Abstract
Advances in the depth and quality of transcriptome sequencing have revealed many new classes of long non-coding RNAs (lncRNAs). lncRNA classification has mushroomed to accommodate these new findings, even though the real dimensions and complexity of the non-coding transcriptome remain unknown. Although evidence of functionality of specific lncRNAs continues to accumulate, conflicting, confusing, and overlapping terminology has fostered ambiguity and lack of clarity in the field in general. The lack of fundamental conceptual un-ambiguous classification framework results in a number of challenges in the annotation and interpretation of non-coding transcriptome data. It also might undermine integration of the new genomic methods and datasets in an effort to unravel function of lncRNA. Here, we review existing lncRNA classifications, nomenclature, and terminology. Then we describe the conceptual guidelines that have emerged for their classification and functional annotation based on expanding and more comprehensive use of large systems biology-based datasets.
The non-coding RNA universe
The classic view of the transcriptome landscape and its mRNA-centric paradigm for transcript annotation has undergone a fundamental change [1, 2]. The ENCODE project estimates that (mostly non-coding) transcripts cover 62–75% of our genome [3], and contribute greatly to the overall estimate of 80% potentially functional sequence in our DNA [4]. Similarly, RNAseq studies show that transcripts from these non-coding regions dominate the population of non-ribosomal non-mitochondrial RNAs in a human cell [5]. Non-coding RNAs (ncRNAs) have emerged as a major source of biomarkers [6–10], targets for therapeutics [8, 11], and potential explanations for the function of non-coding GWAS variants [12]. Constant expansion of the evidence for broad functionality of ncRNAs [13], in processes ranging from heritable epigenetic change [14] to species-specific changes in cognition [15], may finally answer the long standing question of the role of non-coding DNA in eukaryotic biology [16, 17].
Although there has been an emphasis on the annotation and classification of ncRNAs with properties similar to those of protein-coding mRNAs, the vast majority of genomic space used for RNA production remains under-explored. Moreover, although some ncRNAs share properties with coding mRNAs, such as splicing and polyadenylation [18, 19], sequence conservation [18, 19], and export to the cytosol [20], many others do not, highlighting the differences in the functionalities of coding versus ncRNAs [5, 11, 13, 21–24].
The state of non-coding annotation is still in its early days, but the field now has sufficient perspective to establish a logical conceptual framework for the classification of the universe of transcripts that emanate from non-coding genomic regions. New methodologies for integrated classification have accompanied a massive expansion of global transcriptome datasets, particularly from genomics consortiums such as FANTOM [25], ENCODE [3], and GTEx [26]. Methods for grouping RNA sequencing (RNA-seq) (Box 1) reads into a single transcribed region have improved rapidly [27–30]. Progress has also been made in machine learning approaches aimed at the integration and biological interpretation of diverse datasets [29, 31, 32]. All this has culminated in widely used sets of annotations of lncRNAs, such as the one provided by the GENCODE consortium [33], and others [31, 34–37].
Box 1. Overview of high-throughput technologies used to detect and quantify ncRNAs.
RNA sequencing (RNA-seq): currently, one of the most commonly used procedures in transcriptome profiling. Typically, RNA is converted into cDNA using random hexamers followed by massive random sequencing of the resulting cDNAs using next-generation sequencing technologies. As a result, millions of short sequence tags can be generated per experiment. Subsequent mapping of the tags reveals the genomic position encoding the RNA and its relative mass in the cell. The procedure is suitable for various aspects of transcriptome research: RNA mapping, quantitation, alternative splicing analysis etc.
Messenger RNA sequencing (mRNA-seq): RNA-seq on polyA+ fraction of RNA, often synonymous with RNA-seq.
Direct RNA sequencing (DRS): sequencing of native RNA, without library preparation including cDNA conversion step [143], has been successfully used to sequence native polyA+ and identify alternative polyadenylation sites. DRS is particularly useful in applications where artifacts of reverse transcription are undesirable, such as precise strand of origin determination, and in applications that deal with minute amounts of nucleic acids such as single-cell applications. Theoretically, it can provide multiple tags per molecule, however so far it has been used in applications that provide a single tag per molecule at the polyadenylation site.
Cap assisted gene expression (CAGE): a transcriptome profiling procedure that targets RNAs with a 5′-cap [144]. CAGE generates short (typically 27 nt) sequence tags from 5′ ends of such RNAs, with one tag per RNA molecule. It enables accurate mapping of 5′ ends this subset of RNAs.
Serial analysis of gene expression (SAGE): targets polyadenylated messages and generates a single internal (typically close to the 3′ end) tag per RNA molecule [145].
Paired-end tag (PET): also targets polyadenylated RNAs and generates a tag that combines information on 5′ and 3′ ends of the same RNA molecule [146].
Rapid amplification of cDNA ends (RACE): an ‘outward’ PCR-based method designed to identify sequences connected to a given region, which can be used in conjunction with NGS or microarrays, for deep transcriptome profiling of a specific locus [44].
Targeted RNA sequencing: selection of RNAs from a locus of interest using tiling microarrays followed by RNA-seq to achieve the same goal [45].
GRO-seq: A typical RNA profiling experiment measures steady-state levels of RNA. On the other hand, GRO-seq [134] combines nuclear run-on experiments and NGS analysis to provide information on transcription competent RNA polymerase complexes.
Here, we review existing lncRNA classes and then describe the conceptual guidelines that have emerged for their classification and functional annotation based on the expanding and more efficient use of large systems biology-based datasets. This framework endeavours to guide researchers in the classification of ncRNAs and interpretation of next-generation sequencing (NGS) data, especially in non-coding portions of the genome.
Criteria and features of existing classes and categories of lncRNAs
The classification of the great majority of lncRNAs relies on the empirical attributes originally used to detect them (Table 1, Figure 1). This reflects their short history relative to protein-coding genes, and provides a convenient basis by which to classify these un-characterized RNA species.
Table 1.
