Background: Small nucleolar RNAs (snoRNAs) are thought to be exclusively nuclear.
Results: Doxorubicin stress results in significant accumulation of cytosolic snoRNAs via a pathway involving Nox4D and superoxide.
Conclusion: snoRNAs are present in the cytoplasm, and their cytosolic abundance is dynamically regulated.
Significance: Previously unexplained snoRNA biology may be the result of snoRNA interactions with cytoplasmic targets.
Keywords: Intracellular Trafficking, Molecular Cell Biology, NADPH Oxidase, Reactive Oxygen Species (ROS), Small Nucleolar RNA (snoRNA), Subcellular Fractionation, Doxorubicin
Abstract
Small nucleolar RNAs (snoRNAs) guide nucleotide modifications of cellular RNAs in the nucleus. We previously showed that box C/D snoRNAs from the Rpl13a locus are unexpected mediators of physiologic oxidative stress, independent of their predicted ribosomal RNA modifications. Here we demonstrate that oxidative stress induced by doxorubicin causes rapid cytoplasmic accumulation of the Rpl13a snoRNAs through a mechanism that requires superoxide and a nuclear splice variant of NADPH oxidase. RNA-sequencing analysis reveals that box C/D snoRNAs as a class are present in the cytoplasm, where their levels are dynamically regulated by NADPH oxidase. These findings suggest that snoRNAs may orchestrate the response to environmental stress through molecular interactions outside of the nucleus.
Introduction
snoRNAs2 are a diverse class of short, non-coding RNAs (ncRNAs) that localize to the nucleus and guide nucleotide modifications of other RNAs. In vertebrates, most snoRNAs are encoded by the introns of pre-mRNAs, and the majority are predicted to guide nucleotide modifications of ribosomal RNAs (rRNAs) or small nuclear RNAs (snRNAs) (1, 2). snoRNAs nucleate snoRNA-ribonucleoprotein complexes that bring together an RNA-modifying enzyme and a target RNA through antisense base pairing with the snoRNA. snoRNAs localize to the nucleus, in the nucleolus or Cajal body, where most box C/D snoRNAs guide 2′-O-methylation by fibrillarin and most box H/ACA snoRNAs guide pseudouridylation by dyskerin (3–5). However, evidence is also emerging that some snoRNAs have non-canonical functions in RNA editing, alternative splicing, maintenance of chromatin structure, and miRNA-like post-transcriptional regulation of gene expression (6–8). The mechanistic details of these non-canonical functions are largely unknown.
Our laboratory has shown that box C/D snoRNAs encoded by introns of the ribosomal protein L13a (Rpl13a) locus are critical mediators of cell death in response to oxidative stress (9). Haploinsufficiency of these ncRNAs protects from oxidative stress but does not change the 2′-O-methylation status of predicted ribosomal RNA targets (9), suggesting a non-canonical mechanism. Unexpectedly, we have also found that RpL13a snoRNAs accumulate in the cytoplasm of cells during lipotoxic or oxidative stress (9).
The present study was designed to elucidate the relationship between oxidative stress and the cytoplasmic localization of snoRNAs. As a model system we used doxorubicin (dox), a potent inducer of superoxide in cardiomyocytes (10). The production of superoxide by dox is catalyzed by NADPH oxidase (Nox) enzymes, which are important mediators of dox cardiotoxicity in vivo (11). In this report we show that the cytoplasmic localization of Rpl13a snoRNAs is regulated by Nox-dependent oxidative tone. Moreover, RNA-seq reveals that Nox activity broadly regulates cytosolic snoRNA localization.
EXPERIMENTAL PROCEDURES
Materials, Cell Culture, Drug Treatment, Cytotoxicity, and Viability Assays
Dulbecco's modified Eagle's medium (DMEM), FBS, and digitonin were from Sigma. Silencer Select siRNAs, RNAiMAX, SUPERase-In, TRIzol LS, and Power SYBR Master Mix were from Life Technologies. Dox, Mn(III)TMPyP (MnT), and diphenyleneiodonium chloride (DPI) were from Cayman Chemical. H9c2 cells (ATCC) were maintained in DMEM + 10% FBS. Dox or inhibitors were added as indicated. Cytotoxicity and cell viability were determined using the CytoTox 96 Non-radioactive Cytotoxicity Assay and CellTiter-Glo Luminescent Cell Viability Assay (Promega).
Subcellular Fractionation
All manipulations were at 4 °C. Cells were collected by trypsinization and washed. For fractionation by detergent extraction, cell pellets were incubated in digitonin buffer (150 mm NaCl, 50 mm HEPES, pH 7.4, digitonin 100 μg/ml, EDTA 5 mm, and SUPERase-In RNase inhibitor 0.1 units/μl) for 10 min and centrifuged for 10 min at 2000 × g to yield a cytosolic supernatant and nuclear pellet (modified from (12)). The nuclear pellet was washed with PBS before RNA isolation. For fractionation by hypotonic lysis, cells were swelled in disruption buffer (10 mm KCl, 1.5 mm MgCl2, 20 mm Tris-HCl, 5 mm EDTA, and 0.1 unit/μl SUPERase-In RNase inhibitor), homogenized by Dounce (glass-glass), and centrifuged for 5 min at 1500 × g to yield crude cytosolic and nuclear fractions. The cytosol was further cleared by centrifugation for 5 min at 20,000 × g. The nuclear fraction was washed in disruption buffer before RNA isolation (modified from Rio et al. (13)).
