Summary
Linking the molecular effects of mutations to fitness is central to understanding evolutionary dynamics. Here we establish a quantitative relation between the global effect of mutations on the E. coli proteome and bacterial fitness. We created E. coli strains with specific destabilizing mutations in the chromosomal folA gene encoding dihydrofolate reductase (DHFR) and quantified the ensuing changes in the abundances of 2000+ E. coli proteins in mutant strains using tandem mass tags with subsequent LC-MS/MS. mRNA abundances in the same E. coli strains were also quantified. The proteomic effects of mutations in DHFR are quantitatively linked to phenotype: the standard deviations of the distributions of logarithms of relative (to wild-type) protein abundances anti-correlate with bacterial growth rates. Proteomes hierarchically cluster first by media conditions, and within each condition, by the severity of the perturbation to DHFR function. These results highlight the importance of a systems-level layer in the genotype-phenotype relationship.
Keywords: DHFR, genotype-phenotype relation, fitness effect of mutations, proteome, transcriptome
Introduction
Understanding the genotype-phenotype relationship requires vantage points from multiple scales, ranging from the molecular, through the systems, to the cellular/organismal (Lehner, 2013). Several studies demonstrated that mutations in metabolic enzymes have local effects on fitness through changes in metabolic flux (Applebee et al., 2011; Dean et al., 1986; Soskine and Tawfik, 2010). Mutations that change protein stability can also affect fitness through modulation of the number of folded (active) proteins (Bershtein et al., 2006; Firnberg et al., 2014; Wylie and Shakhnovich, 2011), or by affecting the number of toxic unfolded species (Dobson, 2003; Drummond and Wilke, 2008).
However, in most cases a direct link between the mutational effects on protein function and organismal phenotype is not obvious due to pleiotropic effects, such as protein aggregation (Drummond and Wilke, 2008) and formation of functional and non-functional multimers (Bershtein et al., 2012; Lynch, 2013; Zhang et al., 2008). Furthermore, recent studies have shown that partial inhibition of an enzyme can cause broad changes in the metabolic profile of the cell, extending far beyond the immediate products of enzymes in question (Kwon et al., 2010; Kwon et al., 2008).
The systems-level proteomic response to a genetic variation is an important missing link in the multiscale genotype-phenotype relationship. Earlier studies showed that bulk characteristics of the macromolecular composition in the cell cytoplasm, e.g., the total protein concentration or the ratio of proteins to RNA, are sensitive to changes in growth conditions, such as the availability of nutrients (Ehrenberg et al., 2013; Klumpp et al., 2009). However, the effect of mutations or changed growth conditions on the abundances of individual proteins in the cytoplasm is not known. The key objective of the present study is to understand to what extent point mutations in a metabolic enzyme and/or variations in the media affect the proteome composition in the bacterial cytoplasm and how these changes are related to the fitness effects of such mutations.
We used isobaric tandem mass tag (TMT) proteome labeling with subsequent LC-MS/MS to analyze changes in the E. coli proteome in response to a selected set of destabilizing mutations in the chromosomal copy of the folA gene (encoding the core metabolic enzyme DHFR) and found that these mutations reproducibly change the abundances of most detected E. coli proteins. Furthermore, we established that the proteome-level changes are directly related to the fitness effects of these mutations and/or media variation during the growth of the E. coli strains.
Results
Effect of DHFR mutations and media variations on E. coli fitness
folA is an optimal target for studying the genotype-phenotype relationship. First, its product is an important metabolic enzyme. DHFR catalyzes the electron transfer reaction to form tetrahydrofolate, a carrier of single-carbon functional groups utilized in biochemical reactions of the central metabolism, including the de-novo synthesis of purine, pyrimidine, methionine, and glycine (Schnell et al., 2004). Hence, DHFR is an essential enzyme whose function is directly linked to organismal fitness. Second, since DHFR is present at a low copy number (only 40 copies/cell) (Taniguchi et al., 2010), its mutants are less likely to cause aggregation-associated toxicity. Finally, DHFR is a well-established antibiotic target with a competitive inhibitor, trimethoprim, readily available (Toprak et al., 2012). Recently we introduced a set of chromosomal missense point mutations in the open reading frame of the E. coli folA gene and simultaneously evaluated their effects on the biophysical and biochemical properties of the encoded DHFR and on E. coli’s fitness (Bershtein et al., 2013; Bershtein et al., 2012). The mutations were selected to include both conserved and variable loci and to cover a broad range of molecular effects on the stability of the protein (Bershtein et al., 2012). Whereas many destabilizing DHFR mutants escaped aggregation or degradation by forming soluble oligomers and, as a result, were not detrimental, a subset of mutations did cause a noticeable loss of fitness (Bershtein et al., 2012). In the present study, we focused on this latter subset of DHFR mutations. Specifically, we selected four mutant strains carrying single and multiple destabilizing mutations with estimated ΔΔG values (based on the assumption of additivity of stability effects of single point mutations) ranging from 2.8 to 6.4 kcal/mol. Growth rates of the strains were used as a proxy for their fitness (Brauer et al., 2008; Geiler-Samerotte et al., 2011; Geiler-Samerotte et al., 2013). Mutants W133V and V75H+I155A showed a slight drop in growth rates, while the growth of V75H+I91L+I155A and I91L+W133V strains was severely compromised (Figure 1). We determined that the observed loss of fitness stems primarily from the loss-of-function effect of the destabilizing mutation that renders DHFR molecules susceptible to rampant aggregation or degradation in the cell (Bershtein et al., 2013). Indeed, the fitness levels of the DHFR mutant strains can be almost fully restored to the wild-type (WT) levels either metabolically or functionally. Metabolic complementation was achieved by supplementing the growth media with the “folA mix” – a combination of purine, thymidine, pantothenate, glycine and methionine known to sustain the growth of E. coli lacking the folA gene (see Experimental Procedures and (Singer et al., 1985)). Addition of the “folA mix” equalizes the growth rates between WT and the mutants by decreasing the growth rate for WT and significantly increasing it for the mutants (Figure 1). Functional complementation was achieved by plasmid expression of WT DHFR. Addition of WT DHFR fully restores the fitness of all mutants, except for V75H+I91L+I155A (Figure 1), indicating that for this mutant alone, secondary effects, such as misfolding-related toxicity (Drummond and Wilke, 2008; Geiler-Samerotte et al., 2011), might play a more noticeable role.
