Background: Katanin p60 is a protein that actively severs microtubules.
Results: Mutations within the AAA+ pore of katanin p60 and in the C-terminal regions of tubulins perturb efficient microtubule severing.
Conclusion: Interactions between the conserved residues in the katanin p60 pore and the acidic tails of both tubulins may be important.
Significance: Both tubulin molecules are essential for microtubule severing by katanin.
Keywords: ATPases Associated with Diverse Cellular Activities (AAA), Microtubule, Sea Urchin, Tubulin, Yeast, Katanin
Abstract
The microtubule (MT) network is highly dynamic and undergoes dramatic reorganizations during the cell cycle. Dimers of α- and β-tubulins rapidly polymerize to and depolymerize from the end of MT fibrils in an intrinsic GTP-dependent manner. MT severing by ATP-driven enzymes such as katanin and spastin contributes significantly to microtubule dynamics, and it has been shown that katanin p60, a AAA+ family protein, has ATPase and MT-severing activities. The mechanism of MT severing by katanin p60 is poorly understood, and the residues in katanin p60 and tubulins important for severing activity were therefore explored in this study. MT-severing activity, but not ATPase activity, was inhibited by mutations of the conserved aromatic residue and the flanking basic residues in the pore region of the katanin p60 hexameric ring. When the acidic residue-rich C-terminal unstructured segment of either α- or β-tubulin was removed, polymerized MTs were resistant to katanin p60 treatment. Interactions between katanin p60 and the mutant MTs, on the other hand, were unaffected. Taken together, these findings led us to propose that the interactions between the positively charged residues of katanin p60 and the acidic tails of both tubulins are essential for efficient severing of MTs.
Introduction
Microtubules (MTs)4 represent an essential component of the intracellular architecture; they function to separate chromosomes during mitosis and as rails of intracellular transport, and act as the cytoskeleton of the cell. MTs typically consist of 13 protofilaments, each of which is a polymer of α-tubulin and β-tubulin dimers (1). MTs are not static structures because they dramatically change in length, especially during mitosis. Tubulin dimers attach to and are released from the ends of MT filaments in an intrinsic GTPase-dependent manner. Various MT-associated proteins, furthermore, significantly contribute to the reorganization of MT networks (2). Three AAA+ family proteins (ATPases associated with diverse cellular activities), katanin, spastin, and fidgetin, have been identified as MT-severing enzymes (3, 4). These ATP-dependent MT-severing enzymes are found in many eukaryotes such as Tetrahymena, nematodes, fruit flies, and humans, but are absent in yeasts. Mutations in the human spastin gene have also been identified as the most common causes of autosomal dominant hereditary spastic paraplegia (5). It has been shown that during mitosis spastin and fidgetin sever MTs and facilitate depolymerization of the tubulin dimers from the minus-end of MTs, although the Caenorhabditis elegans homolog of fidgetin may not have MT-severing activity (6–8). Katanin is also required for control of spindle length during mitosis and meiosis by severing MTs and facilitating depolymerization from the plus-end of MTs (8–11). In neurons, both katanin and spastin play important roles in reconstitution of axonal branches (12). Katanin has also been shown to be essential for cilia biogenesis in Tetrahymena (13) and for flagella biogenesis in Chlamydomonas (14, 15). Katanin also plays a role in the cell motility of Drosophila cells by regulating the MT networks (16). Although the role of MT-severing enzymes in cells has been investigated extensively, the molecular mechanisms of MT severing are poorly understood.
Katanin is composed of a 60-kDa enzymatic subunit (p60) and an 80-kDa regulatory subunit (p80). Katanin p60 is a AAA+ protein and exhibits MT-severing activity in an ATPase-dependent manner (17, 18), whereas katanin p80 regulates the MT-severing activity of katanin p60 and determines the localization of the protein complex (19). Katanin p60 contains an N-terminal domain followed by a AAA+ domain. A recent NMR analysis revealed that the solution structure of the N-terminal region of katanin p60 strikingly resembles the structures of MT interacting and trafficking domains (Fig. 1A) (20). Although it has been shown that the N-terminal domain alone is able to bind to MT, the whole katanin p60 protein more efficiently interacts with MTs, indicating that multiple regions in katanin p60 are responsible for the interaction with MTs (21). AAA+ domains typically oligomerize into hexameric rings with a central cavity termed a pore (22, 23). It has been suggested that katanin p60 can form a hexameric ring in the presence of ATP (18, 21), although the detailed structure of this ring has not been elucidated. In contrast, an x-ray crystal structure of the AAA+ domain of Drosophila spastin reveals that it possesses three loops exposed to the central pore (24). The most N-terminal loop, referred to as the pore loop, contains an aromatic residue that is highly conserved among AAA+ proteins. This conserved aromatic residue has been shown to be essential for the function of various AAA+ proteins (25–27). In the case of spastin, not only the conserved aromatic residue but also some basic residues in the pore loop play essential roles in the recognition and severing of MTs (28, 29). The corresponding aromatic residue is also conserved in katanin p60 but its significance has not yet been described. In this study, the functional interactions between katanin p60 and MTs were analyzed in detail, especially with regard to the residues in katanin p60 and tubulins thought to be important for MT severing.
FIGURE 1.
