Abstract
The apicomplexan family of pathogens, which includes Plasmodium spp. and Toxoplasma gondii, are primarily obligate intracellular parasites and invade multiple cell types. These parasites express extracellular membrane protein receptors, adhesins, to form specific pathogen-host cell interaction complexes. Various adhesins are used to invade a variety of cell types. The receptors are linked to an actomyosin motor, which is part of a complex comprised of many proteins known as the invasion machinery or glideosome. To date, reviews on invasion have focused primarily on the molecular pathways and signals of invasion, with little or no structural information presented. Over 75 structures of parasite receptors and glideosome proteins have been deposited with the Protein Data Bank. These structures include adhesins, motor proteins, bridging proteins, inner membrane complex and cytoskeletal proteins, as well as co-crystal structures with peptides and antibodies. These structures provide information regarding key interactions necessary for target receptor engagement, machinery complex formation, how force is transmitted, and the basis of inhibitory antibodies. Additionally, these structures can provide starting points for the development of antibodies and inhibitory molecules targeting protein-protein interactions, with the aim to inhibit invasion. This review provides an overview of the parasite adhesin protein families, the glideosome components, glideosome architecture, and discuss recent work regarding alternative models.
Keywords: apicomplexa, invasion machinery, glideosome, adhesins, Toxoplasma, Plasmodium, Cryptosporidium, malaria
1. Introduction
Apicomplexans are primarily obligate, intracellular parasites that can cause devastating human diseases such as malaria (Plasmodium spp.), toxoplasmosis (Toxoplasma gondii), and gastrointestinal illness (Cryptosporidium spp.). These parasites must invade host cells to continue their lifecycle. For example, in Plasmodium spp., the parasite must invade the human host’s liver and red blood cells, as well as the mosquito vector’s midgut. To accomplish this task, the parasite employs an actomyosin motor at the core of a larger complex known as the invasion machinery, or glideosome. The mechanical force generated by this motor is used to invade a host cell, to which parasites attach via a variety of adhesive proteins. In this review, a structural survey of proteins involved in this critical invasion process, focusing on Toxoplasma gondii and Plasmodium spp., where the most research has been done, is presented. If the readers are interested in the cellular and molecular mechanisms of attachment and invasion, they are directed to a review of Plasmodium spp. merozoite invasion, by Cowman et al. (2012). The first part of this structural review focuses on the extracellular interaction of the parasite with the host cell, covering crystal structures that identify mechanisms of binding. The second section covers the intracellular proteins that compose the core invasion machinery and discusses the key protein-protein interactions stabilizing the complex. The third section highlights new findings that call into question the essentiality of machinery components, and discusses compensatory glideosomes, alternative models, and questions that arise from this data.
2. Parasite interaction with the host cell
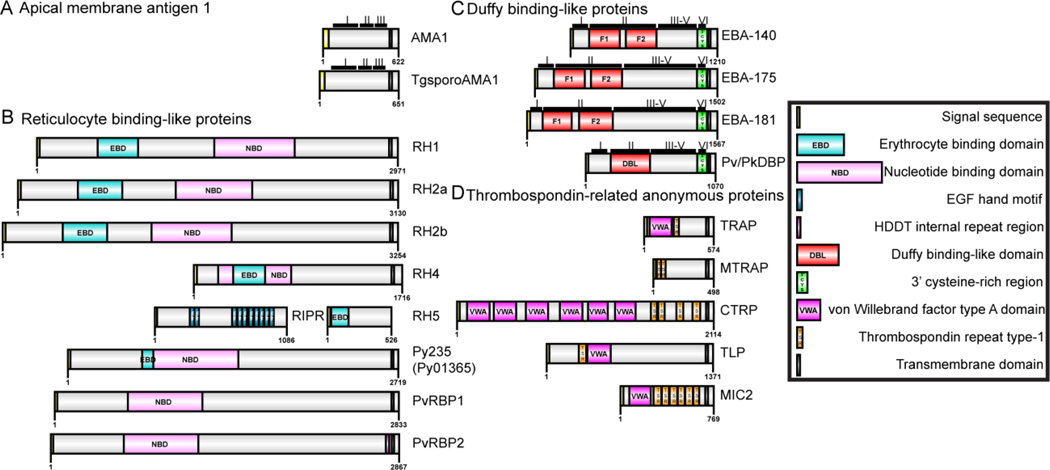
Apicomplexans employ extracellular transmembrane protein receptors to interact with receptors present on the host cell surface to attach and reorient, form a tight junction, and invade successfully. There are four primary families of adhesin proteins into which receptors from Plasmodium spp. and T. gondii can be grouped based on conserved protein domains or localization: the apical membrane antigen (AMA) family, the Duffy binding-like (DBL) family, the reticulocyte binding-like (RBL) family, and the thrombospondin related anonymous protein (TRAP) family. T. gondii and Plasmodium spp. both express proteins in the AMA and TRAP families, while the DBL and RBL proteins are specific to Plasmodium spp. A list of the receptors and their EuPathDB codes (Aurrecoechea et al., 2007), their known interaction partners with references, and any related structures that have been described and deposited in the PDB, are listed in Table 1. Additionally, Figure 1A depicts the domain organization and family grouping of the different receptor proteins. A recently published review by Malpede et al. (2013), following the publication of their structure of the P. vivax Duffy Binding Protein in complex with the Duffy Antigen/Receptor for Chemokines (DARC), provides an introduction to Plasmodium spp. adhesins, including some that are not covered in this review.
Table 1.
Parasite and host cell receptors of invasion.
| Parasite Receptor |
EuPathDB | Host Cell Receptor | Structure |
|---|---|---|---|
| Duffy binding-like (DBL) family and erythrocyte binding-like (EBL) proteins | |||
| PfEBA-140 | PF3D7_1301600 | Glycophorin C | 4GF2 (Lin et al., 2012) (Region II) |
| 4JNO (Malpede et al., 2013) (Region II + sialyllactose) | |||
| PfEBA-175 (BAEBL) | PF3D7_0731500 | Glycophorin A | 1ZRL (Tolia et al., 2005) (Region II) |
| 1ZRO (Tolia et al., 2005) (Region II in presence of sialyllactose) | |||
| 2RJI (Withers-Martinez et al., 2008) (Region VI) | |||
| 4QEX, 4K4M, 4K2U (Chen et al., 2013) (Region II + Fab) | |||
| PfEBA-181 (JESEBL) | PF3D7_0102500 | unknown | -- |
| PvDBP | PVX_110810 | DARC | 3RRC (Batchelor et al., 2011) (Region II) |
| 4NUU (Batchelor et al., 2014) (Region II + DARC dimer) | |||
| 4NUV (Batchelor et al., 2014) (Region II dimer + DARC dimer) | |||
| PkαDBP | PKH_062300 | DARC | 2C6J (Singh et al., 2006) (Region II) |
| Reticulocyte (homolog) binding-like (RBL/RH) family | |||
| PfRH1 | PF3D7_0402300 | sialic-acid containing putative erythrocyte receptor Y | -- |
| PfRH2a | PF3D7_1335400 | chymotrypsin sensitive erythrocyte receptor Z | -- |
| PfRH2b | PF3D7_1335300 | chymotrypsin sensitive erythrocyte receptor Z | -- |
| PfRH4 | PF3D7_0424200 | Complement Receptor 1 (CD35) | 2Q7Z (Furtado et al., 2008) (30 SCR domains of HsCR1) |
| 2MCZ (Park et al., 2014) (SCR domains 1, 2 of HsCR1) | |||
| 2MCY (Park et al., 2014) (SCR domains 2, 3 of HsCR1) | |||
| PfRH5/PfRIPR | PF3D7_0424100 | Basigin (CD147/EMMPRIN) | 4U0Q (Wright et al., 2014a) (Region II + Basigin Domain Ig1&Ig2) |
| 4U0R, 4U1G (Wright et al., 2014a) (Region II + Fab) | |||
| 3QR2 (Redzic et al., 2011) (Basigin Ig0) | |||
| PvRBP1/2 | PVX_098585 | unknown | -- |
| Py235 | PY17X_1468100 | unknown | 3HGF (Gruber et al., 2010) (NBD94) |
| Thrombospondin related anonymous protein (TRAP) family | |||
| PfTRAP | PF3D7_1335900 | unknown | 4HQF, 4HQK (Song et al., 2012) (Closed form VWA + TSR domain) |
| 2BBX (Tossavainen et al., 2006) (TSR domain) | |||
| PvTRAP | PVX_082735 | unknown | 4HQL, 4HQN (Song et al., 2012) (Open form w/ Mg2+/Mn2+) |
| 4HQO (Song et al., 2012) (VWA + TSR) | |||
| 4F1K (Pihlajamaa et al., 2013) (VWA domain) | |||
| 4F1J (Pihlajamaa et al., 2013) (VWA domain+ Mg2+) | |||
| TgMIC2 | TGME49_201780 | unknown | 4OKR, 4OKU (Song and Springer, 2014) |
| 2XGG (Tonkin et al., 2010) (VWA domain) | |||
| PfMTRAP | PF3D7_1028700 | Semaphorin-7a (CD108) | 3NVQ (Liu et al., 2010) (Semaphorin-7a in complex with Plexin-C1) |
| PfTLP | PF3D7_0616500 | unknown | -- |
| PfCTRP | PF3D7_0315200 | unknown | -- |
| Apical membrane antigen (AMA) | |||
| PfAMA1 | PF3D7_1133400 | PfRON2* | 1Z40 (Bai et al., 2005) (Domains I&II) |
| 1YXE (Feng et al., 2005) (Domain II) | |||
| 1HN6 (Nair et al., 2002) (Domain III) | |||
| 3ZWZ (Vulliez-Le Normand et al., 2012) (Domains I&II + RON2 39 aa peptide) | |||
| 3SRI (Vulliez-Le Normand et al., 2012) (Domains I&II + RON2 29 aa peptide) | |||
| 3SRJ (Vulliez-Le Normand et al., 2012) (Domains I&II + inhibitory peptide) | |||
| 2Z8W, 2Z8V (Henderson et al., 2007) (Domains I&II + IgNAR) | |||
| 2Q8B, 2Q8A (Coley et al., 2007) (Domains I&II + Fab) | |||
| 2J5L (Igonet et al., 2007) (Fab + AMA1 region III peptide) | |||
| PvAMA1 | PVX_092275 | PvRON2* | 1W81, 1W8K (Pizarro et al., 2005) (Domains I,II,III) |
| 2J4W (Igonet et al., 2007) (Fab + AMA1 region III peptide) | |||
| 4UXKTBP(+RON2 29 aa peptide) | |||
| TgAMA1 | TGME49_255260 | TgRON2* | 2Y8R (Tonkin et al., 2011) (Y230A Domains I,II,III) |
| 2X2Z (Crawford et al., 2010) (Domains I,II,III) | |||
| 2Y8T (Tonkin et al., 2011) (Domains I,II,III + RON2 37 aa peptide) | |||
| 2Y8S (Tonkin et al., 2011) (Y230A Domains I,II,III + RON2 37 aa peptide) | |||
| TgsporoAMA1 | TGME49_115730 | TgsporoRON2* | 3ZLE (Poukchanski et al., 2013) (Domains I,II,III) |
| 3ZLD (Poukchanski et al., 2013) (Domains I,II,III + RON2 36 aa peptide) | |||
| BdAMA1 | BBOV_IV011230 | BdRON2 | 4APM (Tonkin et al., 2013) (Domains I,II,III) |
| NcAMA1 | NCLIV_028680 | NcRON2 | 4APL (Tonkin et al., 2013) (Domains I,II,III) |
RON2 is inserted into host cell membrane along with RON4, RON5, and RON8 (RON8 is not needed for Plasmodium spp.) forming a receptor for the parasite. RON2 is the only extracellular exposed protein.
TBPDeposited, Hold for Publication
Figure 1. Domain organization of parasite receptors.
(A) Apical membrane antigen family of receptors including AMA1 from P. falciparum and the sporozoite specific AMA1 from T. gondii. (B) Reticulocyte binding-like proteins including the RH proteins from P. falciparum and adapter proteins, RBPs from P. vivax, and Py235 from P. yoelii. (C) Duffy binding-like proteins including the EBAs from P. falciparum and DBPs from P. vivax and P. knowlesi. (D) Thrombospondin-related anonymous protein family including the TRAPs from P. falciparum and MIC2 from T. gondii. A key is provided for the various key features determined from sequence homology or from biochemical and structural information, with region and domain boundaries are marked according to literature reports.
2.1 Apical membrane antigen (AMA) and the rhoptry neck (RON) proteins
Unlike other parasite receptor families, which are shared only amongst the species of a particular genera of apicomplexans, the apical membrane antigen 1 (AMA1) protein is shared by both T. gondii and Plasmodium spp., as well as Babesia, Neospora and Theileria (Peterson et al., 1989; Taylor et al., 1990); Cryptosporidium does not have an AMA1 ortholog. AMA1 is involved in the tight-junction formation that occurs after reorientation of the parasite (Mitchell et al., 2004); however, there have been conflicting reports regarding the exact role of AMA1 and its necessity for invasion (Bargieri et al., 2013; Mital et al., 2005; Richard et al., 2010; Yap et al., 2014).
2.1.1 Apical membrane antigen 1 (AMA1)
The domain organization of AMA1 includes a signal sequence, a microneme targeting sequence, an ectodomain divided into three structural domains as defined by a pattern of eight disulfide bonds (Hodder et al., 1996), and a transmembrane helix followed by a cytoplasmic C-terminal tail (Figure 1A). Domains I and II are similar and belong to the PAN (plasminogen, apple, nematode) superfamily, a protein fold noted to be involved in receptor binding (Tordai et al., 1999).
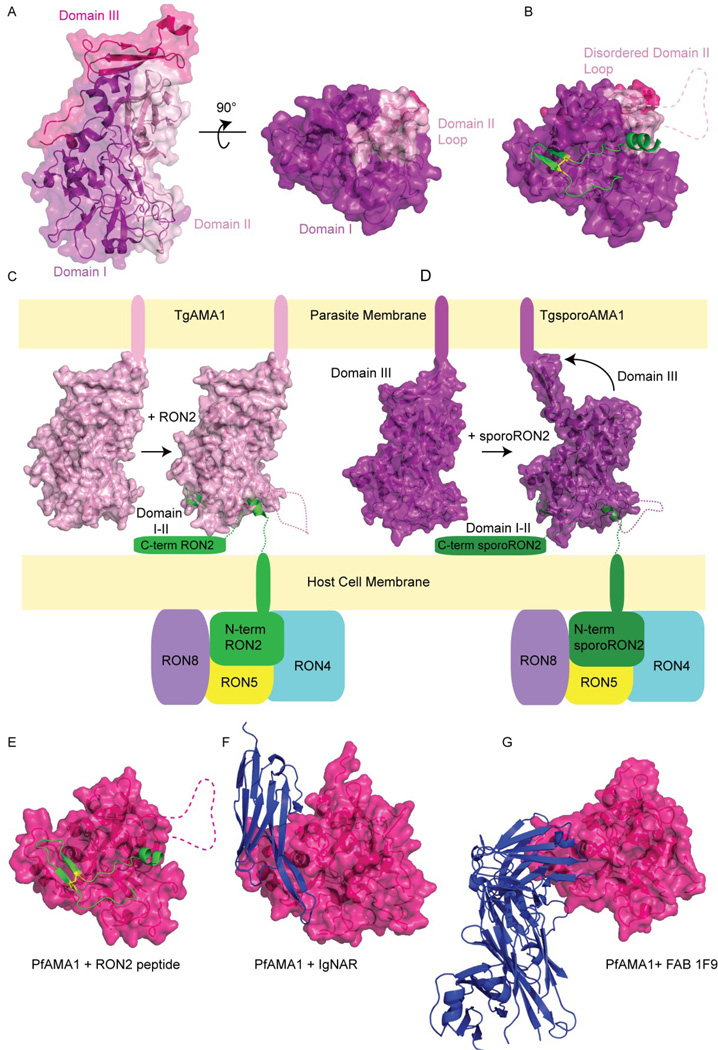
The structure of TgAMA1 is the most complete of the AMA1 structures, with ~98% of the ectodomain modeled (Crawford et al., 2010) and the domain packing is shown in Figure 2A. Domain I is at the distal end of the protein with respect to the assumed orientation on the parasite membrane. Domain I packs alongside Domain II, which contains a loop that extends down to the distal end of the protein and occludes a portion of the receptor binding pocket. Domain III is membrane proximal and contains a stable cysteine knot comprised of 3 disulfide bonds.
Figure 2. Apical membrane antigen 1 structure.
(A) TgAMA1 (surface and cartoon) depicted in two orientations rotated 90° from the other about the x-axis. The three domains of TgAMA1 are highlighted in purple, light pink, and magenta to distinguish Domains I, II and III, respectively. (B) TgAMA1 (surface) bound to the TgRON2 peptide (green, cartoon). The disulfide bond that stabilizes the cysteine loop is shown as yellow sticks. The dotted pink line represents the disordered loop from Domain II that is displaced from its contact with Domain I upon RON2 binding. Depiction of the orientation of (C) TgAMA1 (light pink, surface) and (D) TgsporoAMA1 (purple, surface) in the unbound and RON2 peptide-bound (green, cartoon) state on the parasite membrane surface. The parasite derived RON2/4/5/8 complex is shown inserted into the host cell membrane. An arrow indicates the Domain III movement in sporoAMA1 seen in the crystal structure when it is unpacked from Domains I and II upon binding of sporoRON2. (E) PfAMA1 (magenta, surface) bound to RON2 peptide with disulfide bond (yellow, sticks), and disordered loop (dashed). Structure of PfAMA1 in complex with inhibitory (F) IgNAR and (G) Fab 1F9, shown binding to Domain I where the RON2 cysteine loop binds.
2.1.2 Unique parasite derived, host cell receptor, rhoptry neck proteins (RONs)
AMA1 is a unique receptor because it does not bind to a host cell derived membrane receptor. Proteins secreted from the rhoptries, known as the rhoptry neck proteins (RONs) (Bradley et al., 2005), localize to the moving junction, and form a complex of RON2/4/5 that is inserted into the host cell, to which AMA1 then binds (Alexander et al., 2005; Lebrun et al., 2005). In T. gondii, RON8 is a key component of the complex (Besteiro et al., 2009), but is not required in Plasmodium spp. invasion. It was further shown that AMA1 binds only to RON2 directly (Cao et al., 2009). The RON2/4/5/8 complex is targeted to the host cell membrane, with RON4/5/8 exposed to the host cell cytoplasm, and RON2, predicted to contain at least one transmembrane helix and a large ectodomain, providing a receptor for AMA1 to bind to on the extracellular side (Besteiro et al., 2009) (Figure 2C).
To further investigate the RON2-AMA1 interaction and RON2 topology in the membrane, specific antisera were developed against different regions of RON2 (Lamarque et al., 2011). Experiments using the antisera to detect regions of RON2 exposed to either the host cell cytoplasm or extracellular environment, showed that RON2 is inserted in the host cell membrane as an integral membrane protein, with a cytoplasmic N-terminal domain followed by a transmembrane domain and ectodomain. It remains unclear whether the well-conserved C-terminal domain is exposed to the extracellular space or is located in the cytoplasm, based on these studies. Peptides corresponding to the predicted RON2 ectodomain were generated and co-crystallized with AMA1 (Tonkin et al., 2011; Vulliez-Le Normand et al., 2012). Both the P. falciparum and T. gondii AMA1 structures in complex with their respective RON2 peptides show that the peptide adopts a U-conformation, with an N-terminal helix and a cysteine loop, binding to a hydrophobic groove after displacement of the Domain II loop (Figures 2B, 2E).