Figure 1. The effect of mutations, media composition, and DHFR overexpression on bacterial growth.
Growth rates of all studied strains under various conditions. Growth rates were determined from the exponential phase of growth using the three parameter fit of ln(OD/OD0) vs time curves proposed in (Zwietering et al., 1990). Both metabolic (“folA mix”) and functional (pBAD WT DHFR overexpression) complementation brought the growth rates of the mutant strains very close to the WT levels. Error bars represent the standard deviation of three independent growth measurements.
The extent of proteome variation is anti-correlated with E. coli fitness
To determine the relationship between the fitness of the selected mutant strains and the systems-level response to the DHFR mutations, we quantified changes in the protein abundances in the E. coli proteome. To this end, we applied chemical labeling based on isobaric TMT technology with subsequent LC-MS/MS quantification (Altelaar et al., 2013; Slavov et al., 2014; Thompson et al., 2003). This method allowed us to obtain relative protein abundances (RPA) between each strain/condition in question and a reference strain. As a reference, we chose WT E. coli in our standard growth media (M9 supplemented with amino acids; see Experimental Procedures). We obtained RPA for about half of the E. coli proteome (~2000 proteins, see Table 1) for each mutant strain and media condition (standard M9 and M9 supplemented with the “folA mix”) (see Experimental Procedures, and Table S1 for RPA of each individual protein). In addition, we determined RPA in the WT strain in the presence of trimethoprim (TMP), an antibiotic that inhibits the DHFR activity (Table S1). In total, we quantified 11 proteomes that included all conditions listed in Figure 1, except the functional complementation of DHFR activity (plasmid expression). To control for natural biological variation at different stages of growth, we also collected the RPA data for WT strains grown to different optical density (OD) levels (Table S1). We were able to detect and quantify close to 2,000 proteins available for direct comparison between all 11 proteomes.
Table 1.
Number of genes and protein products detected and quantified in the quantitative proteomics and transcriptomics experiments.
| Proteome (− folA mix) | Proteome (+ folA mix) | Transcriptome | |||
|---|---|---|---|---|---|
| W133V | 2195 | WT | 1849 | W133V | 4192 |
| V75H+I155A | 2194 | W133V | 1850 | V75H+I155A | 4189 |
| TMP 0.5 µg/mL | 2195 | V75H+I155A | 1850 | TMP 0.5 µg/mL | 4189 |
| V75H+I91L+I155A | 2194 | V75H+I91L+I155A | 1849 | V75H+I91L+I155A | 4191 |
| I91L+W133V | 2195 | I91L+W133V | 1847 | I91L+W133V | 4189 |
To assess the relationship of the proteome changes to the transcriptome, we obtained, under identical experimental conditions, transcripts of the folA mutant strains and the WT strain treated with 0.5 µg/mL of TMP (see Experimental Procedures and Supplemental Information). The complete transcriptomics data are provided in Table S2.
We plotted the distributions of logarithms of RPA (LRPA) and found that their standard deviations (S.D.) vary widely from strain to strain (Figures 2A and S1). The logarithms of mRNA abundances relative to WT (LRMA) are distributed qualitatively similar to LRPA (Figure 2B). (Note that the means of the LRPA distributions may vary from sample to sample due to slight variation of final OD of samples, so cannot be a reliable measure of the systems-level response.) The S.D. of LRPA distributions are directly correlated with the key biophysical property of the mutant DHFR variants – their thermodynamic stability (Figure 2C). More strikingly, there exists a robust and highly statistically significant anti-correlation between the S.D. of LRPA and the growth rates (Figure 2D). Generally, the S.D. of LRMA are about twice as big as the S.D. of LRPA (Figure 2E), suggesting that mRNA abundances are more sensitive to genetic variation, probably due to the lower copy numbers of mRNAs compared to the proteins that they encode.
Figure 2. Global statistical properties of proteomes correlate strongly with fitness.
A. Distribution of LRPA. (Data shown are for W133V strain. LRPA distributions for all strains and conditions can be found in related Figure S1). B. Same as A for LRMA. C. S.D. of LRPA are directly correlated to the destabilizing effects of mutations. The change (relative to WT DHFR) in folding free energy (ΔΔG) for multiple mutants is calculated by the additive approximation using empirical measurements obtained for single DHFR mutants in (Bershtein et al., 2012). D. S.D. of LRPA is anti-correlated with growth rate. E. S.D. of LRMA is correlated with S.D. of LRPA. The slope is close to 2, suggesting that transcriptomes are more readily perturbed than proteomes. (See related Figures S1 and S2)
Importantly, the variation of S.D. of LRPA between strains and conditions is not a mere consequence of natural biological variation between growth stages: the S.D. of LRPA for the WT strain grown to different OD remain remarkably constant (Figure S2). In addition, when comparing two proteomes extracted independently from the WT strain grown up to entrance into stationary phase under identical conditions (biological repeats), the correlation of LRPA between them is very high (R = 0.94) (Figure S4), indicating that the TMT-labeling based proteome quantification technique is highly reproducible.