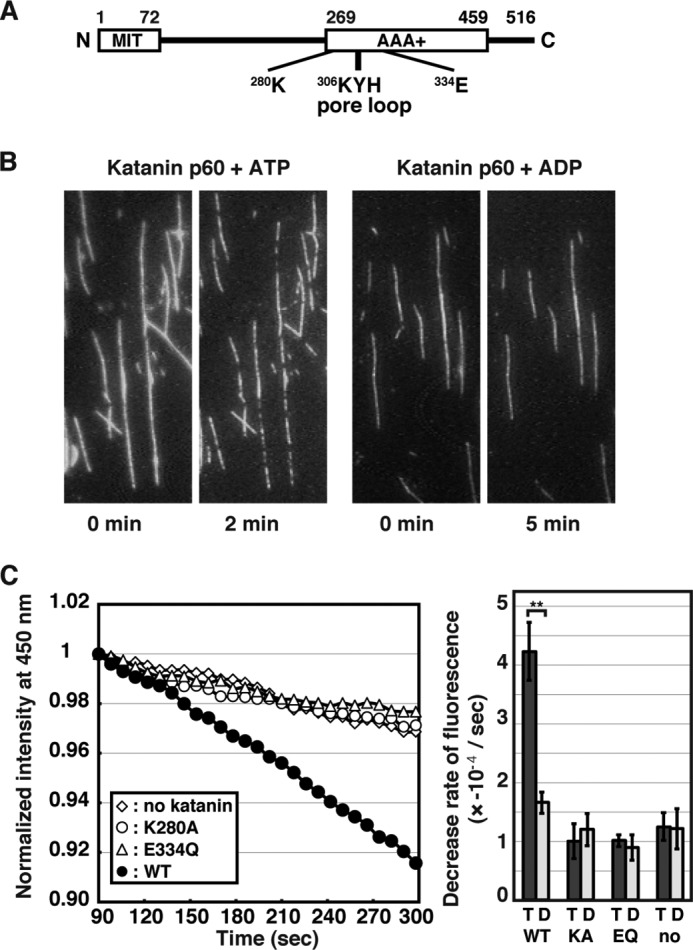
MT-severing activity of katanin p60. A, a schematic representation of the structure of sea urchin katanin p60. The N-terminal domain resembling the MT interacting and trafficking domain (MIT), the AAA+ domain, and residues focused on in this study are highlighted. B, taxol-stabilized rhodamine-labeled bovine MTs (1 μm) were applied onto a microscope slide coated with a mutant kinesin subunit. After washing off excess MTs, the attached MTs were incubated with 0.1 μm katanin p60 in the presence of 1 mm ATP (left panels) or 1 mm ADP (right panels) for the indicated times at room temperature. MTs were visualized by fluorescence microscopy. C, left panel: 1 μm porcine MTs were incubated without katanin p60 (diamonds) or together with 0.02 μm wild type katanin p60 (closed circles), K280A mutant (open circles), or E334Q mutant (triangles) in the presence of 1 mm ATP at 25 °C. The DAPI fluorescence at 450 nm (excitation at 370 nm) was monitored over time and normalized such that the fluorescence at 90 s after the start of incubation was set to 1. Representative results are shown. Right panel, statistics of the decrease in DAPI fluorescence. The rates of decrease in DAPI fluorescence from at least three independent measurements were analyzed and shown as mean ± S.E. T and D indicate measurements in the presence of ATP and ADP, respectively. **, p < 0.01.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant Katanin p60
Sea urchin katanin p60 cDNA (a gift from the McNally lab) was amplified by PCR and inserted into pAcHLT-B, generating a plasmid expressing an N-terminally His6-tagged katanin p60 via the baculovirus-insect protein expression system. Plasmids expressing mutant versions of katanin p60 were generated by oligonucleotide-directed mutagenesis. All constructs were verified by DNA sequencing.
Sf9 insect cells were transfected with the katanin p60-expressing plasmids and BD Baculogold DNA, and grown in Grace medium (45.7 g/liter of Grace medium; BD Biosciences) containing 0.35 g/liter of NaHCO3, 3.33 g/liter of BactoTC Lactalbumin Hydrolysate (BD Biosciences), 3.33 g/liter of BactoTC Yeastolate (BD Biosciences) at pH 6.1–6.2 (adjusted with NaOH) and supplemented with 10% fetal bovine serum (Sigma) and 1% antibiotics (Nacalai tesque) in 15-cm dishes. The cells were harvested at ∼72 h post-infection by low speed centrifugation. The purification procedure was based on a previously described procedure (18) with minor modifications. Cells were lysed in 50 mm Tris-HCl, pH 8.5, 500 mm NaCl, 2 mm MgCl2, 20 mm imidazole, 10 mm 2-mercaptoethanol, 0.02% Nonidet P-40, 0.1 mm ATP, and 20% glycerol. Proteins were bound to a HisTrap HP column (GE Healthcare) and eluted with 50 mm Tris-HCl, pH 8.5, 100 mm NaCl, 2 mm MgCl2, 200 mm imidazole, 10 mm 2-mercaptoethanol, 0.07% Nonidet P-40, 0.1 mm ATP, and 20% glycerol.
In Vitro Assays
ATPase activity assays and DAPI MT disassembly assays were performed according to previously published procedures (18), with modified ATPase assay (20 mm K-HEPES, pH 7.4, 2 mm MgCl2, 0.5 mm EGTA, 5% glycerol) and DAPI MT disassembly assay (20 mm K-HEPES, pH 7.4, 2 mm MgCl2, 2 mm 2-mercaptoethanol) buffers. MT-severing activity was assessed by fluorescence microscopy of rhodamine-labeled porcine MT (Cytoskeleton) performed as described previously (17, 18).
Purification of Recombinant Yeast Tubulins
The SUY57 yeast strain (30), a gift from Dr. Etsuko Muto, RIKEN Brain Science Institute, was used as a parental strain. Briefly, the SUY57 strain has chromosomal deletions of the TUB1, TUB2, and TUB3 genes but is rescued by a single copy plasmid harboring the TUB1 and TUB2 genes and the URA3 gene as a selection marker. In this study, single-copy plasmids expressing N-terminally His-tagged wild type Tub1p, His-tagged Tub1p lacking the five C-terminal amino acid residues, Tub2p, and Tub2p lacking the 24 C-terminal amino acid residues under the control of their own promoters were constructed. The genes for Tub2p have five additional mutations generating A19K, T23V, G26D, N227H, and Y270F, which equip Tub2p with the ability to interact with taxol (30, 31). These plasmids were used to transform the SUY57 yeast strain, and strains expressing modified tubulins were generated by plasmid shuffling. Yeast cells were cultured for 12–16 h in a rich glucose medium, and each tubulin dimer was purified using homemade TOG columns (32). The procedure generated approximately ∼0.7 mg competent tubulins from 9 liters of culture.