2.1.3 sporoAMA1 and sporoRON2
Recently, sporozoite-specific, novel T. gondii paralogs of AMA1 and RON2 were identified and designated sporoAMA1 and sporoRON2 (Poukchanski et al., 2013). Neospora and Eimeria also have homologs of sporoAMA1 and sporoRON2 in addition to the generic AMA1 and RON2, while Plasmodium, Babesia, and Theileria only have homologs related to the generic forms of AMA1 and RON2. There is no detectable cross-interaction between the generic AMA1 and sporoRON2 and vice versa, due to different specific interactions in the hydrophobic groove, especially in the cysteine loop region (Poukchanski et al., 2013). In the crystal structure of sporoAMA1 bound to sporoAMA1, the position of Domain III is altered upon binding of RON2 in comparison to the apo structure (Figure 2D). In the apo AMA1 structures, as well as the generic Pf/Pv/TgAMA1-RON2 structures, Domain III lays along the base of Domains I and II (Figure 2C). In the sporoAMA1-sporoRON2 structure, Domain III rotates more than 90°, extending away from Domains I and II (Figure 2D). Whether this is physiologically relevant or can trigger a signaling event is still an open question. This movement, as previously mentioned, is not observed in the binding of RON2 peptides in the generic Pf/Pv/TgAMA1-RON2 structures. Additionally, Domain III in the sporoAMA1-sporoRON2 crystal structure is making a crystal contact with Domain III of a symmetry mate, like shaking hands, so it may be nothing more than a crystallization artifact.
2.1.4 Inhibition of the AMA1-RON complex
Recognition of this binding motif and further co-crystallization of PfAMA1 with antibodies shed light on their inhibitory mode of action. An IgNAR single-domain antibody was mutated to select for AMA1 from three P. falciparum strains (Henderson et al., 2007). It was shown in a crystal structure that the CD3 loops bind the hydrophobic groove where the RON2 peptide sits, and two β-strands of the IgNAR align with the two strands of RON2 stabilized by the cysteine loop (Figure 2F). Another growth and invasion inhibitory fragment antibody, 1F9, binds to the same end of the hydrophobic groove where the cysteine loop of the RON2 peptide binds (Figure 2G) (Coley et al., 2007). The binding of this antibody can be competed against using serum from individuals exposed to P. falciparum, indicating that acquired antibodies target the same epitope as 1F9. While these antibodies show growth inhibitory effects against laboratory strains, sequencing of lab and field strains has generated hundreds of non-redundant AMA1 sequences, with most polymorphisms concentrated in Domain I (Cortes et al., 2003; Escalante et al., 2001); this variability could pose a problem for vaccine development against this antigen.
A small molecule inhibitor of the AMA1-RON2 interaction in P. falciparum, was determined using a high throughput screen of a 21,000 compound library (Srinivasan et al., 2013). Addition of the compound reduced tight junction formation and reduced invasion levels. The compound was also shown to be effective against multiple lab strains, including the DD2 drug resistant strain, with an IC50 of 10–14 µM. The ultimate effectiveness of antibodies and compounds may rely on how essential the AMA1-RON2 interaction is to the parasite, which has been called into question, as is discussed later in section 4.2.
2.2 Reticulocyte binding-like homolog (RBL/RH) protein family
The reticulocyte binding-like protein family is specific to Plasmodium spp., and is composed of receptors needed for direct interaction with the reticulocyte and erythrocyte surface receptors. There are several subfamilies of reticulocyte binding-like proteins across Plasmodium spp. Most proteins in these families contain a signal sequence, N-terminal ectodomain, comprised of a nucleotide binding domain (Ramalingam et al., 2008) and erythrocyte binding domain (Gao et al., 2008; Gruber et al., 2011; Sahar et al., 2011), a transmembrane helix, and cytoplasmic C-terminal tail (Figure 1B).
2.2.1 Reticulocyte binding-like proteins (RBPs)
There has been little reported regarding the structure and function of the reticulocyte binding-like proteins (RBPs) from P. vivax, PvRBP1 and PvRBP2 (Galinski et al., 1992) or the P. yoelii homolog Py235 (Holder et al., 1991). PvRBP receptors have yet to be identified, in part because it is not possible to culture blood stages of P. vivax; however, PvRBP1 peptides that bind to reticulocytes, have been identified (Cantor et al., 2001; Urquiza et al., 2002). A short review by Li and Han (2012) describes the RBPs in further detail.
2.2.2 Reticulocyte binding-like homolog proteins (RH)
In P. falciparum, five variants of reticulocyte binding-like homolog proteins (RH) are involved in the invasion process: RH1, RH2a, RH2b, RH4, and RH5 (Kaneko et al., 2002; Rayner et al., 2000; Rayner et al., 2001; Triglia et al., 2001). Tham et al. (2012) review the function of P. falciparum RH proteins, and the potential of raising antibodies against these antigens to inhibit invasion. RH proteins 1–4 possess erythrocyte binding domains (EBD), nucleotide binding domains (NBD), and a C-terminal transmembrane helix and cytoplasmic tail.
Receptors for RH1, 2a and 2b have yet to be identified. The RH4 receptor interacts in a sialic acid-independent manner with complement receptor 1 (CR1/CD35) (Tham et al., 2010). Biochemical and biophysical analysis has narrowed down the RH4 interaction site to the N-terminal, complement control protein modules 1–3 (CCP 1–3) of CR1, which is the most membrane-distal CCP region (Park et al., 2014). Furthermore, all residues necessary to bind RH4 are contained solely in CCP-1.
Diverging from the typical architecture of RH proteins, is RH5, which lacks both an identified NBD and a transmembrane helix. To form a complex joining the parasite with the red blood cell, RH5 interacts with RIPR (RH5 interacting protein) (Chen et al., 2011), which also lacks a transmembrane domain. It is assumed that there is another binding partner with a transmembrane domain with which RIPR or RH5 forms a complex, anchoring RH5 to the parasite. RH5 interacts with basigin (CD147/EMMPRIN) (Crosnier et al., 2011) and a crystal structure of RH5 in complex with basigin has been determined (Wright et al., 2014a).
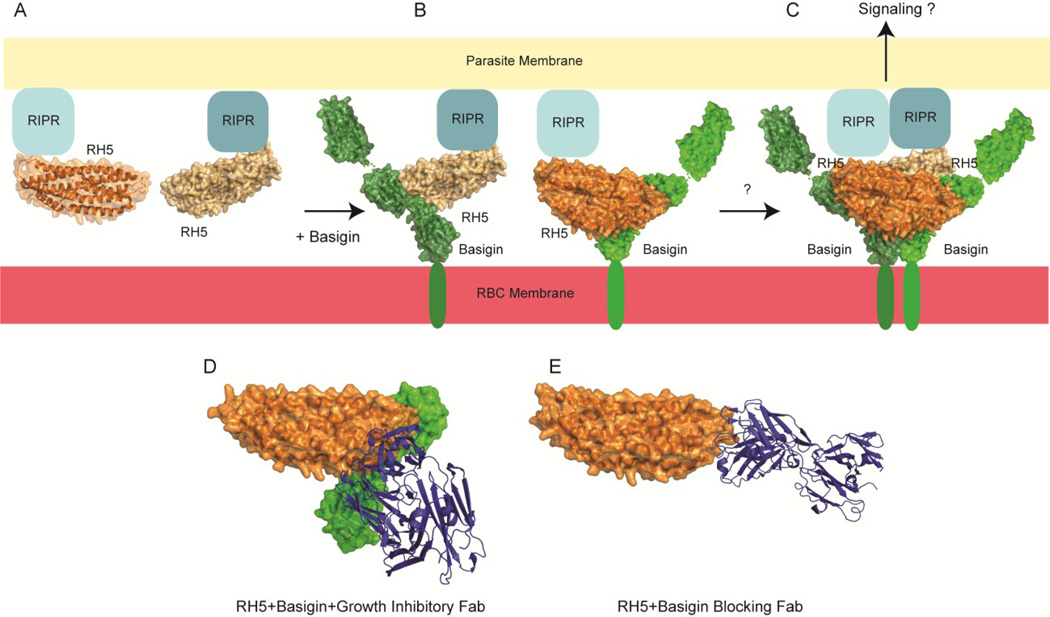
RH5 consists of two bundles of three α-helices each (Figure 3A). In the basigin-RH5 crystal structure, the asymmetric unit contains a 2:2 stoichiometry of basigin and RH5, forming a tetramer consisting of two hetero-dimers. One dimer of basigin-RH5 is formed by the interaction of one tip of the RH5 helix bundle with Regions II and III of basigin (Figure 3B). The tetramer is then formed by the other helical bundle tip interacting with Regions II and III of the other basigin molecule, on the opposite side of the second RH5 molecule (Figure 3C). There is no direct interaction between the two RH5 molecules. Region III of the basigin molecules, which are the most membrane-proximal, stabilize the dimer. This complex leaves an accessible surface for RIPR binding to RH5. SAXS (small angle X-ray scattering) and analytical ultracentrifugation (AUC) data indicate that the basigin-RH5 complex has a 1:1 stoichiometry (Wright et al., 2014a); however, the local effective concentrations at the membrane surface may make the dimer of dimers observed in the crystal structure physiologically possible, and one can speculate that this dimerization event could lead to a signaling event.
Figure 3. RH5 in complex with human receptor basigin.
Schematic of (A) RH5 (orange and light yellow, surface and cartoon) interaction with the parasite membrane via RIPR (blue squares). Addition of basigin (green, surface) results in a (B) 1:1 complex, as confirmed in SAXS and AUC experiments. The crystal structure of the basigin-RH5 complex shows a (C) 2:2 stoichiometry in the ASU. RH5 in complex with a (D) growth inhibitory antibody (blue, cartoon) that does not prevent basigin binding. Basigin is modeled, not present in the crystal structure, to indicate that the binding sites do not overlap. RH5 in complex with a (E) growth inhibitory antibody (blue, cartoon) that does block basigin binding.
RH5 was also crystallized in the presence of two growth inhibitory Fabs that act through different mechanisms (Wright et al., 2014a). Fab 9AD4 is potent (Douglas et al., 2014), however, it does not block binding of basigin in vitro. In the co-crystal structures it is observed that the antibody does not bind to the tip of RH5’s helical bundle, which would thereby prevent basigin binding, but instead binds to another interface (Figure 3D). This binding may present access problems for RH5 to interact with basigin due to steric hindrance at the membrane or it may interfere with RIPR binding to RH5. Monoclonal QA1, however, does disrupt basigin binding, and this effect is due to QA1 binding at the tip of the RH5 helical bundle (Figure 3E), blocking the basigin interaction, as observed in vitro.
2.3 Duffy binding-like proteins (DBP) and the erythrocyte binding-like (EBL) proteins
Another family of proteins involved in erythrocyte and reticulocyte invasion by Plasmodium spp. is the Duffy binding-like family, which consists of the Duffy-binding-like proteins (DBPs) and erythrocyte binding-like proteins (EBLs). DBP and EBL proteins contain a signal sequence, N-terminal ectodomain, a transmembrane helix, and a short C-terminal tail. The ectodomain is divided into 6 regions based on sequence homology between the DBPs and EBLs. Region II is cysteine rich and contains the Duffy binding-like (DBL) domains, responsible for binding to red blood cell receptors, followed by Regions III–V and Region VI, a second conserved cysteine rich domain (Adams et al., 1992) (Figure 1C). A review discussing the discovery and role of DBLs/EBLs is provided by Iyer et al. (2007) and the proteins are further explored by Li and Han (2012).
P. falciparum has multiple receptors for erythrocyte invasion including a subfamily of DBLs known as the erythrocyte binding-like (EBL) proteins, or erythrocyte binding antigens (EBA). This family consists of several proteins implicated in invasion including EBA-140 (Thompson et al., 2001), EBA-175 (Adams et al., 1992) and EBA-181 (Gilberger et al., 2003). The EBLs bind in a sialic acid-dependent manner to the erythrocyte. While the receptor for EBA-181 is unknown, EBA-175 binds to Glycophorin A (Orlandi et al., 1992) and EBA-140 binds Glycophorin C (Lobo et al., 2003).
2.3.1 EBA-175 (BAEBL)
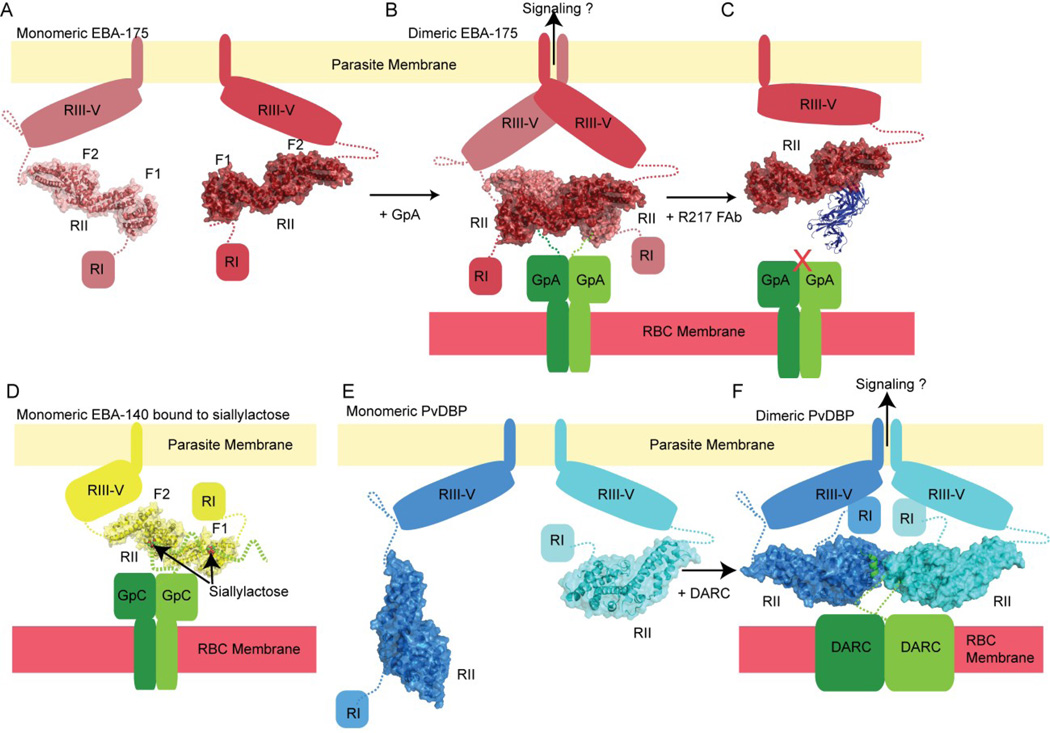
Region II of EBA-175 was crystallized in the presence and absence of sialyllactose as a dimer (Tolia et al., 2005). Region II consists of two DBL domains, F1 and F2, each composed primarily of α-helices and a β-hairpin, and the domains are linked by three α-helices (Figure 4A). The two monomers interact along their entire extended length forming a “handshake”, with the F1 domain of one monomer interacting with the F2 domain of the other and vice versa (Figure 4B). The dimer observed in the crystal is consistent with in vitro data where dynamic light scattering (DLS) indicates a dimer at high concentrations; however, AUC experiments at lower concentrations indicate that EBA-175 is primarily monomeric. It is tempting to speculate that on the membrane, the higher effective concentrations of EBA-175 promote dimerization with help from the obligate dimer of glycophorin A recruiting two monomers of EBA-175 and stabilizing the interaction, which could lead to a signaling event.
Figure 4. Duffy binding-like protein structures.
Schematic depicting orientation of EBA-175 Region II (red, surface and cartoon) (A) before receptor engagement and (B) and dimerization after engagement of dimeric glycophorin A on the red blood cell surface. (C) An inhibitory Fab (blue, cartoon) blocking the sialyllactose binding pocket as well as contacts needed for EBA-175 dimerization. (D) Crystal structure of monomeric EBA-140 with a sialyllactose molecule bound in both the F1 and F2 domains of Region II, which engage the dimeric glycophorin C receptor on the red blood cell surface. Schematic of (E) PvDBP (blue, cartoon and surface) (F) dimerization upon binding of the DARC peptide, part of the dimeric DARC receptor (green) on the red blood cell surface.
While EBA-175 was crystallized in the presence of sialyllactose, poor density meant that the authors could not model the glycans (Tolia et al., 2005). Even though the glycans could not be unambiguously modeled, it was observed that all the binding sites lie along the dimer interface and that the glycan binding involved making contacts with both monomers, indicating that dimerization is likely necessary for glycan/receptor engagement. An inhibitory Fab, R217 (Chen et al., 2013), contacts proposed glycan binding residues as well as the F2 β-finger (Figure 4C). This structural information allows one to rationalize the method of inhibition in which the antibody prevents binding to glycophorin A by two mechanisms. The antibody blocks the glycan binding site directly. Second, the antibody prevents dimerization of EBA-175 by interacting with the F2 β-finger, which promotes dimerization by inserting into a cavity on F1 of the other monomer. By blocking dimerization, glycan binding would also be affected, as the glycan sites are formed between dimers.
2.3.2 EBA-140
A structure of the binding Region II of EBA-140 in the presence and absence of sialyllactose was determined (Lin et al., 2012; Malpede et al., 2013). Unlike the dimer form observed for EBA-175, EBA-140 in the presence and absence of sialyllactose crystalizes as a monomer in the asymmetric unit, consistent with SAXS data indicating that it is a monomer in solution. The two DBL domains, composed of α-helices, are connected via a short helical linker (Figure 4D). The EBA-140-sialyllactose co-crystal structure determined two unique glycan-binding pockets, one in each of the DBL domains, F1 and F2 (Malpede et al., 2013). Mutations of sialic acid binding residues in F1 led to a severely compromised binding to erythrocytes while similar mutations in F2 led to only a 10% binding reduction. Other mutations in F2 resulted in more severe binding defects, but not to the same level as mutations of F1. The authors concluded that while the F2 binding site is functional, that it may not be essential for invasion. Additionally, only four polymorphisms have been reported in EBA-140, and these differences all localize to the F1 domain (Maier et al., 2009) with one found in the glycan binding pocket (Malpede et al., 2013).
2.3.3 Duffy binding-like proteins (DBPs)
P. vivax which primarily invades reticulocytes, and P. knowlesi, which invades normocytes, express PvDBP1/2 (Wertheimer and Barnwell, 1989) and multiple PkDBPs (Haynes et al., 1988), respectively. The proteins share 70% homology and a conserved DBL domain. Recently a structure was published of PvDBP with a peptide from its host cell receptor, the Duffy antigen/chemokine receptor (DARC) (Batchelor et al., 2014; Wertheimer and Barnwell, 1989). PvDBP consists of two α-helical bundles and a β-hairpin near the N-terminus (Batchelor et al., 2011) (Figure 4E). A dimer of PvDBP with one DARC peptide bound forming a heterotrimer, and a heterotetramer of 2 PvDBPs and 2 DARC peptides (Figure 4F), were characterized. The DARC peptide binds at the edge of the PvDBP dimer interface. Authors proposed that the multiple states observed in the crystal structures were representative of physiologically relevant states. SAXS experiments indicate that PvDBP is a monomer at low concentrations in the absence of ligand/DARC peptide, whereas at higher PvDBP concentrations, data suggests that there is a mixture of monomers and dimers present (Batchelor et al., 2011). Additionally, addition of DARC peptide drove PvDBP dimerization at both low and high concentrations. Using ITC (isothermal titration calorimetry), it was confirmed that the PvDBP dimer first binds one DARC peptide and then a second peptide, in agreement with the two crystal states observed (Batchelor et al., 2014).
