SUMMARY
Telomere-led rapid prophase movements (RPMs) in meiotic prophase have been observed in diverse eukaryote species. A shared feature of RPMs is that the force that drives the chromosomal movements is transmitted from the cytoskeleton, through the nuclear envelope, to the telomeres. Studies in mice suggested that dynein movement along microtubules is transmitted to telomeres through SUN1/KASH5 nuclear envelope bridges to generate RPMs. We monitored RPMs in mouse seminiferous tubules using four-dimensional fluorescence imaging and quantitative motion analysis to characterize patterns of movement in the RPM process. We find that RPMs reflect a combination of nuclear rotation and individual chromosome movements. The telomeres move along microtubule tracks which are apparently continuous with the cytoskeletal network, and exhibit characteristic arrangements at different stages of prophase. Quantitative measurements confirmed that SUN1/KASH5, microtubules, and dynein but not actin were necessary for RPMs and that defects in meiotic recombination and synapsis resulted in altered RPMs.
INTRODUCTION
Proper segregation of chromosomes during meiosis requires that homologous chromosomes be physically connected by a mechanical link. This requires the homologs to pair, synapse, form chiasmata that link the homologs, and avoid ectopic connections with non-homologous chromosomes. How chromosome mechanics are coordinated with recombination and how homologous chromosome interactions are regulated are central questions in meiosis. Telomereled rapid prophase movements of the chromosomes (RPMs) have been proposed to move chromosomes relative to one another, helping establish homologous interactions during pairing, resolve chromosome entanglements and regulate chiasma placement (reviewed in (Koszul and Kleckner, 2009)). Since the first identification of dramatic prophase movements in rat spermatocytes (Parvinen and Soderstrom, 1976) RPMs have been observed in a wide range of organisms (Chikashige et al., 1994; Conrad et al., 2008; Ding et al., 1998; Koszul et al., 2008; Labrador et al., 2013; Rickards, 1975; Scherthan et al., 2007; Sheehan and Pawlowski, 2009; Wynne et al., 2012), including mouse (Morelli et al., 2008; Morimoto et al., 2012b; Parvinen and Soderstrom, 1976; Shibuya et al., 2014a; Shibuya et al., 2014b; Yao and Ellingson, 1969).
Work from organisms so far analyzed has revealed a conserved general mechanism supporting active prophase chromosome movements (reviewed in (Hiraoka and Dernburg, 2009; Koszul and Kleckner, 2009). This involves cytoskeletal components that originate the forces generating the movements which are transduced to chromosome telomeres through protein complexes located at the nuclear envelope. However, the mechanism operating the machinery that support chromosome movements vary in different organisms and the specific variations in components of the system in different organisms are not well understood. For example, during fission yeast meiosis, nuclear envelope associated telomeres cluster at the spindle pole body, after which the entire nucleus is dragged by microtubules and associated motors back and forth along the length of the cell (Chikashige et al., 1994). In contrast, in Saccharomyces cerevisiae telomeres become associated transiently through the nuclear envelope to nucleus-hugging actin cables that are continuous with the actin cytoskeleton. In this case chromosome movement may to occur via a passive process as chromosome ends are transiently associated with dynamic actin cables (Koszul et al., 2008).
The participation of microtubules or actin in generating RPMs is a documented difference in model organisms. With the exception of S. cerevisiae in which chromosome movement seems associated with dynamic actin cables (Koszul et al., 2008; Trelles-Sticken et al., 2005), microtubule and dynein have been suggested to be the main components of the force generating RPMs in rat (Salonen et al., 1982), Schizosaccharomyces pombe (Chikashige et al., 2007; Vogel et al., 2009; Yamamoto and Hiraoka, 2003), Caenorhabditis elegans (Wynne et al., 2012) and mouse ((Morimoto et al., 2012b), and this work); however, this aspect seems to be controversial in maize (Sheehan and Pawlowski, 2009).
A particularly conserved aspect of chromosome movements is the protein complexes that bridge telomeres to the cytoskeleton (Hiraoka and Dernburg, 2009; Koszul and Kleckner, 2009) and provide the molecular connections that can transduce forces generated in the cytoplasm to the end of the chromosomes. In the mouse, the SUN1 and KASH5 proteins are localized to the inner and outer nuclear membrane of the nuclear envelope, respectively, and physically interact with each other connecting the internal regions of the nuclear envelope with the cytoskeleton (Horn et al., 2013; Morimoto et al., 2012b). The recent discovery of KASH5, a meiosis-specific protein that physically interacts with both SUN1 in the inner membrane and dynein in the cytoplasm, shed light on the components of the system that link the cytoplasmic force-generating mechanism with the intra-nuclear cargo in mammals. The functional importance of SUN1, KASH5, and dynein in quality control by preventing non-homologous pairing was first proposed in C. elegans in which dynein acts as a licensing factor for the formation of the synaptonemal complex, most likely by overcoming the inhibition imposed by the chromosome-nuclear envelope connection acting through SUN1 and KASH5 (Sato et al., 2009). In this model non-homologous chromosomes are readily separated by the RPM-generated forces, but homologous chromosomes have sufficient affinity to resist.
The finding of molecular components involved in the RPM process raises a number of interesting issues: How do the components work together to generate RPMs? What is the basis by which RPMs are regulated? What are the roles of RPMs in the meiotic process? As a first step towards addressing these questions in a mammalian model we sought to develop a quantitative methodology that would have the resolution to provide a detailed analysis of RPMs. In this paper, we employed 3D fluorescence time-lapse microscopy in intact seminiferous tubules supplemented with customized visualization, and in silico analysis and simulation. This approach proved to have the sensitivity to reveal changes in RPM characteristics during meiotic progression, and differences in RPMs brought about by defects in recombination and synapsis. This study reveals that in mouse spermatocytes RPMs are result of a combination of nuclear rotation and chromosome autonomous movements. The chromosome autonomous movements are consistent with a mechanism by which telomeres move along microtubule tracks and can change directions at intersections of the tracks. The ability to carry out live imaging, combined with quantitative image analysis, offers a tool to investigate the regulation of RPMs, chromosome reorganizations that precede dynamic mid-prophase events, and their contribution to faithful transmission of genetic information.
