Background: Activating mutations of the receptor tyrosine kinase RET are associated with oncogenic function in medullary thyroid cancer.
Results: RET is a dual specificity kinase, phosphorylates ATF4, and inhibits expression of the ATF4 target proapoptotic genes.
Conclusion: RET prevents apoptosis through inhibition of ATF4 activity.
Significance: Simultaneous targeting of RET and ATF4 may provide clinical benefit in cancers with RET abnormalities.
Keywords: Apoptosis, Thyroid, Transcription Regulation, Transcription Repressor, Tyrosine-Protein Kinase (Tyrosine Kinase), Ubiquitylation (Ubiquitination)
Abstract
The RET proto-oncogene, a tyrosine kinase receptor, is widely known for its essential role in cell survival. Germ line missense mutations, which give rise to constitutively active oncogenic RET, were found to cause multiple endocrine neoplasia type 2, a dominant inherited cancer syndrome that affects neuroendocrine organs. However, the mechanisms by which RET promotes cell survival and prevents cell death remain elusive. We demonstrate that in addition to cytoplasmic localization, RET is localized in the nucleus and functions as a tyrosine-threonine dual specificity kinase. Knockdown of RET by shRNA in medullary thyroid cancer-derived cells stimulated expression of activating transcription factor 4 (ATF4), a master transcription factor for stress-induced apoptosis, through activation of its target proapoptotic genes NOXA and PUMA. RET knockdown also increased sensitivity to cisplatin-induced apoptosis. We observed that RET physically interacted with and phosphorylated ATF4 at tyrosine and threonine residues. Indeed, RET kinase activity was required to inhibit the ATF4-dependent activation of the NOXA gene because the site-specific substitution mutations that block threonine phosphorylation increased ATF4 stability and activated its targets NOXA and PUMA. Moreover, chromatin immunoprecipitation assays revealed that ATF4 occupancy increased at the NOXA promoter in TT cells treated with tyrosine kinase inhibitors or the ATF4 inducer eeyarestatin as well as in RET-depleted TT cells. Together these findings reveal RET as a novel dual kinase with nuclear localization and provide mechanisms by which RET represses the proapoptotic genes through direct interaction with and phosphorylation-dependent inactivation of ATF4 during the pathogenesis of medullary thyroid cancer.
Introduction
The RET proto-oncogene, which encodes a receptor tyrosine kinase and regulates a complex network of signal transduction pathways, is often aberrantly activated through mutations or oncogenic fusion. RET oncogenic mutations contribute to cell transformation in medullary thyroid cancer (MTC)2 (1). MTC occurs in sporadic (75%) or hereditary (25%) forms with three clinical subtypes: multiple endocrine neoplasia types 2A and 2B and familial MTC. The MEN2A subtype harbors RET mutations affecting the cysteine residues (609, 611, 618, 620, or 634). These mutations lead to the formation of permanent receptor dimers with constitutive autophosphorylation activity, stimulate downstream signaling pathways, and thus promote transformation. The most aggressive form of MTC is associated with the MEN2B subtype with RET mutations identified in the tyrosine kinase domain of RET (M918T) and causes conformational change and alters substrate specificity.
Furthermore, RET overexpression has been observed in pancreatic cancer (2), melanoma (3), and leukemia (4). RET copy number gains, mutations, and rearrangements have been observed in breast cancer (5). Oncogenic fusions of RET that drive misexpression of the tyrosine kinase domain are found in papillary thyroid carcinoma (6), chronic myelomonocytic leukemia (7), and lung adenocarcinomas (8, 9). Conversely, disruption of RET signaling by a dominant negative truncated form of RET reduces cell viability, abolishes phosphorylation of downstream signaling molecules, reduces cell cycle progression, and stimulates apoptosis (10). RET inhibition also decreases the growth and metastatic potential of estrogen receptor-positive breast cancer cells (11). The direct transforming impact of RET as an oncogenic driver is further supported by studies in which RET transgenic mice developed a variety of malignancies (12, 13). Despite these studies of RET-activated oncogenic signaling pathways, the molecular mechanism by which RET prevents cell death remains unclear.
Cell survival involves suppression of the intrinsic apoptosis pathway through complex interactions between members of the BCL-2 family (14). The BH3-only proapoptotic family members, including NOXA and PUMA, are responsible for relaying various environmental stresses to promote cell death (14). Both NOXA and PUMA are direct transcriptional targets of p53 (15), but they can be regulated by other transcription factors, including ATF4 (16, 17), cMYC (18), FOXO3a (19), Sp1 (20), and E2F1 (21).
The transcription factor ATF4 plays a central role in the activation of the integrated stress response pathway (22). In response to stressors, eIF2α phosphorylation induces ATF4 translation, which then activates expression of its downstream target genes. Some studies indicate that ATF4 plays a prosurvival role (23), whereas others indicate a proapoptotic role, suggesting that the function of ATF4 is context-dependent (16, 17, 24, 25). The fact that p53 expression is lost in MTC cells (26) and the central role of ATF4 in promoting apoptosis led us to hypothesize that RET might prevent the induction of apoptosis via regulating ATF4. Here, we demonstrate that RET represses expression of the ATF4 target proapoptotic genes NOXA and PUMA in an MTC-derived cell line through phosphorylation-dependent degradation of ATF4.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Eeyarestatin and sunitinib were purchased from Tocris Bioscience, and cisplatin was from Sigma-Aldrich. The sources of antibodies are as follows: PUMA, MCL-1, BAD, BIM, BAX, and BCL-XL (Cell Signaling Technology); NOXA (Calbiochem); ATF4 (C-20), RET (C-19), RET (C-20), RET (H300), and ubiquitin (P4D1) (Santa Cruz Biotechnology, Inc.); ATF4 (D4B8) (Cell Signaling). Peptides containing amino acids 102–125 of ATF4 were custom made from Abgent (San Diego, CA).
Cell Lines
The TT, H1993, HCC2935, and HEK293T cell lines were purchased from ATCC. MZCRC1 cells were kindly provided by Dr. Alex Knuth (University Hospital Zürich, Zurich, Switzerland) and were described previously (27–29). The thyroid cell lines were verified by sequencing; TT cells harbor a codon 634 cysteine to tryptophan (C634W) exon 11 RET mutation, and MZCRC1 cells harbor a codon 918 methionine to threonine (M918T) exon 16 RET mutation.
Plasmid Construction and Lentiviral Transduction
Plasmid constructs containing NOXA-luc were purchased from Addgene (30). WT RET, RET-C634W, and RET-M918T long isoform (RET-51) constructs were described previously (31). RET-K758, FLAG-tagged ATF4-T107, ATF4-T114, ATF4-T115, ATF4-T119, and ATF4–4TA constructs were generated with a site-directed mutagenesis kit (Stratagene). Lentiviral vectors (pLKO.1) containing RET and ATF4-specific shRNAs were purchased from Sigma-Aldrich. Lentiviral RET shRNA plasmids were co-transfected into HEK293T cells along with packaging (VPR8.9) and envelope (VSV-G) plasmids using X-tremeGENE (Roche Applied Science) for 2 days. The virus particles containing RET shRNA or control shRNA were used to infect TT cells. Transfected cells were selected in media containing 2 μg/ml puromycin (Clontech). ATF4-WT and ATF4–4TA were cloned in lentiviral vectors (OriGene). The RET kinase domain spanning amino acids 657–1114 for WT and RET-M918T was cloned in pEF4-MYC (Life Technologies, Inc.). GFP-RET and RFP-ATF4 were cloned in pcDNA3.1. RET siRNAs were purchased from Sigma-Aldrich.
Cell Viability, Anchorage-independent Growth, Proliferation, Cell Cycle, and Apoptosis Analyses
Cell viability was measured using an MTT assay with 40,000 cells in a 96-well plate. Cells were treated as indicated in figures and incubated in 200 μl of 0.6 mg/ml MTT in serum-free medium for 4 h and then were solubilized in dimethyl sulfoxide for 30 min following quantification with a spectrophotometer (VICTOR3, PerkinElmer Life Sciences) at 595 nm. Anchorage-independent colony formation assays were performed as described previously (32). Apoptotic cell death was determined using the BD ApoAlert annexin V-FITC apoptosis kit (BD Biosciences) according to the manufacturer's instructions, and cells were analyzed on FACScan system (BD Biosciences). In addition, cells were fixed and stained with antibodies against cleaved caspase-3 and poly(ADP-ribose) polymerase (PARP) (BD Biosciences) and visualized by flow cytometry as described previously (33).
Immunofluorescence Staining and Fluorescence Lifetime Imaging Microscopy (FLIM) Analysis
Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) (pH 7) for 20 min, permeabilized, and blocked in 5% bovine serum albumin. Incubation with the indicated antibodies was carried out for 1 h at 37 °C followed by incubation with a secondary antibody for 1 h at room temperature. DAPI was used to visualize nuclei. FLIM analysis was performed as described previously (34, 35). Cells were imaged using a Leica SP5 II confocal microscope. FLIM samples were excited with a tunable femtosecond titanium-sapphire pumped laser (Mai Tai BB, Spectra-Physics). The wavelength used for two-photon excitation was 900 nm. Images were obtained with an oil immersion objective (numerical aperture 1.4) and a line scan speed of 400 Hz, with image size of 512 × 512 pixels. For FLIM analysis, the pixels were reduced to 256 × 256. FLIM data were collected using an SPC830 data and image acquisition card (Becker & Hickl) for time-correlated single photon counting (TCSPC). The fluorescence decays were fitted with a single exponential decay model using SPC Image software (Becker & Hickl), and the GFP fluorescence lifetimes were displayed in a false color map.