2.4 Thrombospondin related anonymous protein (TRAP) family and MIC2 protein
The last family of receptors discussed here are the thrombospondin related anonymous proteins (TRAPs) (Robson et al., 1988). The TRAP protein is a pan-apicomplexan protein including Plasmodium spp. TRAP, T. gondii MIC2 (microneme-associated protein 2) (Brossier and David Sibley, 2005; Wan et al., 1997), Cryptosporidium spp. TRAPC1 (Spano et al., 1998a; Spano et al., 1998b), Eimeria spp. Etp100 (Tomley et al., 1991), and Babesia spp. TRAP (Gaffar et al., 2004). Plasmodium spp. have multiple TRAP variants necessary for motility and invasion at different stages (Sultan et al., 1997), including TRAP for liver invasion (Muller et al., 1993), CTRP for mosquito midgut invasion (Trottein et al., 1995), MTRAP for erythrocyte invasion (Baum et al., 2006), and TLP (TRAP-like protein) for gliding in the skin (Moreira et al., 2008). The typical domain organization is a signal sequence followed by an N-terminal ectodomain, containing at least one thrombospondin repeat type 1 (TSR) domain and a von Willebrand factor type-A (VWA) motif, a transmembrane helix, and a C-terminal tail (Figure 1D).
2.4.1 TSR and VWA domains and structures
Structures of PfTRAP, PvTRAP and TgMIC2 have been characterized and have provided insight into the mechanisms of TRAP function. In looking at the numerous structures of the VWA and TSR domains, it was observed that TRAP exists in two states, either an open or closed conformation, which is dependent on metal binding to a MIDAS motif (metal ion dependent adhesion sites) (Song and Springer, 2014; Song et al., 2012). Other VWA domains, specifically those in integrin I domains, also contain MIDAS motifs and can adopt open and closed conformations based on magnesium and ligand binding.
A PfTRAP construct containing the VWA and TSR domain was crystallized (Song et al., 2012), however, only the VWA domain could be modeled, even though it was confirmed via SDS-PAGE that the crystals contained the full-length construct. The VWA adopted an α/β Rossmann fold (Figure 5A). A void in the crystal packing at the C-terminus of the VWA domain was large enough to accommodate multiple conformations of the TSR domain, which was apparently disordered given the lack of electron density.
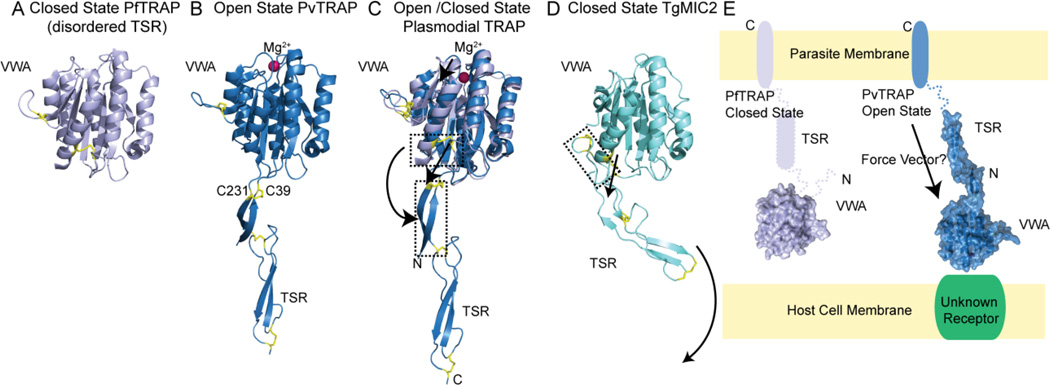
Figure 5. Open and closed state of TRAP proteins.
(A) VWA domain of PfTRAP (grey, cartoon) in the closed state, with key disulfide bonds in yellow. (B) Open state of PvTRAP (blue, cartoon) depicting the VWA with Mg2+ (magenta, sphere) in the MIDAS site, and TSR domain connected by an extended β-strand formed by residues up- and downstream of the VWA domain. (C) Alignment of the open and closed TRAP state showing the unwinding of the helix allowing the disulfide bond between C231 and C39 (PvTRAP) to shift down and away from the VWA domain to stabilize the extended β-strand before the TSR domain. (D) Closed state of TgMIC2 depicting an unextended, ordered TSR domain, however, the helix has not unwound allowing the disulfide bond to shift down and away from the VWA domain and forcing an extension of the TSR domain into the open state as indicated by the arrow. (E) Schematic of the orientation of TRAP relative to the parasite membrane with the N-terminal VWA domain farthest from the membrane followed by a disordered TSR and C-terminal transmembrane domain. Binding of ligand in the membrane distal VWA domain induces a switch to the open state with an extended TSR. This open state may allow force (arrow) to be transmitted along the more rigid TSR domain to the host cell attachment at the unknown receptor.
PfTRAP adopted a closed state conformation in the absence of magnesium and was structurally different, compared to the open state of PvTRAP with magnesium bound (Figure5A and 5B) (Song et al., 2012). In the closed, apo structure (Figure 5A), the TSR domain was disordered; however, in the open metal bound state, an extended β-ribbon is formed and the TSR domain becomes ordered. An extended β-ribbon is formed between the N-terminal residues before the VWA domain start and residues that unwind from the last α-helix in the VWA domain (Figure 5B). This movement and extension can be seen in the positioning of the disulfide bond between C39 and C231 in the PvTRAP open state versus the position of the disulfide bond between C43 and C235 in the PfTRAP closed state (Figure 5C). Three disulfide bonds between C39 and C321, C240 and C261, and C253 and C289, stabilize the structure of the ordered TSR domain in the PvTRAP open state.
The homolog of PfTRAP, TgMIC2, was also crystallized with its VWA and TSR domains modeled (Song and Springer, 2014). The protein was modeled in the closed state, as can be seen by the positioning of the previously noted disulfide bond, which has not extended away from the VWA domain, preventing MIC2 from adopting an extended structure similar to that of the PvTRAP (Figure 5D). While the three disulfide bonds present in MIC2 correspond to those in the extended β-strand and TSR domains in the plasmodial proteins, there is an additional loop stabilized by a disulfide bond that may extend away from the VWA domain in the open state (Figure 5D). A magnesium bound, open state of TgMIC2 was not crystallized, but it is thought that binding to the MIDAS motif would cause an unwinding of the VWA helix and extension of the β-strand and TSR domain to adopt a conformation similar to that of PvTRAP.
It has been proposed by the authors that binding of a ligand/receptor to the VWA domain induces a conformational change, causing the extension of the TSR domain, creating a stable arm through which force generated by the actomyosin motor of the invasion machinery is transmitted to the host cell surface (Figure 5E) (Juliana et al., 2012). One caveat to this model is that a structure of PfTRAP with a magnesium bound to the MIDAS motif was crystallized in the closed state, indicating that this open/closed mechanism may not be conserved between PfTRAP and PvTRAP (Pihlajamaa et al., 2013). However, the construct used in crystallization did not contain the TSR domain and lacked the N-terminal residues, which form the β-strand preceding the VWA domain and forms part of the extended β-ribbon observed in the PvTRAP structure.
2.4.2 TRAP receptors
Receptors for the TRAP proteins have yet to be identified, except for PfMTRAP. The Semaphorin-7a (CD108) receptor was identified and characterization of the interaction between MTRAP and Semaphorin-7a ectodomains was performed via size exclusion chromatography and surface plasmon resonance (Bartholdson et al., 2012). Semaphorin-7a is a homodimer in solution according to SEC (size exclusion chromatography) and MALS (multi-angle light scattering), and MTRAP elutes as a monomer in SEC. Experiments indicate that the binding of MTRAP to Semaphorin-7a occurs in a 2:1 stoichiometry and that MTRAP interacts via its TSR domain, as it lacks the VWA domains that other family members possess.
3. The Invasion Machinery (Glideosome)
For motility and invasion, apicomplexan parasites employ an actomyosin based motor that is part of a larger invasion machinery complex known as the glideosome. Multiple interacting proteins are used to bridge and transmit the locomotive force of the motor to the parasite’s attachment point at the host cell surface and to anchor the motor within the parasite. Most of these proteins are conserved across apicomplexa. The proteins have been sorted into three groups: motor-receptor bridging proteins, motor complex proteins, and inner membrane complex (IMC) proteins and anchors. A list of the machinery components, accessory proteins and anchors, their known interaction partners with references, EuPathDB codes, and any structures that have been deposited with the PDB, are noted in Table 2. A schematic of the glideosome can be found in Figure 9A.
Table 2.
Proteins of the invasion machinery complex.
| Protein | EuPathDB | Interacting Proteins | Structure |
|---|---|---|---|
| Motor-Receptor Bridge | |||
| GAPDH | PF3D7_1462800 | PfRH1, PfRH4, EBA-175, EBA-181 (Pal-Bhowmick et al., 2012) | 1YWG (Satchell et al., 2005) (Pf tetramer) |
| TGME49_289690 | 3STH (SSGCID et al., 2011) (Tg dimer) | ||
| TGME49_269190 | Actin | 2B4R, 2B4T (Robien et al., 2006) (Pf tetramer + protease inhibitor near NAD+ site) | |
| Aldolase | PF3D7_1444800 | PfTRAP/PvTRAP/PbTRAP/TgMIC2 (Bosch et al., 2007b; Boucher and Bosch, 2014; Buscaglia et al., 2003; Jewett and Sibley, 2003) | 1A5C (Kim et al., 1998) (apo Pf tetramer) |
| TGME49_236040 | 2PC4, 2EPH (Bosch et al., 2007b) (Pf tetramer + PbTRAP peptide) | ||
| TGME49_321900 | |||
| Actin (Buscaglia et al., 2003) | 4TU1 (Boucher and Bosch, 2014) (Tg tetramer) | ||
| PfMTRAP (Baum et al., 2006) | 4D2J (Tonkin et al., 2014b) | ||
| PfCTRP & PfTLP (Heiss et al., 2008) | 4TR9NR (Pf tetramer + PbTRAP peptide + small molecule) | ||
| TgMIC6 & TgMIC12 (Sheiner et al., 2010) | |||
| PfAMA1/TgAMA1 (Sheiner et al., 2010; Srinivasan et al., 2011) | |||
| PfRH1, PfRH2b, PfRH4 (Pal-Bhowmick et al., 2012) | |||
| Motor Complex | |||
| Actin | PF3D7_1246200 | Aldolase (Buscaglia et al., 2003) | 4CBU (Vahokoski et al., 2014) (PfActin1) |
| PF3D7_1412500 | Myosin A (Bergman et al., 2003; Heintzelman and Schwartzman, 1999; Herm-Gotz et al., 2002) | 4CBX (Vahokoski et al., 2014) (PbActin2) | |
| TGME49_209030 | 4CBW (Vahokoski et al., 2014) (PbActin1 + muscle actin D-loop*) | ||
| Profilin (Kursula et al., 2008; Plattner et al., 2008) | |||
| ADF1/2 (Allen et al., 1997; Schuler et al., 2005a) | |||
| Capping Proteins (Ganter et al., 2009; Tardieux et al., 1998) | |||
| Formin (Baum et al., 2008; Daher et al., 2010) | |||
| Myosin A | PF3D7_1342600 | Actin (Bergman et al., 2003; Heintzelman and Schwartzman, 1999; Herm-Gotz et al., 2002) | |
| TGME49_235470 | |||
| PfMTIP/TgMLC1 (Bergman et al., 2003; Herm-Gotz et al., 2002) | |||
| PfMTIP/TgMLC1 | PF3D7_1246400 | Myosin A (Bergman et al., 2003; Herm-Gotz et al., 2002) | 4AOM (Douse et al., 2012) (PfMTIP + PfMyoA peptide) |
| TGME49_257680 | |||
| GAP40/45/50 (Baum et al., 2006; Frenal et al., 2010; Gaskins et al., 2004) | 2QAC (Bosch et al., 2007a) (PfMTIP + PfMyoA peptide, closed form) | ||
| 2AUC (Bosch et al., 2006) (PkMTIP + PyMyoA peptide, open form) | |||
| 4GGN (Turley et al., 2013) (PkMTIP + PyMyoA peptide, closed form) | |||
| 4MZK, 4MZJ (Douse et al., 2013) (PfMTIP + MyoA stapled tail peptide) | |||
| 4MZL (Douse et al., 2013) (PfMTIP + HBS mimetic peptide) | |||
| 4GFT (Khamrui et al., 2013) (PfMTIP + nanobody) | |||
| 4R1E (Douse et al., 2014) | |||
| Inner Membrane Complex (IMC) | |||
| GAP40/45/50 | PF3D7_0515700 | MTIP/MyoA (Baum et al., 2006; Frenal et al., 2010; Gaskins et al., 2004) | 3TGH (Bosch et al., 2012) (PfGAP50) |
| PF3D7_1222700 | |||
| PF3D7_0918000 | Alveolin (Bullen et al., 2009) | ||
| TGME49_249850 | |||
| TGME49_223940 | |||
| TGME49_219320 | |||
| GAPM1/2/3 | PF3D7_1323700 | Alveolin & GAP45/50 (Bullen et al., 2009) | -- |
| PF3D7_0423500 | |||
| PF3D7_1406800 | |||
| IMC Sub-compartment | TGME49_260820 | Unknown | 4CHM (Tonkin et al., 2014a) (TgISP1) |
| TGME49_237820 | 4CHJ (Tonkin et al., 2014a) (TgISP3) | ||
| Proteins (ISPs) 1–3 | TGME49_316540 | ||
| Subpellicular Network (SPN) | |||
| Alveolin | PF3D7_0304000 | GAPM1/2/3 (Bullen et al., 2009) | -- |
| PF3D7_1221400 | |||
| PF3D7_1141900 | |||
| TGME49_231640 | |||
| Actin polymerization factors | |||
| Profilin | PF3D7_0932200 | Actin (Kursula et al., 2008; Plattner et al., 2008) | 2JKF (Kursula et al., 2008) (Pf) |
| TGME49_293690 | 2JKG (Kursula et al., 2008) (Pf + Mg2+) | ||
| 3NEC (Kucera et al., 2010) (Tg) | |||
| Formin | PF3D7_0530900 | Actin (Baum et al., 2008; Daher et al., 2010) | -- |
| PF3D7_1219000 | |||
| TGME49_206430 | |||
| TGME49_206580 | |||
| TGME49_213370 | |||
| ADF/cofilin | PF3D7_0503400 | Actin (Allen et al., 1997; Schuler et al., 2005a) | 3Q2B (Wong et al., 2011), 2XF1 (Singh et al., 2011)(PfADF1) |
| PF3D7_1361400 | |||
| TGME49_220400 | 2XFA (Singh et al., 2011) (PbADF2) | ||
| 2L72 (Yadav et al., 2011) (TgADF) | |||
| CAP | PF3D7_0105300 | Actin | 2B0R (Hliscs et al., 2010) (CpC-CAP) |
TBPDeposited, Held for Publication.
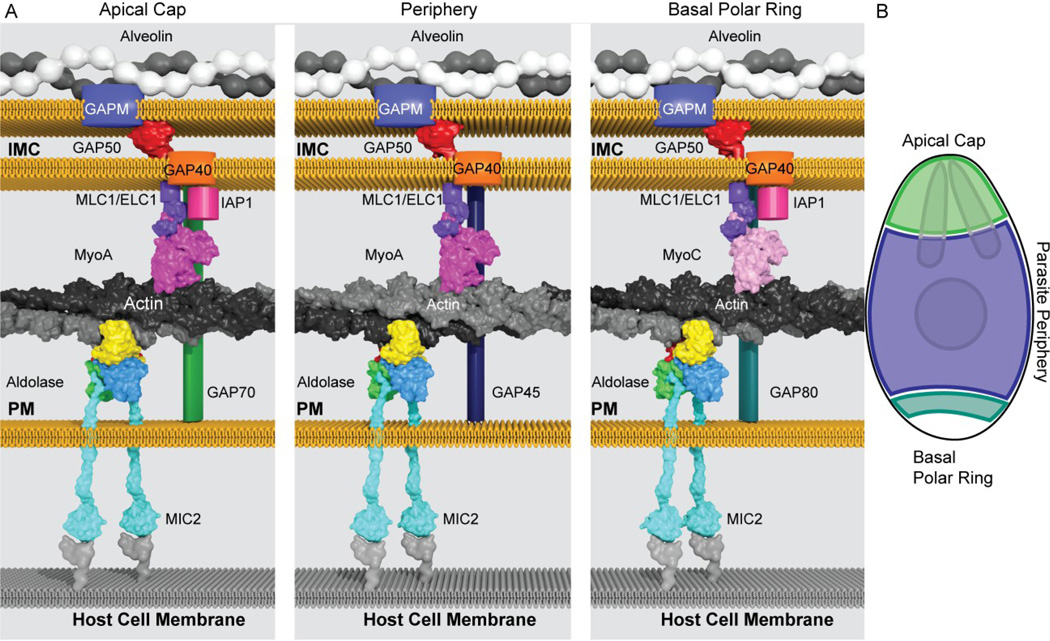
Figure 9. Schematic of apicomplexan glideosomes.
Protein structures solved from both T. gondii and Plasmodium spp. and homologous protein structures were combined with the results of biochemical and genetic data to depict the (A) glideosome model. The three panels illustrate the motor complexes present in the apical cap (left), along the parasite periphery (middle), and at the basal polar ring (right). The actin-myosin motor (grey/black and magenta/light pink) is bridged by tetrameric aldolase (multicolor) to the extracellular adhesins (cyan), which connects to unknown cellular receptors on the host cell membrane (grey). The motor is anchored to the IMC via its interaction with MTIP (Plasmodium spp.) or MLC1/ELC1 (T. gondii) (purple). In T. gondii, the MyoA (magenta) motor is located in the apical cap and periphery of the parasite while the MyoC (light pink) motor acts at the basal polar ring. The myosin-MLC1 motors interact with either lipidated GAP70 (green), GAP45 (dark blue), or GAP80 (teal), which span the supra-alveolar space between the plasma and IMC membranes, at the apical cap, periphery or basal polar ring, respectively. At the apical cap and basal polar ring, IAP1 (hot pink) interacts with the MyoA/C-GAP70/80-MLC1-ELC1 complex. These motor complexes interact with GAP40 (orange) and GAP50 (red) located in the IMC. GAPMs (blue) are located on the cytosolic side of the IMC membrane and interact with the alveolins (white/grey). (B) Schematic of T. gondii tachyzoite highlighting the apical cap (green), periphery (blue), and basal polar ring (teal) with nucleus and rhoptries depicted.