Wireless-electrodeless Quartz-crystal Microbalance (Wireless QCM)
A wireless QCM system, the details of which appear elsewhere (33), was used in this study. The sensor chips used were ∼30-μm thick AT-cut quartz plates with an area of 1.7 × 2.5 mm2. The fundamental resonance frequencies of the chips are ∼54 and 56 MHz. For the immobilization of organic substances, a 2-nm thin film of chromium and then an 18-nm thin film of gold were deposited on both chip surfaces. The quartz crystals were cleaned in a piranha solution (98% H2SO4, 33% H2O2; 4:1) for 30 min, rinsed several times with ultrapure water, treated with oxygen plasma for 30 min, and then immersed in a solution of 10 mm 10-carboxy-1-decanethiol/ethanol for 24 h at 4 °C. The sensor surfaces were then activated using a 100 mm 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride solution for 1 h at room temperature. The quartz crystals were immersed in the 1 μm MT-containing solution, and after rinsing with the buffer, they were immersed in 10 mg/ml of bovine serum albumin to block the remaining activated sites. Purified katanin p60 was then injected into the system at 1, 5, 10, 25, and 50 nm. The Kd (and Ka) values were calculated from the relationship between exponential coefficient α of the binding curve and katanin p60 concentration (34).
RESULTS
Purified Katanin p60 Severs MTs in Vitro in a AAA+ ATPase-dependent Manner
Because it has been suggested that purified recombinant katanin is susceptible to instability (11), we first verified that the preparation of recombinant katanin p60 used in this study exhibit the activities required for MT severing. N-terminally His-tagged sea urchin katanin p60 was purified using the baculovirus-insect expression system. Two katanin p60 mutants, K280A and E334Q, which each have a single replacement in the Walker A and B motifs, respectively, were also produced. Because the Walker A and B motifs in AAA+ domains are responsible for ATP binding and hydrolysis, respectively, these mutants were predicted to have no ATPase activity. The ATPase activities of the purified recombinant katanin p60s were measured using the malachite green assay and it was found that even the wild type katanin p60 showed low ATPase activity (Table 1), probably because of the sensitivity limit of the assay used. In contrast, the wild type katanin p60, but not the inactive mutants, showed significant ATPase activity in the presence of MTs (Table 1), as shown previously (17). Next, the MT-severing activity of the recombinant katanin p60 was assessed by two independent assays. As shown in Fig. 1B, wild type katanin p60 actively severed polymerized rhodamine-labeled bovine MTs in vitro in the presence of ATP, but not in the presence of ADP (Fig. 1B) (18). A quantitative DAPI MT disassembly assay was performed (18), because DAPI bound to polymerized MTs has been shown to emit higher fluorescence than free DAPI (35). When taxol-stabilized MTs were incubated alone, the fluorescence was slightly decreased over time due to spontaneous dissociation of tubulins as well as DAPI fluorescence decay (Fig. 1C). When the taxol-stabilized MTs were incubated with wild type katanin p60 in the presence of ATP, on the other hand, DAPI fluorescence intensity was significantly decreased over time (Fig. 1C), although the decrease rate was slightly slower than that observed previously (18). The linear decreases in DAPI fluorescence with time may indicate that the MT-severing activity of katanin p60 was saturated with MTs. The data were therefore simply fit to a linear model to compare the efficiencies of the reactions (Fig. 1C). DAPI fluorescence intensities from MTs incubated with the K280A and E334Q katanin p60 mutants were indistinguishable from that measured for MTs incubated in the absence of katanin p60 (Fig. 1C), clearly indicating that purified katanin p60 retains AAA+ ATPase-dependent MT-severing activity.
TABLE 1.
ATPase activity
ATPase activity of 0.1 μm katanin p60 with 1 mm ATP at 25 °C was measured in the absence or presence of 1 μm MTs by a malachite green assay (18). The mean ATPase activities ± S.E. were determined from at least three independent experiments. ATPase activities of ∼5 nmol/nmol/min were considered background levels (*) under the experimental conditions used since ATPase-negative mutants were also measured to have these levels of activity.
| Katanin p60 | MT | ATPase activity |
|---|---|---|
| nmol/nmol/min | ||
| Wild type | − | *4.36 ± 0.57 |
| Wild type | + | 39.7 ± 3.95 |
| K280A | + | *5.04 ± 0.67 |
| E334Q | + | *5.61 ± 3.87 |
| K306A | − | *5.65 ± 0.29 |
| K306A | + | 80.6 ± 3.63 |
| Y307A | − | *5.68 ± 0.63 |
| Y307A | + | 11.7 ± 3.42 |
| H308A | − | *5.46 ± 1.22 |
| H308A | + | 66.1 ± 14.36 |
Conserved Residues in the Pore Loop Region Are Required for MT Severing
An aromatic amino acid residue is highly conserved in the essential pore loop of most bacterial and eukaryotic AAA+ proteins (Fig. 2A) (36). Based on the sequence similarity among the homologs of katanin p60, basic amino acid residues flanking the conserved aromatic residue are also conserved (Fig. 2A). Because some basic amino acid residues in the pore loop are required for MT severing by spastin (28), the conserved basic amino acid residues of katanin p60 may also be essential for function. To gain insight into the functional importance of the conserved residues in the pore loop region of katanin p60, each residue was replaced with alanine and the resulting katanin p60 mutants (K306A, Y307A, and H308A) were purified. All three mutants were found to have significant ATPase activity in the presence of MTs, although the Y307A mutation was found to moderately lower the ATPase activity (Table 1). In contrast, in the DAPI MT disassembly assay with the pore loop mutants, the fluorescence decline over time in the presence of ATP was indistinguishable from that in the presence of ADP (Fig. 2, C–F), indicating that MT-severing activity is perturbed by the pore loop mutations.
FIGURE 2.
