Abstract
Our understanding of cell biology and its integration with materials science has led to technological innovations in the bioengineering of tissue-mimicking grafts that can be utilized in clinical and pharmaceutical applications. Bio-engineering of native-like multiscale building blocks provides refined control over the cellular microenvironment, thus enabling functional tissues. In this review, we focus on assembling building blocks from the biomolecular level to the millimeter scale. We also provide an overview of techniques for assembling molecules, cells, spheroids, and microgels and achieving bottom-up tissue engineering. Additionally, we discuss driving mechanisms for self- and guided assembly to create micro-to-macro scale tissue structures.
Starting from bottom to top
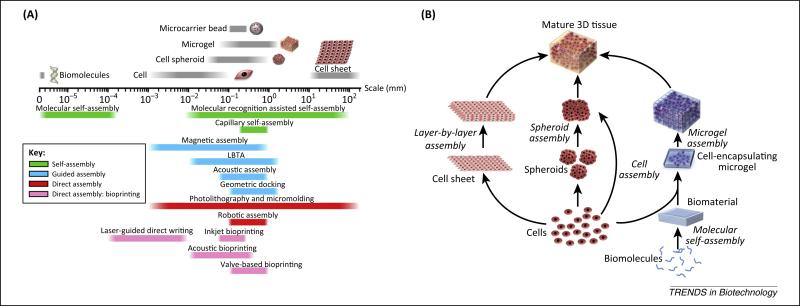
In nature, biological systems are highly hierarchical in structure, spanning a broad scale ranging from the molecular to macroscopic level. This complex hierarchy in tissues is formed by building blocks that enable and regulate the system function. To engineer such complex tissues, biomanufacturing tools that can generate and manipulate multi-scale building blocks are needed. Bottom-up tissue engineering represents a promising strategy to create tissues by assembling heterogeneous building blocks such as peptides, cells, and cell-encapsulating microscale hydro-gels in a multiscale fashion [1–3]. In this review, we highlight the principles of engineering and assembling complex tissues using emerging biomanufacturing techniques (Figure 1A). We also focus on applicable assembly techniques for different scales of building blocks. At the molecular level, self-assembly dominates the formation of complex structures comprising biomolecules such as peptides, oligosaccharides, and nucleic acids, and is driven by charge interactions and/or biochemical binding affinities. Assembly approaches including magnetic assembly, liquid-based template assembly (LBTA), molecular/geometric recognition, and bioprinting are used to assemble building blocks such as cells and cell spheroids [2,4–6]. By combining these biomanufacturing tools, 3D tissues can be assembled using biomolecules, biomaterials, cells, cell spheroids, microgels, and cell sheets (Figure 1B). In vivo, cells are embedded in a 3D niche comprising extracellular matrix (ECM) and neighboring cells with a well-defined spatial distribution. Biomaterials mimicking the native ECM are critically important to form cell-encapsulating building blocks for the bioengineering of artificial tissues (Box 1). Finally, we explore future perspectives on assembly approaches for bottom-up tissue engineering in high-throughput systems.
Figure 1.
(A) Corresponding technologies for assembling building blocks at different scales. The size of each biological entity is shown above the scale axis, while the sample size that each assembly technology can manipulate is shown below the scale axis. (B) Schematic of multiscale assembly strategies from bottom to top for engineering 3D tissue constructs. The assembly strategies can follow paths starting with biomolecules or cells and can be integrated in the engineering of the final 3D tissue constructs.
Biomolecular assembly strategies
Assembly of biomolecules is a spontaneous or directed process employing electrostatic interactions, chemical bonds (e.g., covalent and noncovalent bonds), and affinity-based binding. These assembly approaches incorporate biomolecules such as proteins [7], nucleic acids [8], carbohydrates [9], and lipids [10] to form scalable functional biomaterials that can be used in 3D cell culture.
Peptides for self-assembly
Amino acids can be utilized as building blocks to form scalable bioconstructs by tuning and altering specific functional groups. The most well-examined self-assembling peptides in tissue engineering are the derivatives of arginine– alanine–aspartate (RAD) sequences, which generate nanofibers enhancing cell adhesion, growth, and function [7]. For example, the length of neuronal axons can be extended up to several hundred microns and formation of active synapses can be assisted on peptide-derived biomaterials [11]. Further, P11, Q11, β hairpins, and peptide amphiphiles can be listed as additional peptide sequences providing alignment of supramolecular fibrils over macroscopic scales [12–18].
Carbohydrates for self-assembly and molecular conjugation
Self-assembly of carbohydrates is driven by intermolecular, noncovalent interactions such as hydrogen bonding, π–π stacking, or electrostatic interactions. Carbohydrates such as oligosaccharides and glycopolypeptides have highly specific molecular recognition properties that can also form structurally complex and functional units for biomedicine [9,19]. For example, cyclodextrins are set of natural water-soluble oligosaccharides that can be held together and form cavities that can accommodate cells and therapeutics [9,20]. Combinations of polymers and cyclodextrins can form highly specific biomaterials with enhanced selectivity for loading and delivering their cargo (e.g., drugs, genes) to the target tissue or organ [9].
Nucleic acids for self-assembly and molecular conjugation
Nucleic acids have been used to create self-assembled biomotifs [21], nanopores/nanochannels [22], and responsive ‘smart’ structures (e.g., i-motif structures) [23]. Self-assembly of such structures is driven by pH and electrolyte changes and controlled by mass load and alterations in molecular structure [24,25]. Recently, the DNA molecule has been employed as a biomaterial to build diverse 2D and 3D nanostructures due to the ease of its manipulation by enzymatic reactions [26,27]. Assembly structures featuring nucleic acids can be generated with the ‘DNA origami’ method, creating various geometries comprising designed DNA sequences [8,28,29]. In tissue engineering applications [30,31], branched, complementary (palindromic) sticky-end sequences were earlier constructed to produce large-scale, 3D, cell-encapsulating DNA polymer structures [27]. DNA self-assembly methods can facilitate affinity- and label-based strategies for the selective immobilization of multiple cell types on DNA nanoarrays [32].
Biomolecular assembly strategies combined with microscale technologies can be useful in the development of specific controlled-release mechanisms. Such assembly strategies can also be used to functionalize biomaterials for cell adhesion, for cell patterning to provide a controlled microenvironment for cell fate maintenance of specific niches. Assembled biomolecular elements can also be strategically used for selective biodegradation of matrix and provide control over biodegradation. Deeper understanding of cell–material interactions specific to tissue types and the biocompatible assembly procedures will define the future applications of biomolecular assembly constructs.
Cell assembly
Tissues in the human body such as liver, brain, and pancreatic islets comprise densely packed cells with a minimal ECM fraction [33]. Scaffold- and microgel-based tissue engineering approaches are unable to provide a high cell-packing density. As an alternative, cell sheets have emerged as high-density monolayer cultures that can be piled together forming multilayer cellular constructs [34]. The culture of a single-layer cell sheet can take up to 1 week and an automated system is then required to create the multilayer structure from single-layer building blocks. This approach has been utilized for the bioengineering of several tissues including myocardial [35], periodontal [36], corneal [37], and articular cartilage [38].
Surface topography may assist the alignment and patterning of cells on diverse biomaterials [39,40]. The specific alignment of cells and the ECM can affect cell function, physiology, and the differentiation of stem cells [41–43]. Cell sheets with engineered cellular patterns generated on tissue-specific surface topographies can more readily mimic native cell assembly and may accelerate progression to the final tissue-engineered construct.
Given that cell sheets comprise a single cell type, introducing complexity into the architecture is possible by assembling layers of other cell types. Cell sheets comprising multiple cell types can be utilized by patterning approaches to bring complexity to a single layer [44,45].
