Background: In peripheral nerves, the initial immune response to injury influences regeneration.
Results: S100a8 and S100a9 are the top induced genes in nerves post-injury. S100A8/A9 activate the chemotactic genes and pathways in Schwann cells and stimulate myeloid cell infiltration into the nerve.
Conclusion: S100A8/A9 initiate immune cell transmigration into the nerve.
Significance: S100A8/A9 are novel modulators of peripheral nerve injury.
Keywords: Cellular Immune Response, Chemokine, Cytokine, Gene Expression, S100 Proteins, Schwann Cells, Nerve Regeneration, Peripheral Nerve
Abstract
To shed light on the early immune response processes in severed peripheral nerves, we performed genome-wide transcriptional profiling and bioinformatics analyses of the proximal (P, regenerating) and distal (D, degenerating) nerve stumps on day 1 in the sciatic nerve axotomy model in rats. Multiple cell death-related pathways were activated in the degenerating D stump, whereas activation of the cytoskeletal motility and gluconeogenesis/glycolysis pathways was most prominent in the P stump of the axotomized nerve. Our bioinformatics analyses also identified the specific immunomodulatory genes of the chemokine, IL, TNF, MHC, immunoglobulin-binding Fc receptor, calcium-binding S100, matrix metalloproteinase, tissue inhibitor of metalloproteinase, and ion channel families affected in both the P and D segments. S100a8 and S100a9 were the top up-regulated genes in both the P and D segments. Stimulation of cultured Schwann cells using the purified S100A8/A9 heterodimer recapitulated activation of the myeloid cell and phagocyte chemotactic genes and pathways, which we initially observed in injured nerves. S100A8/A9 heterodimer injection into the intact nerve stimulated macrophage infiltration. We conclude that, following peripheral nerve injury, an immediate acute immune response occurs both distal and proximal to the lesion site and that the rapid transcriptional activation of the S100a8 and S100a9 genes results in S100A8/A9 hetero- and homodimers, which stimulate the release of chemokines and cytokines by activated Schwann cells and generate the initial chemotactic gradient that guides the transmigration of hematogenous immune cells into the injured nerve.
Introduction
In general, peripheral nerves display a strong regenerative potential. Relative to other injury types, complete nerve transection (axotomy) severely damages the endoneurial tube and entails inefficient regenerative growth (1–4). Since the original findings of nerve fiber breakdown distal to transection injury (5), substantial knowledge has been accumulated using sciatic nerve injury models in rodents. This knowledge recognizes that the dramatic endoneurial remodeling in the proximal (P,3 regenerating) and distal (D, degenerating) nerve stumps during the first hours post-transection set the course for a long-term “staggered” axonal growth (up to 4 weeks in rats) and, consequently, incomplete functional (motor and sensory) recovery (3, 4, 6).
Wallerian degeneration is a well orchestrated process initiated by an instant influx of extracellular calcium that activates the calpain- and ubiquitin-proteasome-dependent disintegration of the axonal cytoskeleton in the D stump (7). Concomitantly, the P axons die back to the next node of Ranvier (4, 8) and form axonal end bulbs (9).Although a rigorous, actin-supported process, an end bulb transforms into a regenerating P axonal sprout and forms a growth cone (4, 10, 11). Schwann cells, the main cell population in the peripheral nerve, carefully guide the growing axons toward the end organ. However, disruption of endoneurial tubes caused by transection injury results in a disorganized extracellular matrix that obscures the Schwann cell alignment into a column of proliferating cells, the bands of Büngner, and their ability to deposit the substrata favorable to axonal growth (4, 6).
Denervated Schwann cells rapidly trans-differentiate into Büngner cells post-axotomy. This process involves silencing of the genes coding for myelin proteins and the induction of glial fibrillary acidic protein, p75NGF, neuregulin, and other genes that are required to support Schwann cell dedifferentiation, mitosis, and partnership with regenerating axons (7, 12–14). Schwann cells also play a key role in secreting inflammatory chemokines, cytokines, and matrix metalloproteinases (MMPs), which work in concert to stimulate the development of chemotactic gradients and the directed immune cell migration across the blood-nerve barrier and into the damaged nerve (12, 15–17). Various hematogenous immune cell types, including granulocytes (neutrophils and mast cells) and agranulocytes (monocytes/macrophages and lymphocytes), infiltrate the nerve in the course of Wallerian degeneration (12, 15–17).
Calprotectin (S100A8/A9) is a heterodimeric, non-covalent complex of acidic calcium-binding S100A8 and S100A9. S100A8/A9 induces chemotaxis (18, 24), cytoskeletal reorganization (19), calcium signaling (20), and cytokine expression (21) in immune cells, particularly phagocytes. In peripheral nerves, the phagocytic clearance of axonal and myelin debris is initiated by Schwann cells, which extrude myelin sheaths to create ovoid “digestion chambers” (22, 23). This clearance is completed by macrophages infiltrating the nerve on days 2–14 post-injury (16, 17, 22, 24). Because the S100a8 and S100a9 genes were highly up-regulated on day 1 post-injury in murine nerves (25), we hypothesized that the initial positive chemotactic gradient is a result of Schwann cell stimulation by S100A8/A9 and that this gradient arises shortly after peripheral nerve injury and then stimulates the infiltration of the immune cells toward the trauma site.
Here we first confirmed and expanded the transcriptional profiling studies in peripheral nerves (25–33). Using comparative genome-wide transcriptional profiling and subsequent bioinformatics (Ingenuity, NextBio) analyses in the P and D stumps on day 1 post-axotomy, we recorded early gene expression changes in the severed sciatic nerve in rats. Stimulation of cultured Schwann cells by the purified S100A8/A9 heterodimer recapitulated those transcriptional events, which supported chemotaxis of myeloid cells toward both the P and D segments post-injury in vivo. In addition, S100A8/A9 injection stimulated macrophage infiltration into the nerve in vivo. Our findings suggest that the rapid injury-induced up-regulation of S100A8/A9 in Schwann cells then stimulates the release of cytokines and chemokines, which, combined, provide the initial chemotactic gradient that attracts hematogenous immune cells to the injured nerve.
MATERIALS AND METHODS
Reagents and Antibodies
Routine reagents were purchased from Sigma unless indicated otherwise. The following antibodies were used in our experiments: rabbit polyclonal anti-S100A9 (Novus, catalog no. NB110-89726) and murine monoclonal anti-S100B (Sigma, catalog no. S2532) and anti-CD68 (Serotec, catalog no. MCA341R). We also used the following antibodies from Cell Signaling Technology: a rabbit polyclonal antibody to phospho (p)ERK (catalog no. 9101), to pPI3K (catalog no. 4228), to pJNK (catalog no. 4668), to ERK (catalog no. 9102), to PI3K (catalog no. 4292), and to JNK (catalog no. 9258). A murine monoclonal antibody to β-actin was from Sigma (catalog no. A53166).
S100A8/A9
Purification of endotoxin-free murine S100A8/A9 proteins was performed as described for human S100A8/A9 in Ref. 34. The levels of homodimers were below 10%, as determined by size exclusion chromatography and ELISAs employing all antibody combinations (A8/A8, A9/A9, A8/A9, and A9/A8).4 The levels of LPS were below 2 pg/μg of the complex, as determined by the limulous amoebocyte lysate test.
Animal Procedures
Adult female Sprague-Dawley rats weighing 200–225 g (Harlan) were maintained at 22 °C on 12 h light/12 h dark cycle. Animals had access to food and water ad libitum. Anesthesia was induced by a 5% isoflurane/air mixture (Forane, Butler-Schein) and maintained using a 2.5% mixture. The common sciatic nerve was exposed unilaterally at mid-thigh level and transected using surgical scissors or injected into the fascicle with purified S100A8/A9 (10 μg in 5 μl of PBS) or an equal volume of a vehicle (PBS alone) using a 33-gauge needle and a Hamilton syringe. On day 1 or week 1 after injury or injection, nerve segments (proximal, distal, and contralateral (normal) to the axotomy/injection site) were collected for subsequent analysis. Animals were sacrificed using an overdose of a rodent anesthesia mixture (50 mg/ml Nembutal (Abbott Laboratories) and 5 mg/ml diazepam (Steris Laboratories) in 0.9% PBS (Steris Laboratories)), followed by an intraperitoneal injection of Beuthanasia (100–150 mg/ml, Schering-Plough Animal Health). All animal procedures were done in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and protocols approved by the Institutional Animal Care and Use Committee at the Veterans Affairs San Diego Healthcare System.