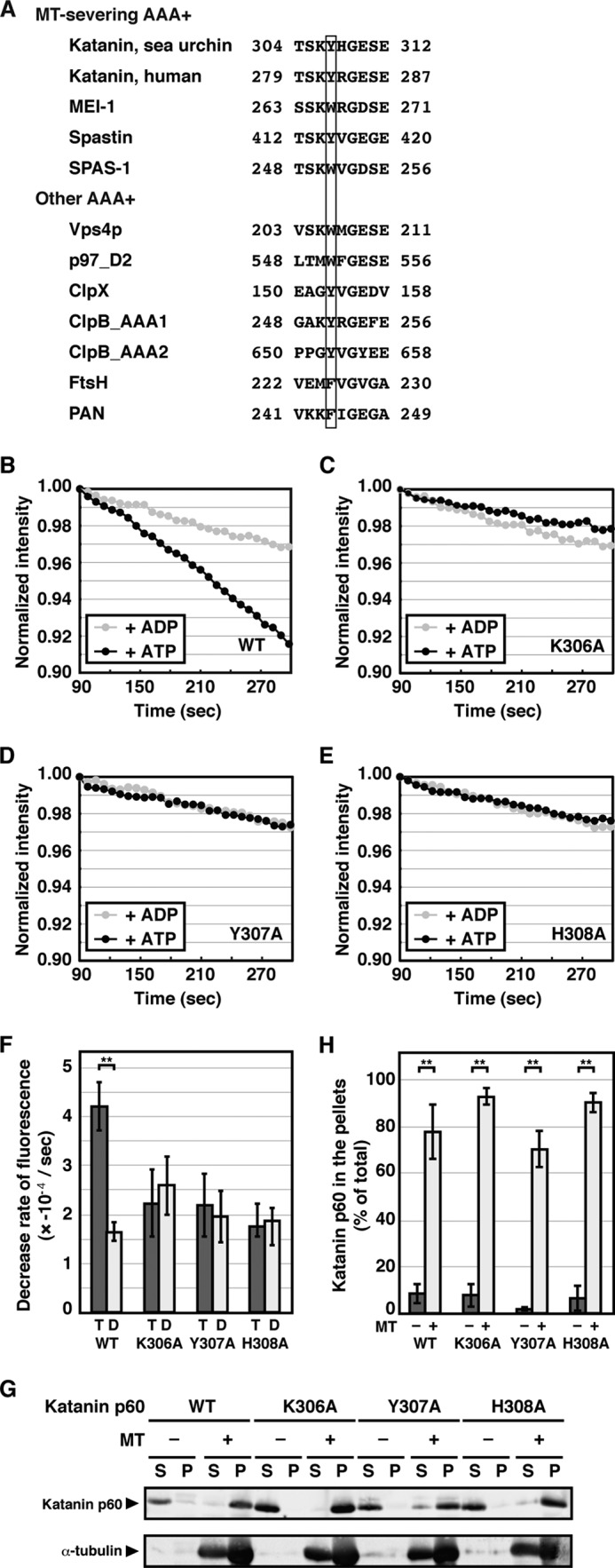
MT-severing activity of the katanin p60 pore loop mutants. A, sequence alignment of AAA+ proteins. The protein sequence in the pore loop region of sea urchin katanin was aligned with other AAA+ proteins including human katanin p60, katanin p60 homolog MEI-1 from C. elegans, human spastin, spastin homolog SPAS-1 from C. elegans, Vps4p from S. cerevisiae, the C-terminal AAA+ domain of human p97, ClpX from Escherichia coli, the first and second AAA+ domains of ClpB from E. coli, FtsH from E. coli, and PAN from Methanococcus jannaschii. The box indicates the conserved aromatic amino acid residues. B–E, MT-severing activity of the pore loop mutant. Taxol-stabilized porcine MTs (1 μm) were incubated with 0.02 μm wild type katanin p60 (B), K306A mutant (C), Y307A mutant (D), or H308A mutant (E) in the presence of 1 mm ATP (black circles) or 1 mm ADP (gray circles) at 25 °C. Disassembly of the MTs was monitored using DAPI as described in the legend to Fig. 1C. Representative results are shown. F, statistics of the decrease in DAPI fluorescence. The rates of decrease in DAPI fluorescence from four independent measurements were analyzed and shown as mean ± S.E. **, p < 0.01. G, interactions between MTs and katanin p60 pore loop mutants. Taxol-stabilized porcine MTs (1 μm) were incubated with 0.4 μm katanin p60 in ATPase assay buffer supplemented with 1 mm ATP at room temperature for 15 min. The MTs were precipitated by centrifugation at 100,000 × g for 10 min at 25 °C and the resulting supernatants and pellets were subjected to SDS-PAGE followed by immunoblotting using anti-His tag (upper panel) and anti-α-tubulin (lower panel) antibodies. H, quantification of katanin p60 bound to MTs. Band intensities of katanin p60 collected in the supernatants and pellets were quantified and mean ± S.E. of three independent experiments are shown.
The ATPase activities of the pore loop mutants as well as that of wild type katanin p60 were stimulated by the addition of MTs (Table 1), suggesting that these mutants have an ability to interact with MTs. To confirm these interactions, recombinant katanin p60 was incubated with MTs in the presence of ATP, after which MTs were precipitated by ultracentrifugation. All three pore loop mutants as well as the wild type katanin p60 co-precipitated with MTs, whereas they were collected in soluble fractions in the absence of MTs (Fig. 2G, 2H), indicating that the conserved pore loop mutants do in fact interact with MTs.
C-terminal Acidic Tails of Both Tubulins Are Required for Katanin p60 to Sever MTs, but Not for Binding to MTs
A previous study showed that the katanin p60/p80 heterodimer binds to subtilisin-treated MTs, but fails to sever these modified MTs (17). It has also been shown that katanin p60 interacts with wild type MTs more efficiently than subtilisin-treated MTs (37). Subtilisin is suggested to remove the α-tubulin and β-tubulin C-terminal regions, which are disordered clusters of acidic amino acid residues (38). Therefore, katanin p60 may recognize the acidic tail of either α-tubulin or β-tubulin or both. To determine which C-terminal tail is necessary for MT severing by katanin p60, we tested in this study whether katanin p60 severs mutant MTs lacking each acidic tail of tubulin. Recombinant tubulins have not been successfully prepared using bacterial expression systems mostly due to the fact that folding of tubulins requires a complex chaperone system (39). Tubulins have therefore been purified only from eukaryotic cells; however, because vertebrate MTs consist of tubulin isoforms encoded by multiple genes, preparation of MTs composed of tubulins lacking the C-terminal tails requires substantial efforts to mutate all of the tubulin genes. In contrast, the budding yeast Saccharomyces cerevisiae has only two α-tubulin genes (TUB1 and TUB3) and a single β-tubulin gene (TUB2), of which the TUB3 gene is dispensable for yeast cell growth. Overall, there is a high degree of sequence conservation between vertebrate and yeast tubulins, except in the disordered acidic tails (Fig. 3), and the budding yeast is therefore a suitable organism for the production of engineered tubulins (30). In this study, yeast strains with disrupted TUB1, TUB2, and TUB3 genes were used and engineered to express Tub1p and Tub2p from single copy plasmids. Because Tub2p lacks the key amino acid residues for interaction with taxol, which is often used to stabilize vertebrate MTs, five mutations were introduced into Tub2p (see “Experimental Procedures”). The taxol-interacting Tub2p will henceforth be referred to as “wild type” to focus on the acidic tails. The engineered strains have a combination of wild type or C-terminally deleted mutants of Tub1p and Tub2p: the five and 24 C-terminal amino acid residues in Tub1p and Tub2p, respectively, were deleted because these regions may be disordered regions based on sequence similarity data (Fig. 3). Purified yeast tubulins were shown to form filaments upon incubation with GTP (data not shown).
