Abstract
Background and Objective
Inhibitors of apoptosis proteins (IAPs) have been well investigated in human cancers, where they are frequently overexpressed and associated with poor prognosis. Here we explored the role of baculoviral IAP repeat containing 6 (BIRC6), a member of IAPs, in human colorectal cancer (CRC).
Methods
We used Western blotting and immunohistochemistry to examine BIRC6 expression in 7 CRC cell lines and 126 CRC clinical samples. We determined the biological significance of BIRC6 in CRC cell lines by a lentivirus-mediated silencing method.
Results
We reported that BIRC6 was overexpressed in CRC cell lines and clinical CRC tissues. BIRC6 overexpression was correlated with tumor size and invasion depth of CRC. BIRC6 overexpression is associated with worse overall survival (OS) (P = 0.001) and shorter disease-free survival (DFS) (P = 0.010). BIRC6 knockdown inhibited cell proliferation, arrested cell cycle at S phase, downregulated cyclin A2, B1, D1 and E1 levels, and sensitized CRC cells to chemotherapy in vitro and in vivo.
Conclusions
Taken together, these data suggests that BIRC6 overexpression is a predictor of poor prognosis in colorectal cancer and BIRC6 could be a potential target of CRC therapy.
Introduction
Colorectal cancer (CRC) is the third most frequently diagnosed malignancy and the fourth leading cause of cancer death worldwide[1]. Due to screening and removal of premalignant polyps, incidence rates have declined over the last 3 decades[2]. The use of new drugs such as oxaliplatin, bevacizumab, cetuximab and panitumumab allows patients with metastatic colorectal cancer survive longer[3,4,5,6]. Despite the progress in the diagnosis and treatment of CRC, the mortality from this disease remains high. Given the poor prognosis of CRC, novel prognostic markers and therapeutic strategies need to be developed to further improve the outcome of colorectal cancer.
The inhibitors of apoptosis protein (IAP) family has been demonstrated to be crucial in apoptosis resistance in a wide range of malignancy[7,8,9,10]. These proteins are distinguished by the presence of up to three copies of Baculoviral IAP Repeat (BIR) domain. The IAPs bind to and suppress a variety of pro-apoptotic factors, thereby effectively inhibit apoptosis in cancer cells[11]. Baculoviral inhibition of apoptosis protein repeat containing 6 (BIRC6), also known as Apollon or Bruce, is the largest member of the IAP family with a single BIR domain at its N-terminal and an active ubiquitin-conjugating (UBC) enzyme domain at its C-terminal[12]. In addition to its IAP activity, BIRC6 has the distinctive property of activity as a chimeric E2/E3 ubiquitin ligase in mammals[13]. With its UBC domain, BIRC6 promotes the degradation of Smac and inhibits the activity of caspase-9, which play key roles in the initiation of apoptosis[14]. While BIRC6 itself is regulated by ubiquitination and proteasomal degradation mediated by E2, UbcH5c and Nrdp1[15].
A large amount of evidence showed that BIRC6 was highly expressed in several types of cancer. It has been reported that brain cancer cells with high levels of BIRC6 were resistant to various anticancer drugs[12]. BIRC6 overexpression was associated with unfavorable prognosis in childhood de novo acute myeloid leukemia[16]. Moreover, it has been reported that p53 is a downstream effector of BIRC6[17,18]. These findings suggest that BIRC6 maybe a new therapeutic target for malignant tumor.
Qiu et al. [14] first proved that siRNAs targeting BIRC6 promoted apoptotic cell death. Subsequently, several studies confirmed that silencing BIRC6 caused elevated apoptosis[17,18,19]. In addition, preliminary investigation demonstrated that BIRC6 expression was more abundant in CRC tissues than in non-neoplastic tissues using cDNA microarrays[20]. Similar result was obtained in the comparative proteomics study of colon cancer stem cells[21]. However, there have been no reports of its prognostic relevance based on clinical data. In the present study, we explored the prognostic significance and biologic features of BIRC6 in colorectal cancer. Our data showed a strong correlation between the BIRC6 overexpression and the unfavorable clinical features. In addition, BIRC6 upregulation in CRC predicted poor prognosis of patients. Furthermore, We found that BIRC6 knockdown inhibited CRC cell proliferation, arrested CRC cell cycle at S phase, downregulated cyclin A2, B1, D1 and E1 levels, and sensitized CRC cells to chemotherapy.
Materials and Methods
Patients, tissue specimens and follow-up
The present study was predominantly conducted by a retrospective design. CRC samples (n = 126) were obtained from patients who received surgery at the Zhongshan Hospital, Fudan University between January 2008 and August 2008. None of them received radiotherapy. Among 126 patients, 42 Patients receieved oxaliplatin-based chemotherapy (FOLFOX4; oxaliplatin, 5-fluorouracil and leucovorin) after curative resection. The follow-up information of all participants was updated every 3 months by telephone. All patients were monitored prospectively by serum CEA, ultrasound examination, endoscope, computed tomography every 3–6 months. The diagnosis of recurrence was based on the imaging method and biopsy (if possible). Patients were followed until death or April 1, 2013, with a mean postoperative follow-up duration of 59 months. Overall survival (OS) was defined as the time from the date of surgery to death by any cause, patients alive being censored at the last follow-up. Disease-free survival (DFS) was defined as the time elapsed from surgery to the first occurrence of any of the following events: colorectal cancer distant metastasis; recurrence of colorectal cancer; development of second non-colorectal malignancy excluding basal cell carcinomas of the skin and carcinoma in situ of the cervix; or death from any cause without documentation of a cancer-related event. Patients with TNM stage IV tumors were excluded when analyzing DFS. Thirty paired fresh resection CRC tissues and the paired adjacent non-cancerous tissues were collected for Western blotting. The study was approved by the Research Ethics Committee of Zhongshan Hospital. All patients provided written consent forms for this study.
Microsatellite instability (MSI) analysis and KRAS mutation analysis
MSI analysis was examined using three mononucleotide repeat markers: BAT25, BAT26, and CAT25 as described previously[22]. KRAS mutation analysis was performed by PCR amplification of genomic DNA using the following primers: sense 5’-AAGGCCTGCTGAAAATGACTG-3’ and antisense 5’-AGAATGGTCCTGCACCAGTAA-3’.
Immunohistochemistry and evaluation
Paraffin-embedded sections of normal and tumor tissues were deparaffinized in xylene and rehydrated in a decreasing ethanol series diluted in distilled water. Following antigen retrieval with a 10 mM citrate buffer, CRC sections were incubated overnight at 4°C with the primary anti-BIRC6 polyclonal antibody (Abcam, Cambridge, MA). Following 30 min incubation with secondary antibody against HRP-conjugated-rabbit Ig, sections were developed in 3, 3’-diaminobenzidine solution under microscopic observation and counterstained with hematoxylin.
The sections were observed under a light microscopy, for a histological review, to examine tumor microheterogeneity in antigen distribution. Five randomized microscopic views of 400-fold magnification of each section were observed and scored. Both the intensity of immunohistochemical staining (0, negative; 1, weak; 2, intermediate; 3, strong) and the percentage of positive cells (0, 0% positive cells; 1, 1–10% positive cells; 2, 11–50% positive cells; 3, >50% positive cells) were evaluated. The final score of each sample was obtained by multiplying the scores of staining intensity and percentage of positive cells. Samples were classified as negative when the final scores were 0 to 3 and positive when 4 to 9 [23]. The BIRC6 staining was scored independently by two pathologists blinded to the clinical characteristics of the patients. The intra-observer reproducibility was tested by obtaining the widely used statistical κ-scores that grade the strength of agreement to six categories [poor (κ-score, <0.00), slight (0.00–0.20), fair (0.21–0.40), moderate (0.41–0.60), substantial (0.61–0.80) and almost perfect (0.81–1.00)][24].