3.1 Motor-receptor bridging proteins
As has been discussed previously, apicomplexans use a wide range of extracellular receptors to attach to a host cell membrane for invasion. In order to transmit the force of the cytoplasmic actomyosin motor to the extracellular attachments, bridging proteins are required. In apicomplexans, two bridging proteins have been identified: aldolase and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). In the Plasmodium spp. genomes there are single copies of both aldolase and GAPDH, while in T. gondii, there are annotations of the genome indicating two copies of each enzyme.
3.1.1 Aldolase
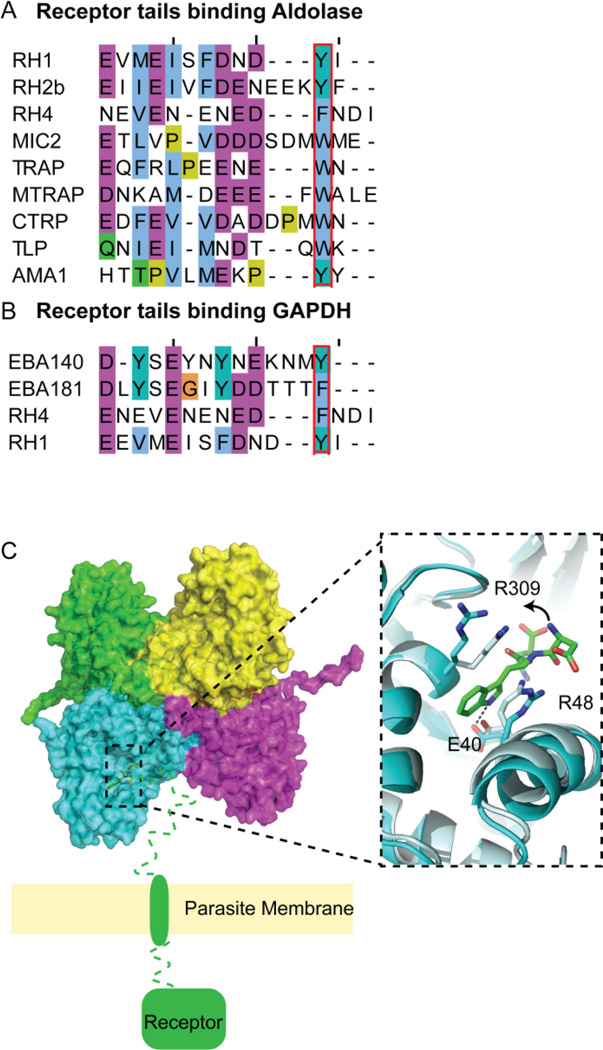
A key bridging protein is the enzyme, fructose 1,6 bisphosphate aldolase. In addition to catalyzing a reaction in the glycolytic pathway, this protein has a second non-enzymatic function; the aldolase tetramer interacts with actin on one face (Buscaglia et al., 2003), and the C-terminal tails of multiple receptor proteins via its active site. In Plasmodium spp., aldolase interacts with multiple receptors responsible for invasion: the TRAPs, AMA1, and RH1/2b/4, and in T. gondii, AMA1 and MIC2/6/12 (Baum et al., 2006; Bosch et al., 2007b; Boucher and Bosch, 2013; Buscaglia et al., 2003; Heiss et al., 2008; Jewett and Sibley, 2003; Pal-Bhowmick et al., 2012; Sheiner et al., 2010; Srinivasan et al., 2011). Alignment of the receptor tails (Figure 6A), mutational studies, and a PfAldolase-PbTRAP crystal structure (Bosch et al., 2007b) indicate that a conserved tryptophan residue is key to the binding (Kappe et al., 1999). A C-terminal PbTRAP peptide (EDNDWN) was co-crystallized with aldolase, with the last three residues modeled in the aldolase active site (Bosch et al., 2007b). To accommodate the peptide, there is a helix shift and R48 and R309 move to sandwich and stack with the key tryptophan (W605), which makes and additional hydrogen bond contact with E40 (Figure 6C). The structure of T. gondii aldolase 1 has been determined (Boucher and Bosch, 2014; Tonkin et al., 2014b). It was noted that the sequence identity in the active site/peptide binding site was 100% conserved and surface plasmon resonance experiments indicate that TgAldolase is competent to bind P. falciparum TRAP tails (data not shown).
Figure 6. Binding of adhesin tails to aldolase and GAPDH.
Alignment of C-terminal tail sequence of receptors known to bind (A) aldolase and (B) GAPDH, highlighting a key residue for binding in red. (C) Tetrameric PfAldolase (multicolored chains, surface) with PbTRAP peptide residues, EWN, bound and modeled in the active site, and an overlay of the binding site of apo (grey) and bound (cyan) aldolase (cartoon). The key tryptophan of the TRAP tail is sandwiched between R48 and R309, after R309 swings out of the active site and the helix containing R48 shifts to allow binding. This positioning allows the tryptophan to make a hydrogen bond to E40.
In addition to aldolase-TRAP co-crystal structures, the ternary co-crystal structure, with a small molecule stabilizing the TRAP-peptide in the aldolase active site, has been deposited and will be described elsewhere (PDB: 4TR9, PCT/US2012/061875) (Cardozo et al., 2013). Briefly, when tested in parasite cultures, the stabilizing compound results in a gliding and invasion phenotype that is in agreement with inhibition of the glideosome at the aldolase-TRAP interface.
3.1.2 GAPDH
A second enzyme used as a connection between actin and the receptors is GAPDH. While the tetrameric structure of PfGAPDH (Robien et al., 2006; Satchell et al., 2005) and dimer of TgGAPDH (SSGCID et al., 2011) have been deposited, to date no co-crystals of GAPDH and a peptide have been structurally characterized. It has been demonstrated via biochemical and biophysical assays that GAPDH can bind the C-terminal peptide tails (Figure 6B) of RH1, RH4, EBA-175 and EBA-181 (Pal-Bhowmick et al., 2012).
3.1.3 Unknown bridging molecules
While interactions between aldolase/GAPDH and the C-terminal tails of multiple receptors has been demonstrated, there are some receptors that do not interact with these bridging partners. EBA-140, RH2a, RH5/RIPR, Pv/PkDBP, and PvRBPs have not been shown to interact with either aldolase or GAPDH. Lack of an apparent interaction partner may indicate that there is either an undiscovered bridging molecule or that the proteins are able to interact with the motor in the absence of a bridging molecule. Another possibility is that the primary function of the receptor is for attachment, and not motility, and therefore, it does not interact with the motor.
3.2 Motor-complex proteins
The locomotive force for invasion is generated by the actomyosin motor. Forward movement of the parasite is generated by myosin pulling back on short actin polymers, to which the extracellular receptors are bridged via aldolase, GAPDH, or yet to be identified components.
3.2.1 Actin
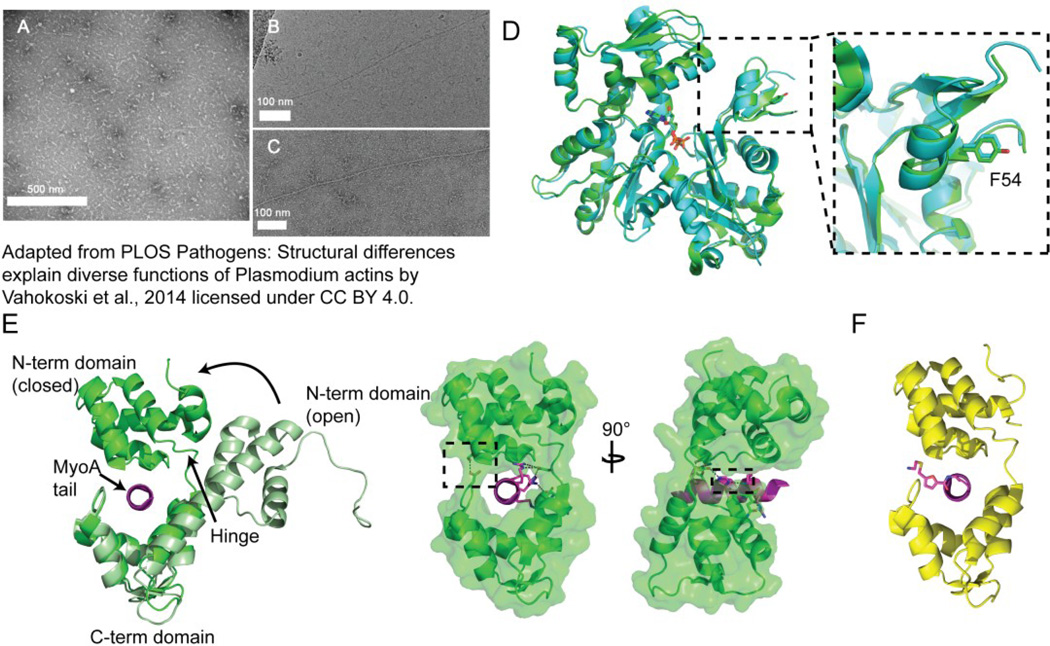
Actin forms the linear track of the glideosome motor. Typically, actin polymerizes in a head-to-tail fashion forming helical, parallel strands that are highly stable and range from several hundred nanometers to microns in length. However, both Plasmodium spp. and T. gondii form unstable actin polymers around 100 nm in length (Sahoo et al., 2006; Schmitz et al., 2005; Schuler et al., 2005b; Vahokoski et al., 2014). Plasmodium spp. express two variants of actin, actin 1 and actin 2 (Wesseling et al., 1988), whereas most apicomplexans, including T. gondii, express only one (Dobrowolski et al., 1997). Actin 2 forms filaments similar to canonical F-actin (Figure7B and 7C), whereas actin 1 is responsible for the short, fragile filaments observed in apicomplexans (Figure 7A) (Vahokoski et al., 2014). The dynamics of apicomplexan actins are unique. Plasmodium spp. actins 1 and 2 can hydrolyze ATP more efficiently and are able to oligomerize in response to ADP (Vahokoski et al., 2014), while TgActin 1 can form filaments at critical concentration 3–4 times lower than F-actin (Sahoo et al., 2006). Observations that TgActin 1 could polymerize at lower critical concentrations as well as without a lag phase, is the result of isodesmic polymerization, where the initial nucleation and polymerization steps have the same rate constants (Skillman et al., 2011).
Figure 7. Motor components actin and MTIP bound to MyoA tail.
EM images of (A) PfActin 1 forming short 100 nm fibers and (B and C) PfActin 2 forming long filaments, adapted from PLOS Pathogens “Structural differences explain diverse functions of Plasmodium actins” by Vahokoski et al. (2014) licensed under a Creative Commons Attribution 4.0 International License. (D) Overlay of the canonical actin (green, cartoon) with PfActin 1 (cyan, cartoon), and ATP and a nonconserved F54 depicted as sticks. (E) MTIP (cartoon) in the open (light green) and closed (dark green) states with the MyoA peptide tail (magenta, cartoon) bound. The hinge, which does not have secondary structure in the closed state but is part of the long central helix in the open state is marked. Two views of the closed MTIP structure, separated by 90° rotation about the y-axis, show the serine-aspartate latch (sticks and boxed) that stabilizes the closed structure. The second orientation depicts the hydrogen bonding network (sticks and dashes), stabilizing the MTIP-MyoA complex, formed between the aspartate residue in the hinge, the arginine, histidine, and lysine along the MyoA helix, the asparagine carboxy terminus of MTIP, and the last arginine on the MyoA peptide. (F) MyoA-tail chimera (magenta, cartoon and stick) bound to MTIP (yellow, cartoon).
Why do these actins not behave like canonical actins? Structures of plasmodial actins 1 and 2 have been determined, shedding some light on this question (Vahokoski et al., 2014). Structural alignment of actin 1, responsible for the short filaments and for forming the linear track of the glideosome, with a canonical actin shows that the overall structures do not differ radically (Figure 7D). In general, structural rearrangements of the plasmodial actins are found in regions responsible for contacts between monomers in a filament, as well as known interfaces for regulatory protein binding. It was demonstrated that substitution of the DNase I (D-)loop from canonical F-actin into actin 1 could “rescue” polymerization and induce formation of longer filaments, similar to those formed by actin 2 (Vahokoski et al., 2014). However, the sequences of the D-loops of actin 1 and actin 2 are not very different, and yet actin 2 is able to form longer filaments, indicating that actin 2 has managed to develop a mechanism for more robust polymerization through other means. Additionally, the Y54F substitution at the base of the D-loop in actin 1, which differentiates it from actin 2 and canonical actins, could explain the lower levels of polymerization; however, mutation of this residue to tyrosine did not rescue polymerization.
3.2.2 Actin accessory proteins
In general, organisms have evolved a set of actin accessory proteins that aid in the nucleation of actin, stimulate polymerization, stabilize filaments, cap filaments, and sever filaments. Apicomplexans have a limited subset of the actin accessory proteins that are present in higher eukaryotes (Schuler and Matuschewski, 2006). For a detailed structure/function review of these actin binding proteins can be found in the review by Olshina et al. (2012). While these proteins are not discussed here, but have been listed in Table 2.
3.2.3 Myosin A
Myosin A (MyoA) pulls on the actin filament to move the parasite forward by generating mechanochemical force through the hydrolysis of ATP. TgMyoA and Plasmodium spp. MyoA are part of the Class XIV myosin family. This class of myosins possesses a head domain, but lack a neck composed of IQ motifs, and a tail. Apicomplexan MyoA diverges from this architecture as it does possess a tail domain (Heintzelman and Schwartzman, 1997; Heintzelman and Schwartzman, 1999; Pinder et al., 2000; Pinder et al., 1998). PfMyoA does not contain a canonical IQ-motif (IQxxxRGxxxR) present in other myosins, but does have a modified motif VQxxxRKxxx(A/V). It has been shown that TgMyoA moves at speeds of ~5 µm/s in vitro, despite lacking features of typical myosins such as a conserved glycine in the motor domain, a conserved residue in an actin binding loop, and possession of a degenerate IQ-motif.
Unfortunately, structures of the MyoA motor domain do not exist due to difficulty in expression, but recently the successful expression of TgMyoA in Sf9 insect cells was accomplished via co-expression with TgMLC1 (myosin light chain 1), TgELC1 (essential light chain 1), and a chaperone, TgUNC, a myosin-specific co-chaperone from the UCS family, Unc45b (Bookwalter et al., 2014). This strategy will hopefully enable structural characterization of the protein in the future.
3.2.4 MTIP/MLC1
To anchor the actomyosin motor to the inner membrane complex, T. gondii encodes for the myosin light chain (MLC) protein (Herm-Gotz et al., 2002) and Plasmodium spp. express the ortholog, myosin-A tail interacting protein (MTIP) (Bergman et al., 2003). These proteins are distantly related to the myosin light chain proteins, but lack the calcium binding motif common to the family. Unique to the apicomplexan interaction of MyoA with MTIP/TgMLC1 is that the interaction is mediated via the MyoA tail containing a single degenerate IQ motif, instead of by a neck domain containing multiple, canonical IQ motifs (Bergman et al., 2003; Herm-Gotz et al., 2002). An N-terminal deletion mutant, PkMTIPΔ78, was first crystallized at a low, non-physiological pH with a helical MyoA peptide containing the modified IQ motif (Bosch et al., 2006) (Figure 7E). The PkMTIPΔ78 structure possesses a central helix that connects the N- and C-terminal domains, forming one helix in each domain. The N-terminal domain of the truncated construct consists of four helices, forming two degenerate EF hands (no Ca2+ binding was observed), while the C-terminal domain is composed of four helices, forming two degenerate EF hands, as well as a small anti-parallel β-sheet. The MyoA peptide binds to the C-terminal domain via residues extending into hydrophobic pockets and via complementary electrostatic interactions.
A later structure of PfMTIPΔ60-MyoA was crystallized at physiological pH and a different conformation was observed (Bosch et al., 2007a). The long, continuous central helix previously observed at low pH is kinked at physiological pH, resulting in a closed MTIP structure, where the N- and C-terminal domains latch via an electrostatic interaction. This state enables a tight interaction of the MyoA peptide with both the C- and N-terminal domains and hinge region. This kink, or compacted structure, is facilitated by the deprotonation of MyoA residue H810, allowing hydrogen bonding interactions with an aspartate in the MTIP hinge and the MyoA lysine (K813) at the canonical glycine position of the IQ motif. This closed conformation is believed to be the functionally competent state.
Phosphorylation of MTIP was thought to play a role in regulation of the MTIP-MyoA interaction when studies determined that MTIP could be phosphorylated in vitro via CDPK1, and a phospho-proteomics study identified a phosphorylated S108 (Green et al., 2008; Treeck et al., 2011). To test whether phosphorylation affected MyoA binding, the MTIP S108E mutation was made. The S108E mutation affected the stability of the MyoA-MTIP complex as measured via ITC and DSF (differential scanning fluorimetry) (Douse et al., 2012). In the crystal structure it is seen that S108 of the N-terminal domain forms a latch with D173 of the C-terminal domain (Figure 7E). Phosphorylation of this residue would alter the charge state and disrupt the latch, which prevents formation of the closed state. This result indicates that phosphorylation may be regulating motor assembly.
As the MTIP-MyoA interaction is necessary to anchor the motor to the IMC, some work has been done to identify small molecules that disrupt the protein-protein interaction. Using the crystal structure of the MyoA-MTIPΔ60 complex, a hybrid virtual screening approach was implemented, first building a template pharmacophore based on key interactions of MyoA with MTIP, and then screening a large library for compounds matching the pharmacophore (Kortagere et al., 2010). Identified molecules were then docked against the crystal structure and several compounds tested for growth inhibition of P. falciparum cultures exhibited IC50 < 25 µM, leading to the identification of a pyrazole-urea compound that led to decreased growth and altered gliding phenotypes. However, proof of a direct interaction of the compound with MTIP was not established.
A more recent study used a fragment-based approach to screen molecules against the protein-protein interaction using DSF to screen an initial library, followed by cross-validation with ITC and NMR (Douse et al., 2014). Unfortunately, crystal structures of the compounds complexed with MTIP alone were not able to be obtained, likely due to the low binding affinities (KD ~ mM) typical of fragments. To overcome the low binding affinity, a MTIP-stabilizing fragment, which was demonstrated to interact with residues implicated in MyoA binding, was linked to the N-terminus of a truncated MyoA peptide to form a chimera. A crystal structure of MTIP in complex with the chimera was solved providing further insight into how the compound interacts with MTIP (Figure 7F). This structure may allow for rational expansion of the fragment to target other regions and residues of MTIP and development of a tighter binding compound.
3.2.5 ELC1
Calcium signaling has been demonstrated to be important for egress and invasion in apicomplexans, with CDPK1 phosphorylating multiple glideosome proteins (Green et al., 2008). Myosin activity is often regulated by calcium through calcium-dependent phosphorylation or calcium binding to EF hands of myosin regulatory elements (Himmel et al., 2009). As was previously mentioned, MTIP contains degenerate EF hands, and does not bind calcium. In the course of a phospho-proteomics study to identify calcium-dependent phosphorylation targets in T. gondii during invasion, a previously uncharacterized, putative calmodulin-like protein was identified (Nebl et al., 2011). Sequence searches revealed that the identified protein was most similar to the essential light chain of scallop myosin II. This protein was therefore termed the essential light chain 1 (ELC1). It was demonstrated that ELC1 co-purified with the MyoAMLC1-GAP40/45/50 glideosome and more strongly associated with the complex during calcium signaling. This light chain element may be responsible for direct calcium regulation of the myosin, as opposed to MLC1 phosphorylation by CDPK1.