Point mutations in the folA gene deterministically affect abundances of most proteins
The broad distributions of LRPA and LRMA might indicate that variations in protein and mRNA abundances are just a consequence of stochastic sample-to-sample variation between colony founder cells. If this were the case, we could not see strong reproducibility from sample to sample and/or between strains. Another possibility is that broad distributions of LRPA and LRMA are due to long-time intrinsic stochasticity in gene expression (Elowitz et al., 2002), which extends beyond the cell-to-cell variation to affect the total abundances in the bulk. In that case, we might still find that the overall statistical properties of the proteome response to mutations, such as S.D. of LRPA/LRMA, are robust, i.e., reproducible, between samples in biological repeats. An extreme scenario of this case is that each protein abundance varies deterministically in response to genetic or media variation. By a “deterministic” response, we mean that the LRPA/LRMA of each protein is reproducible (apart from the experimental noise) from sample to sample at the identical conditions. We note that the mere analysis of the distribution LRPA or LRMA from individual experiments does not allow us to distinguish between stochastically and deterministically varying quantities since the LRPA or LRMA for all genes, whether stochastic or deterministic, appear to be drawn from the same distributions, as shown in Figures 2 and S1. Therefore, only comparison of LRPA/LRMA between biological repeats can reveal the degrees of stochasticity and determinism in the proteomics and transcriptomics responses to folA mutations.
For further analysis, we separated the strain-to-strain variation of global statistical properties -- average LRMA/LRPA and its S.D. -- from the variation of the abundances of individual proteins. To that end we normalized LRPA and LRMA for each gene in each strain to obtain z-scores:
| (1) |
where index i refers to gene, is the LRPA or LRMA for gene i, 〈Ystrain〉 denotes an average quantity Yi over all genes for a given strain or condition in corresponding experiments, and is the S.D. of , a quantity already plotted on Figure 2B.
Next, we estimated how many proteins change their abundances deterministically in response to a mutation and/or media variation. Specifically, we assumed that the LRPA or LRMA in a proteome of total K proteins separate into two groups: N proteins, whose relative-to-WT variation is deterministic, and the remaining (K-N), whose variation is stochastic. We also assumed that the LRPA or LRMA of individual genes (and therefore their corresponding z-scores) obtained in a single experiment (as shown in Figures 2 and S1) are drawn from the same distribution so that it is not possible to decompose this distribution into distinct distributions corresponding to stochastically and deterministically varying genes or protein abundances. Therefore, we turned to the comparison of biological repeats in order to determine the fraction of deterministically changing genes.
For N “deterministic” genes, the z-scores of LRPA obtained from different biological repeats A and B for the same strain s are identical, up to the experimental noise:
| (2) |
where ηi is the experimental noise and is the LRPA z-score for particular gene i of strain s in the biological repeat experiment A. The z-scores of the remaining K-N “stochastic” genes are statistically independent between biological repeats.
A simple statistical analysis based on the application of the central limit theorem (see Supplementary Methods) establishes the relationship between the number of deterministically varying genes, N, to the Pearson correlation, r, between the sets of LRPA or LRMA z-scores and determined for biological repeats A and B:
| (3) |
The data (Figure S3) show that the Pearson correlation between z-score sets for biological repeats for both LRPA and LRMA is high, in the range 0.56–0.95 (overall higher for LRMA than for LRPA), suggesting that most of the observed LRMA and LRPA in the mutant strains are not just simple manifestation of a noisy gene expression, or an epigenetic sample-to-sample variation in the founder clones. Rather, we observed that in each case more than 1,000 genes vary their mRNA and protein abundances in a deterministic manner in response to point mutations in the folA gene. It is important to note that this conclusion does not depend on the assumptions about the amplitude of the experimental noise. Eq. 3 still holds with significant accuracy even if the experimental noise in the LRMA or LRPA measurements is comparable to the amplitude of abundance changes. As shown in Supplementary Methods, the reason for that conclusion is that the Pearson correlation is evaluated over a very large number of genes, i.e. K≈2000≫1, whereas the relative error in Eq. 3 is of the order of .
A possible confounding factor is that the observed deterministic variation of LRPA is due to variation between the growth stages and culture densities for different strains. To explore this possibility, we again compared the proteomes of the folA mutant strains to the proteomes of WT grown to different OD. Low correlations between the WT and mutant proteomes at all OD (Figure 3A) indicate that the variation of proteomes at different growth stages does not account for the LRPA in the mutant strains.
Figure 3. Correlation between proteomes and transcriptomes.
A. Scatter plots of z-scores of LRPA between proteomes of the wild-type strain grown to various OD levels, and proteomes of mutant strains, and TMP-treated WT strain. Low correlations indicate that perturbations observed in response to DHFR mutations or functional inhibition by TMP is largely unrelated to natural variation rooted in different stages of the growth cycle. B. Scatter plots and correlation coefficients between proteomes of all strains under standard growth conditions (right panel), and in presence of the “folA mix” (left panel). Addition of the “folA mix” minimizes the variation between different proteomes. C. Transcriptomics data obtained for strains grown under standard conditions (identical to A). Correlations are overall higher for mRNA abundances, but similar classes of transcriptomes are discernible. (See related Figures S3 and S4 for correlation in biological repeats).
We conclude that the E. coli proteome and transcriptome are highly sensitive to point mutations in the metabolic enzyme DHFR; a vast number (in the range of 1000–2000) of genes vary their transcription levels and abundances in response to mutations in the folA gene.
Growth rate is not the sole determinant of the proteomes of mutant strains
Next, we determined the Pearson correlation coefficient between the LRPA z-scores for all strains and conditions. There is a remarkable pattern in the correlations between proteomes of different strains. Proteomes that show a moderate decrease in growth (W133V, V75H+I155A, and WT treated with 0.5 µg/mL of TMP) are closely correlated between themselves, as are the proteomes of strains with a severe decrease in growth rates (I91L+W133V, V75H+ I91L +I155A, and WT treated with 1 µg/mL of TMP) (Figure 3B, top panel). The correlation between members of these two groups is considerably weaker, albeit still highly statistically significant. Addition of the “folA mix,” which nearly equalizes the growth between WT and even the most detrimental mutants (Figure 1), significantly reduces this separation into two classes, making correlations between all proteomes uniformly high (Figure 3B, left panel). A similar, but less pronounced pattern of correlations is observed for LRMA (Figure 3C).