Cell culture
LoVo, SW620, DLD-1, HT-29, HCT116, SW480 and SW1116 were purchased from the Type Culture Collection of Chinese Academy of Sciences (Shanghai, China), human colon healthy cell line NCM460 were purchased from INCELL corporation. The SW620, HT-29, SW480 and HCT116 cells were routinely maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT). LoVo, DLD-1 and SW1116 were maintained in RPMI 1640 medium plus 10% fetal bovine serum. NCM460 were grown in M3:10 medium. Cells were maintained at 37°C in a humidified incubator under 5% CO2.
Establishment of stable BIRC6-knockdown clones
Lentiviral transduction was used to establish BIRC6 knockdown stable clones. A panel of shRNA (short hairpin RNA) lentiviral vectors differed in BIRC6-targeting sequences (TRCN0000041-57~61) and pLKO.1-puro empty vector were purchased from Sigma Aldrich (St Louis, MO, USA). Cells were transfected at a MOI of 1 for 24h, screened by puromycin. The inhibition efficiency was identified by Western blotting.
Western blotting
Colorectum tissue or harvested cells were homogenized in SDS lysis buffer (40 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% SDS, 1 mM aprotinin, 1 mM PMSF and 10 mg/ml leupeptin) and then centrifuged for 15 min at 4°C. Total protein lysates extracted from samples were quantitated with BCA Protein Assay Kit (Pierce, Rockford, IL, USA). An aliquot of protein was boiled with protein loading buffer for 5 min, and was loaded on SDS polyacrylamide gel. Equal amounts of protein were separated by 8% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA) using a mini trans-blot apparatus (Bio-Rad Laboratories, Hercules, CA, USA). Membranes were blocked with PBS-0.05% Tween 20 containing 5% nonfat dry milk for 1 h, followed by incubation with antibody against BIRC6 or glyceraldehyde-3-phosphate dehydrogenase antibody at 4°C overnight. The membrane was then incubated with secondary horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse antibody (Jackson Immune Research Laboratories Inc, West Grove, PA). Blots were developed using an enhanced chemiluminescence kit (Tiangen, China). Each experiment was repeated at least three times.
Cell proliferation assay
Cell proliferation was measured using the Cell Counting Kit-8 (CCK-8) (Dojindo Co., Kumamoto, Japan) according to the instructions of the supplier. Cells were incubated with CCK-8 for 1 hour with 3 multiples and proliferation rate was assessed by measuring the absorbance at 450 nm with the Universal Micro-plate Reader. Each experiment was repeated three times[25].
Colony formation assay
Anchorage-independent growth ability was measured using soft agar colony formation assay and plate colony formation assay respectively. For soft agar colony formation, control or stable BIRC6-knockdown cells (1 × 103 cells/well) were resuspended in DMEM or RPMI 1640 containing 0.3% noble agarose (Takara Shuzo Co., Tokyo, Japan) in six-well plates. The suspension was laid over DMEM or RPMI 1640 containing 0.6% noble agarose and further covered with 0.5 ml DMEM or RPMI 1640. The plates were then incubated in a 5% CO2 incubator at 37°C for 14 days, with replenishment of medium every other day. Colonies were imaged using Nikon ECLIPSE TE300 and macroscopically visible colonies in 3 randomly chosen fields per well were counted for quantification. For plate colony formation assay, cells from different treated groups were seeded into six-well plates (800 cells/well) for 2 weeks. Colonies were stained with 0.1% crystal violet in 20% methanol and colonies containing more than 50 cells were counted. Plating efficiency was calculated as: plating efficiency = (colony number/plating cell number) × 100%, in triplicate.
Cell cycle and apoptosis assay
The effect of BIRC6 knockdown on the cell cycle was analyzed by flow cytometry. Cells were trypsinized and fixed with ice-cold 70% ethanol at 4°C overnight. For DNA content analysis, cells were incubated with 50 μg/mL propidium iodide (Sigma, St. Louis, MO) in the presence of 100 μg/mL RNase and 0.2% Triton X-100 for 30 minutes at 37°C. DNA content was determined in the BD FACSAriaII. The distribution of cells in each phase of the cell cycle was calculated using the Flowjo Program. Each experiment was performed in triplicate.
For apoptosis analysis by flow cytometry, cells were treated with 5-FU, CDDP, oxaliplatin or CPT-11 (All from Sigma, St. Louis, MO)[26]. Drug-induced apoptosis was analyzed using Annexin V assay kit (BD Biosciences, San Jose, CA) according to the instructions of the manufacturer.
Tumor xenograft models
Twenty male Balb/c nude mice (4 weeks of age, 12–14 g) were purchased from Experimental Animals Center of Shanghai Institute of Life Science (Shanghai, China) and were raised under specific pathogen-free conditions. All surgery was performed under anesthesia with sodium pentobarbital. DLD-1 cells (stable BIRC6 knockdown 59 and the control clones, 106) in 0.1 ml of PBS were injected subcutaneously into the right flank of each mouse. Mice were sacrificed at 4 weeks post-injection; tumors were excised and weighed. Tumor volume was calculated by the formula: 0.5 × L × W2 (L = length of tumor; W = width of tumor). Animal experiments were performed according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals, prepared by the National Academy of Sciences and published by the National Institutes of Health, and also approved by the ethics committee of Fudan University.
Statistical analysis
Statistical analysis was performed using IBM SPSS statistical software (version 17.0). Categorical variables were compared by χ2 test or Continuity Correction or Fisher’s exact tests as appropriate. Continuous variables were compared using Wilcoxon ranksum test and independent two sample t-test. Univariate analysis were performed to identify the factors that affect survival of CRC. Multivariate analysis were done using the Cox multivariate proportional hazard regression model. Survival curves were estimated using the Kaplan-Meier method (the log-rank test). All data were presented as mean ± standard error of the mean (SEM) from three independent experiments and P-values were determined from 2-sided tests. Statistical significance was displayed as *, P < 0.05 or **, P < 0.01.