RNA Isolation and Quantitative Real-time PCR (qPCR)
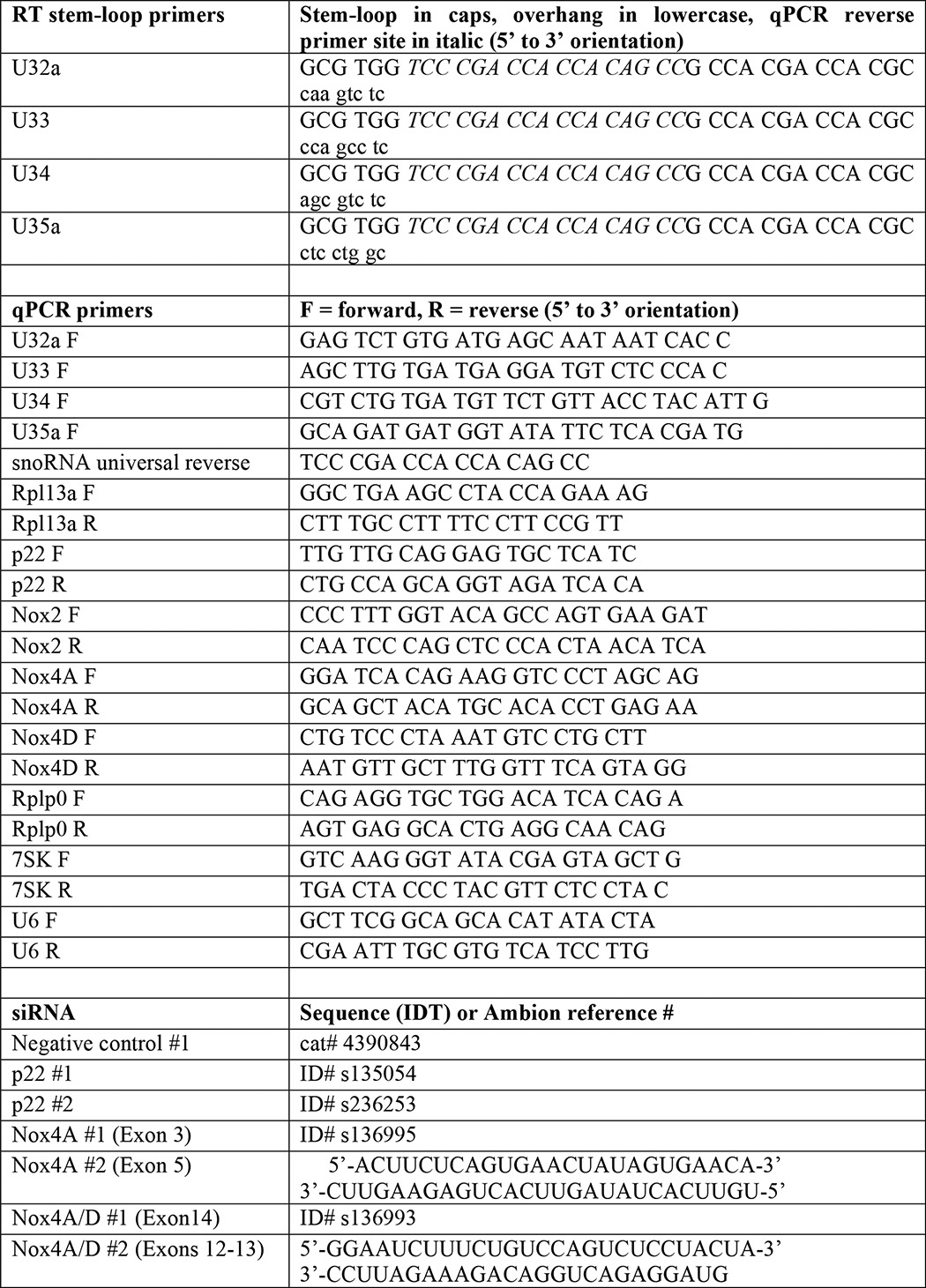
Total RNA was isolated from cytoplasmic or nuclear fractions using TRIzol LS. cDNA was synthesized with oligo-dT for mRNAs, random hexamer primers for U6 and 7SK, and stem-loop primers specific for snoRNAs followed by qPCR as previously described (9). Primer sequences are shown in Table 1. Relative quantitation of target transcript expression was calculated using the ddCT method using Rplp0 (36B4) as an endogenous control on an ABI 7500 Fast Real-time PCR System.
TABLE 1.
Primers and siRNA
Immunoblots
Protein from whole cell or nuclear radioimmune precipitation assay buffer (RIPA) lysate (RIPA buffer: 150 mm NaCl, 50 mm HEPES, pH 7.4, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate) or fractionated cytosol was quantified, separated by SDS-PAGE, immunoblotted, and detected by ECL. Antibodies were heat shock protein 90 (Hsp90) at 1:2000 (Enzo SPA-846), α-tubulin at 1:500 (Sigma; Thr-6199), nucleophosmin (NPM) at 1:500 (Abcam ab15440), histone H3 at 1:5000 (Abcam ab1791), and Nox4 (Abcam ab109225). Chemiluminescent detection was performed with horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories).
Immunofluorescence
Cells were grown on glass coverslips, fixed with 4% paraformaldehyde, permeabilized with Nonidet P-40 or Triton X-100, and blocked in 200 μg/ml ChromPure IgG corresponding to secondary antibody species (Jackson Immuno Research Laboratories). Primary antibody for detection of nucleophosmin (Life Technologies 325200) was used at 1:1000, and secondary Alexa Fluor® 350 donkey anti-mouse IgG (Life Technologies A10035) was used at 5 μg/ml. Primary antibody to detect p22 (Santa Cruz sc-20781) was used at 1:50 followed by secondary Alexa Fluor 488 goat anti-rabbit IgG at 5 μg/ml and counterstained with Hoechst 33342, 1 μg/ml. Images were obtained using an Axioskop 2 MOT Plus microscope (Zeiss). Fluorescence intensity for p22 was measured from 10 random fields of cells using ImageJ (National Institutes of Health).
siRNA
siRNA (Silencer Select or synthesized by Integrated DNA Technology) were introduced into cells using RNAiMax per the manufacturer's protocol. Details for siRNAs are shown in Table 1.
