonin-mediated folding in vivo. Mol. Cell 2, 93–99 [DOI] [PubMed] [Google Scholar]
- 48. Levy-Rimler G., Viitanen P., Weiss C., Sharkia R., Greenberg A., Niv A., Lustig A., Delarea Y., Azem A. (2001) The effect of nucleotides and mitochondrial chaperonin 10 on the structure and chaperone activity of mitochondrial chaperonin 60. Eur. J. Biochem. 268, 3465–3472 [DOI] [PubMed] [Google Scholar]
- 49. Rospert S., Glick B. S., Jenö P., Schatz G., Todd M. J., Lorimer G. H., Viitanen P. V. (1993) Identification and functional analysis of chaperonin 10, the groES homolog from yeast mitochondria. Proc. Natl. Acad. Sci. U.S.A. 90, 10967–10971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Höhfeld J., Hartl F. U. (1994) Role of the chaperonin cofactor Hsp10 in protein folding and sorting in yeast mitochondria. J. Cell Biol. 126, 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cheng M. Y., Hartl F. U., Martin J., Pollock R. A., Kalousek F., Neupert W., Hallberg E. M., Hallberg R. L., Horwich A. L. (1989) Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature 337, 620–625 [DOI] [PubMed] [Google Scholar]
- 52. Cheng M. Y., Hartl F. U., Horwich A. L. (1990) The mitochondrial chaperonin hsp60 is required for its own assembly. Nature 348, 455–458 [DOI] [PubMed] [Google Scholar]
- 53. Hallberg E. M., Shu Y., Hallberg R. L. (1993) Loss of mitochondrial hsp60 function: nonequivalent effects on matrix-targeted and intermembrane-targeted proteins. Mol. Cell Biol. 13, 3050–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wenz L. S., Opaliński L., Schuler M. H., Ellenrieder L., Ieva R., Böttinger L., Qiu J., van der Laan M., Wiedemann N., Guiard B., Pfanner N., Becker T. (2014) The presequence pathway is involved in protein sorting to the mitochondrial outer membrane. EMBO Rep. 15, 678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ryan M. T., Voos W., Pfanner N. (2001) Assaying protein import into mitochondria. Methods Cell Biol. 65, 189–215 [DOI] [PubMed] [Google Scholar]
- 56. Lytovchenko O., Naumenko N., Oeljeklaus S., Schmidt B., von der Malsburg K., Deckers M., Warscheid B., van der Laan M., Rehling P. (2014) The INA complex facilitates assembly of the peripheral stalk of the mitochondrial F1Fo-ATP synthase. EMBO J. 33, 1624–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 58. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 59. Issel-Tarver L., Christie K. R., Dolinski K., Andrada R., Balakrishnan R., Ball C. A., Binkley G., Dong S., Dwight S. S., Fisk D. G., Harris M., Schroeder M., Sethuraman A., Tse K., Weng S., Botstein D., Cherry J. M. (2002) Saccharomyces genome database. Methods Enzymol. 350, 329–346 [DOI] [PubMed] [Google Scholar]
- 60. Becker T., Pfannschmidt S., Guiard B., Stojanovski D., Milenkovic D., Kutik S., Pfanner N., Meisinger C., Wiedemann N. (2008) Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal anchored receptors. J. Biol. Chem. 283, 120–127 [DOI] [PubMed] [Google Scholar]
- 61. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 62. Schneider H.-C., Westermann B., Neupert W., Brunner M. (1996) The nucleotide exchange factor MGE exerts a key function in the ATP-dependent cycle of mt-Hsp70-Tim44 interaction driving mitochondrial protein import. EMBO J. 15, 5796–5803 [PMC free article] [PubMed] [Google Scholar]
- 63. Mapa K., Sikor M., Kudryavtsev V., Waegemann K., Kalinin S., Seidel C. A., Neupert W., Lamb D. C., Mokranjac D. (2010) The conformational dynamics of the mitochondrial Hsp70 chaperone. Mol. Cell 38, 89–100 [DOI] [PubMed] [Google Scholar]
- 64. Qiu J., Wenz L. S., Zerbes R. M., Oeljeklaus S., Bohnert M., Stroud D. A., Wirth C., Ellenrieder L., Thornton N., Kutik S., Wiese S., Schulze-Specking A., Zufall N., Chacinska A., Guiard B., Hunte C., Warscheid B., van der Laan M., Pfanner N., Wiedemann N., Becker T. (2013) Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell 154, 596–608 [DOI] [PubMed] [Google Scholar]
- 65. Kerner M. J., Naylor D. J., Ishihama Y., Maier T., Chang H. C., Stines A. P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Hartl F. U. (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122, 209–220 [DOI] [PubMed] [Google Scholar]
- 66. Ryan M. T., Naylor D. J., Hoogenraad N. J., Høj P. B. (1995) Affinity purification, overexpression, and characterization of chaperonin 10 homologues synthesized with and without N-terminal acetylation. J. Biol. Chem. 270, 22037–22043 [DOI] [PubMed] [Google Scholar]
- 67. Dubaquié Y., Looser R., Rospert S. (1997) Significance of chaperonin 10-mediated inhibition of ATP hydrolysis by chaperonin 60. Proc. Natl. Acad. Sci. U.S.A. 94, 9011–9016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wittig I., Beckhaus T., Wumaier Z., Karas M., Schägger H. (2010) Mass estimation of native proteins by blue native electrophoresis: principles and practical hints. Mol. Cell Proteomics 9, 2149–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.