Results
Enhanced BIRC6 expression in CRC cells lines and clinic CRC tissues
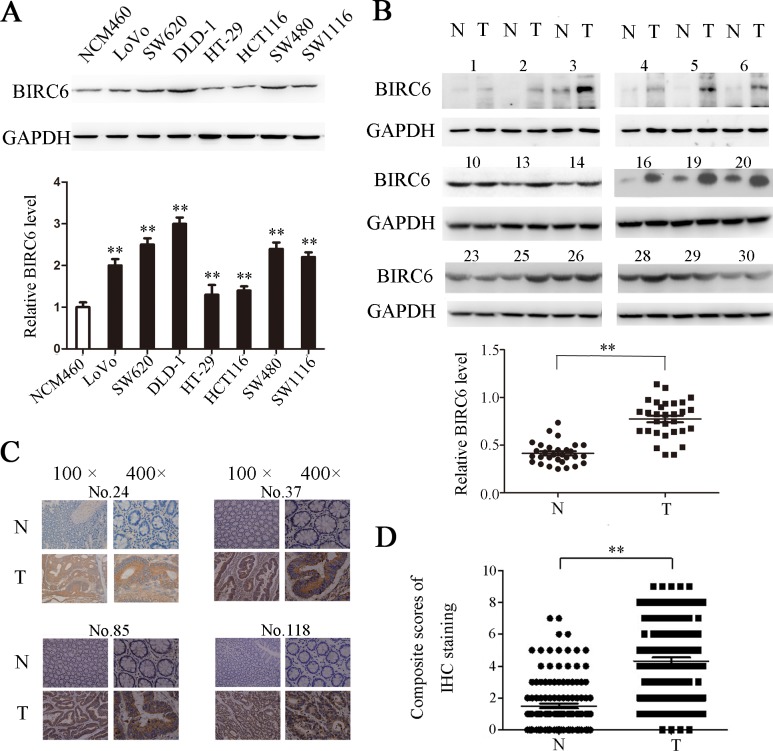
We first detected the BIRC6 expression in 7 CRC cells. Western blotting showed that BIRC6 was overexpressed in LoVo, SW620, DLD-1, HT-29, HCT116, SW480 and SW1116, whereas it was weakly detected in normal colonic epithelial cell line NCM460 (Fig 1A). We next performed Western blotting to examine the BIRC6 expression in 30 paired CRC tissues and adjacent nontumorous tissues. The data implied that BIRC6 was elevated in tumor tissues (Fig 1B). We further assessed the BIRC6 expression in 126 CRC patients by immunohistochemistry. As a result, significant BIRC6 staining was detected in CRC tissues (positive, 73 of 126), whereas the staining in corresponding normal tissues was much weaker (positive, 17 of 126) (Fig 1C and 1D). Notably, the reproducibility of our classification of BIRC6 expression was found to be ‘almost perfect’ (κ-value, 0.816) when the 126 slides of CRC tissues were assessed by two independent observers. The results above indicated that BIRC6 was significantly upregulated in CRC cells lines and clinic CRC tissues.
Fig 1. BIRC6 was overexpressed in CRC cell lines and tumor tissues of CRC patients.
(A) Expression of BIRC6 in 1 human colon healthy cell line and 7 CRC cell lines. Upper panel: Typical patterns of BIRC6 expression in NCM460 and CRC cell lines. Lower panel: Relative intensity of BIRC6 normalized to GAPDH. Data present Mean ± SEM from three independent experiments. (B) Expression of BIRC6 in 30 paired CRC tissues (T) and adjacent nontumorous tissues (N). Upper panel: Typical patterns of BIRC6 expression in CRC tissues. Lower panel: Relative intensity of BIRC6 normalized to GAPDH. (C) Immunohistochemistry staining of BIRC6 in tumor tissues and self-paired adjacent non-tumorous tissues from four representative CRC patients. (D) Scores of immunohistochemistry staining of BIRC6 in 126 cases. **, P < 0.01.
Correlation between BIRC6 expression and clinical pathological data
We investigated the correlation of BIRC6 expression with clinicopathologic features in 126 CRC patients. Patient clinical characteristics are listed in S1 Table. There was no significant correlation between BIRC6 expression and age, gender, tumor location, lymph node metastasis (N stage), distant metastasis (M stage), histology type, degree of differentiation, KRAS status and MSI status. However, BIRC6 expression positively correlated with tumor size (P = 0.044) and invasion depth (T stage) (P = 0.013) (Table 1).
Table 1. Correlation between BIRC6 expression and clinicopathologic characteristics.
| characteristics | BIRC6 expression in tumor tissues | |||
|---|---|---|---|---|
| Negative (n = 53) | Positive (n = 73) | P | ||
| Age (years) | ≤50 | 16 | 18 | 0.490 |
| >50 | 37 | 55 | ||
| Gender | male | 30 | 41 | 0.961 |
| female | 23 | 32 | ||
| Tumor location | colon | 27 | 45 | 0.231 |
| rectal | 26 | 28 | ||
| Tumor size (cm) | ≤5cm | 49 | 58 | 0.044 |
| >5cm | 4 | 15 | ||
| Invasion depth | T1-T2 | 15 | 8 | 0.013 |
| T3-T4 | 38 | 65 | ||
| Lymph node metastasis | N0 | 30 | 42 | 0.917 |
| N1-N2 | 23 | 31 | ||
| Distant metastasis | M0 | 44 | 58 | 0.615 |
| M1 | 9 | 15 | ||
| TNM stage | I-II | 26 | 36 | 0.977 |
| III-IV | 27 | 37 | ||
| Tumor histology type | adenocarcinoma | 47 | 61 | 0.418 |
| mucinous/signet-ring | 6 | 12 | ||
| Tumor degree of differentiation | well/moderate | 38 | 44 | 0.184 |
| poor | 15 | 29 | ||
| KRAS status | Wild type | 39 | 50 | 0.54 |
| Mutation | 14 | 23 | ||
| MSI status | MSS | 45 | 59 | 0.55 |
| MSI | 8 | 14 | ||
Pearson χ2 tests for all analyses. Bold items have been considered statistically significant. Abbreviations: TNM, tumor-node-metastasis; MSI, microsatellite instability; MSS, microsatellite stable.
Prognostic value of enhanced BIRC6 expression
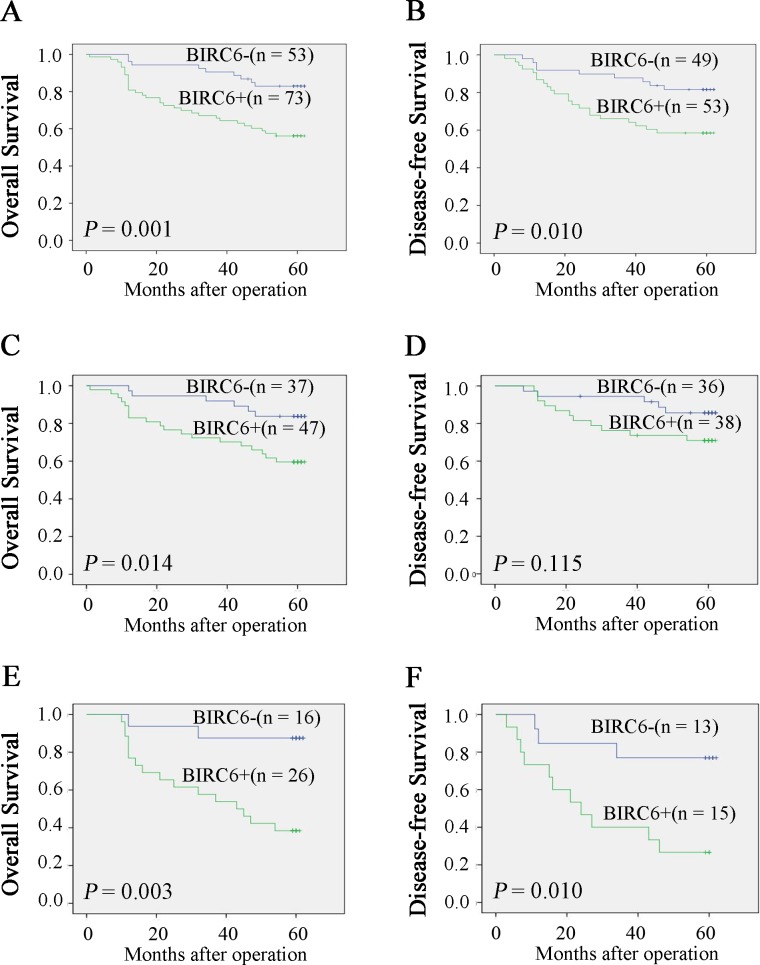
Kaplan-Meier analysis and log-rank test were used to determine the relationship between BIRC6 expression and prognosis. CRC patients with positive BIRC6 expression tended to have shorter overall survival (OS) and disease-free survival (DFS) (P = 0.001 and P = 0.010, respectively) (Fig 2A and 2B). We next divided patients into two groups: no chemotherapy group and chemotherapy group. As it showed in Fig 2C and 2D, BIRC6 expression was correlated with OS (P = 0.038) and DFS (P = 0.041) in no chemotherapy group. Similar results were observed in chemotherapy group (P = 0.003 and P = 0.010) (Fig 2E and 2F). Univariate analysis demonstrated that positive BIRC6 expression was associated with worse OS (P = 0.002) and DFS (P = 0.013) (Table 2). Other factors correlated with OS were T stage, N stage, M stage and tumor degree of differentiation. Factors affecting DFS included T stage, N stage, KRAS status and MSI status. In addition, multivariate analysis identified enhanced BIRC6 level a risk factor for both OS (P = 0.045) and DFS (P = 0.026) (Table 3).
