Abstract
Examining the function of individual human hippocampal subfields remains challenging due to their small sizes and convoluted structures. Previous human functional magnetic resonance (fMRI) studies at 3 Tesla (T) have successfully detected differences in activation between hippocampal cornu ammonis (CA) field CA1, combined CA2, 3 and dentate gyrus (DG) region (CA23DG), and the subiculum during associative memory tasks. In this study we investigated hippocampal subfield activity in healthy participants using an associative memory paradigm during high-resolution functional magnetic resonance imaging (fMRI) scanning at 7T. We were able to localize fMRI activity to anterior CA2 and CA3 during learning, and to the posterior CA2 field, the CA1, and the posterior subiculum during retrieval of novel associations. These results provide insight into more specific human hippocampal subfield functions underlying learning and memory and a unique opportunity for future investigations of hippocampal subfield function in healthy individuals as well as those suffering from neurodegenerative diseases.
Keywords: Hippocampus, memory, fMRI, MRI, high-resolution imaging
Introduction
Numerous studies have confirmed a crucial role for the hippocampus in declarative memory, or the memory of previously experienced events and learned facts (Squire 1992, 2004). Subfields of the hippocampus (Cornu ammonis [CA] fields 1–3, dentate gyrus [DG], and subiculum) differ in both structure and function (Leutgeb et al., 2007; Lee et al., 2004; Sharp, 2006; Taube et al., 1990; Boccara et al., 2010; Duvernoy, 2005; Carr et al., 2010a). There have been numerous studies in humans and animal models suggesting different roles for these subregions, but the results are still not currently clear. Computational models suggest the hippocampal CA3 region is involved in the successful formation of new associations (Treves & Rolls, 1994; Marr, 1971). Specifically these models have posited that the recurrent collaterals in CA3 play a role in binding together elements from episodes or associations. Thus, one might predict that this region might be particularly active during learning new associations. Furthermore, rodent electrophysiological studies have found that both CA3 and DG regions may be involved in pattern separation processes (Leutgeb et al., 2007), or the orthogonalization of overlapping information, that may be necessary for accurate learning of similar items in memory. Neunuebel & Knierim (2014) have recently presented evidence that the CA3 performs pattern completion in that it showed relatively coherent activity in the face of distortions of the environment and variations in input from the DG.
While DG and CA3 are thought to perform different functions based on computational models of the hippocampus (Marr, 1971; O’Reilly & McClelland, 1994), most work with humans has not been able to separate the DG and CA3 regions due to difficulty with defining borders between these regions using standard MRI techniques. Nevertheless, a number of studies have shown evidence of encoding related activity and pattern separation in an ROI including DG, CA2 and CA3 (CA23DG; Zeineh et al., 2003; Eldridge et al., 2005; Suthana et al., 2011; Bakker et al., 2008; Stokes et al., 2014; Yassa & Stark, 2011). However, there are some inconsistencies across studies with respect to hippocampal regions CA1 and CA23DG (Azab et al., 2014; Lacy et al., 2011).
The limited number of available studies and possible species-specific differences (O’Keefe, 1999) both in terms of the memory tasks used in different species and inherent differences in circuitry illustrate the need for advances in human hippocampus subregion functional analyses in vivo. In humans, functional magnetic resonance (fMRI) studies at 3 Tesla (T) have successfully detected differences in activation between CA1, CA23DG, and the subiculum during various memory tasks (for review see Carr et al., 2010a). For example, Zeineh and colleagues (2003) demonstrated that encoding and retrieval processes are associated with distinct hippocampal subfields involved during a face-name association task. Specifically, the authors revealed an association between neural activity in the combined CA23DG region during learning and in the subiculum during recall. While the CA23DG region has been consistently shown to be active during the learning of novel paired associates (Zeineh et al., 2003; Eldridge et al., 2005; Suthana et al., 2011), it remains unknown whether CA2, CA3 or the DG within the human hippocampus contribute to the increase in learning related activity. Previous limitations of these studies only allowed for separation of the CA23DG area from neighboring CA1 and subiculum regions (Carr et al., 2010a). Separating CA1 from the other hippocampal CA fields in the anterior hippocampus has also been previously challenging (Zeineh et al., 2003; Suthana et al., 2009, 2011). More recently, using 3T MRI scanning and multi-voxel pattern analysis (MVPA), Bonnici and colleagues (2012) were able to identify differential hippocampal subfield contributions in a decision-making paradigm. The authors attempted to distinguish CA1, CA3 and the dentate gyrus (DG) using structural and functional scanning with 0.52 × 0.52mm and 1.5 × 1.5mm in-plane resolution respectively. A potential strength of the approach by Bonnici and colleagues (2012) was the acquisition of isotropic voxels, although this in turn will typically reduce in-plane resolution somewhat. The method we use focuses on higher in-plane resolution on both functional and structural scans (0.35 × 0.35 structural, and 1 × 1mm functional) and uses a subsequent interpolation procedure to achieve isotropy. However, in contrast to our 7T MRI data and specific analysis technique, Bonnici and colleagues (2012) acknowledge that 3T MRI scanning does not allow the delineation of the hippocampal CA2 field, which would be part of CA3 in their segmentation approach. Another potential limitation with the Bonnici et al. study is that the lower signal to noise ratio afforded at 3T likely made accurate delineation of CA3 vs. DG challenging and potentially more arbitrary. Consistent with this potential concern, even advanced high-resolution protocols at 3T/4T do not discriminate CA3 vs. DG (i.e., Muller et al, 2007; Zeineh et al., 2000). Although Yushkevich et al. (2010, 2014) were able to apply separate DG and CA3 labels using a fully automated segmentation technique, we believe that the higher field strength afforded at 7T, as well as our higher resolution structural and EPI scans, likely made this discrimination more precise both anatomically and functionally.
Limited data on the functional significance of hippocampal area CA2 also illustrates the need for better resolution in the hippocampus in human fMRI. Once believed to be a transitional zone between CA1 and CA3, rodent data suggest unique molecular characteristics for the CA2 field that imply a functional distinction from other CA regions (Zhao et al., 2007, Mercer et al., 2012). Although the CA2 region has been suggested to mediate hippocampal-dependent memory through its synaptic connections between CA1 and CA3 (Chevaleyre and Siegelbaum, 2010), there is evidence for disproportionate atrophy in CA2 in schizophrenia (Benes et al., 1998) and relative sparing in temporal lobe epilepsy (Sloviter, 1983). Thus, localizing blood oxygen level dependent (BOLD) activation separately to the hippocampal regions CA2, CA3, or the DG would be an advance for in-vivo investigations of the human hippocampus with regards to understanding memory processes per se as well as the state and progression of neurological diseases.
In the current study, we combined 7T magnetic resonance imaging (MRI) and fMRI with cortical unfolding analyses (Zeineh et al., 2001, 2003; Suthana et al., 2011) to create two-dimensional (2D) maps yielding in-plane resolutions of 0.35 mm (structural) and 1.0 mm (functional). Additionally, in a complementary analysis we created 3D regions of interest (ROIs) from which percent signal changes in BOLD activation were measured. These techniques enabled us to measure BOLD signal changes in separate subfields of the hippocampus (CA1, CA2, CA3, and DG) and adjacent medial temporal lobe (MTL) regions during a hippocampal-dependent paired-associates memory task. We tested the hypothesis that learning related activity would be localized to the anterior CA3 subregion, based on computational models indicating engagement of CA3 in the successful formation of new associations (Treves & Rolls, 1994; Marr, 1971) and previous findings of encoding related activity within the anterior hippocampus (Schacter & Wagner, 1999; Prince et al., 2005) whereas we expected retrieval-related activity in the posterior subiculum, based on previous data demonstrating activation in this area during memory retrieval (Zeineh et al., 2003; Eldridge 2005).
Materials and methods
Subjects
Subjects in the current study were fourteen (seven female) right-handed (mean age ± S.E. = 24.7 ± 0.73; range = [19–29]) participants with no history of neurological disease. Six subjects were excluded (out of a total of 20) from the analysis due to excessive relative head motion (4 subjects excluded) or failure to complete the behavioral task (2 subjects excluded). For calculating head motion in structural scans, we estimated according to Gedamu et al. (2008), the ratio of two ROI's (one in the phase encoding direction and one in the read direction) of the image). Subjects with a ratio of greater than 2 for structural scans and a relative motion of greater than 3mm in the fMRI data were excluded. The study was performed at the Max Planck Institute (MPI) for Human Cognitive and Brain Sciences in Leipzig, Germany under the MPI and UCLA Institutional Review Board testing protocols and approved by the UCLA Human Subjects Protection Committee. All subjects gave written informed consent to participate in this study.