The observation that strains having similar growth rates tend to have similar proteomes might suggest that the growth rate is the single determinant of the proteome composition. However, a more careful analysis shows that this is not the case: the growth rate is not the sole determinant of the proteome composition. We clustered the LRPA z-scores using the Ward clustering algorithm (Ward, 1963) (see Supplemental Information) and found that proteomes cluster hierarchically in a systematic, biologically meaningful manner (Figure 4A). At the first level of the hierarchy, proteomes separate into two classes depending on the growth media: strains grown in the presence of the “folA mix” tend to cluster together as do the strains grown in supplemented M9 without the “folA mix.” At the next levels of the hierarchy, i.e. at each media condition, strains cluster according to their growth rates (Figure 4A).
Figure 4. Proteomes cluster hierarchically according to media conditions and growth rates.
A. Ward hierarchical clustering of proteomes. Values of the horizontal axis at split points indicate Ward distances between corresponding clusters (see Supplementary Methods). B. Same as A for null model proteomes generated to correlate according to growth rates. C. Correlation between Ward distance at branch point of a cluster and number of members of the cluster for real proteomes. D. Control: same as C for NMPs.
Hierarchical clustering of proteomes suggests a peculiar interplay of media conditions and the internal state of the cells (growth rate) in sculpting their proteomes. To evaluate the significance of this finding, we generated hypothetical null model proteomes (NMPs) whose correlations are determined exclusively by their assigned growth rates (see Supplemental Information), and clustered them by applying the same Ward algorithm. We stochastically generated numerous NMPs (as described in Supplemental Information) and found, for each realization, the same tree (Figure 4B). The NMP tree in Figure 4B is qualitatively different from the real data (Figure 4A), thereby rejecting the null hypothesis that the growth rate is the sole determinant of the correlation between the proteomes. The differences between real and null model proteomes are further highlighted by the observation that real proteomes cluster hierarchically while NMPs do not. Each branch point on the tree represents the root of a cluster, which has two properties, the Ward distance at the branch point (i.e., branch point on the x-axis coordinate) and the number of members – proteomes that belong to it (Figure 4). For hierarchical clustering these two properties are correlated, while for simple trees they are not. Indeed, the analysis shows that real proteomes cluster hierarchically while NMPs do not (Figures 4C and 4D).
folA expression is up-regulated but DHFR abundances drop in the mutant strains
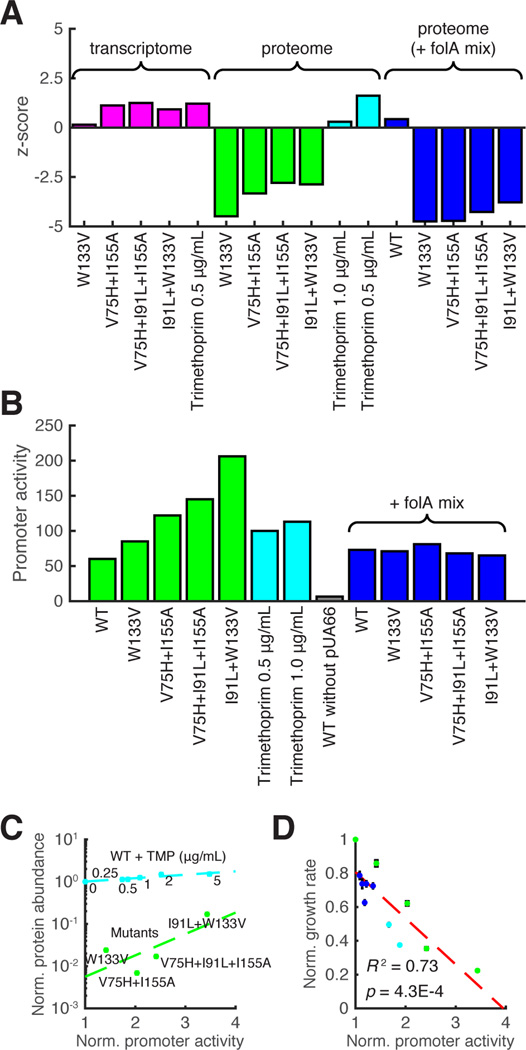
Transcriptomics data show that expression of the folA gene is up-regulated in all the mutants, and, as noted before (Bollenbach et al., 2009), in the WT strain exposed to TMP (Figure 5A). However, the increase in DHFR abundance can be detected only in the TMP-treated WT strain. All mutant strains show a significant loss of DHFR abundance (Figure 5A), presumably due to degradation and/or aggregation inside the cell.
Figure 5. DHFR protein and mRNA abundances A.
Changes in folA mRNA and DHFR protein abundances (relative to WT, in z-score), as detected by the high-throughput techniques for all strains. Expression of both TMP-treated WT and mutant folA genes is upregulated (transcriptome data, blue columns). The abundance of WT DHFR is also elevated with TMP addition. Conversely, all mutant DHFR proteins show a substantial drop in abundances under standard growth conditions (green columns), and upon addition of the “folA mix.” B. Direct measurements of the folA promoter activity using GFP reporter plasmid for all strains and for all conditions studied. Metabolic complementation (addition of the “folA mix”) eliminates the up-regulation of expression of the mutant folA genes, despite the drop in abundances of the mutant DHFR proteins (Fig.4). C. DHFR abundance determined by the western blot correlates with promoter activity but dramatically differs between mutant strains and TMP-treated WT. Variation in abundance measurements by Western Blot (standard deviation obtained in 4 independent experiments) constitutes app. 8%. D. The level of folA promoter activation is strongly anti-correlated with the bacterial growth rate. The color code of strains and conditions is the same as in Figure 1 and Figure 4.