Fig 2. High expression of BIRC6 correlated with poor survival rate.
Overall survival (A) and disease-free survival (B) between patients with positive and negative expression of BIRC6 were estimated using the Kaplan-Meier method and compared by the log rank test. Overall survival (C) and disease-free survival (D) were estimated in no chemotherapy group. Overall survival (E) and disease-free survival (F) were estimated in chemotherapy group. Patients with TNM stage IV tumors were excluded when analyzing disease-free survival.
Table 2. Univariate analysis of factors associated with survival and recurrence.
| Variables | OS | DFS | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | P | Hazard ratio (95% CI) | P | |
| Age, y (>50 vs. ≤50) | 0.985 (0.502–1.931) | 0.964 | 0.762 (0.359–1.619) | 0.479 |
| Gender (female vs. male) | 1.405 (0.761–2.594) | 0.276 | 1.531 (0.755–3.107) | 0.238 |
| Location (rectal vs. colon) | 1.039 (0.561–1.925) | 0.903 | 1.182 (0.584–2.391) | 0.642 |
| Tumor size, cm (>5 vs. ≤5) | 0.596 (0.212–1.672) | 0.325 | 0.487 (0.148–1.601) | 0.236 |
| Invasion depth (T3-T4 vs. T1-T2) | 3.174 (0.979–10.285) | 0.054 | 2.084 (0.729–5.959) | 0.171 |
| Lymph node metastasis (N1-N2 vs. N0) | 3.162 (1.656–6.039) | 0.000 | 2.585 (1.265–5.283) | 0.009 |
| Distant metastasis (M1 vs. M0) | 5.977 (3.216–11.106) | 0.000 | ||
| TNM stage (III-IV vs. I-II) | 7.573 (3.178–18.048) | 0.000 | ||
| Tumor histology type (adenocarcinoma vs. mucinous/signet-ring) | 1.263 (0.560–2.850) | 0.574 | 0.451 (0.108–1.890) | 0.276 |
| Tumor degree of differentiation (poor vs. well/moderate) | 1.923 (1.041–3.554) | 0.037 | 1.122 (0.528–2.382) | 0.765 |
| KRAS status (mutation vs. wild type) | 1.128 (0.584–2.178) | 0.720 | 2.037 (0.997–4.160) | 0.051 |
| MSI status (MSI vs. MSS) | 1.774 | 0.115 | 2.724 | 0.028 |
| (0.869–3.620) | (1.114–6.657) | |||
| Expression of BIRC6 (positive vs. negative) | 3.170 (1.512–6.647) | 0.002 | 2.673 (1.230–5.812) | 0.013 |
Bold items have been considered statistically significant. Cox proportional hazards regression model was used in univariate analysis. Patients with TNM stage IV tumors were excluded when analyzing disease-free survival. Abbreviations: 95% CI, 95% confidence interval; OS, overall survival; DFS, disease-free survival; TNM, tumor-node-metastasis; MSI, microsatellite instability; MSS, microsatellite stable.
Table 3. Multivariate analysis of factors associated with OS and DFS.
| Hazard ratio (95% CI) | P | |
|---|---|---|
| OS | ||
| Invasion depth (T3-T4 vs. T1-T2) | 0.965 (0.270–3.451) | 0.956 |
| Lymph node metastasis (N1-N2 vs. N0) | 2.406 (1.234–4.692) | 0.010 |
| Distant metastasis (M1 vs. M0) | 4.288 (2.243–8.196) | 0.000 |
| Tumor degree of differentiation (poor vs. well/moderate) | 1.171 (0.621–2.208) | 0.625 |
| Expression of BIRC6 (positive vs. negative) | 2.214 (1.019–4.810) | 0.045 |
| DFS | ||
| Invasion depth (T3-T4 vs. T1-T2) | 1.028 (0.324–3.260) | 0.963 |
| Lymph node metastasis (N1-N2 vs. N0) KRAS status (mutation vs. wild type) MSI status (MSI vs. MSS) | 2.198 (1.036–4.666) 1.743 (0.840–3.619) 3.138 (1.259–7.825) | 0.040 0.136 0.014 |
| Expression of BIRC6 (positive vs. negative) | 2.564 (1.117–5.886) | 0.026 |
Bold items have been considered statistically significant. Multivariate analysis and Cox proportional hazards regression model were used. Variables were adopted for their prognostic significance by univariate analysis (P < 0.2). Abbreviations: 95% CI, 95% confidence interval; OS, overall survival; DFS, disease-free survival; MSI, microsatellite instability; MSS, microsatellite stable.
BIRC6 knockdown inhibited CRC cell proliferation
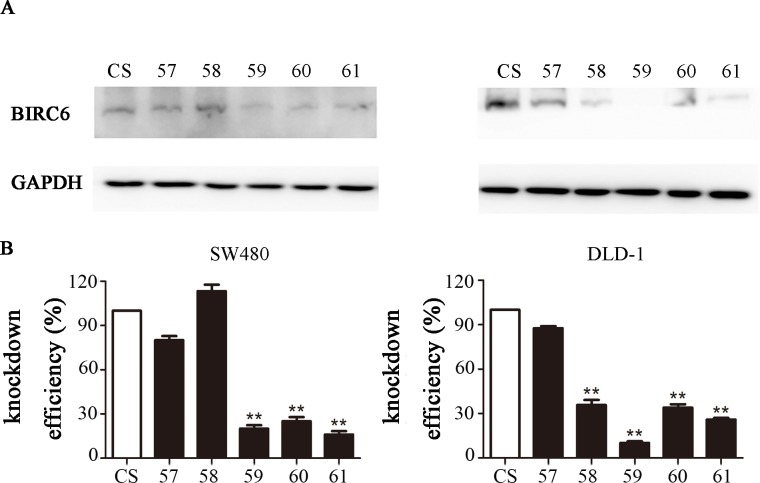
Since the full-length cDNA of BIRC6 extends for 14.5 kb, it is difficult to overexpress BIRC6 in a cell line. Instead, we used lentiviral transduction to establish BIRC6 knockdown stable clones in two CRC cell lines: SW480 and DLD-1. The down-regulated BIRC6 expression was observed significantly in two BIRC6-knockdown cell lines (59 and 61), as shown by Western blotting (Fig 3). These two clones were used in the subsequent analysis.