Experimental design
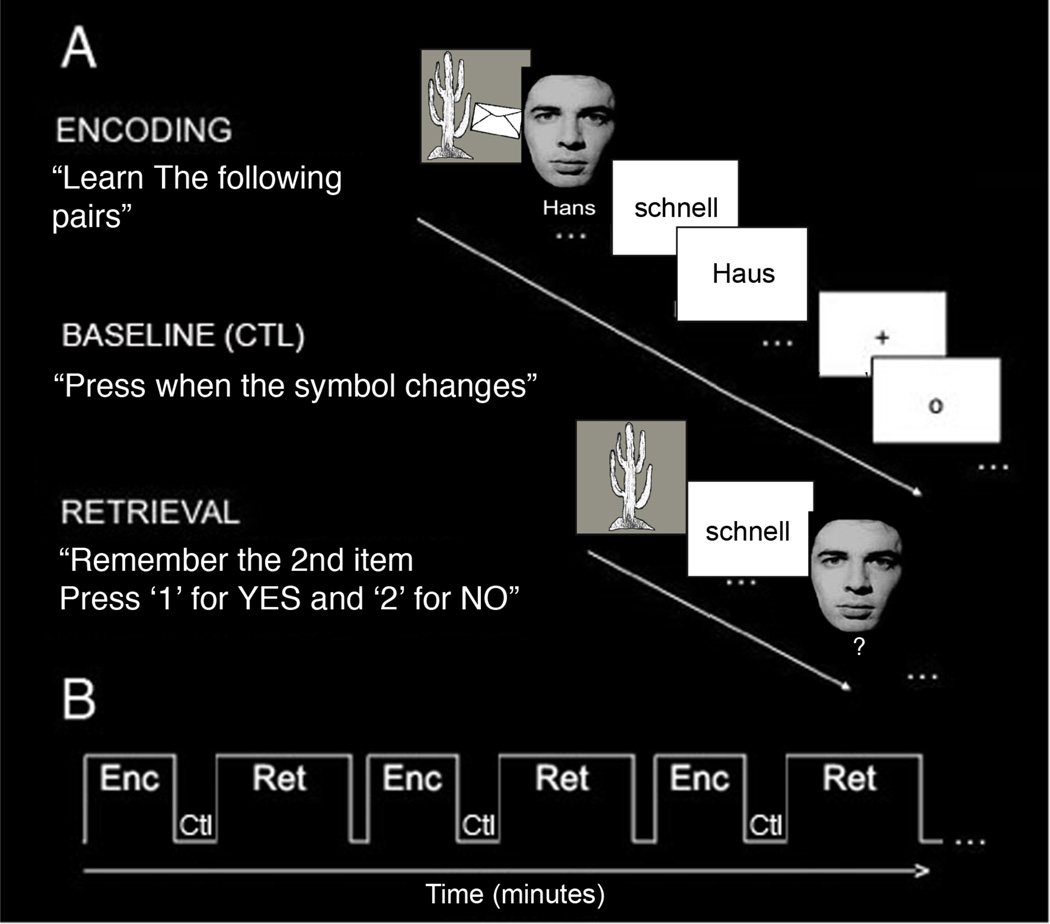
Subjects learned (encoded) and recalled (retrieved) unrelated novel pairs of face-name, word-word, and object-object associations during fMRI scanning (Fig. 1A). The task consisted of six blocks of alternating encoding and retrieval separated by a control (baseline) condition (Fig. 1). During the encoding blocks, subjects saw the same twelve pairs of unrelated associations visually shown on the screen (4 face-name, 4 word-word, 4 object-object) and were instructed to learn the associated pairs. During encoding, face-name and object-object visual pairs were each presented on the screen for duration of 3 seconds. Word-word pairs were also presented visually for a total of 3 seconds with 1.5-second duration for each word. During recall of pairs, the first item of the pair was presented visually as the cue for subsequent recall of the associated learned item. We used varying types of associations in order to look at MTL involvement during associative learning more generally. Previous studies examining specific types of associative learning separately (i.e. face-name, word-word, or object-object) have already shown CA23DG to be involved during all three types of learning (Zeineh et al., 2003; Eldridge et al., 2005; Suthana et al., 2010). In this study, we were interested in determining which CA field would be recruited during associative learning in general. Immediately following each encoding block, subjects completed a baseline control task in which they were instructed to fixate on a symbol (“+” or “o”) centered on the screen and press a button each time it changed. We chose this baseline to provide a more accurate and active baseline level; higher cognitive non-mnemonic baselines have been shown to activate the hippocampus to a lesser extent than simple fixation (Stark and Squire, 2001). Immediately following the 30-second baseline condition, subjects completed the retrieval block in which they saw the first item of the pair and were asked whether or not they were able to silently recall the second associated item. The subject pressed one of two keys corresponding to successful recall or unsuccessful recall. To prevent head motion while scanning, subjects were asked to silently recall the second item of the pair, but were then immediately tested again verbally in the scanner following the final block of the task in order to ensure accurate recall. Prior to the scanning session, all subjects completed an identical task using alternate pairs in order to ensure normal memory performance levels and to familiarize them with the task.
Fig. 1. Behavioral Task.
(A) Subjects learned (encoding) and recalled (retrieval) unrelated-pairs of words (example shown is schnell-Haus [fast-house]), objects, and face-names. During the encoding block, subjects saw twelve pairs of unrelated items and were instructed to learn the pairs. During the baseline (ctl) block, subjects were instructed to fixate on a symbol (“+” or “o”) and press a button when the symbol changed. During the retrieval block, subjects saw the first item of the pairs and were asked to remember the second paired item. (B) The task consisted of six blocks of alternating encoding (Enc) and retrieval (Ret) separated by the baseline control (Ctl) task.
During fMRI acquisition, word pairs were presented visually using a projection screen from a PC (Windows XP) computer using Presentation software (Neurobehavioral Systems, Inc, San Francisco, CA). Key presses were recorded for behavioral analysis.
Image Acquisition
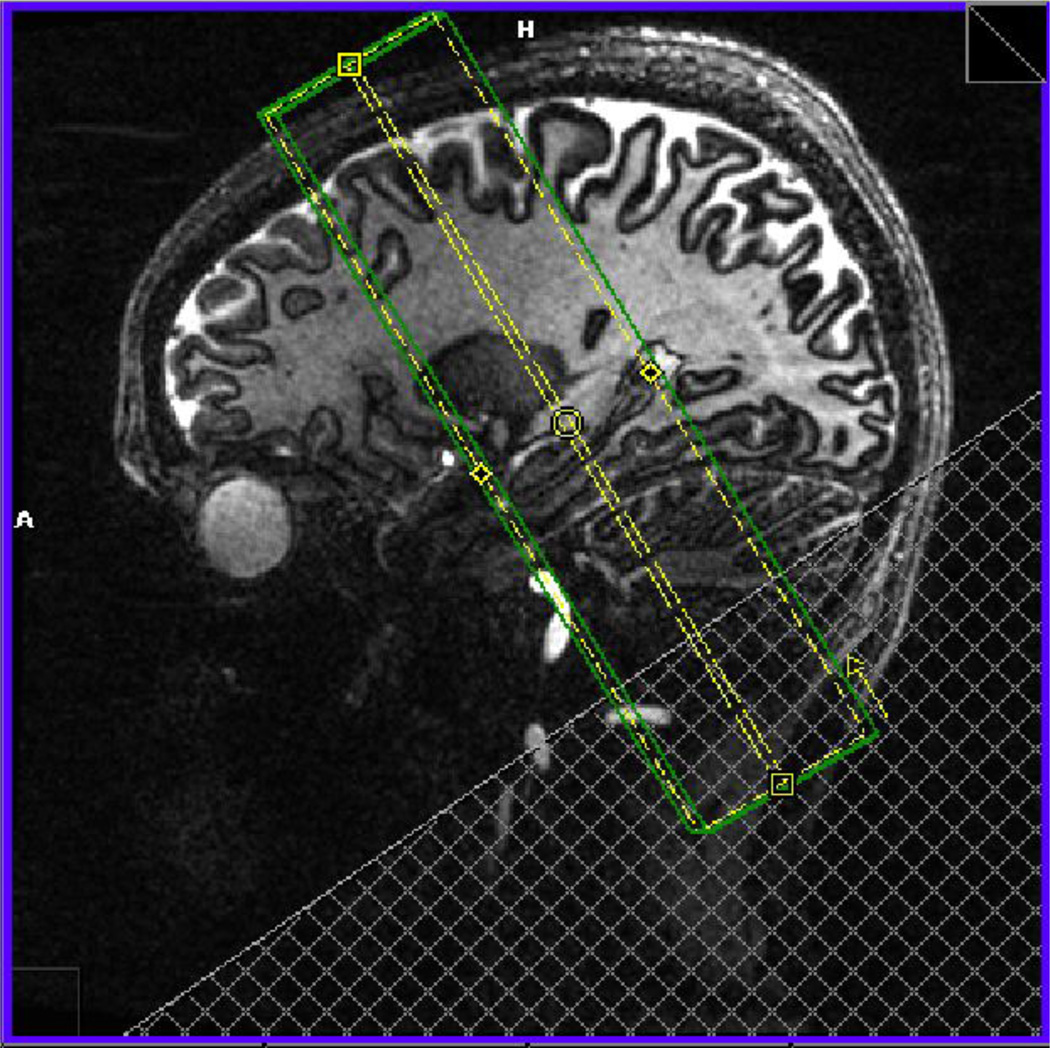
Using a 7T Siemens MR scanner (Siemens Healthcare Sector, Erlangen, Germany) with a 24-channel head coil (Nova Medical, Wilmington, MA, USA), we acquired turbo spin echo images (hippocampal high-resolution [HHR] structural scan, matrix size 640 × 640, TR = 4800 ms, TE = 22 ms, 21 slices without gap, voxel size = 0.35 × 0.35 × 2.0 mm) and echo planar images (EPI) with zoomed GRAPPA (Heidemann et al., 2012; HHR functional scan, matrix size of 200 × 200, TR = 3000 ms, TE = 19 ms, 21 slices without gap, field of view = 200 mm, total acceleration factor 3.33, voxel size = 1.0 × 1.0 × 2.0 mm). To register the EPI images to the HHR structural scan, we used a structural scan with matched resolution to the functional ([HHR coplanar scan]; matrix size 192 × 192, TR = 5930 ms, TE = 20 ms, 21 slices without gap, voxel size = 1.0 × 1.0 × 2.0 mm). For a list of scanning parameters, see table 1. All of the images were acquired in the coronal oblique plane perpendicular to the long-axis of the hippocampus (Fig. 2) because of the homogeneity of the structure along the long axis but variability in–plane; thus we maximized in-plane resolution.
Table 1.
Shown are the scanning parameters for the hippocampal high-resolution (HHR) structural, low resolution matched to functional (HHR Coplanar) and the functional scans (HHR Functional).
| Parameter | HHR Structural | HHR Coplanar | HHR Functional |
|---|---|---|---|
| TR (ms) | 4800 | 5930 | 3000 |
| TE (ms) | 22 | 30 | 19 |
| Number of slices | 21 | 21 | 21 |
| Slice thickness (mm) | 2 | 2 | 2 |
| Distance factor (%) | 0 | 0 | 0 |
| Phase Encoding | |||
| Direction | F >>H | F >>H | F >>H |
| FoV read | 224 | 192 | 200 |
| FoV phase | 100 | 100 | 90 |
| Flip angle | 60 | 60 | 90 |
| Concatenations | 1 | 1 | 1 |
| Fat suppression | None | None | None |
| Measurements | 1 | 1 | 202 |
| PAT mode | GRAPPA | GRAPPA | GRAPPA |
| Reference lines PE | 32 | 32 | 41 |
| Acceleration factor | 2 | 2 | 3 |
| Voxel size | 0.3 × 0.3 × 2 mm | 1.0 × 1.0 × 2 mm | 1.0 × 1.0 × 2 mm |
| Relative SNR | 1 | 1 | 1 |
| BW | 81 | 80 | 926 |
| Echo spacing (ms) | 21.8 | 20.1 | 1.2 |
| Time (min) | 7:38 | 4:16 | 10:23 |
Fig. 2. Orientation of Image Acquisition.
Shown is the area where high-resolution structural and zoomed EPI images are acquired in the coronal oblique plane perpendicular to the long axis of the hippocampus.