3.3 Inner membrane complex (IMC)
The actomyosin motor is attached to the inner membrane complex (IMC) comprised of alveoli. These alveoli underlay the entirety of the parasite’s plasma membrane, except at the base and apex, and are connected to the parasite’s cytoskeleton. Several proteins connect the actomyosin motor to the IMC and are involved in attachment to the cytoskeleton. In this section, the structural features of the glideosome associated proteins (GAP), glideosome associated proteins with multiple membrane spans (GAPM), IMC sub-compartment proteins (ISPs), and the anchoring alveolins are discussed. A table with predicted and experimentally derived post-translational modifications that may affect assembly, localization, and the function of the motor, are listed in Table 3. An in depth review of the apicomplexan IMC architecture and involved proteins, has been published by Kono et al. (2013).
Table 3.
Confirmed and predicted post-translational modifications of the invasion machinery components.
| Protein | Myristoylation | Palmitoylation | Phosphorylation |
|---|---|---|---|
| Aldolase | - | + Pf1 | + Pf/Tg2,3,4 |
| GAPDH | - | - | + Pf2,4 |
| Actin 1 | - | - | + Pf/Tg2,3,4 |
| MyoA | - | - | + Pf/Tg2,3,4,5 |
| MyoB | - | - | + Pf3,4,5 |
| MyoC | - | - | + Pf/Tg2,4 |
| MyoD | - | - | |
| MyoE | - | - | + Pf2,3,4,5 |
| MTIP | - | + Pf1 | + Pf4,6 |
| ELC1 | - | + Tg predicted* | |
| MLC1 | - | + Tg13 | +Tg4 |
| MLC2 | - | +Tg12 | +Tg4 |
| GAP40 | - | + Pf1 | + Pf2,3 |
| GAP45 | + Pf7 | + Pf1,8,13 | + Pf2,3,5 |
| GAP50 | - | + Pf1 | + Tg4 |
| GAP70 | +Tg predicted15 | +Tg predicted15 | + Tg4 |
| GAP80 | +Tg predicted15 | +Tg predicted15 | + Tg4 |
| GAPM1 | - | + Pf predicted* | + Pf4 |
| GAPM2 | - | + Pf1 | + Pf2,3,4 |
| GAPM3 | - | + Pf1 | + Pf5 |
| IAP1 | - | +Tg15 | +Tg4 |
| ISP1 | + Tg9+ Pf10 | + Tg9+ Pf10 | + Pf10 |
| ISP2 | + Tg9 | + Tg9 | + Tg4 |
| ISP3 | + Tg9+ Pf10 | + Tg9+ Pf10 | + Tg9+ Pf10 |
| ISP4 | - | + Tg11 | |
| TgIMC1, 4, 6, 9, 10, 11, 12, 13, 14, 15 | + Tg predicted14 | + Tg4 | |
| IMC1-c (ALV5) | + Pf1 | + Pf2,3,4,5 | |
| IMC1-e (ALV2) | + Pf1 | + Pf4,5 | |
| IMC1-f (ALV6) | +Pf predicted* | + Pf3 | |
| IMC1-g (ALV4) | + Pf1 | + Pf2,3,4,5 |
3.3.1 Glideosome associated proteins (GAP)
Three glideosome associated proteins (GAPs) are conserved between T. gondii and Plasmodium spp. (Frenal et al., 2010; Gaskins et al., 2004; Rees-Channer et al., 2006) and are notated by their apparent molecular weight, as determined by SDS-PAGE, as GAP40, GAP45, and GAP50. It has been demonstrated that GAP40, GAP45 and GAP50 co-localize with the MyoA-MTIP/MLC1 complex (Gaskins et al., 2004).
GAP45, MyoA, and MTIP/MLC1 form a precomplex mediated by the N-terminal domain of MTIP/MLC1 interacting with the C-terminus of the peripheral membrane protein GAP45 (Gaskins et al., 2004; Johnson et al., 2007; Rees-Channer et al., 2006). The structure of GAP45 is unknown, however, it is predicted that GAP45 consists primarily of a coiled-coil domain. At the N-terminus, GAP45 is myristoylated and palmitoylated (Rees-Channer et al., 2006; Wright et al., 2014b). This acylation targets GAP45 preferentially to the plasma membrane and serves to prevent GAP45 from associating with the nascent IMCs (Frenal et al., 2010). GAP45 has additional predicted palmitoylation sites at its C-terminus, and this modification is expected to facilitate GAP45 localization to the IMC, allowing the MyoA-MTIP/MLC1 complex to also associate with the IMC (Rees-Channer et al., 2006). Phosphorylation is the final step that controls association of the complex with the IMC (Gilk et al., 2009; Ridzuan et al., 2012). Phosphorylation of residues in the C-terminal domain of GAP45, controls association with the IMC. In addition to GAP45, two related proteins, present only in coccidians (e.g. T. gondii and Cryptosporidium spp.), GAP70 (Frenal et al., 2010) and GAP80 (Frenal et al., 2014b), have been identified and have been shown to interact with MyoA-MLC1 and MyoC-MLC1, respectively, in a manner similar to the interaction of GAP45 and MyoA-MLC1.
There still remains a question whether or not GAP45 is contacting both the IMC and plasma membrane simultaneously, spanning the supra-alveolar space. Measurements of the distance between the plasma membrane and IMC via cryo-EM, have ranged from 23–30 nm in P. berghei sporozoites (Kudryashev et al., 2010). An I-TASSER (Roy et al., 2010) homology model of PfGAP45 predicts a coiled-coil with a 180° bend in the middle (Figure 8A). This structure is approximately 15 nm in length when folded, and if extended, could potentially span the supraalveolar space to make contact with both membranes simultaneously.
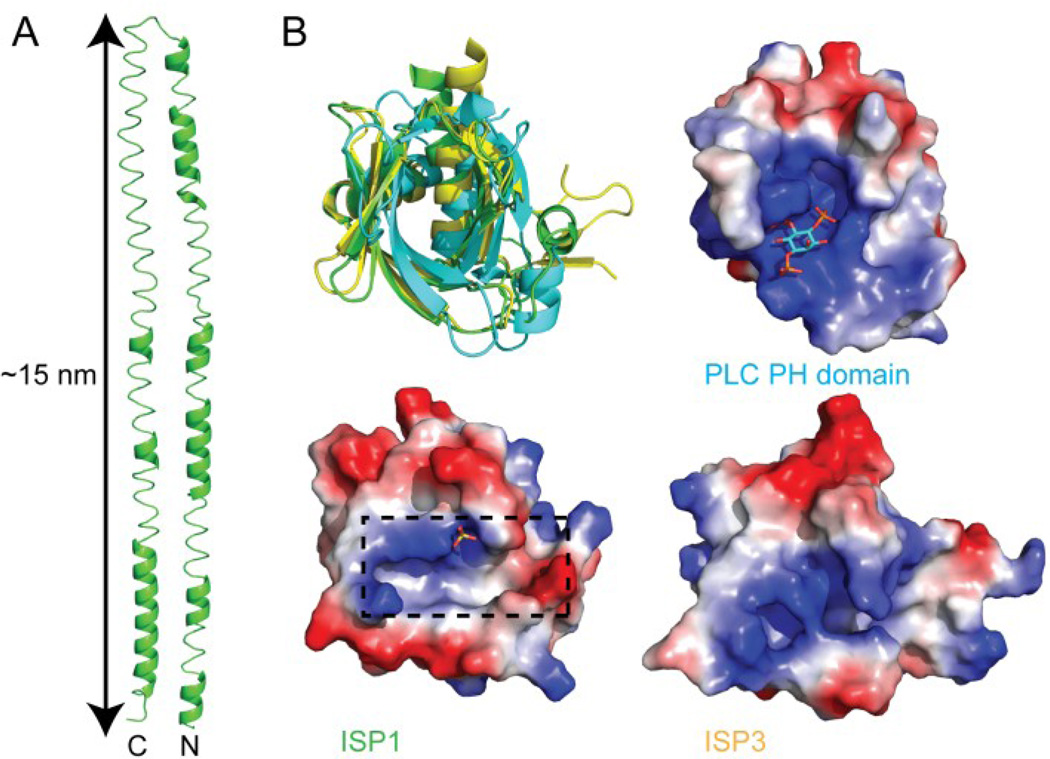
Figure 8. IMC associated proteins GAP45 and ISP1 and 3.
(A) I-TASSER homology model of PfGAP45 (green, cartoon) composed of coiled-coils spanning 15 nm. (B) Structures of the Pleckstrin homology domain of ISP1 (green, cartoon), ISP3 (yellow, cartoon), and structurally similar phospholipase C (green, cartoon). Calculated electrostatic surface potential for PLC and ISP1 and 3 with phospholipid bound to PLC and sulfate to ISP1 (sticks). Box indicates a potential protein binding site.
GAP45 also interacts with GAP50, a protein bound to the IMC (Gaskins et al., 2004). As previously mentioned, GAP50 contains a signal sequence for targeted secretion to the IMC, as well as a C-terminal transmembrane helix for insertion into the IMC vesicle. The likely orientation of GAP50 is with the N-terminal domain located within the lumen of the IMC (Johnson et al., 2007). The structure of GAP50 (Bosch et al., 2012) is most closely related to the human purple acid phosphatase; however, the recombinant protein shows no protein phosphatase activity in vitro, but does have activity against small molecules such as ATP and IPP (isopentenyl pyrophosphate), with a preference for di- and tri-phosphates (Müller et al., 2010). Due to the conserved nature of the protein and the lack of direct interaction other than via co-immunoprecipitation/ purification (co-IP) of GAP45 and GAP50, it is possible that there are other binding partners that are yet to be discovered.
The last member of the GAP family to be discussed is GAP40, a polytopic membrane protein (Frenal et al., 2010). GAP40 co-purifies with the MyoA-MTIP/MLC1-GAP45/50 complex. GAP40 is comprised of nine transmembrane spanning domains, and could act as an anchor of the motor to the IMC. The direct interactions of the GAP proteins with each other is still under investigation, but it has been noted that the glycosylation and phosphorylation states of the proteins play a role in their interactions with the other IMC components (Fauquenoy et al., 2011; Gilk et al., 2009).
3.3.2 Glideosome associated proteins with multiple membrane spans (GAPM)
The actomyosin motor is attached to the outermost side of the IMC alveoli membranes. In cryo-EM freeze fracture studies, 9 nm particles on the innermost, cytoplasmic side of the membrane of the IMC were observed (Raibaud et al., 2001). Investigation of the makeup of these particles led to the discovery of the glideosome associated proteins with multiple membrane spans (GAPMs) (Bullen et al., 2009). There are three GAPM variants (GAPM 1–3), which possess six transmembrane domains and are present at the IMC in large, detergent resistant oligomeric complexes. GAPM interacts with the alveolins, which form part of the parasite’s cytoskeletal network. The proteins also co-IP with both GAP50 and GAP45 of the IMC, indicating that GAPM may interact with other IMC associated proteins across the lumenal space of the IMC.
3.3.3 IMC sub-compartment proteins (ISPs)
The IMC sub-compartment proteins (ISP) are conserved across apicomplexans and are unique because the proteins are localized to different parts of the IMC along the parasite (Beck et al., 2010; Fung et al., 2012; Poulin et al., 2013). T. gondii encodes for ISPs 1–4, while only orthologs of ISP1 and ISP3 have been identified in Plasmodium spp. In T. gondii, ISP1 localizes to the apical end of the parasite, ISP2 localizes along the central region of the IMC, and ISP3 is able to localize to both the basal and central regions.
The ISPs do not possess signal sequences or transmembrane sequences; however, all ISPs are predicted to have palmitoylation and myristoylation sites near the N-terminus. The working theory is that targeting of the ISPs relies on recognition by an IMC bound palmitoyl acyl transferases (PAT) (Beck et al., 2010). Myristoylation of ISP1–3 would lead to nonspecific membrane association. If an ISP comes in contact with a PAT, it would be palmitoylated and bound to that membrane. PATs located at different parts of the IMC would control the specific localization of each ISP by only palmitoylating their specific substrate protein. In T. gondii, there are three putative membrane-bound O-acyl transferases, which typically palmitoylate secreted proteins, and 18 DHHC-CRD PATs, which typically palmitoylate non-secreted proteins for targeting to membranes (Beck et al., 2013; Resh, 2006). This information raises the questions as to whether there are multiple PATs capable of recognizing specific substrates, and how in turn the PATs would be localized to different sub-compartments of the IMC. A recent review by Frenal et al. (2014a) discusses palmitoylation and depalmitoylation cycles in T. gondii, and their role in protein localization.
Crystal structures of TgISP1 and TgISP3 (Tonkin et al., 2014a) reveal that the proteins adopt a Pleckstrin homology (PH) fold (Figure 8B), that is typically involved in phospholipid binding; however, there was no evidence of phospholipid binding. This result is likely due to a mutation of the residues that bind the phospholipid. Analysis of surface electrostatics show that the binding site in the ISP proteins is more hydrophobic and less positively charged than the phospholipid binding site from phospholipase C (Figure 8B). Analysis of the surface of ISP1 reveals a hydrophobic groove and a sulfate molecule bound near the groove leading to speculation that the protein has adapted to bind a protein partner possibly mediated by phosphorylation.
3.4 Subpellicular Network (SPN)
The inner membrane complex, made up of flattened membranes and associated with multiple, acylated peripheral membrane proteins and integral membrane proteins, interacts with the parasite’s subpellicular network (SPN) (Mann and Beckers, 2001). It was found in T. gondii that a network of 8–10 nm fibers were associated with the cytoplasmic face of the IMC.
3.4.1 Alveolins
The alveolin protein, TgIMC1 (ALV1), was identified as the main protein present in the 2D lattice of the SPN (Mann and Beckers, 2001). IMC1 shares homology with articulins, which are cytoskeleton proteins found in protists. The TgIMC1 orthologs for Plasmodium spp. were annotated as IMC1a-h (Khater et al., 2004). The alveolins, which vary in size, contain multiple repeat domains with a common motif of (E/D)(K/R)ΦΦ(E/D)ΦPx (Φ=hydrophobic and x=any amino acid), flanked by nonconserved regions on either side, and are predicted to have extended coiled-coil domains (Gould et al., 2008). It has been shown that knockout of IMC1a, IMC1b and IMC1h negatively affect the gliding motility of sporozoites and ookinetes and reduce infectivity (Khater et al., 2004; Tremp and Dessens, 2011; Volkmann et al., 2012). Connecting this cytoskeleton to the actomyosin motor, gliding motility and invasion/infectivity is likely accomplished via a connection with the IMC and GAPM1–3 proteins. The GAPMs co-purify with the alveolins, forming detergent-resistant oligomeric complexes, likely providing an anchoring point for the IMC to the subpellicular network (Bullen et al., 2009).
4. Exploring the essentiality of the adhesins and glideosome components
In the past year, several groups have published data regarding conditional knockouts of proteins key to the glideosome model in T. gondii. Conditional knockouts, using a DiCre-recombinase system, of ama1, aldolase, mic2, act1, gap45 and myoA have been generated in T. gondii and it has been shown that invasion still progresses, albeit at severely reduced levels (Andenmatten et al., 2013; Egarter et al., 2014; Frenal et al., 2014b; Shen and Sibley, 2014). Discussed below are these experiments and what the results mean in our understanding of the actomyosin motor based invasion by T. gondii, how this work translates to other apicomplexans, specifically Plasmodium spp., and alternative models that have been proposed for invasion. A summary of the conditional knockouts and the observed phenotypes, is presented in Table 4.
Table 4.
DiCre-recombinase inducible knockouts in T. gondii effects on egress and invasion phenotypes.
| Induced Knockout |
Egress | Invasion | Gliding | Compensatory Mechanism | Reference |
|---|---|---|---|---|---|
| MyoA | 2% | 16% | 37% | MyoC glideosome | Egarter et al., 2014 |
| MyoB/C | 100% | 100% | 100% | MyoA glideosome | Egarter et al., 2014 |
| MyoA/B/C | 2% | 5% | N.D. | MyoD? MyoE? (untested) | Egarter et al., 2014 |
| MLC1 | 5% | 28% | 42% | MyoD-MLC2? (untested) | Egarter et al., 2014 |
| GAP45 | 4% | 6% | 100% | GAP80 over-expression Frenal et al., 2014b | Egarter et al., 2014 |
| GAP80* | 85% | 80% | N.D. | GAP45 | Frenal et al., 2014b |
| GAP40 | N.D. | N.D. | N.D. | essential | Egarter et al., 2014 |
| GAP50 | N.D. | N.D. | N.D. | essential | Egarter et al., 2014 |
| Actin 1 | 2% | 10% | 10% | ? | Egarter et al., 2014 |
| Aldolase** | N.D. | 100% | 100% | GAPDH? | Shen and Sibley, 2014 |
| AMA1 | N.D. | ~25% | 100% | Other adhesins | Shen and Sibley, 2014 |
Phenotype from GAP80 clonal KO line with GAP45 iKO in absence of ATc. All other clones are inducible knockouts via the DiCre-recombinase system established in T. gondii.
Performed in low glucose conditions to alleviate toxicity of glycolysis intermediates.
4.1 AMA1 knockouts
A clonal knockout of ama1 in T. gondii tachyzoites and non-clonal knockout in P. berghei merozoites, were produced, which negatively affected host cell attachment (Bargieri et al., 2013); however, these parasite lines were still able to form a tight junction and invade cells in a smooth, one-step fashion. The merozoites and tachyzoites displayed a 3–5 fold lower invasion efficiency overall, but when normalized to the number of merozoites attached to cells, the parasites could invade cells better. It was concluded that AMA1 is dispensable for invasion by both P. berghei merozoites and sporozoites and T. gondii tachyzoites and opened up the possibility that the model of AMA1 and the RON complex as an essential component of the moving junction may be incorrect.
Following the study in T. gondii and P. berghei, an ama1 conditional knockout in P. falciparum was produced (Yap et al., 2014). Authors of the study concluded that AMA1 is not dispensable in merozoite invasion by P. falciparum. The ama1 gene was placed under conditional control in merozoites using the DiCre-recombinase system. Efficiency of deletion was ~80%. Because the deletion occurred after DNA-replication, due to the trophozoite specific promoter used to express the recombinase, and in combination with the imperfect deletion efficiency, there was a resulting mix of genetically null and wild-type (WT) merozoites. However, because micronemes, where AMA1 resides, form prior to segmentation, it was possible for genetically null parasites to have a range of AMA1 levels, which was observed with levels ranging from undetectable to near WT. The invasion efficiency of the PfAMA1 deficient merozoites was very low, and it was explained that there was likely insufficient AMA1 present to form a tight junction or to seal the invasion pore if a merozoite managed to have high enough levels of AMA1. One explanation offered by the authors to reconcile the P. falciparum results with the P. berghei and T. gondii data (Bargieri et al., 2013) is that both T. gondii and P. berghei possess orthologs/paralogs of AMA1, which could act as redundant adhesins; however, no redundancies have been identified in P. falciparum, which would make it essential for the model in which AMA1 plays a role in the moving junction. A more thorough detailing of this controversy is found in a review by Harvey et al. (2014).