Different known classes of lncRNAs
| Category | Abbreviation | Reference | Specific Examples |
|---|---|---|---|
| Classification based on transcript length | |||
| Long non-coding RNA | lncRNA | [38, 39] | |
| Long-intergenic non-coding RNA; large Intervening Non-Coding RNA, long-intervening non-coding RNA; | lincRNA | [18] | ANRIL [117], H19 [149], HOTAIR [18], HOTTIP [150], lincRNA-p21 [151], XIST [152], Paupar [153] |
| Very long intergenic non-coding RNA | vlincRNA | [29] | HELLP transcript [42], Vlinc_21, vlinc_185, vlinc_377, vlinc_500 [29] |
| macroRNA | [28, 154] | Airn, Gtl2lt, KCNQOT1, Lncat, Nespas (reviewed in Ref [154]), STAiR1 [28] | |
| Promoter-associated long RNA | PALR | [38] | |
| Classification based on association with annotated protein-coding genes | |||
| Intronic ncRNA; Stable intronic sequence RNA; totally intronic RNA, partially intronic RNA | sisRNA, TIN, PIN | [49, 50, 54] additional references in the text | |
| Circular intronic RNAs | ciRNAs | [55] | |
| Sense ncRNA | [44] | ||
| Natural antisense ncRNA | asRNA, NAT | [57] | BACE1-AS [155], aHIF [156], Tsix [157] |
| Mirror antisense | [44, 67] | Globin antisense [67] | |
| Exonic circular RNAs | ecircRNAs | [62] | cANRIL [118] |
| Chimeric RNAs, trans-spliced RNAs, exon juxtaposition | [44, 63–65] | ||
| Standalone ncRNAs made from 3′UTRs | uaRNA | [60] | |
| Chromatin-interlinking RNA | ciRNA | [68] | |
| Transcription start site-associated RNAs | TSSa-RNAs | [158] | |
| Classification based on association with other DNA elements of known function | |||
| Enhancer-associated RNA | eRNA | [159] | |
| Promoter-associated long RNA | PALR | [38] | |
| Upstream antisense RNA | uaRNA | [160] | |
| PROMoter uPstream Transcript | PROMPT | [89] | |
| Telomeric repeat-containing RNA | TERRA | [161] | |
| Classification based on protein-coding RNA resemblance | |||
| mRNA-like noncoding RNAs | mlncRNAs | [18] | |
| Long-intergenic non-coding RNA; large Intervening Non-Coding RNA, long-intervening non-coding RNA; | lincRNA | [18] | ANRIL [117], H19 [149], HOTAIR [18], HOTTIP [150], lincRNA-p21 [151], XIST [152] |
| Classification based on association with repeats | |||
| C0T-1 repeat RNA | [162] | ||
| Long interspersed nuclear element | LINE1/2 | [163] | |
| Transcribed endogenous retroviruses | [81] | ||
| Expressed Satellite Repeats | [164] | ||
| Non-coding RNA driven by promoters within repeats | vlincRNAs, NASTs | [29, 76] | Vlinc_21, vlinc_185, vlinc_377, vlinc_500 [29] |
| Polypurine-repeat-containing RNA | GRC-RNA | [165] | |
| transcribed pseudogenes | [83] | PTENP1 and KRASP1 [86] | |
| Classification based on association with a biochemical pathway or stability | |||
| Nrd1-unterminated transcript | NUT | [166] | |
| miRNA primary transcripts | [167] | H19 [168] | |
| piRNA primary transcripts | [169] | ||
| Cryptic unstable transcript | CUT | [88] | |
| PROMoter uPstream Transcript | PROMPT | [89] | |
| Xrn1-sensitive unstable transcript | XUT | [91] | |
| Stable Uncharacterized Transcript, Stable Unannotated Transcript | SUT | [92] | |
| Classification based on sequence and structure conservation | |||
| Transcribed-Ultraconserved Regions | T-UCR | [95] | UCR106 [95] |
| Hypoxia-induced noncoding ultraconserved transcript | HINCUT | [100] | |
| Long-intergenic non-coding RNA; large Intervening Non-Coding RNA, long-intervening non-coding RNA; | lincRNA | [18] | HOTAIR [18], HOTTIP [150] |
| RNA-Z regions | [97] | ||
| EvoFold regions | [98] | ||
| Classification based on expression in different biological states | |||
| Long stress-induced non-coding transcript | LSINCT | [101] | |
| Hypoxia-induced noncoding ultraconserved transcript | HINCUT | [100] | |
| Non-Annotated Stem Transcript | NAST | [76] | |
| Classification based on association with subcellular structures | |||
| Chromatin-associated RNA | CAR | [102] | |
| Chromatin-interlinking RNA | ciRNA | [68] | |
| Nuclear bodies associated RNAs | [170] | ||
| PRC2 associated RNAs | [19, 103] | ||
| Classification based on function | |||
| Long noncoding RNAs with enhancer- like function; ncRNA-activating | ncRNA-a | [108] | ncRNA-a7 [108] |
| miRNA primary transcripts | [167] | H19 [168] | |
| piRNA primary transcripts | [169] | ||
| Competing endogenous RNA | ceRNA | [109] | PTENP1 and KRASP1 [86] |
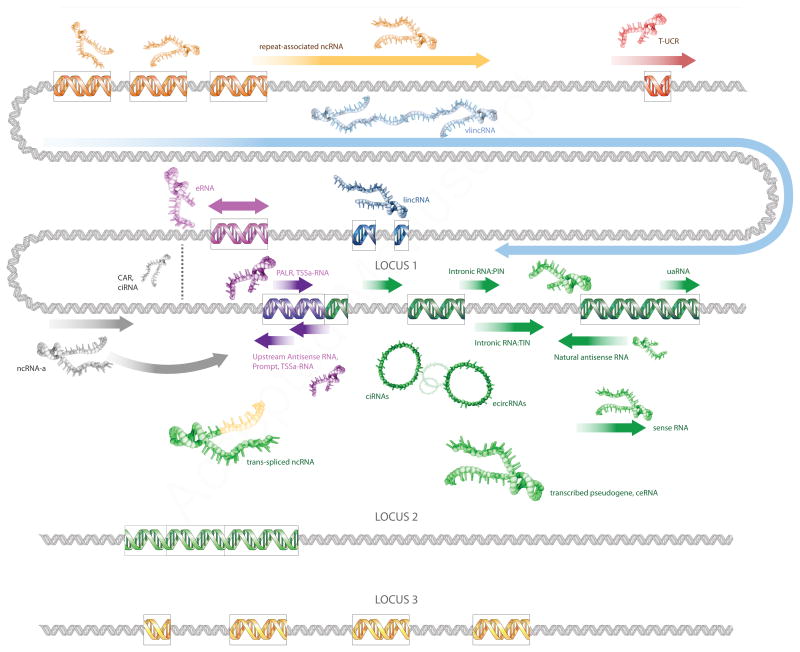
Figure 1. Schematic diagram illustrating various classes of ncRNAs.
Three hypothetical loci are shown. Protein coding exons are shown as green (locus 1) or yellow boxes (locus 3). Locus 2 signifies a pseudogene of locus 1. Regulatory regions of locus 1 are shown in purple (promoter) and magenta (enhancer). Repeats are denoted by brown boxes. Lines with arrows represent ncRNAs. CAR: chromatin-associated RNA. ceRNA: Competing endogenous. RNA ciRNA: chromatin-interlinking RNA (grey) or circular intronic RNA (green). ecircRNA: exonic circular RNA. eRNA: enhancer-associated RNA. lincRNA: long intervening non-coding RNA. ncRNA-a: activating non-coding RNAs. PALR: promoter-associated long RNA. PIN: partially intronic RNA. TIN: totally intronic RNA. TSSa-RNA: transcription start site-associated RNA. T-UCR: Transcribed Ultraconserved Regions. uaRNA: 3′UTR-derived RNAs. vlincRNA: very long intergenic non-coding RNA. The role depicted here for CARs and ciRNAs in stabilizing a chromatin loop is hypothetical.
Classification based on transcript length
The length estimate of ncRNAs serves as the most commonly used attribute for their classification. Typically, a threshold of 200 bases separates long from short ncRNAs [38, 39] (Table 1). Often, our knowledge is limited to sequence reads mapped to a ‘region of transcription’, and even with improvements in NGS read length [40], this will probably continue for the foreseeable future.
Building transcribed regions based on RNA-seq profiling of total RNA (rather than the polyA+ fraction, see below) led to the discovery that intergenic space encodes thousands of very long intergenic non-coding RNAs (vlincRNAs), whose primary transcripts can range in length from 50 kb to 1MB [28, 29, 41]. Spanning at least 10% of human genome [5, 29], vlincRNAs have been implicated in important biological processes such as pluripotency [29], cancer [28, 29], apoptosis [29], cell-cycle progression [28, 42], and cellular senescence [41].