Fig 3. BIRC6 knockdown stable clones were established by lentiviral transduction.
(A) BIRC6 knockdown efficiency was testified by Western blotting. (B) Relative expression of BIRC6 normalized to GAPDH was calculated. **, P < 0.01 vs. control.
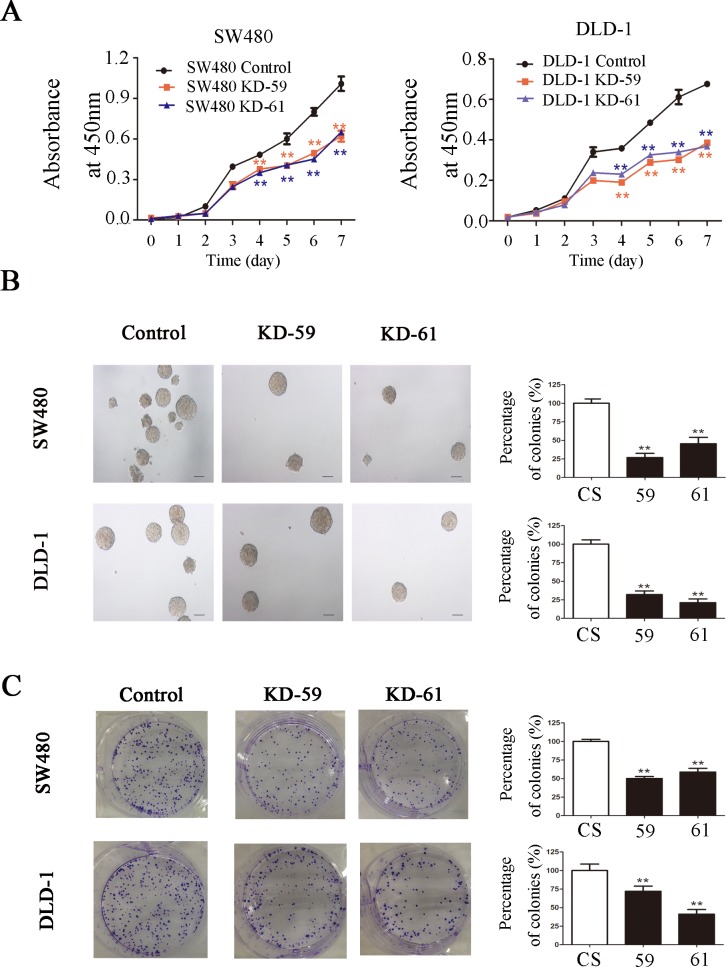
CCK-8 assay showed that BIRC6 knockdown significantly inhibited the proliferation of SW480 and DLD-1 cells in a time-dependent manner in vitro (Fig 4A). Colony formation assay showed that BIRC6 knockdown reduced colony formation in agar in SW480 and DLD-1 cells (Fig 4B). Similar results were observed in colony formation in plates (Fig 4C).
Fig 4. BIRC6 knockdown inhibited CRC cell proliferation.
(A) Cell-growth curves of control (CS) and two BIRC6 knockdown clones (59 and 61). (B) Representative images of soft agar colony formation assay and relative levels of colonies in BIRC6 knockdown clones normalized to the control. (C) Representative images of plate colony formation assay and relative levels of colonies in BIRC6 knockdown clones normalized to the control. Data present Mean ± SEM from three independent experiments. **, P < 0.01 vs. control.
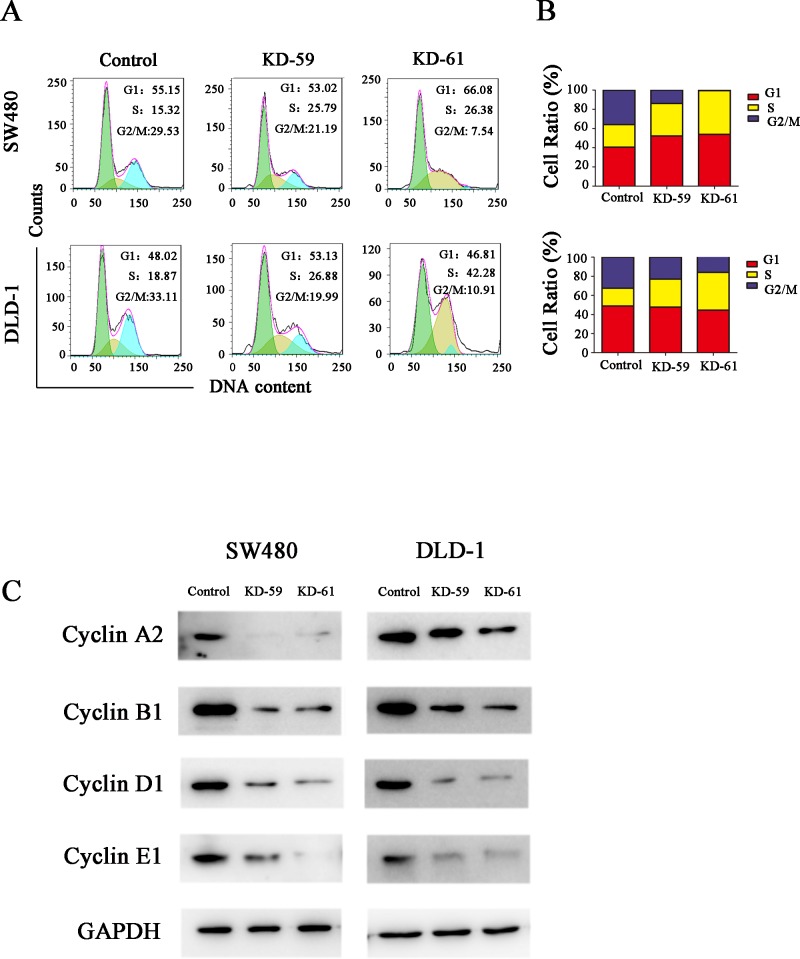
BIRC6 knockdown induced cell cycle arrest at S phase and downregulated cyclin A2, B1, D1 and E1 levels in CRC cells
Cell cycle analysis showed that the percentage of cells in S phase increased while the percentage of cells in G2/M phase decreased after BIRC6 knockdown in both SW480 cells and DLD-1 cells (Fig 5A and 5B), which indicated that BIRC6 knockdown prevented the CRC cells from entering the mitotic phase. To check whether BIRC6 could regulate the genes involved in cell-cycle control, we detected the effect of BIRC6 on the expression of several cyclins. Western blotting indicated that BIRC6 knockdown decreased the levels of cyclin A2, B1, D1 and E1 in both SW480 cells and DLD-1 cells (Fig 5C).
Fig 5. BIRC6 knockdown induced cell cycle arrest at S phase.
(A) Representative images of cell cycle assessed by PI staining. (B) Quantification of the cell cycle distribution. Control cells were cells transfected with pLKO.1-puro empty vector; KD-59 and KD-61 were cells transfected with shRNA lentiviral vectors differed in BIRC6-targeting sequences. (C) Expression of cyclin A2, B1, D1 and E1 were performed by Western blotting in the control and two knockdown clones.