Computational Unfolding
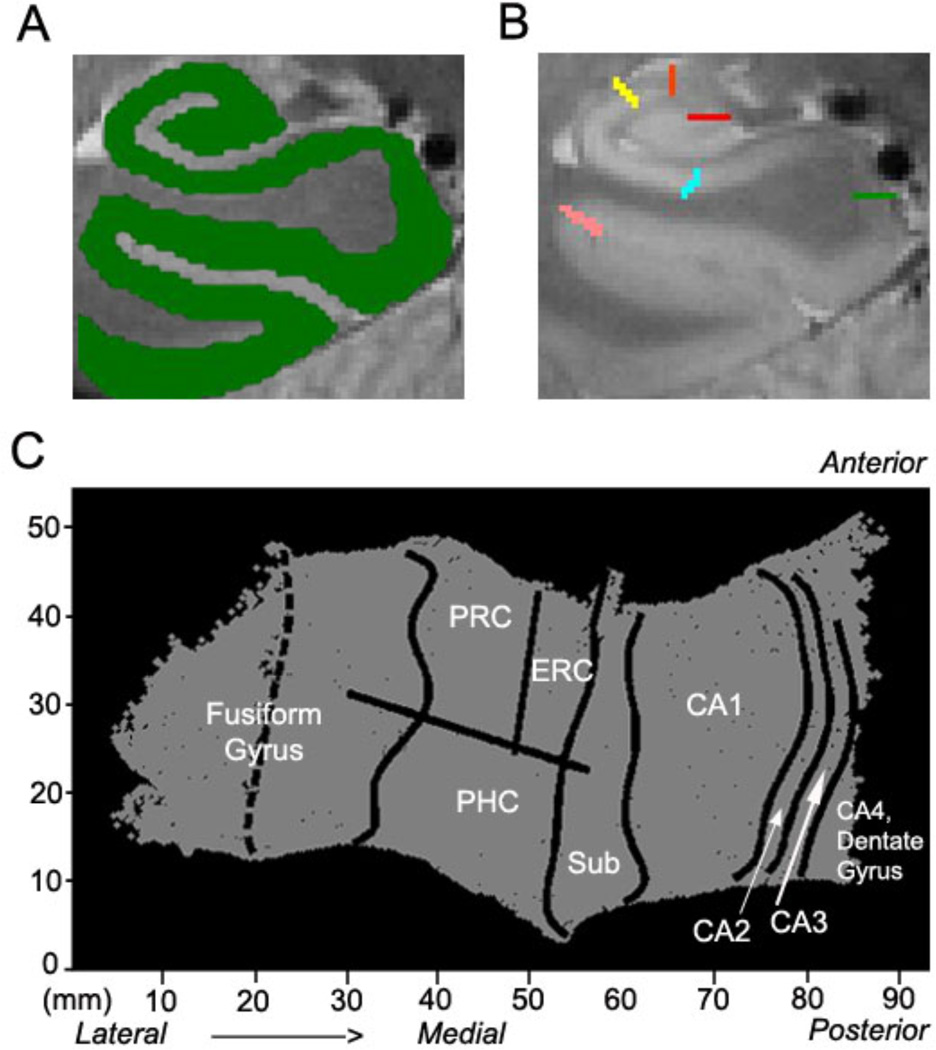
The 3D gray matter of hippocampal and adjacent MTL regions (Fig. 3A) was created by manually segmenting the white matter and CSF and growing the gray matter area between these two segmented areas (Fig. 4; Teo et al., 1997). To improve segmentation along the long-axis of the hippocampus, the entire image (i.e. segmented CSF, gray and white matter) was interpolated by a factor of 6 across slices, yielding a final isotropic voxel size of 0.35 mm3 (Engel et al., 1997). We used an automated interpolation algorithm, which involved linear interpolation along the z-axis, anisotropic curvature smoothing, thresholding, and 3D rendering (Ekstrom et al., 2009). Then connected gray matter layers were grown out (Fig. 4C,G) using a region-expansion algorithm to cover all pixels defined as gray matter, resulting in a gray matter strip containing CA fields 1, 2, and 3, 4/DG, subiculum, ERC, PRC, and PHC. To enable the computational algorithm to begin growing out layers in the DG/CA4 region, the thin sulcus between the dentate gyrus and the CA fields (the stratum radiatum, stratum lacunosum moleculare and vestigial hippocampal sulcus [SRLM-HS] that does not have CSF) must be labeled with a white matter or CSF mask having a width of at least 1 voxel each. This step of labeling the region in this manner enables the application of the unfolding algorithm. Therefore, there could be a small amount of CSF (e.g. Fig. 4, bright signal, panel D) included in the white matter label (e.g. Fig. 4, panel E, blue). Yet, in order to apply the unfolding algorithm, white matter and CSF labels must be continuous masks without any voxels not belonging to the mask (e.g., a few CSF labeled voxels within a white matter label), and thus these regions would for example be labeled as white matter. The resulting volume of connected gray matter layers would then be computationally unfolded (Fig. 3C) with an iterative algorithm based on metric multidimensional scaling, which flattens the entire MTL gray matter volume to 2D space. The position of CA1, subiculum, ERC, PRC, and PHC on the 2D map were found by mapping pixels from known points from the structural images (Fig. 3B) using atlases by Amaral and Insausti (2012), and Duvernoy (2005) similar to previously published studies (Zeineh et al, 2003; Ekstrom et al., 2009; Suthana et al., 2009). The position of CA3 was determined based on the Duvernoy atlas (2005). Specifically, for posterior slices the border between CA3 and CA4/DG was determined by following the overlying boundary line between CA2 and CA4/DG downwards until hitting the hippocampal sulcus thereby creating the rounded enclosed structure of the CA3/DG. For anterior slices, the border between CA3 and CA4/DG was placed by following the first overlying digitation of the hippocampal sulcus downwards to enclose the CA4/DG structure. With this segmentation procedure, anterior CA3 is fully enlarged due to inclusion of the white matter areas of the fimbria. For our fMRI results, inclusion of the fimbria would not present an issue due to the assumed absence of any BOLD signal originating from white matter areas. For the most 2–3 anterior slices, CA1 rather than the CA3 region dominated the hippocampal head as can be seen on the unfolded group map (Fig. 7). Posterior CA3 demarcation was based on the Duvernoy atlas and other similar methods using high-resolution imaging (Wisse et al., 2012). Anterior and posterior CA2 demarcation was determined using two separate anterior and posterior sized rectangles across slices in between CA1 and CA3 similar to a virtual square used in a previous study (Wisse et al., 2012). The CA4 region has not been separable from the DG with human imaging methods used in vivo. However, the separate existence of the CA4 field is debatable and in most cases is no longer considered to be a separate part of the dentate itself (Amaral and Insausti, 2012). We therefore included it within an encompassing region together with the DG itself. Due to improvements in resolution, our current method now treats CA fields 2, 3, and DG as separate entities (CA2, CA3, DG). We also created anatomical ROIs by projecting the bounded regions from 2D space to 3D space and created separate mask ROI images. Initial boundary delineation was done in the 3D image space, which was then projected into 2D space. The gray matter strip (e.g. green area outlined in Fig. 4C) was then broken up into regions according to these boundary lines and projected back into 3D space. The separate segmented regions were then visually inspected and edited manually if needed. These regions included DG, CA3, CA2, CA1, subiculum, ERC, PRC, and PHC and can be seen in an example subject’s MRI (Fig. 5).
Fig. 3. Generation of Flat Maps.
Shown is for one example subject. (A) The gray matter (green) is flattened into a (C) 2D map (right side shown) showing regions CA4DG (CA4, dentate gyrus), CA2, CA3, CA1, subiculum (sub), entorhinal cortex (ERC), perirhinal cortex (PRC), parahippocampal cortex (PHC), and fusiform gyrus. (B) Boundaries between regions are demarcated and projected into 2D space after gray matter is unfolded into a flat map. Shown are the boundaries between dentate gyrus and CA3 (red), CA3 and CA2 (orange), CA2 and CA1 (yellow), CA1 and subiculum (light blue), subiculum and PHC (green), PHC and fusiform (pink). For boundaries between anterior regions (ERC and PRC) see figure 4.
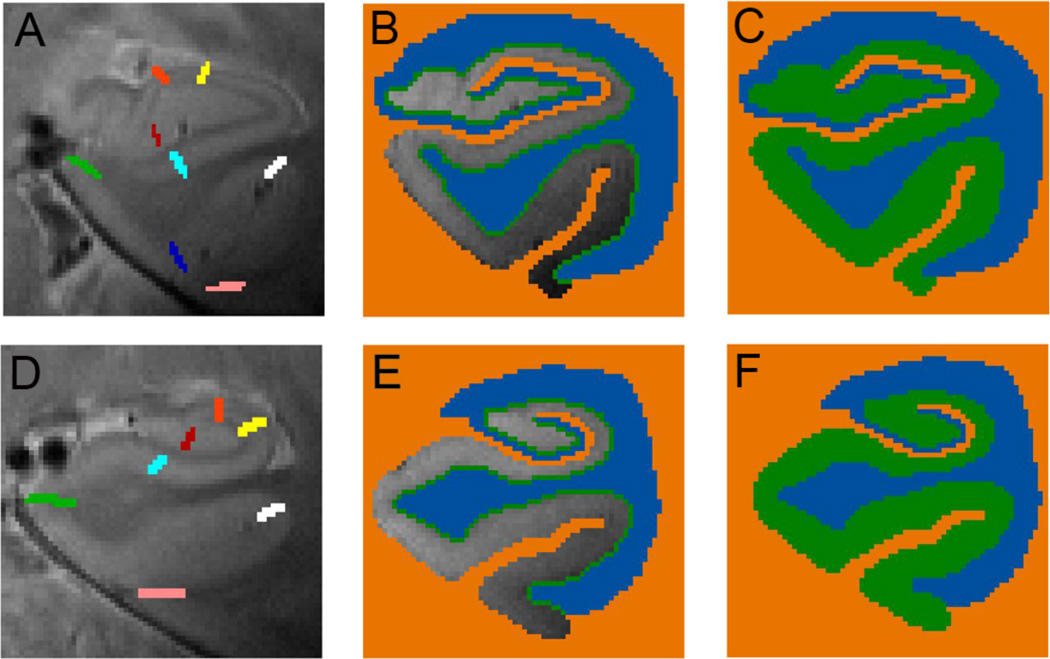
Fig. 4. Unfolding method.
Each subjects’ gray matter (green, C, F) is created by segmenting the lateral and dorsal white matter and CSF areas (blue) and medial non-MTL gray matter and cerebral spinal fluid (orange). The gray matter is grown in layers in between (B, E) from blue to orange voxels. (A, D) Boundaries between regions are demarcated on each slice for each individual subject in 3D space. (A) Shown are the anterior MTL boundaries between dentate gyrus and CA3 (red), CA3 and CA2 (orange), CA2 and CA1 (yellow), CA1 and subiculum (light blue), subiculum and ERC (green), ERC and PRC (dark blue), PRC and fusiform (pink). (D) Additional posterior boundaries are shown including the boundary between subiculum and PHC (green) and between PHC and fusiform (white).