Classification based on association with annotated protein-coding genes
This commonly used attribute (Table 1, Figure 1) serves as the foundation of the GENCODE classification of lncRNAs [33]. It underlies the logical challenge of overlapping non-coding and coding transcripts at a given locus - called “transcriptional forests” by the FANTOM consortium [43]. Targeted methods (Box 1) based on Rapid Amplification of cDNA Ends (RACE) experiments [44] and RNA-seq [45] indicate that transcriptional forests constitute a general feature of the human genome. A prominent category of ncRNAs has emerged from these transcriptional forests composed of sense ncRNAs that overlap coding mRNAs on the same strand and share some sequence with the latter, yet do not encode proteins [44, 46–48]. This category includes unspliced sense partially intronic RNAs (PINs) [49], and spliced transcripts that combine exons from coding and non-coding regions of a gene [47, 48]. GENCODE recognizes the existence of such spliced lncRNAs in their “sense overlapping” biotype [33].
The PIN and “sense overlapping” categories allow for overlap between lncRNAs and exons of a protein-coding gene. However, a protein-coding gene can produce lncRNAs found exclusively in its introns, known as totally intronic RNAs (TINs) [49] (Table 1, Figure 1). TINs make up the majority (~70%) of all non-coding (non-rRNA) nuclear-encoded RNA and 40–50% of all cellular (non-rRNA) RNAs by mass, as established by single-molecule sequencing [50]. Evidence that large numbers of introns encode standalone RNAs originally came from microarray expression profiling [49, 51] and in silico analysis of expressed sequence tag (EST) databases [49, 52]. Even genomes as compact as those of human viruses can encode functional intronic RNAs [53]. Large numbers of standalone intronic RNAs recently found in Xenopus oocytes [54] and mice [50] support the conclusion that introns encode functional ncRNAs on a global scale. Some of these transcripts likely represent circular intronic ncRNAs (ciRNAs) (produced from introns that escape debranching) that can accumulate in cells and regulate expression of their parent genes [55] (Figure 1). Overlap on the opposite DNA strand from their associated protein-coding gene represents another frequently used attribute of lncRNAs. These natural antisense transcripts (NATs) (Figure 1) occur in 50–70% of all protein-coding genes [56, 57].
ncRNAs can also be composed solely of sequences of exons of protein-coding mRNAs (Figure 1, Table 1). For example, transcript cleavage followed by post-transcriptional 5′-cap addition [58, 59] can result in production of stand-alone ncRNAs from various parts of mRNAs [58], notably 3′ UTRs [60]. In fact, the type 0 variant of the cap structure may associate with the post-transcriptionally capped 5′ ends [61]. Additional cellular processes could produce this type of ncRNA, such as the reverse-splicing implicated in the production of circular exonic RNAs [62], trans-splicing leading to production of chimeric RNAs [63, 64], exon juxtaposition [65], and presumed RNA copying, leading to production of “mirror antisense” transcripts [44, 66, 67]. Finally, RNAs whose sequences have features of bona fide coding transcripts may have other roles as revealed by the class of chromatin-interlinking RNAs (ciRNAs). These RNAs participate in maintaining interphase chromatin configuration and mostly include spliced transcripts with long 3′UTRs [68].
Classification based on association with other DNA elements of known function
Notable classes of such RNAs include enhancer- and promoter-associated long RNAs (Table 1, Figure 1). These long RNAs are involved in linking the dynamics of nuclear architecture, chromatin signalling plasticity, and transcriptional regulation [69]. Interestingly, enhancers that give rise to RNA species have greater likelihood of functionality in reporter assays than those that do not [70], arguing for a functional, rather than spurious, link between RNA and this type of genomic element.
Classification based on mRNA resemblance
As mentioned previously, research has focussed on ncRNAs with a spliced structure, conserved sequence, and a polyA tail [18, 19, 34, 35, 71] (Figure 1). In fact, lncRNAs annotated by GENCODE - even those solely confined to intronic sequences - represent primarily spliced transcripts [33]. These features were used to identify thousands of transcripts in mouse [18] and human [19], called long intervening ncRNAs (lincRNAs) [18]. This approach has revealed many important functional lncRNAs, such as HOTAIR, which mediates gene silencing by facilitating localization of the epigenetic repressor Polycomb repressive complex 2 (PRC2) to its target sequence [72]. Expression analysis of ~10,000 human lincRNAs across 1,300 tumour samples using microarrays revealed hundreds non-coding transcripts potentially driving four different cancer types [73]. Numerous other studies have implicated lincRNAs in human development and disease [74]. As an indication of their functionality, expression analysis of 11 tetrapod species found 2,508 lincRNAs expressed in at least three species and originating more than 90 million years ago [71].
Classification based on association with repeats
About half of the human genome consists of repeats of various categories, and many ncRNA-encoding genomic regions overlap these elements (Figure 1). Promoters within repeats drive expression of many ncRNAs [75], especially in pluripotent [29, 76, 77] and cancerous cells [29]. Promoters within the long tandem repeats (LTRs) of endogenous retroviruses specifically associate with ncRNAs from several classes of non-annotated stem transcripts (NASTs) [76] in pluripotent stem cells, including lincRNAs [77] and vlincRNAs [29] (Table 1). In addition, LTR-driven vlincRNAs identify common regulatory architectures between stem and cancer cells [29], an interesting reminder of ideas from the stem cell theory of cancer [78].
Individual copies of repeats are expressed from their own promoters and contribute to the ncRNA transcriptome. For example, RNA polymerase III (Pol III) transcribes non-coding repeated elements, such as Alu, B1, and B2, that can bind to RNA Pol II and affect its activity in response to stress [79]. Long interspersed nuclear elements (LINEs) comprise 20% of the genome and express mostly non-coding transcripts due to 3′ truncation and accumulated mutations [80]. Similarly, expression of non-coding ERVs is a well-documented phenomenon, [81]. Finally, examination of transcripts containing repeat copies continues to reveal additional regulatory functions for these molecules, as exemplified by Alu-mediated inter-molecular interaction of coding and ncRNAs in trans described recently [82].
Transcripts from a specific subset of repeated sequences – non-coding copies of protein-coding genes or mRNAs (pseudogenes) [83] have gained prominence upon realization that they can function in various ways [84], including binding and titrating regulatory molecules that normally interact with the functional copies [85, 86]. Moreover, pseudogenes can be transcribed from the opposite strand thus producing transcripts capable of inter-molecular interaction with the productive copy [87] or its promoter [85].
Classification based on a biochemical pathway or stability
Classification of ncRNA based on their association with substrate pools of different RNA degradation pathways and enzymes has recently gained popularity. Inhibition of components of the exosome (RRP6, RRP40, and RRP44) or nonsense mediated decay (XRN1) has revealed populations of ncRNAs not easily observed in wild-type cells [88–92] (Table 1). This approach also provides information about the pathways of their metabolism. The latter is another attribute used for classification of ncRNAs, as exemplified by XUTs (Xrn1-sensitive unstable transcripts) [91] (Table 1). Most of the pathways analysed so far in this classification involve RNA degradation, and these lncRNAs overlap with classes of NATs [91] and promoter-associated RNAs [89, 90, 92].