Stimulation of Cultured Schwann Cells Using S100A8/A9
Primary Schwann cells were isolated from sciatic nerves of postnatal day 1–3 Sprague-Dawley rats (24). Schwann cells were further purified as described previously (35). The purity of Schwann cell cultures was monitored by S100B immunostaining (24). Schwann cells (>99% purity) were then cultured in 75-cm2 flasks coated with 50 μg/ml poly-d-lysine (Chemicon) in DMEM containing 1 g/liter glucose, 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 21 μg/ml bovine pituitary extract (Life Technologies), and 4 μm forskolin (Calbiotech) at 37 °C and 5.0% CO2. Cells were used in experiments at passages 3–7. For stimulation, Schwann cells were plated in wells of a 6-well plate and allowed to reach a 75% confluence level. Purified S100A8/A9 (5 μg/ml) was added to the cells for 1 or 24 h. Cells were then washed and harvested. Total RNA was isolated using the Direct-zol RNA MiniPrep system (Zymo Research) and used in the follow-up transcription profiling experiments. The RNA purity was estimated by measuring the A260/280 and the A260/230 ratios. RNA integrity was assessed using an Experion automated electrophoresis system (Bio-Rad). The purified RNA was quantified using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific).
Genome-wide Transcriptional Profiling
Nerve segments from 3 rats/group were isolated and stored at −20 °C in RNA-later (Life Technologies). Total RNA was extracted using TRIzol and purified using an RNeasy column (Qiagen). RNA purity, integrity, and quantity were assessed as described above. Total RNA aliquots (50 ng each) were labeled using a LowInput QuickAmp labeling kit and Cy3-CTP (Agilent Technologies). The labeled samples (1500 ng each) were hybridized for 18 h at 65 °C to SurePrint G3 rat GE 8 × 60K microarray chips (Agilent Technologies) and featured over 30,000 individual transcripts. Microarray chips were washed, developed using Cy3-Streptavidin Fluor conjugates (GE Healthcare), and scanned using a C Scanner (Agilent Technologies). Raw data were processed using Feature Extraction software (Agilent Technologies). Normalization to the median was performed using GeneSpring GX software (Agilent Technologies). Differentially expressed mRNAs with the signal intensity over 2-fold relative to the background standard deviation were identified by Student's t test. Statistically significant data (p < 0.05) were analyzed further to calculate the gene expression levels.
Systems Biology Analysis
To determine the affected cellular regulatory and signaling pathways, the individual genes, the expression of which differed at least 2-fold between the respective samples, were analyzed using Ingenuity Pathway Analyses (IPA) (Qiagen) and NextBio software (NextBio). The heatmap charts were generated using GenePattern software (Broad Institute, Cambridge, MA).
RT-PCR
Real-time RT-PCR was conducted using a Mx4000TM Multiplex quantitative PCR system (Agilent Technologies) in 25-μl reactions containing TaqMan Universal PCR Master Mix (Ambion), cDNA (50 ng), specific forward and reverse primers (900 nm each), and probes (200–300 nm). Primers and probes for Il6 and Il1β were from Roche (catalog nos. 04686934001 and 04689011001, respectively), and for Timp1 they were from Applied Biosystems (catalog no. Rn01430873_g1). Primers and probes for Mmp9, Tnfα, and Gapdh were designed as described previously (35, 36) and synthesized by Biosearch Technologies. Relative mRNA levels were quantified using the 2(−ΔΔ C(T)) method (37). Normalization to Gapdh and -fold-change calculation were performed using MxPro software (Agilent Technologies).
Neuropathology
Nerve segments (3 animals/group) were isolated and post-fixed for 48 h at 4 °C with 2.5% glutaraldehyde in 0.1 m phosphate buffer (pH 7.4). Specimens were washed with the phosphate buffer, post-fixed with 1% osmic acid (Ted Pella), dehydrated in graded (30–100%) ethyl alcohol and propylene oxide, and embedded in Araldite resin (Alaldite 502, catalog no. #8060), Eponate 12 resin (catalog no. 18005), dodecenyl succinic anhydride (catalog no. 18022), and DMP-30 (catalog no. 18042) (all obtained from Ted Pella). 1-μm-thick sections were cut using a diamond knife in an automated RM2065 microtome (Leica Microsystems) and then stained using methylene blue/azure II solution as described previously (35).
Immunohistochemistry
Three animals per group were perfused with 4% paraformaldehyde. Nerve segments were isolated, post-fixed in 4% paraformaldehyde, cryoprotected in graded sucrose solution, and embedded in O.C.T. compound (Tissue-Tek) on dry ice (36). Sections (10 μm thick) were blocked for 16 h at 4 °C in PBS containing 5% normal goat serum (Vector Laboratories) and 0.25% Triton X-100. Sections were incubated for 16 h at 4 °C with the respective primary antibodies and washed in PBS, followed by incubation with the corresponding secondary Alexa Fluor 488 or Alexa Fluor 594 antibody conjugates (Invitrogen). Slides were mounted in SlowFade Gold Antifade medium with DAPI (Life Technologies). Alternatively, sections were incubated with the respective biotin-conjugated secondary antibody and stained using the Vectastain Elite ABC system and 3′-3-diaminobenzidine substrate (Vector Laboratories) and Methyl Green (Vector Laboratories) counterstain. Images were acquired using a DMR microscope (Leica Microsystems) and processed using Openlab 4 imaging software (Improvision).
Immunoblotting
Nerve segments (3 animals/group) were collected, snap-frozen in liquid nitrogen, and then used for protein extraction. Nerve segments were homogenized in 50 mm Tris-HCl (pH 7.4) containing 1% Triton X-100, 150 mm NaCl, 10% glycerol, 0.1% SDS, 5 mm EDTA, 1 mm phenylmethylsulphonyl fluoride, and aprotinin and leupeptin (1 μg/ml each). Protein concentration was determined using a bicinchoninic acid assay (Pierce). Proteins (50 μg) were separated using SDS gel electrophoresis in 15% acrylamide gels (Bio-Rad) and transferred onto a nitrocellulose membrane using iBlot (Invitrogen). The membranes were blocked for 16 h at 4 °C in 5% nonfat milk in TBS and incubated with the primary antibodies diluted with 5% BSA in TBS. Membranes were washed in TBS containing 0.1% Tween and incubated for 1 h at ambient temperature with the respective HRP-conjugated secondary antibody (Cell Signaling Technologies) and the enhanced chemiluminescence system (GE Healthcare). For a loading control, the membranes were reprobed for β-actin. The membranes were scanned and digitized.
Statistical Analyses
Statistical analyses were performed using two-tailed, unpaired Student's t test and, when variances were unequal, by an unpaired t test with Welch's correction using either KaleidaGraph 4.03 (Synergy Software) or SPSS 16.0 (SPSS) software. Analyses of variance for repeated measures were employed for comparing three or more groups, followed by Tukey-Kramer post hoc test. p < 0.05 was considered significant.