4.2 Glideosome component knockouts
A conditional DiCre-recombinase system in T. gondii has been established and used to investigate the role of key invasion machinery components (Andenmatten et al., 2013; Egarter et al., 2014). Conditional/induced knockouts (iKO) were generated for mic2, myoA, gap40, gap45, gap50, mlc1, and act1. In the process of creating conditional knockouts, it was possible to isolate clonal knockouts of mic2 and myoA, whereas clonal knockouts could not be obtained of gap40, gap45, gap50, mlc1 or act1. It is important to remind readers that the following experiments were performed with T. gondii tachyzoites, and not Plasmodium spp. parasites.
4.2.1 MyoA knockout
In the myoA knockout, the Meissner group observed that the invasion rate was only 16% that of wild-type (WT) (Andenmatten et al., 2013; Egarter et al., 2014). Gliding motility was reduced by ~65% and tachyzoites moved mostly by circular gliding and in short steps. The myoA iKO parasites displayed different kinetics of invasion, with a few parasites entering smoothly and efficiently like WT in ~25 seconds, while the majority entered in a “stop-and-go” manner and could take up to 10 minutes to invade. The observation that myoA iKO parasites could still invade, albeit more slowly, suggested that MyoA is important for efficient invasion, but that there could also be an alternate force generation model. An explanation for the ability of myoA iKO invasion is that other myosins could act as a redundancy mechanism. The Soldati group has indicated that MLC1 is able to interact with MyoC (Jacot et al., 2014), and may be able to substitute for MyoA. To test this possibility, a triple conditional knockout of myoA/B/C was produced, resulting in a more severe phenotype than the single myoA iKO, however, parasites could still invade. It is still possible that the other myosins such as MyoD and MyoE, which were not knocked out, could also complement myoA/B/C function, providing T. gondii with multiple compensatory myosins.
4.2.2 MLC1 knockout
The mlc1 iKO caused mislocalization of MyoA, as expected, and reduced the ability to egress to 5% that of WT, and led to ~70% reduction in invasion (Egarter et al., 2014). This data suggests that MLC1 is important for host cell egress and MyoA localization, and that other MLCs are unable to fully complement this function. While there may not be another MLC capable of fully complementing the role of MLC1, it was noted by the authors that it was not possible to rule out the possibility that the MyoD-MLC2-motor (Polonais et al., 2011) could complement loss of the MyoA-MLC1-motor.
4.2.3 GAP45 knockout
The gap45 iKO resulted in cytosolic localization of the MyoA/MLC1 complex and affected the parasite morphology, due to separation of the IMC from the plasma membrane (Egarter et al., 2014). Along with a change in localization and morphology, depletion of GAP45 resulted in ~95% reduction in egress as well as invasion. However, gliding remained at 100% that of WT parasites, suggesting that morphology is the cause of invasion defects and not motility.
4.2.4 Actin 1 knockout
To further investigate the requirement of the actomyosin motor, the Meissner group generated a conditional knockout strain of act1 (Andenmatten et al., 2013; Egarter et al., 2014). A clonal act1 KO could not be isolated and conditional knockouts resulted in a defect in apicoplast segregation. Unlike in Plasmodium spp., it is not possible to supplement T. gondii cultures with IPP to determine if the act1 clonal KO is unable to be obtained solely due to an apicoplast defect. The conditional act1 knockout displayed gliding and invasion defects, but was still motile and could invade at ~10% that of WT, It was reasoned that actin 1 is not essential for invasion, only for apicoplast segregation or some other replicative process. As there is only one actin in T. gondii, if this actin is dispensable, an alternative to the glideosome would be needed.
To account for the ability to invade without actin, the authors propose a gelation-solation osmotic motor comprised of actin-like filaments (Egarter et al., 2014). However, if actin 1 is not required for invasion, as suggested by their data, and unlike Plasmodium spp., T. gondii does not possess actin 2, then what is the makeup of the gel? There is a family of actin-related proteins (Arps), also known as actin-like proteins (ALPs), members of the actin superfamily, which may be able to form this gel (Gordon and Sibley, 2005). Apicomplexans have 8–10 of these Arps and knockouts of the associated genes could answer the question of whether or not these proteins are involved in invasion and gliding motility. There is also a question of whether there is any remaining actin around. While it was reported that actin levels were undetectable via Western blot and fluorescence, and that would mean the concentration was lower than the critical concentration needed for filament formation (Egarter et al., 2014), it was noted previously that T. gondii actin 1 can form filaments at a concentration 3–4 times lower than canonical actin (Sahoo et al., 2006). So, it may be possible that filaments could still form from any remaining actin pools. More experiments will hopefully reveal the existence of either complementary motors or evidence for alternative motor models.
4.2.5 Aldolase conditional knockout
In addition to the knockouts performed by the Meissner group, the Sibley group was able to generate an aldolase conditional knockout in T. gondii, that was able to invade fibroblasts under low glucose conditions (Shen and Sibley, 2014). The low glucose conditions prevented accumulation of toxic glycolysis intermediates, enabling the aldolase iKO tachyzoites to invade in the absence of aldolase. The glucose data is interesting, but explanations of possible redundancies, we believe, are not adequately addressed.
Only briefly discussed in the Sibley paper (2014) is the fact that GAPDH, another known bridging protein, could possibly rescue the invasion of the parasite in the absence of aldolase. Reported mutations that can disrupt the AMA1 interaction with GAPDH, did not lead to inhibition of invasion, supporting the case that GAPDH is not necessary for invasion. However, it is known in T. gondii that the AMA1 pathway is nonessential and is likely a redundant pathway to invasion (Bargieri et al., 2013). So, a clone with an AMA1 defective in both aldolase and GAPDH binding that can invade, does not prove the non-essentiality of GAPDH. A conditional knockout of both GAPDH and aldolase would be needed to provide more evidence that bridging to this motor is not necessary. Also, this point recalls the evidence presented earlier that while AMA1 seems to be dispensable for T. gondii and it is possible that paralogs/orthologs to AMA1 are able to compensate for AMA1 loss, that AMA1 is essential for tight junction formation and pore closure in P. falciparum (Yap et al., 2014).
4.2.6 Egress phenotypes
Discussion has focused on the invasion and gliding phenotypes, yet many of these conditional knockouts blocked host cell egress. Host cell egress is blocked in myoA/B/C triple knockouts as well as the act1, mlc1, gap40, gap45, and gap50 conditional knockouts (Andenmatten et al., 2013; Egarter et al., 2014). The question that arises is this: Why is egress blocked when proteins, traditionally associated with the invasion machinery, are knocked out? Clearly, while these proteins could be considered nonessential for invasion based on the ability of the conditional knockouts to invade host cells, it is also likely that either proper localization or expression of the invasion machinery associated proteins, acts as a regulator of egress. It is possible that proper glideosome assembly acts as a checkpoint, so that only parasites that are fully competent for invasion are released.
4.2.7 Discussion of alternative invasion mechanisms and knockouts
It is important to keep in mind that the conditional knockout of glideosome components discussed here has only been performed in T. gondii (Andenmatten et al., 2013; Egarter et al., 2014), and that while it has been presented that apicomplexans share these invasion proteins, that the mechanisms of invasion may be different between the genera. However, it is also important to take these data and begin to look for alternative models and glideosomes to explain a mechanism of host cell invasion.
One possibility to explain conditional knockouts of the MyoA glideosome components in T. gondii, is that the MyoA-MLC1 complex is only one of multiple, redundant motors (e.g. MyoD-MLC2, MyoC-MLC1). Recent work has described a novel glideosome in T. gondii involving MyoC (Frenal et al., 2014b). It has been demonstrated that GAP80, a member of the GAP45 family, is able to recruit MyoC to the basal polar ring with the help of IMC-associated protein 1 (IAP1). Previous work had already shown that MyoC could interact with MLC1 (Polonais et al., 2011). MyoC, like MyoA, also interacts with the IMC embedded proteins, GAP40 and GAP50, and forms the MyoC-MLC1-GAP80-IAP1-GAP40-GAP50 glideosome (Frenal et al., 2014b).
In WT parasites, the MyoA-MLC1-GAP45-GAP40-GAP50 glideosome functions along the parasite periphery, while the MyoA-MLC1-GAP70-GAP40-GAP50 glideosome functions at the apical cap and the MyoC-MLC1-GAP80-IAP1-GAP40-GAP50 glideosome functions at the basal polar ring (Figure 9A, 9B). Depletion of MyoA in a conditional knockout strain leads to a redistribution of the MyoC glideosome along the parasite periphery and into the apical cap, likely accounting for the observed low levels of invasion. In gap80 and gap70 iKOs, GAP45 can assume their roles, but neither GAP80 nor GAP70 could complement GAP45 function. If the conditional knockout work done in T. gondii is replicated in Plasmodium spp. and invasion is observed, these alternative glideosomes with GAP70 and GAP80, would not be a possible explanation, since GAP70 and GAP80 are only present in coccidians. However, the demonstrated plasticity of the glideosomes in T. gondii, and the ability of the MyoC glideosome to redistribute along the parasite in the absence of the MyoA glideosome, would lead one to speculate that there is a similar plasticity and redundancy mechanisms for Plasmodium spp. to allow for invasion.
Alternatively, it has been suggested that there are other modes of invasion (linear glideosome versus gelation-solation model). Perhaps multiple motors exist and have different roles in invasion. It has been suggested that a linear glideosome motor is needed for efficiency in reorientation, tight junction formation, and the start of invasion and another motor completes invasion once it is going. Another possibility is that invasion is regulated by the host cell via a host cell mediated uptake or autophagy/phagocytosis pathway. Regardless, these conditional knockouts of the glideosome components do cause severe phenotypes, with a 95% reduction in invasion in some cases (Andenmatten et al., 2013; Egarter et al., 2014; Frenal et al., 2014b; Shen and Sibley, 2014), and are still worthy of continued study and translational work to develop inhibitors of these glideosomes.
5. Conclusion
When this structural survey was performed, approximately 50 protein structures of apicomplexan adhesin proteins and host cell receptors had been deposited with the PDB. Many of these structures concern key domains that have been previously implicated in receptor interactions or have been targeted by antibodies during the course of vaccine development. Antibody-adhesin co-structures are useful to structurally identify their mechanism of interaction and inhibition, to help guide antibody development to regions of the adhesin that are both accessible and needed for receptor engagement or adhesin function. Structures of receptor domains bound to ligand or peptide have provided insight into assembly and recognition as well as clues as to how to disrupt them, which may lead to new therapeutics. Structural, biochemical, and cellular/genetic investigations are needed in the future to understand how apicomplexan adhesin proteins interact with ligands, as well as to discover their interacting partners.
Around 35 structures related to the glideosome machinery have been deposited, accounting for ~60% of the glideosome complex. Lacking are apicomplexan structures of the unique Myosin A, as well as ISP2, GAP40, GAP45 and GAPM1–3. With successful purification of MyoA, it may not be much longer before a structure is obtained, providing a better understanding of how this unique myosin operates. Structures of GAPM1–3 may be difficult due to the multiple transmembrane passes and GAP45 due to the predicted extended-coiled-coil structure. Additionally, structures can provide key information regarding complex formation that enables the targeting of protein-protein interactions of the invasion machinery with small molecule disruptors or stabilizers.
Multiple papers were released implicating that many components of this machinery were not essential in T. gondii and called for the proposal of alternate mechanisms of invasion. This review has summarized the results of these papers and presented evidence that seems to be contradictory, including the knockout results from the Soldati group, which showed plasticity of the glideosomes and identification of novel glideosome complexes. Further work to use a conditional knockout system in P. falciparum should address whether these components are essential for invasion in other apicomplexans. There are still unanswered questions such as why there is an egress block associated with machinery component knockouts, and what is the composition of an alternative motor. Keeping an open mind to the possibility that the established model may represent only one of several mechanisms of invasion, is important. However, the reports of the invasion machinery’s non-essentiality seem to be premature in light of recent work, meaning that further investigation will be necessary and the glideosome is still a viable area for study to develop novel anti-parasitics.
Supplementary Material
Acknowledgements
We would like to acknowledge the contributions of all the groups working on understanding the cellular, molecular, and structural details of parasite attachment and invasion. We apologize for any studies that we may have overlooked. Additionally, we would like to give our sincere thanks to the referees for this paper, whose thoroughness and insightful critiques have helped enormously in developing, expanding, and refining this review.
This work was supported by funding from The Bloomberg Family Foundation (J.B.), and the NIH Chemistry-Biology Interface Training Grant (T32GM080189) and a Predoctoral Fellowship from the Johns Hopkins Malaria Research Institute (L.E.B.).
Abbreviations
- Pf
Plasmodium falciparum
- Pb
Plasmodium berghei
- Pv
Plasmodium vivax
- Py
Plasmodium yoelii
- Pk
Plasmodium knowlesi
- Tg
Toxoplasma gondii
- ACT1
actin 1
- AMA1
apical membrane antigen 1
- AUC
analytical ultracentrifugation
- DARC
Duffy antigen/chemokine receptor
- DBL
Duffy binding-like domain
- DBP
Duffy binding-like protein
- DLS
dynamic light scattering
- DSF
differential scanning fluorimetry
- EBA
erythrocyte binding antigen
- EBD
erythrocyte binding domain
- EBL
erythrocyte binding-like protein
- EGF
epidermal growth factor
- Fab
fragment antibody
- GAP
glideosome associated protein
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GAPM
glideosome associated proteins with multiple membrane spans
- IgNAR
immunoglobulin new antigen receptor
- IAP1
IMC associated protein 1
- iKO
DiCre-recombinase induced knockout
- IMC
inner membrane complex
- IMC1-x
inner membrane complex protein 1-x
- IPP
isopentenyl pyrophosphate
- ISP
IMC sub-compartment proteins
- ITC
isothermal titration calorimetry
- MALS
multi-angle light scattering
- MIC2
microneme-associated protein 2
- MLC
myosin light chain
- MSP
merozoite surface protein
- MTIP
myosin tail interacting protein
- MTRAP
merozoite thrombospondin related anonymous protein
- MyoA
myosin A
- NBD
nucleotide binding domain
- PAN
plasminogen, apple, nematode
- RBP
reticulocyte binding-like protein
- RH
reticulocyte binding-like homolog
- RIPR
RH5 interacting protein
- RON
rhoptry neck protein
- TSR
thrombospondin repeat domain
- TRAP
thrombospondin related anonymous protein
- SAXS
small angle X-ray scattering
- SEC
size exclusion chromatography
- SPN
subpellicular network
- VWA
von Willebrand factor type A domain
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Lauren E. Boucher, Email: lboucher@jhu.edu.
Jürgen Bosch, Email: jbosch@jhu.edu.