Classification based on sequence or structure conservation
Sequence conservation, though highly informative in predicting functional protein-coding mRNAs, remains a metric with controversial merit in the non-coding space. Its absence - typical for lncRNAs [93] - does not universally imply lack of functionality [22, 24]. Still, many ultra-conserved regions (UCRs) – sequences of DNA 100% conserved in human, rat, and mouse – map to the non-coding space of the genome [94]. A large number of UCRs are transcribed as ncRNAs, and some are associated with malignant states [95]. As secondary RNA structure plays a crucial role in ncRNA function [24, 96], a number of bioinformatics approaches such as RNA-Z [97] and EvoFold [98] leverage structure rather than sequence conservation to predict ncRNA-encoding regions (Table 1) [99].
Classification based on biological states
A number of cancer-associated transcribed UCRs (T-UCRs) encoding ncRNAs were induced by hypoxia and thus further sub-classified as “hypoxia-induced noncoding ultraconserved transcripts” (HINCUTs) [100]. They serve as an example of another attribute used for ncRNA classification: induction after treatment with a stimulus or association with a certain biological state. Another example is “long stress-induced non-coding transcripts” (LSINCTs) [101].
Classification based on subcellular localization
RNA localization can provide important clues to its function. ncRNAs tend to be enriched in the nucleus [38, 56], which suggests their involvement in the temporal-spatial regulation of nuclear architecture. For example, chromatin-associated RNAs (CARs) – both intronic and intergenic – form an integral component of chromatin, with the potential to regulate the expression of nearby genes [102] (Figure 1). The ENCODE consortium performed extensive profiling of three sub-nuclear compartments (chromatin, nucleolus, and nucleoplasm) revealing their RNA compositions [3]. Within the nucleus, ncRNA association with, and targeting of, the gene-silencing PRC2 complex led to the identification of thousands of PRC2-associated ncRNAs in mouse embryonic stem cells [103] and human cell lines [19]. ncRNAs form components of other nuclear sub-compartments such as paraspeckles, the nucleolus, and the nuclear matrix [104]. Presumably, additional classes of ncRNAs associated with these and other compartments likely await discovery. Interestingly, some lncRNAs localize to the cytosol [105] and actually associate with ribosomes. Even the small mitochondrial genome encodes lncRNAs [106], underscoring the variety of different processes in which these transcripts could participate (see below).
Classification based on function
lncRNAs can participate in a plethora of different cellular processes: chromatin remodelling, regulation of transcription and translation, RNA stability, scaffolding, and innate immunity just to name a few. We discuss here only examples of functions used for classification, and direct the reader to other reviews focusing on lncRNA molecular mechanisms [6, 7, 13, 24, 39, 107].
Activating ncRNAs (ncRNA-a), which have enhancer-like properties, represents an example of classification based on function (Table 1). This class is distinguished from enhancer RNAs (eRNAs) [108] by positively regulating nearby genes (Figure 1). One notable member of this class, designated ncRNA-a7, regulates the Snai1 transcription factor. Depleting ncRNA-a7 leads to major phenotypic changes at both cellular and molecular levels [108]. The category of ncRNA-a will probably continue to grow, as the accumulation of high-quality expression datasets identify more lncRNAs that positively correlate with nearby genes (St. Laurent et al, submitted).
Another example is Competing endogenous RNAs (ceRNAs) [109]. They share sequence similarity with protein-coding transcripts and function by competing for regulatory molecules [109]. Any ncRNA sharing a sequence with another (coding or non-coding) RNA could potentially be a ceRNA, such as transcribed pseudogenes, which represent important ceRNAs [86] (Figure 1). Conceivably, the ceRNAs could form part of a complex regulatory matrix driven by differential affinity among many contextually associated RNA molecules [110].
Some lncRNAs serve as precursors for shorter functional RNAs as exemplified by primary transcripts for mi- and piRNAs (Table 1). In fact, the long and short cleavage products could have distinct functions, as evidenced by short non-coding tRNA-like molecule produced during maturation of MALAT1 lncRNA [111]. The ENCODE consortium estimates that approximately 6% of all annotated coding and non-coding transcripts overlap with short RNAs [3]. A recent report suggests that an 18 nt short RNA produced from a coding mRNA regulates translation [112]. Also, ncRNAs derived from 3′ ends of mRNAs associate with Argonaute proteins, suggesting they represent novel regulatory molecules [90]. Short RNAs derived from protein-coding transcripts can also mediate trans-generational silencing of the parent gene [14]. Conceivably, cleavage could also generate functional lncRNAs from a longer precursor ncRNA, where both the precursor and the product could have different functions.
Finally, we note that not every lncRNA transcript functions solely as a non-coding element. Peptide sequencing data revealed presence of 250 novel mouse peptides encoded by presumed lncRNAs [113]. The full extent of the novel mammalian proteome encoded by lncRNAs is not yet clear however. Although many lncRNAs appear to associate with ribosomes [105, 114], this frequently does not result in protein synthesis [115, 116], but instead could reveal lncRNA regulation of translation [105]. Nonetheless, the protein coding potential is currently used as one metric in lncRNA definition [37].
Challenges of current lncRNA classification
As described above, the existing classifications of lncRNAs rest on their descriptive and distinctive properties: from their size, to their localization, to their function. For example, the GENCODE system, as one of the few practical and up-to-date classifications available, also classifies lncRNAs into “Antisense RNA” or “lincRNA”, in addition to intron-associated biotypes [33]. Although logical principles guide these classifications, they have inherited a number of unavoidable shortcomings. First, the existing classes capture a small fraction of lncRNAs present in the cell as illustrated in Figure 2. The various lists of lncRNAs annotated based on resemblance to protein-coding mRNAs account for only 0.05–1.12% of cellular RNA (Figure 2) while functional intronic RNAs could constitute as much as 16% [50]. Second, the overlap between multiple existing annotations of lncRNAs derived by different groups is small [33]. Third, the descriptions of the classes in the current annotation schemes can be vague. For example, a lncRNA could initiate at an enhancer element or initiate a large distance away and merely overlap it, yet currently they would both be classified as eRNAs. Fourth, the existing classes are not mutually exclusive. Thus, a lincRNA could theoretically also classify as an eRNA, and an LSINCT, and a CAR, and a T-UCF, and so on. For example, ANRIL is a lncRNA [117], a NAT [117], and a circular RNA [118]. This point is particularly problematic, as few datasets cover all of these characteristics and therefore many ncRNAs are not comprehensively assessed. Fifth, they lack systematization: following the current schemes, in the future one might expect hundreds of overlapping classes of ncRNAs as new knowledge is incorporated into the classification. There are already at least 50 associations with a multitude of biological or biochemical processes (Table 1). Sixth, an attribute used in the current classifications may decline over time in relevance or utility. Considering the growing role of trans regulation by ncRNAs via intermolecular interactions [42, 82, 119], the fact that a lncRNA associates with an enhancer, promoter, or intron, or is antisense to a known gene may not reflect the actual function of that ncRNA. Instead, the latter could function by interacting with transcripts derived from elsewhere in a genome.
Figure 2. Properties of different published lists of human transcripts representing various classes of ncRNAs.
Sequence conservation was defined by the conserved elements from the Vertebrate Multiz Alignment & Conservation (100 Species) from the UCSC Browser [147]. Relative conservation represents the fraction of conserved bases relative to the total lengths for each list of ncRNAs. Relative mass and expression levels represent averages of several malignant and normal tissues profiled using single-molecule RNA-seq analysis [5, 29]. Only uniquely aligning non-rRNA and non-chrM reads were considered. Relative mass represents proportion of reads mapping to a particular genomic element relative to all reads. The relative expression is the relative mass divided by the total length of each list and normalized to the relative expression of coding exons (defined by UCSC Genes). Promoter-associated RNAs were defined by the regions 3 kb upstream of annotated start sites of UCSC Genes. Given the lack of a comprehensive list of standalone human intronic RNAs, we extrapolated the relative mass of those based on mouse data [50]. The GENCODE annotations [33] are based on v19.