Engineering complex cellular structures
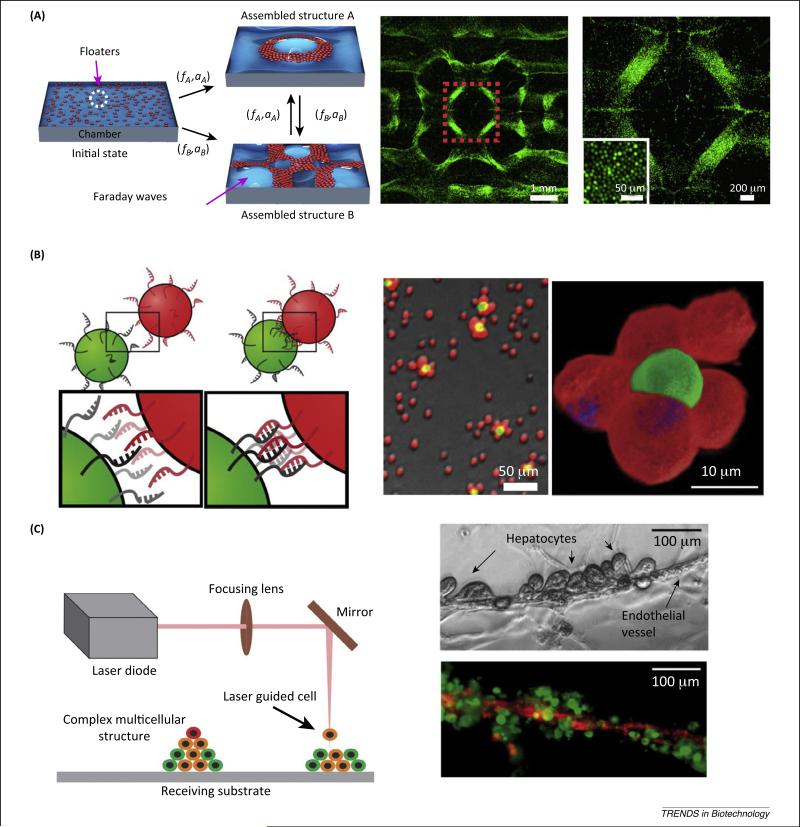
Micromolding methods, using molds fabricated through soft photolithography and rapid prototyping, can enable the engineering of 3D tissue constructs with complex structures [46,47]. Cells that are deposited in nonadhesive molds (e.g., agarose, polyacrylamide) self-organize into 3D microtissue constructs. The resulting cell aggregates can comprise closely packed single or multiple cell populations and retain high cell viability for more than 2 weeks [48]. Unlike micromolding techniques, liquid-based templated assembly enables dynamic reconfiguration of the topography of a pattern template by tuning vibrational frequency and acceleration to assemble floating biological entities (Figure 2A) [49]. Assembly of floaters (cells or microcarriers) is based on their drift on standing waves to either nodes or antinodes depending on wettability and the density of floaters relative to the assembly fluid. By exploring the waveforms of standing waves, diverse symmetric and periodic patterns of floaters can be generated to mimic repeating structural units in native tissues. In addition, assembly of floaters on the liquid-based templates can be performed in a parallel and scalable way with an assembly time of less than 10 s. Simultaneous assembly of millions of cells suspended into different monolayer patterns is demonstrated in a 4 cm2 chamber [49].
Figure 2.
Cell assembly technologies. (A) Liquid-based template assembly (LBTA). Left: Schematics of reconfigurable assembly by LBTA. Right: Assembled cell pattern. The region marked with red dashed lines indicates the magnified region. Cells were stained with cell tracker (green). Reproduced, with permission, from [49]. (B) Molecular recognition-assisted cell assembly. Left: Schematic demonstration. Right: Images of assembled cells and enlarged heterogeneous cell arrangement. Green- and red-stained Jurkat cells marked with the complementary DNA sequences. Adapted from [53]. (C) Sketch showing laser-guided direct writing (LGDW). Left: LGDW. The laser is focused into a cell suspension and the radiation force due to the difference in refractive index moves cells onto a receiving substrate. Right: Phase contrast and immunofluorescence of human hepatocytes and endothelial structure engineered by LGDW. Reproduced, with permission, from [56].
Engineering heterogeneous cell spatial arrangement
Cell–cell and cell–substrate interactions between heterogeneous cell types regulate important cytophysiological processes related to tissue regeneration such as stem cell differentiation, self-organization into tissue-specific architecture, and expression of tissue-specific function [50–52]. Therefore, the ability to achieve predefined heterogeneous cell arrangements is needed in tissue engineering. Molecular recognition-assisted cell assembly enables the generation of heterogeneous multicellular aggregates with designed 3D cell arrangements [53]. Engineering of tissue mimics comprising a functional paracrine signaling pathway was demonstrated using CHO cells and hematopoietic progenitor cells (Figure 2B) [53]. These two cell types were separately conjugated to short oligonucleotides with complementary sequences then assembled into 3D cell aggregates with designed interconnectivity. DNA linkage between cells can be reversed, thereby allowing intercellular interactions and tissue formation. These heterogeneous aggregates can be utilized as building blocks for the engineering of tissue constructs.
Engineering complex tissue structure with single-cell controllability
Laser-guided direct writing (LGDW) can assemble heterogeneous cell populations into complex 3D structures with single-cell control [54,55]. In LGDW, individual suspended cells are trapped and manipulated by optical radiation force (Figure 2C). Hierarchical 3D structures can be formed by picking and placing single cells one by one using a layer-by-layer strategy. For example, sinusoid-like structures were generated by direct 3D bioprinting of endothelial cells and hepatocytes [56]. This technique provides a venue to study cell–cell interactions and cell signaling in synthetic biology and tissue engineering in a highly controlled manner. Despite its high spatial precision, assembly throughput is limited to hundreds of cells per hour per laser beam. Multiple laser beam systems can be further utilized for biological applications and improve the capacity of the system and the time required for the assembly.
Formation of tissue spheroids
Tissue spheroids or aggregates are used not only as tissue-based microphysiological systems for drug discovery but also as building blocks to form complex tissue architectures through tissue fusion [57]. Spheroid-based tissue engineering aims to: (i) generate high-throughput and uniform tissue spheroids; and (ii) assemble tissue spheroids into more complex constructs. Technologies have been developed to generate 3D homogeneous or heterogeneous spheroids, such as hanging drop techniques, rotating wall vessels, magnetic levitation, and suspension culture in low adhesion/attachment plates [58–61]. Multiplex tissue spheroids can be generated in hanging drop networks with predefined intertissue connections, which facilitates ‘human-on-a-chip’ research [62,63]. Recently, microfluidic droplet platforms have been developed for the rapid formation of multicell-type spheroids. Single cells are clustered together in double-emulsion droplets at high-throughput, forming spheroids of uniform size [64–66].
Assembly methods for tissue spheroids
Compared with scaffold-free cell assembly, tissue spheroid assembly allows manipulation of hundreds to thousands of cells per spheroid. Bioprinting technology has been demonstrated to assemble spheroids into close-packed tissue [67,68]. Using this technique, it was possible to bioprint double-layered vascular tubes or 3D liver tissues that retained main liver function for up to 40 days and exhibited dose-dependent responses to several liver toxicants [69]. LBTA has also been reported to assemble thousands of tissue spheroids into periodic symmetry patterns in a few seconds [49]. In another study, magnetized tissue spheroids were patterned with magnetic fields into multiple geometries over several length scales [70].