RESULTS
Characteristic Changes in Nuclear Morphology Mark the Progression through Meiotic Prophase in Living Cells
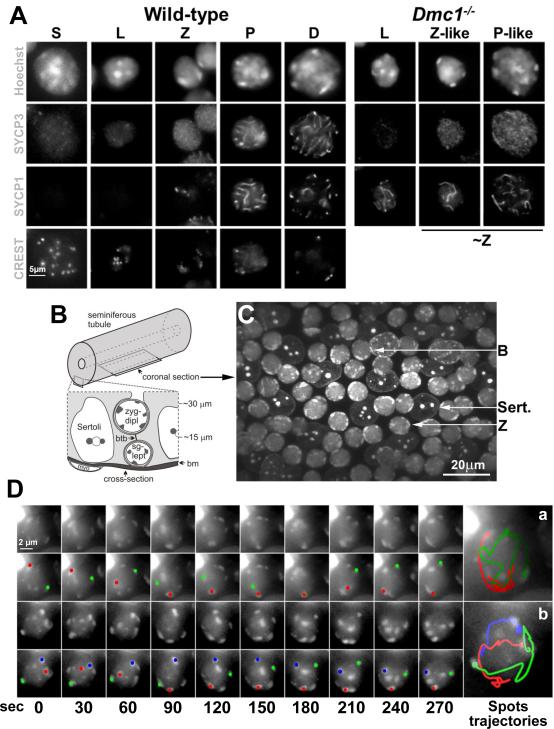
Because meiosis in spermatocytes occurs in cells in the complex cellular milieu of the seminiferous tubule we sought to identify cell staining methods, compatible with live cell imaging in this tissue, that would allow us to track the behaviors of telomeres. We began by analyzing squash preparations of spermatocytes in which we correlated the changes in nuclear size and pericentromeric heterochromatin morphology (revealed by Hoechst 33342 staining) that occur in spermatocyte development with the localization of elements of the synaptonemal complex (SYCP1 and SYCP3) and centromeres (CREST; Figure 1A). Mouse chromosomes are acrocentric (the centromere is very close to one end of the chromosome) and the centromeres are associated with long blocks of pericentric heterochromatin – thus one telomere of each chromosome is marked by this pericentric heterochromatin. These heterochromatin foci (Sheehan and Pawlowski, 2009) indicate the positions of individual or groups of chromosomes that can be tracked in time-lapse images, and here will be referred to simply as spots.
Figure 1. Fluorescent Labeled Peri-Centromeric Heterochromatin Spots Exhibit Rapid, Heterogeneous and Independent Movements.
A. Single-plane fluorescent images from 3D stacks of s-phase (S), leptotene (L), zygotene (Z), pachytene (P), and diplotene (D) nuclei in wild-type and leptotene (L), zygotene-like (Z-like) and pachytene-like (P-like) in Dmc1−/− spermatocytes squash preparations. For comparison Z-like and P-like nucleus were grouped under ~Z. Magnification bar represents 5μm. B. Diagram showing the location of the indicated cell types and their approximate distances from the basement membrane (bm) of the seminiferous tubule as viewed in cross-section. The position of a coronal section, as shown in C and used to acquire time-lapse images, is also indicated. C. Micrograph of a seminiferous tubule stained with Hoechst 33342, showing the morphology of Sertoli (Sert.), bouquet (B) and zygotene (Z) nuclei. Magnification bar represents 20μm. D. Maximum-intensity projections of time-lapse images of wild-type zygotene nuclei. A zygotene nucleus in which two heterochromatin spots merge, remain together for 60 sec and then separate (a) and an example of a nucleus in which three spots moved independently at different speeds (b) is shown. For each nucleus, the peri-centromeric heterochromatin spots (top panel) and the trajectory of selected spots represented in different colors (bottom panel) are shown. Magnification bar represents 2μm.
Hoechst 33342 stain revealed multiple spots of pericentric heterochromatin at pre-meiotic S phase meiocytes (identified as EDU positive cells, not shown) with many of the spots lying in the nuclear interior (Fig. 1A). Leptotene stage is characterized by initial stages of the synaptonemal complex formation when SYCP3 begins to form core structures called axial elements along each sister chromatid pair. In these spermatocytes some spots are still in the interior of the nucleus but heterochromatin moves to the nuclear periphery as leptotene proceeds. Later, marking the onset of zygotene, a tripartite synaptonemal complex is formed which central regions marked by SYCP1. In zygotene spermatocytes all the heterochromatin spots appear bound to the nuclear envelope. This localization is expected, as it is clear that the ends of the chromosome axes, i. e., the telomeres, attach to the nuclear envelope early in meiotic prophase, placing the telomere-adjacent pericentric heterochromatin at the nuclear periphery. During the leptotene-zygotene transition in mouse most telomeres cluster on one side of the nucleus to form the bouquet, which disperses relatively quickly, so that bouquet stage is relatively short (Scherthan, 2001). In pachytene there is increasing compaction of chromatin and the synapsing axes appear as brighter, linear signals.
The characteristic nuclear morphologies we observed in squashed spermatocytes were apparent when we analyzed seminiferous tubule explants stained with Hoechst 33342 (Figures 1B and C). The seminiferous tubule has a stratified organization, with spermatogenic cells developing in columns as they move away from the tubule periphery (Fig. 1B). Sertoli cells form a layer parallel to the tubule wall and provide support to the cells as they develop. Figure 1C show examples of Sertoli cells and meiocytes at different stages of prophase I.
Telomere Movements during Mouse Meiotic Prophase are Rapid and Heterogeneous
To analyze the detail of RPMs in mouse spermatocytes, we measured the movements of the pericentric heterochromatin spots in a cover-glass bottomed dish to allow the collection of 3D image stacks (see supplemental experimental procedures) of coronal sections (Figure 1B). Acquisition of 16 image slices in 1μm steps along the Z dimension (the direction of focus) was required in order to capture multiple nuclei in a single image stack. When RPMs were occurring at the maximal speed, acquisition of stacks as frequently as every 10 seconds was required to enable identification of the same spot in successive time-points. Fluorescence image stacks of the same nuclei were acquired every 10 seconds for a total of 10 minutes (giving a total of 61 time-points). Under these conditions, the RPMs measured at the beginning of a time-course are not different from those measured at the end. These recordings showed that RPMs are rapid and heterogeneous. Figure 1D shows an example of a zygotene nucleus in which two heterochromatin spots merge, remain together for 40 seconds and then separate (panel a). An example of a nucleus in which three spots moved independently at different speeds is also shown (panel b).
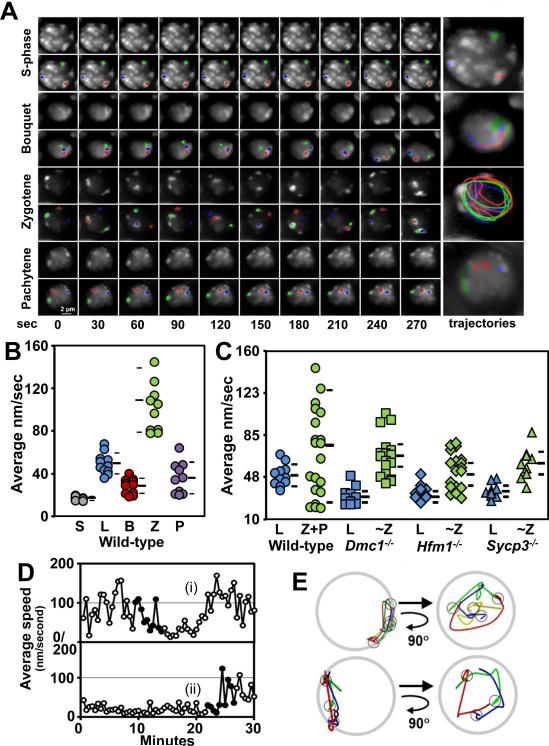
Using the 4D imaging method, we tracked the movements of pericentric spots in nuclei determined to be at different meiotic stages (pre meiotic S-phase, bouquet, zygotene, and pachytene) based on their Hoechst 33342 staining patterns (Fig. 2A). To quantitatively measure RPMs, individual spots in at least 10 meiotic prophase nuclei at each stage were analyzed using software that we previously developed (OMRFQANT;(Conrad et al., 2008; Lee et al., 2012). The analysis of pre-meiotic S-phase spermatocytes revealed that in these nuclei, the average RPM speed was 18 nm/sec, far below the minimum average speed at any stage of prophase I (Figure 2A and B). Spots moved much faster in leptotene then decreased from leptotene (49.5 ± 9.7 nm/sec, n = 11) to the bouquet stage (28.5 ± 7.0 nm/sec, n = 11), increased sharply in zygotene, (109.2 ± 33.2 nm/sec, n = 11) then fell in pachytene (36.0 ± 14.8 nm/sec, n = 10; Figure 2B). The movements could be divided into two categories. First, spots exhibited autonomous movements that appeared unrelated to the movements of other spots (e.g. Fig. 2A, the green spot in the pachytene nucleus). But the most prominent movement occurred when the spots moved in concert, appearing to rotate together as a semi-rigid unit (e.g. Fig. 2A, the zygotene nucleus) (supplemental movies 1-4).