Dual-Luciferase Assay
Using X-tremeGENE (Roche Applied Science), the 4X-CRE or pGL3-NOXA-Luc construct was transfected into HEK293T cells, with Renilla luciferase plasmid (Promega) as an internal control. Dual-Luciferase assays were performed at 24 h post-transfection, and reporter activity was normalized as relative luciferase activity (firefly luciferase/Renilla luciferase).
Real-time PCR
Total RNA was extracted with a Qiagen RNeasy kit and treated with DNase. cDNA was prepared using reverse transcriptase and used for quantitative real-time PCR using TaqMan primer probes (Applied Biosystems) and 18 S rRNA or hypoxanthine-guanine phosphoribosyltransferase (HPRT) as control.
ChIP Assay
ChIP was performed as described previously (36). The quantitative PCR was performed on ChIP DNA using FastStart SYBR Green Master (Roche Applied Science). The primer sequences used for the ChIP have been described previously (30), and the primers used for the real-time PCR included the following: primer set 1, CCTACGTCACCAGGGAAGTT (forward) and GATGCTGGGATCGGGTGT (reverse); primer set 2, TAACAGGCTGCTGTCTCTGG (forward) and TTGGGAATTAAGTCGGACCA (reverse).
Biochemical Assays
Cell extracts, Western blot analysis, immunoprecipitation, and cell fractionations were performed as described previously (37).
In Vitro Binding Assay
35S-Labeled RET was prepared by in vitro transcription and translation using the TNT system as described previously (32). Labeled RET was incubated with GST or GST-ATF4 followed by pull-down with glutathione S-transferase-agarose beads and then separation of SDS-PAGE and autoradiography.
In Vitro Kinase Assay and Mass Spectrometry
For the kinase reaction, 0.25 μg of commercial RET active enzyme (catalog no. 14-570, Millipore) or immunopurified FLAG-tagged RET kinase domain from TT cells and transfected HEK293T cells was incubated with 2 μg of GST-ATF4 in presence of 1 mm ATP for 30 min at 30 °C. Samples were separated on SDS-PAGE followed by Western blotting with tyrosine- and threonine-specific phosphoantibodies. Site-specific phosphorylation of GST-ATF4 by commercial RET was identified by mass spectrometry. For dot blot analysis, RET kinase domain wild type was expressed in HEK293T cells and purified with FLAG-agarose beads. FLAG-tagged RET bound with agarose beads was incubated with peptides in kinase buffer at 30 °C for 1 h. Following incubation, supernatant was spotted onto nitrocellulose membrane and then incubated with phospho-specific antibodies.
In Vivo Ubiquitination Assay
Cells were treated with MG132 (10 μm) for 8 h. ATF4 was immunoprecipitated by c-20 antibody followed by Western blot analysis with ubiquitin antibody.
Statistical Analysis
All data were expressed as means ± S.D. Data were analyzed with GraphPad Software using the indicated tests. Statistical significance was indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
RESULTS
RET Inhibition Increases Sensitivity to Cisplatin-mediated Apoptosis
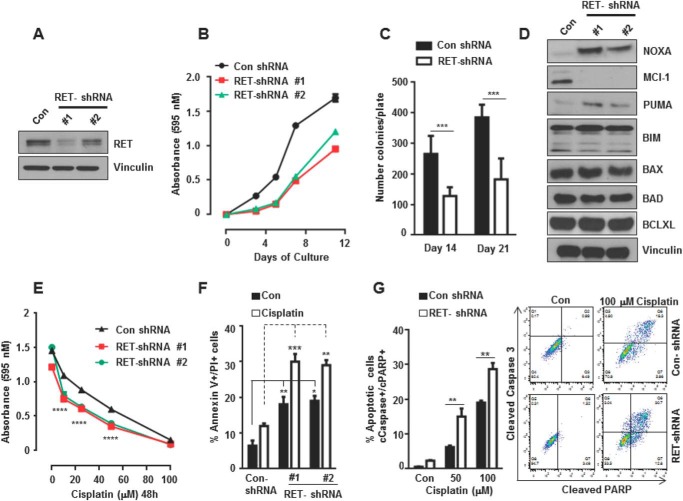
The human MTC cell line TT was derived from a patient with aggressive medullary thyroid carcinomas and carries a frequent RET mutation (C634W) identified in patients with multiple endocrine neoplasia type 2A disease. This mutation confers dominant ligand-independent constitutive activity of the mutant RET protein. Oncogenic RET has been shown to increase antiapoptotic signaling (10). We first examined the mechanism by which RET prevents apoptosis upon stress pathway activation. TT cell lines stably expressing RET-specific shRNA were generated by lentiviral shRNA transduction. Western blot analysis confirmed that RET protein levels were 60–90% lower in the RET-depleted cells than in the controls (Fig. 1A). RET depletion in the TT cells decreased cell survival and inhibited colony formation on soft agar (Fig. 1, B and C). To define the mechanism of decreased survival, we examined the levels of proapoptotic and antiapoptotic regulators in the RET-depleted and control TT cells. A significant increase in levels of the proapoptotic regulators NOXA (i.e. PMAIP1) and PUMA (i.e. BBC3) was observed in RET-depleted TT cells, whereas the level of the antiapoptotic regulator MCL-1 was markedly decreased (Fig. 1D). The levels of the proapoptotic regulators BIM (i.e. BCL2L11), BAX (BCL2L4), and BAD (BBC2, BCL2LB) and the antiapoptotic regulator BCL-XL (i.e. BCL2L1) were not affected by RET depletion.
FIGURE 1.
RET knockdown decreases cell survival and sensitizes cells to cisplatin-induced cell death. A, Western blot analysis showing efficiency of RET knockdown in RET shRNA TT cells (clone 1 and clone 2) compared with control (Con). Vinculin served as a loading control. B, cell viability was measured by an MTT assay in control shRNA and RET shRNA TT cells at the indicated time points. Shown are the mean and S.D. (error bars) of six replicates in a representative experiment. C, RET knockdown inhibits anchorage-independent growth. Control and RET shRNA 1 cells were seeded in soft agar, and colonies were counted after 2 and 3 weeks. Data are presented as mean and S.D. of six replicates in a representative experiment (Student's t test). D, Western blot showing levels of apoptotic and survival proteins in control and RET shRNA TT cells. E, MTT assay for cell viability in RET shRNA and control shRNA cells treated with cisplatin at the indicated dose for 48 h. Shown are mean and S.D. of six replicates in a representative experiment. Two-way analyses of variance and Bonferroni's multiple comparison tests were used. F, quantification of apoptotic and dead cells using an annexin V/propidium iodide assay in control shRNA and RET shRNA TT cells after treatment with cisplatin (20 μm) for 24 h. Two-way analyses of variance and Bonferroni's multiple comparison tests were used. G, cleaved caspase-3/cleaved PARP-positive cells were quantified using flow cytometry after treatment with cisplatin at the indicated concentrations for 24 h (right panels). Data are shown as means ± S.D. from three independent experiments (left panel). The statistical analysis used was two-way analysis of variance, Tukey's multiple comparison test. Asterisk indicates p value, *, p < 0.05; **, p < 0.01; ***, p < 0.001.
It has been shown that NOXA expression is up-regulated in response to cisplatin and related platinum compounds (38). Because knockdown of RET up-regulated NOXA levels (Fig. 1D), we hypothesized that RET knockdown would also enhance the efficacy of cisplatin-induced cell death. We observed that RET-depleted TT cells were more sensitive to cisplatin than control cells, as shown by an MTT assay (Fig. 1E). Quantification of apoptotic cells by annexin V/propidium iodide staining showed increase in cell death in cisplatin treated RET-shRNA cells compared with control shRNA cells (Fig. 1F). The percentage of apoptotic cells was quantified by measuring cleaved caspase-3/cleaved PARP double-positive cells using flow cytometry. We found that RET-depleted cells were more sensitive to cisplatin-induced cell death than control shRNA cells treated with cisplatin (Fig. 1G).