Bibliography
- Adams JH, Sim BK, Dolan SA, Fang X, Kaslow DC, Miller LH. A family of erythrocyte binding proteins of malaria parasites. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DL, Mital J, Ward GE, Bradley P, Boothroyd JC. Identification of the moving junction complex of Toxoplasma gondii: a collaboration between distinct secretory organelles. PLoS pathogens. 2005;1:e17. doi: 10.1371/journal.ppat.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen ML, Dobrowolski JM, Muller H, Sibley LD, Mansour TE. Cloning and characterization of actin depolymerizing factor from Toxoplasma gondii. Molecular and biochemical parasitology. 1997;88:43–52. doi: 10.1016/s0166-6851(97)00069-8. [DOI] [PubMed] [Google Scholar]
- Andenmatten N, Egarter S, Jackson AJ, Jullien N, Herman JP, Meissner M. Conditional genome engineering in Toxoplasma gondii uncovers alternative invasion mechanisms. Nature methods. 2013;10:125–127. doi: 10.1038/nmeth.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson-White BR, Ivey FD, Cheng K, Szatanek T, Lorestani A, Beckers CJ, Ferguson DJ, Sahoo N, Gubbels MJ. A family of intermediate filament-like proteins is sequentially assembled into the cytoskeleton of Toxoplasma gondii. Cellular microbiology. 2011;13:18–31. doi: 10.1111/j.1462-5822.2010.01514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C, Heiges M, Wang H, Wang Z, Fischer S, Rhodes P, Miller J, Kraemer E, Stoeckert CJ, Jr, Roos DS, Kissinger JC. ApiDB: integrated resources for the apicomplexan bioinformatics resource center. Nucleic acids research. 2007;35:D427–D430. doi: 10.1093/nar/gkl880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai T, Becker M, Gupta A, Strike P, Murphy VJ, Anders RF, Batchelor AH. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12736–12741. doi: 10.1073/pnas.0501808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargieri DY, Andenmatten N, Lagal V, Thiberge S, Whitelaw JA, Tardieux I, Meissner M, Menard R. Apical membrane antigen 1 mediates apicomplexan parasite attachment but is dispensable for host cell invasion. Nature communications. 2013;4:2552. doi: 10.1038/ncomms3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholdson SJ, Bustamante LY, Crosnier C, Johnson S, Lea S, Rayner JC, Wright GJ. Semaphorin-7A is an erythrocyte receptor for P. falciparum merozoite-specific TRAP homolog, MTRAP. PLoS pathogens. 2012;8:e1003031. doi: 10.1371/journal.ppat.1003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor JD, Zahm JA, Tolia NH. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nature structural & molecular biology. 2011;18:908–914. doi: 10.1038/nsmb.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor JD, Malpede BM, Omattage NS, Dekoster GT, Henzler-Wildman KA, Tolia NH. Red Blood Cell Invasion by Plasmodium vivax: Structural Basis for DBP Engagement of DARC. PLoS pathogens. 2014;10:e1003869. doi: 10.1371/journal.ppat.1003869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J, Richard D, Healer J, Rug M, Krnajski Z, Gilberger TW, Green JL, Holder AA, Cowman AF. A conserved molecular motor drives cell invasion and gliding motility across malaria life cycle stages and other apicomplexan parasites. The Journal of biological chemistry. 2006;281:5197–5208. doi: 10.1074/jbc.M509807200. [DOI] [PubMed] [Google Scholar]
- Baum J, Tonkin CJ, Paul AS, Rug M, Smith BJ, Gould SB, Richard D, Pollard TD, Cowman AF. A malaria parasite formin regulates actin polymerization and localizes to the parasite-erythrocyte moving junction during invasion. Cell host & microbe. 2008;3:188–198. doi: 10.1016/j.chom.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Beck JR, Rodriguez-Fernandez IA, de Leon JC, Huynh MH, Carruthers VB, Morrissette NS, Bradley PJ. A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS pathogens. 2010;6:e1001094. doi: 10.1371/journal.ppat.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JR, Fung C, Straub KW, Coppens I, Vashisht AA, Wohlschlegel JA, Bradley PJ. A Toxoplasma palmitoyl acyl transferase and the palmitoylated armadillo repeat protein TgARO govern apical rhoptry tethering and reveal a critical role for the rhoptries in host cell invasion but not egress. PLoS pathogens. 2013;9:e1003162. doi: 10.1371/journal.ppat.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman LW, Kaiser K, Fujioka H, Coppens I, Daly TM, Fox S, Matuschewski K, Nussenzweig V, Kappe SH. Myosin A tail domain interacting protein (MTIP) localizes to the inner membrane complex of Plasmodium sporozoites. Journal of cell science. 2003;116:39–49. doi: 10.1242/jcs.00194. [DOI] [PubMed] [Google Scholar]
- Besteiro S, Michelin A, Poncet J, Dubremetz JF, Lebrun M. Export of a Toxoplasma gondii rhoptry neck protein complex at the host cell membrane to form the moving junction during invasion. PLoS pathogens. 2009;5:e1000309. doi: 10.1371/journal.ppat.1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookwalter CS, Kelsen A, Leung JM, Ward GE, Trybus KM. A Toxoplasma gondii Class XIV Myosin, Expressed in Sf9 Cells with a Parasite Co-chaperone, Requires Two Light Chains for Fast Motility. The Journal of biological chemistry. 2014;289:30832–30841. doi: 10.1074/jbc.M114.572453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch J, Paige MH, Vaidya AB, Bergman LW, Hol WG. Crystal structure of GAP50, the anchor of the invasion machinery in the inner membrane complex of Plasmodium falciparum. Journal of structural biology. 2012;178:61–73. doi: 10.1016/j.jsb.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch J, Turley S, Roach CM, Daly TM, Bergman LW, Hol WG. The closed MTIP-myosin A-tail complex from the malaria parasite invasion machinery. Journal of molecular biology. 2007a;372:77–88. doi: 10.1016/j.jmb.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch J, Buscaglia CA, Krumm B, Ingason BP, Lucas R, Roach C, Cardozo T, Nussenzweig V, Hol WG. Aldolase provides an unusual binding site for thrombospondin-related anonymous protein in the invasion machinery of the malaria parasite. Proceedings of the National Academy of Sciences of the United States of America. 2007b;104:7015–7020. doi: 10.1073/pnas.0605301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch J, Turley S, Daly TM, Bogh SM, Villasmil ML, Roach C, Zhou N, Morrisey JM, Vaidya AB, Bergman LW, Hol WG. Structure of the MTIP-MyoA complex, a key component of the malaria parasite invasion motor. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4852–4857. doi: 10.1073/pnas.0510907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher LE, Bosch J. Development of a multifunctional tool for drug screening against plasmodial protein-protein interactions via surface plasmon resonance. Journal of molecular recognition : JMR. 2013;26:496–500. doi: 10.1002/jmr.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher LE, Bosch J. Structure of Toxoplasma gondii fructose-1,6-bisphosphate aldolase. Acta Crystallographica. Section F, Structural Biology Communications. 2014;70:1186–1192. doi: 10.1107/S2053230X14017087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley PJ, Ward C, Cheng SJ, Alexander DL, Coller S, Coombs GH, Dunn JD, Ferguson DJ, Sanderson SJ, Wastling JM, Boothroyd JC. Proteomic analysis of rhoptry organelles reveals many novel constituents for host-parasite interactions in Toxoplasma gondii. The Journal of biological chemistry. 2005;280:34245–34258. doi: 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- Brossier F, David Sibley L. Toxoplasma gondii: microneme protein MIC2. The international journal of biochemistry & cell biology. 2005;37:2266–2272. doi: 10.1016/j.biocel.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Bullen HE, Tonkin CJ, O'Donnell RA, Tham WH, Papenfuss AT, Gould S, Cowman AF, Crabb BS, Gilson PR. A novel family of Apicomplexan glideosome-associated proteins with an inner membrane-anchoring role. The Journal of biological chemistry. 2009;284:25353–25363. doi: 10.1074/jbc.M109.036772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaglia CA, Coppens I, Hol WG, Nussenzweig V. Sites of interaction between aldolase and thrombospondin-related anonymous protein in plasmodium. Molecular biology of the cell. 2003;14:4947–4957. doi: 10.1091/mbc.E03-06-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor EM, Lombo TB, Cepeda A, Espinosa AM, Barrero CA, Guzman F, Gunturiz ML, Urquiza M, Ocampo M, Patarroyo ME, Patarroyo MA. Plasmodium vivax: functional analysis of a highly conserved PvRBP-1 protein region. Molecular and biochemical parasitology. 2001;117:229–234. doi: 10.1016/s0166-6851(01)00355-3. [DOI] [PubMed] [Google Scholar]
- Cao J, Kaneko O, Thongkukiatkul A, Tachibana M, Otsuki H, Gao Q, Tsuboi T, Torii M. Rhoptry neck protein RON2 forms a complex with microneme protein AMA1 in Plasmodium falciparum merozoites. Parasitology international. 2009;58:29–35. doi: 10.1016/j.parint.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Cardozo TJ, Bosch J, Nemetski SM. Small molecule malarial aldolase-trap enhancers and glideosome inhibitors. 2013 Vol. PCT/US2012/061875, A61K 31/166 (2006.01); A61K 31/16 (2006.01); A61K 38/16 (2006.01); A61K 38/17 (2006.01) ed. [Google Scholar]
- Chen E, Paing MM, Salinas N, Sim BK, Tolia NH. Structural and functional basis for inhibition of erythrocyte invasion by antibodies that target Plasmodium falciparum EBA-175. PLoS pathogens. 2013;9:e1003390. doi: 10.1371/journal.ppat.1003390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lopaticki S, Riglar DT, Dekiwadia C, Uboldi AD, Tham WH, O'Neill MT, Richard D, Baum J, Ralph SA, Cowman AF. An EGF-like protein forms a complex with PfRh5 and is required for invasion of human erythrocytes by Plasmodium falciparum. PLoS pathogens. 2011;7:e1002199. doi: 10.1371/journal.ppat.1002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley AM, Gupta A, Murphy VJ, Bai T, Kim H, Foley M, Anders RF, Batchelor AH. Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody. PLoS pathogens. 2007;3:1308–1319. doi: 10.1371/journal.ppat.0030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MO, Wright JC, Jones M, Rayner JC, Choudhary JS. Confident and sensitive phosphoproteomics using combinations of collision induced dissociation and electron transfer dissociation. Journal of proteomics. 2014;103:1–14. doi: 10.1016/j.jprot.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes A, Mellombo M, Mueller I, Benet A, Reeder JC, Anders RF. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infection and immunity. 2003;71:1416–1426. doi: 10.1128/IAI.71.3.1416-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman AF, Berry D, Baum J. The cellular and molecular basis for malaria parasite invasion of the human red blood cell. The Journal of cell biology. 2012;198:961–971. doi: 10.1083/jcb.201206112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford J, Tonkin ML, Grujic O, Boulanger MJ. Structural characterization of apical membrane antigen 1 (AMA1) from Toxoplasma gondii. The Journal of biological chemistry. 2010;285:15644–15652. doi: 10.1074/jbc.M109.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C, Bustamante LY, Bartholdson SJ, Bei AK, Theron M, Uchikawa M, Mboup S, Ndir O, Kwiatkowski DP, Duraisingh MT, Rayner JC, Wright GJ. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature. 2011;480:534–537. doi: 10.1038/nature10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher W, Plattner F, Carlier MF, Soldati-Favre D. Concerted action of two formins in gliding motility and host cell invasion by Toxoplasma gondii. PLoS pathogens. 2010;6:e1001132. doi: 10.1371/journal.ppat.1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowolski JM, Niesman IR, Sibley LD. Actin in the parasite Toxoplasma gondii is encoded by a single copy gene, ACT1 and exists primarily in a globular form. Cell motility and the cytoskeleton. 1997;37:253–262. doi: 10.1002/(SICI)1097-0169(1997)37:3<253::AID-CM7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Douglas AD, Williams AR, Knuepfer E, Illingworth JJ, Furze JM, Crosnier C, Choudhary P, Bustamante LY, Zakutansky SE, Awuah DK, Alanine DG, Theron M, Worth A, Shimkets R, Rayner JC, Holder AA, Wright GJ, Draper SJ. Neutralization of Plasmodium falciparum merozoites by antibodies against PfRH5. Journal of immunology (Baltimore, Md. : 1950) 2014;192:245–258. doi: 10.4049/jimmunol.1302045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douse CH, Vrielink N, Wenlin Z, Cota E, Tate EW. Targeting a Dynamic Protein-Protein Interaction: Fragment Screening against the Malaria Myosin A Motor Complex. ChemMedChem. 2014 doi: 10.1002/cmdc.201402357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douse CH, Mass SJ, Thomas JC, Garnett JA, Cota E, Tate EW. Structural examination of constrained Plasmodium falciparum myosin A peptide helices using pentenyl glycine stapling and hydrogen bond surrogate approaches. Protein Data Bank. 2013 [Google Scholar]
- Douse CH, Green JL, Salgado PS, Simpson PJ, Thomas JC, Langsley G, Holder AA, Tate EW, Cota E. Regulation of the Plasmodium motor complex: phosphorylation of myosin A tail-interacting protein (MTIP) loosens its grip on MyoA. The Journal of biological chemistry. 2012;287:36968–36977. doi: 10.1074/jbc.M112.379842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egarter S, Andenmatten N, Jackson AJ, Pall G, Black JA, Ferguson DJP, Tardieux I, Mogilner A, Meissner M. The Toxoplasma Acto-MyoA motor complex is important but not essential for gliding motility and host cell invasion. bioRxiv. 2014 doi: 10.1371/journal.pone.0091819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Grebert HM, Chaiyaroj SC, Magris M, Biswas S, Nahlen BL, Lal AA. Polymorphism in the gene encoding the apical membrane antigen-1 (AMA-1) of Plasmodium falciparum. X. Asembo Bay Cohort Project. Molecular and biochemical parasitology. 2001;113:279–287. doi: 10.1016/s0166-6851(01)00229-8. [DOI] [PubMed] [Google Scholar]
- Fauquenoy S, Hovasse A, Sloves PJ, Morelle W, Dilezitoko Alayi T, Slomianny C, Werkmeister E, Schaeffer C, Van Dorsselaer A, Tomavo S. Unusual N-glycan structures required for trafficking Toxoplasma gondii GAP50 to the inner membrane complex regulate host cell entry through parasite motility. Molecular & cellular proteomics : MCP. 2011;10 doi: 10.1074/mcp.M111.008953. M111 008953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng ZP, Keizer DW, Stevenson RA, Yao S, Babon JJ, Murphy VJ, Anders RF, Norton RS. Structure and inter-domain interactions of domain II from the blood-stage malarial protein, apical membrane antigen 1. Journal of molecular biology. 2005;350:641–656. doi: 10.1016/j.jmb.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Frenal K, Kemp LE, Soldati-Favre D. Emerging roles for protein S-palmitoylation in Toxoplasma biology. International journal for parasitology. 2014a;44:121–131. doi: 10.1016/j.ijpara.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Frenal K, Marq JB, Jacot D, Polonais V, Soldati-Favre D. Plasticity between MyoC- and MyoA-Glideosomes: An Example of Functional Compensation in Toxoplasma gondii Invasion. PLoS pathogens. 2014b;10:e1004504. doi: 10.1371/journal.ppat.1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenal K, Polonais V, Marq JB, Stratmann R, Limenitakis J, Soldati-Favre D. Functional dissection of the apicomplexan glideosome molecular architecture. Cell host & microbe. 2010;8:343–357. doi: 10.1016/j.chom.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Fung C, Beck JR, Robertson SD, Gubbels MJ, Bradley PJ. Toxoplasma ISP4 is a central IMC sub-compartment protein whose localization depends on palmitoylation but not myristoylation. Molecular and biochemical parasitology. 2012;184:99–108. doi: 10.1016/j.molbiopara.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtado PB, Huang CY, Ihyembe D, Hammond RA, Marsh HC, Perkins SJ. The partly folded back solution structure arrangement of the 30 SCR domains in human complement receptor type 1 (CR1) permits access to its C3b and C4b ligands. Journal of molecular biology. 2008;375:102–118. doi: 10.1016/j.jmb.2007.09.085. [DOI] [PubMed] [Google Scholar]
- Gaffar FR, Yatsuda AP, Franssen FF, de Vries E. A Babesia bovis merozoite protein with a domain architecture highly similar to the thrombospondin-related anonymous protein (TRAP) present in Plasmodium sporozoites. Molecular and biochemical parasitology. 2004;136:25–34. doi: 10.1016/j.molbiopara.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Galinski MR, Medina CC, Ingravallo P, Barnwell JW. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell. 1992;69:1213–1226. doi: 10.1016/0092-8674(92)90642-p. [DOI] [PubMed] [Google Scholar]
- Ganter M, Schuler H, Matuschewski K. Vital role for the Plasmodium actin capping protein (CP) beta-subunit in motility of malaria sporozoites. Molecular microbiology. 2009;74:1356–1367. doi: 10.1111/j.1365-2958.2009.06828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Yeo KP, Aw SS, Kuss C, Iyer JK, Genesan S, Rajamanonmani R, Lescar J, Bozdech Z, Preiser PR. Antibodies targeting the PfRH1 binding domain inhibit invasion of Plasmodium falciparum merozoites. PLoS pathogens. 2008;4:e1000104. doi: 10.1371/journal.ppat.1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskins E, Gilk S, DeVore N, Mann T, Ward G, Beckers C. Identification of the membrane receptor of a class XIV myosin in Toxoplasma gondii. The Journal of cell biology. 2004;165:383–393. doi: 10.1083/jcb.200311137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilberger TW, Thompson JK, Triglia T, Good RT, Duraisingh MT, Cowman AF. A novel erythrocyte binding antigen-175 paralogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. The Journal of biological chemistry. 2003;278:14480–14486. doi: 10.1074/jbc.M211446200. [DOI] [PubMed] [Google Scholar]
- Gilk SD, Gaskins E, Ward GE, Beckers CJ. GAP45 phosphorylation controls assembly of the Toxoplasma myosin XIV complex. Eukaryotic cell. 2009;8:190–196. doi: 10.1128/EC.00201-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JL, Sibley LD. Comparative genome analysis reveals a conserved family of actin-like proteins in apicomplexan parasites. BMC genomics. 2005;6:179. doi: 10.1186/1471-2164-6-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Tham WH, Cowman AF, McFadden GI, Waller RF. Alveolins, a new family of cortical proteins that define the protist infrakingdom Alveolata. Molecular biology and evolution. 2008;25:1219–1230. doi: 10.1093/molbev/msn070. [DOI] [PubMed] [Google Scholar]
- Green JL, Rees-Channer RR, Howell SA, Martin SR, Knuepfer E, Taylor HM, Grainger M, Holder AA. The motor complex of Plasmodium falciparum: phosphorylation by a calcium-dependent protein kinase. The Journal of biological chemistry. 2008;283:30980–30989. doi: 10.1074/jbc.M803129200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A, Gunalan K, Ramalingam JK, Manimekalai MS, Gruber G, Preiser PR. Structural characterization of the erythrocyte binding domain of the reticulocyte binding protein homologue family of Plasmodium yoelii. Infection and immunity. 2011;79:2880–2888. doi: 10.1128/IAI.01326-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A, Manimekalai MS, Balakrishna AM, Hunke C, Jeyakanthan J, Preiser PR, Gruber G. Structural determination of functional units of the nucleotide binding domain (NBD94) of the reticulocyte binding protein Py235 of Plasmodium yoelii. PloS one. 2010;5:e9146. doi: 10.1371/journal.pone.0009146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KL, Yap A, Gilson PR, Cowman AF, Crabb BS. Insights and controversies into the role of the key apicomplexan invasion ligand, Apical Membrane Antigen 1. International journal for parasitology. 2014;44:853–857. doi: 10.1016/j.ijpara.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Haynes JD, Dalton JP, Klotz FW, McGinniss MH, Hadley TJ, Hudson DE, Miller LH. Receptor-like specificity of a Plasmodium knowlesi malarial protein that binds to Duffy antigen ligands on erythrocytes. The Journal of experimental medicine. 1988;167:1873–1881. doi: 10.1084/jem.167.6.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzelman MB, Schwartzman JD. A novel class of unconventional myosins from Toxoplasma gondii. Journal of molecular biology. 1997;271:139–146. doi: 10.1006/jmbi.1997.1167. [DOI] [PubMed] [Google Scholar]