RNA Sequencing and Bioinformatics
Cells from n = 3 experiments were treated as indicated, cytosol was recovered by detergent extraction, and total RNA was isolated using TRIzol LS followed by Qiagen RNeasy miRNA clean-up. Indexed RNA-seq libraries were made using the Illumina TruSeq small RNA kit. Libraries were amplified with 14 cycles of PCR, pooled, and separated by PAGE. Products from small RNA with input sizes of 35–350 nucleotides were excised, collected, and sequenced using an Illumina HiSeq 2500. Demultiplexed data were uploaded to Galaxy. Sequences were evaluated for quality by FASTQC, trimming was applied to reject scores of <20, and sequences were clipped to remove adapter sequence. Sequences were aligned to the rat genome (rn4, Bowtie for Illumina, default settings) and aligned reads were exported to Partek Genomics Suite (v.6.6) for annotation and quantitative analysis using the most recent rat genome build that contains snoRNAs (Ensembl Build e69, RGSC3.4.69). Note that tRNA was not annotated in this build and was, therefore, not included in our analysis. Annotated reads were normalized as reads per million aligned reads. Analysis was limited to genes with >1 aligned read per sample (GEO: GSE67050).
Statistical Methods
Results and 95% confidence intervals for qPCR data were calculated using 7500 Software v.2.0.6 (Life Technologies). Pairwise comparisons are significant at p < 0.05 where error bars do not overlap. For RNA-seq analysis, Partek Genomics Suite was used for hierarchical clustering, and identification of differentially expressed genes was based on reads/million aligned reads-normalized data. All other statistics were calculated in Prism v6 using analysis of variance for multiple comparisons or t test for pairwise comparisons. Statistical significance for these tests was defined using α = 0.05 and p < 0.05.
RESULTS
Doxorubicin Stimulates Rapid Cytosolic Accumulation of Rpl13a snoRNAs
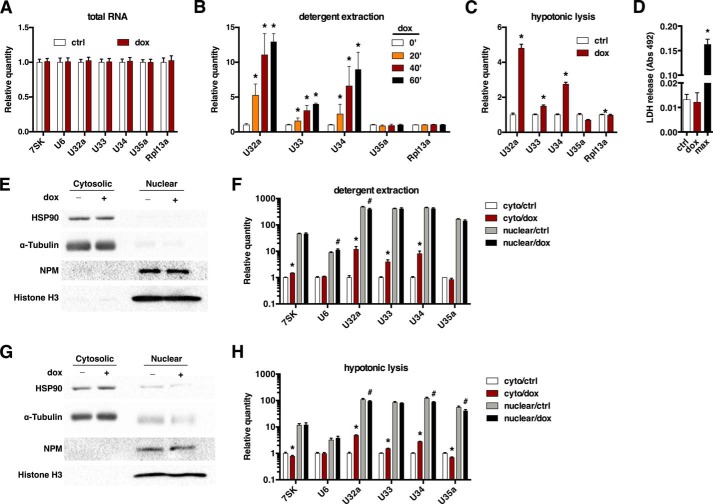
To understand how the cytoplasmic localization of snoRNAs is regulated by oxidative stress, we first treated H9c2 rat cardiomyoblasts with dox to induce reactive oxygen species and quantified total Rpl13a snoRNAs U32a, U33, U34, and U35a by qPCR. We found no change in total snoRNA levels, Rpl13a mRNA, or other small ncRNAs 7SK and U6 after 1 h of dox treatment (Fig. 1A). We then repeated the treatment and isolated cytosol by selective detergent extraction. Rpl13a snoRNAs were easily detected in the cytoplasm of untreated cells, and levels of U32a, U33, and U34 increased after dox treatment, without a change in Rpl13a mRNA expression (Fig. 1B). Similar results were obtained using detergent-free hypotonic cell lysis to isolate cytosolic RNA (Fig. 1C). To ensure that this rapid and robust response did not simply reflect drug cytotoxicity, we assayed for cellular lactate dehydrogenase (LDH) release, which showed no toxicity at 1 h (Fig. 1D).
FIGURE 1.
Dox stimulates rapid cytosolic accumulation of Rpl13a snoRNAs. A, H9c2 cells were treated with 20 μm dox for 1 h, and total cellular RNA was assayed by qPCR. Relative quantification is shown, with normalization to the control transcript Rplp0. Error bars show 95% confidence interval for n = 3 experiments. ctrl, control. B, H9c2 cells were treated with 20 μm dox for the times shown. Cytosol was isolated by selective permeabilization of the plasma membrane with digitonin buffer (detergent extraction) and quantified as in A. *, p < 0.05 versus control. C, cells were treated with dox for 1 h as in B or untreated (ctrl), and cytosol was isolated by detergent-free hypotonic lysis. Cytosolic snoRNA was quantified as in B. D, cytotoxicity was assayed by lactate dehydrogenase (LDH) release into the media after 1 h of treatment with 20 μm dox. Cell lysis to release total cellular lactate dehydrogenase served as maximal cytotoxicity (max). The graph shows mean absorbance at 492 nm + S.E. for n = 3 experiments. *, p < 0.05 versus control. E–H, cytosolic and nuclear extracts were prepared from control and 1-h dox-treated cells by detergent extraction (E and F) and by hypotonic lysis (G and H). Fractions were analyzed by Western blot for protein markers of cytoplasm (Hsp90, α-tubulin), nucleus (histone H3), and nucleolus (NPM) (E and G). qPCR quantification of nuclear ncRNAs (7SK and U6) and the Rpl13a snoRNAs shows the relative amounts of RNA from cytosolic and nuclear fractions (F and H, note the log scale; *, p < 0.05 versus control for cytoplasm; #, p < 0.05 versus control for nuclear).