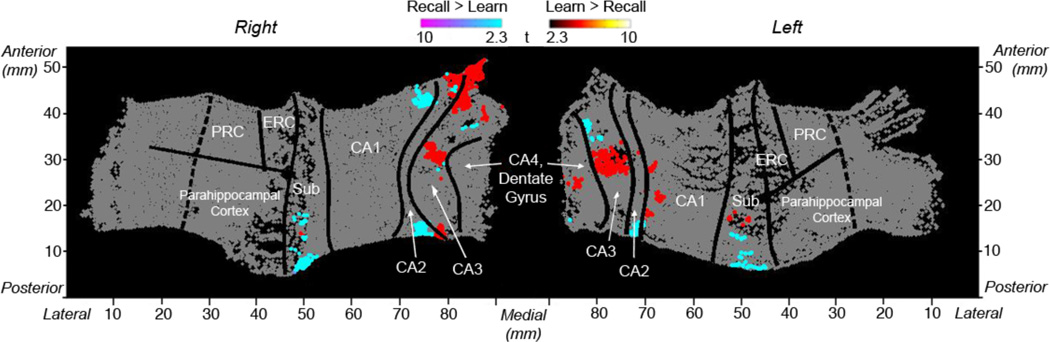
Fig. 7. Group Voxel-wise Analysis.
Group voxel based mixed-effects unfolded t-test maps (statistical maps of significantly activated and deactivated regions; recall > learn: 2.3 ≥ t ≥ 10 and learn > recall: 2.3 < t < 10, p < 0.05 cluster corrected). Shown are significant increases and decreases within the left MTL regions during learning and recall of associative pairs. Regions shown include CA4 and dentate gyrus, CA3, CA2, CA1, subiculum (sub), entorhinal cortex (ERC), perirhinal cortex (PRC), and parahippocampal cortex.
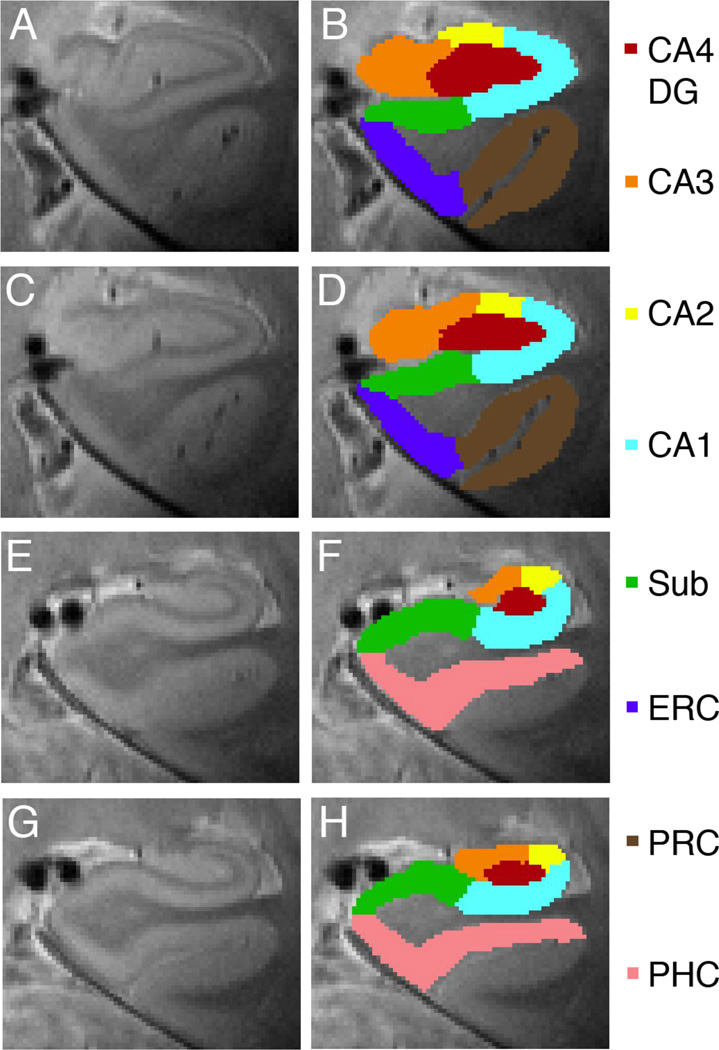
Fig. 5. Anatomical Regions of Interest.
Voxels in 2D space are projected into 3D space to create anatomical regions of interests showing anterior (A–D) and posterior regions (E–H): CA4 and dentate gyrus (CA4DG [red]), CA3 including fimbria (orange), CA2 (yellow), CA1 (light blue), subiculum (green), entorhinal cortex (blue), perirhinal cortex (brown) and parahippocampal cortex (pink).
Functional Imaging and statistical analysis
To investigate differences in overall activity in contrasts of interest fMRI analysis was completed using the FEAT (fMRI Expert Analysis Tool, version 5.98) tool of FMRIB Software Analysis (FSL version 4.0, www.fmrib.ox.ac.uk/fsl). Preprocessing included skull stripping, retrospective head motion correction and image quality analysis before the data were entered into statistical analysis. Motion correction to the medial volume was applied to functional images using MCFLIRT (FMRIB's motion correction linear image registration tool; Jenkinson et al., 2002); a normalized correlation ratio cost function and linear interpolation was used. Skulls were removed from brain images using BET (brain extraction tool; Smith et al., 2002). Spatial smoothing was applied to images using a Gaussian kernel of full-width-half-maximum at 2mm across subregional boundaries for our ROI analysis and within subregional boundaries for unfolding analyses. However, since smoothing did not change our overall results, we present ROI results and example fMRI data using unsmoothed data. Images were high-pass temporal filtered using a Gaussian-weighted least-squares straight-line fitting (sigma = 100.0s). High-resolution functional images were aligned using FLIRT (FMRIB’s Linear Image Registration Tool) to high-resolution coplanar images with a rigid-body transformation with 6 degrees of freedom. The high-resolution coplanar images were then registered to the subject’s high-resolution structural images with 6 degrees of freedom. For all subjects, registration was improved by using a weighted mask of each individual subjects’ MTL regions to optimize FLIRT registration to these structures. To do this, we created a mask of bilateral MTL regions for each individual subject and used these images as a reference volume to weight each individual’s registration allowing for focused registration of MTL regions. We also manually checked each subjects’ registration using landmarks visible on both structural and functional images (e.g. blood vessels). FILM (FMRIB’s improved linear model) with local autocorrelation correction (Woolrich et al., 2001) was used for all time-series statistical analysis. To create regressors of interest a delta function with trial onset times was convolved with a canonical (double gamma) hemodynamic response function, along with the temporal derivative. Prior to the ROI analysis only, a cluster corrected threshold at Z > 2.3 and p < 0.05 was applied to all contrasts of interest. Contrasts of interests included Learn versus Baseline, Learn versus Recall, Recall versus Baseline, and Recall versus Learn. For the Learn versus Recall contrast we were interested in looking at areas of significant BOLD activation that were greater during learning compared to recall. Conversely, for the Recall versus Learn contrast, we were interested in areas of BOLD activation that were significantly greater during the recall compared to the learn condition.
Each subject’s individual high-resolution functional activations were warped into an unfolded 2D group template, which was created based on the individual subject anatomical images and boundaries. For each voxel, the fit between each individual subject’s activation (e.g., beta values) for contrasts of interest were then compared across subjects using a mixed-effects t-test (t ≥ 2.3, p < 0.05, cluster corrected). See Thompson et al. (2000), Zeineh et al. (2001), and Ekstrom et al. (2009) for details on these methods.
A priori, anatomical ROIs were defined in 2D space and then projected into 3D space (Fig. 5) using reverse transformation parameters that were used for projecting 3D to 2D (Ekstrom et al., 2010). Masks for each ROI were created using the demarcated boundaries that were drawn in 3D space. Using the 3D ROI anatomical masks, FSL Featquery (FEAT version 5.98) was then used to calculate the average percent signal change within each ROI. A two (condition) by seven (region) repeated measures analysis of variance (ANOVA) was conducted to compare the effect of condition on percent signal change during learning compared to baseline and recall compared to baseline. Percent signal change was also computed for the six blocks of the task separately for CA3 and subiculum. For the ROI analysis, post hoc t-test comparisons were made only if global ANOVA indicated a statistically significant (p < 0.05) effect of condition (learn vs. baseline and recall vs. baseline).
Results
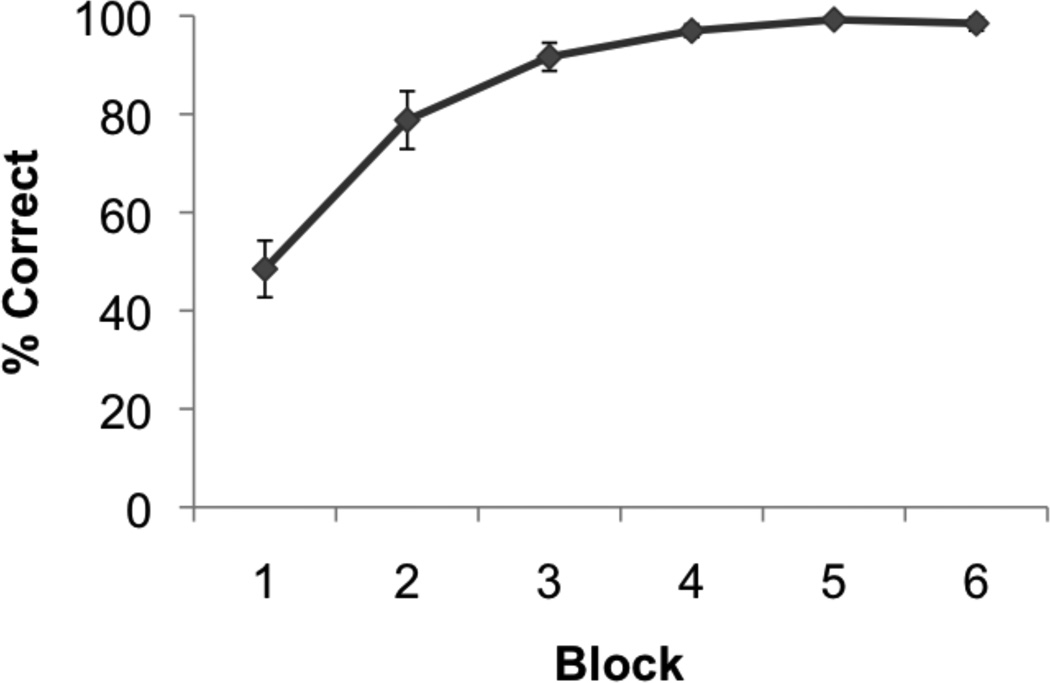
Participants successfully learned and recalled novel unrelated paired-associates (N = 14, percent correct: mean ± s.e.m., 89.09 ± 3.08; Fig. 6) during the hippocampal-dependent memory task (Fig. 1). Performance increased across blocks of the task (F(5,65) = 43.322; p < 0.001). Figure 3C shows an example of a flat map of an individual’s right MTL regions. The 3D ROIs generated from the flat maps are shown on anterior (Fig. 5A–D) and posterior (Fig. 5E–H) coronal images showing the hippocampus (Fig. 5, CA4DG [red], CA3 [orange], CA2 [yellow], CA1 [light blue], subiculum [green],) and adjacent MTL regions (Fig. 5, ERC [blue], PRC [brown], and Fig. 5D, PHC [pink]).