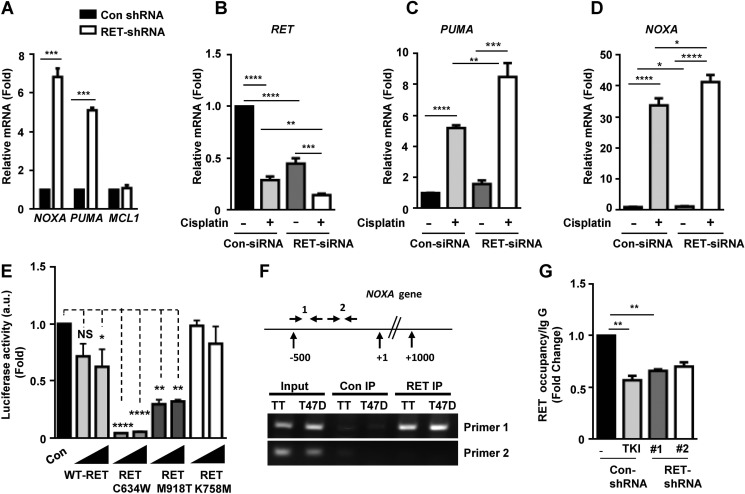
To address the mechanisms of RET-mediated regulation in expression of NOXA and PUMA, we analyzed their transcript levels in control and RET-depleted TT cells. We observed that the levels of NOXA and PUMA mRNAs were strongly stimulated in RET-depleted shRNA TT cells compared with control shRNA cells, whereas there was no change in MCL-1 mRNA levels observed, indicating that RET mediates transcriptional repression of the NOXA and PUMA genes in MTC cells (Fig. 2A). Similar results were reproduced in another MTC cell line, MZCRC1, in which knockdown of RET by siRNA increased the levels of NOXA and PUMA mRNAs (Fig. 2, C and D). Cisplatin-induced stimulation in NOXA and PUMA mRNA levels were higher in RET siRNA-transfected cells compared with control siRNA cells (Fig. 2, C and D). However, RET expression was reduced by cisplatin in both RET siRNA and control siRNA cells (Fig. 2B). These findings suggest that RET is an essential regulator of survival in MTC cells, and its specific inhibition leads to the induction of proapoptotic effector genes, causing sensitivity to genotoxic stress-induced cell death.
FIGURE 2.
RET represses NOXA and PUMA expression and NOXA promoter activity. A, quantitative real-time PCR showing mRNA levels of NOXA, PUMA, and MCL-1 in control shRNA and RET shRNA 1 TT cells using S18 mRNA as internal control. Data are shown as means ± S.D. (error bars) performed in triplicate from three independent experiments (Asterisk indicates p value, ***, p = 0.0006, Student's t test). B–D, MZCRC1 cells were transfected with control and RET siRNAs for 48 h and treated with cisplatin (100 μm) for 24 h. Quantitative real-time PCR shows mRNA levels of RET (B), PUMA (C), and NOXA (D) in control siRNA and RET siRNA cells. Values were normalized against HPRT mRNA levels. Shown are means ± S.D. of triplicates from two independent experiments (unpaired Student's t test). E, RET kinase activity is required for repression of NOXA promoter activity. Reporter assays in transfected HEK293T cells with the NOXA reporter and increasing amounts of WT RET, active mutant RET (RETC634W, RET-M918T), or kinase-dead mutant RET (RET-K758A) plasmids. Normalized luciferase enzyme activity (a.u. indicates arbitrary units) is presented as mean ± S.D. of triplicate samples from one representative experiment. F, schematic of the NOXA locus and the primers used in ChIP experiments. Shown is the ChIP assay for RET occupancy at the NOXA gene in TT cells and breast cancer T47D cells. RET occupancy was observed within a region close to the proximal NOXA promoter as amplified by primer set 1 (primer set 1 amplifies a genomic region −500 bp from the transcription start site; primer set 2 amplifies a genomic region +500 bp from the transcription start site. G, ChIP assays indicating RET occupancy at the NOXA promoter in TT cells treated with TKI (10 μm vandetanib for 8 h) and RET shRNA TT cells. Data are shown as means ± S.D. from two independent experiments. The statistical analysis used was unpaired Student's t test; **, p = 0.0048.
RET Kinase Activity Is Required for Inhibition of the NOXA Promoter Activity
Using reporter assays (NOXA-luciferase) in HEK293T cells, we showed that ectopic expression of both wild type and mutants (M918T and C634W mutations) RET long isoform (RET 51) repressed the NOXA promoter activity in a dose-dependent manner (Fig. 2E). Interestingly, NOXA promoter inhibition by activated RET-C634W and RET-M918T were more robust than inhibition by RET-WT. However, co-expression of a kinase-dead RET mutant (RET-K758M) did not inhibit NOXA promoter activity, indicating that RET kinase activity was required for repression of NOXA transcription (Fig. 2E). To gain further insight into the mechanisms of RET-dependent repression of the apoptotic genes, we performed a chromatin immunoprecipitation assay in MTC TT cells and breast cancer T47D cells using RET antibodies. Our results indicate that RET interacted with the chromatin fragments of the NOXA gene in TT and breast cancer T47D cell lines. The RET occupancy was observed at the proximal promoter within −500 bp upstream of the transcriptional start site of NOXA gene (primer 1, Fig. 2F). Furthermore, RET occupancy at the NOXA promoter was appreciably reduced in TT cells treated with tyrosine kinase inhibitor vandetanib and in RET shRNA TT cells as well (Fig. 2G). These results support our hypothesis that RET has a nuclear function in regulation of gene expression. These data further prompted us to examine localization of RET in the nucleus, wherein this mediates nuclear function in regulation of NOXA and PUMA genes.
Receptor Tyrosine Kinase RET Is Localized in the Nucleus
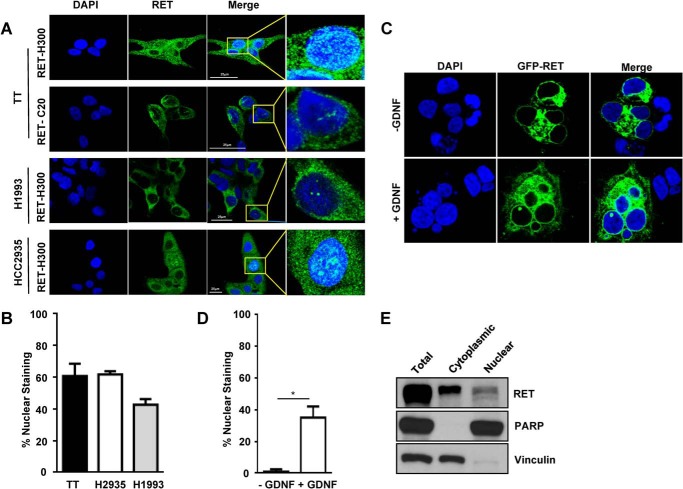
The nuclear localization of other receptor tyrosine kinases is well documented (39). Therefore, a direct assessment of nuclear localization was performed through immunostaining of TT cells and lung cancer cells using antibodies specific to the N and C termini of RET. These studies showed that although RET was localized predominantly in the plasma membrane and cytoplasm, RET was also localized in the nucleus, indicating that intact RET can translocate into the nucleus (Fig. 3, A and B). We also observed RET in the nuclei of the non-small lung cancer cell lines H1993 and HCC2935, suggesting that nuclear localization of RET is not restricted to MTC cells (Fig. 3, A and B). Next, we investigated whether RET activation was required for its translocation to the nucleus. Green fluorescent protein GFP-RET was ectopically expressed in HEK293T cells, which were then stimulated with glial cell-derived neurotropic factor and its soluble coreceptor GFRα1. Nuclear localization of GFP-RET was only observed in glial cell-derived neurotropic factor-treated cells, which confirmed that activation was essential for translocation (Fig. 3, C and D). Further, cellular fractionation followed by Western blot studies detected intact full-length RET protein in the nuclear fractions of TT cells (Fig. 3E).
FIGURE 3.
RET nuclear localization in cancer cells. A, immunostaining of RET in TT cells and non-small lung cancer cell lines with the indicated RET antibodies. Scale bars are indicated by the white lines. B, quantification of RET-positive cells with nuclear staining. Approximately 200–300 cells were counted at ×20 magnification in five different fields for each cell line. Data are means ± S.D. (error bars) from two independent experiments. C, HEK 293T cells were transfected with WT GFP-RET and treated with soluble coreceptor GFRα1 and glial cell-derived neurotropic factor (GDNF) (100 ng/ml) for 15 min and stained with DAPI. D, quantification of cells showing nuclear RET as performed in B. E, Western blot analysis for RET in nuclear and cytoplasmic fractions of TT cells. Vinculin and PARP were used as cytoplasmic and nuclear markers, respectively. Asterisks indicate p value, *, p < 0.05.