FIGURE 3.
Sequence alignments of tubulins in yeast and human. The protein sequences of α-tubulins (Tub1p from S. cerevisiae and tubulin α1b from Homo sapiens; A) and β-tubulins (Tub2p from S. cerevisiae and tubulin β chain isoform b from H. sapiens); B, were aligned using the ClustalW algorithm (49). Bars indicate the region missing in a crystal structure of tubulin dimers (50) and closed triangles mark the sites at which the C-terminal tails were removed. Open triangles indicate the residues mutated to convert the yeast β-tubulin to a taxol-interacting form.
Because budding yeast lacks any MT-severing AAA+ proteins including katanin and spastin, we investigated whether sea urchin katanin p60 recognizes yeast MTs in vitro. The ATPase activity of katanin p60 in the presence of MTs reconstituted from yeast wild type tubulins was 24.7 ± 4.37 nmol/nmol/min, indicating that yeast MTs effectively stimulate the ATPase activity of katanin p60, as shown for porcine MTs (Table 1). In the DAPI MT disassembly assay using polymerized yeast wild type MTs, fluorescence was significantly decreased over time in the presence of katanin p60 and ATP, but not in the presence of ADP (Fig. 4, A and E). The efficiency of severing yeast MTs by katanin p60 was slightly less efficient than that of severing porcine MTs (Figs. 2F and 4E) although it was not to a statistically significant degree (p = 0.087). These findings indicate that katanin p60 effectively recognizes and severs yeast MTs. In contrast, when MTs were prepared from tubulins lacking either one C-terminal tail or both, the DAPI fluorescence in the presence of katanin p60 and ATP was indistinguishable from that measured for MTs incubated with ADP (Fig. 4, B–E).
FIGURE 4.
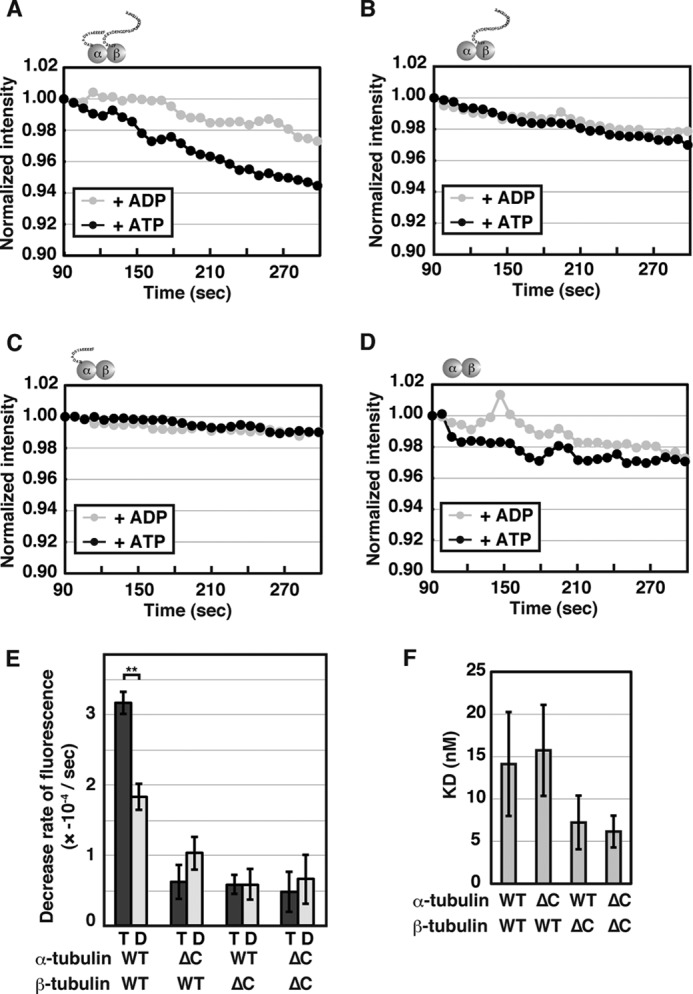
Severing of recombinant yeast MTs by katanin p60. A–D, taxol-stabilized yeast MTs (1 μm) composed of wild type tubulins (A), C-terminally deleted α-tubulin and wild type β-tubulin (B), wild type α-tubulin and C-terminally deleted β-tubulin (C), or C-terminally deleted α- and β-tubulins (D) were incubated with 0.02 μm katanin p60 in the presence of 1 mm ATP or ADP at 25 °C. DAPI fluorescence was monitored as described in Fig. 1C. E, statistics of the decrease in DAPI fluorescence. The rates of decrease in DAPI fluorescence from at least three independent measurements were analyzed and shown as mean ± S.E. T and D indicate the results in the presence of ATP and ADP, respectively. **, p < 0.01. F, dissociation constants of the interactions between katanin p60 and yeast MTs. Interactions of katanin p60 and yeast MTs in the presence of 1 mm ATPγS were measured by the wireless QCM system at room temperature and mean ± S.E. from three or four independent measurements are shown.
To examine whether katanin p60 recognizes MTs lacking C-terminal tails of each tubulin, a quantitative analysis of the potential interactions was performed using the wireless QCM system (33). A major advantage of this method is that quantitative interactions of large structures such as MTs, intracellular organelles, and even cells can be measured, unlike with other conventional methods such as surface plasmon resonance, with which such interactions would be difficult to assess. Katanin p60 was shown to interact with porcine and yeast wild type MTs with dissociation constants of 49 ± 17 and 14 ± 6 nm, respectively (p = 0.10). Relatively large standard errors were obtained for this data, and can be mostly attributed to variable and less controlled attachment of MTs on the sensor surface. The interactions between katanin p60 and the MTs composed of tubulins with deletions of the C-terminal tails were similar to the interaction between katanin p60 and wild type MTs (p = 0.33–0.86; Fig. 4F). These findings indicate that the C-terminal acidic tails of both α- and β-tubulins are essential for severing of MTs by katanin p60, but not for efficient interaction of katanin p60 with MTs.