Fig. 6. Behavioral Performance.
The task consisted of 6 blocks of encoding (learn) and retrieval (recall). Shown is the average percent correct across subjects (N=14) during blocks 1 thru 6 of the paired associates task.
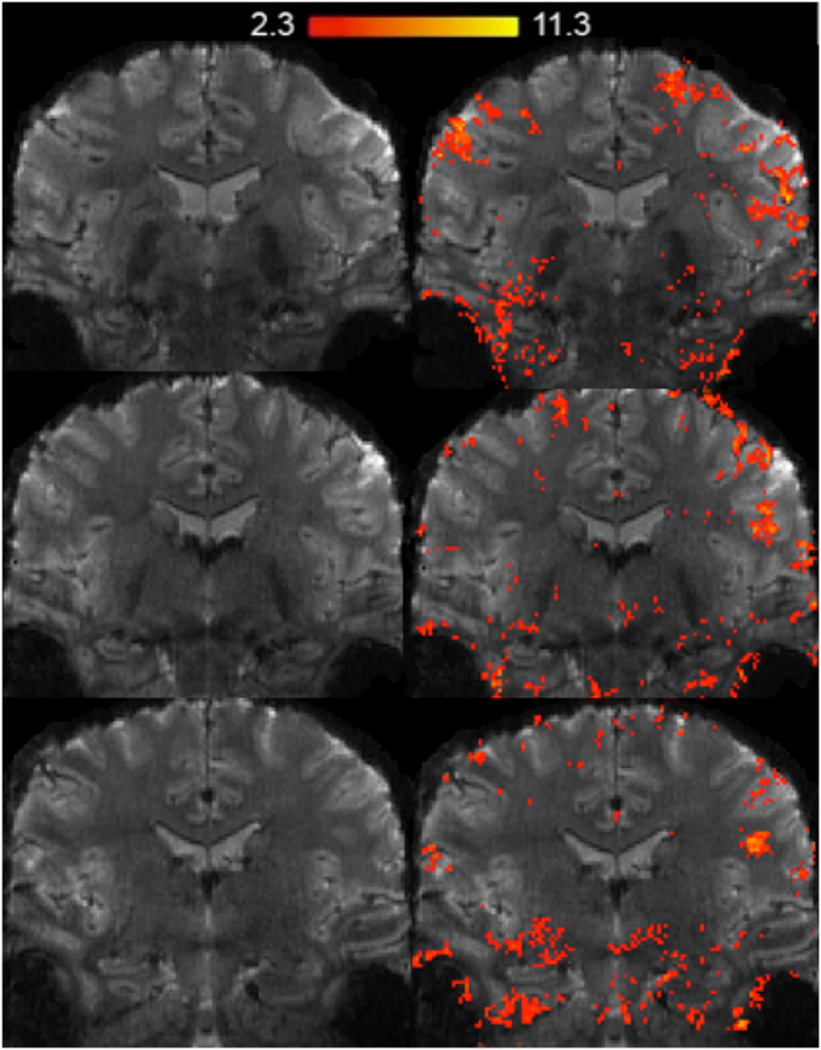
A voxel-wise analysis (Fig. 7) across subjects showed significant areas of activation within hippocampal subfields during learning versus recall of unrelated paired associates. Specifically, activation during learning was significantly higher compared to recall within the anterior hippocampal CA2 and CA3 subfields. In contrast, we found significant clusters of activation within posterior bilateral CA2 and subiculum during recall compared to learning. For an example subject’s significant BOLD activations (learning and recall compared to baseline) superimposed onto their fMRI image see figure 8.
Fig. 8. fMRI example data.
An example subject’s activation map superimposed on an oblique-coronal high-resolution fMRI image during the learning and recall of paired associates compared to baseline. Shown are clusters of significantly activated regions (statistical maps of significantly activated regions; z ≥ 2.4, p < 0.05 corrected).
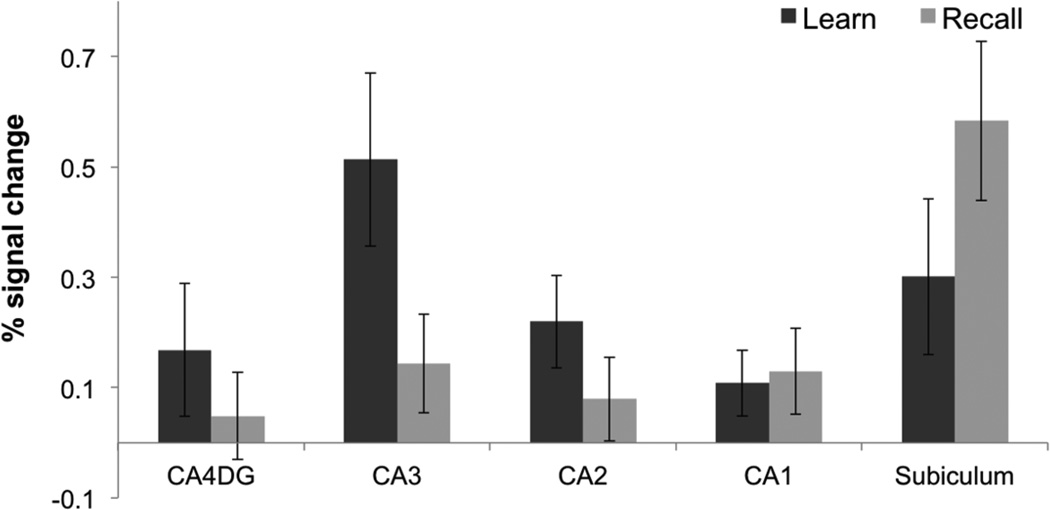
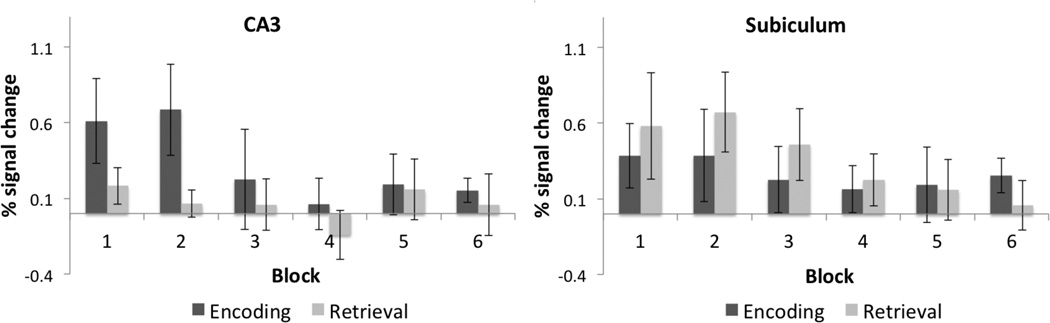
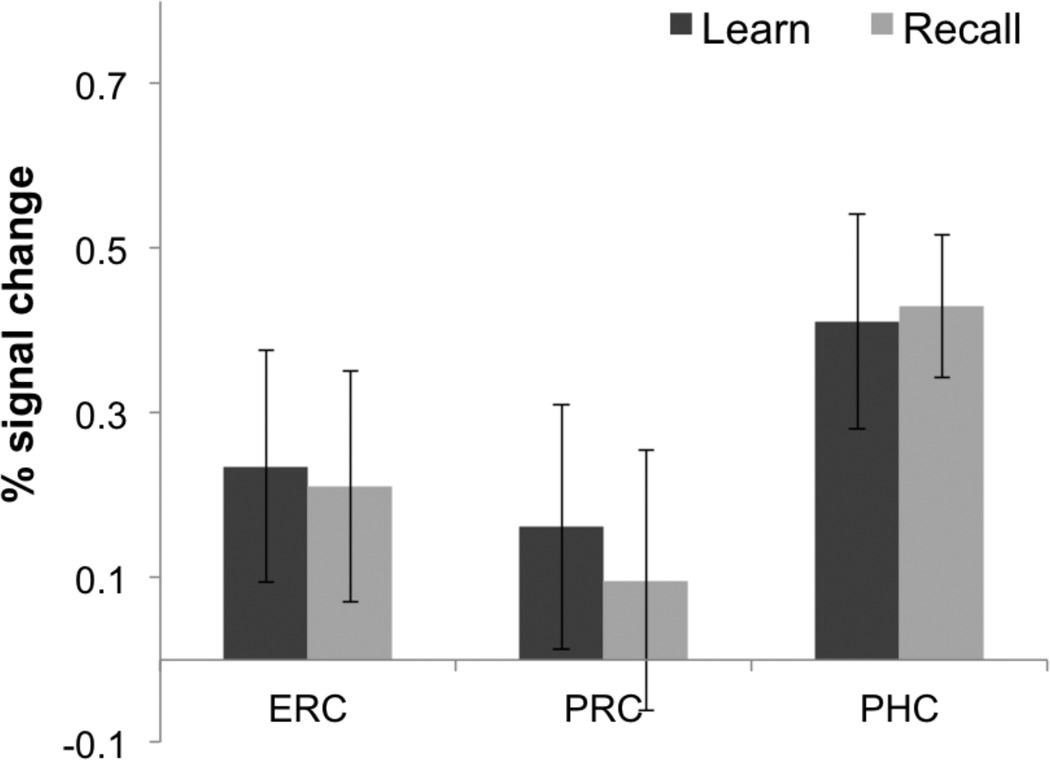
As a complementary analysis, we also created 3D ROIs and calculated percent signal change during learning and recall compared to baseline separately during blocks 1–3 where most learning occurred (Fig. 9). A repeated measures ANOVA demonstrated a significant effect of condition (Learn vs. Baseline, Recall vs. Baseline) on percent signal change (F(1,83) = 5.37, p < 0.05). Post-hoc comparisons showed a significant increase in BOLD activation during learning within the CA3 (Learn > Baseline, t(13) = 3.39; p < 0.05), CA2 (Learn > Baseline, t(13) = 2.72; p < 0.05), subiculum (Learn > Baseline, t(13) = 2.21; p < 0.05), and PHC (Learn > Baseline, t(13) = 3.27; p < 0.05). Furthermore, we found significantly higher activation during learning compared to recall in the hippocampal CA3 region (Learn > recall, t(13) = 2.3; p < 0.05) consistent with our voxel wise analysis (Fig. 7). Consistent with previous findings within the CA23DG region (Zeineh et al., 2003) encoding related activity within the CA3 decreased across learning blocks (F(1,83) = 2.32, p < 0.05; Fig. 10A). Our results also show a trend for decreasing subicular activation during learning (F(1,83) = 2.23, p = 0.053; Fig. 10B), which is consistent with prior findings (Preston et al., 2010; Zeineh et al., 2003). However, subicular BOLD activation in some of these studies was significantly greater during recall compared to learning (Zeineh et al., 2003; Suthana et al., 2009, 2011). In the current study, we found a significant increase in BOLD activation within the posterior subiculum during recall (Recall > Baseline, t(13) = 3.73; p < 0.05) and a trend for activation higher during recall compared to learning (Recall > Learn, t(13) = 1.26; p = 0.19) with a decrease across learning blocks (Fig. 10B). There was a significant increase in BOLD activation within the PHC (Fig. 11) during learning and recall separately compared to baseline (Learn > Baseline, t(13) = 2.55; p < 0.05; Recall > Baseline, t(13) = 3.41; p < 0.05). However, there were no significant differences between learning and recall in any other CA fields (DG, CA1, and CA2) or extra-hippocampal ROIs (ERC, PRC, and PHC; Fig. 11). A higher signal dropout associated with large susceptibility variations nearby extra-hippocampal ROIs, however, make it challenging to reliably detect BOLD activation changes within anterior fusiform and perirhinal cortices.