RET Represses ATF4-mediated Apoptotic Gene Transcription
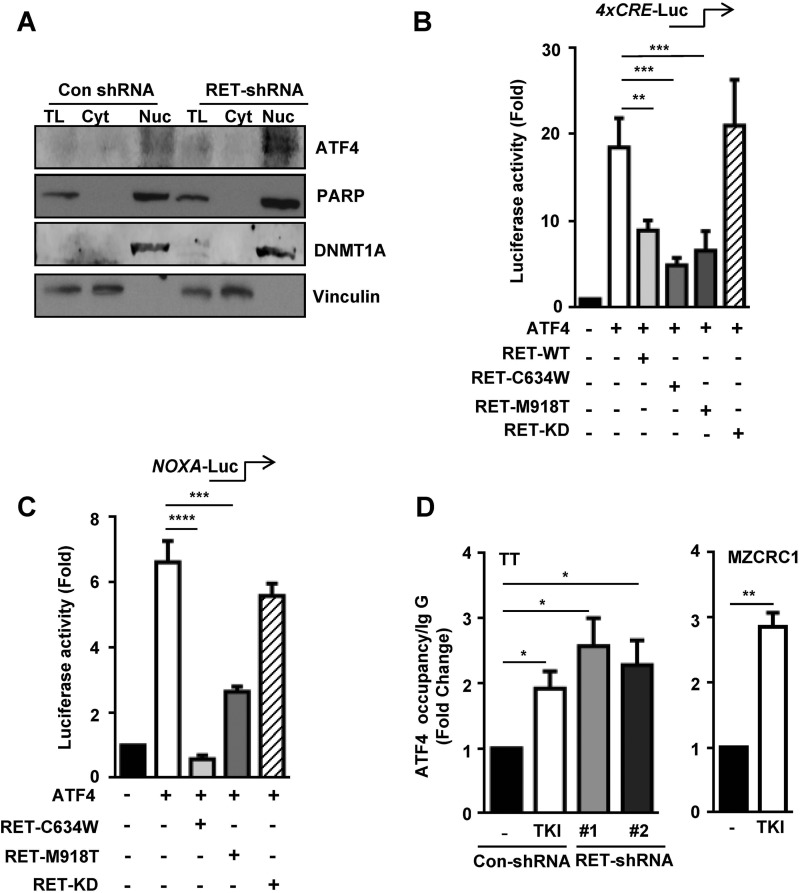
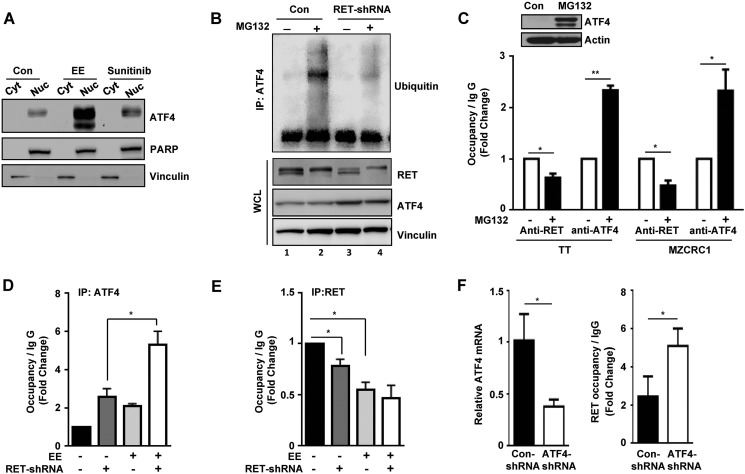
In silico analysis of the promoter sequence bound by RET identified several cis-acting elements, including consensus binding sites for p53 (15), c-Myc (18), and ATF4 (30). Because no expression of a p53 transcript is detected in MTC cells (26), we chose to focus on ATF4 because of its well defined role in the activation of apoptotic genes, including NOXA and PUMA (16, 30). Interestingly, ATF4 levels were significantly increased in the nuclear fraction of RET-depleted TT cells compared with control cells (Fig. 4A), in parallel with levels of NOXA and PUMA (Fig. 1D), which are both targets of ATF4. These observations led us to hypothesize that RET regulates the ATF4 activity.
FIGURE 4.
RET negatively regulates the ATF4 transcriptional activity. A, nuclear retention of ATF4 in RET shRNA TT cells. PARP and DNMT1A (DNA methyl transferase) were used as nuclear markers, and vinculin was used as a cytoplasmic marker. TL, total lysates; Cyt, cytoplasmic fraction; Nuc, nuclear fraction. B and C, RET inhibits ATF4-dependent gene activation. Shown are reporter assays with HEK293T cells transfected with the 4xCRE-Luc reporter (B) and the NOXA-Luc reporter (C) along with expression vectors encoding ATF4-WT, RET-WT, mutant RET (RET-C634W, RET-M918T), and RET kinase-dead mutant (RET-KD). D, ChIP assays indicating ATF4 occupancy at the NOXA promoter in TT cells treated with TKI (10 μm vandetanib for 8 h) and RET shRNA TT cells (left); MZCRC1 cells were treated with 10 μm sunitinib for 8 h (right). Data are shown as means ± S.D. (error bars) from two independent experiments. The statistical analysis used was unpaired Student's t test. Asterisks indicate p value, *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
ATF4 is known to activate its target gene through the cyclic AMP-response element (CRE). Therefore, we tested whether RET regulates ATF4-mediated activation of CRE. Consistent with a previous report (40), overexpression of ATF4 in HEK293T cells stimulated an ∼20-fold increase in the CRE-luc reporter activity (Fig. 4B). This ATF4-stimulated activation of CRE was significantly inhibited by co-expression of RET-WT, RETC634W, and RET-M918T but was not repressed by kinase-dead mutant RET (RET-K758M) (Fig. 4B). To examine the effect of RET on ATF4 transcriptional activity, we transfected the NOXA-luc reporter into HEK293T cells. Co-expression of ATF4 stimulated a 6-fold increase in the NOXA-luc reporter activity, whereas co-expression of RETC634W or RET-M918T significantly repressed the ATF4-stimulated NOXA reporter activity (Fig. 4C). Indeed, kinase-dead RET was unable to repress the NOXA reporter activity, further supporting our notion that kinase activity of RET is required for repression of ATF4-dependent NOXA reporter activity.
To examine the effect of RET on occupancy of ATF4 at its target genes, we performed ChIP assays using ATF4 antibodies in tyrosine kinase inhibitor (TKI)-treated TT cells and in the RET shRNA TT cell lines. Under these conditions, the levels of ATF4 occupancy at the NOXA promoter increased both in the TKI-treated cells and in the RET-depleted cells compared with control cells (Fig. 4D, left). Similar results were observed using the MZCRC1 cell line (Fig. 4D, right). These results indicate that ATF4 occupancy at NOXA is inversely correlated with the level and activity of RET. We conclude that increased RET activity and levels in MTC are favorable for maintaining suppression of ATF4 and the proapoptotic genes.
Stabilized ATF4 Promotes Its Recruitment to the NOXA Promoter
We have demonstrated that in MTC cells, RET is highly expressed, whereas the stress-induced transcription factor ATF4 and its target proapoptotic genes NOXA and PUMA are weakly expressed (Figs. 1D, 2A, and 4A), thus providing a mechanistic clue that RET should prevent apoptosis by antagonizing ATF4 stability. Reports indicate that eeyarestatins are ERAD-specific inhibitors that elicit an integrated stress response program at the endoplasmic reticulum, which activates ATF4-dependent induction of cell death (30, 41). Consistent with these reports, we showed that ATF4 levels were significantly higher in nuclear fractions of TT cells treated with eeyarestatin (EE) than in nuclear fraction of control cells (Fig. 5A). Levels of nuclear ATF4 were increased in TKI-treated TT cells (sunitinib) compared with untreated control cells. These data imply that elevated RET levels in MTC cells should affect ATF4 stability. To determine the effect of RET in ATF4 stability, we performed ubiquitination assay in control and RET shRNA TT cells treated with the proteasome inhibitor MG132. In the presence of MG132, a high molecular mass species due to polyubiquitination of ATF4 was observed in control TT cells. In contrast, the levels of polyubiquitinated ATF4 were reduced in RET-depleted cells (Fig. 5B, compare lane 2 with lane 4). These data suggest that ATF4 is prone to degradation in MTC cells. We speculate that RET should sequester ATF4 for ubiquitination and degradation, thus maintaining low levels of ATF4 in MTC cells.
FIGURE 5.
RET mediates suppression of apoptotic target genes through ubiquitination and degradation of ATF4. A, nuclear retention of ATF4 after eeyarestatin (EE) (10 μm for 8 h) or TKI treatment (10 μm sunitinib for 8 h) compared with untreated controls (Con). PARP and vinculin were used as nuclear and cytoplasmic markers, respectively. B, ubiquitination of ATF4 in control shRNA and RET shRNA 1 TT cells in the presence or absence of 10 μm MG132 for 8 h. Western blot showing polyubiquitinated ATF4 with ubiquitin and levels of RET and ATF4 in whole cell lysates (WCL). C, stabilization of ATF4 decreased the RET occupancy but increased the ATF4 occupancy in the NOXA promoter. Top, Western blot showing levels of ATF4 in lysate of TT cells treated with MG132 (5 μm) for 16 h. TT and MZCRC1 cells were treated with MG132 (5 μm) for 16 h and subjected to ChIP assays using anti-RET and anti-ATF4 antibodies. DMSO-treated cells were used as control. -Fold changes shown are the means ± S.D. (error bars) of two independent experiments (Student's t test). D and E, depletion of RET by shRNA or treatment with eeyarestatin increased ATF4 recruitment to the NOXA promoter. Shown are ChIP assays for occupancies of ATF4 (D) and RET (E) in control shRNA-TT cells or RET shRNA TT cells with or without treatment with eeyarestatin (10 μm for 8 h). Data are shown as means ± S.D. from two independent experiments (Student's t test). F, left, levels of ATF4 mRNA in control and ATF4 shRNA TT cells measured by quantitative PCR. Right, ChIP assays indicating RET occupancy at the NOXA promoter in control and ATF4 shRNA TT cells. Data are means ± S.D. from two independent experiments (Student's t test). *, p < 0.05; **, p < 0.01.