In scaffold-free cell assembly techniques, optimization of the assembly methods and monitoring cell health is necessary to enable broad applications in tissue engineering. It is essential to create experimental platforms and designs that minimize cell damage due to the exposure to multiple stress factors such as shear forces, osmotic shock, thermal fluctuations, pH changes, and toxic chemicals that may alter cell physiology or be lethal.
Assembly of cell-encapsulating microgels
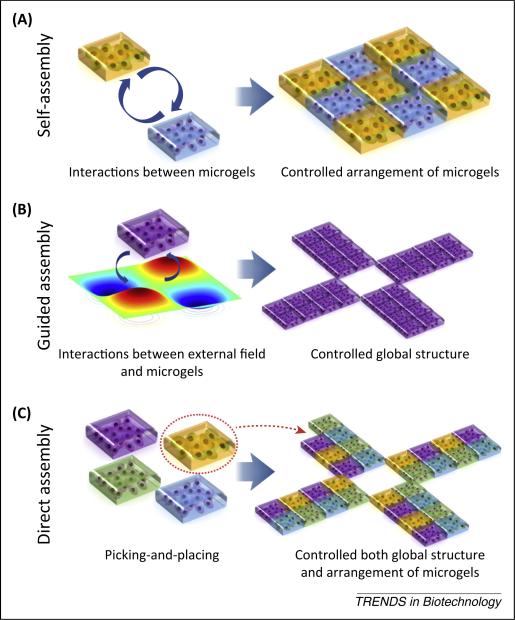
The engineering and assembly of tissue-mimicking 3D structures with cell-specific niches is important for tissue regeneration, maturation, and functionalization. Various technologies were developed to assemble microgels into designed architectures and spatial organizations. These assembly technologies can be classified into three categories based on interaction modes between microgels and guiding forces: (i) self-assembly; (ii) guided assembly; and (iii) direct assembly (Figure 3).
Figure 3.
Illustration of mechanisms for assembly technologies. (A) Self-assembly explores intrinsic interactions among building blocks to generate ordered structures. (B) Guided self-assembly explores interactions between building blocks and external forces to generate controlled global structures. (C) Guided assembly with a pick-and-place strategy builds complex structures with controlled global geometry and arrangement of cell carriers piece by piece.
Self-assembly
Self-assembly describes a process in which an ordered system or structure is spontaneously formed from building blocks driven only by their intrinsic interactions [71] (Figure 3A). Self-assembly is characterized by the minimization of free energy and the formation of ordered, periodic arrangements of building blocks within a resulting structure. Self-assembly mechanisms include capillary self-assembly and molecular recognition-assisted self-assembly [25,72–78].
Capillary self-assembly
Self-assembly by capillary force is driven by minimizing the interfacial free energy at the fluid–fluid interface. By tuning the geometric shape, size, and wettability of building blocks, the formation of periodic patterns can be engineered through capillary self-assembly [72–74]. Capillary self-assembly has been utilized to create centimeter-scale hydrogel assemblies with controlled micrometer-scale biological features at the air– liquid interface [75] and the aqueous–oil interface [76].
Molecular recognition-assisted self-assembly
Molecular recognition has been explored to assemble microgels into predesigned 3D spatial interactions by adjusting complementary molecules such as oligonucleotide pairs [25,78,79]. Orthogonal DNA coding-assisted assembly of multiple polyethylene glycol (PEG)-based microtissues has been reported [78]. In this strategy, microgels were modified with single-stranded DNA and were self-assembled by sequence-specific hybridization onto patterned DNA microarrays. Similarly, by exploring face-specific DNA modification on cubic microgels, self-assembly of diverse structures was demonstrated in interfacial agitation systems [25]. In another strategy, host and guest gels are designed to interact with each other by utilizing corresponding molecular recognition of cyclodextrins and hydrocarbon groups [79]. By increasing temperature, these strong molecular interactions were dissociated, thus demonstrating reversible binding of the blocks.
Guided assembly
This approach describes a process in which an ordered system or structure is spontaneously formed from disordered building blocks driven by their interactions with extrinsic fields such as magnetic or acoustic field forces (Figure 3B). Guided assembly is characterized by the minimization of the free energy of the entire interactive system and the formation of organized building blocks that is globally defined by the spatiotemporal distribution of the extrinsic field. Guided assembly can be based on magnetic fields, acoustic fields, geometric recognition, or liquid-based templates [49,80–89,91,92].
Magnetic field-guided assembly
Guided assembly using magnetic forces exploits interactions between microgels and magnetic fields that can be either static [80] or alternating fields established by a magnet [81] or electromagnet [82]. Magnetic assembly of cell-encapsulating microgels into 3D structures has been recently reported for tissue engineering [83–85]. Microgels incorporating magnetic nanoparticles (MNPs) can be assembled into a heterogeneous, multilayered structure by a magnetic field [83]. The controlled release of MNPs can be critical for potential clinical applications [93]. In this regard, cell-encapsulating MNP-free hydrogels were explored for the assembly of 3D constructs using their paramagnetic free radicals when placed in a magnetic field [84,85]. MNP-free microgels with tunable magnetic properties were levitated in a magnetic field and precisely formed various patterns of 3D heterogeneous tissue assemblies of multiple cell and biomaterial types. There is a need to evaluate various types of para-magnetic media to enable long-term cell culture and to control the chemical microenvironment minimizing any detrimental effects on cells and assembled tissue constructs.
Acoustic field-guided assembly
Guided assembly using acoustic fields exploits interactions between microgels and acoustic fields, which are usually generated by bulk acoustic waves [86] or surface acoustic waves [87]. Recently, acoustic technologies in combination with microfluidics have been utilized for diverse biological applications such as cell sorting [88] and cell patterning [89]. Furthermore, acoustic fields were used in tissue engineering, for which microgels were initially dispersed in droplets and then closely packed into a monolayer structure in less than 10 s by applying acoustic waves [91]. This acoustic assembly method can be potentially further expanded into 3D assembly methods, going beyond 2D assembled constructs.
Geometric recognition-guided assembly
This technique exploits geometric matching between shape-coded micro-gels and congruent wells on the substrate that organize these microgels into a designed spatial configuration. This approach is promising for the creation of microenvironments with various cell types and biochemical factors. For example, shape-coded cell niches were docked into predesigned receiving wells by gravity sedimentation for the study of cell–microenvironment and cell–cell interactions [92].
Liquid-based template assembly
Guided assembly using standing waves has been demonstrated with LBTA, where neuron-seeded carrier beads were assembled into 3D neural networks with controlled architectures [49]. These neuronal networks are significant for the generation of in vitro brain models to understand the wiring and mapping of neuron circuits. In addition, assembly of microgels into various patterns was demonstrated with hydrogels ranging from 500 μm to 2 mm in size. This method requires biocompatible aqueous media that meet the specific criteria for viscosity, density, and the facilitation of a stable final construct.
Direct assembly
Direct assembly utilizes a pick-and-place strategy to create an ordered architecture by picking and placing building blocks piece by piece (Figure 3C). Assembly time is proportional to the number of building blocks required in the fully assembled structure. Direct assembly is a deterministic process and the assembled structure can be highly complex by virtue of having spatial and temporal control over a single building block. Guided assembly methods include bioprinting, digital patterning, and robotic assembly [2,4,94,95].