RESULTS
Early Post-injury Response Genes in the Proximal and Distal Nerve Stumps
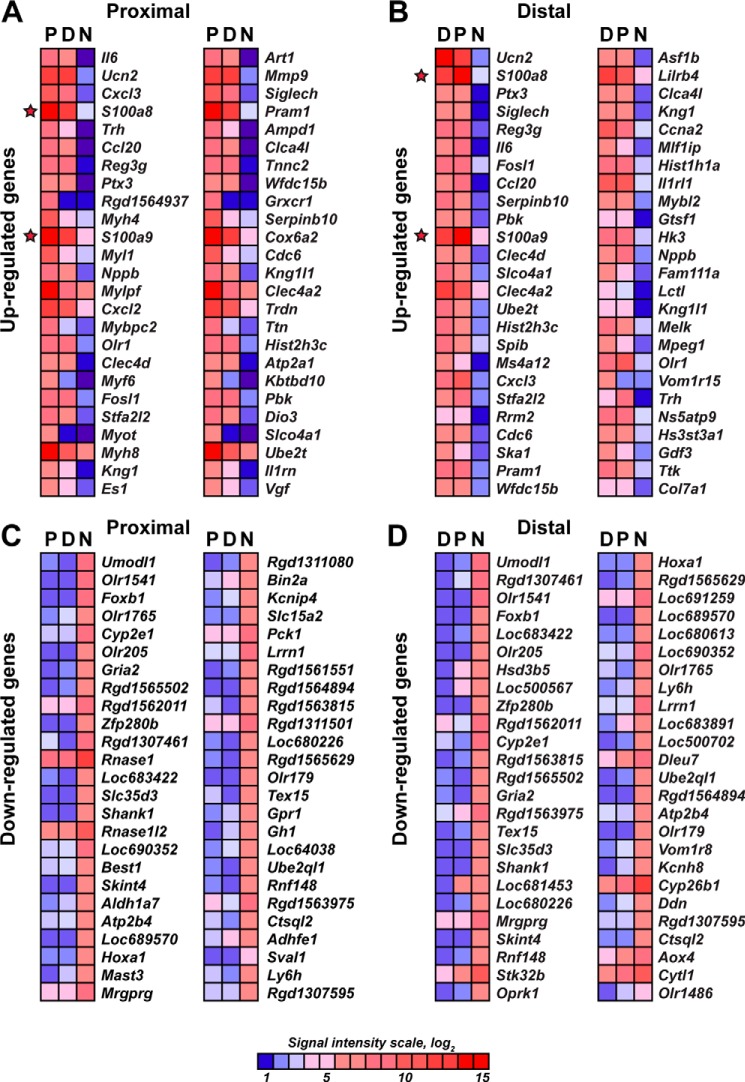
The early endoneurial changes in axotomized peripheral nerve are believed to predispose the regenerating axons for their long-term staggered growth (3, 4, 6). To identify genes whose expression was affected by axotomy in the sciatic nerve, we performed comparative genome-wide transcriptional profiling of the rat sciatic nerve P and D axotomized segments and a contralateral normal (N) nerve as a control on day 1 post-transection. More than 3831 individual affected genes (1816 up-regulated and 2015 down-regulated) in the P segment and more than 4540 individual genes (2165 up-regulated and 2375 down-regulated) in the D segment were identified relative to normal nerves. A heatmap of the 50 most affected up- and down-regulated genes in the P and D segments is shown in Fig. 1. In agreement with the earlier observations, the documented cell populations in sciatic nerves on day 1 post-injury included Schwann cells (∼80% of the total cell count), fibroblasts, resident macrophages, mast cells, endothelial cells, and infiltrating neutrophils (15, 16, 38).
FIGURE 1.
A–D, heatmaps of the genome-wide transcriptional profiling data of the axotomized (day 1) rat sciatic nerve. Red and blue correspond to high and the low expression levels, respectively. The color map shows the signal intensity scale. The top 50 up-regulated (A and B) and top 50 down-regulated (C and D) genes in the proximal and distal segments are shown. Only statistically significant data (p < 0.05) were used in our analysis. The S100a8 and S100a9 genes are highlighted with stars.
In general, the early injury-induced transcriptional changes demonstrated a level of similarity in the P and D stumps. Therefore, in both stumps, we recorded the up-regulation of the genes that are directly involved in the immune response (Fig. 1A), including interleukin (Il6), chemokine (CXC motif) ligand 3 (Cxcl3), chemokine (CC motif) ligand 20 (Ccl20), and the pentraxin-related protein Ptx3. Intriguingly, the calcium-binding protein S100a8 and S100a9 genes were highly up-regulated in both the P and D stumps.
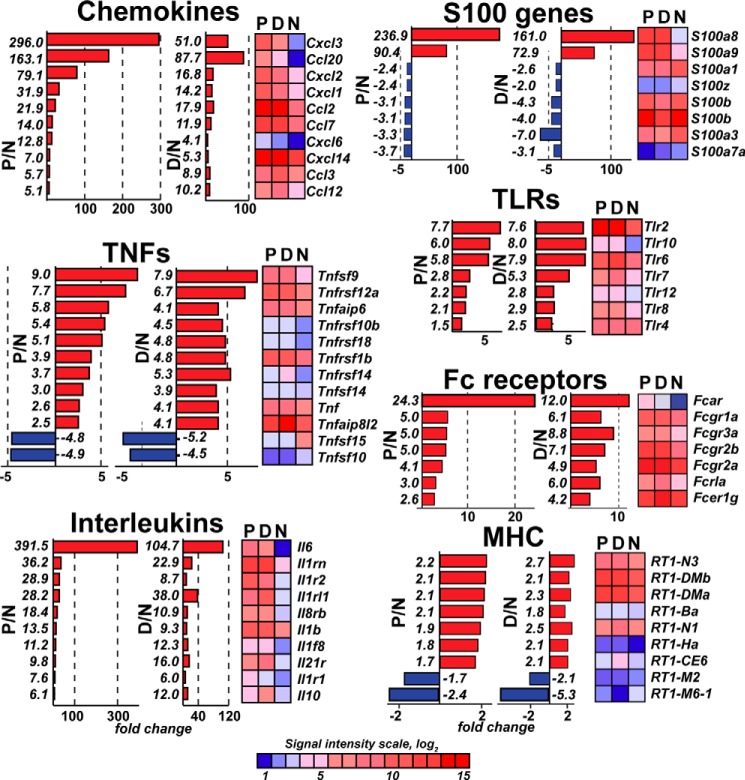
Further detailed analyses of the data demonstrated a significant similarity in the immunomodulatory gene expression patterns between in the P and D stumps, although the expression levels of the certain genes were distinct (Figs. 2 and 3). Therefore, the expression of Il6, Cxcl3, and Ccl20 was enhanced 390-fold, 300-fold, and 160-fold in the P samples, respectively, and 105-fold, 50-fold, and 88-fold in the D stumps, respectively (Fig. 2). Similarly, the expression of S100a8, S100a9, and Mmp9 (a typical proinflammatory MMP type) was enhanced 237-fold, 90-fold, and 55-fold in the P stumps and 161-fold, 73-fold, and 24-fold in the D stumps, respectively (Figs. 2 and 3A). On the other hand, the enhanced expression of Toll-like receptors (TLRs), TNF superfamily, antibody-binding Fc receptor Fcgr1–3 genes, and RT1 genes encoding the MHC cell surface proteins in rats (Fig. 2); tissue inhibitor of metalloproteinase-1 (Timp1); Mmp12 (macrophage metalloelastase); and many ion channels was roughly similar in the P and D stumps (Fig. 3, A and B). Using real-time RT-PCR, the induction of Il6, Il1β, Tnfα, Timp1, and Mmp9 was confirmed in the injured nerve segments (Fig. 3C). Taken together, these results indicate that nerve axotomy induced an early and robust transcriptional activity related to immune response genes in both the P and D segments.
FIGURE 2.
Immunomodulatory gene families affected in the proximal and distal segments on day 1 post-axotomy. The chemokine, S100, TLR, TNF, interleukin, Fc receptor, and MHC families are shown. -Fold-change values were calculated relative to the normal nerve using the normalized intensity values (p < 0.05). Red and blue correspond to up- and down-regulated genes, respectively. Heatmaps for the affected genes (p < 0.05) are shown. Genes with more than 2-fold changes (5-fold for chemokines and interleukins) in 3 rats/group were included in the analysis. The color map shows the signal intensity scale.
FIGURE 3.
A–C, specific gene families affected in the proximal and distal segments at day 1. The MMP and TIMP (A) and ion channel (B) gene families are shown. -Fold-change values were calculated relative to the normal nerve using the normalized intensity values (p < 0.05). The ion channel genes with a -fold-change of more than 3 are displayed. Red and blue correspond to up- and down-regulated genes, respectively. Heatmaps of the affected genes (p < 0.05) are shown. The color map shows the signal intensity scale. C, real-time RT-PCR of the Il6, Il1β, Tnfα, Timp1, and Mmp9 genes in rat sciatic nerves at day 1 post-axotomy in the D sample. The mean relative mRNA levels of 4 rats/group were normalized to Gapdh. The columns indicate the -fold-change relative to the normal nerve samples (*, p < 0.05; **, p < 0.01). Data are mean ± S.E.