Furthermore stabilization of ATF4 by MG132 in both MTC cell lines, TT and MZCRC1, allowed a significant increase in ATF4 occupancy at the NOXA promoter, but RET occupancy decreased (Fig. 5C, top panel for TT cells). Moreover, ATF4 occupancy was also significantly enhanced at the NOXA promoter in RET shRNA TT cells that were treated with eeyarestatin compared with RET shRNA or eeyarestatin-treated control cells, and RET occupancy was decreased (Fig. 5, D and E), suggesting that ATF4 prevents interactions of RET with the NOXA gene. These results were further confirmed in ATF4 shRNA TT cells, which indicated that occupancy of RET at the NOXA promoter increased in ATF4-depleted TT cells compared with control TT cells (Fig. 5F, right). These findings consistently indicate that there is an inverse relationship between ATF4 and RET occupancy at the target NOXA gene. Overall, these data strongly suggest that aberrantly activated RET decreases ATF4 levels and consequently inhibits activation of the proapoptotic NOXA and PUMA genes.
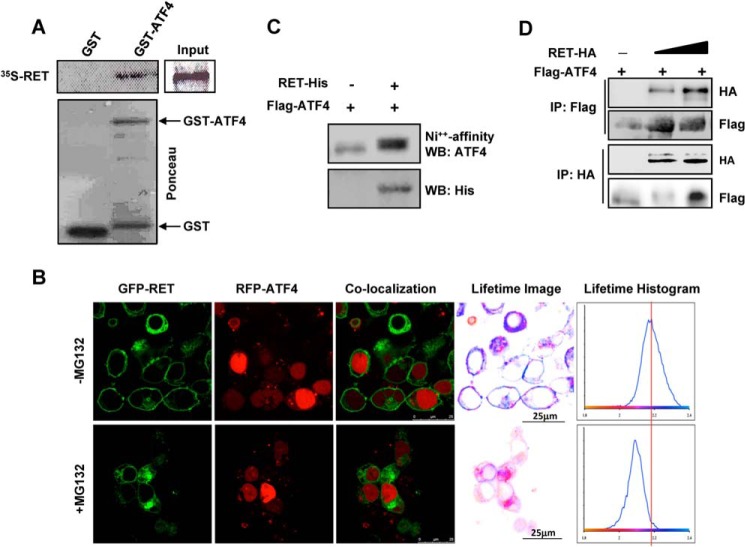
RET Physically Interacts with ATF4 in Vitro and in Vivo
The kinase-dependent inhibition of ATF4 activity by RET led us to examine their physical interactions and the phosphorylation of ATF4 by RET. A GST pull-down assay demonstrated that in vitro-translated [35S]methionine-labeled RET-C634W specifically interacted with recombinant GST-ATF4 but not with GST alone (Fig. 6A), indicating a physical interaction between ATF4 and RET in vitro.
FIGURE 6.
RET interacts with ATF4. A, in vitro GST pull-down assay with 35S-labeled RET-C634W proteins in the presence of GST and GST-ATF4. B–D, interactions between RET and ATF4 in cells; B, FRET between RET and ATF4 was measured using FLIM after transfection in HEK293T cells treated with 5 μm MG132 for 16 h. Confocal images and co-localization of the same field are shown (left). The histograms as shown (right) indicate the average fluorescence lifetime distributions corresponding to cells in the field of view. Lifetime images were generated by pixel-by-pixel mapping of the lifetime data and are represented as false color images adjacent to histograms. Scale bars are indicated by the white line. C, ATF4 was co-purified with RET. HEK293T cells were transfected with His-RET- kinase domain (residues 657–1114) and FLAG-ATF4 followed by purification of RET with nickel beads and then Western blot (WB) with ATF4 or His tag antibodies. D, RET was co-immunoprecipitated (IP) with ATF4. FLAG-ATF4 and HA-tagged full-length WT RET were expressed in HEK293T cells followed by reversed immunoprecipitation and Western blot with the indicated antibodies.
Direct interactions between RET and ATF4 in vivo were studied by FLIM after co-expression of wild type GFP-RET and RFP-ATF4 in HEK293T cells treated with MG132. Representative confocal images and co-localization of the same field of view are also shown (Fig. 6B). FLIM analysis showed that in the presence of MG132, GFP-RET is within 10 nm of RFP-ATF4, supporting a direct physical interaction of the two proteins. The specific interaction is observed by a peak shift to the left of the vertical line (i.e. to shorter lifetimes resulting from FRET between GFP and RFP) (Fig. 6B). The histograms on the right show the average fluorescence lifetime distributions corresponding to cells in the field of view. Lifetime images were generated by pixel-by-pixel mapping of the lifetime data and represented as false color images. The interactions between RET and ATF4 were only observed in cells treated with proteasome inhibitor. In cells expressing the RET receptor, the average lifetime is centered on ∼2.2 ns. This is in contrast to the proteasome inhibitor-treated cells, where the average lifetime shortens to 2.1 ns, seen as a left shift in the peak. The 100-ps shortening of the average lifetime is the result of FRET between GFP-RET and RFP-ATF4, indicating direct interactions between RET and ATF4.
In addition, using affinity purification of RET with Ni2+ beads in the lysates of HEK293T expressed with His-tagged RET(657–1114) and FLAG-tagged ATF4, we found that ATF4 was co-purified with RET, further confirming in vivo interaction of these proteins (Fig. 6C). In co-immunoprecipitation (IP) and reverse co-immunoprecipitation assays, we further demonstrated that interactions between RET and ATF4 after transfections in HEK293T cells (Fig. 6D).
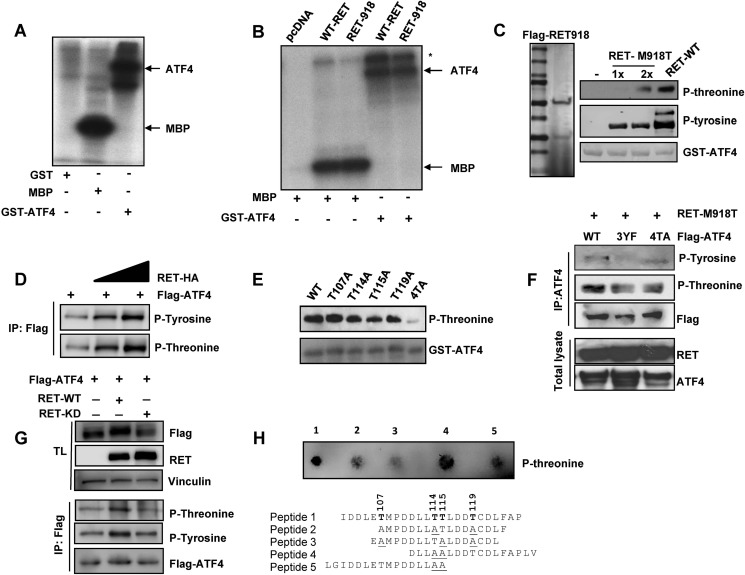
RET Exhibits a Novel Dual Kinase Activity and Phosphorylates ATF4
RET interactions with ATF4 implies that ATF4 may serve as a kinase substrate. This assumption is further supported by the observation that RET kinase activity is required for inhibition of ATF4-dependent transcription. Therefore, we hypothesized that RET inhibits ATF4 activity through phosphorylation, which antagonizes its maximal transcriptional activity for target genes.
To demonstrate that RET phosphorylates ATF4, we carried out three independent approaches. First, we showed that immunopurified mutant RET (C634W) from TT cells phosphorylated recombinant GST-ATF4 and a known RET substrate, myelin basic protein, in the presence of [32P]ATP (Fig. 7A). Second, immunopurified RET from transfected HEK293T cells was able to phosphorylate GST-ATF4 and myelin basic protein (Fig. 7B). Third, we expressed the kinase domain of RET-M918T as FLAG-RET(657–1114)-His-tagged protein in HEK293T cells, purified by a two-tandem immunoaffinity approach using first Ni2+ beads and then FLAG antibody-conjugated beads (Fig. 7C, left). Recombinant GST-ATF4 was phosphorylated by the purified RET(657–1114) at both threonine and tyrosine residues, as indicated by Western blot with phosphothreonine and phosphotyrosine antibodies (Fig. 7C, right). A commercially available active RET enzyme served as a control for RET activity. To determine ATF4 phosphorylation in vivo, FLAG-ATF4 and full-length RET-HA were expressed in HEK293T cells, followed by immunoprecipitation of ATF4 and then Western blot with phospho-specific antibodies. Fig. 7D shows that ectopically expressed RET in HEK293T cells increased ATF4 phosphorylation at both threonine and tyrosine.
FIGURE 7.