Given the surprising nature of our results, we performed additional experiments to validate the integrity of our cytosolic isolation. For both cytosolic isolation methods, immunoblotting showed enrichment of cytosolic proteins and depletion of nuclear/nucleolar proteins in cytosolic fractions, whereas qPCR demonstrated that the small ncRNAs 7SK and U6 were predominantly in the nuclear fraction (Fig. 1, E–H). Overall, the partitioning of these nuclear RNA markers to the nuclear fraction was 4–5-fold higher with the detergent extraction method than with the hypotonic lysis method (compare Figs. 1, F and H), demonstrating the superior performance of the former method for obtaining cytoplasm that is free of contaminating nuclear material. Importantly, neither 7SK nor U6 control RNA demonstrated the marked increase in cytosolic levels with dox treatment that we observed for the RpL13a snoRNAs.
Using the detergent extraction method, we found U32a, U33, and U34 were 400–500× more abundant in the nucleus than the cytosol (99.4–99.8% nuclear), consistent with their known nucleolar localization (Fig. 1F). Although brief treatment with dox did not increase the nuclear content of these snoRNAs, the drug induced 12-, 4-, and 8-fold increases in the cytosolic abundance of U32a, U33, and U34, lowering their nuclear-to-cytosolic ratios to 33, 104, and 52, respectively. Cytosolic isolation by hypotonic lysis showed a similar pattern of results with somewhat blunted effect sizes (Fig. 1H). Thus, based on two independent methods, we conclude that dox induces accumulation of these snoRNAs in the cytosol. Because total cellular levels of Rpl13a snoRNAs did not change with treatment, levels of cytoplasmic snoRNAs in these and subsequent experiments are presented without normalization to total cellular snoRNA content.
Superoxide Is Necessary for Cytosolic Accumulation of Rpl13a snoRNAs in Response to Doxorubicin
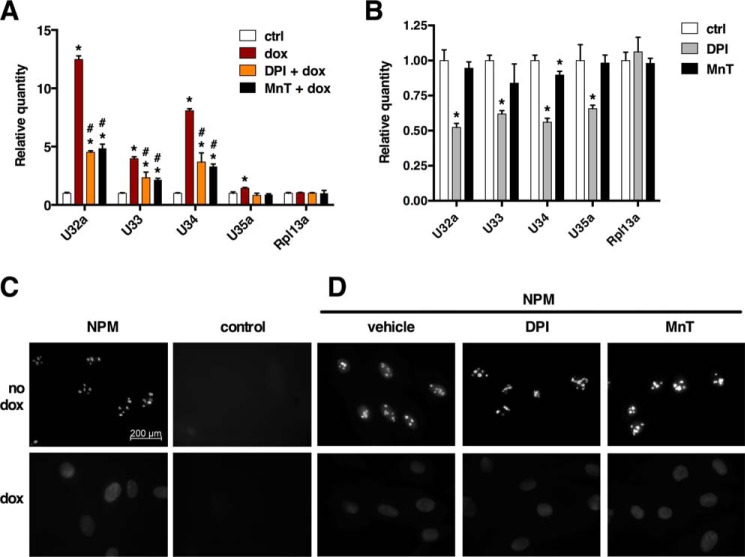
Given that dox induces superoxide through Nox enzymes, we tested whether the superoxide scavenger Mn(III)TMPyP [MnT] or the Nox inhibitor DPI could inhibit cytosolic accumulation of Rpl13a snoRNAs. As shown in Fig. 2A, co-treatment of cells with MnT or DPI significantly blunted the accumulation of cytosolic snoRNAs after treatment with dox for 1 h. This observation suggests that production of superoxide via Nox is required for Rpl13a snoRNA accumulation in the cytosol after treatment with dox. Interestingly, treatment with DPI alone resulted in lower levels of cytoplasmic snoRNAs, providing evidence for the role of Nox enzymes in the subcellular localization of these snoRNAs that is independent of the dox effect (Fig. 2B).
FIGURE 2.
Superoxide production is necessary for cytosolic accumulation of Rpl13a snoRNAs in response to dox. A, H9c2 cells were treated for 1 h with either vehicle alone (ctrl) or combinations of 20 μm dox, 500 nm DPI, and 200 μm MnT as shown. Cytosolic RNA was isolated by detergent extraction and quantified relative to Rplp0 by qPCR. Error bars show 95% confidence interval for n = 3 experiments. B, H9c2 cells were treated with DPI or MnT alone. Cytosol was isolated and analyzed for cytosolic snoRNAs as in A. C and D, H9c2 cells were analyzed by immunofluorescence microscopy for detection of NPM under control and 1 h dox-treated conditions and also in the presence of DPI or MnT as in A. Micrographs show representative fields. Scale bar, 200 μm. *, p < 0.05 versus control; #, p < 0.05 versus dox alone.
We further tested whether the inhibitory effects of MnT and DPI on cytosolic snoRNA accumulation simply reflected inhibition of nucleolar stress that is known to be induced by dox (14). Immunofluorescence staining showed dox-induced dissolution of NPM nucleolar punctae (Fig. 2C), but NPM remained confined to the nucleus. Although MnT and DPI blocked the cytosolic accumulation of snoRNAs after dox treatment, pretreatment with MnT or DPI had no effect on the dox-induced nucleolar stress, suggesting that nucleolar dissolution is not dependent on superoxide or Nox activity (Fig. 2D). Our findings are consistent with a model in which Rpl13a snoRNA accumulation in the cytosol is a ROS-regulated event and not simply the result of generalized nucleolar stress.