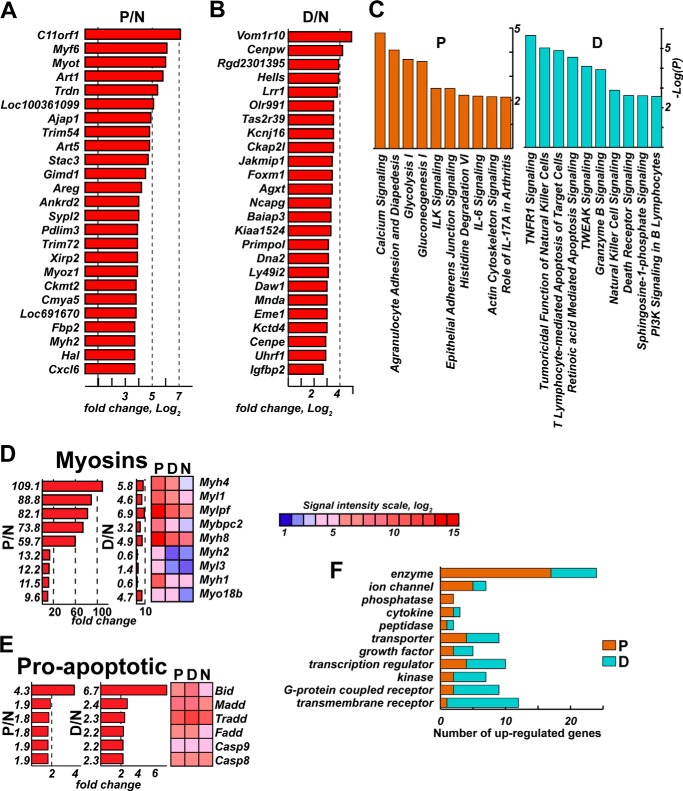
To discriminate transcriptional changes in the P and D segments, we excluded genes with similar expression pattern changes in both segments and then identified genes that were significantly and selectively up-regulated in the single respective segment (Fig. 4, A and B). Subsequent bioinformatics analysis revealed the specific pathways that were selectively affected in the P versus D samples. Specifically, multiple signaling pathways, including myosin, calcium, integrin-linked kinase, and gluconeogenesis/glycolysis signaling were selectively up-regulated in the P stumps (Fig. 4C). The most dramatic difference was observed in the myosin cytoskeleton-related genes, which were 50- to 100-fold up-regulated in the regenerating P segments and only enhanced severalfold in the D segment (Fig. 4D). In contrast, the signaling pathways that are largely focused on cell death, including apoptosis, TNF receptor 1 (TNFR1) signaling, and natural killer cell and death receptor signaling, were selectively stimulated in the D stumps (Fig. 4C). Therefore, transcriptional activation of the TNFα signaling network in the degenerating segment was due to the selective up-regulation of multiple proapoptotic genes, including MADD, FADD, BID, and caspases (Fig. 4E).
FIGURE 4.
A–D, the top 25 up-regulated genes that are selectively up-regulated on day 1 post-axotomy in the proximal (A) or distal (B) nerve segments. The horizontal axis represents the signal intensity ratio (log2). C, canonical pathway analysis (IPA) of the differentially expressed genes in the P (orange) and D (blue) nerve segments. D and E, the up-regulated myosin (D) and proapoptotic (E) gene families affected in the proximal and distal segments on day 1. -Fold-change values were calculated relative to the normal nerve using the normalized intensity values (p < 0.05). Red and blue correspond to up- and down-regulated genes, respectively. Heatmaps of the affected genes (p < 0.05) are shown. The color map shows the signal intensity scale. F, protein types up-regulated in the P (orange) and D (blue) nerve segments. Genes with a –fold change of more than 3 were included in this analysis. The p values were calculated using right-tailed Fisher's exact test.
Likewise, there was a bias in the P segment versus the D segment related to the affected protein types. The injury predominantly affected the genes coding for enzymes and ion channels in the P stumps, whereas the transmembrane G-coupled receptors were selectively up-regulated in the D segments (Fig. 4F). Overall, these findings support and extend the observations made by others, which focused mainly on either the individual P or D changes or on later time points post-axotomy (25–33).
Schwann Cell S100A9 Expression and Differential Activation of Kinase Pathways in Nerves Post-injury
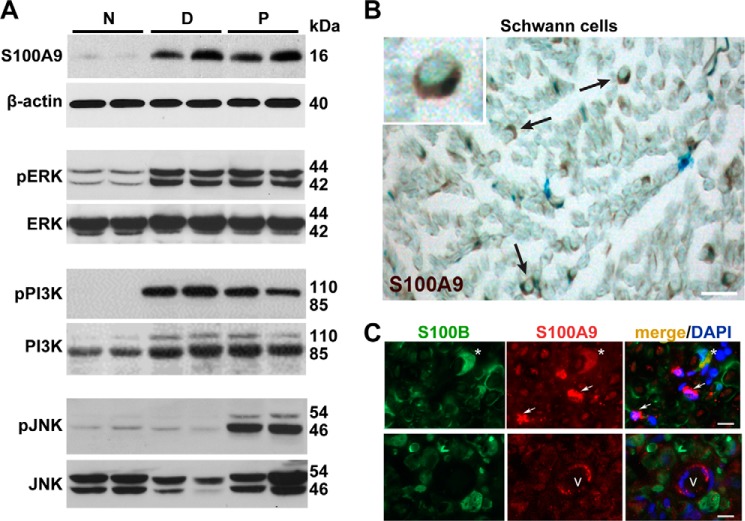
Next we assessed the levels and the source for S100A8/A9 protein in the injured nerve. S100 protein levels in the normal nerve were low and increased dramatically on day 1 post-axotomy in both the P and D segments, shown exemplarily for S100A9 (Fig. 5A).
FIGURE 5.
A–C, S100A9 and activation of the ERK, JNK, and PI3K kinases. A, immunoblotting for S100A9, β-actin, phosphorylated and total ERK, PI3K, and JNK in the proximal and distal sciatic nerve segments on day 1 post-axotomy. Normal samples represent contralateral nerve control. Data from 2 rats/group are shown. B, immunostaining of S100A9 (2′2-diaminobenzidine, brown) in the distal stump on day 1 post-axotomy. Schwann cells are indicated by arrows. The inset shows a single Schwann cell. Scale bar = to 25 μm. C, immunostaining of S100B (green) and S100A9 (red) in the injured nerve. S100B/S100A9-positive Schwann cells are indicated by asterisks. S100A9-positive, S100B-negative non-Schwann cells and vessel cells are indicated by arrows and V, respectively. Nuclei were stained with DAPI (blue). Scale bars = to 15 μm.
Rapid gene induction post-injury is normally followed by activation of the signaling kinase pathways. Specifically, ERK, JNK, and PI3K regulate a wide variety of functions relating to Schwann and immune cell functions in nerves post-injury (39–43) and S100A8/A9-induced cell signaling (18–21, 44, 45). In agreement with these earlier data, a significant activation of pERK1/2 was observed in the P and D stumps on day 1 post-transection compared with the N nerve (Fig. 5A). The main phosphorylated isoform in both the P and D samples was pPI3K 85 kDa, whereas the PI3K 110-kDa isoform was the dominant species in the total PI3K pool. In turn, pJNK1/2 was activated in the P stumps on day 1 post-transection compared with the N nerve. There was no similar increase in pJNK in the D stump samples. The S100A9 protein predominantly localized to crescent-shaped Schwann cells of both the P and D segments (for simplicity, only D is shown), as confirmed by colocalization of S100A9 with a phenotypic Schwann cell marker, S100B (Fig. 5, B and C). In addition, insignificant S100A9 immunoreactivity was occasionally observed in non-S100B-reactive vessel endothelial and other endoneurial cells of axotomized nerves (Fig. 5C).
S100A8/A9 Stimulates Cultured Schwann Cells
Calprotectin (S100A8/A9) induces immune cell chemotaxis (18, 46) and cytokine expression (21). Because S100a8 and S100a9 were among the top-induced genes in both the P and D segments, we hypothesized that the S100A8/A9 heterodimer was implicated in the initial positive chemotactic gradient in the denervated Schwann cells.