The consolidated conceptual framework of lncRNA classification
The concepts driving lncRNA classification have begun to benefit from recent developments in the annotation of non-coding transcripts and the dramatically improved techniques for measuring them. Below, we review the conceptual components that provide the basis for these ongoing improvements (illustrated in Figure 3).
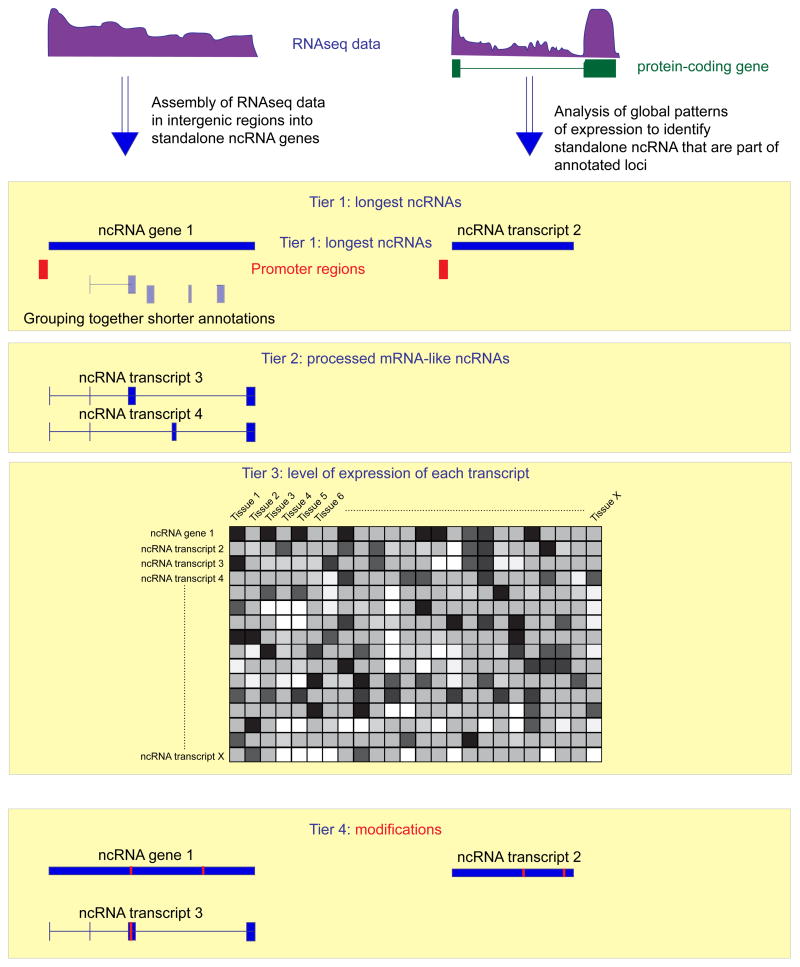
Figure 3. Outline of the consolidated conceptual framework of ncRNA classification.
Highly accurate empirical RNA-seq data drives both annotation and quantification of the longest ncRNA (Tier 1) and of processed ncRNA species (Tier 2) across the entire genome. The quantitation data serves as the basis for the combined global matrix of knowledge of expression of each (coding and non-coding) RNA gene and transcript across multiple biological sources (Tier 3). This information provides the input for the functional annotation of non-coding transcripts using systems biology approaches. Mapping of RNA modifications provides the final layer of knowledge in this scheme.
Tier 1: mapping the longest unprocessed transcript
The fact that the existing lists of lncRNAs miss a large fraction of ncRNA mass (Figure 2) argues that the annotation process has to start at a higher level. The first logical step in this effort is mapping the longest non-coding transcript (Figure 3).
Subdividing the intergenic space into standalone ncRNA loci (genes) has obvious benefits. First, it allows for consolidation of disparate and often incomplete ncRNAs represented by ESTs, lincRNAs, and mRNAs, into a single locus. As an illustration, the clinically important 8q24 region upstream of the MYC gene contains a number of different lncRNA elements [12] (Figure 4). Given the distances that separate them, it may not be obvious that these annotations are part of the same transcript, yet the RNA-seq signal clearly groups them together into one locus associated with a specific regulatory region (Figure 4). Second, such a grouping would allow experiments to focus on the locus rather than its many different genomic elements, allowing for seamless integration of the data from independent experiments. Third, it would clarify the issue of RNA association with different genomic features, for example enhancers, by showing whether the transcripts originate from these DNA elements, or merely overlap them. Overall, the longest transcripts would serve as scaffolds to bring together all the disparate annotations into gene-like structures with their own specific transcription regulatory regions, helping to resolve the problem of overlap. In this case, promoter information and CAGE tag data (Box 1) [25] would help in both assessing the quality of the map and understanding the regulation of such genes.
Figure 4. A genomic view of the 8q24 region upstream of the human MYC gene.
This clinically-important locus containing many GWAS hits associated with several cancers represents an example of a genomic region that could clearly benefit from the new annotation scheme. The RNAseq analysis reveals fairly strong signal on both strands covering most of this >1Mbp region. Yet, the known lncRNA annotations represent only a small fraction of this locus and judging by the distribution of the RNAseq signal and known promoters, are likely part of much larger transcript units (for example vlincRNAs shown on the figure). Transcriptome RNA-seq data is represented by the polyA- nuclear RNA from normal epidermal keratinocytes (NHEK) and embryonic stem cells (H1) generated by the ENCODE consortium [3]. In addition, vlincRNAs [29], promoters [32], and disease-associated variants from genome-wide association studies [148] (GWAS) are shown. Reproduced with permission from [12].
A lncRNA may not always be produced from its own dedicated promoter, as exemplified by circular intronic RNAs [55]. Such standalone functional intronic RNAs would however have certain features, such as low correlation with other exons or introns of the same gene, relatively high expression levels with low variance, and occasionally, differential expression in a biological time course [50]. These properties can now be measured by highly quantitative analysis of RNA levels across multiple diverse samples. Thus, defining standalone transcripts would require an additional dimension – quantitative measurement to allow for analysis co-expression with multiple neighbouring transcripts.
Tier 2: defining processed transcripts
The transcriptional forest concept implies that multiple RNA species share the same genomic space, either transcribed independently or derived by processing of longer precursors [38, 43, 120]. Mapping sites of polyadenylation [121, 122] provides additional information on completeness of such maps. Application of highly-sensitive methods targeted to specific regions using RACE [47] or capture-sequencing [45] would increase the discovery of processed species. Multiple levels of processing exist, such as A to I editing [123, 124] and others [125] each under their own regulatory control.
Tier 3: the additional dimension of expression levels
In the past, genomic coordinates alone determined genomic annotation. However, in the case of overlapping transcripts it cannot predict which isoforms likely function in a particular tissue. Thus, progress of our understanding of the complexities of the transcriptome (Box 1) argues for an additional dimension – expression of each RNA produced at a given locus (Figure 3). The pioneering efforts by the FANTOM Consortium [25] make this undertaking possible.