Direct assembly with digital patterning
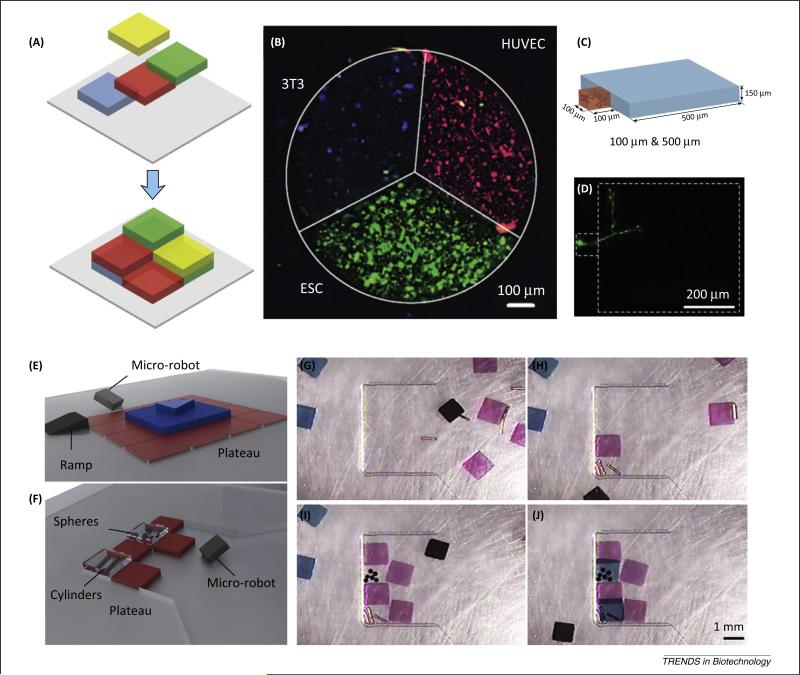
Photolithography has been used in multiple fields such as semiconductor technologies and bioengineering to pattern 3D hydrogels. In this approach, digitally designed photo masks are used to create patterns, providing a precise and easy technique to create otherwise structurally complex models. This strategy has also been incorporated into a simple alignment system allowing precise placement of 3D hydrogels in heterogeneous patterns encapsulating multiple cell types (Figure 4A–D) [94]. Use of this system to mimic the cellular composition of native cortical brain tissue has been reported and it was demonstrated that the ratio of inhibitory neurons to excitatory neurons was preserved in the engineered in vitro model (Figure 4D). The precise heterogeneous placement of multiple cell types is important for the creation of tissue samples closer in composition to the native tissue and also allows the investigation and analysis of interactions between different cell types (Figure 4B). In addition, control over the cellular microenvironment and spatial position facilitates investigations of cell responses to different materials. This system provides a high-throughput, easy-to-use method that yields engineered tissue structures to mimic native tissues and provide the means for in-depth analyses of processes such as cell–cell interactions.
Figure 4.
Digital patterning. (A) Schematic of digital patterning of heterogeneous structures. (B) Patterning of three cell types [ESCs in green, 3T3 cells in blue, human umbilical vein endothelial cells (HUVECs) in red] in a single tissue structure. (C,D) Digital patterning of a 100-μm hydrogel next to a 500-μm hydrogel, where only the 100-μm hydrogel includes a single neuron. Reproduced, with permission, from [94]. (E–J) 3D patterning of living units via magnetic microrobots. (E,F) Microrobotic assembly of heterogeneous objects. (G–J) Heterogeneous assembly of soft hydrogels and rigid objects. Reproduced, with permission, from [95].
Direct assembly of microgels with microrobotics
Robotic assembly utilizes a pick-and-place approach to engineer 3D constructs from microgels. Recently, a versatile method has been presented to encode functional building blocks into complex 3D tissue constructs using a microrobot [95]. The microrobot is controlled by an operator through real-time computer monitoring with spatiotemporal control in fluid microenvironments while pushing microgels along the way. This approach provides the capacity to achieve high-resolution patterning of microgels into viable complex tissue constructs. For instance, a three-layer heterogeneous pyramidal structure comprising building blocks on each layer was fabricated (Figure 4E–J). Micro-robotic assembly can also be used in building heterogeneous structures comprising various materials such as PEG hydrogels, copper cylinders, and polystyrene spheres.
Bioprinting
Bioprinting is a direct assembly approach used to engineer arbitrarily shaped 3D tissue constructs by depositing cell-encapsulating hydrogel prepolymer droplets onto a receiving substrate on demand [4–6,96–101,115]. Engineering of 3D tissue-like droplet networks by printing picoliter aqueous droplets in bulk oil or within oil drops has been reported [102,103]. In this work, printed droplet networks were functionalized with membrane proteins, enabling instant electrical communication through a specific pathway. These networks can also be designed using stimulus-responsive osmolytes to achieve self-folding in a predictable way. Direct assembly with bioprinting can generate custom shapes for personalized medicine. For instance, a 3D bionic ear was bioprinted from cell-loaded hydrogel matrix with the anatomical architecture of a human ear and an electronic component acting as an antenna to receive signals [104].
Assembling cell-encapsulating building blocks provides the basis of bottom-up tissue engineering, through which complex microtissues can be generated. The approaches described above provide a high degree of control over the final structure within a customized microenvironment. The properties of cell-encapsulating materials affect cell physiology, metabolism, and stem cell differentiation. Bioengineering the chemical and physical features (e.g., biocompatibility, porosity, stiffness, diffusion of gas and nutrients) of these materials will enhance the function of the encapsulated cells and enable the broad applicability of the construct. Moreover, tissue-specific designed biomaterials can address bottlenecks specific to the assembly method such as mechanical stability and ease of manipulation.
Evaluation of multiscale assembly systems
Our ability to build constructs that mimic the complex architecture of native tissues is limited by our scientific knowledge and the tools that we have to bioengineer and construct complexity. The size scale of the materials to be assembled are diverse, as are the technologies that are needed to handle this diversity. Overall, the assembly technologies are bound by the main design parameters and limitations that need to be overcome (Table 1). (i) Viability and function: The introduction of advanced technologies to the 3D assembly and biomanufacturing process should not come at the cost of the viability or function of the assembled cells. (ii) Time consumed during assembly and throughput: To allow large sets of data to be generated for statistical analysis, and to enable high-throughput creation of constructs, the assembly time and being able to create constructs in parallel are significant. For instance, in bioprinting, using a single-nozzle robot is a limiting factor (Table 2). (iii) Scalability and control over construct size: Scalable assembly of thousands of building blocks such as microgels or extrusions in bioprinting can be challenging, for larger 3D constructs or many identical constructs. (iv) Ease-of-use, repeatability, and broad access to the assembly technology: Some of the technology platforms require either expensive equipment or sophisticated set-ups that are not commercially available and need to be designed or custom manufactured as prototypes (e.g., bioprinting, laser printing requiring lasers, robotics, microscopy systems). It would be desirable for these assembly technologies to be easily adapted broadly to laboratories around the globe with minimal investment, minimizing operator-to-operator variability and enabling repeatable outcomes. (v) Mechanical stability and cellular ECM remodeling: Depending on the application of interest, the assembled constructs may need to be stabilized during culture to maintain a specific structure during the post-assembly and post-analysis stages. For instance, in cell-encapsulating constructs, it is well known that cells remodel their microenvironment and secrete their own ECM. This opens additional optimization steps for materials that are being selected especially for biodegradability and their degradation rate and interactions with other cell types in cocultures. (vi) Dynamic temporal and spatial control over precision and 3D complexity: Spatial control over a single building block enables building complexity in architecture and facilitates the establishment of tissue hierarchy. However, temporal resolution makes it possible to build things on demand as constructs pass through various phases of development and maturation [94,95]. (vii) Handling diverse materials and multiple cell types: Using the directed and guided methods to tune the dynamically spatial coding of materials enables creating heterogeneity in the assembled structures (e.g., magnetically assembling building blocks of different materials and content) [85]. (viii) Cutting across multiple scales: Assembly methods can be harmonized in a sequential or parallel manner, in which capabilities to assemble nano-scale and macroscale building units can be interfaced to build complexity across a wide dynamic range of scales (molecules, cells, microgels) to work in combination simultaneously or interchangeably (e.g., integrating DNA templates into assembly microgels [25]).