DISCUSSION
AAA+ proteins typically form hexameric ring structures, the formation of which is required for effective ATP hydrolysis, because ATP binds to the interface of neighboring protomers and residues of the two protomers are required for hydrolysis of a single ATP molecule (40). Electron microscopy has revealed that katanin p60 also forms a ring structure (18) and it has been suggested that MTs facilitate oligomer formation of katanin p60, thereby stimulating the ATPase activity of katanin p60 (21). On the other hand, it has also been suggested that the ATPase activity of katanin p60 may be stimulated by conformational changes upon binding to MTs rather than by oligomer formation (41). In either case, the conserved pore residues of katanin p60 have less impact on the stimulation of the ATPase activity by MTs (Fig. 2). The Y307A mutant showed lower ATPase activity compared with the other katanin p60 proteins and such a reduction in ATPase activity by mutation of the conserved pore residue was previously also observed in other AAA+ proteins such as FtsH and ClpX (25, 42).
AAA+ proteins typically use energy generated from ATP hydrolysis to induce conformational changes of substrate proteins, thereby leading to unfolding, disassembly, and disaggregation of substrates. It has been shown that the conserved aromatic residue in the pore loop of the AAA+ protein ClpB directly interacts with a substrate protein (26). Nucleotide-dependent conformational changes of the pore loop of ClpX may pull substrates into the narrow pore, facilitating unfolding of the substrates (43). Here, we show that not only the conserved aromatic residue but also the flanking basic residues in katanin p60 are required for MT severing. In addition to this, we found that the C-terminal acidic tails of both tubulins are essential for MT severing. The AAA+ domain of spastin has been suggested to interact with the C-terminal tails of tubulins, and this interaction was shown to be perturbed by a mutation in the pore loop of spastin (24, 29). Taking into account the analogy with these highly homologous proteins, it is conceivable that the conserved pore residues of katanin p60 interacts with the C-terminal tails of both tubulins through electrostatic interactions as well as by a means conserved among the AAA+ proteins. Deletion of these residues did not affect the overall interactions between katanin p60 and MTs, suggesting that other regions, such as the core bodies of tubulins and the N-terminal domain of katanin p60, participate as major interaction sites.
Are the mechanisms of MT severing by two AAA+ proteins, katanin p60 and spastin, the same? Oligomer formations of katanin p60 and spastin are facilitated by MTs (21, 29). Aromatic and basic residues in the pore loop of both proteins are essential for the MT-severing activity (Fig. 2) (29, 44). We found that the C-terminal tails of both tubulins are required for MT severing by katanin p60 (Fig. 4). Both tails are recognized by spastin (29), although it has not been clear whether either or both tubulin tails are required for severing by spastin. Both AAA+ proteins possess MT-interacting regions in addition to the AAA+ domains (20, 29). Thus, interactions of katanin p60 and spastin with MTs may occur in a similar manner. However, it has been shown that Tau, a MT-associating protein, protects MTs against severing by katanin p60 more efficiently than that by spastin in neurons (45). It was also shown that Tau could not interact with subtilisin-treated MTs (46), indicating that Tau binds to either C-terminal end of tubulins. A peptide composed of the tail of β-tubulin, but not that of α-tubulin, perturb efficient severing of MTs by spastin (24). Taking these results together, we propose that katanin requires C-terminal tails of both tubulins for MT severing, whereas spastin may require that of either tubulin.
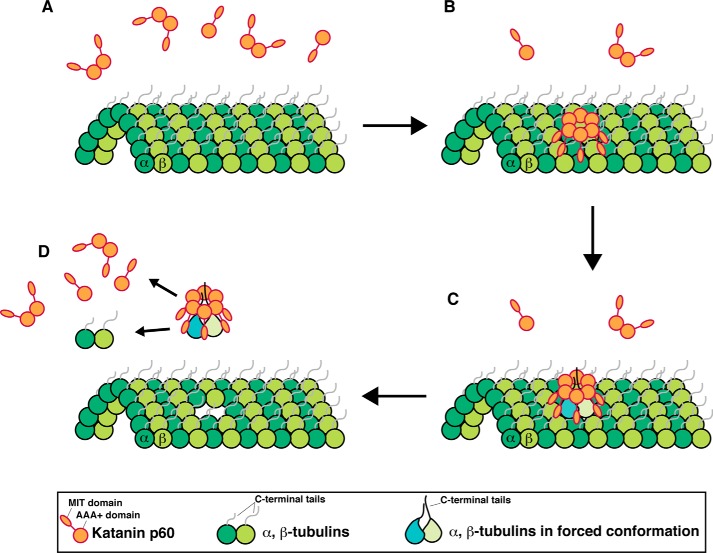
We propose possible mechanisms for the requirement of the C-terminal tails of both tubulins for MT severing by katanin p60 (Fig. 5). Katanin p60 tends to oligomerize, possibly hexamerize, on MTs (Fig. 5, A and B). The pore loop of katanin p60 may coincidentally capture C-terminal segments of both α- and β-tubulins (Fig. 5B). The simultaneous pulling of both tails may generate mechanical tension in the tubulin dimer, thereby leading to destabilization of tubulin-tubulin contacts in the lattice (Fig. 5C). The resulting defective interaction may promote the release of the tubulin dimer from the MT (Fig. 5D). In this case, it would not be necessary for the tubulins to traverse the katanin p60 pore entirely, because subtle conformational changes may be enough for the release. This proposed mechanism is compatible with previous observations (17), in which released tubulins are competent to repolymerize, and may also be supported by the observation that katanin preferentially acts on defective sites that are generated by changing protofilament numbers or tubulin loss from the filaments within MTs in vitro (47). Alternatively, a C-terminal tubulin tail may be required in a different step from the one requiring the tail of another tubulin. For example, it would be possible that katanin p60 pulls a tubulin tail, whereas another tubulin tail is required to position the katanin hexamer for effective function. In this case, a tubulin monomer may be released in each reaction. Future studies investigating how many tubulin molecules are released by katanin p60 will distinguish the two possible mechanisms. High-speed atomic force microcopy (48) is a cutting edge tool that can be used to visualize even single tubulin molecules in MTs5 and thus may reveal the mechanisms of MT-severing by AAA+ proteins. Furthermore, because lengths and net charges differ significantly between α- and β-tubulin and among species, it would be interesting to investigate what features of the C-terminal tails of tubulins are required for MT severing by AAA+ ATPases. The engineered MTs used in this study will be a powerful tool for analyzing MT-associated biological processes.