Fig. 9. Hippocampal Region of Interest Analysis.
Average percent (%) signal changes during learning and recall separately compared to baseline during learning blocks 1–3 in CA4 and dentate gyrus (CA4DG), CA3, CA2, CA1, and subiculum. Error bars correspond to the standard error across subjects.
Fig. 10. CA3 and Subiculum activity during each task block.
Average percent (%) signal changes during learning and recall separately compared to baseline in CA3 and subiculum for each of the six task blocks. Error bars correspond to the standard error across subjects.
Fig. 11. Extra-hippocampal Region of Interest Analysis.
Average percent (%) signal changes during learning and recall separately compared to baseline in entorhinal cortex (ERC), perirhinal cortex (PRC), and parahippocampal cortex (PHC). Error bars correspond to the standard error across subjects.
Discussion
We present data from high-resolution in vivo fMRI of human hippocampal subfields acquired at 7T. Utilizing a paired-associates task (Zeineh et al., 2003; Eldridge et al., 2005), we show that BOLD activation during learning reflects activity generated from the anterior CA3 and the anterior CA2 subfield, whereas BOLD activation during recall reflects activity from the posterior CA2 field, the CA1, and the posterior subiculum. Prior fMRI studies have consistently found activation in a combined CA23DG region during learning and in the subiculum during recall (Zeineh et al., 2003; Eldridge et al., 2005; Suthana et al., 2011; Viskontas et al., 2009, Carr et al., 2010b). However, extracting BOLD signal changes from the DG and the CA fields 2–3 separately was not possible in these past fMRI studies. More recently, Bonnici and colleagues (2012) combined MVPA and high-resolution fMRI to demonstrate differential patterns of activation in subjects discriminating between scenes under perceptual certainty vs. ambiguity in hippocampal CA3 and DG subregions. Specifically, pattern classifiers in all hippocampal subregions were similarly effective under perceptual certainty, but under perceptual ambiguity, classification was superior for CA3 and CA1 compared to other regions. Due to scanning resolution limits the authors were unable to distinguish CA3 and CA2. Because we collected functional data at 7T we were able to distinguish these regions and reveal a differential engagement of these regions in encoding and retrieval. Furthermore, we were able to demonstrate different patterns of activation for encoding and retrieval across the anterior-posterior axis of region CA2.
Our data demonstrate that high-resolution fMRI at 7T can be used to investigate theories of human hippocampal subfield functional established using computational or animal models. Rodent electrophysiology and modeling studies suggest that the CA3 subfield, given its recurrent collateral connectivity, is optimized to encode novel associative information coming in from the entorhinal cortex (Marr, 1971, Rolls & Kesner, 2006). Human fMRI studies have shown that the combined DG/CA2/CA3 region may be preferentially implicated in pattern separation, illustrating its substantial susceptibility to novel stimuli (Carr 2010a, Bakker et al., 2008, Azab et al., 2014; Stokes et al., 2014). In the current study, using an associative learning procedure, we found learning related activity within the CA3 area but not DG. These results are consistent with models suggesting that region CA3 is an autoassociative network that play a role in binding (Treves & Rolls, 1994; Marr, 1971). Our lack of learning-related activation in DG is possibly due to the sparse firing properties of the human DG subfield. It may also be that our associative learning task did not require pattern separation processes that may come into play in tasks in which highly similar items must be orthogonalized.
We found activation in different CA2 regions during learning and recall. The CA2 field of the hippocampus is a small zone connecting CA3 and CA1; the activity we measure in CA2 may reflect CA3 synaptic BOLD activity given that the hippocampal BOLD fMRI signal has been shown to more strongly correlate with synaptic activity rather than intrinsic firing rate changes per se (Ekstrom et al., 2010). Greater sensitivity to BOLD signal changes at 7T field strength when compared with 3T (Theysohn et al., 2013) may enhance the detection of brain activity within smaller hippocampal regions such as CA2. Interestingly, the association of anterior CA2 with learning and posterior CA2 with retrieval as seen in our data is consistent with the idea of an encoding/retrieval gradient along the longitudinal axis of the hippocampus that has been reported in associative memory (Prince et al., 2005). It may be that this gradient reflects differences in function of CA3 output in anterior and posterior hippocampus.
There are a growing number of studies investigating human hippocampal subregion functioning during learning and memory and there is some variability in BOLD signal findings across studies. This may reflect technological advances that allow the possibility of detecting neural activity in a small region’s functionally heterogeneous substructures. Differential hippocampal subregion activity also could be modulated by factors such as the motivational context (Wolosin et al., 2013), or goal states, e.g., whether the participants’ attention is specifically directed to item distinctiveness or similarity (Carr et al., 2013). Such top-down factors may have major influence on the engagement of circuits within the hippocampus.
Our findings are also consistent with prior studies with respect to subicular activation during recall of previously learned paired associates (Zeineh et al., 2003; Eldridge et al., 2005; Suthana et al., 2011). Although these studies found higher subiculum activation during recall compared to learning, they also showed some subicular activation during learning alone (Zeineh et al., 2003; Preston et al., 2010). Here, we also found increased activation within the subiculum during both learning and recall, with a trend leaning towards higher activation during recall, not inconsistent with prior findings. We also found that CA3, which showed learning-related changes did show decreases in learning-related activation across blocks, corresponding to less new learning occurring. We also found that retrieval-related activation in the posterior subiculum increased from learning to recall blocks, corresponding to increasing retrieval success across learning blocks. As the primary output of the hippocampus, the subiculum may show particularly high levels of activity after successful retrieval involving engagement of the trisynaptic hippocampal circuit.
In addition to high-resolution fMRI data collection, measuring BOLD activation in separate hippocampal subfields requires innovative data analyses techniques. We used two independent approaches; a cortical unfolding approach and an ROI approach both used in previous high-resolution hippocampal studies (Zeineh et al., 2003; Suthana et al., 2009). Cortical unfolding has been used on the hippocampus to computationally unfold the small and highly convoluted subfields into a flattened map and analyze them in 2D, both using voxel-wise and ROI comparisons. Flat maps visualize the entire hippocampus and adjacent MTL regions on one single image, allowing for the viewing of complex patterns of increasing and decreasing activation within and across slices that are otherwise not fully discernable or may be masked using an ROI approach. For example there are combinations of positive and negative activations during spatial navigation (Aguirre et al., 1996; Shipman and Astur, 2008; Suthana et al., 2009, 2011) and reproducible anterior/posterior distinctions along the MTL (Zeineh et al., 2003; Suthana et al., 2011) that are easily viewable on the flattened image. Flat maps also allow greater resolution through interpolation of the consistent long-axis structure of the hippocampus. Furthermore, unlike an ROI analysis, the unfolding technique is not necessarily dependent on subregional boundary placement since activity is visualized altogether with boundaries superimposed onto one map. Additionally, the cortical unfolding technique allows for specific measurements of cortical thickness in all subfields separately, which has been shown to be more sensitive than standard volumetry in detecting early changes associated with genetic risk for Alzheimer’s disease (Burggren et al., 2008; Donix et al., 2010). Flat maps have also been used to provide an overall picture of structural patterns of hippocampal electrode placement across patients with temporal-lobe epilepsy (Ekstrom et al., 2008).
One challenge with any transformation of data, such as from 3D to 2D, is that clusters that span the medial-lateral axis may be separated when flattened. Here, the statistical thresholding of individual beta-weight values generated for each voxel before mapping to 2D is performed after the unfolding procedure is complete. Thus clusters crossing subregional boundaries in the original 3D format will not be maintained. However, we also present the alternative and complementary 3D analysis of our results using a ROI-based approach, which extracts percent signal change at each voxel and then altogether averaged across an ROI. Our ROI-based results are consistent with our flat mapping voxel-wise results showing activation differences between learning and recall conditions within hippocampal subfields CA2, CA3 and subiculum. We do see clusters of significant activation within the CA1 and CA2 region during recall that is not reflected in our ROI analysis. However, it is possible that this activation is masked when averaging across the entire anatomical regions. Thus, because ROI analyses can themselves suffer from masking out subtle areas of significant activation due to the averaging across an entire anatomical ROI, future studies are likely to benefit from the use of both unfolding or voxel-wise and ROI-based methods.
Previously, at 3T, high-resolution fMRI scans provided resolution ranging from 1.5 – 1.6 mm (Zeineh et al., 2000; Kirwan et al., 2007; Ekstrom et al., 2009; Bonnici et al., 2012). At 7T, with its much higher image signal to noise ratio (SNR), we have improved our in-plane fMRI voxel size to 1.0 × 1.0 mm with a slice thickness of 2.0 mm. For many purposes and due to the highly convoluted structure of the human brain, isotropic voxels are beneficial for fMRI (e.g. Hyde et al., 2001) because this implies no directional bias. Although our final interpolated resolution is isotropic, our raw data are not, so some discussion is warranted. Since the SNR is proportional to the voxel volume, a high spatial resolution in general results in a low SNR. To increase the BOLD contrast to noise ratio, which depends on the SNR, we decided to maximize the in-plane resolution at the cost of acquiring non-isotropic voxels with thicker slices. To minimize partial volume effects, we placed the slices orthogonal to the hippocampus. However, like for previous studies, which acquire images perpendicular to the plane of the hippocampus, increased B0 inhomogeneities in the temporal lobes may cause image distortions in phase encoding direction (Ojemann et al., 1997; Olman et al., 2009). Enhanced signal displacement along the superior-inferior axis (phase-encoding direction in our fMRI experiments), however, was attenuated by a combination of zooming and GRAPPA resulting in high acceleration factors and, hence, short EPI readouts. This can be appreciated in Figure 8. However, future high-resolution studies at 7T using the current methodologies should additionally collect B0 maps to minimize distortion effects. We chose the oblique coronal acquisition direction on the grounds that the hippocampal structure is quite homogenous through plane, while variable in-plane. In order to accurately delineate boundaries between CA fields, in-plane resolution must be high enough for their anatomical boundaries to be clearly visualized. For example, the boundary between anterior CA1 and DG is better visible at higher in-plane resolutions. The choice of voxel dimensions will clearly depend on the objectives of the particular study.