Table 1.
Advantages and limitations of assembly methods
| Assembly method | Advantages | Limitations | Refs |
|---|---|---|---|
| Peptide self-assembly | Tunable material properties | Stability of assembly construct | [12,18] |
| Carbohydrate self-assembly | Selective geometry to carry cargo | Material limitation | [9] |
| Nucleic acid self-assembly | High affinity, selectivity | Rapid degradation | [21,27] |
| LGDW | High cell packing density Spatial control over heterogeneous cell arrangement; Scaffold free |
Low throughput | [54-56] |
| Cell sheet assembly | Large size Scaffold free |
Limited cell types; without control over cell arrangement within each layer | [34] |
| Micromolding | Rapid assembly of high numbers of cells High cell packing density; Scaffold free |
No control over spatial organization of heterogeneous cell types | [46,47] |
| Capillary self-assembly | Minimal external manipulation | No control over spatial organization of heterogeneous building units No global control over final structure |
[70-75] |
| Molecular recognition | Highly specific | Requires surface modification | [27] |
| Magnetic assembly | High speed | Potential cytotoxicity No control over spatial organization of heterogeneous building units |
[81-83] |
| Acoustic assembly | High speed | No control over spatial organization of heterogeneous building units | [89] |
| Geometric recognition | High heterogeneity | Low throughput | [90] |
| LBTA | Allows scalable and parallel manipulation Allows formation of complex structure High speed |
Limited control over spatial organization of heterogeneous building units | [49] |
| Photolithography | High precision High speed Scalable |
Potential cell damage due to UV exposure and free radicals | [94] |
| Microrobotic assembly | High precision Allows assembly of heterogeneous materials |
Low throughput | [95] |
| Bioprinting | Allows formation of computer designed arbitrary structure | High mechanical stress on cells Random control over cell number and types in individual droplets |
[3,102] |
Table 2.
Assembly time for selected guided and direct assembly methods
| Category | Method | Assembly time | Refs |
|---|---|---|---|
| Guided assembly | Geometric docking | ~20 min/batch | [92] |
| Surface tension | ~60 s/batch | [76] | |
| Molecular recognition-assisted self-assembly | 20-120 s/batch | [79] | |
| LBTA | ~5 s/batch | [49] | |
| Acoustic assembly | 5-30 s/batch | [91] | |
| Magnetic assembly | <3 s/batch | [83] | |
| Direct assembly | Photolithography | 30-120 s | [94] |
| LGDW | A single beam to hundreds of cells/h | [54-56] | |
| Bioprinting | 1000-1000 000 droplets/s | [3] | |
| Microrobotic assembly | 1-10 min/building unit depending on operator's skill | [95] |
The selection of a specific assembly technique should be made based on parameters such as the scale range of the building units and the size of the final construct, the complexity of the construct (e.g., biomaterial and cell types), and the throughput. When building complex architectures with high precision, resolution, and control, bioengineers will face trade-offs in design based on the choice of assembly technique. Despite the limitations of each assembly approach (Table 1), they all have unique advantages that can meet the criteria for specific applications or address well-defined questions in biotechnology. For instance, the relatively slow speed of direct assembly compared with other techniques may limit it to applications for which more time can be taken to generate 3D tissue engineered constructs. Regarding utilizing these techniques to address specific questions such as how bioengineered microtissues using different materials encapsulating diverse cell types interact with each other, many valuable insights can be gained from such investigations.
Toward high-throughput systems
Automation in biotechnological assays has progressed rapidly over the past decade. For instance, automated assay kits and gene analyzers provide drug discovery and cell analysis platforms that are easy to use and have high accuracy. Researchers are able to perform experiments with many variables using low quantities of sample and reagents and obtain results in a timely manner. The automated systems are difficult to adapt to 3D assembly, despite the promising outcomes they have provided for 2D platforms [98]. The biotechnology industry has growing interest in personalized medicine tools with custom solutions that meet the demands of clients [105–107] and pharmacotoxicology [108]. Such directions bring the need for more agile and precise automated assembly platforms to handle complexity and increase the applicability of these manufacturing and assembly processes. The technological platforms developed in laboratories need to be evaluated and translated to industrial high-throughput production. Standardization of the assembly process will enable reproducibility of outcomes, minimize operator variations, optimize the use of biological materials, standardize process steps, and reduce waste [109]. Automated high-throughput systems will also make the products more accessible, inexpensive, and efficient.
Concluding remarks and future perspectives
Multiscale assembly strategies from the molecular level to the macroscale promise to generate complex tissues with multiple building blocks in various spatial organizations using various materials. In the past two decades, multiple methods and technologies have been developed to assemble biomolecules, cells, tissue spheroids, and cell-encapsulating microgels. Currently, most of these methods remain in developmental stages and are enabling progress in addressing the existing bottlenecks such as assembly efficiency, speed, and mechanical stability. In addition to finding solutions for these challenges, the next steps need to focus on novel biomaterials, integrating developed assembly strategies to establish valid solutions to existing challenges in tissue engineering, and regenerative medicine for industrial and clinical applications. The need for multiscale assembly will push the development of high-throughput microtissue platforms for diagnostics, drug screening, and cell biology research. In addition, precision and reproducibility in assembly approaches will potentially enable on-demand patient-specific tissues for regenerative medicine, creating a new direction in personalized medicine.
Box 1. Microenvironments and biomanufacturing.
Hydrogels such as fibrin, collagen, alginate, and PEG-based biomaterials are used extensively in tissue engineering due to their biocompatibility, porous structure, moldability, and the tunability of their biological, chemical and mechanical properties [110]. Biomaterial synthesis focuses on capturing cell-specific ECM features by incorporating cell-specific signaling molecules, matrix stiffness, and porosity. The biological properties of synthetic hydrogels can be tuned by incorporating peptides that enhance cell adhesion [e.g., arginine–glycine–aspartic acid (RGD) and glutamic acid peptide] [111]. Microenvironments can be engineered with photopatterning, where ECM proteins or soluble factors can be spatially organized within 3D PEG-based hydrogels, thereby controlling the distribution of encapsulated mesenchymal stem cells [112]. Moreover, technological advances enable reversible patterning of biologically active molecules in hydrogels using light [113]. In another approach, hybrid biomaterials can merge the biological complexity of the natural ECM with the mechanical strength of engineered constructs. In such an approach, PEG- and poly(L-lactic acid) (PLLA)-based scaffolds have been combined with fibrin and poly(lactic-co-glycolic acid) (PLGA) microspheres loaded with fibroblast growth factor (FGF) and platelet-derived growth factor (PDGF)-BB. These mechanically stable biomimetic scaffolds enhance both in vivo engraftment and vascularization with released angiogenic factors [114].
Acknowledgments
U.D. acknowledges that this material is based in part upon work supported by the National Science Foundation under NSF CAREER Award Number 1150733, R15HL115556, and R01EB015776.
Footnotes
Disclaimer statement
U.D. is a founder of, and has an equity interest in: (i) DxNow Inc., a company that is developing microfluidic and imaging technologies for point-of-care diagnostic solutions, and (ii) Koek Biotech, a company that is developing microfluidic IVF technologies for clinical solutions; his interests were reviewed and are managed by the Brigham and Women's Hospital and Partners HealthCare in accordance with their conflict of interest policies. The other authors indicated no potential conflicts of interest.