Figure 2. RPMs are Regulated by Meiotic Progression and Affected in Recombination Mutants.
A. Maximum-intensity projections of time-lapse images of wild-type prophase nuclei. Magnification bar represents 2μm. B. Quantitation of the movement of spots for wild-type premeiotic S-phase (S), leptotene (L), bouquet (B), zygotene (Z), and pachytene (P) spermatocytes. Each circle represents the average of all spots analyzed in a single spermatocyte; horizontal lines indicate the median and standard deviation values. C. Quantitation of spot movements for wild-type and recombination and synapsis mutants. Each symbol represents the average of all spots analyzed in a single spermatocyte; horizontal lines indicate the median and standard deviation values. D. Plot of average speed changes during bouquet formation and resolution in wild-type zygotene nuclei. The duration of the bouquet for cell (i) was ~8 min and >22 min for cell (ii). In panel D, filled dots indicate the periods that are shown as marker traces in E where the cells enter (i) and exit (ii) the bouquet stage. E. Traces of spot movements for the periods shown in D. The small circles mark the spot positions when they were in the bouquet stage, i.e., at the end of the trace (top panel) and the beginning of the trace (bottom panel). Each nucleus is viewed as in the original movie (top diagrams) and was then rotated to show the view down the arrows i.e., from the center of the nucleus towards the bouquet cluster (bottom diagrams). The latter views demonstrate that the bouquet clusters lie on the axes of the clockwise rotations. See also Figure S1.
In summary, in mouse spermatocytes RPM characteristics vary with the stage of meiotic prophase, as has been reported earlier in S. cerevisiae and C. elegans (Conrad et al., 2008; Wynne et al., 2012). The restricted chromosome movements in pre-meiotic S-phase nuclei and the characteristic changes in the speed of RPMs during prophase progression indicate that chromosome movements are highly regulated during the meiotic program.
RPMs are Defective in Recombination and Synapsis Mutants
Work in several model organisms has shown that mutations that block meiotic progression also reduce chromosome movement (Conrad et al., 2008; Hiraoka et al., 2000; Labrador et al., 2013; Wynne et al., 2012). To test whether this is true in mouse spermatocytes we examined the effects on RPMs of mutations in three genes that are involved in prophase chromosome behavior. DMC1 is a meiotic-specific recombinase required for strand invasion (Neale and Keeney, 2006), HFM1 helicase is required for stabilization of strand invasion intermediates and crossover formation (Lynn et al., 2007), and SYCP3 is required for synaptonemal complex assembly and homologous chromosome synapsis (Page and Hawley, 2004).
Mutations in mouse DMC1 and SYCP3 result in a zygotene/pachytene-like arrest and deletion of HFM1 lead to arrest at the end of prophase I in diakinesis. In this study, imaged nuclei of mutants (Dmc1−/−, Sycp3−/− and Sun1−/−) undergoing early prophase arrest that possessed a similar heterochromatin morphology to that of wild-type zygotene (Z) and pachytene (P) nuclei were classified as ~Z (zygotene-like plus pachytene-like) in order to compare RPMs in the mutants and wild-type (Fig. 1A).
Compared to wild-type, the RPMs in Dmc1−/− spermatocytes were slower during early prophase (leptotene, 31.1 ± 8.0 nm/sec, n = 10; Kolmorogov-Smirnov test, p = 0.001) and occurred at almost the same rate during late prophase (~Z, 67.0 ± 19.1 nm/sec, p = 0.2, n = 17; Figure 2C); these changes are similar to the changes to RPMs observed in Dmc1−/− budding yeast (Conrad et al., 2008).
Most mouse Hfm1−/− spermatocytes undergo normal recombination initiation and homologous chromosome pairing. However, the intermediate and late stages of recombination appear to be aberrant and are accompanied by incomplete synapsis (Guiraldelli et al., 2013). RPMs in early (leptotene, 34.1 ± 7.1 nm/sec, n = 12, p = 0.004) and late prophase (~Z, 53.8 ± 15.7 nm/sec, n = 20, p = 0.03) in Hfm1−/− spermatocytes were defective compared to wild-type spermatocytes (Figure 2C).
Sycp3−/− spermatocytes fail to form axial/lateral elements of the synaptonemal complex, which is accompanied by an absence of synapsis and defective DNA repair (Yuan et al., 2000). Compared to wild-type nuclei, RPMs were significantly reduced in early (leptotene, 31.1 ± 8.0 nm/sec, n = 10, p = 0.005) and late (~Z, 67.0 ± 19.2 nm/sec, n= 10, p = 0.08) Sycp3−/− nuclei (Figure 2C).
The maximum speed of RPMs occurred at early prophase (zygotene) (Figure 2B) and apparently required DMC1 and HFM1. This indicates that maximum RPMs occur at approximately the time of and in the presence of early-mid recombination intermediates that are expected to be present when homologous chromosomes are actively pairing. Therefore, it is possible that RPMs are modulated by recombination and may facilitate homologous chromosome pairing and/or reduce unproductive interactions between unrelated chromosomes. However, the mutations in genes affecting recombination and synaptonemal complex analyzed in this study may also affect other important meiotic processes and ultimately meiotic progression, which may indirectly turn in deficient RPMs.
Our evidence that RPMs are dependent on meiotic progression and are affected in recombination and synaptonemal complex mutants raises the question of whether RPMs are influenced by the extent of homologous chromosome pairing. To test this possibility, we compared the average speed of RPMs in leptotene and pachytene spermatocytes. In contrast to pachytene spermatocytes in which all the chromosome complement is engaged in synapsis, leptotene cells show very limited chromosome pairing and synapsis have not been yet achieved. Average speeds of RPMs in leptotene and pachytene nuclei were not significantly different (Figure 2B). This suggests that the extent of pairing and synapsis per se are not important factors that regulate RPMs.