Nox Mediates Cytosolic Accumulation of Rpl13a snoRNAs in Response to Doxorubicin
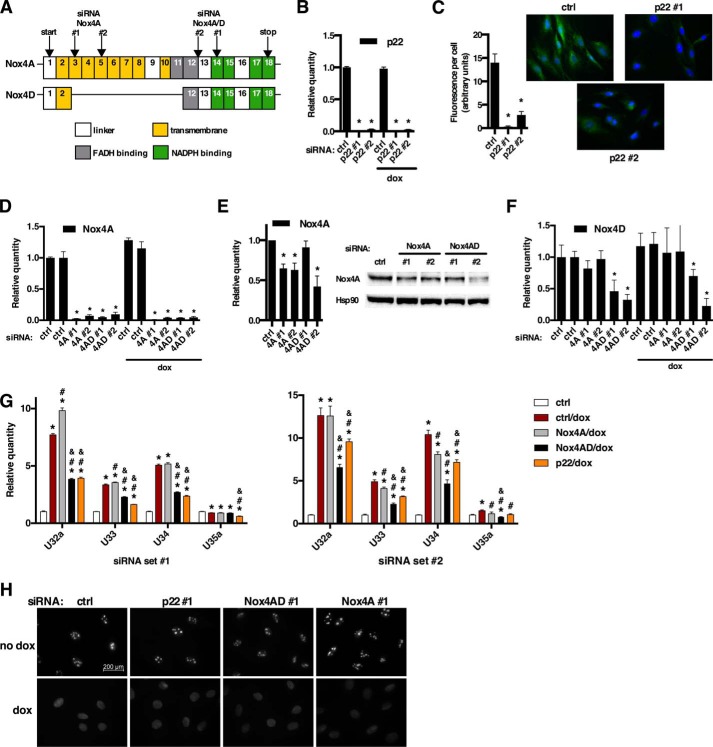
The ability of MnT and DPI to inhibit cytosolic snoRNA accumulation after dox strongly suggested a mechanism involving a member of the Nox enzyme family of multisubunit enzyme complexes. qPCR revealed constitutive expression of Nox4 but not Nox2 in H9c2 cells.3 Nox4D is an enzymatically active splice variant of Nox4 that localizes to the nucleus and nucleolus, and we hypothesized that this was a likely source of nuclear ROS that could influence Rpl13a snoRNA localization (15, 16). Using a previously validated siRNA knockdown approach for Nox4 isoforms (15), we used two independent siRNAs for each target: membrane-associated Nox4A at exons 3 or 5 (siRNA Nox4A), both the Nox4A and Nox4D isoforms at exons 12–13 or 14 (siRNA Nox4A/D), or the regulatory p22 subunit (Fig. 3A). Knockdown of p22, Nox4A, and Nox4D mRNAs was confirmed by qPCR (Figs. 3, B, D, and F). Knockdown of p22 and Nox4A proteins was confirmed by immunofluorescence and immunoblot, respectively (Fig. 3, C and E); however, commercially available antibodies that recognize rat Nox4D are not available. siRNA knockdown of p22 or Nox4A/D recapitulated the effects of MnT and DPI, with markedly blunted cytosolic accumulation of Rpl13a snoRNAs in response to dox (Fig. 3G). In contrast, siRNA to Nox4A alone had little effect, implicating Nox4D as an important mediator of cytosolic snoRNA accumulation in response to dox. As with DPI and MnT, none of the siRNA treatments altered the pattern of nucleolar dissolution induced by dox (Fig. 3H).
FIGURE 3.
NAPDH oxidase mediates cytosolic accumulation of Rpl13a snoRNAs in response to dox. A, the exonic structures of Nox4A and alternatively-spliced nuclear form Nox4D are depicted, with localization of siRNAs that target Nox4A only (exons 3 and 5) or both Nox4A and D (exons 12–13 and 14). B–G, H9c2 cells were transfected for 24 h with siRNA targeting transcripts as shown: control, p22, Nox4A/D, or Nox4A. Cells were then treated with 20 μm dox for 1 h where indicated. RNAs were quantified by qPCR and are reported as mean values relative to Rplp0 for n = 3 experiments. Error bars show 95% confidence interval. Total RNA was assessed for target knockdown with and without dox treatment (B, D, and F). Cells were analyzed by immunofluorescence microscopy to confirm knockdown of p22 protein (C). Fluorescence intensity under identical imaging conditions was quantified, and the graph shows average intensity per cell (>50 cells per condition). Representative micrographs are shown; green = p22; blue = Hoechst. Cell lysates were analyzed by Western blot to confirm knockdown of the 67kD Nox4A isoform (E). Rat Nox4D was not detected by any commercially available antibodies. The graph shows densitometry quantification of n = 3 independent experiments with normalization to Hsp90. A representative blot is shown. Cytosolic RNA was isolated by detergent extraction, and Rpl13a snoRNAs were quantified by qPCR (G). H, cells were treated as above and analyzed by immunofluorescence microscopy for localization of NPM. Micrographs show representative fields. Scale bar, 200 μm. *, p < 0.05 versus control; #, p < 0.05 versus control/dox; &, p < 0.05 versus Nox4A/dox.