Tier 4: RNA modifications
A map of all (>100) RNA modifications [125] constitutes the final layer of annotation (Figure 3). These patterns could represent an information rich source for distinguishing RNA molecules, thus assisting in their classification. So far technological limitations prevent us from efficient genome-wide mapping of most RNA modifications, and assaying technically accessible modifications is fraught with pitfalls [124] such as false discovery due to technological noise [126]. Hopefully, existing [127] and emerging technologies [128] will enable progress in cataloguing RNA modifications.
From consolidated conceptual framework to function
The first key of the new framework is consolidation, achieved by grouping disparate lncRNA transcripts into genes or standalone Tier 1 transcripts. The second aspect moves from phenomenological description of lncRNAs to their genomic coordinates, which parallels the evolution of the concept of the gene [129]. The third aspect uses empirical data to unravel the layers of overlapping transcripts, as exemplified by the study of intronic RNAs in mice [50].
The fourth aspect assigns functional weight to a lncRNA by integrating information from diverse high throughput methods [130]. Among others, these methods include CLIP-seq [131] for detection and measurement of RNA – protein interactions, SHAPE-seq [132] for analysis of RNA secondary structure, ChIRP [133] for measurement of RNA–chromatin interactions, and GRO-seq (Box 1) to measure transcription [134]. This multi-faceted approach combines independent sources of evidence for the functionality of an RNA molecule, underscoring the complexities of lncRNA involvement in the flow of biological information [24, 135]. Fortunately new machine learning methods have emerged to identify and decipher complex patterns in the data, yielding probabilistic evaluation of ncRNA function across large populations of transcripts [71, 136].
The evolution of the conceptual framework of ncRNA classification described above provides a roadmap for the analysis of an RNA-seq experiment and its integration into a broader knowledge base of high-throughput multi-dimensional information. The availability of a common set of genomic coordinates for various stages of ncRNA processing (Figure 3, Tiers 1 and 2) provides the key resource that enables the integration of data from multiple experiments. Tiers 3 and 4 assist in refinement of the classification by separating overlapping transcripts that have different patterns of gene expression and modification.
For effective data integration, systems biology approaches require expression datasets that cover large numbers of biological sources with extraordinary precision [137]. Small yet biologically important effects [138] can be lost in technological noise [139]. Similarly, loss of ncRNAs and transcriptome complexity can occur during library preparation [140] and RNA isolation [141]. Processing of NGS data also presents a number of challenges. For example, algorithms building transcripts (Tier 1) should account for regions of the genome that have poor alignability due to repetitive regions [142]. NGS reads unassigned after these steps can then serve as input into ab initio algorithms such as the genomic binning approach [50, 139] to detect differentially expressed regions [27–30]. Without a doubt, cycles of iterations consisting of annotation, expression measurement, and addition of new transcripts and transcribed regions from global RNA measurement experiments will illuminate the puzzle of pervasive transcription.
Concluding remarks
Assigning functions to the mass of lncRNAs produced in the cell requires novel thinking and approaches. Many of the classic reductionist methods that worked well for coding genes have proven less useful to the challenges of deciphering the elaborate populations of transcripts generated by pervasive ncRNA transcription. Instead, global, systems-biology and genomics-driven approaches have emerged, which rely on an integrative framework of annotation and classification. This framework increases emphasis on the quality of genome-wide RNA measurements to allow for the ready integration of data from multiple types of experiments. It facilitates the development of improved tools for the integration of the highly multi-dimensional data from these experiments into the classification framework, thereby revealing associations between both coding and non-coding transcripts. Finally, it supports the rational and structured selection of subsets of these predictions for biological follow-up using reductionist methods.
Highlights.
Non-coding RNAs constitute the majority of transcriptional output of the human genome.
Lack of functional knowledge of most of ncRNAs has led to classification with a number of issues.
A new classification is emerging rooted in unbiased whole-genome surveys of RNA.
The new classification should allow for easy data integration across multiple experiments.
Acknowledgments
We wish to thank Maxim Ri, Denis Antonets and Dmitry Shtokalo for help with the bioinformatics analysis and Mark Mazaitis for expert assistance with the figure preparations. Studies on long noncoding RNAs in the Wahlestedt laboratory are currently supported by the US National Institute of Health awards DA035592, MH084880 and NS071674.
Glossary
- 5′-cap
an altered nucleotide present at 5′ ends of a eukaryotic RNA and vital for its functioning
- ENCODE project
the Encyclopedia Of DNA Elements, a public research consortium launched in September 2003 by the National Human Genome Research Institute. The goal of the project is to identify all functional elements in the human genome sequence
- Endogenous retrovirus (ERV)
a genomic element that was traced back to a retrovirus integrated into an ancestral genome and since retained. ERV sequences comprise ~8% of the human genome
- Expressed Sequence Tag (EST)
a relatively short and typically partial sequence of a longer RNA molecule
- FANTOM consortium
an international research consortium established by scientists at RIKEN, Japan in 2000, initially to assign functional annotations to the full-length cDNAs collected during the Mouse Encyclopedia Project. FANTOM has since developed and expanded over time to encompass different fields of transcriptome analysis
- Genomic bin approach
an approach designed to detect differentially-expressed regions of the genome in the regions where no annotation is available
- Long tandem repeat (LTR)
identical pieces of DNA found at the ends of retroviruses and critical for viral life cycle. LTRs contain elements required for viral gene expression. LTRs of ERVs often retain these elements and thus can initiate or control expression of host transcripts
- Paraspeckle
a subcellular compartment that could be identified in nuclear interchromatin space
- Polycomb repressive complex 2 (PRC2)
a multi-protein complex that reversibly modifies chromatin structure and silences target genes
- Tiling microarray
a microarray design (typically oligonucleotide-based) where probes interrogate an entire genomic region of interest at regular intervals agnostic of genomic annotations. This design differs from other microarrays that target only specific genomic features of interest, like exons of known genes
Footnotes
Online linking.