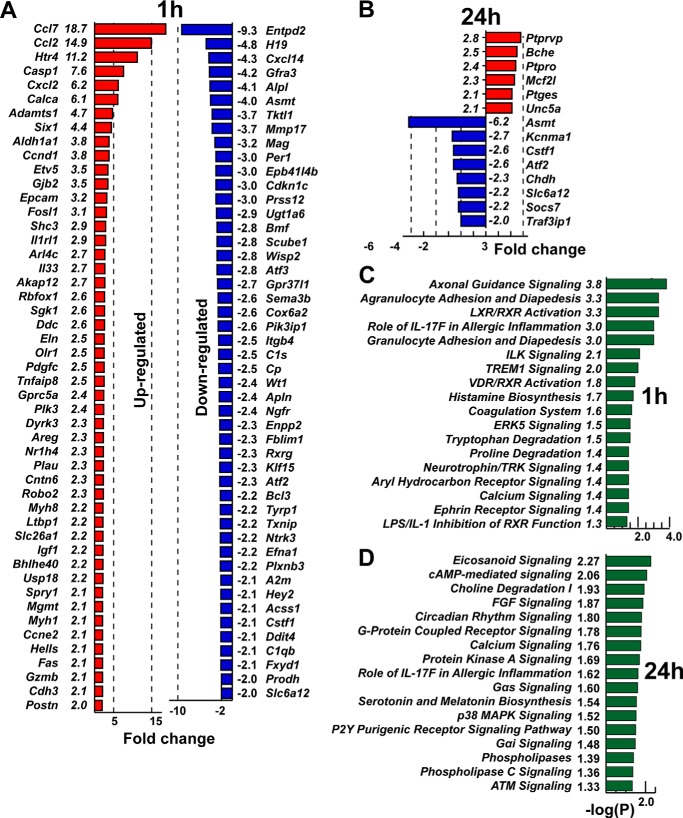
To test this hypothesis, we performed genome-wide transcriptional profiling of cultured Schwann cells coincubated for 1 and 24 h with the purified S100A8/A9 protein complex. As a result of a short-term, 1-h coincubation with S100A8/A9, a number of the genes, especially chemokine genes, including Ccl7, Ccl2, and Cxcl2, were up-regulated severalfold in Schwann cells. Our further IPA suggested that S100A8/A9 affected multiple cell adhesion and movement, chemotaxis, and signaling pathways in Schwann cells (Fig. 6, A and C). In sharp contrast, only a few genes were up-regulated in Schwann cells after long-term, 24-h coincubation with S100A8/A9 (Fig. 6, B and D). S100A8/A9 did not significantly regulate the expression of many other inflammatory genes of the IL, TNF, MMP, TLR, and S100 families in Schwann cells in either sample (Fig. 6, A–D). Taken together, these results indicate that the affected pathways were largely dissimilar in the 1-h versus the 24-h cell samples and that the most significant effect of S100A8/A9 takes place in Schwann cells shortly after their stimulation with S100A8/A9.
FIGURE 6.
A–D, the top up- and down-regulated genes in cultured Schwann cells stimulated with S100A8/A9 for 1 h (A) and 24 h (B). –Fold change values were calculated relative to the intact Schwann cell control (p < 0.05). Red and blue indicate up- and down-regulated genes, respectively. C and D, canonical pathway analysis (IPA) of the differentially expressed genes in Schwann cells following 1-h (C) and 24-h (D) stimulation with S100A8/A9. The p values (green bars) were calculated using right-tailed Fisher's exact test.
Similarity of the Inflammatory Gene Network in Injured Nerve and S100A8/A9-treated Schwann Cells
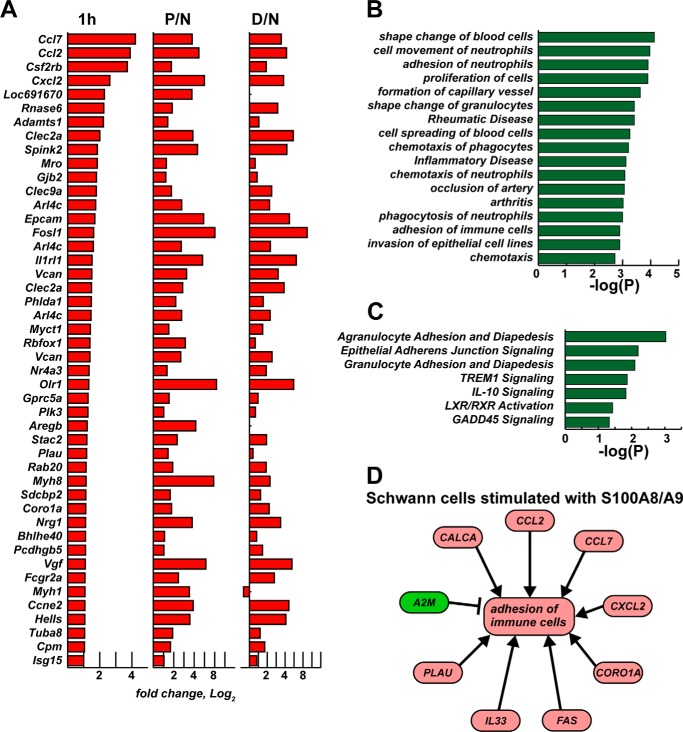
We next determined whether the individual genes up-regulated in Schwann cells treated with S100A8/A9 for 1 h were similar to those affected in the P and D segments of the axotomized sciatic nerve (Fig. 7A). The follow-up IPA pointed to these similarly affected biological functions, diseases, and canonical pathways (Fig. 7, B and C), which were characteristic for stimulating the chemotaxis, adhesion, and motility of immune cells, including neutrophils and phagocytes. Furthermore, in Schwann cells treated with S100A8/A9, adhesion of immune cells was directly related to up-regulation of chemokine (Ccl2, Ccl7, Cxcl2), calcitonin CALCA, Fas, Il33, and urokinase-type plasminogen activator PLAU genes and down-regulation of a protease inhibitor and cytokine transporter, A2m (α-2-macroglobulin) (Fig. 7D).
FIGURE 7.
A–D, up-regulated gene network overlap in S100A8/A9-stimulated cultured Schwann cells and injured nerve. A, up-regulated genes in Schwann cells after 1 h of stimulation with S100A8/A9 relative to the intact cell control and in the proximal (P/N) and distal (D/N) nerve segments relative to the normal nerve control. The bars correspond to -fold change (log2) of normalized signal intensity (p < 0.05). B, biological function and disease analysis (IPA) of the genes in A. C, canonical pathway analysis (IPA) of the genes in A. The bars indicate p values, which were calculated using right-tailed Fisher's exact test. D, activation of the immune cell adhesion network following 1-h stimulation of Schwann cells with S100A8/A9. Red and green correspond to up- and down-regulated genes, respectively. Arrows indicate stimulation.
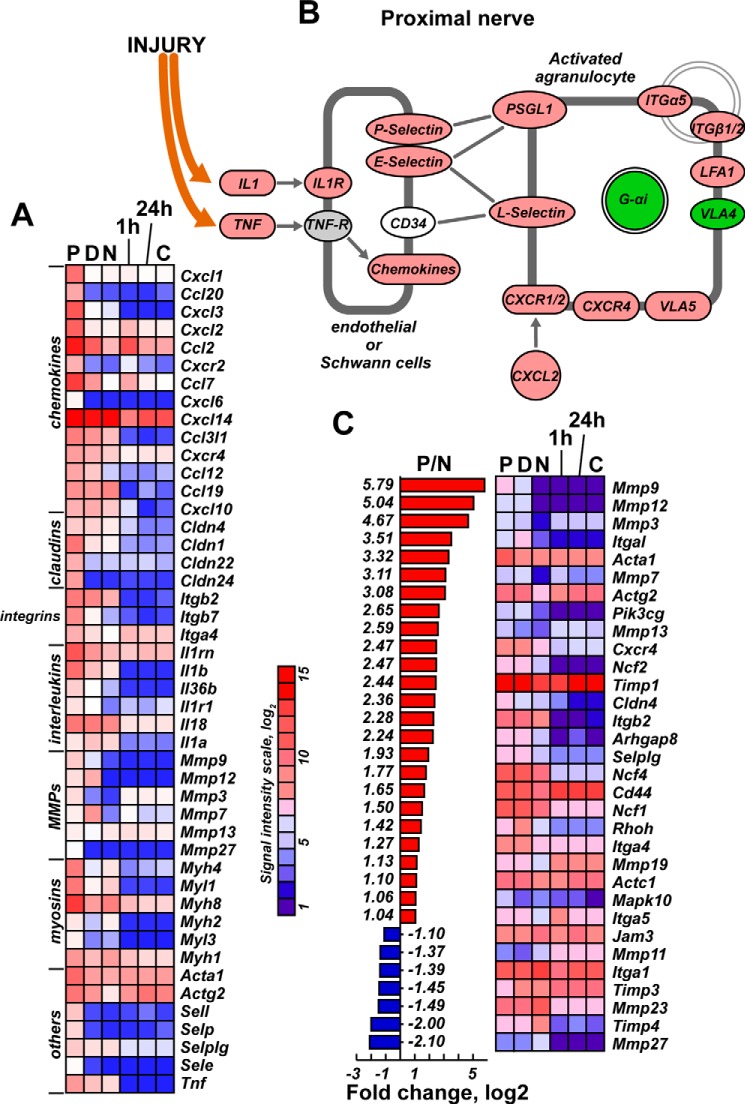
The role of the agranulocyte/granulocyte activation and adhesion pathways in injured nerves was recapitulated by S100A8/A9 stimulation of the cultured Schwann cells and confirmed by our additional bioinformatics analysis of the transcriptional profiling data. Therefore, transcriptional activity of multiple genes, including P-, E- and L-selectins and Lfa1, was enhanced in both the P segment and in stimulated Schwann cells (Fig. 8).