We sought to explore this observation further using targeted analysis of the folA promoter activity and intracellular DHFR abundance. To that end, we used a reporter plasmid in which the folA promoter is fused to the green fluorescent protein (GFP) (Zaslaver et al., 2006) and quantified the DHFR abundance with the western blot using custom-raised antibodies (see Experimental Procedures). The measure of the promoter activation -- GFP fluorescence normalized by biomass (OD) -- is shown in Figure 5B for all strains. Consistent with the transcriptomics data, the loss of DHFR function causes activation of the folA promoter proportionally to the degree of functional loss, as can be seen from the effect of varying the TMP concentration. Conversely, the abundances of the mutant DHFR proteins remain very low, despite the comparable levels of promoter activation (Figure 5C). The addition of the “folA mix” brought promoter activity of the mutant strains close to the WT level (Figure 5B). This result clearly indicates that the cause of activation of the folA promoter is metabolic in all cases. Overall, we observed a strong anti-correlation between growth rates and promoter activation across all strains and conditions (Figure 5D), consistent with the view that the metabolome rearrangement is the master cause of both effects - fitness loss and folA promoter activation.
Major transcriptome and proteome effects of folA mutations extend pleiotropically beyond the folate pathway
Combined, the proteomics and transcriptomics data provide a significant resource for understanding the mechanistic aspects of the cell response to mutations and media variation. The complete data sets are presented in Tables S1 and S2 in the Excel format to allow an interactive analysis of specific genes whose expression and abundances are affected by the folA mutations.
To focus on specific biological processes rather than individual genes, we grouped the genes into 480 overlapping functional classes introduced by Sangurdekar and coworkers (Sangurdekar et al., 2011). For each functional class, we evaluated the cumulative z-score as an average among all proteins belonging to a functional class (Table S3) at a specific experimental condition (mutant strain and media composition). A large absolute value of indicates that LRPA or LRMA for all proteins within a functional class shift up or down in concert.
Figures 6A and S5 show the relationship between transcriptomic and proteomic cumulative z-scores for all gene groups defined in (Sangurdekar et al., 2011). While the overall correlation is statistically significant, the spread indicates that for many gene groups their LRMA and LRPA change in different directions. The lower left quarter on Figures 6A and S5 is especially noteworthy, as it shows several groups of genes whose transcription is clearly up-regulated in the mutant strains whereas the corresponding protein abundance drops, indicating that protein turnover plays a crucial role in regulating such genes. Note that inverse situations – when transcription is significantly down-regulated but protein abundances increase – are much less common for all strains. Interestingly, this finding is in contrast with observations in yeast where induced genes show high correlation between changes in mRNA and protein abundances (Lee et al., 2011).
Figure 6. Pleiotropic biological effects of DHFR mutation and DHFR inhibition by trimethoprim.

A. Scatter plot showing the relative variation of cumulative average z-scores for transcriptomic changes (LRMA) and proteomic changes (LRPA) for overlapping groups of genes defined in (Sangurdekar et al., 2011). Data points corresponding to groups of genes whose cumulative average LRMA z-scores are positive (up-regulation) are shown in red, while transcriptionally down-regulated groups are shown in blue. The data shown are for the W133V strain (see related Figure S5 for the remaining mutant strains and TMP-treated WT). B. Variation for specific gene groups. Clustering global variation in mRNA and protein abundances as belonging to functional classes (upper panel) (see (Sangurdekar et al., 2011)) or co-regulated by a specific operon (lower panel) reveals the highly statistically significant variation of several functional groups. The color code indicates the direction of change (blue – down-regulation, red – up-regulation) and the color depth codes for the logarithm of p-values against the null model of independent variation within a group of genes (see Supplemental Information).
As a next step in the analysis, we focused on several interesting functional groups of genes, especially the ones that show opposite trends in LRMA and LRPA. The statistical significance p-values that show whether a group of genes is significantly up- or down-regulated, either in the proteome or the transcriptome or both, can be estimated based on a simple null model of independence of LRPA or LRMA of genes within a class, as explained in Supplemental Information. Figure 6B shows the p-values for variation of LRPA/LRMA for genes grouped by function (upper panel) and by operon (lower panel).
Besides shifts in folA expression and DHFR abundances, significant variations were found for many important functional groups of genes (Figure 6B, upper panel; due to the overall large dynamic range of p-values, some statistically significant changes may be difficult to discern in the figure. See Table S3 for actual p-values.). First, the genes responsible for motility shut down across the mutant strains with a concomitant drop in their protein abundances (see the fliA operon in Figure 6B, lower panel). Interestingly, addition of the “folA mix” completely reverses this trend (except for only partial reversal for the I91V+W133V mutant). Also, while a broad set of SOS response genes is transcriptionally up-regulated (in contrast to the RpoS-regulated subset of stress-induced genes), the protein abundances of these gene products are highly elevated only in the slowest growing strains, I91L+W133V and V75H+I91V+I155A. Addition of the “folA mix” alleviates the SOS response in all strains. Moreover, TMP does not trigger the SOS response at either 0.5 nor 1.0 µg/mL, nor does it trigger DNA repair genes. Possibly, the depletion of precursor purines and pyrimidines might not lead to overall DNA damage that triggers the SOS response. Expression of genes belonging to the pyrimidine biosynthesis pathway is significantly up-regulated, but the abundances of their protein products drop in all strains, with most significant impact on the slower growing I91L+W133V and V75H+I91V+I155A strains and WT treated with a high concentration of TMP. Addition of the “folA mix” again reverses this proteomic trend, giving rise to increased abundances of all the gene products belonging to this pathway.
folA mutations cause a wide-spread transcriptional rewiring in E. coli
Additional systematic insights come from the analysis of the variation of genes grouped by common transcriptional units regulated by operons. For example, the genes responsible for the uptake of ferric ions (under the Fur regulator) exhibit major transcriptional down-regulation and a concomitant drop in protein abundance. For some genes, however, variations of transcript numbers and protein abundances do not exactly go hand in hand. For example, arginine catabolism genes (ArgR operon) are transcriptionally up-regulated (Figure 6B, lower panel). However, their protein abundances significantly drop in the mutant strains in the M9 medium and slightly drop in the presence of the “folA mix.” This effect is probably common to the genes in the nitrogen metabolism pathway, as seen for the RpoN and NtrC operons. Other pathways like catabolite activation (CRP) and fumarate/nitrate reduction (FNR) show concerted transcriptome and proteome changes (up-regulation in both cases) for the folA mutants that moderately affect growth rates (W133V and V75H+I155A). However, there is a reversal of this trend for the mutants that exhibit severely compromised growth (V75H+I91L+I155A, I91L+W133V), and the abundances of CRP- and FNR-regulated proteins drop significantly. An interesting insight comes from the analysis of RpoS-dependent genes. It has been shown that the phosphorylated response regulator ArcA is a direct suppressor of RpoS transcription (Mika and Hengge, 2005). Indeed, we observed transcriptional up-regulation of ArcA and down-regulation of RpoS. However, at the proteome level there is a down-regulation of ArcA for the V75H+I91L+I155A and I91L+W133V strains, while a small but noticeable increase in the abundance of proteins controlled by RpoS for the same mutants. This also holds true for the WT at high concentration of TMP.