RET phosphorylates ATF4. A, kinase assay was performed using immunopurified RET from TT cell lysates with GST-ATF4 and myelin basic protein (MBP) as substrates. [32P]ATP served as a phosphate donor in the kinase assay. SDS-PAGE followed by autoradiography demonstrates specific phosphorylation of both proteins by RET. B, kinase assays were performed as in A with immunopurified full-length WT-RET and M918T mutant RET derived from transfected HEK293T cells. *, RET autophosphorylation. C, a tandem affinity-purified RET phosphorylates both tyrosine and threonine residues. Purification of RET-M918T enzyme (residues 657–1114) from transfected HEK293T cells was carried out with Ni2+ beads followed by a second affinity selection with FLAG antibody-agarose beads to yield a highly purified RET polypeptide on SDS-PAGE (left). A kinase assay was performed with purified RET-M918T and commercial wild type RET (Millipore) along with GST-ATF4 as a substrate and cold ATP, followed by Western blot analysis with phosphothreonine and phosphotyrosine antibodies (right panels). A gel stained with Ponceau indicates the levels of GST-ATF4. D, RET phosphorylates ATF4 in vivo. FLAG-ATF4 was expressed in HEK293T cells along with increasing amounts of full-length HA-RET. ATF4 phosphorylation was analyzed after immunoprecipitation (IP) and then Western blot with phospho-specific antibodies as indicated. E, mutation of targeted threonine residues reduces ATF4 phosphorylation. Shown is an in vitro kinase assay with GST-ATF4 and mutant GST-ATF4 in the presence of commercially obtained RET active enzyme. GST antibody was used as a control. F, in vivo phosphorylation of mutant ATF4. HEK293T cells were transfected with ATF4–4TA and ATF4–3YF along with RET-M918T (residues 657–1114) followed by immunoprecipitation with ATF4 antibody and Western blot with the indicated antibodies. G, decreased ATF4 phosphorylation in RET-KD-expressing cells. HEK293T cells were transfected with FLAG-ATF4 along with full-length RET-WT and RET-KD followed by immunoprecipitation with FLAG antibody and Western blot with the indicated antibodies. H, RET phosphorylates threonine residue. Peptides after incubation with purified RET wild type with the kinase domain (residues 657–1114) were spotted on nitrocellulose and then incubated with phosphothreonine antibody.
Identification of Threonine Phosphorylation in ATF4 by Mass Spectrometry
Site-specific phosphorylation of GST-ATF4 by commercial RET enzyme in vitro assays was identified by high sensitivity LC-MS/MS in an Orbitrap Elite high resolution mass spectrometer that revealed threonine phosphorylation at Thr-107, -114, -115, and -119 (supplemental Figs. 1 and 2). To confirm the specificity of RET-mediated ATF4 phosphorylation, we generated ATF4 mutants in which threonine was substituted with alanine and tyrosine with phenylalanine. We observed a moderate decrease in GST-ATF4 phosphorylation with targeted substitution of T114A, T115A, and T119A by RET kinase in vitro, but simultaneous mutation of all four threonines (T107A, T114A, T115A, and T119A; denoted 4TA) abrogated phosphorylation (Fig. 7E). For in vivo phosphorylation, wild type ATF4, ATF4–4TA, and ATF4–3YF (Y197F, Y228F, and Y261F) were expressed together with the RET (M918T) kinase domain in HEK293T cells. Immunoprecipitation of ATF4 followed by Western blot indicated that although tyrosine phosphorylation was completely abolished in ATF4–3YF, threonine phosphorylation was moderately decreased in ATF4–4TA, possibly due to the presence of other phosphorylatable threonines by other kinases present in HEK293T cells (Fig. 7F). Moreover, we further showed that ATF4 phosphorylation levels at both tyrosine and threonine residues were decreased in RET kinase-dead-expressing cells compared with wild type RET kinase-expressing cells (Fig. 7G). These data strongly indicate that RET phosphorylates ATF4 in cells.
To further confirm the RET as dual specificity tyrosine/threonine kinase, we used ATF4 peptides as substrate for an RET kinase activity in vitro assay. These peptides were identified by a mass spectrometry approach to contain phosphorylated threonine (Thr-107, -114, -115, and -119) by RET enzyme. Threonines were substituted with alanine to identify preferential substrate for RET. Dot blot analysis followed by phosphothreonine antibody clearly indicates that there were reduced levels of phosphorylation in peptide 2 (with one threonine at 115), peptide 3 (at Thr-114), and peptide 5 (at Thr-107) compared with wild type peptide 1, which had all four threonines. Indeed, RET was able to phosphorylate peptide 4 (at Thr-119) to the same extent as it did peptide 1 (Fig. 7H), indicating that RET utilized these sites for phosphorylation with relative specificity. Together, these data suggest that RET exhibits threonine kinase activity and phosphorylates ATF4 at threonine and tyrosines.
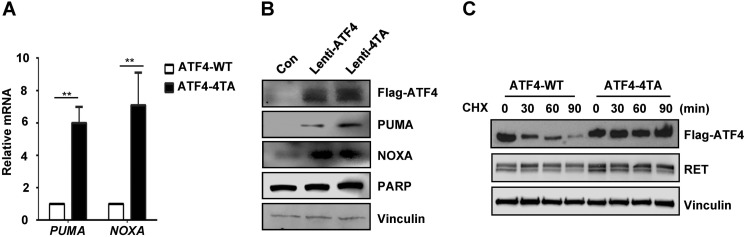
Threonine Phosphorylation Regulates Stability and Activity of ATF4
To examine the effect of ATF4 phosphorylation on activation of its target genes, wild type ATF4 and ATF4–4TA were expressed in TT cells using lentiviral constructs. Quantitative RT-PCR measurement indicates that levels of NOXA and PUMA mRNAs were stimulated to a level 6–9 times higher in TT cells with expression of mutant ATF4–4TA than in those with wild type ATF4 (Fig. 8A). Levels of the PUMA protein were also higher in TT cells with ATF4–4TA expression than in those with wild type ATF4 (Fig. 8B). Although NOXA mRNA levels were increased in ATF4–4TA expressing cells, NOXA protein levels remained unchanged, suggesting a possible post-translational control in regulation of NOXA protein. Furthermore, cycloheximide treatment of these cells overexpressing mutant ATF4–4TA or wild type ATF4 indicated that the phosphodefective ATF4–4TA protein was rather much more stable than wild type ATF4 without any change in RET levels (Fig. 8C). Together, these studies strongly indicate that RET regulates ATF stability and its transcriptional activity through phosphorylation and ubiquitination.
FIGURE 8.
Threonine phosphorylation of ATF4 regulates its activity. A, quantitative RT-PCR shows mRNA levels of ATF4 targets NOXA and PUMA in TT cells expressing lentiviral ATF4-WT or ATF4–4TA. Error bars, S.D. values from two independent experiments. **, p < 0.01. B, Western blot analysis shows levels of indicated proteins in TT cells expressing lentiviral ATF4-WT or ATF4–4TA for 24 h. Vinculin served as a loading control. C, Western blot analysis indicates the levels of lentivirus-mediated expression of ATF4-WT and ATF4–4TA in TT cells after treatment with cycloheximide (CHX) (100 μg/ml) for the indicated periods using anti-FLAG, anti-RET, and anti-vinculin antibodies.
DISCUSSION
This study provides several lines of evidence for a novel nuclear role of RET tyrosine kinase receptor to attenuate apoptosis. RET functions to regulate both ATF4 stability and DNA binding, through phosphorylation-dependent mechanisms. Specific targeting of this pathway by stabilization of ATF4 in combination with inhibition of RET offers a possible new therapeutic approach because it renders tumor cells more susceptible to genotoxic-induced apoptosis.
During the past several decades, 58 receptor tyrosine kinases have been identified and classified into 20 different subfamilies (39). 18 of these 58 receptor tyrosine kinases have been detected in the nucleus and are directly involved in regulating gene expression that is related to cell proliferation, survival, and insensitivity to therapeutic agents (37, 39, 42). Although RET is widely reported as a survival factor (43) and cytoplasmic signaling molecule (44), our data reveal that intact activated RET tyrosine kinase receptor also exists in the nuclei of MTC cells, where its function is probably to control gene transcription. The interaction of RET with the chromatin of NOXA and most likely that of other proapoptotic genes explains the functional relevance of RET nuclear localization. We demonstrated that, as with other receptor tyrosine kinases, such as ErbB-2 (45), RET kinase activity is required for its nuclear function. Our finding that RET also is localized to the nuclei of non-small lung cancer cells suggests that an activated form of RET is expressed in lung cancer cells and might be an attractive and novel alternative therapeutic target in other RET-dependent cancers.
ATF4 is a universal stress-responsive gene and an important effector of the integrated stress response pathway (25). In the presence of stress stimuli, ATF4 levels increase to activate expression of its primary target CHOP, which mediates oxidative stress and cell death (25). ATF4 is known to activate several other proapoptotic genes, including NOXA, PUMA, and TRB3, during cell death (16, 46). Small molecules that elevate ATF4 activity inhibit autochthonous murine neuroblastoma tumor growth in xenograft models (17). It has been previously shown that phosphorylation of ATF4 occurs at several sites (Thr-212, Ser-215, Ser-218, Ser-223, Ser-230, Ser-234, and Ser-247) and regulates both stability and transcriptional activity. For example, serine 215 phosphorylation by casein kinase 2 increases ATF4 transcriptional activity (47, 48).