References
- 1.Elbert DL. Bottom-up tissue engineering. Curr. Opin. Biotechnol. 2011;22:674–680. doi: 10.1016/j.copbio.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurkan UA, et al. Emerging technologies for assembly of microscale hydrogels. Adv. Healthc. Mater. 2012;1:149–158. doi: 10.1002/adhm.201200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tasoglu S, et al. Manipulating biological agents and cells in micro-scale volumes for applications in medicine. Chem. Soc. Rev. 2013;42:5788–5808. doi: 10.1039/c3cs60042d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tasoglu S, Demirci U. Bioprinting for stem cell research. Trends Biotechnol. 2013;31:10–19. doi: 10.1016/j.tibtech.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon S, et al. Drop-on-demand single cell isolation and total RNA analysis. PLoS ONE. 2011;6:e17455. doi: 10.1371/journal.pone.0017455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demirci U, Montesano G. Single cell epitaxy by acoustic picolitre droplets. Lab Chip. 2007;7:1139–1145. doi: 10.1039/b704965j. [DOI] [PubMed] [Google Scholar]
- 7.Maude S, et al. Biomimetic self-assembling peptides as scaffolds for soft tissue engineering. Nanomedicine. 2013;8:823–847. doi: 10.2217/nnm.13.65. [DOI] [PubMed] [Google Scholar]
- 8.Andersen ES, et al. DNA origami design of dolphin-shaped structures with flexible tails. ACS Nano. 2008;2:1213–1218. doi: 10.1021/nn800215j. [DOI] [PubMed] [Google Scholar]
- 9.Zhao F, et al. Supramolecular self-assembly forming a multifunctional synergistic system for targeted co-delivery of gene and drug. Biomaterials. 2014;35:1050–1062. doi: 10.1016/j.biomaterials.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 10.Arslan Yildiz A, et al. Biomimetic membrane platform: fabrication, characterization and applications. Colloids Surf. B Biointerfaces. 2013;103:510–516. doi: 10.1016/j.colsurfb.2012.10.066. [DOI] [PubMed] [Google Scholar]
- 11.Holmes TC, et al. Extensive neurite outgrowth and active synapse formation on self-assembling peptide scaffolds. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6728–6733. doi: 10.1073/pnas.97.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kyle S, et al. Recombinant self-assembling peptides as biomaterials for tissue engineering. Biomaterials. 2010;31:9395–9405. doi: 10.1016/j.biomaterials.2010.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prakash A, et al. Recombinant production of self-assembling b-structured peptides using SUMO as a fusion partner. Microb. Cell Fact. 2012;11:92. doi: 10.1186/1475-2859-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung JP, et al. Co-assembling peptides as defined matrices for endothelial cells. Biomaterials. 2009;30:2400–2410. doi: 10.1016/j.biomaterials.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leonard SR, et al. Solid-state NMR evidence for b-hairpin structure within MAX8 designer peptide nanofibers. Biophys. J. 2013;105:222–230. doi: 10.1016/j.bpj.2013.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rughani RV, Schneider JP. Molecular design of b-hairpin peptides for material construction. MRS Bull. 2008;33:530–535. doi: 10.1557/mrs2008.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shroff K, et al. Fibronectin-mimetic peptide–amphiphile nanofiber gels support increased cell adhesion and promote ECM production. Soft Matter. 2010;6:5064–5072. [Google Scholar]
- 18.Zhang S, et al. A self-assembly pathway to aligned monodomain gels. Nat. Mater. 2010;9:594–601. doi: 10.1038/nmat2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaikh AY, et al. Cationic charged helical glycopolypeptide using ring opening polymerization of 6-deoxy-6-azido-glyco-N-carboxyanhydride. Biomacromolecules. 2014;15:3679–3686. doi: 10.1021/bm5009537. [DOI] [PubMed] [Google Scholar]
- 20.Li J. Self-assembled supramolecular hydrogels based on polymer–cyclodextrin inclusion complexes for drug delivery. NPG Asia Mater. 2010;2:112–118. [Google Scholar]
- 21.Wang S, et al. Enthalpy-driven three-state switching of a superhydrophilic/superhydrophobic surface. Angew. Chem. Int. Ed. Engl. 2007;46:3915–3917. doi: 10.1002/anie.200700439. [DOI] [PubMed] [Google Scholar]
- 22.Xia F, et al. Gating of single synthetic nanopores by proton-driven DNA molecular motors. J. Am. Chem. Soc. 2008;130:8345–8350. doi: 10.1021/ja800266p. [DOI] [PubMed] [Google Scholar]
- 23.Chen C, et al. Stimuli-responsive controlled-release system using quadruplex DNA-capped silica nanocontainers. Nucleic Acids Res. 2011;39:1638–1644. doi: 10.1093/nar/gkq893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shu W, et al. DNA molecular motor driven micromechanical cantilever arrays. J. Am. Chem. Soc. 2005;127:17054–17060. doi: 10.1021/ja0554514. [DOI] [PubMed] [Google Scholar]
- 25.Qi H, et al. DNA-directed self-assembly of shape-controlled hydrogels. Nat. Commun. 2013;4:2275. doi: 10.1038/ncomms3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu D, et al. DNA-based switchable devices and materials. NPG Asia Mater. 2011;3:109–114. [Google Scholar]
- 27.Um SH, et al. Enzyme-catalysed assembly of DNA hydrogel. Nat. Mater. 2006;5:797–801. doi: 10.1038/nmat1741. [DOI] [PubMed] [Google Scholar]
- 28.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 29.Andersen ES, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–75. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 30.Lee JB, et al. A mechanical metamaterial made from a DNA hydrogel. Nat. Nanotechnol. 2012;7:816–820. doi: 10.1038/nnano.2012.211. [DOI] [PubMed] [Google Scholar]
- 31.Cheng E, et al. A pH-triggered, fast-responding DNA hydrogel. Angew. Chem. Int. Ed. Engl. 2009;121:7796–7799. doi: 10.1002/anie.200902538. [DOI] [PubMed] [Google Scholar]
- 32.Koyfman AY, et al. Cell-targeted self-assembled DNA nanostructures. J. Am. Chem. Soc. 2009;131:14237–14239. doi: 10.1021/ja9015638. [DOI] [PubMed] [Google Scholar]
- 33.Bhatia SN. Microfabrication in Tissue Engineering and Bioartificial Organs. Springer; 1999. [Google Scholar]
- 34.Owaki T, et al. Cell sheet engineering for regenerative medicine: current challenges and strategies. Biotechnol. J. 2014;9:904–914. doi: 10.1002/biot.201300432. [DOI] [PubMed] [Google Scholar]
- 35.Matsuura K, et al. Toward the development of bioengineered human three-dimensional vascularized cardiac tissue using cell sheet technology. Int. Heart J. 2014;55:1–7. doi: 10.1536/ihj.13-337. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, et al. Recent advances in cell sheet technology for periodontal regeneration. Curr. Stem Cell Res. Ther. 2014;9:162–173. doi: 10.2174/1574888x09666140213150218. [DOI] [PubMed] [Google Scholar]
- 37.Soma T, et al. Maintenance and distribution of epithelial stem/ progenitor cells after corneal reconstruction using oral mucosal epithelial cell sheets. PLoS ONE. 2014;9:e110987. doi: 10.1371/journal.pone.0110987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato M, et al. Articular cartilage regeneration using cell sheet technology. Anat. Rec. (Hoboken) 2014;297:36–43. doi: 10.1002/ar.22829. [DOI] [PubMed] [Google Scholar]