FANTOM Consortium: http://fantom.gsc.riken.jp/
ENCODE Consortium: http://www.genome.gov/encode/
St. Laurent Institute: http://www.stlaurentinstitute.com/
Database of RNA modifications: http://mods.rna.albany.edu/
NIH Roadmap Epigenomics project: http://www.roadmapepigenomics.org/
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kapranov P, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 2.Okazaki Y, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 3.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapranov P, et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. 2010;8:149. doi: 10.1186/1741-7007-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark BS, Blackshaw S. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front Genet. 2014;5:164. doi: 10.3389/fgene.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibb EA, et al. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qureshi IA, Mehler MF. Long non-coding RNAs: novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics: the journal of the American Society for Experimental NeuroTherapeutics. 2013;10:632–646. doi: 10.1007/s13311-013-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reis EM, Verjovski-Almeida S. Perspectives of Long Non-Coding RNAs in Cancer Diagnostics. Front Genet. 2012;3:32. doi: 10.3389/fgene.2012.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vergara IA, et al. Genomic “Dark Matter” in Prostate Cancer: Exploring the Clinical Utility of ncRNA as Biomarkers. Front Genet. 2012;3:23. doi: 10.3389/fgene.2012.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 12.St Laurent G, et al. Dark matter RNA illuminates the puzzle of genome-wide association studies. BMC medicine. 2014;12:97. doi: 10.1186/1741-7015-12-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark MB, et al. The dark matter rises: the expanding world of regulatory RNAs. Essays in biochemistry. 2013;54:1–16. doi: 10.1042/bse0540001. [DOI] [PubMed] [Google Scholar]
- 14.Liebers R, et al. Epigenetic regulation by heritable RNA. PLoS Genet. 2014;10:e1004296. doi: 10.1371/journal.pgen.1004296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smalheiser NR. The RNA-centred view of the synapse: non-coding RNAs and synaptic plasticity. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2014:369. doi: 10.1098/rstb.2013.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penman S. If genes just make proteins and our proteins are the same, then why are we so different? Journal of cellular biochemistry. 1991;47:95–98. doi: 10.1002/jcb.240470202. [DOI] [PubMed] [Google Scholar]
- 17.Zuckerkandl E. A general function of noncoding polynucleotide sequences. Mass binding of transconformational proteins. Molecular biology reports. 1981;7:149–158. doi: 10.1007/BF00778746. [DOI] [PubMed] [Google Scholar]
- 18.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang K, et al. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene. 2014;547:1–9. doi: 10.1016/j.gene.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 21.Kapranov P, St Laurent G. Dark Matter RNA: Existence, Function, and Controversy. Front Genet. 2012;3:60. doi: 10.3389/fgene.2012.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.St Laurent G, 3rd, Wahlestedt C. Noncoding RNAs: couplers of analog and digital information in nervous system function? Trends Neurosci. 2007;30:612–621. doi: 10.1016/j.tins.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 25.FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lonsdale J, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hackermuller J, et al. Cell cycle, oncogenic and tumor suppressor pathways regulate numerous long and macro non-protein coding RNAs. Genome Biol. 2014;15:R48. doi: 10.1186/gb-2014-15-3-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.St Laurent G, et al. VlincRNAs controlled by retroviral elements are a hallmark of pluripotency and cancer. Genome Biology. 2013;14:R73. doi: 10.1186/gb-2013-14-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boley N, et al. Genome-guided transcript assembly by integrative analysis of RNA sequence data. Nat Biotechnol. 2014;32:341–346. doi: 10.1038/nbt.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia H, et al. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478–1487. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amaral PP, et al. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:D146–151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volders PJ, et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:D174–180. doi: 10.1093/nar/gku1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 39.Wang KC, Chang HY. Molecular Mechanisms of Long Noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharon D, et al. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol. 2013;31:1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazorthes S, et al. A vlincRNA participates in senescence maintenance by relieving H2AZ-mediated repression at the INK4 locus. Nature communications. 2015;6:5971. doi: 10.1038/ncomms6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Dijk M, et al. HELLP babies link a novel lincRNA to the trophoblast cell cycle. The Journal of clinical investigation. 2012;122:4003–4011. doi: 10.1172/JCI65171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 44.Kapranov P, et al. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mercer TR, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denoeud F, et al. Prominent use of distal 5′ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res. 2007;17:746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Djebali S, et al. Efficient targeted transcript discovery via array-based normalization of RACE libraries. Nat Methods. 2008;5:629–635. doi: 10.1038/nmeth.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Makrythanasis P, et al. Variation in novel exons (RACEfrags) of the MECP2 gene in Rett syndrome patients and controls. Human mutation. 2009;30:E866–879. doi: 10.1002/humu.21073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakaya HI, et al. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007;8:R43. doi: 10.1186/gb-2007-8-3-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.St Laurent G, 3rd, et al. Intronic RNAs constitute the major fraction of the non-coding RNA in mammalian cells. BMC Genomics. 2012;13:504. doi: 10.1186/1471-2164-13-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fachel AA, et al. Expression analysis and in silico characterization of intronic long noncoding RNAs in renal cell carcinoma: emerging functional associations. Mol Cancer. 2013;12:140. doi: 10.1186/1476-4598-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engelhardt J, Stadler PF. Hidden treasures in unspliced EST data. Theory in biosciences = Theorie in den Biowissenschaften. 2012;131:49–57. doi: 10.1007/s12064-012-0151-6. [DOI] [PubMed] [Google Scholar]
- 53.Moss WN, Steitz JA. Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA. BMC Genomics. 2013;14:543. doi: 10.1186/1471-2164-14-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gardner EJ, et al. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 2012;26:2550–2559. doi: 10.1101/gad.202184.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 56.Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 57.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 58.Affymetrix ENCODE Transcriptome Project; Cold Spring Harbor Laboratory ENCODE Transcriptome Project. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Otsuka Y, et al. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol. 2009;29:2155–2167. doi: 10.1128/MCB.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mercer TR, et al. Expression of distinct RNAs from 3′ untranslated regions. Nucleic Acids Res. 2011;39:2393–2403. doi: 10.1093/nar/gkq1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abdelhamid RF, et al. Multiplicity of 5′ Cap Structures Present on Short RNAs. PLoS ONE. 2014;9:e102895. doi: 10.1371/journal.pone.0102895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Djebali S, et al. Evidence for transcript networks composed of chimeric RNAs in human cells. PLoS ONE. 2012;7:e28213. doi: 10.1371/journal.pone.0028213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finta C, Zaphiropoulos PG. Intergenic mRNA molecules resulting from trans-splicing. The Journal of biological chemistry. 2002;277:5882–5890. doi: 10.1074/jbc.M109175200. [DOI] [PubMed] [Google Scholar]
- 65.Zaphiropoulos PG. RNA molecules containing exons originating from different members of the cytochrome P450 2C gene subfamily (CYP2C) in human epidermis and liver. Nucleic Acids Res. 1999;27:2585–2590. doi: 10.1093/nar/27.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kapranov P, et al. New class of gene-termini-associated human RNAs suggests a novel RNA copying mechanism. Nature. 2010;466:642–646. doi: 10.1038/nature09190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Volloch V, et al. Antisense globin RNA in mouse erythroid tissues: structure, origin, and possible function. Proc Natl Acad Sci U S A. 1996;93:2476–2481. doi: 10.1073/pnas.93.6.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caudron-Herger M, et al. Coding RNAs with a non-coding function: maintenance of open chromatin structure. Nucleus. 2011;2:410–424. doi: 10.4161/nucl.2.5.17736. [DOI] [PubMed] [Google Scholar]
- 69.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Necsulea A, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 72.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Du Z, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harries LW. Long non-coding RNAs and human disease. Biochemical Society transactions. 2012;40:902–906. doi: 10.1042/BST20120020. [DOI] [PubMed] [Google Scholar]
- 75.Faulkner GJ, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 76.Fort A, et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet. 2014;46:558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- 77.Kelley DR, Rinn JL. Transposable elements reveal a stem cell specific class of long noncoding RNAs. Genome Biol. 2012;13:R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sell S. On the stem cell origin of cancer. The American journal of pathology. 2010;176:2584–2494. doi: 10.2353/ajpath.2010.091064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mariner PD, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 80.Belancio VP, et al. LINE dancing in the human genome: transposable elements and disease. Genome Med. 2009;1:97. doi: 10.1186/gm97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Flockerzi A, et al. Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV Transcriptome Project. BMC Genomics. 2008;9:354. doi: 10.1186/1471-2164-9-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang J, et al. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013;27:793–804. doi: 10.1101/gad.212639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zheng D, et al. Pseudogenes in the ENCODE regions: consensus annotation, analysis of transcription, and evolution. Genome Res. 2007;17:839–851. doi: 10.1101/gr.5586307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pink RC, et al. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. 2011;17:792–798. doi: 10.1261/rna.2658311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Johnsson P, et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013;20:440–446. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kerin T, et al. A noncoding RNA antisense to moesin at 5p14.1 in autism. Science translational medicine. 2012;4:128ra140. doi: 10.1126/scitranslmed.3003479. [DOI] [PubMed] [Google Scholar]