In our study, we provide compelling evidence using mass spectrometry and other biochemical studies that RET phosphorylates ATF4 at threonines, revealing RET to be a dual specificity kinase. We found that ATF4 phosphorylation at threonines (Thr-107, -114, -115, and -119) destabilized ATF4 and decreased its ability to activate NOXA and PUMA transcription. These findings strongly suggest that RET may regulate the expression of a wide range of ATF4 target genes. This novel phosphorylation function of RET could be used to identify other key targets whose functions are regulated by RET threonine kinase activity and to reveal other roles of aberrantly activated RET in tumor progression.
NOXA is one of nine major BH3-only proteins that are required for proteasome and endoplasmic reticulum stress-induced apoptosis (49, 50). We observed that RET negatively regulates expression of the proapoptotic genes NOXA and PUMA but not that of other members of the BCL2 family, such as BIM, BAX, and BCL-XL. The levels of the prosurvival protein MCL-1, but not its mRNA, were significantly decreased in RET-depleted cells, suggesting that RET regulates MCL-1 synthesis or turnover, acting as a positive regulator of this survival gene (51, 52). Because NOXA regulates the localization and stability of MCL-1 in small cell lung cancer cells (53), we speculate that RET regulates MCL-1 expression through repression of NOXA transcription.
In summary, our data provide evidence for a key role of RET in cell survival through negative regulation of proapoptotic gene transcription. These functions are propagated through regulation of ATF4 expression, a central mediator of stress-induced apoptosis. The ability of RET to destabilize ATF4 renders MTC cells resistant to chemotherapy and TKIs.
Supplementary Material
Acknowledgments
We thank Dr. Mien-Chie Hung (University of Texas M.D. Anderson Cancer Center) for insightful comments and suggestions. We thank Yihong Ye (National Institutes of Health) for the NOXA reporter construct and Tsowin Hai (Ohio State University) for the 4X-CRE construct.
This work was supported by an American Thyroid Association research grant (to R. B.Y.) and by a Kosberg Foundation grant (to R. F. G.). This work was also supported, in whole or in part, by National Institutes of Health Cancer Center Support Grant CA016672 (to the University of Texas M.D. Anderson Cancer Center).

This article contains supplemental Figs. 1 and 2.
- MTC
- medullary thyroid cancer
- FLIM
- fluorescence lifetime imaging microscopy
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PARP
- poly(ADP-ribose) polymerase
- CRE
- cAMP-response element
- TKI
- tyrosine kinase inhibitor.
REFERENCES
- 1. Wells S. A., Jr., Santoro M. (2009) Targeting the RET pathway in thyroid cancer. Clin. Cancer Res. 15, 7119–7123 [DOI] [PubMed] [Google Scholar]
- 2. Zeng Q., Cheng Y., Zhu Q., Yu Z., Wu X., Huang K., Zhou M., Han S., Zhang Q. (2008) The relationship between overexpression of glial cell-derived neurotrophic factor and its RET receptor with progression and prognosis of human pancreatic cancer. J. Int. Med. Res. 36, 656–664 [DOI] [PubMed] [Google Scholar]
- 3. Ohshima Y., Yajima I., Takeda K., Iida M., Kumasaka M., Matsumoto Y., Kato M. (2010) c-RET molecule in malignant melanoma from oncogenic RET-carrying transgenic mice and human cell lines. PLoS One 5, e10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Díaz-Beyá M., Navarro A., Ferrer G., Díaz T., Gel B., Camós M., Pratcorona M., Torrebadell M., Rozman M., Colomer D., Monzo M., Esteve J. (2013) Acute myeloid leukemia with translocation (8;16)(p11;p13) and MYST3-CREBBP rearrangement harbors a distinctive microRNA signature targeting RET proto-oncogene. Leukemia 27, 595–603 [DOI] [PubMed] [Google Scholar]
- 5. Nikolsky Y., Sviridov E., Yao J., Dosymbekov D., Ustyansky V., Kaznacheev V., Dezso Z., Mulvey L., Macconaill L. E., Winckler W., Serebryiskaya T., Nikolskaya T., Polyak K. (2008) Genome-wide functional synergy between amplified and mutated genes in human breast cancer. Cancer Res. 68, 9532–9540 [DOI] [PubMed] [Google Scholar]
- 6. Nikiforov Y. E., Rowland J. M., Bove K. E., Monforte-Munoz H., Fagin J. A. (1997) Distinct pattern of ret oncogene rearrangements in morphological variants of radiation-induced and sporadic thyroid papillary carcinomas in children. Cancer Res. 57, 1690–1694 [PubMed] [Google Scholar]
- 7. Ballerini P., Struski S., Cresson C., Prade N., Toujani S., Deswarte C., Dobbelstein S., Petit A., Lapillonne H., Gautier E. F., Demur C., Lippert E., Pages P., Mansat-De Mas V., Donadieu J., Huguet F., Dastugue N., Broccardo C., Perot C., Delabesse E. (2012) RET fusion genes are associated with chronic myelomonocytic leukemia and enhance monocytic differentiation. Leukemia 26, 2384–2389 [DOI] [PubMed] [Google Scholar]
- 8. Kohno T., Ichikawa H., Totoki Y., Yasuda K., Hiramoto M., Nammo T., Sakamoto H., Tsuta K., Furuta K., Shimada Y., Iwakawa R., Ogiwara H., Oike T., Enari M., Schetter A. J., Okayama H., Haugen A., Skaug V., Chiku S., Yamanaka I., Arai Y., Watanabe S., Sekine I., Ogawa S., Harris C. C., Tsuda H., Yoshida T., Yokota J., Shibata T. (2012) KIF5B-RET fusions in lung adenocarcinoma. Nat. Med. 18, 375–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takeuchi K., Soda M., Togashi Y., Suzuki R., Sakata S., Hatano S., Asaka R., Hamanaka W., Ninomiya H., Uehara H., Lim Choi Y., Satoh Y., Okumura S., Nakagawa K., Mano H., Ishikawa Y. (2012) RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 18, 378–381 [DOI] [PubMed] [Google Scholar]
- 10. Drosten M., Hilken G., Böckmann M., Rödicker F., Mise N., Cranston A. N., Dahmen U., Ponder B. A., Pützer B. M. (2004) Role of MEN2A-derived RET in maintenance and proliferation of medullary thyroid carcinoma. J. Natl. Cancer Inst. 96, 1231–1239 [DOI] [PubMed] [Google Scholar]
- 11. Gattelli A., Nalvarte I., Boulay A., Roloff T. C., Schreiber M., Carragher N., Macleod K. K., Schlederer M., Lienhard S., Kenner L., Torres-Arzayus M. I., Hynes N. E. (2013) Ret inhibition decreases growth and metastatic potential of estrogen receptor positive breast cancer cells. EMBO Mol. Med. 5, 1335–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Portella G., Salvatore D., Botti G., Cerrato A., Zhang L., Mineo A., Chiappetta G., Santelli G., Pozzi L., Vecchio G., Fusco A., Santoro M. (1996) Development of mammary and cutaneous gland tumors in transgenic mice carrying the RET/PTC1 oncogene. Oncogene 13, 2021–2026 [PubMed] [Google Scholar]
- 13. Kawai K., Iwashita T., Murakami H., Hiraiwa N., Yoshiki A., Kusakabe M., Ono K., Iida K., Nakayama A., Takahashi M. (2000) Tissue-specific carcinogenesis in transgenic mice expressing the RET proto-oncogene with a multiple endocrine neoplasia type 2A mutation. Cancer Res. 60, 5254–5260 [PubMed] [Google Scholar]
- 14. Chipuk J. E., Moldoveanu T., Llambi F., Parsons M. J., Green D. R. (2010) The BCL-2 family reunion. Mol. Cell 37, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oda E., Ohki R., Murasawa H., Nemoto J., Shibue T., Yamashita T., Tokino T., Taniguchi T., Tanaka N. (2000) Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288, 1053–1058 [DOI] [PubMed] [Google Scholar]
- 16. Galehdar Z., Swan P., Fuerth B., Callaghan S. M., Park D. S., Cregan S. P. (2010) Neuronal apoptosis induced by endoplasmic reticulum stress is regulated by ATF4-CHOP-mediated induction of the Bcl-2 homology 3-only member PUMA. J. Neurosci. 30, 16938–16948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qing G., Li B., Vu A., Skuli N., Walton Z. E., Liu X., Mayes P. A., Wise D. R., Thompson C. B., Maris J. M., Hogarty M. D., Simon M. C. (2012) ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell 22, 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nikiforov M. A., Riblett M., Tang W. H., Gratchouck V., Zhuang D., Fernandez Y., Verhaegen M., Varambally S., Chinnaiyan A. M., Jakubowiak A. J., Soengas M. S. (2007) Tumor cell-selective regulation of NOXA by c-MYC in response to proteasome inhibition. Proc. Natl. Acad. Sci. U.S.A. 104, 19488–19493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. You H., Pellegrini M., Tsuchihara K., Yamamoto K., Hacker G., Erlacher M., Villunger A., Mak T. W. (2006) FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J. Exp. Med. 203, 1657–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ming L., Sakaida T., Yue W., Jha A., Zhang L., Yu J. (2008) Sp1 and p73 activate PUMA following serum starvation. Carcinogenesis 29, 1878–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hershko T., Ginsberg D. (2004) Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J. Biol. Chem. 279, 8627–8634 [DOI] [PubMed] [Google Scholar]