The determination of boundaries between areas may be improved in the future, as it currently relies on visualization of MRI landmarks that reflect underlying cellular histology from postmortem studies. Current segmentation of MTL subfields is affected by variability across protocols. For example, the CA2/CA3 ROIs defined in our study may be larger than those in other protocols because white matter areas such as the fimbria are included. Nevertheless, this segmentation should not affect fMRI signals, which only reflect the gray matter areas of these subfields. In addition, a benefit to our computational unfolding approach is that it allows for visualization of the MTL regions altogether and does not suffer from segmentation biases. However, other approaches may also have advantages. Our previous studies have shown that if intra-rater reliability for boundary demarcation is kept high (Burggren et al., 2008), and methods are kept consistent across different studies, errors resulting from manual MRI segmentation biases can be minimized. Wisse and colleagues (2012) demonstrated that 7T MRI data allow manual segmentation of hippocampal CA fields with excellent inter-rater agreement, which highlights that high field strength, improves this challenging task. Integration of data from myelin-stained (Geyer et al., 2011) sections of postmortem hippocampus into probabilistic atlases may greatly improve the reliability of hippocampal subregional boundary determinations. Future studies may benefit from the combination of computational unfolding, ROI analysis, MVPA (Bonnici et al., 2012), histology guided segmentation approaches (Adler et al., 2014), cytoarchitectural postmortem atlases (Yushkevich et al., 2009), high-field structural imaging (Mueller et al., 2007; Zeineh et al., 2014), and automated segmentation procedures (Van Leemput et al., 2009, Yushkevich et al., 2010, 2014) in order to provide a complete, accurate, and fast analysis of structural and functional imaging studies of human hippocampal subfields during normal memory and disorders such as Alzheimer’s disease or temporal-lobe epilepsy. Our results summarize our recent improvements in high-resolution imaging and computational unfolding methods of the human hippocampus in vivo. With these novel advancements we are able to localize BOLD activation to separate hippocampal dentate and CA subfields. Our findings are consistent with animal and computational models implicating a crucial role for CA3 in learning. Overall, we believe that these techniques will improve investigations of hippocampal subregion characteristics associated with learning, memory and neurodegenerative processes affecting hippocampal circuitry.
ACKNOWLEDGEMENTS
This work was supported by the NIMH T90 431587-BH-29793, NSF GK-12 0742410. We thank Domenica Wilfling, Elizabeth Wladimirow, and Enrico Reimer for their help in subject recruitment, data management, and study coordination. Finally, we also wish to thank all of the subjects for their participation in this study.
Footnotes
FINANCIAL DISCLOSURE: Adam Bazih has significant equity and financial interest with K_space LLC, myfMRI, Inc., and Boombang, Inc.
References
- Adler DH, Pluta J, Kadivar S, Craige C, Gee JC, Avants BB, et al. Histology-derived volumetric annotation of the human hippocampal subfields in postmortem MRI. Neuroimage. 2014;84:505–523. doi: 10.1016/j.neuroimage.2013.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre GK, Detre JA, Alsop DC, D’Esposito M. The parahippocampus subserves topographical learning in man. Cereb Cortex. 1996;6:823–829. doi: 10.1093/cercor/6.6.823. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Insausti R. The human nervous system. San Diego: Academic Press; 2012. The hippocampal formation. [Google Scholar]
- Azab M, Stark SM, Stark CE. Contributions of human hippocampal subfields to spatial and temporal pattern separation. Hippocampus. 2014;24(3):293–302. doi: 10.1002/hipo.22223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker A, Kirwan CB, Miller M, Stark CE. Pattern separation in the human hippocampal CA3 and dentate gyrus. Science. 2008;319:1640–1642. doi: 10.1126/science.1152882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, Kwok EW, Vincent SL, Todtenkopf MS. A reduction of nonpyramidal cells in sector CA2 of schizophrenics and manic depressives. Biol Psychiatry. 1998;44:88–97. doi: 10.1016/s0006-3223(98)00138-3. [DOI] [PubMed] [Google Scholar]
- Boccara CN, Sargolini F, Thoresen VH, Solstad T, Witter MP, Moser EI, Moser MB. Grid cells in pre- and parasubiculum. Nat Neurosci. 2010;13:987–994. doi: 10.1038/nn.2602. [DOI] [PubMed] [Google Scholar]
- Bonnici HM, Chadwick MJ, Kumaran D, Hassabis D, Weiskopf N, Maguire EA. Multi-voxel pattern analysis in human hippocampal subfields. Frontiers Human Neurosci. 2012;6:290. doi: 10.3389/fnhum.2012.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggren AC, Zeineh MM, Ekstrom AD, Braskie MN, Thompson PM, Small GW, et al. Reduced cortical thickness in hippocampal subregions among cognitively normal apolipoprotein E e4 carriers. NeuroImage. 2008;41:1177–1183. doi: 10.1016/j.neuroimage.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr VA, Rissman J, Wagner AD. Imaging the human medial temporal lobe with high-resolution fMRI. Neuron. 2010a;65:298–308. doi: 10.1016/j.neuron.2009.12.022. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr VA, Viskontas IV, Engel SA, Knowlton BJ. Neural activity in the hippocampus and perirhinal cortex during encoding is associated with the durability of episodic memory. J. Cogn. Neurosci. 2010b;22:2652–2662. doi: 10.1162/jocn.2009.21381. [DOI] [PubMed] [Google Scholar]
- Carr VA, Engel SA, Knowlton BJ. Top-down modulation of hippocampal encoding activity as measured by high-resolution functional MRI. Neuropsychologia. 2013;51(10):1829–1837. doi: 10.1016/j.neuropsychologia.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevaleyre V, Siegelbaum SA. Strong CA2 pyramidal neuron synapses define a powerful disynaptic cortico-hippocampal loop. Neuron. 2010;66:560–572. doi: 10.1016/j.neuron.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donix M, Burggren AB, Suthana NA, Siddarth P, Ekstrom AD, Krupa A, Jones M, Rao A, Martin-Harris L, Ercoli LM, Miller KJ, Small GW, Bookheimer SY. Longitudinal Changes in Medial Temporal Cortical Thickness in Normal Subjects with the APOE-4 polymorphism. Neuroimage. 2010;53:37–43. doi: 10.1016/j.neuroimage.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvernoy HM. The human hippocampus: Functional Anatomy, Vascularization, and Serial Sections with MRI. Berlin: Springer; 2005. [Google Scholar]
- Ekstrom A, Suthana NA, Salamon N, Behnke E, Bookheimer SY, Fried I. High-Resolution Depth Electrode Localization and Imaging in Patients with Pharmacologically Intractable Epilepsy. Journal of Neurosurgery. 2008;108:812–815. doi: 10.3171/JNS/2008/108/4/0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrom AD, Bazih AJ, Suthana NA, Al-Hakim R, Ogura K, Zeineh M, Burggren AC, Bookheimer SY. Advances in high-resolution imaging and computational unfolding of the human hippocampus. Neuroimage. 2009;47:42–49. doi: 10.1016/j.neuroimage.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrom A. How and when the fMRI BOLD signal relates to underlying neural activity: the danger in dissociation. Brain Res Rev. 2010;62:233–244. doi: 10.1016/j.brainresrev.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge LL, Engel SA, Zeineh MM, Bookheimer SY, Knowlton BJ. A dissociation of encoding and retrieval processes in the human hippocampus. J Neurosci. 2005;25:3280–3286. doi: 10.1523/JNEUROSCI.3420-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel SA, Glover GH, Wandell BA. Retinotopic organization in human visual cortex and the spatial precision of functional MRI. Cerebral Cortex. 1997;7:181–192. doi: 10.1093/cercor/7.2.181. [DOI] [PubMed] [Google Scholar]
- Gedamu EL, Collins DL, Arnold DL. Automated quality control of brain MR images. J Magn Reson Imaging. 2008;28:308–319. doi: 10.1002/jmri.21434. [DOI] [PubMed] [Google Scholar]
- Geyer S, Weiss M, Reimann K, Lohmann G, Turner R. Microstructural parcellation of the human cerebral cortex - from Brodmann's post-mortem map to in vivo mapping with high-field magnetic resonance imaging. Front Hum Neurosci. 2011;5:19. doi: 10.3389/fnhum.2011.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann RM, Ivanov D, Trampel R, Fasano F, Meyer H, Pfeuffer J, Turner R. Isotropic Sub-Millimeter fMRI in the Human Brain at 7 Tesla: Combining Reduced Field-of-View Imaging and Partially Parallel Acquisitions. Magn Reson Med. 2012;68:1506–1516. doi: 10.1002/mrm.24156. [DOI] [PubMed] [Google Scholar]
- Hyde JS, Biswal BB, Jesmanowicz A. High-resolution fMRI using multislice partial k-space GR-EPI with cubic voxels. Magn Reson Med. 2001;46:114–125. doi: 10.1002/mrm.1166. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Kirwan CB, Jones CK, Miller MI, Stark CE. High-resolution fMRI investigation of the medial temporal lobe. Hum Brain Mapp. 2007;28:959–966. doi: 10.1002/hbm.20331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy JW, Yassa MA, Stark SM, Muftuler LT, Stark CE. Distinct pattern separation related transfer functions in human CA3/dentate and CA1 revealed using high-resolution fMRI and variable mnemonic similarity. Learning and Memory. 2011;18(1):15–18. doi: 10.1101/lm.1971111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Yoganarasimha D, Rao G, Knierim JJ. Comparison of population coherence of place cells in hippocampal subfields CA1 and CA3. Nature. 2004;430:456–459. doi: 10.1038/nature02739. [DOI] [PubMed] [Google Scholar]
- Leutgeb JK, Leutgeb S, Moser MB, Moser EI. Pattern separation in the dentate gyrus and CA3 of the hippocampus. Science. 2007;315:961–966. doi: 10.1126/science.1135801. [DOI] [PubMed] [Google Scholar]
- Marr D. Simple memory: a theory for archicortex. Phil Trans. R. Soc Lond. B. 1971;262:23–81. doi: 10.1098/rstb.1971.0078. [DOI] [PubMed] [Google Scholar]