Discussion
Quantitative transcriptomics and, more recently, proteomics are powerful tools in systems biology. They have been widely used to analyze systems-level changes associated with disease phenotypes in mammalian cells (Vogel and Marcotte, 2012). Other applications include the study of the systems-level response to major perturbations such as whole genome duplication (de Godoy et al., 2008), osmolarity and oxidative stresses (Maier et al., 2011; Vogel and Marcotte, 2012), and loss of function mutations in the RNA degradosome in E. coli, which affect global RNA turnover and regulation (Bernstein et al., 2004; Zhou et al., 2013). Also, quantitative proteomics was used to explore the general relationship between cells proteome and growth rates (Brauer et al., 2008; Geiler-Samerotte et al., 2013; Slavov and Botstein, 2011). In particular, Drummond and colleagues established the relationship between growth rates and the total numbers of soluble and insoluble proteins in yeast (Geiler-Samerotte et al., 2013). In contrast to earlier studies, the focus of the present work is on the systems-level proteome and transcriptome response to the minimal and most fundamental genetic perturbations – missense point mutations introduced through genome editing into a core metabolic enzyme.
A popular conceptual view in systems biology postulates that modularity and stability of transcriptional networks had evolved to confer robustness to biological systems (Bornholdt and Sneppen, 2000; Wagner and Wright, 2007). In particular, an effect of a point mutation in a robust biological system should be limited to genes and their protein products that physically, genetically or metabolically interact with a mutated protein. Here we found that local perturbations of DHFR function reproducibly affect transcription and protein abundances of a huge number of genes apparently unrelated to the folate pathway, highlighting a highly pleiotropic systems-level effect of mutations in DHFR. A detailed analysis of groups of genes provided a rationale for some but not all of these shifts. All mutant and TMP-treated WT strains shut down motility, presumably as a way to conserve resources. However, for many pathways an intuitive explanation of the changes is not obvious. For example, genes responsible for nitrogen metabolism and ferric ion uptake are significantly affected. Moreover, for these genes, mRNA and protein abundances change in the opposite directions in a statistically significant way, indicating the importance of regulation at the level of protein turnover. Another striking example of the turnover effect is DHFR itself. Both destabilizing DHFR mutations and TMP treatment caused activation of the folA promoter. However, the abundance of DHFR proteins increases only upon TMP treatment. Up-regulation of the gene does not save the destabilized mutants. This effect can be attributed to protein quality control (PQC), which detects and degrades partly folded mutant DHFR (Bershtein et al., 2013). It should be noted that the overall increase in DHFR abundance upon TMP treatment cannot alleviate the detrimental fitness effect of TMP; the number of active DHFR molecules would still decrease upon addition of TMP due to the inhibition of DHFR by the antibiotic.
The key finding of this study is that point mutations in an essential enzyme have a profound pleiotropic effect extending to the level of the whole proteome and transcriptome. Moreover, the S.D. of the LRPA or LRMA appears to provide a reliable global quantification of the degree of the pleiotropic effects associated with a given mutation. “Narrow” (low S.D.) distributions indicate that the mutations do not induce widespread systems-level perturbations and their fitness effects are minimal, whereas “wide” distributions (high S.D.) reveal a comprehensive systems-level response with ensuing pronounced fitness effects. While we do not have a full mechanistic explanation for this finding, some reasons can be speculated. In particular, we note that partial loss of DHFR function has a profound effect on the pool of cell metabolites (Kwon et al., 2010). Such a global change may affect biophysical properties (such as stability, or Kd of interaction) and the ensuing degradation rates of multiple proteins, thus causing changes in the protein turnover balance. Indirect support for this view comes from the hierarchical clustering of proteomes, which shows that media composition rather than mere growth rate determines the crucial segregation between proteomes at the top of the hierarchy. Mutations in DHFR cause a domino-like effect leading to transcriptional activation of the folA gene, the changes in abundance for the whole E. coli proteome, and finally, changes of fitness of the mutant strains. The quantitative measures of these effects on all scales strongly correlate, suggesting the existence of a common underlying cause that drives these changes. Future studies will reveal the existence and exact nature of this cause.
Experimental Procedures
Promoter activity
Strains were transformed with pUA66 plasmid carrying folA promoter fused to GFP coding gene (Zaslaver et al., 2006). Promoter activity is defined by a ratio between fluorescent signal (excitation 495 nm, emission 510 nm) and biomass production (measured as OD at 600nm)
Intracellular protein abundance
Cells were grown in supplemented M9 medium for 4 hours at 37°C, chilled on ice for 30 min and lysed with BugBuster (Novagen). DHFR amounts in the soluble fraction were determined by SDS-PAGE followed by Western Blot using Rabbit-anti E.coli’s DHFR polyclonal antibodies (custom raised by Pacific Immunology).
Preparation of E. coli strains with chromosomal mutations in folA gene
The genome editing approach to create E. coli strains with chromosomal mutations in folA gene is based on homologous recombination as reported previously (Bershtein et al., 2012).