Bouquet Formation is Associated with Temporarily Reduced Rates of Movement and Spiraling Movements of the Chromosome Ends
Accumulation of telomeres in a limited region of the nuclear envelope periphery defines bouquet formation. Although this is a conserved phenomenon observed from yeast to humans (Scherthan, 2001), the characteristics of chromosome movements during bouquet formation and dissolution in a mammal have not been studied. Time-lapse images were taken over a total capture time of 30 min, which enabled the visualization of the formation and/or dissolution of nine bouquets. Maximal clustering which was often adjacent to the centrosome (Figure S1A and (Liebe et al., 2004)), was found to persist for an average period of 15 ±7 minutes. During this time RPMs were minimal or temporarily cease. In all analyzed cells chromosome movements during the bouquet stage apparently involve spiraling of the chromosome ends into place followed by subsequent spiraling away from the cluster, which dissolves the bouquet several minutes later (Figure 2D). In eight out of nine instances, as viewed from the middle of each nucleus, the spots rotated counterclockwise (clockwise in the ninth scored cell) on entry into and/or exit from the bouquet (Figure 2D and E and supplemental movie 3). During the period in which movies were acquired, some nuclei exhibited periods of telomere clustering followed by dissolution and re-clustering of telomeres (not shown). This suggests that bouquet stage is dynamic, with cycles of telomere clustering and dissolution.
RPMs in the Mouse Are a Mixture of Nuclear Rotation and Autonomous Movements
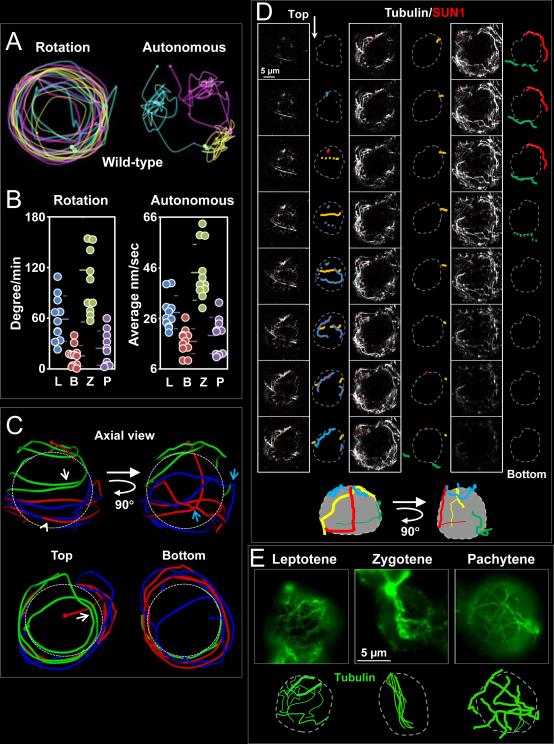
Our analysis of chromosome movements showed that the RPMs in mouse spermatocytes are composed of two types of movement: nuclear rotation in which individual spots in the nucleus do not move relative to one another but the entire set moves relative to extra-nuclear material (e.g. surrounding cells) and autonomous chromosome movements in which spots move relative to other spots in the nucleus (Figure 3A). We developed an in silico approach to separately evaluate these two types of movement (see Supplemental Information for details). Graphical depictions of the “unwinding” of wild-type datasets are shown in supplemental movie 5. We observed the appearance of multi-axial spinning in which the nucleus rotated concurrently with autonomous movements of individual spots. Furthermore, we observed two modes of autonomous motion. In one mode, a spot exhibited constant changes in direction and remained close to its origin (we referred to this area as a domain, yellow trajectory, upper right of Figure 3A). In the second mode, spots exhibited continuous movement in one direction, which brought the spot to a new domain (the pink marker moves to the yellow domain three times during the 10 min, Figure 3A). The sum of the degrees each nucleus rotated is reported in degrees per minute (leptotene, 59.0 ± 26.8; bouquet, 15.3 ± 12.1; zygotene, 116.8 ± 61.7; and pachytene, 24.4 ± 20.2) and the residual, autonomous movement made by each spot, calculated by subtracting the movement attributable to nuclear rotation, is reported as the average speed in nm per second (leptotene, 28.2 ± 6.5; bouquet, 16.9 ± 11.9; zygotene, 44.2 ± 11.0; and pachytene 19.1 ± 7.2; Figure 3B). In terms of raw movements, both rotation and autonomous movements followed similar patterns of variation during prophase and reached maxima at zygotene.
Figure 3. Identification of a Cytoplasmic Microtubule Network with Defined Stationary Tracks Associated with the Nuclear Envelope.
A. RPMs appear to be composed of nuclear rotation and independent chromosome movements. The positions and trajectories of three individual heterochromatin spots in wild-type zygotene nuclei are marked in different colors with a small sphere indicating the initial position and a large sphere indicating the final position. Rotation (left panel) and autonomous (right panel) movements of individual markers were detected in the same nucleus. B. Quantification of autonomous and rotational movements. For all datasets, the mean and standard deviation values are indicated by horizontal lines. Autonomous movements during each stage were significantly different to that of the neighboring stage (all p < 0.0001), except for the comparison of leptotene and pachytene (p = 0.11). Rotational movements in zygotene were significantly different to that of all other stages (all p < 0.03). C. Trajectory of three heterochromatin independent spots in a zygotene spermatocyte nucleus. D. Microtubule cables and their relationship with the nuclear envelope. Consecutive optical sections (0.24 μm) spanning an entire fixed squashed spermatocyte nucleus reveal a complex microtubule cable network arrangement in 3D. The nuclear envelope defined by SUN1 localization is represented by a grey dotted line and microtubules (outlined in colors) are followed through successive sections. The schematic representations (bottom) represent 3D reconstructions of microtubule cable disposition along the nucleus. Magnification bar represents 5μm. E. Single slice image of wild-type spermatocytes at different stages of prophase immunostained using anti α-tubulin antibodies. The cartoons are schematic representations of the representative microtubule patterns at each stage. Magnification bar represents 5μm. See also Figure S1. See also Figure S2.
We observed a strong correlation between rotational and autonomous movements in wild-type spermatocytes at every stage of prophase I (r2 of 0.85; Figure S2A). This suggests that either a single mechanism propels both types of movements or, if there are different mechanisms, that they are regulated in a similar manner.
In summary, observation of the movements, combined with computational subtraction of the whole-nucleus rotation-type movements indicated that two types of spot motion occur in mouse spermatocytes, nuclear rotation and independent autonomous movements. We speculate that both types of chromosome movements may depend upon cytoskeletal microtubule tracks.
Chromosome Ends Move Along Stationary Tracks
In order to determine whether or not the spots move along designated tracks we dissected the trajectory of individual or groups of spots in zygotene nuclei. Figure 3C shows the trajectory of three different spots in one nucleus. Observation of spot trajectories in several zygotene nuclei revealed that: (1) a spot can move back and forth along a particular trajectory (white arrows, green trajectory); (2) different spots can move along the same path, separated by many seconds or minutes (arrowhead, blue and red trajectory) (see supplemental movie 5); (3) spots frequently paused followed by sharp-angle turns (blue arrows, blue and red trajectory).
These observations suggest the presence of relative stable tracks or closely adjacent paths/tracks that can be used by one or multiple spots simultaneously or separated in time. The pauses in movement may be the result of spots moving to a differently-directed track or reaching the end of a track. Resumption of movement in a different direction at a sharp angle may indicate the spot has changed tracks. Sometimes spots resume movement when another spot seems to collide with it, and the pair moves together, suggesting passive spot movement. The simulation in supplemental movie 6 shows how, in combination, these behaviors can describe the observed behaviors of the spots.