- 39.Isenberg BC, et al. A thermoresponsive, microtextured substrate for cell sheet engineering with defined structural organization. Biomaterials. 2008;29:2565–2572. doi: 10.1016/j.biomaterials.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahn EH, et al. Spatial control of adult stem cell fate using nanotopographic cues. Biomaterials. 2014;35:2401–2410. doi: 10.1016/j.biomaterials.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim DH, et al. Biomimetic nanopatterns as enabling tools for analysis and control of live cells. Adv. Mater. 2010;22:4551–4566. doi: 10.1002/adma.201000468. [DOI] [PubMed] [Google Scholar]
- 42.Kim DH, et al. Matrix nanotopography as a regulator of cell function. J. Cell Biol. 2012;197:351–360. doi: 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moraes C, et al. Defined topologically-complex protein matrices to manipulate cell shape via three-dimensional fiber-like patterns. Lab Chip. 2014;14:2191–2201. doi: 10.1039/c4lc00122b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haraguchi Y, et al. Fabrication of functional three-dimensional tissues by stacking cell sheets in vitro. Nat. Protoc. 2012;7:850–858. doi: 10.1038/nprot.2012.027. [DOI] [PubMed] [Google Scholar]
- 45.Tavana H, Takayama S. Aqueous biphasic microprinting approach to tissue engineering. Biomicrofluidics. 2011;5:13404. doi: 10.1063/1.3516658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duffy DC, et al. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal. Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 47.Xia Y, Whitesides GM. Soft lithography. Annu. Rev. Mater. Sci. 1998;28:153–184. [Google Scholar]
- 48.Napolitano A, et al. Scaffold-free three-dimensional cell culture utilizing micromolded nonadhesive hydrogels. Biotechniques. 2007;43:494–500. doi: 10.2144/000112591. [DOI] [PubMed] [Google Scholar]
- 49.Chen P, et al. Microscale assembly directed by liquid-based template. Adv. Mater. 2014;26:5936–5941. doi: 10.1002/adma.201402079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasai Y. Cytosystems dynamics in self-organization of tissue architecture. Nature. 2013;493:318–326. doi: 10.1038/nature11859. [DOI] [PubMed] [Google Scholar]
- 51.Woodford C, Zandstra PW. Tissue engineering 2.0: guiding self-organization during pluripotent stem cell differentiation. Curr. Opin. Biotechnol. 2012;23:810–819. doi: 10.1016/j.copbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li L, Xie T. Stem cell niche: structure and function. Annu. Rev. Cell Dev. Biol. 2005;21:605–631. doi: 10.1146/annurev.cellbio.21.012704.131525. [DOI] [PubMed] [Google Scholar]
- 53.Gartner ZJ, Bertozzi CR. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4606–4610. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Odde DJ, Renn MJ. Laser-guided direct writing for applications in biotechnology. Trends Biotechnol. 1999;17:385–389. doi: 10.1016/s0167-7799(99)01355-4. [DOI] [PubMed] [Google Scholar]
- 55.Akselrod GM, et al. Laser-guided assembly of heterotypic three-dimensional living cell microarrays. Biophys. J. 2006;91:3465–3473. doi: 10.1529/biophysj.106.084079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nahmias Y, Odde DJ. Micropatterning of living cells by laser-guided direct writing: application to fabrication of hepatic–endothelial sinusoid-like structures. Nat. Protoc. 2006;1:2288–2296. doi: 10.1038/nprot.2006.386. [DOI] [PubMed] [Google Scholar]
- 57.Jakab K, et al. Engineering biological structures of prescribed shape using self-assembling multicellular systems. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2864–2869. doi: 10.1073/pnas.0400164101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Asghar W, et al. In vitro three-dimensional cancer culture models. In: Bae YH, et al., editors. Cancer Targeted Drug Delivery. Springer; 2013. pp. 635–665. [Google Scholar]
- 59.Souza GR, et al. Three-dimensional tissue culture based on magnetic cell levitation. Nat. Nanotechnol. 2010;5:291–296. doi: 10.1038/nnano.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsiao AY, et al. 384 Hanging drop arrays give excellent Z-factors and allow versatile formation of co-culture spheroids. Biotechnol. Bioeng. 2012;109:1293–1304. doi: 10.1002/bit.24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barrila J, et al. Organotypic 3D cell culture models: using the rotating wall vessel to study host–pathogen interactions. Nat. Rev. Microbiol. 2010;8:791–801. doi: 10.1038/nrmicro2423. [DOI] [PubMed] [Google Scholar]
- 62.Frey O, et al. Reconfigurable microfluidic hanging drop network for multi-tissue interaction and analysis. Nat. Commun. 2014;5:4250. doi: 10.1038/ncomms5250. [DOI] [PubMed] [Google Scholar]
- 63.Guven S, et al. Functional maintenance of differentiated embryoid bodies in microfluidic systems: a platform for personalized medicine. Stem Cells Transl. Med. 2015 doi: 10.5966/sctm.2014-0119. http://dx.doi.org/10.5966/sctm.2014-0119. [DOI] [PMC free article] [PubMed]
- 64.Chan HF, et al. Rapid formation of multicellular spheroids in double-emulsion droplets with controllable microenvironment. Sci. Rep. 2013;3:3462. doi: 10.1038/srep03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu L, et al. Encapsulation of single cells on a microfluidic device integrating droplet generation with fluorescence-activated droplet sorting. Biomed. Microdevices. 2013;15:553–560. doi: 10.1007/s10544-013-9754-z. [DOI] [PubMed] [Google Scholar]
- 66.Torisawa YS, et al. Efficient formation of uniform-sized embryoid bodies using a compartmentalized microchannel device. Lab Chip. 2007;7:770–776. doi: 10.1039/b618439a. [DOI] [PubMed] [Google Scholar]
- 67.Mironov V, et al. Organ printing: from bioprinter to organ biofabrication line. Curr. Opin. Biotechnol. 2011;22:667–673. doi: 10.1016/j.copbio.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 68.Mironov V, et al. Organ printing: tissue spheroids as building blocks. Biomaterials. 2009;30:2164–2174. doi: 10.1016/j.biomaterials.2008.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Norotte C, et al. Scaffold-free vascular tissue engineering using bioprinting. Biomaterials. 2009;30:5910–5917. doi: 10.1016/j.biomaterials.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bratt-Leal AM, et al. Magnetic manipulation and spatial patterning of multi-cellular stem cell aggregates. Integr. Biol. (Camb.) 2011;3:1224–1232. doi: 10.1039/c1ib00064k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grzybowski BA, et al. Self-assembly: from crystals to cells. Soft Matter. 2009;5:1110–1128. [Google Scholar]
- 72.Bowden N, et al. Self-assembly of mesoscale objects into ordered two-dimensional arrays. Science. 1997;276:233–235. doi: 10.1126/science.276.5310.233. [DOI] [PubMed] [Google Scholar]
- 73.Bowden N, et al. Mesoscale self-assembly of hexagonal plates using lateral capillary forces: synthesis using the ‘capillary bond’. J. Am. Chem. Soc. 1999;121:5373–5391. [Google Scholar]
- 74.Wolfe DB, et al. Mesoscale self-assembly: capillary interactions when positive and negative menisci have similar amplitudes. Langmuir. 2003;19:2206–2214. [Google Scholar]
- 75.Zamanian B, et al. Interface-directed self-assembly of cell-laden microgels. Small. 2010;6:937–944. doi: 10.1002/smll.200902326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Du Y, et al. Directed assembly of cell-laden microgels for fabrication of 3D tissue constructs. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9522–9527. doi: 10.1073/pnas.0801866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singh P, Joseph DD. Fluid dynamics of floating particles. J. Fluid Mech. 2005;530:31–80. [Google Scholar]