Broad Regulation of snoRNAs in the Cytoplasm
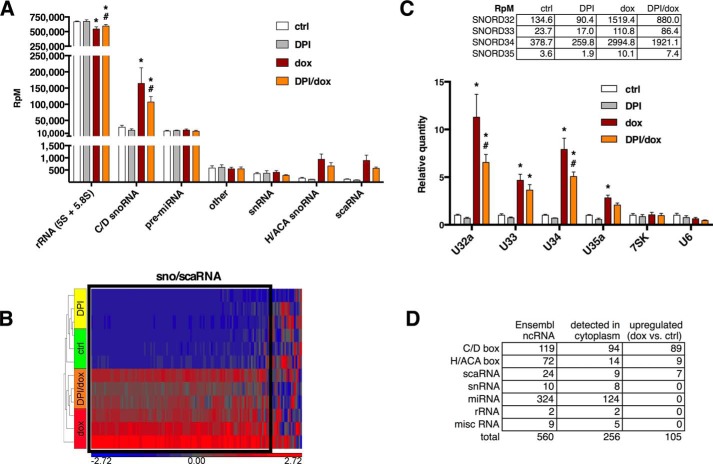
To determine whether regulated accumulation of snoRNAs in the cytoplasm is restricted to the Rpl13a locus or a more general phenomenon, we treated H9c2 cells with dox and/or DPI and prepared small RNA-sequencing libraries from RNA of 35–350 nucleotides in order to capture snoRNAs while minimizing overlap with miRNAs. Aligned reads were annotated using Ensembl Rat Build e69, which specifies 560 small ncRNA (including 215 sno/scaRNAs). Total raw reads per condition were similar, as were the number of reads aligned to the genome, and >99% of annotated reads corresponded to small ncRNA genes (Table 2). Dox treatment resulted in a significant increase in both the number and proportion of annotated reads that mapped to box C/D snoRNA (Table 2 and Fig. 4A). Box C/D snoRNAs were surprisingly abundant, accounting for 4% of annotated reads at baseline and 22% after dox treatment. Treatment with DPI significantly lowered the amount of cytoplasmic box C/D snoRNAs after dox treatment, mirroring our findings for the Rpl13a snoRNAs. Cytoplasmic box H/ACA snoRNA and scaRNA reads followed the same pattern as box C/D snoRNAs, but these changes did not reach statistical significance. In contrast, reads for rRNA (5 S and 5.8 S), pre-miRNA, snRNA, and miscellaneous other small RNA did not increase in response to dox treatment. We also detected miRNA sequences that likely represent incompletely processed miRNA precursors or reflect incomplete gel separation, but these did not change with treatment. Unsupervised hierarchical clustering showed the expected segregation by treatment group (Fig. 4B). Furthermore, the majority of sno/scaRNAs clustered tightly together (within black box), whereas snRNA (including U6), rRNA, and miscellaneous other small RNAs (including U6, 7SK, RNase P, Y-RNA, and Vault) did not (shown to the right of black box).
TABLE 2.
Pooled RNA-seq read counts from cytoplasm are shown for the analysis pipeline
Data are the cytoplasmic RNA-seq reads (total per condition).
| Control | DPI | dox | DPI/dox | |
|---|---|---|---|---|
| Raw | 15,777,864 | 14,321,914 | 16,038,254 | 16,539,780 |
| Aligned | 15,297,385 | 13,838,068 | 15,282,623 | 15,844,700 |
| Annotated | 11,026,233 | 9,976,388 | 11,264,042 | 11,475,445 |
| Annotated ncRNA | 10,972,911 | 9,931,129 | 11,201,347 | 11,418,071 |
| sn/sno/scaRNA | 453,102 | 285,808 | 2,472,756 | 1,709,804 |
| miRNA | 265,594 | 259,443 | 305,213 | 266,277 |
| rRNA (5 S + 5.8 S) | 10,243,718 | 9,376,317 | 8,413,350 | 9,431,538 |
| Miscellaneous | 8,746 | 8,247 | 8,236 | 8,667 |
| snRNA | 5,275 | 4,959 | 6,105 | 4,469 |
| scaRNA | 1,828 | 1,221 | 13,340 | 8,989 |
| C/D snoRNA | 443,462 | 278,003 | 2,439,357 | 1,685,855 |
| H/ACA snoRNA | 2,537 | 1,625 | 13,954 | 10,491 |
FIGURE 4.
snoRNAs as a class are dynamically regulated in the cytoplasm. H9c2 cells were treated with dox (20 μm) and/or DPI (500 nm) for 1 h. A, normalized RNA-seq read counts for annotated cytoplasmic small ncRNA classes. Annotated reads were normalized as reads per million raw reads (RpM) and shown as the mean ± S.E. for n = 3 experiments. ctrl, control. B, unsupervised hierarchical clustering of 132 annotated ncRNA used reads from A to generate a heat map showing relative changes in cytoplasmic small ncRNA levels with the treatments. The black box highlights clustering of snoRNAs and scaRNAs. Reads mapping to miRNAs were omitted for clarity. C, RNA-seq expression data for cytosolic U6, 7SK, and Rpl13a snoRNAs in response to treatment with DPI, dox, or DPI/dox. Read data are shown as reads/million aligned reads to demonstrate the relative abundance of different Rpl13a snoRNA species (upper). These values were then further normalized to the control sample and graphed to show relative expression by treatment (lower; mean values + S.E.; n = 3 samples). D, 105 small ncRNA genes in the cytoplasm were differentially expressed after dox treatment. The number of genes annotated for the rat genome (Ensembl ncRNA), number of genes found experimentally by cytoplasmic RNA-seq (detected in cytoplasm), and number of genes detected in the cytoplasm and up-regulated by dox (versus control) are shown by ncRNA subclass. *, p < 0.05 for treatment versus control; #, p < 0.05 for DPI/dox versus dox alone.
Although all four Rpl13a snoRNAs are transcribed in a single pre-mRNA, normalized read counts from cytoplasmic small ncRNA showed a 100-fold difference between the least abundant (U35a) and the most abundant Rpl13a snoRNA (U34) (Fig. 4C, upper). Changes in cytosolic abundance for U32a, U33, and U34 after treatment with DPI and/or dox matched extremely well with data obtained by qPCR, further validating that method of quantitation (Fig. 4C, lower). The RNA-seq method detected a 3-fold increase in cytosolic U35a after dox treatment, which was not previously detected by qPCR. This could relate to relative sensitivities of these methods, as U35a is the least abundant of the Rpl13a snoRNAs. The range of abundance among the four Rpl13a snoRNAs processed from the same pre-mRNA may reflect different post-transcriptional mechanisms that regulate the production and/or degradation of specific snoRNAs. Sequence coverage for the Rpl13a snoRNAs (66–82 nucleotides long) extended across the entire length of the snoRNAs, suggesting that these are intact snoRNA species and not miRNA-like cleavage products.