Media and growth conditions
Cells were grown from a single colony overnight at 30°C in M9 minimal medium supplemented with 0.2% glucose, 1 mM MgSO4, 0.1% casamino acids, and 0.5 µg/mL thiamine. Overnight cultures were diluted 1/100 and grown at 37°C. For proteomics and transcriptomics analysis (see below and Supplementary Methods) cultures were harvested after 4 hours of growth. Growth rate measurements were conducted for 16 hours in Bioscreen C system (Growth Curves USA). OD data were collected at 600nm at 15 min intervals. The resulting growth curves were fit to a bacterial growth model to obtain growth rate parameters (Zwietering et al., 1990). For metabolic complementation (Singer et al., 1985), growth medium was supplemented with 1 mM adenine, 1 mM thymine, 1 µg/mL D-panthothenate, 0.5 mM glycine, and 0.5 mM methionine (the “folA mix”). For functional complementation strains were transformed with pBAD plasmid [EMBL] carrying WT folA gene and grown in presence of 100 µg/mL ampicillin and 0.002% arabinose.
Transcriptomics
Total RNA was extracted using RNeasy® Protect Bacteria Mini Kit following the manufacturer’s instructions. Library construction and sequencing were performed at Genewiz, Inc (South Plainfield, NJ) on the Illumina HiSeq2000 platform in a 1×100bp single-read (SR) configuration, with a total of at least 120 million reads per lane. The reads were aligned to the E. coli MG1655 reference genome (NC_000913) using Rockhopper (McClure et al., 2013) to get transcript levels.
Proteomics
For global proteome analysis, E. coli cells were lysed into 50 mM NaH2PO4 buffer (pH8) supplemented with BugBuster extraction reagent and benzonase (Novagen), following the manufacturer’s instruction. Soluble cell lysates were trypsinized overnight by Promega (Madison, WI) Trypsin/Lys-C enzyme mixture with ratio 1:30 enzyme to protein and labeled with TMT reagent (TMT®, Thermo, San Jose, CA) followed by nanoLC-MS/MS separation and analysis (see SI). Tryptic peptides mixtures were separated on ERLIC chromatography using earlier published protocol (Ma et al., 2014). After separation, each fraction was submitted to 90min LC-MS/MS run to Orbitrap Elite (Thermo, Bremen) mass spectrometer. Eluted from LC peptides were submitted to MS/MS in Orbitrap Elite for a High Collision Dissociation (HCD) and in iontrap instrument for Collision Induced Dissociation (CID) using “Top 20 method with dynamic exclusion”. Briefly, “Top 20 methods” allow mass spectrometer instrument to submit peaks that elute from nanoLC at any given time point to further dissociation process called MS/MS either by HCD or by CID methods and putting already MS/MSed peaks in an exclusion list for next 30 sec to avoid same peaks been peaked up twice for same procedure. This method allow instrument to go deep into proteome and identify majority of peaks that are eluting from nanoLC separation independent from their absolute intensities. Data were searched on Proteome Discoverer 1.4.1.14 (Thermo, San Jose, CA) search engine against E. coli database added with common contaminants and sequences of mutated versions of DHFR protein. All results were filtered by use of Percolator v2.05 (Kall et al., 2007) to 1 % False Discovery Rate (FDR) on protein level. To address the co-isolation interference effect reported for TMT® labeling in MS2 mode (Wuhr et al., 2012), all data were filtered to allow a maximum of 40% of ions co-isolation. Such a threshold was shown to preserve a large body of data without forfeiting the quality of protein quantitation, with exception of ratios >10, for which some level of underestimation was observed (Slavov et al., 2014).
Supplementary Material
Acknowledgements
This work is supported by NIH grant GM068670 (to ES), long-term postdoctoral fellowship from the Human Frontier Science Program (to SB), and NSF grant MCB-1243837. We are grateful to Adrian Serohijos for discussions and help, Bharat V. Adkar for analysis of the transcriptomics data and Will Jacobs and Amy I. Gilson for critical reading of the manuscript and helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers. Transcriptomic sequences were deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra). The experiment accession numbers for the raw reads deposit are as follows: WT -- SRX951916, DHFR mutant M133V -- SRX951945, DHFR mutant V75H+I155A -- SRX951946, DHFR mutant I91L+W133V -- SRX951948, DHFR mutant V75H+I91L+I155A -- SRX951950, and WT with TMP 0.5 µg/mL -- SRX951951. The BioProject accession number for the whole project is PRJNA277706, and the BioSample accession number is SAMN03393359.
Author Contributions. SB and ES designed research, SB and Sanchari Bhattacharyya carried out experiments, JMC and ES adopted statistical tools for quantitative analysis of the data, JMC carried out quantitative analysis, SB, ES and Sanchari Bhattacharyya analyzed the data, and ES wrote the paper.