- Mercer A, Botcher NA, Eastlake K, Thomson AM. SP-SR interneurones: a novel class of neurones of the CA2 region of the hippocampus. Hippocampus. 2012;22(8):1758–1769. doi: 10.1002/hipo.22010. [DOI] [PubMed] [Google Scholar]
- Mueller SG, Stables L, Du AT, Schuff N, Truran D, Cashdollar N, Weiner MW. Measurement of hippocampal subfields and age-related changes with high resolution MRI at 4T. Neurobiol Aging. 2007;28:719–726. doi: 10.1016/j.neurobiolaging.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neunuebel JP, Knierim JJ. CA3 retrieves coherent representations from degraded input: direct evidence for CA3 pattern completion and dentate gyrus pattern separation. Neuron. 2014;81(2):416–427. doi: 10.1016/j.neuron.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojemann JG, Akbudak E, Snyder AZ, McKinstry RC, Raichle ME, Conturo TE. Anatomic localization and quantitative analysis of gradient refocused echo-planar fMRI susceptibility artifacts. Neuroimage. 1997;6:156–167. doi: 10.1006/nimg.1997.0289. [DOI] [PubMed] [Google Scholar]
- O'Keefe J. Do hippocampal pyramidal cells signal non-spatial as well as spatial information? Hippocampus. 1999;9(4):352–364. doi: 10.1002/(SICI)1098-1063(1999)9:4<352::AID-HIPO3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Olman CA, Davachi L, Inati S. Distortion and signal loss in medial temporal lobe. PLoS One. 2009;4:e8160. doi: 10.1371/journal.pone.0008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly RC, McClelland JL. Hippocampal conjunctive encoding, storage, and recall: avoiding a trade-off. Hippocampus. 1994;4(6):661–682. doi: 10.1002/hipo.450040605. [DOI] [PubMed] [Google Scholar]
- Preston AR, Bornstein AM, Hutchinson JB, Gaare ME, Glover GH, Wagner AD. High-resolution fMRI of content-sensitive subsequent memory responses in human medial temporal lobe. J Cogn Neurosci. 2010;22:156–173. doi: 10.1162/jocn.2009.21195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince SE, Daselaar SM, Cabeza R. Neural correlates of relational memory: successful encoding and retrieval of semantic and perceptual associations. J Neurosci. 2005;25(5):1203–1210. doi: 10.1523/JNEUROSCI.2540-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls ET, Kesner RP. A computational theory of hippocampal function, and empirical tests of the theory. Progress in Neurobiology. 2006;79(1):1–48. doi: 10.1016/j.pneurobio.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Schacter DL, Wagner AD. Medial temporal lobe activations in fMRI and PET studies of episodic encoding and retrieval. Hippocampus. 1999;9(1):7–24. doi: 10.1002/(SICI)1098-1063(1999)9:1<7::AID-HIPO2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Sharp PE. Subicular place cells generate the same "map" for different environments: comparison with hippocampal cells. Behav Brain Res. 2006;174:206–214. doi: 10.1016/j.bbr.2006.05.034. Review. [DOI] [PubMed] [Google Scholar]
- Shipman SL, Astur RS. Factors affecting the hippocampal BOLD response during spatial memory. Behav Brain Res. 2008;187:433–441. doi: 10.1016/j.bbr.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Sloviter RS. “Epileptic” brain damage in rats induced by sustained electrical stimulation of the perforant path. I. Acute electrophysiological and light microscopic studies. Brain Res Bull. 1983;10:675–697. doi: 10.1016/0361-9230(83)90037-0. [DOI] [PubMed] [Google Scholar]
- Smith CD, Andersen AH, Kryscio RJ, Schmitt FA, Kindy MS, Blonder LX, et al. Women at risk for AD show increased parietal activation during a fluency task. Neurology. 2002;58:1197–1202. doi: 10.1212/wnl.58.8.1197. [DOI] [PubMed] [Google Scholar]
- Squire LR. Memory and the hippocampus: a synthesis from findings with rats, monkeys, and humans. Psychol. Rev. 1992;99:195–231. doi: 10.1037/0033-295x.99.2.195. [DOI] [PubMed] [Google Scholar]
- Squire LR, Stark CE, Clark RE. The medial temporal lobe. Annu. Rev. Neurosci. 2004;27:279–306. doi: 10.1146/annurev.neuro.27.070203.144130. [DOI] [PubMed] [Google Scholar]
- Stark CE, Squire LR. When zero is not zero: the problem of ambiguous baseline conditions in fMRI. Proceedings of the National Academy of Sciences. 2001;98:12760–12766. doi: 10.1073/pnas.221462998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes J, Kyle C, Ekstrom AD. Complementary Roles of Human Hippocampal Subfields in Differentiation and Integration of Spatial Context. J Cogn Neurosci. 2014;30:1–14. doi: 10.1162/jocn_a_00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthana NA, Ekstrom AD, Moshirvaziri S, Knowlton B, Bookheimer SY. Human hippocampal CA1 involvement during allocentric encoding of spatial information. Journal of Neuroscience. 2009;29:10512–10519. doi: 10.1523/JNEUROSCI.0621-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthana NA, Ekstrom A, Moshirvaziri S, Knowlton B, Bookheimer SY. Dissociations within Human Hippocampal Subregions during Encoding and Retrieval of Spatial Information. Hippocampus. 2011;21:694–701. doi: 10.1002/hipo.20833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthana N, Krupa A, Donix M, Burggren AB, Ekstrom AD, Jones, et al. Reduced hippocampal CA2, CA3, and dentate gyrus activity in asymptomatic people at genetic risk for Alzheimer's disease. Neuroimage. 2010;53:1077–1084. doi: 10.1016/j.neuroimage.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube JS, Muller RU, Ranck JB., Jr Head-direction cells recorded from the postsubiculum in freely moving rats. I. Description and quantitative analysis. J. Neurosci. 1990;10:420–435. doi: 10.1523/JNEUROSCI.10-02-00420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo PC, Sapiro G, Wandell BA. Creating connected representations of cortical gray matter for functional MRI visualization. IEEE Transactions on Medical Imaging. 1997;16:852–863. doi: 10.1109/42.650881. [DOI] [PubMed] [Google Scholar]
- Theysohn N, Qin S, Maderwald S, Poser BA, Theysohn JM, Ladd ME, et al. Memory-related hippocampal activity can be measured robustly using FMRI at 7 tesla. Journal of Neuroimaging. 2013;23(4):445–451. doi: 10.1111/jon.12036. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Woods RP, Mega MS, Toga AW. Mathematical/computational challenges in creating deformable and probabilistic atlases of the human brain. Human Brain Mapping. 2000;9:81–92. doi: 10.1002/(SICI)1097-0193(200002)9:2<81::AID-HBM3>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treves A, Rolls ET. Computational analysis of the role of the hippocampus in memory. Hippocampus. 1994;4(3):374–391. doi: 10.1002/hipo.450040319. Review. [DOI] [PubMed] [Google Scholar]
- Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinack J, Dickerson BC, Golland P, Fischl B. Automated segmentation of hippocampal subfields from ultra-high resolution in vivo MRI. Hippocampus. 2009;19:549–557. doi: 10.1002/hipo.20615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viskontas IV, Carr VA, Engel SA, Knowlton BJ. The neural correlates of recollection: hippocampal activation declines as episodic memory fades. Hippocampus. 2009;19:265–272. doi: 10.1002/hipo.20503. [DOI] [PubMed] [Google Scholar]
- Wisse LE, Gerritsen L, Zwanenburg JJ, Kuijf HJ, Luijten PR, Biessels GJ, et al. Subfields of the hippocampal formation at 7 T MRI: in vivo volumetric assessment. Neuroimage. 2012;61(4):1043–1049. doi: 10.1016/j.neuroimage.2012.03.023. [DOI] [PubMed] [Google Scholar]
- Wolosin SM, Zeithamova D, Preston AR. Distributed hippocampal patterns that discriminate reward context are associated with enhanced associative binding. Journal of Experimental Psychology: General. 2013;142(4):1264–1276. doi: 10.1037/a0033609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolrich MW, Ripley BD, Brady M, Smith SM. Temporal autocorrelation in univariate linear modeling of FMRI data. NeuroImage. 2001;14:1370–1386. doi: 10.1006/nimg.2001.0931. [DOI] [PubMed] [Google Scholar]
- Yassa MA, Stark CE. Pattern separation in the hippocampus. Trends in Neurosciences. 2011;34:515–525. doi: 10.1016/j.tins.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Avants BB, Pluta J, Das S, Minkoff D, Mechanic-Hamilton D, Glynn S, Pickup S, Liu W, Gee JC, Grossman M, Detre JA. A high-resolution computational atlas of the human hippocampus from postmortem magnetic resonance imaging at 9.4 T. Neuroimage. 2009;44:385–398. doi: 10.1016/j.neuroimage.2008.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Wang H, Pluta J, Das SR, Craige C, Avants BB, Weiner MW, Mueller S. Nearly automatic segmentation of hippocampal subfields in in vivo focal T2-weighted MRI. Neuroimage. 2010;53:1208–1224. doi: 10.1016/j.neuroimage.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Pluta JB, Wang H, Xie L, Ding SL, Gertje EC, Mancuso L, Kliot D, Das SR, Wolk DA. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum Brain Mapp. 2015;36:258–287. doi: 10.1002/hbm.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeineh MM, Engel SA, Thompson PM, Bookheimer SY. Dynamics of the hippocampus during encoding and retrieval of face-name pairs. Science. 2003;299:577–580. doi: 10.1126/science.1077775. [DOI] [PubMed] [Google Scholar]
- Zeineh MM, Engel SE, Bookheimer SY. Application of cortical unfolding techniques to functional MRI of the human hippocampal region. Neuroimage. 2000;11:668–668. doi: 10.1006/nimg.2000.0561. [DOI] [PubMed] [Google Scholar]
- Zeineh MM, Enge SA, Thompson PM, Bookheimer SY. Unfolding the human hippocampus with high resolution structural and functional MRI. The Anatomical Record. 2001;265:111–120. doi: 10.1002/ar.1061. [DOI] [PubMed] [Google Scholar]
- Zeineh MM, Parekh MB, Zaharchuk G, Su JH, Rosenberg J, Fischbein NJ, et al. Ultrahigh-resolution imaging of the human brain with phase-cycled balanced steady-state free precession at 7 T. Investigative Radiology. 2014;49(5):278–289. doi: 10.1097/RLI.0000000000000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Choi YS, Obrietan K, Dudek SM. Synaptic plasticity (and the lack thereof) in hippocampal CA2 neurons. J Neurosci. 2007;27:12025–12032. doi: 10.1523/JNEUROSCI.4094-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]