Identification of Microtubules Associated with the Nuclear Envelope that Extend Outward into the Cytoskeletal Network
Recent work showed that RPMs depend on microtubule (Morimoto et al., 2012a). Here, we examined the features of the cytoplasmic microtubule network which could help to explain the observed tracking behavior of the spots. We performed immunolocalization of α-tubulin and SUN1 and used confocal microscopy to visualize microtubules associated with the nuclear envelope in lightly-fixed mouse spermatocyte squash preparations (Figure 3D). This approach revealed several features of microtubule cables: (1) The nucleus is semi-spherical and no microtubule cables are detected in this space. (2) In all cells, a few microtubules were associated with the nuclear envelope, with their ends extending outwards beyond the nucleus into the general cytoskeletal network. (3) Microtubule bundles with apparent different diameters could be observed within the network. (4) The microtubule network did not show any particular relationship with the microtubule organizing center (Figure S1B). None of the 20 spermatocytes studied for any of the stages analyzed (leptotene, zygotene and pachytene) showed a α-tubulin immunosignal as the center for microtubule aggregation.
A comparative visual analysis revealed that spermatocytes at certain stages had its own characteristic pattern of microtubule distribution (Figure 3E). The most contrasting patterns of microtubule arrangement were observed at leptotene (70% of scored cells exhibit similar microtubule pattern, n = 30), zygotene (74%, n = 34), and pachytene (90%, n = 25). Pachytene nuclei exhibited a complex, interconnected network of cytoplasmic microtubules bundles with larger diameter than were typical of the other stages (pachytene: 0.30 ± 0.05 μm, n = 15 cells versus leptotene: 0.12 μm ± 0.03, n = 15 cells). Most microtubules in zygotene spermatocytes were characteristically aggregated within a defined area. These observations raise the possibility that microtubule arrangements may influence the type of movement and speed of RPMs.
RPMs Are Dependent on Microtubules and Dynein but Less Responsive to Actin Inhibition
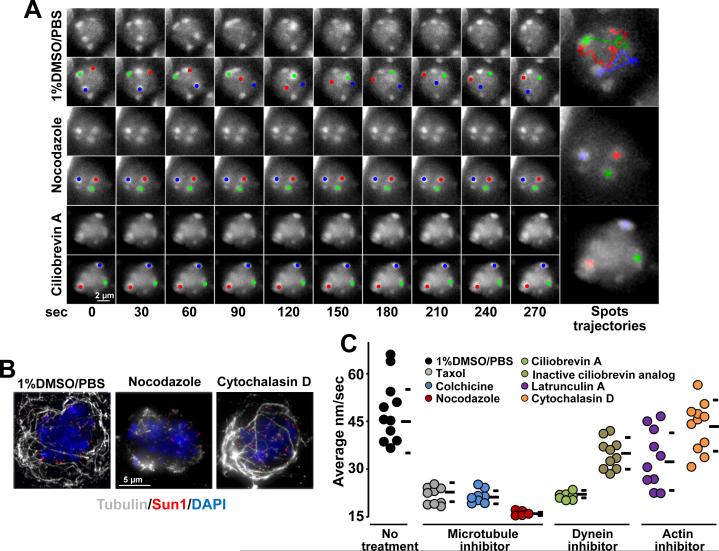
As suggested in recent work (Morimoto et al., 2012a) and the aforementioned results, chromosome movements in mouse spermatocyte nuclei displayed characteristics that may be explained by motor-driven motion along cytoskeletal microtubules. Consistent with these findings, we observed that microtubule destabilization in spermatocytes induced by intraperitoneal injection of 0.1 mg/g nocodazole (Sigma) for 3 h (Figure 4A-C) (or microtubules explants incubation in presence of nocodazole, Figure S3B) reduced RPMs in zygotene nuclei to background levels (average speed 15.7 ± 1.0 nm/sec, n = 15), which is significantly lower than zygotene nuclei in mice injected with 1% DMSO-PBS (45.3 ± 8.9 nm/sec, t-test p < 0.0001, n = 11). We observed that cessation of chromosome movement in nocodazole-treated mice was accompanied by disruption of the microtubule network (Figure 4B).
Figure 4. RPMs are Dependent on Microtubules and dynein.
A. Maximum-intensity projections of time-lapse images of wild-type zygotene spermatocytes after the indicated treatments. Magnification bar represents 2μm.B. Maximum-intensity projections of representative pachytene spermatocytes immunostained for α-tubulin and SUN1 after the indicated treatments. Magnification bar represents 5μm.C. Quantitation of RPMs in zygotene spermatocytes after intraperitoneal injection of 1% DMSO or microtubule, dynein or actin inhibitors; horizontal lines indicate the median and standard deviation values. See also Figure S3.
Significant reductions in the speed of RPMs were also observed in mice injected with colchicine (9.3 mg/g for 3 h; 21.1 ± 2.0 nm/sec, n = 10, p = 0.0004) and the microtubule stabilizing agent taxol (Sigma) (0.9 mg/g for 3 h; 22.8 ± 3.7 nm/sec, n = 10, p = 0.0005; Figure 4C and Figure S3A). RPM speed reduction after taxol treatment may be explained by disruption of the microtubule network (Figure S3C). Taken together, these results indicate that microtubules are an essential component of the system that supports chromosome movement in mouse prophase spermatocytes.
In contrast to budding yeast in which chromosome movements seems to be directed by actin (Koszul et al., 2008), RPMs in C. elegans and rat spermatocytes are less affected by actin poisons compared to microtubules (Salonen et al., 1982; Wynne et al., 2012). RPMs continued to certain extent after intraperitoneal injection with latrunculin A (Enzo life Sciences) (0.84 mg/g, 3 h; average speed of 32.3 ± 9.1 nm/sec, n = 10, p = 0.06) and cytochalasin D (Sigma) (0.6 mg/g, 3 h; 43.4 ± 8.3 nm/sec, n = 11, p = 0.5; Figure 4B and C, Figure S3A). In agreement, in experiments where seminiferous microtubules explants were incubated with varying amounts of small molecule inhibitors, although slowed to varying degrees, RPMs were markedly less sensitive to actin poisons (Figure S3B).
Recent reports demonstrate that telomere attachment to the nuclear envelope must be coupled to cytoplasmic dynein on cytoplasmic microtubules to ensure meiotic progression (Ding et al., 2007; Horn et al., 2013; Morimoto et al., 2012b). In agreement with a requirement for dynein in RPMs, intraperitoneal injection of 0.72 mg/g of ciliobrevin A (HPI-4, Sigma (Firestone et al., 2012)), reduced RPMs in zygotene spermatocytes (21.5 ± 1.0 nm/sec; t-test p = 0.0004). RPMs were markedly less sensitive to an inactive ciliobrevin analog (#2, Shengjie Bio-Tech (Firestone et al., 2012) (Figure 4C). Inhibitory effect on RPMs was also observed after microtubules explants incubation in presence of ciliobrevin A, Figure S3B). The pattern of the microtubule network remained unaltered after dynein inhibitor treatment (data not shown). This indicates that dynein per se, and not a secondary effect on microtubule network integrity (i.e., microtubule stability) account for the effects of ciliobrevin A on RPMs.
Taken together, our data indicate that RPMs in mouse spermatocytes are mainly dependent on the microtubule cytoskeleton and associated dynein motors. We should note that although RPMs are less responsive to actin filament inhibitors, cytochalasin D still had a significant effect on RPMs. This indicates a possible direct or indirect, but relative minor, role for actin in generating RPMs.