Overall, we detected 256 of 560 annotated small ncRNA, including 94 of 119 annotated box C/D snoRNAs in the cytoplasm of H9c2 cells, with lower yields of box H/ACA snoRNAs and scaRNAs (Fig. 4D). We then determined which of these small ncRNA were differentially represented in the cytoplasm after dox treatment using normalized read counts and parameters of >2-fold change with false discovery rate <0.05. The 105 genes that passed these criteria were all sno/scaRNA with up-regulated cytoplasmic expression after dox treatment, and the bulk of these were box C/D snoRNAs (Fig. 4D). The 89 up-regulated box C/D snoRNAs accounted for 95% of the detectable cytoplasmic box C/D snoRNAs in these cells, suggesting a strong class effect. Although a much smaller fraction of annotated box H/ACA snoRNAs and scaRNAs were detected in the cytoplasm, more than half of those that could be detected also showed significant accumulation after dox treatment. Consistent with our qPCR data, 7SK and U6 control RNAs were detected in the cytoplasm (Fig. 4C), but these RNAs did not accumulate with dox treatment. Overall, these data provide evidence that dox causes cytoplasmic accumulation of a wide range of snoRNAs via a Nox-dependent oxidative stress mechanism.
DISCUSSION
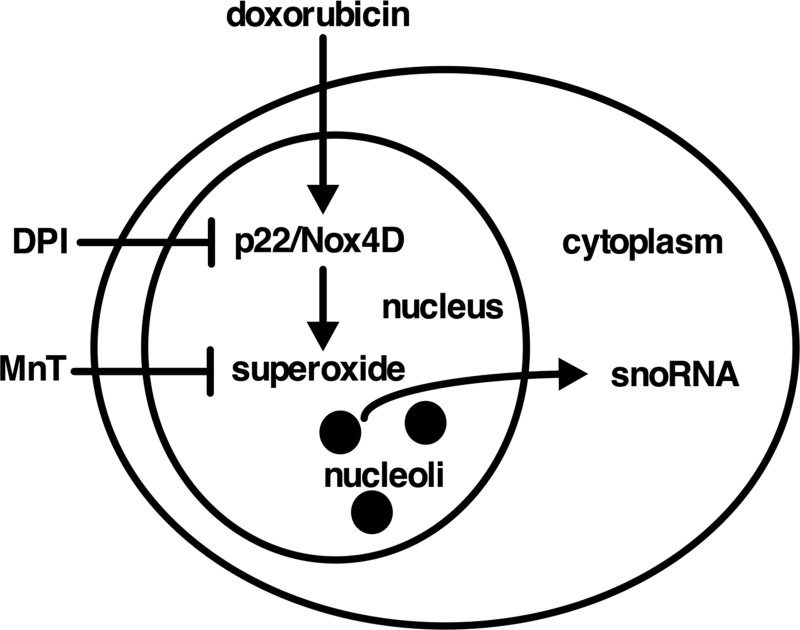
In this report we have shown by qPCR and RNA-seq analyses that box C/D snoRNAs are not exclusively localized to the nucleus but are also present in the cytoplasm, where their cytoplasmic abundance as a class is dynamically regulated by oxidative tone. Our results support a model in which dox localizes to the nucleus and interacts with Nox4D to generate nuclear superoxide, which is required for release of snoRNAs to the cytoplasm (Fig. 5).
FIGURE 5.
Proposed model showing regulation of cytosolic snoRNA abundance by Nox4D and superoxide in response to doxorubicin.
Prior studies using less sensitive methods of in situ hybridization and northern blotting have shown that snoRNAs reside in the nucleus and are concentrated in nucleoli and Cajal bodies, a distribution that supports their canonical function in the maturation of rRNA and snRNAs (17, 18). Our data showing that >99% of Rpl13a snoRNAs are nuclear in unstressed cells are consistent with this (Fig. 1F). However, by using the high sensitivity methods of qPCR and RNA-seq, we show for the first time that a broad range of snoRNAs are easily detectable in the cytoplasm. The accumulation of Rpl13a snoRNAs in the cytoplasm after oxidative stress is rapid and significant. Previous studies provided evidence of independently transcribed and 5′-trimethylguanosine-capped snoRNAs U3, U8, and U13 traffic between the nucleus and cytoplasm (19). We now demonstrate localization of intronically encoded, uncapped box C/D snoRNAs in the cytoplasm and provide insights into the regulation of this distribution.
Our data demonstrate that the cytoplasmic abundance of box C/D snoRNAs is regulated at least in part by Nox and superoxide. Our knockdown studies implicate Nox4D as a key regulator for the nuclear-cytoplasmic partitioning of Rpl13a snoRNAs, and this isoform is well positioned to catalyze redox cycling of dox to produce superoxide within the nucleus (15). The rapid accumulation of snoRNAs in the cytoplasm in the setting of oxidative stress is consistent with a model in which superoxide regulates the traffic of pre-existing nucleolar snoRNAs to the cytoplasm, although we cannot rule out contributions of regulated degradation of these species in the cytoplasm. Because the nuclear snoRNAs are in vast excess to the cytoplasmic snoRNAs, even small changes in the nuclear pool would be expected to create large relative changes in the cytoplasmic pool. A model of tight regulation of cytoplasmic snoRNA levels is further supported by our prior observation that mutant cells with 50% reduction of nuclear Rpl13a snoRNAs maintain wild-type cytoplasmic levels of these snoRNAs (20). This suggests that cells have robust mechanisms for maintaining the cytoplasmic levels of snoRNAs and that detection of snoRNAs in the cytoplasm is not simply a “mass effect” in which some proportion of snoRNAs spill over into the cytoplasm.
The regulated presence of snoRNAs in the cytoplasm furthers our understanding of these ncRNAs and provides a new context in which they may serve important cellular functions. In the case of the Rpl13a snoRNAs, we previously showed that methylation of predicted rRNA target sites does not correlate with their function in the oxidative stress response (9). The presence of these and other box C/D snoRNAs in the cytoplasm raises the possibility that snoRNAs might target cytoplasmic RNAs such as mRNAs. Antisense interactions with such targets could lead to nucleotide modifications or other processing events, analogous to the well characterized role for snoRNAs in modification of nascent rRNAs in the nucleus. A recent publication showing that yeast H/ACA snoRNAs are responsible for pseudouridylation of certain mRNA species suggests that the same could be true for box C/D snoRNAs (21). Given that the number and diversity of snoRNAs rivals that of miRNAs, snoRNAs as a class could exert broad effects on cytoplasmic functions.