FIGURE 8.
A–C, agranulocyte/granulocyte activation and adhesion pathways in S100A8/A9-stimulated Schwann cells and the injured nerve. A, chemokine, claudin, integrin, interleukin, Mmp, myosine, and other genes regulated in both proximal/distal nerve segments and in cultured Schwann cells after 1-h and 24-h stimulation with S100A8/A9. A signal intensity scale is shown. B, the agranulocyte activation pathway indicates up-regulated (red) and down-regulated (green) genes observed in the proximal nerve segment. C, regulation of agranulocyte-specific genes in the injured nerve compared with cultured Schwann cells after 1-h and 24-h stimulation with S100A8/A9. P/N, -fold-change (log2) of the signal intensity in the proximal nerve segment relative to the normal nerve (p < 0.05). Red and blue correspond to up- and down-regulated genes, respectively. Also shown is a heatmap of the affected genes (p < 0.05) in post-injury nerve segments and in cultured Schwann cells after 1-h and 24-h stimulation with S100A8/A9.
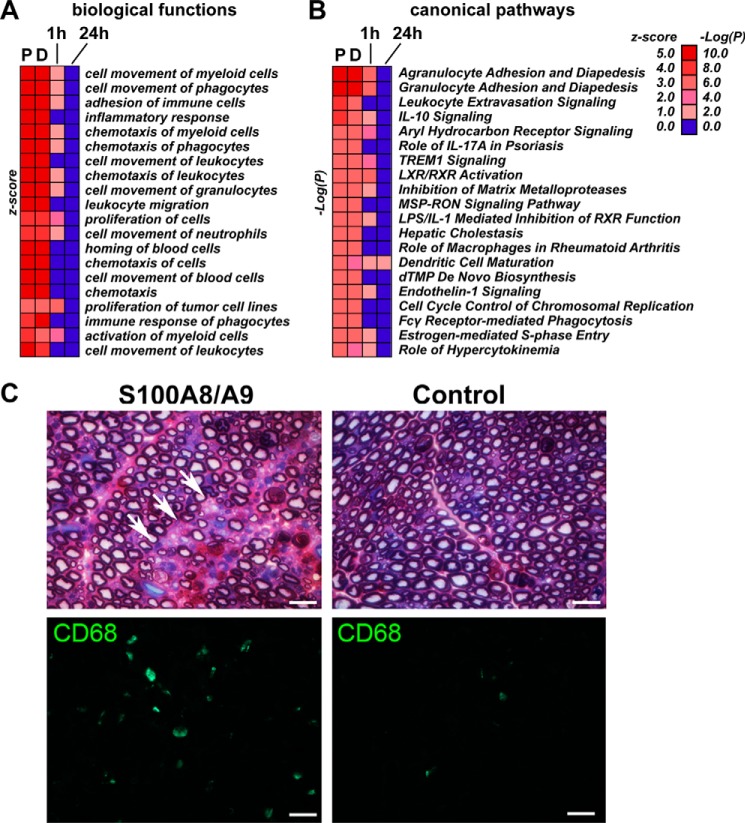
Intriguingly, some key proinflammatory factors in the axotomized nerve, including Il1β, Tnfα, Mmp9, and Mmp12, were not similarly induced in isolated Schwann cells stimulated with S100A8/A9 (Fig. 8, A and C). These data imply the presence of immune mechanisms in the injured nerve that are not fully recapitulated in the purified Schwann cell cultures or are independent of S100A8/A9 activity. Conversely, multiple other biological functions and canonical pathways supporting myeloid, phagocyte, and leukocyte cell movement, adhesion, and chemotaxis were induced in the P and D nerve segments in a similar way as in S100A8/A9-stimulated Schwann cells (Fig. 9A).
FIGURE 9.
A–C, S100A8/A9 stimulates macrophage migration into the nerve. A and B, IPA biological functions (A) and canonical pathways (B) in injured nerve and Schwann cells stimulated for 1 h and 24 h with S100A8/A9. The color intensity map shows the IPA prediction confidence z scores (biological functions) and −log(P) values (canonical pathways). The p values were calculated using right-tailed Fisher's exact test. C, ultrastructure and immunostaining analyses of the naïve nerve after injection of S100A8/A9. Top left panel, methylene blue/azure II staining in 1-μm-thick sciatic nerve sections of S100A8/A9-injected nerves display the areas of endoneurial edema with infiltrating phagocytes (arrows). Top right panel, uncompromised axons surrounded by a compact rim of myelin sheath in the control nerve. Bottom panels, CD68 immunostaining of macrophages (green) in S100A8/A9-injected and control 10-μm-thick nerve sections. Scale bars = 25 μm.
S100A8/A9 Stimulates Immune Cell Recruitment into Nerves
Extravasation of myeloid cells into the nerve, specifically of hematogenous CD68+ macrophages, is a critical event of Wallerian degeneration between days 2 and 14 post-injury (12, 15–17). Therefore, we tested the IPA prediction for myeloid cell migration after S100A8/A9 stimulation (Fig. 9A) using direct intraneural injection of the purified S100A8/A9 heterodimer into the intact nerve, followed by ultrastructural and immunohistochemical CD68 analyses of the injection site on day 7 post-injection (Fig. 9B). The nerves exposed to S100A8/A9 displayed areas of endoneurial edema with clusters of infiltrating immune cells, including phagocytes (Fig. 9B). In contrast, the nerve bundles injected with control PBS maintained normal morphology, displaying uncompromised axons surrounded by a compact rim of myelin sheath. The significant increase in the macrophage numbers after S100A8/A9 injection was confirmed using an antibody to CD68 (Fig. 9B). These data confirm the key role of S100A8/A9 in creating a functional chemotactic gradient that guides myeloid cell migration into peripheral nerves.
DISCUSSION
Peripheral nerve injury that involves complete transection/axotomy of the nerve trunk can be broadly characterized as a clearance Wallerian degeneration process within the D stump and the regeneration of the surviving axons in the P stump, which remain connected to the neuronal cell body in the ganglia. Although both the degenerative and regenerative processes generally begin immediately after most types of nerve injury, axotomy entails a prolonged lag period in the regenerative process and staggered neurite growth (for up to 4 weeks in rats) (3, 4, 6). Overall, our data confirm and extend the findings by us and others of the early transcriptional response to peripheral nerve injury, characterized by disintegration of the axonal cytoskeleton, immune response, and cell death unfolding in the D segment (26, 29, 31, 32) and cell proliferation, migration, axon guidance, and regeneration in the P segment proximal to nerve injury (33, 47). Our results offer additional comparative insights into the specific rapid cell responses and activation of gene families and pathways that are favorable for chemotaxis, adhesion, and extravasation of myeloid cells in the D and P segments within the first day of peripheral nerve axotomy.
Multiple immune response genes from the chemokine, IL, TNF, TLR, S100, and MMP families were induced in both the P and D stumps. These data indicate that molecular programs facilitating the acute inflammatory or degenerative changes exist in both the D and P segments. Indeed, calpain-dependent acute axonal degeneration of proximal axons occurs in the spinal cord within minutes to hours after injury (48). Likewise, an increase in immediate-early immune response genes (e.g. Mmp9, Ccl20, Cxcl2, and Il6) and the calcium, agranulocyte, and granulocyte signaling pathways was observed in the P segment within day 1 post-axotomy in this study and also by us and others earlier (33, 47).
Schwann cells remain the main cell population in peripheral nerves 1 day post-injury. Accordingly, the predominant changes described here mainly represent features of injury-induced Schwann cell activation and trans-differentiation into Büngner cells. In addition, resident reactive fibroblasts may regulate genes involved in epineurial scarring, as endothelial cells of the nerve vasculature, and a small population of the resident macrophages and mast cells may contribute, although insignificantly, to the transcriptional profiles in nerves on day 1 post-injury.
Infiltrating cell populations in nerves within day 1 post-injury include neutrophils and patrolling lymphocytes (15, 17, 38, 49). Consistently, activation of granulocyte and agranulocyte signaling is recorded on day 1 post-axotomy. Hematogenous macrophages generally infiltrate the D or P segments after day 2 post-injury (2) to complete debris clearance processes (22, 23).