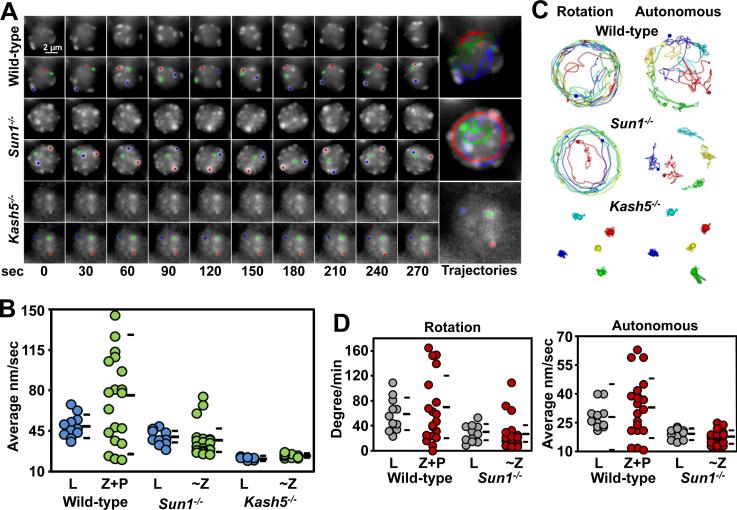
Deficient RPMs in Sun1−/− and Kash5−/− Spermatocytes
Previous works have shown that mouse SUN1 and KASH5 proteins form a complex that spans both nuclear membranes to couple telomeres to dynein on cytoplasmic microtubules (reviewed in (Shibuya and Watanabe, 2014; Stewart and Burke, 2014)). Based on the suggested requirements for these proteins in prophase telomere movements, we used 4D imaging to define the effect that deficiencies in SUN1 and KASH5 have on the tracking of telomere spots in meiotic prophase.
We observed that the RPMs are severely defective, but not absent, in Sun1−/− spermatocytes (Figure 5A and B and supplemental movie 7). The average speed of raw movement (39.1 ± 7.0 nm/sec; Kolmorogov-Smirnov test, p = 0.001), rotation (29.4 ± 13.8 degrees/min; Kolmorogov-Smirnov test, p = 0.01) and autonomous movements in leptotene nuclei (19.2 ± 2.9 nm/sec; t-test, p = 0.0006) and ~Z nuclei (average raw movements 38.0 ± 16.1 nm/sec, p = 0.003; rotation 27.5 ± 26.1 degrees/min, p = 0.002; and autonomous movements 17.8 ± 3.3 nm/sec, p < 0.0001) were significantly different from wild-type, confirming the visual impression of deficient rotation and near absent autonomous movements (Figure 5C and D and supplemental movie 8). These observations are consistent with a conserved role for SUN1 orthologs (i.e. SUN2) in chromosome movements in other species (Conrad et al., 2008; Hiraoka and Dernburg, 2009; Wynne et al., 2012).
Figure 5. SUN1 and KASH5 are Required for RPMs.
A. Maximum-intensity projections of time-lapse images of wild-type, Sun1−/− and Kash5−/− zygotene spermatocyte nuclei. Magnification bar represents 2μm.B. Quantitation of spot movements in the seminiferous tubules of wild-type, Sun1−/− and Kash5−/− mice. C. Autonomous and rotational movements are defective in Sun1−/− and Kash5−/− nuclei. Movement of individual markers highlighted in color are viewed down the axis of rotation. D. Quantification of autonomous and rotational movements in wild-type and Sun1−/− nuclei. For comparison wild-type, zygotene (Z), and pachytene (P) nuclei were grouped. See also Figure S2.
The strong correlations between the speed of rotation and autonomous movements in wild-type and Dmc1−/− mice, and reduced correlations between rotation and autonomous movements in Sun1−/− mice (Figure S2B) suggest that a close mechanistic relationship exists between rotation and autonomous movements.
We measured RPMs in spermatocyte nuclei from Kash5−/− mice. The average speeds for raw movements in leptotene (23.6 ± 1.3 nm/sec, p < 0.0001) and ~Z (25.6 ± 1.8 nm/sec, p < 0.0001) indicated that RPMs were absent (Figure 5B). Spermatocyte nuclei in Kash5−/− mice exhibited no residual rotation or autonomous movements (Figure 5C) which is a more severe phenotype compared to Sun1−/− spermatocytes.
In sum, quantitative measurement of RPMs reveals that KASH5 is uniquely required for normal RPMs levels. However, in the absence of SUN1, chromosome movements continue, albeit at a very much reduced level. The implication here is that there must be an additional SUN protein expressed in spermatocytes that can tether at least some KASH5 at the nuclear periphery. An obvious candidate is SUN2, a protein recently documented at the spermatocyte nuclear envelope and proposed to have a relative minor role in RPMs respect to SUN1.
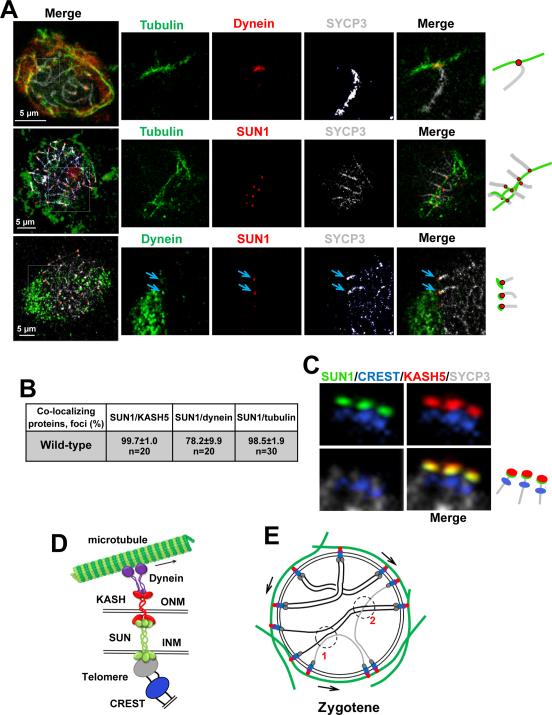
We ask whether one or several SUN1/KASH5 complexes contact microtubule track associated to the nuclear envelope. We used structurally preserved spermatocytes and confocal microscopy to visualize the association of telomeres to cytoskeletal components via SUN1-KASH5 nuclear envelope bridges. Co-immunolabelling experiments in pachytene wild-type spermatocytes revealed high levels of co-localization of SUN1/KASH5 (99.7 ± 1.0%, n = 20), SUN1/dynein (78.2 ± 9.9%, n = 20), and SUN1/tubulin (98.5 ± 1.9%, n = 30), and demonstrated that SUN1/KASH5/dynein connect the ends of the telomeres to a section of microtubule cable that is close to the nuclear envelope (Figure 6A-D). Notably, several SUN1-associated telomeres were attached to the same microtubule cable, which formed a continuous network with other cables extending in divergent and parallel directions (Figure 6A, middle panel). This indicates that multiple telomeres may use the same or closely associated microtubules/tracks simultaneously.
Figure 6. Visualization of KASH5-SUN1 Complexes Coupling Telomeres to dynein on Cytoskeletal Microtubules.