FIGURE 5.
A proposed mechanism for MT severing by katanin p60. Katanin p60 seems to oligomerize on MTs A and B, simultaneous pulling of both C-terminal tails of α- and β-tubulins may generate mechanical tensions in the tubulin dimer (C), leading to destabilization of tubulin-tubulin contacts. The resulting defective interaction may promote the release the tubulin dimer (D). The released tubulins are competent to repolymerize.
Acknowledgments
We thank Dr. Francis J. McNally for the katanin p60-expressing plasmid and Dr. Etsuko Muto for the yeast SUY57 strain. We also thank members of the Ogura Lab for helpful discussion and staffs of LILA at Kumamoto University for technical assistances.
This work was supported in part by grants-in-aid from Japan Society for the Promotion of Science, from MEXT of Japan, and from the CREST program of JST.
Dr. T. Uchihashi, personal communication.
- MT
- microtubule
- AAA+
- ATPases associated with diverse cellular activities
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1. Downing K. H., Nogales E. (1998) Tubulin and microtubule structure. Curr. Opin. Cell Biol. 10, 16–22 [DOI] [PubMed] [Google Scholar]
- 2. Walczak C. E. (2000) Microtubule dynamics and tubulin interacting proteins. Curr. Opin. Cell Biol. 12, 52–56 [DOI] [PubMed] [Google Scholar]
- 3. Roll-Mecak A., McNally F. J. (2010) Microtubule-severing enzymes. Curr. Opin. Cell Biol. 22, 96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharp D. J., Ross J. L. (2012) Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 125, 2561–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hazan J., Fonknechten N., Mavel D., Paternotte C., Samson D., Artiguenave F., Davoine C.-S., Cruaud C., Dürr A., Wincker P., Brottier P., Cattolico L., Barbe V., Burgunder J.-M., Prud'homme J.-F., Brice A., Fontaine B., Heilig B., Weissenbach J. (1999) Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat. Genet. 23, 296–303 [DOI] [PubMed] [Google Scholar]
- 6. Mukherjee S., Diaz Valencia J. D., Stewman S., Metz J., Monnier S., Rath U., Asenjo A. B., Charafeddine R. A., Sosa H. J., Ross J. L., Ma A., Sharp D. J. (2012) Human fidgetin is a microtubule severing enzyme and minus-end depolymerase that regulates mitosis. Cell Cycle 11, 2359–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Onitake A., Yamanaka K., Esaki M., Ogura T. (2012) Caenorhabditis elegans fidgetin homolog FIGL-1, a nuclear-localized AAA ATPase, binds to SUMO. J. Struct. Biol. 179, 143–151 [DOI] [PubMed] [Google Scholar]
- 8. Zhang D., Rogers G. C., Buster D. W., Sharp D. J. (2007) Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J. Cell Biol. 177, 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Srayko M., Buster D. W., Bazirgan O. A., McNally F. J., Mains P. E. (2000) MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14, 1072–1084 [PMC free article] [PubMed] [Google Scholar]
- 10. McNally K., Audhya A., Oegema K., McNally F. J. (2006) Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175, 881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Díaz-Valencia J. D., Morelli M. M., Bailey M., Zhang D., Sharp D. J., Ross J. L. (2011) Drosophila katanin-60 depolymerizes and severs at microtubule defects. Biophys. J. 100, 2440–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu W., Solowska J. M., Qiang L., Karabay A., Baird D., Baas P. W. (2005) Regulation of microtubule severing by katanin subunits during neuronal development. J. Neurosci. 25, 5573–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharma N., Bryant J., Wloga D., Donaldson R., Davis R. C., Jerka-Dziadosz M., Gaertig J. (2007) Katanin regulates dynamics of microtubules and biogenesis of motile cilia. J. Cell Biol. 178, 1065–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dymek E. E., Smith E. F. (2012) PF19 encodes the p60 catalytic subunit of katanin and is required for assembly of the flagellar central apparatus in Chlamydomonas. J. Cell Sci. 125, 3357–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kannegaard E., Rego E. H., Schuck S., Feldman J. L., Marshall W. F. (2014) Quantitative analysis and modeling of katanin function in flagellar length control. Mol. Biol. Cell. 25, 3686–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang D., Grode K. D., Stewman S. F., Diaz-Valencia J. D., Liebling E., Rath U., Riera T., Currie J. D., Buster D. W., Asenjo A. B., Sosa H. J., Ross J. L., Ma A., Rogers S. L., Sharp D. J. (2011) Drosophila katanin is a microtubule depolymerase that regulates cortical-microtubule plus-end interactions and cell migration. Nat. Cell Biol. 13, 361–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McNally F. J., Vale R. D. (1993) Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 75, 419–429 [DOI] [PubMed] [Google Scholar]
- 18. Hartman J. J., Mahr J., McNally K., Okawa K., Iwamatsu A., Thomas S., Cheesman S., Heuser J., Vale R. D., McNally F. J. (1998) Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 93, 277–287 [DOI] [PubMed] [Google Scholar]
- 19. McNally K. P., Bazirgan O. A., McNally F. J. (2000) Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. J. Cell Sci. 113, 1623–1633 [DOI] [PubMed] [Google Scholar]
- 20. Iwaya N., Kuwahara Y., Fujiwara Y., Goda N., Tenno T., Akiyama K., Mase S., Tochio H., Ikegami T., Shirakawa M., Hiroaki H. (2010) A common substrate recognition mode conserved between katanin p60 and VPS4 governs microtubule severing and membrane skeleton reorganization. J. Biol. Chem. 285, 16822–16829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hartman J. J., Vale R. D. (1999) Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 286, 782–785 [DOI] [PubMed] [Google Scholar]
- 22. Ogura T., Wilkinson A. J. (2001) AAA+ superfamily ATPases: common structure-diverse function. Genes Cells. 6, 575–597 [DOI] [PubMed] [Google Scholar]