- 22. Ron D., Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 23. Pike L. R., Singleton D. C., Buffa F., Abramczyk O., Phadwal K., Li J. L., Simon A. K., Murray J. T., Harris A. L. (2013) Transcriptional up-regulation of ULK1 by ATF4 contributes to cancer cell survival. Biochem. J. 449, 389–400 [DOI] [PubMed] [Google Scholar]
- 24. Lange P. S., Chavez J. C., Pinto J. T., Coppola G., Sun C. W., Townes T. M., Geschwind D. H., Ratan R. R. (2008) ATF4 is an oxidative stress-inducible, prodeath transcription factor in neurons in vitro and in vivo. J. Exp. Med. 205, 1227–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han J., Back S. H., Hur J., Lin Y. H., Gildersleeve R., Shan J., Yuan C. L., Krokowski D., Wang S., Hatzoglou M., Kilberg M. S., Sartor M. A., Kaufman R. J. (2013) ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Velasco J. A., Medina D. L., Romero J., Mato M. E., Santisteban P. (1997) Introduction of p53 induces cell-cycle arrest in p53-deficient human medullary-thyroid-carcinoma cells. Int. J. Cancer 73, 449–455 [DOI] [PubMed] [Google Scholar]
- 27. Zhu W., Hai T., Ye L., Cote G. J. (2010) Medullary thyroid carcinoma cell lines contain a self-renewing CD133+ population that is dependent on ret proto-oncogene activity. J Clin. Endocrinol. Metab. 95, 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vitagliano D., De Falco V., Tamburrino A., Coluzzi S., Troncone G., Chiappetta G., Ciardiello F., Tortora G., Fagin J. A., Ryan A. J., Carlomagno F., Santoro M. (2011) The tyrosine kinase inhibitor ZD6474 blocks proliferation of RET mutant medullary thyroid carcinoma cells. Endocr. Relat. Cancer 18, 1–11 [DOI] [PubMed] [Google Scholar]
- 29. Cooley L. D., Elder F. F., Knuth A., Gagel R. F. (1995) Cytogenetic characterization of three human and three rat medullary thyroid carcinoma cell lines. Cancer Genet. Cytogenet. 80, 138–149 [DOI] [PubMed] [Google Scholar]
- 30. Wang Q., Mora-Jensen H., Weniger M. A., Perez-Galan P., Wolford C., Hai T., Ron D., Chen W., Trenkle W., Wiestner A., Ye Y. (2009) ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 106, 2200–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parthasarathy R., Cote G. J., Gagel R. F. (1999) Hammerhead ribozyme-mediated inactivation of mutant RET in medullary thyroid carcinoma. Cancer Res. 59, 3911–3914 [PubMed] [Google Scholar]
- 32. Bagheri-Yarmand R., Mandal M., Taludker A. H., Wang R. A., Vadlamudi R. K., Kung H. J., Kumar R. (2001) Etk/Bmx tyrosine kinase activates Pak1 and regulates tumorigenicity of breast cancer cells. J. Biol. Chem. 276, 29403–29409 [DOI] [PubMed] [Google Scholar]
- 33. Lee M. J., Ye A. S., Gardino A. K., Heijink A. M., Sorger P. K., MacBeath G., Yaffe M. B. (2012) Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell 149, 780–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ahmed Z., George R., Lin C. C., Suen K. M., Levitt J. A., Suhling K., Ladbury J. E. (2010) Direct binding of Grb2 SH3 domain to FGFR2 regulates SHP2 function. Cell. Signal. 22, 23–33 [DOI] [PubMed] [Google Scholar]
- 35. Ahmed Z., Lin C. C., Suen K. M., Melo F. A., Levitt J. A., Suhling K., Ladbury J. E. (2013) Grb2 controls phosphorylation of FGFR2 by inhibiting receptor kinase and Shp2 phosphatase activity. J. Cell Biol. 200, 493–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gururaj A. E., Singh R. R., Rayala S. K., Holm C., den Hollander P., Zhang H., Balasenthil S., Talukder A. H., Landberg G., Kumar R. (2006) MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proc. Natl. Acad. Sci. U.S.A. 103, 6670–6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hsu S. C., Hung M. C. (2007) Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J. Biol. Chem. 282, 10432–10440 [DOI] [PubMed] [Google Scholar]
- 38. Sheridan C., Brumatti G., Elgendy M., Brunet M., Martin S. J. (2010) An ERK-dependent pathway to Noxa expression regulates apoptosis by platinum-based chemotherapeutic drugs. Oncogene 29, 6428–6441 [DOI] [PubMed] [Google Scholar]
- 39. Lemmon M. A., Schlessinger J. (2010) Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koyanagi S., Hamdan A. M., Horiguchi M., Kusunose N., Okamoto A., Matsunaga N., Ohdo S. (2011) cAMP-response element (CRE)-mediated transcription by activating transcription factor-4 (ATF4) is essential for circadian expression of the Period2 gene. J. Biol. Chem. 286, 32416–32423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Q., Shinkre B. A., Lee J. G., Weniger M. A., Liu Y., Chen W., Wiestner A., Trenkle W. C., Ye Y. (2010) The ERAD inhibitor Eeyarestatin I is a bifunctional compound with a membrane-binding domain and a p97/VCP inhibitory group. PLoS One 5, e15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gschwind A., Fischer O. M., Ullrich A. (2004) The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat. Rev. Cancer 4, 361–370 [DOI] [PubMed] [Google Scholar]
- 43. Fonseca-Pereira D., Arroz-Madeira S., Rodrigues-Campos M., Barbosa I. A., Domingues R. G., Bento T., Almeida A. R., Ribeiro H., Potocnik A. J., Enomoto H., Veiga-Fernandes H. (2014) The neurotrophic factor receptor RET drives haematopoietic stem cell survival and function. Nature 514, 98–101 [DOI] [PubMed] [Google Scholar]
- 44. Richardson D. S., Rodrigues D. M., Hyndman B. D., Crupi M. J., Nicolescu A. C., Mulligan L. M. (2012) Alternative splicing results in RET isoforms with distinct trafficking properties. Mol. Biol. Cell 23, 3838–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang S. C., Lien H. C., Xia W., Chen I. F., Lo H. W., Wang Z., Ali-Seyed M., Lee D. F., Bartholomeusz G., Ou-Yang F., Giri D. K., Hung M. C. (2004) Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2. Cancer Cell 6, 251–261 [DOI] [PubMed] [Google Scholar]
- 46. Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H. (2005) TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 24, 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ampofo E., Sokolowsky T., Götz C., Montenarh M. (2013) Functional interaction of protein kinase CK2 and activating transcription factor 4 (ATF4), a key player in the cellular stress response. Biochim. Biophys. Acta 1833, 439–451 [DOI] [PubMed] [Google Scholar]
- 48. Frank C. L., Ge X., Xie Z., Zhou Y., Tsai L. H. (2010) Control of activating transcription factor 4 (ATF4) persistence by multisite phosphorylation impacts cell cycle progression and neurogenesis. J. Biol. Chem. 285, 33324–33337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang L., Lopez H., George N. M., Liu X., Pang X., Luo X. (2011) Selective involvement of BH3-only proteins and differential targets of Noxa in diverse apoptotic pathways. Cell Death Differ. 18, 864–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Armstrong J. L., Flockhart R., Veal G. J., Lovat P. E., Redfern C. P. (2010) Regulation of endoplasmic reticulum stress-induced cell death by ATF4 in neuroectodermal tumor cells. J. Biol. Chem. 285, 6091–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ding Q., He X., Xia W., Hsu J. M., Chen C. T., Li L. Y., Lee D. F., Yang J. Y., Xie X., Liu J. C., Hung M. C. (2007) Myeloid cell leukemia-1 inversely correlates with glycogen synthase kinase-3β activity and associates with poor prognosis in human breast cancer. Cancer Res. 67, 4564–4571 [DOI] [PubMed] [Google Scholar]
- 52. Ding Q., He X., Hsu J. M., Xia W., Chen C. T., Li L. Y., Lee D. F., Liu J. C., Zhong Q., Wang X., Hung M. C. (2007) Degradation of Mcl-1 by β-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol. Cell. Biol. 27, 4006–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nakajima W., Hicks M. A., Tanaka N., Krystal G. W., Harada H. (2014) Noxa determines localization and stability of MCL-1 and consequently ABT-737 sensitivity in small cell lung cancer. Cell Death Dis. 5, e1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.