References
- Altelaar AF, Frese CK, Preisinger C, Hennrich ML, Schram AW, Timmers HT, Heck AJ, Mohammed S. Benchmarking stable isotope labeling based quantitative proteomics. Journal of proteomics. 2013;88:14–26. doi: 10.1016/j.jprot.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Applebee MK, Joyce AR, Conrad TM, Pettigrew DW, Palsson BO. Functional and metabolic effects of adaptive glycerol kinase (GLPK) mutants in Escherichia coli. J Biol Chem. 2011;286:23150–23159. doi: 10.1074/jbc.M110.195305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein JA, Lin PH, Cohen SN, Lin-Chao S. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc Natl Acad Sci U S A. 2004;101:2758–2763. doi: 10.1073/pnas.0308747101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Mu W, Serohijos AW, Zhou J, Shakhnovich EI. Protein quality control acts on folding intermediates to shape the effects of mutations on organismal fitness. Molecular cell. 2013;49:133–144. doi: 10.1016/j.molcel.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Mu W, Shakhnovich EI. Soluble oligomerization provides a beneficial fitness effect on destabilizing mutations. Proc Natl Acad Sci U S A. 2012;109:4857–4862. doi: 10.1073/pnas.1118157109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- Bollenbach T, Quan S, Chait R, Kishony R. Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell. 2009;139:707–718. doi: 10.1016/j.cell.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornholdt S, Sneppen K. Robustness as an evolutionary principle. Proc Biol Sci. 2000;267:2281–2286. doi: 10.1098/rspb.2000.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- Dean AM, Dykhuizen DE, Hartl DL. Fitness as a function of beta-galactosidase activity in Escherichia coli. Genet Res. 1986;48:1–8. doi: 10.1017/s0016672300024587. [DOI] [PubMed] [Google Scholar]
- Dobson CM. Protein folding and disease: a view from the first Horizon Symposium. Nat Rev Drug Discov. 2003;2:154–160. doi: 10.1038/nrd1013. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg M, Bremer H, Dennis PP. Medium-dependent control of the bacterial growth rate. Biochimie. 2013;95:643–658. doi: 10.1016/j.biochi.2012.11.012. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Firnberg E, Labonte JW, Gray JJ, Ostermeier M. A comprehensive, high-resolution map of a gene's fitness landscape. Mol Biol Evol. 2014;31:1581–1592. doi: 10.1093/molbev/msu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SM, Hartl DL, Drummond DA. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:680–685. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Hashimoto T, Dion MF, Budnik BA, Airoldi EM, Drummond DA. Quantifying condition-dependent intracellular protein levels enables high-precision fitness estimates. PloS one. 2013;8:e75320. doi: 10.1371/journal.pone.0075320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- Klumpp S, Zhang Z, Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139:1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YK, Higgins MB, Rabinowitz JD. Antifolate-induced depletion of intracellular glycine and purines inhibits thymineless death in E. coli. ACS Chem Biol. 2010;5:787–795. doi: 10.1021/cb100096f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YK, Lu W, Melamud E, Khanam N, Bognar A, Rabinowitz JD. A domino effect in antifolate drug action in Escherichia coli. Nature chemical biology. 2008;4:602–608. doi: 10.1038/nchembio.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MV, Topper SE, Hubler SL, Hose J, Wenger CD, Coon JJ, Gasch AP. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Molecular systems biology. 2011;7:514. doi: 10.1038/msb.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B. Genotype to phenotype: lessons from model organisms for human genetics. Nat Rev Genet. 2013;14:168–178. doi: 10.1038/nrg3404. [DOI] [PubMed] [Google Scholar]
- Lynch M. Evolutionary diversification of the multimeric states of proteins. Proc Natl Acad Sci U S A. 2013;110:E2821–E2828. doi: 10.1073/pnas.1310980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Ward CC, Jungreis I, Slavoff SA, Schwaid AG, Neveu J, Budnik BA, Kellis M, Saghatelian A. Discovery of human sORF-encoded polypeptides (SEPs) in cell lines and tissue. J Proteome Res. 2014;13:1757–1765. doi: 10.1021/pr401280w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T, Schmidt A, Guell M, Kuhner S, Gavin AC, Aebersold R, Serrano L. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Molecular systems biology. 2011;7:511. doi: 10.1038/msb.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F, Hengge R. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigmaS (RpoS) in E. coli. Genes Dev. 2005;19:2770–2781. doi: 10.1101/gad.353705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangurdekar DP, Zhang Z, Khodursky AB. The association of DNA damage response and nucleotide level modulation with the antibacterial mechanism of the anti-folate drug trimethoprim. BMC Genomics. 2011;12:583. doi: 10.1186/1471-2164-12-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell JR, Dyson HJ, Wright PE. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu Rev Biophys Biomol Struct. 2004;33:119–140. doi: 10.1146/annurev.biophys.33.110502.133613. [DOI] [PubMed] [Google Scholar]
- Singer S, Ferone R, Walton L, Elwell L. Isolation of a dihydrofolate reductase-deficient mutant of Escherichia coli. J Bacteriol. 1985;164:470–472. doi: 10.1128/jb.164.1.470-472.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Botstein D. Coupling among growth rate response, metabolic cycle, and cell division cycle in yeast. Mol Biol Cell. 2011;22:1997–2009. doi: 10.1091/mbc.E11-02-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Budnik BA, Schwab D, Airoldi EM, van Oudenaarden A. Constant growth rate can be supported by decreasing energy flux and increasing aerobic glycolysis. Cell reports. 2014;7:705–714. doi: 10.1016/j.celrep.2014.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soskine M, Tawfik DS. Mutational effects and the evolution of new protein functions. Nat Rev Genet. 2010;11:572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Analytical chemistry. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- Toprak E, Veres A, Michel JB, Chait R, Hartl DL, Kishony R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2012;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Wright J. Alternative routes and mutational robustness in complex regulatory networks. Bio Systems. 2007;88:163–172. doi: 10.1016/j.biosystems.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Ward JH. Hierarchical Grouping to Optimize an Objective Function. J Am Stat Assoc. 1963;58 236-&. [Google Scholar]
- Wuhr M, Haas W, McAlister GC, Peshkin L, Rad R, Kirschner MW, Gygi SP. Accurate multiplexed proteomics at the MS2 level using the complement reporter ion cluster. Analytical chemistry. 2012;84:9214–9221. doi: 10.1021/ac301962s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie CS, Shakhnovich EI. A biophysical protein folding model accounts for most mutational fitness effects in viruses. Proc Natl Acad Sci U S A. 2011;108:9916–9921. doi: 10.1073/pnas.1017572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- Zhang J, Maslov S, Shakhnovich EI. Constraints imposed by non-functional protein-protein interactions on gene expression and proteome size. Molecular systems biology. 2008;4:210. doi: 10.1038/msb.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zhang AB, Wang R, Marcotte EM, Vogel C. The proteomic response to mutants of the Escherichia coli RNA degradosome. Mol Biosyst. 2013;9:750–757. doi: 10.1039/c3mb25513a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwietering MH, Jongenburger I, Rombouts FM, van 't Riet K. Modeling of the bacterial growth curve. Applied and environmental microbiology. 1990;56:1875–1881. doi: 10.1128/aem.56.6.1875-1881.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.