A. Example of wild-type pachytene spermatocytes showing co-localization of components of the system that supports RPMs. The magnified areas highlight the association of the chromosome ends and associated protein complexes with microtubules. Arrows indicate dynein-SUN1 co-localization. Magnification bar represents 5μm.B. Quantitation of the co-localization of protein immunosignals in wild-type spermatocytes. C. Example of pachytene chromosome proximal telomeric ends connected to SUN1/KASH5 nuclear envelope bridges. SYCP3 was used to visualize chromosome cores and CREST marks the proximal telomeric ends. D. Proposed model for meiotic chromosome telomere-nuclear envelope attachment and connection to dynein on microtubules. E. Schematic representation of zygotene nuclei. The proposed model in D and E summarizes observations of this and previous studies. In E arrows represent the direction of telomere movements. At 1, RPMs disrupt a non-homologous unproductive interaction. At 2, RPMs facilitate interlock resolution. See also Figure S4.
The number of telomeres connected to SUN1/KASH5 complexes and the stability of the interactions of these bridges with dynein and microtubule cables may regulate the speed of RPMs. We then examined telomere connections with nuclear envelope bridges by analyzing co-localization of CREST (as a marker of centromere-proximal telomeres) with SUN1 and SUN1/tubulin in pre-meiotic and prophase spermatocytes. As cells entered meiosis, a high number of telomeres associated with SUN1 (Figure S4 A and B). As prophase I progressed, eventually all telomeres associate with nuclear envelope bridges. Similarly, co-localization of SUN1 and tubulin revealed that a high number of nuclear envelope bridges associated with microtubule cables as the cells entered meiosis, and reached a maximum in zygotene and pachytene. Taken together, these results indicate that the number of telomeres connected to the system may influence characteristic RPMs speed during meiotic progression.
DISCUSSION
In this study we explored the complex movements of the telomeres of meiotic prophase cells in their natural environment in seminiferous tubules. Our imaging methods allowed us to resolve the timing and spatial resolution of chromosome movements in cells in various meiotic stages. This analysis shows that RPMs are activated by meiotic entrance, regulated during meiotic progression, and are sensitive to mutations affecting recombination and chromosome synapsis. Together with an in silico analysis, this methodology allowed dissection of chromosome movements during bouquet formation and resolution, and revealed unique characteristics of chromosome movements - most notably that RPMs in mouse spermatocytes are the result of a combination of nuclear rotation and chromosome autonomous movements.
Previous work in mice has demonstrated that treatment with the microtubule poison nocodazole altered telomere movements (Morimoto et al., 2012b; Shibuya et al., 2014a; Shibuya et al., 2014b) and colchicine treatment induced defective chromosome pairing, synapsis and segregation in vivo (Tepperberg et al., 1997, 1999). This work led to the model that telomere movement in mouse depends upon force generation by dynein to move the telomeres along microtubules – similar to what has been deduced from studies of telomere movements in other organisms (Chikashige et al., 2007; Salonen et al., 1982; Vogel et al., 2009; Wynne et al., 2012; Yamamoto and Hiraoka, 2003). Our studies reveal that microtubule bundles lie along the nuclear surface. These microtubule bundles extend into a complex, interconnected cytoskeletal network. Notably, we find that the speed of telomere movement changes as cells proceed through meiotic prophase. These changes in chromosome behavior were found to be accompanied by dramatic changes in the organization of the microtubule network. We observed that microtubule bundles at the nuclear surface interact with one or several SUN1/KASH5 complexes and chromosome ends apparently attached to the nuclear envelope. The linear movements of telomeres along the nuclear envelop and the pauses and sharp angled turns made by telomeres are consistent with the model that microtubule bundles are used as tracks to guide chromosome movement. This model suggests the possibility that microtubule reorganizations may regulate the speed and type of chromosome movement characteristic of different stages of prophase.
Perhaps surprisingly, at the present time little consensus has been achieved about the function of meiotic chromosome motion. Previous work and our findings suggest a model in which telomere-led RPMs, in coordination with changes in cytoskeletal-generating forces - the force-transducing system, and the mechanical properties of chromosomes result in highly-regulated chromosome dynamics (reviewed in (Hiraoka and Dernburg, 2009; Koszul and Kleckner, 2009)). We imagine that the primary biological significance of these mutually-dependent processes is regulation of inter-homolog interactions that operate during chromosome pairing to guarantee that the homologous chromosome pairs are prepared for segregation. Therefore, at different stages of meiotic prophase, RPMs may: (1) allow free movement to promote chromosome interactions and facilitate homologue pairing; (2) reduce unproductive interactions between unrelated chromosomes, such as local non-homologous pairing and synapsis that may result in ectopic crossing over (Figure 6E); (3) resolve chromosome entanglements (Figure 6E); (4) direct the formation and resolution of chromosome bouquet conformation; (5) reduce crossing over at centromeric regions by reducing early inter-homolog telomeric interactions; and (6) regulate the number and position of crossovers.
The use of intact cultured mouse seminiferous tubules has proved to be an advantageous model system for high-resolution observation of chromosome structures. The live imaging methodology and analysis developed here provide a platform for future studies focused to understand unresolved aspects of RPMs such as the molecular basis for their regulation and the role(s) of RPMs in meiotic progression. Furthermore, live imaging in tissue explants that can be cultured for a relative long period of time (Sato et al., 2011) may be used to study other poorly understood meiotic processes such as homologous chromosome pairing and synapsis, which together allow meiotic chromosomes to segregate faithfully.
EXPERIMENTAL PROCEDURES
Mouse Strains
The Oklahoma Medical Research Foundation Animal Care and Use Committee approved all animal protocols. The mice used in this study were as follows: C57BL/6, Dmc1−/− (Pittman et al., 1998), Hfm1−/− (Guiraldelli et al., 2013), Sycp3−/− (Yuan et al., 2000), Kash5−/− (Horn et al., 2013) and Sun1−/− (Chi et al., 2009).
Fluorescent Microscopy and Imaging
Four-dimensional movie stack images (16 slices with 1 μm steps) were acquired every 10 sec for the indicated times using a Zeiss Axioplan 2ie fitted with a 63X 1.4NA objective, Roper CoolSnap camera and custom acquisition software.
Images of fixed spermatocytes were analyzed using AxioVision software (Zeiss). Confocal IF images of fixed spermatocytes were acquired with a Zeiss LSM-710 system.
Full description of procedures for movies acquisition, small molecule inhibitor treatment, and imaging of fixed spermatocytes are available in Supplemental Experimental Procedures.
Algorithms to distinguish whole-nucleus rotational and autonomous spot movement
Full details of the procedure to distinguish rotational and autonomous movements are described in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Direct visualization of chromosome movements in live mouse seminiferous tubules.
Quantitative analysis revealed characteristics and regulation of RPMs.
SUN1/KASH5 connects telomere to dynein on cytoskeletal microtubules to generate RPMs.
Etoc blurb.
Lee et al. show that prophase movement of mouse meiotic chromosomes are regulated in a stage-specific manner and altered in recombination and synapsis mutants. Collectively, their results show that telomeres move along microtubule tracks associated to the nuclear membrane with transduction of force through the nuclear envelope bridges.