- 88.Arigo JT, et al. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 89.Ntini E, et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol. 2013;20:923–928. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- 90.Valen E, et al. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat Struct Mol Biol. 2011;18:1075–1082. doi: 10.1038/nsmb.2091. [DOI] [PubMed] [Google Scholar]
- 91.van Dijk EL, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 92.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 95.Hudson RS, et al. Transcription signatures encoded by ultraconserved genomic regions in human prostate cancer. Mol Cancer. 2013;12:13. doi: 10.1186/1476-4598-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mauger DM, et al. The genetic code as expressed through relationships between mRNA structure and protein function. FEBS Lett. 2013;587:1180–1188. doi: 10.1016/j.febslet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Washietl S, et al. Fast and reliable prediction of noncoding RNAs. Proc Natl Acad Sci U S A. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pedersen JS, et al. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Washietl S, et al. Structured RNAs in the ENCODE selected regions of the human genome. Genome Res. 2007;17:852–864. doi: 10.1101/gr.5650707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ferdin J, et al. HINCUTs in cancer: hypoxia-induced noncoding ultraconserved transcripts. Cell death and differentiation. 2013;20:1675–1687. doi: 10.1038/cdd.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Silva JM, et al. Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics. 2010;95:355–362. doi: 10.1016/j.ygeno.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 102.Mondal T, et al. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol. 2014;26:10–18. doi: 10.1016/j.ceb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van Heesch S, et al. Extensive localization of long noncoding RNAs to the cytosol and mono-and polyribosomal complexes. Genome Biol. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rackham O, et al. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA. 2011;17:2085–2093. doi: 10.1261/rna.029405.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kung JT, et al. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tay Y, et al. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.St Laurent G, et al. Dark matter RNA: an intelligent scaffold for the dynamic regulation of the nuclear information landscape. Front Genet. 2012;3:57. doi: 10.3389/fgene.2012.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wilusz JE, et al. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pircher A, et al. An mRNA-derived noncoding RNA targets and regulates the ribosome. Mol Cell. 2014;54:147–155. doi: 10.1016/j.molcel.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Prabakaran S, et al. Quantitative profiling of peptides from RNAs classified as noncoding. Nature communications. 2014;5:5429. doi: 10.1038/ncomms6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ingolia NT, et al. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Banfai B, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guttman M, et al. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pasmant E, et al. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 118.Burd CE, et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Holdt LM, et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9:e1003588. doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kapranov P, et al. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 121.Jan CH, et al. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ozsolak F, et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peng Z, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 124.St Laurent G, et al. Genome-wide analysis of A-to-I RNA editing by single-molecule sequencing in Drosophila. Nat Struct Mol Biol. 2013;20:1333–1339. doi: 10.1038/nsmb.2675. [DOI] [PubMed] [Google Scholar]
- 125.Grosjean H. Nucleic Acids Are Not Boring Long Polymers of Only Four Types of Nucleotides: A Guided Tour. In: Grosjean H, editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. 7. Landes Bioscience; 2009. pp. 1–18. [Google Scholar]
- 126.Kleinman CL, Majewski J. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1209658. author reply 1302. [DOI] [PubMed] [Google Scholar]
- 127.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barhoumi A, Halas NJ. Detecting Chemically Modified DNA Bases Using Surface-Enhanced Raman Spectroscopy. The Journal of Physical Chemistry Letters. 2011;2:3118–3123. doi: 10.1021/jz201423b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gerstein MB, et al. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 130.Mudge JM, et al. Functional transcriptomics in the post-ENCODE era. Genome Res. 2013;23:1961–1973. doi: 10.1101/gr.161315.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Siegfried NA, et al. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat Methods. 2014;11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chu C, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hah N, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mattick JS. A new paradigm for developmental biology. J Exp Biol. 2007;210:1526–1547. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- 136.Dozmorov MG, et al. Systematic classification of non-coding RNAs by epigenomic similarity. BMC Bioinformatics. 2013;14(Suppl 14):S2. doi: 10.1186/1471-2105-14-S14-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wan YW, et al. On the reproducibility of TCGA ovarian cancer microRNA profiles. PLoS ONE. 2014;9:e87782. doi: 10.1371/journal.pone.0087782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.St Laurent G, et al. On the importance of small changes in RNA expression. Methods. 2013;63:18–24. doi: 10.1016/j.ymeth.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 139.Raz T, et al. Protocol dependence of sequencing-based gene expression measurements. PLoS ONE. 2011;6:e19287. doi: 10.1371/journal.pone.0019287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fu GK, et al. Molecular indexing enables quantitative targeted RNA sequencing and reveals poor efficiencies in standard library preparations. Proc Natl Acad Sci U S A. 2014;111:1891–1896. doi: 10.1073/pnas.1323732111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sultan M, et al. Influence of RNA extraction methods and library selection schemes on RNA-seq data. BMC Genomics. 2014;15:675. doi: 10.1186/1471-2164-15-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Derrien T, et al. Fast computation and applications of genome mappability. PLoS ONE. 2012;7:e30377. doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ozsolak F, et al. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 144.Kodzius R, et al. CAGE: cap analysis of gene expression. Nat Methods. 2006;3:211–222. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- 145.Philippe N, et al. Combining DGE and RNA-sequencing data to identify new polyA+ non-coding transcripts in the human genome. Nucleic Acids Res. 2014;42:2820–2832. doi: 10.1093/nar/gkt1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fullwood MJ, et al. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19:521–532. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Blanchette M, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pachnis V, et al. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc Natl Acad Sci U S A. 1984;81:5523–5527. doi: 10.1073/pnas.81.17.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Brown CJ, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 153.Vance KW, et al. The long non-coding RNA Paupar regulates the expression of both local and distal genes. The EMBO journal. 2014;33:296–311. doi: 10.1002/embj.201386225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Koerner MV, et al. The function of non-coding RNAs in genomic imprinting. Development. 2009;136:1771–1783. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Faghihi MA, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nature medicine. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. Journal of the National Cancer Institute. 1999;91:143–151. doi: 10.1093/jnci/91.2.143. [DOI] [PubMed] [Google Scholar]
- 157.Lee JT, et al. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 158.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Flynn RA, et al. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci U S A. 2011;108:10460–10465. doi: 10.1073/pnas.1106630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. The EMBO journal. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hall LL, et al. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell. 2014;156:907–919. doi: 10.1016/j.cell.2014.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rangwala SH, et al. Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol. 2009;10:R100. doi: 10.1186/gb-2009-10-9-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ting DT, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zheng R, et al. Polypurine-repeat-containing RNAs: a novel class of long non-coding RNA in mammalian cells. Journal of cell science. 2010;123:3734–3744. doi: 10.1242/jcs.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schulz D, et al. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell. 2013;155:1075–1087. doi: 10.1016/j.cell.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 167.Saini HK, et al. Annotation of mammalian primary microRNAs. BMC Genomics. 2008;9:564. doi: 10.1186/1471-2164-9-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li XZ, et al. Defining piRNA primary transcripts. Cell Cycle. 2013;12:1657–1658. doi: 10.4161/cc.24989. [DOI] [PubMed] [Google Scholar]
- 170.Mao YS, et al. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]