- 78.Li CY, et al. DNA-templated assembly of droplet-derived PEG microtissues. Lab Chip. 2011;11:2967–2975. doi: 10.1039/c1lc20318e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harada A, et al. Macroscopic self-assembly through molecular recognition. Nat. Chem. 2011;3:34–37. doi: 10.1038/nchem.893. [DOI] [PubMed] [Google Scholar]
- 80.Demirors AF, et al. Colloidal assembly directed by virtual magnetic moulds. Nature. 2013;503:99–103. doi: 10.1038/nature12591. [DOI] [PubMed] [Google Scholar]
- 81.Grzybowski BA, et al. Dynamic self-assembly of magnetized, millimetre-sized objects rotating at a liquid–air interface. Nature. 2000;405:1033–1036. doi: 10.1038/35016528. [DOI] [PubMed] [Google Scholar]
- 82.Snezhko A, Aranson IS. Magnetic manipulation of self-assembled colloidal asters. Nat. Mater. 2011;10:698–703. doi: 10.1038/nmat3083. [DOI] [PubMed] [Google Scholar]
- 83.Xu F, et al. Three-dimensional magnetic assembly of microscale hydrogels. Adv. Mater. 2011;23:4254–4260. doi: 10.1002/adma.201101962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tasoglu S, et al. Paramagnetic levitational assembly of hydrogels. Adv. Mater. 2013;25:1137–1143. doi: 10.1002/adma.201200285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tasoglu S, et al. Guided and magnetic self-assembly of tunable magnetoceptive gels. Nat. Commun. 2014;5:4702. doi: 10.1038/ncomms5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Onfelt B, et al. Ultrasound-controlled cell aggregation in a multi-well chip. Lab Chip. 2010;10:2727–2732. doi: 10.1039/c004707d. [DOI] [PubMed] [Google Scholar]
- 87.Guldiken R, et al. Sheathless size-based acoustic particle separation. Sensors. 2012;12:905–922. doi: 10.3390/s120100905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen P, et al. Microfluidic chips for cell sorting. Front. Biosci. 2008;13:2464–2483. doi: 10.2741/2859. [DOI] [PubMed] [Google Scholar]
- 89.Huang TJ, et al. Acoustic tweezers: patterning cells and microparticles using standing surface acoustic waves (SSAW). Lab Chip. 2009;9:2890–2895. doi: 10.1039/b910595f. [DOI] [PubMed] [Google Scholar]
- 90.Eng G, et al. Assembly of complex cell microenvironments using geometrically docked hydrogel shapes. Proc. Natl. Acad. Sci. U.S.A. 2013;110:4551–4556. doi: 10.1073/pnas.1300569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu F, et al. The assembly of cell-encapsulating microscale hydrogels using acoustic waves. Biomaterials. 2011;32:7847–7855. doi: 10.1016/j.biomaterials.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eng G, et al. Assembly of complex cell microenvironments using geometrically docked hydrogel shapes. Proc. Natl. Acad. Sci. U.S.A. 2013;110:4551–4556. doi: 10.1073/pnas.1300569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu F, et al. Release of magnetic nanoparticles from cell-encapsulating biodegradable nanobiomaterials. ACS Nano. 2012;6:6640–6649. doi: 10.1021/nn300902w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gurkan UA, et al. Simple precision creation of digitally specified, spatially heterogeneous, engineered tissue architectures. Adv. Mater. 2013;25:1192–1198. doi: 10.1002/adma.201203261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tasoglu S, et al. Untethered micro-robotic coding of three-dimensional material composition. Nat. Commun. 2014;5:3124. doi: 10.1038/ncomms4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu F, et al. In. 2009 IEEE Sensors. IEEE; 2009. Cell bioprinting as a potential high-throughput method for fabricating cell-based biosensors (CBBs). pp. 387–391. [Google Scholar]
- 97.Moon S, et al. Layer by layer three-dimensional tissue epitaxy by cell-laden hydrogel droplets. Tissue Eng. Part C Methods. 2009;16:157–166. doi: 10.1089/ten.TEC.2009.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014;32:773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 99.Gurkan UA, et al. Engineering anisotropic biomimetic fibrocartilage microenvironment by bioprinting mesenchymal stem cells in nanoliter gel droplets. Mol. Pharm. 2014;11:2151–2159. doi: 10.1021/mp400573g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu F, et al. A three-dimensional in vitro ovarian cancer coculture model using a high-throughput cell patterning platform. Biotechnol. J. 2011;6:204–212. doi: 10.1002/biot.201000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu F, et al. A droplet-based building block approach for bladder smooth muscle cell (SMC) proliferation. Biofabrication. 2010;2:014105. doi: 10.1088/1758-5082/2/1/014105. [DOI] [PubMed] [Google Scholar]
- 102.Villar G, et al. A tissue-like printed material. Science. 2013;340:48–52. doi: 10.1126/science.1229495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Durmus NG, et al. Bioprinting: functional droplet networks. Nat. Mater. 2013;12:478–479. doi: 10.1038/nmat3665. [DOI] [PubMed] [Google Scholar]
- 104.Mannoor MS, et al. 3D printed bionic ears. Nano Lett. 2014;13:2634–2639. doi: 10.1021/nl4007744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sinha G. Cell presses. Nat. Biotechnol. 2014;32:716–719. doi: 10.1038/nbt.2983. [DOI] [PubMed] [Google Scholar]
- 106.Reiffel AJ, et al. High-fidelity tissue engineering of patient-specific auricles for reconstruction of pediatric microtia and other auricular deformities. PLoS ONE. 2013;8:e56506. doi: 10.1371/journal.pone.0056506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zopf DA, et al. Bioresorbable airway splint created with a three-dimensional printer. N. Engl. J. Med. 2013;368:2043–2045. doi: 10.1056/NEJMc1206319. [DOI] [PubMed] [Google Scholar]
- 108.Mueller D, et al. 3D organotypic HepaRG cultures as in vitro model for acute and repeated dose toxicity studies. Toxicol. In Vitro. 2014;28:104–112. doi: 10.1016/j.tiv.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 109.Marga F, et al. Toward engineering functional organ modules by additive manufacturing. Biofabrication. 2012;4:022001. doi: 10.1088/1758-5082/4/2/022001. [DOI] [PubMed] [Google Scholar]
- 110.Song YS, et al. Engineered 3D tissue models for cell-laden microfluidic channels. Anal. Bioanal. Chem. 2009;395:185–193. doi: 10.1007/s00216-009-2935-1. [DOI] [PubMed] [Google Scholar]
- 111.Karaman O, et al. Effect of surface modification of nanofibres with glutamic acid peptide on calcium phosphate nucleation and osteogenic differentiation of marrow stromal cells. J. Tissue Eng. Regen. Med. 2013 doi: 10.1002/term.1775. Published online July 30, 2013. http://dx.doi.org/10.1002/term.1775. [DOI] [PubMed]
- 112.Mosiewicz KA, et al. In situ cell manipulation through enzymatic hydrogel photopatterning. Nat. Mater. 2013;12:1072–1078. doi: 10.1038/nmat3766. [DOI] [PubMed] [Google Scholar]
- 113.DeForest CA, Anseth KS. Photoreversible patterning of biomolecules within click-based hydrogels. Angew. Chem. Int. Ed. Engl. 2012;51:1816–1819. doi: 10.1002/anie.201106463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jiang B, et al. Design of a composite biomaterial system for tissue engineering applications. Acta. Biomater. 2014;10:1177–1186. doi: 10.1016/j.actbio.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 115.Tasoglu S, et al. Impact of a compound droplet on a flat surface: A model for single cell epitaxy. Phys. Fluids. 2010;22:082103. doi: 10.1063/1.3475527. [DOI] [PMC free article] [PubMed] [Google Scholar]