Here we identified S100a8 and S100a9 among the top induced genes in peripheral nerve post-injury and, by employing the purified S100A8/A9 heterodimer (calprotectin), established its important role in stimulating the chemokine-cytokine network and the initial chemotactic gradient in Schwann cells. This gradient attracts hematogenous immune cells to the nerve injury site. In addition, this gradient may control the migration of resident cells, including Schwann cells. Schwann cells migrate from both the P and D stumps into the nerve gap resulting from transection (2). Specifically, stimulation of cultured Schwann cells with S100A8/A9 recapitulated a significant portion of the proinflammatory gene network activation we observed in the axotomized nerve, including chemokine (Ccl7, Ccl2, and Cxcl2), inflammatory cytokine (Il1r1 and Il33) and MMPs (Mmp3, Mmp7, and Mmp13), and agranulocyte and granulocyte activation signaling pathways.
S100A8/A9, also known as myeloid-related proteins MRP8/14, initiated myeloid (CD68+ macrophage) migration into the intact nerve. Schwann cells were the main cell source for S100A9 on day 1 post-axotomy. Future studies will need to decipher the S100A8 and S100A9 source and roles in the orchestration of complex immune cell migration patterns and functions in the course of Wallerian degeneration (15, 17, 38, 49).
The S100A8 and S100A9 homodimers replicate, and, under some circumstances exceed, the activities of the S100A8/A9 heterodimer. This study does not rule out the possibility that some effects observed here relate to homodimer formation in heterodimer preparations or homodimer formation in injured nerves expressing high levels of both S100a8 and S100a9 transcripts. Future investigation is required to distinguish between the effects of hetero- and homodimers in modulating the inflammatory program in peripheral nerve injury and repair.
S100A8/A9 are endogenous ligands of TLR4 (44) and RAGE (the receptor for advanced glycation endproducts) (45), both of which are expressed in Schwann cells (50, 51). The S100B protein, a phenotypic marker of Schwann cells, is distinct structurally and functionally from S100A8/A9. Interestingly, S100A8/A9 stimulated other immune response genes in Schwann cells, including the Itga4 gene, coding for integrin α4, which forms the VLA4/α4β1 lymphocyte/monocyte homing receptor. S100A8/A9 also induced the axonal guidance signaling pathway Htr4 gene encoding the G-protein-coupled serotonin 5-HT4 receptor and the inhibition of the MMP pathway in Schwann cells.
S100A8/A9 stimulation was, however, ineffective in regulating Schwann cell expression of the top induced inflammatory genes expressed in nerves shortly after axotomy, including Il6, Il1β, Tnfα, Timp1, and Mmp9. These data corroborate the presence of the S100A8/A9-independent immune activation mechanisms in injured nerves in vivo. In addition, S100A8/A9 stimulation did not activate lymphocyte migration signaling, a late response immune activation event in damaged nerves (38). We conclude that S100A8/A9 may initiate the acute phase response signaling and chemotactic gradient preceding the major inflammatory response in the damaged nerve.
Worth noting are the differences in transcriptional programs observed between the murine and rat nerve injury models. Timp1 was among the top 10 up-regulated genes in axotomized murine nerves (25). In turn, because of the high pre-existing expression of Timp1 in normal rat sciatic nerves, only a 2-fold induction of Timp1 was recorded post-axotomy, suggesting the presence of species-specific mechanisms of transcriptional regulation.
In sum, we determined that S100A8/A9 are potent initiators of the immune response in stimulated Schwann cells of injured peripheral nerves. Up-regulation of S100A8/A9 in Schwann cells shortly post-injury contributes to the activation of the chemokine-cytokine network and the initial chemotactic gradient that guides hematogenous immune cells toward the injury site.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1DE022757 (to V. I. S. and A. Y. S.). This work was also supported by Department of Veterans Affairs Merit Review Award 5I01BX000638 (to V. I. S.).
The normalized microarray data reported in this paper have been submitted to the Gene Expression Omnibus Repository with accession number GSE62282.
A. V. Chernov, J. Dolkas, K. Hoang, M. Angert, G. Srikrishna, T. Vogl, S. Baranovskaya, A. Y. Strongin, and V. I. Shubayev, unpublished data.
- P
- proximal
- D
- distal
- N
- normal
- MMP
- matrix metalloproteinase
- TLR
- Toll-like receptor.
REFERENCES
- 1. Gordon T., Sulaiman O., Boyd J. G. (2003) Experimental strategies to promote functional recovery after peripheral nerve injuries. J. Peripher. Nerv. Syst. 8, 236–250 [DOI] [PubMed] [Google Scholar]
- 2. Zochodne D. W. (2012) The challenges and beauty of peripheral nerve regrowth. J. Peripher. Nerv. Syst. 17, 1–18 [DOI] [PubMed] [Google Scholar]
- 3. McDonald D., Cheng C., Chen Y., Zochodne D. (2006) Early events of peripheral nerve regeneration. Neuron Glia Biol. 2, 139–147 [DOI] [PubMed] [Google Scholar]
- 4. Wood M. D., Kemp S. W., Weber C., Borschel G. H., Gordon T. (2011) Outcome measures of peripheral nerve regeneration. Ann. Anat. 193, 321–333 [DOI] [PubMed] [Google Scholar]
- 5. Waller A. (1850) Experiments on the section of the glossopharyngeal and hypoglossal nerves of the frog and observations of the alterations produced thereby in the structure of their primitive fibers. Philos. Trans. R Soc. Lond. B Biol. Sci. 140, 423–429 [Google Scholar]
- 6. Gordon T., Tyreman N., Raji M. A. (2011) The basis for diminished functional recovery after delayed peripheral nerve repair. J. Neurosci. 31, 5325–5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vargas M. E., Barres B. A. (2007) Why is Wallerian degeneration in the CNS so slow? Annu. Rev. Neurosci. 30, 153–179 [DOI] [PubMed] [Google Scholar]
- 8. Chen Y. Y., McDonald D., Cheng C., Magnowski B., Durand J., Zochodne D. W. (2005) Axon and Schwann cell partnership during nerve regrowth. J. Neuropathol. Exp. Neurol. 64, 613–622 [DOI] [PubMed] [Google Scholar]
- 9. Ramon y Cajal S. (1991) Cajal's Degeneration and Regeneration of the Nervous System, (translated by May R. M., DeFelipe J., Jones E. G, eds), pp. 144–166, Oxford University Press, New York [Google Scholar]
- 10. Nix P., Hisamoto N., Matsumoto K., Bastiani M. (2011) Axon regeneration requires coordinate activation of p38 and JNK MAPK pathways. Proc. Natl. Acad. Sci. U.S.A. 108, 10738–10743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kenney A. M., Kocsis J. D. (1998) Peripheral axotomy induces long-term c-Jun amino-terminal kinase-1 activation and activator protein-1 binding activity by c-Jun and junD in adult rat dorsal root ganglia in vivo. J. Neurosci. 18, 1318–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Z. L., Yu W. M., Strickland S. (2007) Peripheral regeneration. Annu. Rev. Neurosci. 30, 209–233 [DOI] [PubMed] [Google Scholar]
- 13. Allodi I., Udina E., Navarro X. (2012) Specificity of peripheral nerve regeneration: interactions at the axon level. Prog. Neurobiol. 98, 16–37 [DOI] [PubMed] [Google Scholar]
- 14. Jessen K. R., Mirsky R. (2008) Negative regulation of myelination: relevance for development, injury, and demyelinating disease. Glia 56, 1552–1565 [DOI] [PubMed] [Google Scholar]
- 15. Kieseier B. C., Hartung H. P., Wiendl H. (2006) Immune circuitry in the peripheral nervous system. Curr. Opin. Neurol. 19, 437–445 [DOI] [PubMed] [Google Scholar]
- 16. Myers R. R., Campana W. M., Shubayev V. I. (2006) The role of neuroinflammation in neuropathic pain: mechanisms and therapeutic targets. Drug Discov. Today 11, 8–20 [DOI] [PubMed] [Google Scholar]
- 17. Thacker M. A., Clark A. K., Marchand F., McMahon S. B. (2007) Pathophysiology of peripheral neuropathic pain: immune cells and molecules. Anesth. Analg. 105, 838–847 [DOI] [PubMed] [Google Scholar]