- 23. White S. R., Lauring B. (2007) AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic 8, 1657–1667 [DOI] [PubMed] [Google Scholar]
- 24. Roll-Mecak A., Vale R. D. (2008) Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature 451, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamada-Inagawa T., Okuno T., Karata K., Yamanaka K., Ogura T. (2003) Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J. Biol. Chem. 278, 50182–50187 [DOI] [PubMed] [Google Scholar]
- 26. Schlieker C., Weibezahn J., Patzelt H., Tessarz P., Strub C., Zeth K., Erbse A., Schneider-Mergener J., Chin J. W., Schultz P. G., Bukau B., Mogk A. (2004) Substrate recognition by the AAA+ chaperone ClpB. Nat. Struct. Mol. Biol. 11, 607–615 [DOI] [PubMed] [Google Scholar]
- 27. Scott A., Chung H.-Y., Gonciarz-Swiatek M., Hill G. C., Whitby F. G., Gaspar J., Holton J. M., Viswanathan R., Ghaffarian S., Hill C. P., Sundquist W. I. (2005) Structural and mechanistic studies of VPS4 proteins. EMBO J. 24, 3658–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matsushita-Ishiodori Y., Yamanaka K., Hashimoto H., Esaki M., Ogura T. (2009) Conserved aromatic and basic amino acid residues in the pore region of Caenorhabditis elegans spastin play critical roles in microtubule severing. Genes Cells. 14, 925–940 [DOI] [PubMed] [Google Scholar]
- 29. White S. R., Evans K. J., Lary J., Cole J. L., Lauring B. (2007) Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J. Cell Biol. 176, 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Uchimura S., Oguchi Y., Katsuki M., Usui T., Osada H., Nikawa J., Ishiwata S., Muto E. (2006) Identification of a strong binding site for kinesin on the microtubule using mutant analysis of tubulin. EMBO J. 25, 5932–5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gupta M. L., Jr., Bode C. J., Georg G. I., Himes R. H. (2003) Understanding tubulin-Taxol interactions: mutations that impart taxol binding to yeast tubulin. Proc. Natl. Acad. Sci. U.S.A. 100, 6394–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Widlund P. O., Podolski M., Reber S., Alper J., Storch M., Hyman A. A., Howard J., Drechsel D. N. (2012) One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Mol. Biol. Cell. 23, 4393–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ogi H., Fukunishi Y., Omori T., Hatanaka K., Hirao M., Nishiyama M. (2008) Effects of flow rate on sensitivity and affinity in flow injection biosensor systems studied by 55-MHz wireless quartz crystal microbalance. Anal. Chem. 80, 5494–5500 [DOI] [PubMed] [Google Scholar]
- 34. Ogi H., Okamoto K., Nagai H., Fukunishi Y., Hirao M. (2009) Replacement-free electrodeless quartz crystal microbalance biosensor using nonspecific-adsorption of streptavidin on quartz. Anal. Chem. 81, 4015–4020 [DOI] [PubMed] [Google Scholar]
- 35. Heusele C., Bonne D., Carlier M. F. (1987) Is microtubule assembly a biphasic process? a fluorimetric study using 4′,6-diamidino-2-phenylindole as a probe. Eur. J. Biochem. 165, 613–620 [DOI] [PubMed] [Google Scholar]
- 36. Confalonieri F., Duguet M. (1995) A 200-amino acid ATPase module in search of a basic function. BioEssays. 17, 639–850 [DOI] [PubMed] [Google Scholar]
- 37. Eckert T., Le D. T., Link S., Friedmann L., Woehlke G. (2012) Spastin's microtubule-binding properties and comparison to katanin. PLoS One 7, e50161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shiina N., Gotoh Y., Nishida E. (1995) Microtubule-severing activity in M phase. Trends Cell Biol. 5, 283–286 [DOI] [PubMed] [Google Scholar]
- 39. Llorca O., Martín-Benito J., Ritco-Vonsovici M., Grantham J., Hynes G. M., Willison K. R., Carrascosa J. L., Valpuesta J. M. (2000) Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J. 19, 5971–5979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Snider J., Houry W. A. (2008) AAA+ proteins: diversity in function, similarity in structure. Biochem. Soc. Trans. 36, 72–77 [DOI] [PubMed] [Google Scholar]
- 41. Iwaya N., Akiyama K., Goda N., Tenno T., Fujiwara Y., Hamada D., Ikura T., Shirakawa M., Hiroaki H. (2012) Effect of Ca2+ on the microtubule-severing enzyme p60-katanin: insight into the substrate-dependent activation mechanism. FEBS J. 279, 1339–1352 [DOI] [PubMed] [Google Scholar]
- 42. Siddiqui S. M., Sauer R. T., Baker T. A. (2004) Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 18, 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martin A., Baker T. A., Sauer R. T. (2008) Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat. Struct. Mol. Biol. 15, 1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matsushita-Ishiodori Y., Yamanaka K., Ogura T. (2007) The C. elegans homologue of the spastic paraplegia protein, spastin, disassembles microtubules. Biochem. Biophys. Res. Commun. 359, 157–162 [DOI] [PubMed] [Google Scholar]
- 45. Yu W., Qiang L., Solowska J. M., Karabay A., Korulu S., Baas P. W. (2008) The microtubule-severing proteins spastin and katanin participate differently in the formation of axonal branches. Mol. Biol. Cell. 19, 1485–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marya P. K., Syed Z., Fraylich P. E., Eagles P. A. (1994) Kinesin and Tau bind to distinct sites on microtubules. J. Cell Sci. 107, 339–344 [DOI] [PubMed] [Google Scholar]
- 47. Davis L. J., Odde D. J., Block S. M., Gross S. P. (2002) The importance of lattice defects in katanin-mediated microtubule severing in vitro. Biophys. J. 82, 2916–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ando T. (2014) High-speed AFM imaging. Curr. Opin. Struct. Biol. 28, 63–68 [DOI] [PubMed] [Google Scholar]
- 49. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Löwe J., Li H., Downing K. H., Nogales E. (2001) Refined structure of αβ-tubulin at 3.5-Å resolution. J. Mol. Biol. 313, 1045–1057 [DOI] [PubMed] [Google Scholar]