Acknowledgments
We are grateful to John Edwards for critical reading of this manuscript and to Xiaowen Wang for assistance with Partek analysis. RNA-seq was performed by the Genome Technology Access Center at Washington University School of Medicine, supported by National Institutes of Health Grants P30 CA91842, UL1 TR000448, and P30 DK052574.
This work was supported, in whole or in part, by National Institutes of Health Grants T32 HL07081, K08 HL114889, and R01 DK064989.
RNA-seq data are available in Gene Expression Omnibus (GEO) under accession number GSE67050.
C. L. Holley, B. S. Scruggs, and J. E. Schaffer, unpublished observations.
- snoRNA
- small nucleolar RNA
- ncRNA
- non-coding RNA
- DPI
- diphenyleneiodonium chloride
- dox
- doxorubicin
- MnT
- Mn(III)TMPyP
- Nox
- NADPH oxidase
- NPM
- nucleophosmin
- qPCR
- quantitative real-time PCR
- Rpl13a
- ribosomal protein L13a
- Hsp90
- heat shock protein 90
- miRNA
- microRNA
- scaRNA
- small Cajal body-specific RNA.
REFERENCES
- 1. Kiss T., Fayet E., Jády B. E., Richard P., Weber M. (2006) Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 71, 407–417 [DOI] [PubMed] [Google Scholar]
- 2. Matera A. G., Terns R. M., Terns M. P. (2007) Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8, 209–220 [DOI] [PubMed] [Google Scholar]
- 3. Kiss-László Z., Henry Y., Bachellerie J. P., Caizergues-Ferrer M., Kiss T. (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 85, 1077–1088 [DOI] [PubMed] [Google Scholar]
- 4. Ganot P., Bortolin M. L., Kiss T. (1997) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 89, 799–809 [DOI] [PubMed] [Google Scholar]
- 5. Tollervey D., Kiss T. (1997) Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9, 337–342 [DOI] [PubMed] [Google Scholar]
- 6. Kishore S., Stamm S. (2006) The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 311, 230–232 [DOI] [PubMed] [Google Scholar]
- 7. Schubert T., Pusch M. C., Diermeier S., Benes V., Kremmer E., Imhof A., Längst G. (2012) Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin. Mol. Cell. 48, 434–444 [DOI] [PubMed] [Google Scholar]
- 8. Vitali P., Basyuk E., Le Meur E., Bertrand E., Muscatelli F., Cavaillé J., Huttenhofer A. (2005) ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol. 169, 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Michel C. I., Holley C. L., Scruggs B. S., Sidhu R., Brookheart R. T., Listenberger L. L., Behlke M. A., Ory D. S., Schaffer J. E. (2011) Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 14, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deng S., Kruger A., Kleschyov A. L., Kalinowski L., Daiber A., Wojnowski L. (2007) Gp91phox-containing NAD(P)H oxidase increases superoxide formation by doxorubicin and NADPH. Free Radic. Biol. Med. 42, 466–473 [DOI] [PubMed] [Google Scholar]
- 11. Zhao Y., McLaughlin D., Robinson E., Harvey A. P., Hookham M. B., Shah A. M., McDermott B. J., Grieve D. J. (2010) Nox2 NADPH oxidase promotes pathologic cardiac remodeling associated with doxorubicin chemotherapy. Cancer Res. 70, 9287–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holden P., Horton W. (2009) Crude subcellular fractionation of cultured mammalian cell lines. BMC Res. Notes 2, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rio D. C., Ares M., Hannon G. J., Nilsen T. W. (2010) Preparation of cytoplasmic and nuclear RNA from tissue culture cells. Cold Spring Harb. Protoc. 10.1101/pdb.prot5441 [DOI] [PubMed] [Google Scholar]
- 14. Avitabile D., Bailey B., Cottage C. T., Sundararaman B., Joyo A., McGregor M., Gude N., Truffa S., Zarrabi A., Konstandin M., Khan M., Mohsin S., Völkers M., Toko H., Mason M., Cheng Z., Din S., Alvarez R., Jr., Fischer K., Sussman M. (2011) Nucleolar stress is an early response to myocardial damage involving nucleolar proteins nucleostemin and nucleophosmin. Proc. Natl. Acad. Sci. U.S.A. 108, 6145–6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anilkumar N., San Jose G., Sawyer I., Santos C. X., Sand C., Brewer A. C., Warren D., Shah A. M. (2013) A 28-kDa splice variant of NADPH oxidase-4 is nuclear-localized and involved in redox signaling in vascular cells. Arterioscler. Thromb. Vasc. Biol. 33, e104–e112 [DOI] [PubMed] [Google Scholar]
- 16. Matsushima S., Kuroda J., Ago T., Zhai P., Park J. Y., Xie L.-H., Tian B., Sadoshima J. (2013) Increased oxidative stress in the nucleus caused by Nox4 mediates oxidation of HDAC4 and cardiac hypertrophy. Circ. Res. 112, 651–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tyc K., Steitz J. A. (1989) U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 8, 3113–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Samarsky D. A., Fournier M. J., Singer R. H., Bertrand E. (1998) The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 17, 3747–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Watkins N. J., Lemm I., Lührmann R. (2007) Involvement of nuclear import and export factors in U8 box C/D snoRNP biogenesis. Mol. Cell. Biol. 27, 7018–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scruggs B. S., Michel C. I., Ory D. S., Schaffer J. E. (2012) SmD3 regulates intronic noncoding RNA biogenesis. Mol. Cell. Biol. 32, 4092–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwartz S., Bernstein D. A., Mumbach M. R., Jovanovic M., Herbst R. H., León-Ricardo B. X., Engreitz J. M., Guttman M., Satija R., Lander E. S., Fink G., Regev A. (2014) Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]