ACKNOWLEDGEMENTS
We thank Kuan-Teh Jeang (National Institutes of Health) for Sun1−/− mice. We also thank Dean Dawson and Gary Gorbsky (OMRF) for comments on the manuscript. This work was supported by NIGMS, National Institutes of Health Award 1P20GM103636, and by March of Dimes FY14-256 and 4340-06-05-0 OCASCR grants (to R. J. P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and 8 movies and can be found with this article online.
REFERENCES
- Chi YH, Cheng LI, Myers T, Ward JM, Williams E, Su Q, Faucette L, Wang JY, Jeang KT. Requirement for Sun1 in the expression of meiotic reproductive genes and piRNA. Development. 2009;136:965–973. doi: 10.1242/dev.029868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Haraguchi T, Hiraoka Y. Another way to move chromosomes. Chromosoma. 2007;116:497–505. doi: 10.1007/s00412-007-0114-8. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 2008;133:1175–1187. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998;111(Pt 6):701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Firestone AJ, Weinger JS, Maldonado M, Barlan K, Langston LD, O'Donnell M, Gelfand VI, Kapoor TM, Chen JK. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 2012;484:125–129. doi: 10.1038/nature10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiraldelli MF, Eyster C, Wilkerson JL, Dresser ME, Pezza RJ. Mouse HFM1/Mer3 is required for crossover formation and complete synapsis of homologous chromosomes during meiosis. PLoS Genet. 2013;9:e1003383. doi: 10.1371/journal.pgen.1003383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell. 2009;17:598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Ding DQ, Yamamoto A, Tsutsumi C, Chikashige Y. Characterization of fission yeast meiotic mutants based on live observation of meiotic prophase nuclear movement. Chromosoma. 2000;109:103–109. doi: 10.1007/s004120050417. [DOI] [PubMed] [Google Scholar]
- Horn HF, Kim DI, Wright GD, Wong ES, Stewart CL, Burke B, Roux KJ. A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J Cell Biol. 2013;202:1023–1039. doi: 10.1083/jcb.201304004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133:1188–1201. doi: 10.1016/j.cell.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Kleckner N. Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol. 2009;19:716–724. doi: 10.1016/j.tcb.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador L, Barroso C, Lightfoot J, Muller-Reichert T, Flibotte S, Taylor J, Moerman DG, Villeneuve AM, Martinez-Perez E. Chromosome movements promoted by the mitochondrial protein SPD-3 are required for homology search during Caenorhabditis elegans meiosis. PLoS Genet. 2013;9:e1003497. doi: 10.1371/journal.pgen.1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Conrad MN, Dresser ME. Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS genetics. 2012;8:e1002730. doi: 10.1371/journal.pgen.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebe B, Alsheimer M, Hoog C, Benavente R, Scherthan H. Telomere attachment, meiotic chromosome condensation, pairing, and bouquet stage duration are modified in spermatocytes lacking axial elements. Mol Biol Cell. 2004;15:827–837. doi: 10.1091/mbc.E03-07-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn A, Soucek R, Borner GV. ZMM proteins during meiosis: crossover artists at work. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- Morelli MA, Werling U, Edelmann W, Roberson MS, Cohen PE. Analysis of meiotic prophase I in live mouse spermatocytes. Chromosome Res. 2008;16:743–760. doi: 10.1007/s10577-008-1224-8. [DOI] [PubMed] [Google Scholar]
- Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, Watanabe Y. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J Cell Biol. 2012a;198:165–172. doi: 10.1083/jcb.201204085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, Watanabe Y. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. The Journal of cell. 2012b;198:165–172. doi: 10.1083/jcb.201204085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- Parvinen M, Soderstrom KO. Chromosome rotation and formation of synapsis. Nature. 1976;260:534–535. doi: 10.1038/260534a0. [DOI] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA, Schimenti JC. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- Rickards GK. Prophase chromosome movements in living house cricket spermatocytes and their relationship to prometaphase, anaphase and granule movements. Chromosoma. 1975;49:407–455. doi: 10.1007/BF00285133. [DOI] [PubMed] [Google Scholar]
- Salonen K, Paranko J, Parvinen M. A colcemid-sensitive mechanism involved in regulation of chromosome movements during meiotic pairing. Chromosoma. 1982;85:611–618. doi: 10.1007/BF00330775. [DOI] [PubMed] [Google Scholar]
- Sato T, Katagiri K, Gohbara A, Inoue K, Ogonuki N, Ogura A, Kubota Y, Ogawa T. In vitro production of functional sperm in cultured neonatal mouse testes. Nature. 2011;471:504–507. doi: 10.1038/nature09850. [DOI] [PubMed] [Google Scholar]
- Scherthan H. A bouquet makes ends meet. Nature reviews Molecular cell biology. 2001;2:621–627. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104:16934–16939. doi: 10.1073/pnas.0704860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan MJ, Pawlowski WP. Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proc Natl Acad Sci U S A. 2009;106:20989–20994. doi: 10.1073/pnas.0906498106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya H, Ishiguro KI, Watanabe Y. The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat Cell Biol. 2014a;16:145–156. doi: 10.1038/ncb2896. [DOI] [PubMed] [Google Scholar]
- Shibuya H, Morimoto A, Watanabe Y. The dissection of meiotic chromosome movement in mice using an in vivo electroporation technique. PLoS Genet. 2014b;10:e1004821. doi: 10.1371/journal.pgen.1004821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya H, Watanabe Y. The meiosis-specific modification of mammalian telomeres. Cell Cycle. 2014;13:2024–2028. doi: 10.4161/cc.29350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CL, Burke B. The missing LINC: a mammalian KASH-domain protein coupling meiotic chromosomes to the cytoskeleton. Nucleus. 2014;5:3–10. doi: 10.4161/nucl.27819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperberg JH, Moses MJ, Nath J. Colchicine effects on meiosis in the male mouse. I. Meiotic prophase: synaptic arrest, univalents, loss of damaged spermatocytes and a possible checkpoint at pachytene. Chromosoma. 1997;106:183–192. doi: 10.1007/s004120050238. [DOI] [PubMed] [Google Scholar]
- Tepperberg JH, Moses MJ, Nath J. Colchicine effects on meiosis in the male mouse. II. Inhibition of synapsis and induction of nondisjunction. Mutat Res. 1999;429:93–105. doi: 10.1016/s0027-5107(99)00102-5. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken E, Adelfalk C, Loidl J, Scherthan H. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J Cell Biol. 2005;170:213–223. doi: 10.1083/jcb.200501042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel SK, Pavin N, Maghelli N, Julicher F, Tolic-Norrelykke IM. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 2009;7:e1000087. doi: 10.1371/journal.pbio.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne DJ, Rog O, Carlton PM, Dernburg AF. Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J Cell Biol. 2012;196:47–64. doi: 10.1083/jcb.201106022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y. Cytoplasmic dynein in fungi: insights from nuclear migration. J Cell Sci. 2003;116:4501–4512. doi: 10.1242/jcs.00835. [DOI] [PubMed] [Google Scholar]
- Yao KT, Ellingson DJ. Observations on nuclear rotation and oscillation in Chinese hamster germinal cells in vitro. Exp Cell Res. 1969;55:39–42. doi: 10.1016/0014-4827(69)90451-0. [DOI] [PubMed] [Google Scholar]
- Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Hoog C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell. 2000;5:73–83. doi: 10.1016/s1097-2765(00)80404-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.