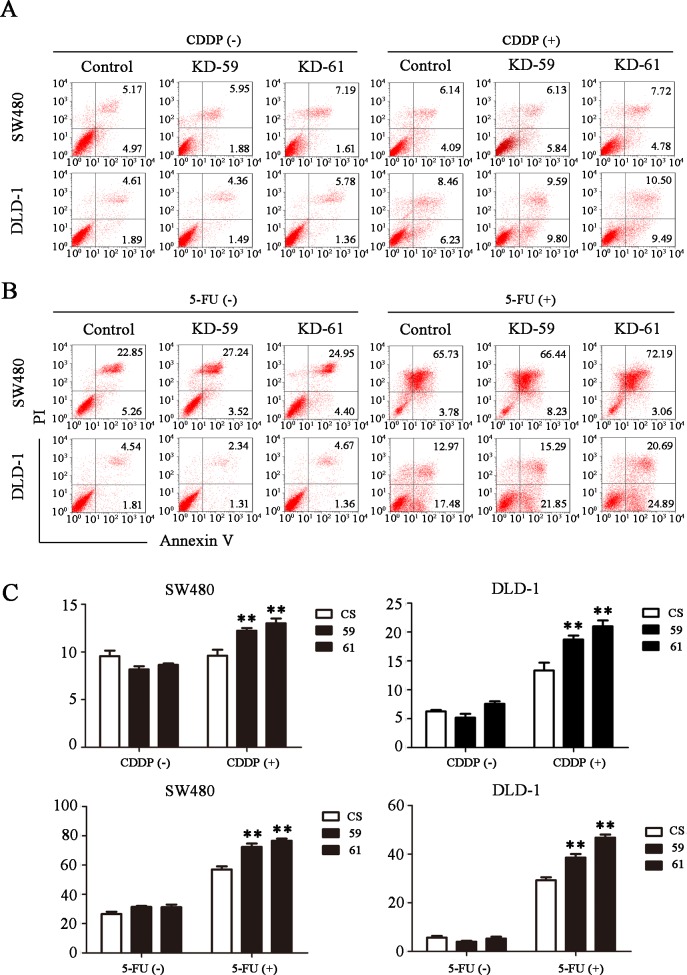
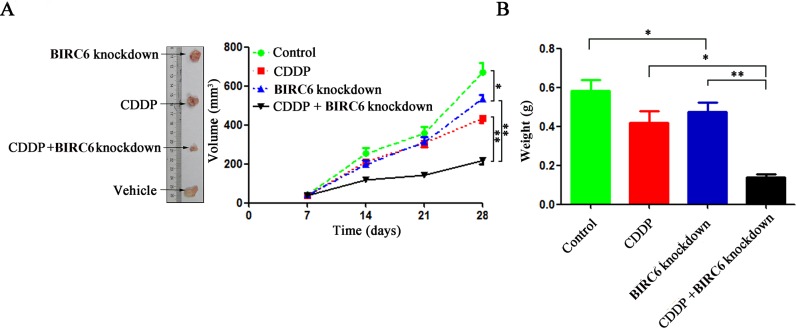
BIRC6 knockdown sensitized CRC cells to chemotherapy in vitro and in vivo
We investigate the role of BIRC6 in chmosensitivity of ESCC cells. Though BIRC6 knockdown alone did not induce apoptosis in SW480 cells and DLD-1 cells, BIRC6 knockdown sensitized SW480 cells and DLD-1 cells to chemo-induced apoptosis including 5-FU, CDDP, oxaliplatin and CPT-11 (Figs 6A and 6B and S2). The finding that BIRC6 knockdown inhibited cell growth and sensitized CRC cells to chemo-induced apoptosis in vitro prompted us to determine whether it exerts a similar effect in vivo. Control or BIRC6 stable knockdown clones of DLD-1 were subcutaneously inoculated into nude mice. When all animals had established tumors averaging from 50 to 100 mm3, tumorbearing mice were treated with CDDP (4 mg/kg; i.p., twice weekly)[27]. The antitumor efficacy was measured by monitoring the tumor volume and weight after treatment. BIRC6 knockdown slightly inhibited tumor growth. Interestingly, combination of BIRC6 knockdown and CDDP treatment significantly inhibited the tumor growth compared with either BIRC6 knockdown or CDDP treatment alone (Fig 7A and 7B).
Fig 6. BIRC6 knockdown sensitized CRC cells to cisplatin (CDDP) or 5-fluorouracil (5-FU)-induced apoptosis.
(A) Representative images of cell apoptosis assessed by Annexin V/PI staining in the control (CS) and BIRC6 knockdown cells treated with CDDP (40 μM for SW480 or 100 μM for DLD-1) for 24 h or not. (B) Representative images of cell apoptosis for the control and BIRC6 knockdown cells treated with 5-FU (75 μg/mL for SW480 or 50 μg/mL for DLD-1) for 48 h or not. (C) Quantification of the apoptosis rate. Data present Mean ± SEM from three independent experiments. **, P < 0.01 vs. control.
Fig 7. BIRC6 knockdown sensitized CRC cells to chemotherapy in a xenograft nude mouse model.
DLD-1 cells stably transfected with control shRNA and BIRC6 knockdown shRNA (59) were injected subcutaneously into nude mice. Tumorbearing mice were treated with CDDP (4 mg/kg; i.p., twice weekly). (A) The tumor size was monitored. (B) Total tumor weight of each group of mice was recorded. Data present Mean ± SEM of 5 mice of each group. *, P < 0.05; **, P < 0.01.
Discussion
Acquired resistance to apoptosis is one of the defining hallmarks of cancer[28]. IAPs, a group of anti-apoptosis proteins that are highly evolutionarily conserved across several species, are key regulators of apoptosis[29]. BIRC6, also named as Apollon, is the largest member of the IAP family with 4830 amino acid residues[12]. It is also the only known IAP member which is membrane associated and localizes to the Golgi compartment and the vesicular system[30]. BIRC6 is highly conserved in different species, suggesting a common biological function. Sequence analysis showed that BIRC6 shared about 92% amino acid identity with Bruce, a member of IAP family from mouse[12]. In addition, there exists the Drosophila homolog of BIRC6 called dBruce[31]. Unlike most of the other mammalian IAPs, BIRC6 have been shown to be essential for viability. Complete inactivation of the Bruce gene led to perinatal lethality and growth retardation in mice embryos after embryonic day 14[32]. Likewise, deletion of the C-terminal half of Bruce caused activation of caspases and apoptosis in the placenta and yolk sac, and resulted in embryonic lethality[17]. Moreover, plenty of evidence indicated that BIRC6 was overexpressed in a variety of cancers including glioma, childhood de novo acute myeloid leukemia, breast cancer, neuroblastoma, prostate cancer and non-small-cell lung cancer, and overexpression of BIRC6 was correlated with carcinogenesis, progression and poor prognosis of malignant tumor[12,16,18,33,34,35]. Although the previous study demonstrated that BIRC6 was up-regulated in colorectal cancer using cDNA microarray and qRT-PCR analysis, the relationship between BIRC6 expression and colorectal cancer progression has not been extensively studied.
In the present study, we observed that the expression of BIRC6 could be detected in both colorectal cancer and adjacent tissues but with different percentage. The level of BIRC6 was obviously upregulated in CRC tissues compared with adjacent nontumorous tissues. We also demonstrated that high expression of BIRC6 was associated with adverse clinical and pathologic features in colorectal cancer. Moreover, patients with positive BIRC6 expression tended to survive shorter and had a higher risk of recurrence than those with negative BIRC6 expression. Univariate and multivariate analysis showed that BIRC6 expression was significantly correlated with OS and DFS, indicating that BIRC6 was a predictor of prognosis.