- 18. Lackmann M., Rajasekariah P., Iismaa S. E., Jones G., Cornish C. J., Hu S., Simpson R. J., Moritz R. L., Geczy C. L. (1993) Identification of a chemotactic domain of the pro-inflammatory S100 protein CP-10. J. Immunol. 150, 2981–2991 [PubMed] [Google Scholar]
- 19. Vogl T., Ludwig S., Goebeler M., Strey A., Thorey I. S., Reichelt R., Foell D., Gerke V., Manitz M. P., Nacken W., Werner S., Sorg C., Roth J. (2004) MRP8 and MRP14 control microtubule reorganization during transendothelial migration of phagocytes. Blood 104, 4260–4268 [DOI] [PubMed] [Google Scholar]
- 20. Hobbs J. A., May R., Tanousis K., McNeill E., Mathies M., Gebhardt C., Henderson R., Robinson M. J., Hogg N. (2003) Myeloid cell function in MRP-14 (S100A9) null mice. Mol. Cell Biol. 23, 2564–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simard J. C., Cesaro A., Chapeton-Montes J., Tardif M., Antoine F., Girard D., Tessier P. A. (2013) S100A8 and S100A9 induce cytokine expression and regulate the NLRP3 inflammasome via ROS-dependent activation of NF-κB(1.). PLoS ONE 8, e72138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fernandez-Valle C., Bunge R. P., Bunge M. B. (1995) Schwann cells degrade myelin and proliferate in the absence of macrophages: evidence from in vitro studies of Wallerian degeneration. J. Neurocytol. 24, 667–679 [DOI] [PubMed] [Google Scholar]
- 23. Perry V. H., Brown M. C., Gordon S. (1987) The macrophage response to central and peripheral nerve injury: A possible role for macrophages in regeneration. J. Exp. Med. 165, 1218–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brockes J. P., Fields K. L., Raff M. C. (1979) Studies on cultured rat Schwann cells: I: establishment of purified populations from cultures of peripheral nerve. Brain Res. 165, 105–118 [DOI] [PubMed] [Google Scholar]
- 25. Kim Y., Remacle A. G., Chernov A. V., Liu H., Shubayev I., Lai C., Dolkas J., Shiryaev S. A., Golubkov V. S., Mizisin A. P., Strongin A. Y., Shubayev V. I. (2012) The MMP-9/TIMP-1 axis controls the status of differentiation and function of myelin-forming Schwann cells in nerve regeneration. PLoS ONE 7, e33664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nagarajan R., Le N., Mahoney H., Araki T., Milbrandt J. (2002) Deciphering peripheral nerve myelination by using Schwann cell expression profiling. Proc. Natl. Acad. Sci. U.S.A. 99, 8998–9003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang N., Li H., Sun Y., Yin D., Zhao Q., Cui S., Yao D. (2014) Differential gene expression in proximal and distal nerve segments of rats with sciatic nerve injury during Wallerian degeneration. Neural Regen. Res. 9, 1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. D'Antonio M., Michalovich D., Paterson M., Droggiti A., Woodhoo A., Mirsky R., Jessen K. R. (2006) Gene profiling and bioinformatic analysis of Schwann cell embryonic development and myelination. Glia 53, 501–515 [DOI] [PubMed] [Google Scholar]
- 29. Bosse F., Hasenpusch-Theil K., Küry P., Müller H. W. (2006) Gene expression profiling reveals that peripheral nerve regeneration is a consequence of both novel injury-dependent and reactivated developmental processes. J. Neurochem. 96, 1441–1457 [DOI] [PubMed] [Google Scholar]
- 30. Kubo T., Yamashita T., Yamaguchi A., Hosokawa K., Tohyama M. (2002) Analysis of genes induced in peripheral nerve after axotomy using cDNA microarrays. J. Neurochem. 82, 1129–1136 [DOI] [PubMed] [Google Scholar]
- 31. Araki T., Sasaki Y., Milbrandt J. (2004) Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 305, 1010–1013 [DOI] [PubMed] [Google Scholar]
- 32. Yao D., Li M., Shen D., Ding F., Lu S., Zhao Q., Gu X. (2013) Expression changes and bioinformatic analysis of Wallerian degeneration after sciatic nerve injury in rat. Neurosci. Bull 29, 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li S., Liu Q., Wang Y., Gu Y., Liu D., Wang C., Ding G., Chen J., Liu J., Gu X. (2013) Differential gene expression profiling and biological process analysis in proximal nerve segments after sciatic nerve transection. PLoS ONE 8, e57000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vogl T., Leukert N., Barczyk K., Strupat K., Roth J. (2006) Biophysical characterization of S100A8 and S100A9 in the absence and presence of bivalent cations. Biochim. Biophys. Acta 1763, 1298–1306 [DOI] [PubMed] [Google Scholar]
- 35. Shubayev V. I., Angert M., Dolkas J., Campana W. M., Palenscar K., Myers R. R. (2006) TNFα-induced MMP-9 promotes macrophage recruitment into injured peripheral nerve. Mol. Cell Neurosci. 31, 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chattopadhyay S., Shubayev V. I. (2009) MMP-9 controls Schwann cell proliferation and phenotypic remodeling via IGF-1 and ErbB receptor-mediated activation of MEK/ERK pathway. Glia 57, 1316–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔ C(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 38. Austin P. J., Moalem-Taylor G. (2010) The neuro-immune balance in neuropathic pain: involvement of inflammatory immune cells, immune-like glial cells and cytokines. J. Neuroimmunol. 229, 26–50 [DOI] [PubMed] [Google Scholar]
- 39. Harrisingh M. C., Perez-Nadales E., Parkinson D. B., Malcolm D. S., Mudge A. W., Lloyd A. C. (2004) The Ras/Raf/ERK signalling pathway drives Schwann cell dedifferentiation. EMBO J. 23, 3061–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sheu J. Y., Kulhanek D. J., Eckenstein F. P. (2000) Differential patterns of ERK and STAT3 phosphorylation after sciatic nerve transection in the rat. Exp. Neurol. 166, 392–402 [DOI] [PubMed] [Google Scholar]
- 41. Maurel P., Salzer J. L. (2000) Axonal regulation of Schwann cell proliferation and survival and the initial events of myelination requires PI 3-kinase activity. J. Neurosci. 20, 4635–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zrouri H., Le Goascogne C., Li W. W., Pierre M., Courtin F. (2004) The role of MAP kinases in rapid gene induction after lesioning of the rat sciatic nerve. Eur. J. Neurosci. 20, 1811–1818 [DOI] [PubMed] [Google Scholar]
- 43. Abe N., Cavalli V. (2008) Nerve injury signaling. Curr. Opin. Neurobiol. 18, 276–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vogl T., Tenbrock K., Ludwig S., Leukert N., Ehrhardt C., van Zoelen M. A., Nacken W., Foell D., van der Poll T., Sorg C., Roth J. (2007) Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat. Med. 13, 1042–1049 [DOI] [PubMed] [Google Scholar]
- 45. Turovskaya O., Foell D., Sinha P., Vogl T., Newlin R., Nayak J., Nguyen M., Olsson A., Nawroth P. P., Bierhaus A., Varki N., Kronenberg M., Freeze H. H., Srikrishna G. (2008) RAGE, carboxylated glycans and S100A8/A9 play essential roles in colitis-associated carcinogenesis. Carcinogenesis 29, 2035–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lackmann M., Cornish C. J., Simpson R. J., Moritz R. L., Geczy C. L. (1992) Purification and structural analysis of a murine chemotactic cytokine (CP-10) with sequence homology to S100 proteins. J. Biol. Chem. 267, 7499–7504 [PubMed] [Google Scholar]
- 47. Wang Y., Tang X., Yu B., Gu Y., Yuan Y., Yao D., Ding F., Gu X. (2012) Gene network revealed involvements of Birc2, Birc3 and Tnfrsf1a in anti-apoptosis of injured peripheral nerves. PLoS ONE 7, e43436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kerschensteiner M., Schwab M. E., Lichtman J. W., Misgeld T. (2005) In vivo imaging of axonal degeneration and regeneration in the injured spinal cord. Nat. Med. 11, 572–577 [DOI] [PubMed] [Google Scholar]
- 49. Ren K., Dubner R. (2010) Interactions between the immune and nervous systems in pain. Nat. Med. 16, 1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Goethals S., Ydens E., Timmerman V., Janssens S. (2010) Toll-like receptor expression in the peripheral nerve. Glia 58, 1701–1709 [DOI] [PubMed] [Google Scholar]
- 51. Perrone L., Peluso G., Melone M. A. (2008) RAGE recycles at the plasma membrane in S100B secretory vesicles and promotes Schwann cells morphological changes. J. Cell Physiol. 217, 60–71 [DOI] [PubMed] [Google Scholar]