Similarly, BIRC6 was also overexpressed in 7 CRC cell lines. We applied RNAi approach to suppress BIRC6 expression and explored the role of BIRC6 in CRC progression. Our results showed that knockdown of BIRC6 reduced CRC cell proliferation, which is consistent with the previous study in several other tumors. The cell cycle analysis indicated that BIRC6 knockdown resulted in S phase block, thus preventing CRC cells from entering into mitosis. Moreover, We found that BIRC6 knockdown decreased cyclin A2, B1, D1 and E1 levels in CRC cells. We proposed that decreased cyclins levels may contribute to BIRC6 knockdown-induced S phase block. Consistent with our hypothesis, Lee et al. [36] found that compound which consist of hinokitiol decreased the levels of cyclin A and cyclin E and led to S phase arrest in HCT116 and SW620 cells. Joe et al. [37] reported that resveratrol decreased the levels of cyclins D1, A, and B1 and resulted in S phase arrest in SW480 cells. However, unlike our results, Kikuchi et al. [38] found that BIRC6 ubiquitylated cyclin A and BIRC6 deficiency accumulated more cyclin A in 293T cells. Accordingly, the effect of BIRC6 on cyclins appears to depend on cell types and target proteins. Finally, we found that BIRC6 knockdown sensitized CRC cells to chemotherapy both in vitro and in vivo, which provides a rationale for combining BIRC6 antagonism with chemotherapy to treat CRC.
The present study has certain limitations. For example, the correlation between BIRC6 and prognosis was determined in a limited number of CRC patients (n = 126), and this correlation required confirmation on a larger-scale basis. In the same sense, the fact that we did not observe a significant correlation between BIRC6 expression and MSI status needs further investigation in larger samples before firmer conclusions can be drawn. It's worth noting that we found that BIRC6 regulated cell cycle progression via modulation of cell-cycle-regulation proteins. While increasing evidence have suggested a biological correlation between MSI and alterations in cell-cycle-regulation proteins in cancer[39,40].
In conclusion, the present study provides evidence that BIRC6 functions as a prognostic factor of human CRC. Moreover, our results suggest that BIRC6 knockdown in combination with chemotherapy may have therapeutic potential in the treatment of human CRC.
Supporting Information
(DOCX)
(DOCX)
Representative images of immunohistochemistry.
(TIF)
(A) Representative images of cell apoptosis assessed by Annexin V/PI staining in the control (CS) and BIRC6 knockdown cells treated with oxaliplatin (10 μM for SW480 and DLD-1) for 48 h or not. (B) Representative images of cell apoptosis for the control and BIRC6 knockdown cells treated with CPT-11 (4 μM for SW480 and DLD-1) for 24 h or not. (C) Quantification of the apoptosis rate. Data present Mean ± SEM from three independent experiments. *, P < 0.05; **, P < 0.01 vs. control.
(TIF)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Shanghai Science and Technology Commission (grant numbers: 10411950100 to XS, 12ZR1405300 to YC), National Natural Science Foundation of China (grant numbers: 81100344, 81173078 to SZ, 81371268 to SC, 81172273, 81272388 to LD, 81472673 to YC, 81272388, 81400628 to XS), the National Clinical Key Special Subject of China (grant number: 371 to XS), and Shanghai Pujiang Program(grant number: No.13PJD008 to GC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 2. Akin O, Brennan SB, Dershaw DD, Ginsberg MS, Gollub MJ, Schöder H, et al. (2012) Advances in oncologic imaging: update on 5 common cancers. CA Cancer J Clin 62: 364–393. 10.3322/caac.21156 [DOI] [PubMed] [Google Scholar]
- 3. Grothey A, Sargent D, Goldberg RM, Schmoll HJ (2004) Survival of patients with advanced colorectal cancer improves with the availability of fluorouracil-leucovorin, irinotecan, and oxaliplatin in the course of treatment. J Clin Oncol 22: 1209–1214. [DOI] [PubMed] [Google Scholar]
- 4. Fuchs CS, Marshall J, Barrueco J (2008) Randomized, controlled trial of irinotecan plus infusional, bolus, or oral fluoropyrimidines in first-line treatment of metastatic colorectal cancer: updated results from the BICC-C study. J Clin Oncol 26: 689–690. 10.1200/JCO.2007.15.5390 [DOI] [PubMed] [Google Scholar]
- 5. Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, Braud FD, et al. (2009) Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol 27: 663–671. 10.1200/JCO.2008.20.8397 [DOI] [PubMed] [Google Scholar]
- 6. Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, et al. (2010) Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol 28: 4697–4705. 10.1200/JCO.2009.27.4860 [DOI] [PubMed] [Google Scholar]
- 7. Srinivasula SM, Ashwell JD (2008) IAPs: what's in a name? Mol Cell 30: 123–135. 10.1016/j.molcel.2008.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Athanasoula K, Gogas H, Polonifi K, Vaiopoulos AG, Polyzos A, Mantzourani M (2014) Survivin beyond physiology: orchestration of multistep carcinogenesis and therapeutic potentials. Cancer Lett 347: 175–182. 10.1016/j.canlet.2014.02.014 [DOI] [PubMed] [Google Scholar]
- 9. Fulda S (2014) Inhibitor of Apoptosis (IAP) proteins in hematological malignancies: molecular mechanisms and therapeutic opportunities. Leukemia 28: 1414–1422. 10.1038/leu.2014.56 [DOI] [PubMed] [Google Scholar]
- 10. Guicciardi ME, Werneburg NW, Bronk SF, Franke A, Yagita H, Thomas G, et al. (2014) Cellular inhibitor of apoptosis (cIAP)-mediated ubiquitination of phosphofurin acidic cluster sorting protein 2 (PACS-2) negatively regulates tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) cytotoxicity. PLOS One 9: e92124 10.1371/journal.pone.0092124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. LaCasse EC, Baird S, Korneluk RG, MacKenzie AE (1998) The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene 17: 3247–3259. [DOI] [PubMed] [Google Scholar]
- 12. Chen Z, Naito M, Hori S, Mashima T, Yamori T, Tsuruo T (1999) A human IAP-family gene, apollon, expressed in human brain cancer cells. Biochem Biophys Res Commun 264: 847–854. [DOI] [PubMed] [Google Scholar]
- 13. Bartke T, Pohl C, Pyrowolakis G, Jentsch S (2004) Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol Cell 14: 801–811. [DOI] [PubMed] [Google Scholar]
- 14. Qiu XB, Goldberg AL (2005) The membrane-associated inhibitor of apoptosis protein, BRUCE/Apollon, antagonizes both the precursor and mature forms of Smac and caspase-9. J Biol Chem 280: 174–182. [DOI] [PubMed] [Google Scholar]
- 15. Qiu XB, Markant SL, Yuan J, Goldberg AL (2004) Nrdp1-mediated degradation of the gigantic IAP, BRUCE, is a novel pathway for triggering apoptosis. EMBO J 23: 800–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sung KW, Choi J, Hwang YK, Lee SJ, Kim HJ, Lee SH, et al. (2007) Overexpression of Apollon, an antiapoptotic protein, is associated with poor prognosis in childhood de novo acute myeloid leukemia. Clin Cancer Res 13: 5109–5114. [DOI] [PubMed] [Google Scholar]
- 17. Ren J, Shi M, Liu R, Yang QH, Johnson T, Skarnes WC, et al. (2005) The Birc6 (Bruce) gene regulates p53 and the mitochondrial pathway of apoptosis and is essential for mouse embryonic development. Proc Natl Acad Sci U S A 102: 565–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lopergolo A, Pennati M, Gandellini P, Orlotti NI, Poma P, Daidone MG, et al. (2009) Apollon gene silencing induces apoptosis in breast cancer cells through p53 stabilisation and caspase-3 activation. Br J Cancer 100: 739–746. 10.1038/sj.bjc.6604927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chu L, Gu J, Sun L, Qian Q, Qian C, Liu X (2008) Oncolytic adenovirus-mediated shRNA against Apollon inhibits tumor cell growth and enhances antitumor effect of 5-fluorouracil. Gene Ther 15: 484–494. 10.1038/gt.2008.6 [DOI] [PubMed] [Google Scholar]
- 20. Bianchini M, Levy E, Zucchini C, Pinski V, Macagno C, Sanctis P, et al. (2006) Comparative study of gene expression by cDNA microarray in human colorectal cancer tissues and normal mucosa. Int J Oncol 29: 83–94. [PubMed] [Google Scholar]
- 21. Van Houdt WJ, Emmink BL, Pham TV, Piersma SR, Verheem A, Vries RG, et al. (2011) Comparative proteomics of colon cancer stem cells and differentiated tumor cells identifies BIRC6 as a potential therapeutic target. Mol Cell Proteomics 10: M111–M11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Findeisen P, Kloor M, Merx S, Sutter C, Woerner SM, Dostmann N, et al. (2005) T25 repeat in the 3' untranslated region of the CASP2 gene: a sensitive and specific marker for microsatellite instability in colorectal cancer. Cancer Res 18:8072–8078. [DOI] [PubMed] [Google Scholar]
- 23. Yang XR, Xu Y, Yu B, Zhou J, Li JC, Qiu SJ, et al. (2009) CD24 is a novel predictor for poor prognosis of hepatocellular carcinoma after surgery. Clin Cancer Res 15: 5518–5527. 10.1158/1078-0432.CCR-09-0151 [DOI] [PubMed] [Google Scholar]
- 24. Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33: 159–174. [PubMed] [Google Scholar]
- 25. Zhao N, Wang X, Zhang Y, Gu Q, Huang F, Zheng W, et al. (2013) Gestational zinc deficiency impairs humoral and cellular immune responses to hepatitis B vaccination in offspring mice. PLOS One 8: e73461 10.1371/journal.pone.0073461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Akiyoshi T, Nakamura M, Yanai K, Nagai S, Wada J, Koga K, et al. (2008) Gamma-secretase inhibitors enhance taxane-induced mitotic arrest and apoptosis in colon cancer cells. Gastroenterology 134: 131–144. 10.1053/j.gastro.2007.10.008 [DOI] [PubMed] [Google Scholar]
- 27. Suzuki K, Kokuryo T, Senga T, Yokoyama Y, Nagino M, Hamaguchi M (2010) Novel combination treatment for colorectal cancer using Nek2 siRNA and cisplatin. Cancer Sci 101: 1163–1169. 10.1111/j.1349-7006.2010.01504.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57–70. [DOI] [PubMed] [Google Scholar]
- 29. LaCasse EC, Mahoney DJ, Cheung HH, Plenchette S, Baird S, Korneluk RG (2008) IAP-targeted therapies for cancer. Oncogene 27: 6252–6275. 10.1038/onc.2008.302 [DOI] [PubMed] [Google Scholar]
- 30. Hauser HP, Bardroff M, Pyrowolakis G, Jentsch S (1998) A giant ubiquitin-conjugating enzyme related to IAP apoptosis inhibitors. J Cell Biol 141: 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vernooy SY, Chow V, Su J, Verbrugghe K, Yang J, Cole S, et al. (2002) Drosophila Bruce can potently suppress Rpr- and Grim-dependent but not Hid-dependent cell death. Curr Biol 12: 1164–1168. [DOI] [PubMed] [Google Scholar]
- 32. Lotz K, Pyrowolakis G, Jentsch S (2004) BRUCE, a giant E2/E3 ubiquitin ligase and inhibitor of apoptosis protein of the trans-Golgi network, is required for normal placenta development and mouse survival. Mol Cell Biol 24: 9339–9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lamers F, Schild L, Koster J, Speleman F, Ora I, Westerhout EM, et al. (2012) Identification of BIRC6 as a novel intervention target for neuroblastoma therapy. BMC Cancer 12: 285 10.1186/1471-2407-12-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Low CG, Luk IS, Lin D, Fazli L, Yang K, Xu Y, et al. (2013) BIRC6 protein, an inhibitor of apoptosis: role in survival of human prostate cancer cells. PLOS One 8: e55837 10.1371/journal.pone.0055837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dong X, Lin D, Low C, Vucic EA, English JC, Yee J, et al. (2013) Elevated expression of BIRC6 protein in non-small-cell lung cancers is associated with cancer recurrence and chemoresistance. J Thorac Oncol 8: 161–170. 10.1097/JTO.0b013e31827d5237 [DOI] [PubMed] [Google Scholar]
- 36. Lee YS, Choi KM, Kim W, Jeon YS, Lee YM, Hong JT, et al. (2013) Hinokitiol inhibits cell growth through induction of S-phase arrest and apoptosis in human colon cancer cells and suppresses tumor growth in a mouse xenograft experiment. J Nat Prod 76: 2195–2202. 10.1021/np4005135 [DOI] [PubMed] [Google Scholar]
- 37. Joe AK, Liu H, Suzui M, Vural ME, Xiao D, Weinstein IB (2002) Resveratrol induces growth inhibition, S-phase arrest, apoptosis, and changes in biomarker expression in several human cancer cell lines. Clin Cancer Res 8: 893–903. [PubMed] [Google Scholar]
- 38. Kikuchi R, Ohata H, Ohoka N, Kawabata A, Naito M (2014) APOLLON protein promotes early mitotic CYCLIN A degradation independent of the spindle assembly checkpoint. J Biol Chem 289: 3457–3467. 10.1074/jbc.M113.514430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morikawa T, Kuchiba A, Liao X, Imamura Y, Yamauchi M, Qian ZR, et al. (2012) Tumor TP53 expression status, body mass index and prognosis in colorectal cancer. Int J Cancer 131: 1169–1178. 10.1002/ijc.26495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zaanan A, Meunier K, Sangar F, Flejou JF, Praz F (2011) Microsatellite instability in colorectal cancer: from molecular oncogenic mechanisms to clinical implications. Cell Oncol (Dordr) 34: 155–176. 10.1007/s13402-011-0024-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Representative images of immunohistochemistry.
(TIF)
(A) Representative images of cell apoptosis assessed by Annexin V/PI staining in the control (CS) and BIRC6 knockdown cells treated with oxaliplatin (10 μM for SW480 and DLD-1) for 48 h or not. (B) Representative images of cell apoptosis for the control and BIRC6 knockdown cells treated with CPT-11 (4 μM for SW480 and DLD-1) for 24 h or not. (C) Quantification of the apoptosis rate. Data present Mean ± SEM from three independent experiments. *, P < 0.05; **, P < 0.01 vs. control.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.