- Heintzelman MB, Schwartzman JD. Characterization of myosin-A and myosin-C: two class XIV unconventional myosins from Toxoplasma gondii. Cell motility and the cytoskeleton. 1999;44:58–67. doi: 10.1002/(SICI)1097-0169(199909)44:1<58::AID-CM5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Heiss K, Nie H, Kumar S, Daly TM, Bergman LW, Matuschewski K. Functional characterization of a redundant Plasmodium TRAP family invasin, TRAP-like protein, by aldolase binding and a genetic complementation test. Eukaryotic cell. 2008;7:1062–1070. doi: 10.1128/EC.00089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KA, Streltsov VA, Coley AM, Dolezal O, Hudson PJ, Batchelor AH, Gupta A, Bai T, Murphy VJ, Anders RF, Foley M, Nuttall SD. Structure of an IgNAR-AMA1 complex: targeting a conserved hydrophobic cleft broadens malarial strain recognition. Structure. 2007;15:1452–1466. doi: 10.1016/j.str.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Herm-Gotz A, Weiss S, Stratmann R, Fujita-Becker S, Ruff C, Meyhofer E, Soldati T, Manstein DJ, Geeves MA, Soldati D. Toxoplasma gondii myosin A and its light chain: a fast, single-headed, plus-end-directed motor. The EMBO journal. 2002;21:2149–2158. doi: 10.1093/emboj/21.9.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel DM, Mui S, O'Neall-Hennessey E, Szent-Gyorgyi AG, Cohen C. The on-off switch in regulated myosins: different triggers but related mechanisms. Journal of molecular biology. 2009;394:496–505. doi: 10.1016/j.jmb.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hliscs M, Sattler JM, Tempel W, Artz JD, Dong A, Hui R, Matuschewski K, Schuler H. Structure and function of a G-actin sequestering protein with a vital role in malaria oocyst development inside the mosquito vector. The Journal of biological chemistry. 2010;285:11572–11583. doi: 10.1074/jbc.M109.054916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodder AN, Crewther PE, Matthew ML, Reid GE, Moritz RL, Simpson RJ, Anders RF. The disulfide bond structure of Plasmodium apical membrane antigen-1. The Journal of biological chemistry. 1996;271:29446–29452. doi: 10.1074/jbc.271.46.29446. [DOI] [PubMed] [Google Scholar]
- Holder AA, Keen JK, Sinha KA, Brown KN. The 235 kD rhoptry protein of Plasmodium yoelii. Acta Leidensia. 1991;60:101–106. [PubMed] [Google Scholar]
- Igonet S, Vulliez-Le Normand B, Faure G, Riottot MM, Kocken CH, Thomas AW, Bentley GA. Cross-reactivity studies of an anti-Plasmodium vivax apical membrane antigen 1 monoclonal antibody: binding and structural characterisation. Journal of molecular biology. 2007;366:1523–1537. doi: 10.1016/j.jmb.2006.12.028. [DOI] [PubMed] [Google Scholar]
- Iyer J, Gruner AC, Renia L, Snounou G, Preiser PR. Invasion of host cells by malaria parasites: a tale of two protein families. Molecular microbiology. 2007;65:231–249. doi: 10.1111/j.1365-2958.2007.05791.x. [DOI] [PubMed] [Google Scholar]
- Jacot D, Frenal K, Marq JB, Sharma P, Soldati-Favre D. Assessment of phosphorylation in Toxoplasma glideosome assembly and function. Cellular microbiology. 2014 doi: 10.1111/cmi.12307. [DOI] [PubMed] [Google Scholar]
- Jewett TJ, Sibley LD. Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Molecular cell. 2003;11:885–894. doi: 10.1016/s1097-2765(03)00113-8. [DOI] [PubMed] [Google Scholar]
- Johnson TM, Rajfur Z, Jacobson K, Beckers CJ. Immobilization of the type XIV myosin complex in Toxoplasma gondii. Molecular biology of the cell. 2007;18:3039–3046. doi: 10.1091/mbc.E07-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ML, Collins MO, Goulding D, Choudhary JS, Rayner JC. Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell host & microbe. 2012;12:246–258. doi: 10.1016/j.chom.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliana C, Fernandes-Alnemri T, Kang S, Farias A, Qin F, Alnemri ES. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. The Journal of biological chemistry. 2012;287:36617–36622. doi: 10.1074/jbc.M112.407130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko O, Mu J, Tsuboi T, Su X, Torii M. Gene structure and expression of a Plasmodium falciparum 220-kDa protein homologous to the Plasmodium vivax reticulocyte binding proteins. Molecular and biochemical parasitology. 2002;121:275–278. doi: 10.1016/s0166-6851(02)00042-7. [DOI] [PubMed] [Google Scholar]
- Kappe S, Bruderer T, Gantt S, Fujioka H, Nussenzweig V, Menard R. Conservation of a gliding motility and cell invasion machinery in Apicomplexan parasites. The Journal of cell biology. 1999;147:937–944. doi: 10.1083/jcb.147.5.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamrui S, Turley S, Pardon E, Steyaert J, Verlinde C, Fan E, Bergman LW, Hol WGJ. Malaria invasion machinery protein-Nanobody complex. Protein Data Bank. 2013 [Google Scholar]
- Khater EI, Sinden RE, Dessens JT. A malaria membrane skeletal protein is essential for normal morphogenesis, motility, and infectivity of sporozoites. The Journal of cell biology. 2004;167:425–432. doi: 10.1083/jcb.200406068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Certa U, Dobeli H, Jakob P, Hol WG. Crystal structure of fructose-1,6-bisphosphate aldolase from the human malaria parasite Plasmodium falciparum. Biochemistry. 1998;37:4388–4396. doi: 10.1021/bi972233h. [DOI] [PubMed] [Google Scholar]
- Kono M, Prusty D, Parkinson J, Gilberger TW. The apicomplexan inner membrane complex. Frontiers in bioscience (Landmark edition) 2013;18:982–992. doi: 10.2741/4157. [DOI] [PubMed] [Google Scholar]
- Kortagere S, Welsh WJ, Morrisey JM, Daly T, Ejigiri I, Sinnis P, Vaidya AB, Bergman LW. Structure-based design of novel small-molecule inhibitors of Plasmodium falciparum. Journal of chemical information and modeling. 2010;50:840–849. doi: 10.1021/ci100039k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera K, Koblansky AA, Saunders LP, Frederick KB, De La Cruz EM, Ghosh S, Modis Y. Structure-based analysis of Toxoplasma gondii profilin: a parasite-specific motif is required for recognition by Toll-like receptor 11. Journal of molecular biology. 2010;403:616–629. doi: 10.1016/j.jmb.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashev M, Lepper S, Baumeister W, Cyrklaff M, Frischknecht F. Geometric constrains for detecting short actin filaments by cryogenic electron tomography. PMC Biophysics. 2010;3:6–6. doi: 10.1186/1757-5036-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursula I, Kursula P, Ganter M, Panjikar S, Matuschewski K, Schuler H. Structural basis for parasite-specific functions of the divergent profilin of Plasmodium falciparum. Structure. 2008;16:1638–1648. doi: 10.1016/j.str.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Lamarque M, Besteiro S, Papoin J, Roques M, Vulliez-Le Normand B, Morlon-Guyot J, Dubremetz JF, Fauquenoy S, Tomavo S, Faber BW, Kocken CH, Thomas AW, Boulanger MJ, Bentley GA, Lebrun M. The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS pathogens. 2011;7:e1001276. doi: 10.1371/journal.ppat.1001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasonder E, Green JL, Camarda G, Talabani H, Holder AA, Langsley G, Alano P. The Plasmodium falciparum schizont phosphoproteome reveals extensive phosphatidylinositol and cAMP-protein kinase A signaling. Journal of proteome research. 2012;11:5323–5337. doi: 10.1021/pr300557m. [DOI] [PubMed] [Google Scholar]
- Lebrun M, Michelin A, El Hajj H, Poncet J, Bradley PJ, Vial H, Dubremetz JF. The rhoptry neck protein RON4 re-localizes at the moving junction during Toxoplasma gondii invasion. Cellular microbiology. 2005;7:1823–1833. doi: 10.1111/j.1462-5822.2005.00646.x. [DOI] [PubMed] [Google Scholar]
- Li J, Han ET. Dissection of the Plasmodium vivax reticulocyte binding-like proteins (PvRBPs) Biochemical and biophysical research communications. 2012;426:1–6. doi: 10.1016/j.bbrc.2012.08.055. [DOI] [PubMed] [Google Scholar]
- Lin DH, Malpede BM, Batchelor JD, Tolia NH. Crystal and solution structures of Plasmodium falciparum erythrocyte-binding antigen 140 reveal determinants of receptor specificity during erythrocyte invasion. The Journal of biological chemistry. 2012;287:36830–36836. doi: 10.1074/jbc.M112.409276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Juo ZS, Shim AH, Focia PJ, Chen X, Garcia KC, He X. Structural basis of semaphorin-plexin recognition and viral mimicry from Sema7A and A39R complexes with PlexinC1. Cell. 2010;142:749–761. doi: 10.1016/j.cell.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo CA, Rodriguez M, Reid M, Lustigman S. Glycophorin C is the receptor for the Plasmodium falciparum erythrocyte binding ligand PfEBP-2 (baebl) Blood. 2003;101:4628–4631. doi: 10.1182/blood-2002-10-3076. [DOI] [PubMed] [Google Scholar]
- Maier AG, Baum J, Smith B, Conway DJ, Cowman AF. Polymorphisms in erythrocyte binding antigens 140 and 181 affect function and binding but not receptor specificity in Plasmodium falciparum. Infection and immunity. 2009;77:1689–1699. doi: 10.1128/IAI.01331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpede BM, Lin DH, Tolia NH. Molecular basis for sialic acid-dependent receptor recognition by the Plasmodium falciparum invasion protein erythrocyte-binding antigen-140/BAEBL. The Journal of biological chemistry. 2013;288:12406–12415. doi: 10.1074/jbc.M113.450643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann T, Beckers C. Characterization of the subpellicular network, a filamentous membrane skeletal component in the parasite Toxoplasma gondii. Molecular and biochemical parasitology. 2001;115:257–268. doi: 10.1016/s0166-6851(01)00289-4. [DOI] [PubMed] [Google Scholar]
- Mital J, Meissner M, Soldati D, Ward GE. Conditional expression of Toxoplasma gondii apical membrane antigen-1 (TgAMA1) demonstrates that TgAMA1 plays a critical role in host cell invasion. Molecular biology of the cell. 2005;16:4341–4349. doi: 10.1091/mbc.E05-04-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell GH, Thomas AW, Margos G, Dluzewski AR, Bannister LH. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infection and immunity. 2004;72:154–158. doi: 10.1128/IAI.72.1.154-158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira CK, Templeton TJ, Lavazec C, Hayward RE, Hobbs CV, Kroeze H, Janse CJ, Waters AP, Sinnis P, Coppi A. The Plasmodium TRAP/MIC2 family member, TRAP-Like Protein (TLP), is involved in tissue traversal by sporozoites. Cellular microbiology. 2008;10:1505–1516. doi: 10.1111/j.1462-5822.2008.01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HM, Reckmann I, Hollingdale MR, Bujard H, Robson KJ, Crisanti A. Thrombospondin related anonymous protein (TRAP) of Plasmodium falciparum binds specifically to sulfated glycoconjugates and to HepG2 hepatoma cells suggesting a role for this molecule in sporozoite invasion of hepatocytes. The EMBO journal. 1993;12:2881–2889. doi: 10.1002/j.1460-2075.1993.tb05950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller IB, Knockel J, Eschbach ML, Bergmann B, Walter RD, Wrenger C. Secretion of an acid phosphatase provides a possible mechanism to acquire host nutrients by Plasmodium falciparum. Cellular microbiology. 2010;12:677–691. doi: 10.1111/j.1462-5822.2010.01426.x. [DOI] [PubMed] [Google Scholar]
- Nair M, Hinds MG, Coley AM, Hodder AN, Foley M, Anders RF, Norton RS. Structure of domain III of the blood-stage malaria vaccine candidate, Plasmodium falciparum apical membrane antigen 1 (AMA1) Journal of molecular biology. 2002;322:741–753. doi: 10.1016/s0022-2836(02)00806-9. [DOI] [PubMed] [Google Scholar]
- Nebl T, Prieto JH, Kapp E, Smith BJ, Williams MJ, Yates JR, 3rd, Cowman AF, Tonkin CJ. Quantitative in vivo analyses reveal calcium-dependent phosphorylation sites and identifies a novel component of the Toxoplasma invasion motor complex. PLoS pathogens. 2011;7:e1002222. doi: 10.1371/journal.ppat.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshina MA, Wong W, Baum J. Holding back the microfilament--structural insights into actin and the actin-monomer-binding proteins of apicomplexan parasites. IUBMB life. 2012;64:370–377. doi: 10.1002/iub.1014. [DOI] [PubMed] [Google Scholar]
- Orlandi PA, Klotz FW, Haynes JD. A malaria invasion receptor, the 175-kilodalton erythrocyte binding antigen of Plasmodium falciparum recognizes the terminal Neu5Ac(alpha 2–3)Gal- sequences of glycophorin A. The Journal of cell biology. 1992;116:901–909. doi: 10.1083/jcb.116.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhowmick I, Andersen J, Srinivasan P, Narum DL, Bosch J, Miller LH. Binding of aldolase and glyceraldehyde-3-phosphate dehydrogenase to the cytoplasmic tails of Plasmodium falciparum merozoite duffy binding-like and reticulocyte homology ligands. mBio 3. 2012 doi: 10.1128/mBio.00292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Guariento M, Maciejewski M, Hauhart R, Tham WH, Cowman AF, Schmidt CQ, Mertens HD, Liszewski MK, Hourcade DE, Barlow PN, Atkinson JP. Using Mutagenesis and Structural Biology to Map the Binding Site for the Plasmodium falciparum Merozoite Protein PfRh4 on the Human Immune Adherence Receptor. The Journal of biological chemistry. 2014;289:450–463. doi: 10.1074/jbc.M113.520346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease BN, Huttlin EL, Jedrychowski MP, Talevich E, Harmon J, Dillman T, Kannan N, Doerig C, Chakrabarti R, Gygi SP, Chakrabarti D. Global analysis of protein expression and phosphorylation of three stages of Plasmodium falciparum intraerythrocytic development. Journal of proteome research. 2013;12:4028–4045. doi: 10.1021/pr400394g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MG, Marshall VM, Smythe JA, Crewther PE, Lew A, Silva A, Anders RF, Kemp DJ. Integral membrane protein located in the apical complex of Plasmodium falciparum. Molecular and cellular biology. 1989;9:3151–3154. doi: 10.1128/mcb.9.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihlajamaa T, Kajander T, Knuuti J, Horkka K, Sharma A, Permi P. Structure of Plasmodium falciparum TRAP (thrombospondin-related anonymous protein) A domain highlights distinct features in apicomplexan von Willebrand factor A homologues. The Biochemical journal. 2013;450:469–476. doi: 10.1042/BJ20121058. [DOI] [PubMed] [Google Scholar]
- Pinder J, Fowler R, Bannister L, Dluzewski A, Mitchell GH. Motile systems in malaria merozoites: how is the red blood cell invaded? Parasitology today (Personal ed.) 2000;16:240–245. doi: 10.1016/s0169-4758(00)01664-1. [DOI] [PubMed] [Google Scholar]
- Pinder JC, Fowler RE, Dluzewski AR, Bannister LH, Lavin FM, Mitchell GH, Wilson RJ, Gratzer WB. Actomyosin motor in the merozoite of the malaria parasite, Plasmodium falciparum: implications for red cell invasion. Journal of cell science. 1998;111(Pt 13):1831–1839. doi: 10.1242/jcs.111.13.1831. [DOI] [PubMed] [Google Scholar]
- Pizarro JC, Vulliez-Le Normand B, Chesne-Seck ML, Collins CR, Withers-Martinez C, Hackett F, Blackman MJ, Faber BW, Remarque EJ, Kocken CH, Thomas AW, Bentley GA. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science (New York, N.Y.) 2005;308:408–411. doi: 10.1126/science.1107449. [DOI] [PubMed] [Google Scholar]
- Plattner F, Yarovinsky F, Romero S, Didry D, Carlier MF, Sher A, Soldati-Favre D. Toxoplasma profilin is essential for host cell invasion and TLR11-dependent induction of an interleukin-12 response. Cell host & microbe. 2008;3:77–87. doi: 10.1016/j.chom.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Polonais V, Javier Foth B, Chinthalapudi K, Marq JB, Manstein DJ, Soldati-Favre D, Frenal K. Unusual anchor of a motor complex (MyoD-MLC2) to the plasma membrane of Toxoplasma gondii. Traffic (Copenhagen, Denmark) 2011;12:287–300. doi: 10.1111/j.1600-0854.2010.01148.x. [DOI] [PubMed] [Google Scholar]
- Poukchanski A, Fritz HM, Tonkin ML, Treeck M, Boulanger MJ, Boothroyd JC. Toxoplasma gondii sporozoites invade host cells using two novel paralogues of RON2 and AMA1. PloS one. 2013;8:e70637. doi: 10.1371/journal.pone.0070637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin B, Patzewitz EM, Brady D, Silvie O, Wright MH, Ferguson DJ, Wall RJ, Whipple S, Guttery DS, Tate EW, Wickstead B, Holder AA, Tewari R. Unique apicomplexan IMC sub-compartment proteins are early markers for apical polarity in the malaria parasite. Biology open. 2013;2:1160–1170. doi: 10.1242/bio.20136163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud A, Lupetti P, Paul RE, Mercati D, Brey PT, Sinden RE, Heuser JE, Dallai R. Cryofracture electron microscopy of the ookinete pellicle of Plasmodium gallinaceum reveals the existence of novel pores in the alveolar membranes. Journal of structural biology. 2001;135:47–57. doi: 10.1006/jsbi.2001.4396. [DOI] [PubMed] [Google Scholar]
- Ramalingam JK, Hunke C, Gao X, Gruber G, Preiser PR. ATP/ADP binding to a novel nucleotide binding domain of the reticulocyte-binding protein Py235 of Plasmodium yoelii. The Journal of biological chemistry. 2008;283:36386–36396. doi: 10.1074/jbc.M803102200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner JC, Galinski MR, Ingravallo P, Barnwell JW. Two Plasmodium falciparum genes express merozoite proteins that are related to Plasmodium vivax and Plasmodium yoelii adhesive proteins involved in host cell selection and invasion. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:9648–9653. doi: 10.1073/pnas.160469097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner JC, Vargas-Serrato E, Huber CS, Galinski MR, Barnwell JW. A Plasmodium falciparum homologue of Plasmodium vivax reticulocyte binding protein (PvRBP1) defines a trypsin-resistant erythrocyte invasion pathway. The Journal of experimental medicine. 2001;194:1571–1581. doi: 10.1084/jem.194.11.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redzic JS, Armstrong GS, Isern NG, Jones DN, Kieft JS, Eisenmesser EZ. The retinal specific CD147 Ig0 domain: from molecular structure to biological activity. Journal of molecular biology. 2011;411:68–82. doi: 10.1016/j.jmb.2011.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees-Channer RR, Martin SR, Green JL, Bowyer PW, Grainger M, Molloy JE, Holder AA. Dual acylation of the 45 kDa gliding-associated protein (GAP45) in Plasmodium falciparum merozoites. Molecular and biochemical parasitology. 2006;149:113–116. doi: 10.1016/j.molbiopara.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein engineering, design & selection : PEDS. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Science's STKE : signal transduction knowledge environment. 2006;2006:re14. doi: 10.1126/stke.3592006re14. [DOI] [PubMed] [Google Scholar]
- Richard D, MacRaild CA, Riglar DT, Chan JA, Foley M, Baum J, Ralph SA, Norton RS, Cowman AF. Interaction between Plasmodium falciparum apical membrane antigen 1 and the rhoptry neck protein complex defines a key step in the erythrocyte invasion process of malaria parasites. The Journal of biological chemistry. 2010;285:14815–14822. doi: 10.1074/jbc.M109.080770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridzuan MA, Moon RW, Knuepfer E, Black S, Holder AA, Green JL. Subcellular location, phosphorylation and assembly into the motor complex of GAP45 during Plasmodium falciparum schizont development. PloS one. 2012;7:e33845. doi: 10.1371/journal.pone.0033845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robien MA, Bosch J, Buckner FS, Van Voorhis WC, Worthey EA, Myler P, Mehlin C, Boni EE, Kalyuzhniy O, Anderson L, Lauricella A, Gulde S, Luft JR, DeTitta G, Caruthers JM, Hodgson KO, Soltis M, Zucker F, Verlinde CL, Merritt EA, Schoenfeld LW, Hol WG. Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 A resolution reveals intriguing extra electron density in the active site. Proteins. 2006;62:570–577. doi: 10.1002/prot.20801. [DOI] [PubMed] [Google Scholar]
- Robson KJ, Hall JR, Jennings MW, Harris TJ, Marsh K, Newbold CI, Tate VE, Weatherall DJ. A highly conserved amino-acid sequence in thrombospondin, properdin and in proteins from sporozoites and blood stages of a human malaria parasite. Nature. 1988;335:79–82. doi: 10.1038/335079a0. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar T, Reddy KS, Bharadwaj M, Pandey AK, Singh S, Chitnis CE, Gaur D. Plasmodium falciparum reticulocyte binding-like homologue protein 2 (PfRH2) is a key adhesive molecule involved in erythrocyte invasion. PloS one. 2011;6:e17102. doi: 10.1371/journal.pone.0017102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo N, Beatty W, Heuser J, Sept D, Sibley LD. Unusual kinetic and structural properties control rapid assembly and turnover of actin in the parasite Toxoplasma gondii. Molecular biology of the cell. 2006;17:895–906. doi: 10.1091/mbc.E05-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchell JF, Malby RL, Luo CS, Adisa A, Alpyurek AE, Klonis N, Smith BJ, Tilley L, Colman PM. Structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum. Acta crystallographica. Section D, Biological crystallography. 2005;61:1213–1221. doi: 10.1107/S0907444905018317. [DOI] [PubMed] [Google Scholar]
- Schmitz S, Grainger M, Howell S, Calder LJ, Gaeb M, Pinder JC, Holder AA, Veigel C. Malaria parasite actin filaments are very short. Journal of molecular biology. 2005;349:113–125. doi: 10.1016/j.jmb.2005.03.056. [DOI] [PubMed] [Google Scholar]
- Schuler H, Matuschewski K. Regulation of apicomplexan microfilament dynamics by a minimal set of actin-binding proteins. Traffic (Copenhagen, Denmark) 2006;7:1433–1439. doi: 10.1111/j.1600-0854.2006.00484.x. [DOI] [PubMed] [Google Scholar]
- Schuler H, Mueller AK, Matuschewski K. A Plasmodium actin-depolymerizing factor that binds exclusively to actin monomers. Molecular biology of the cell. 2005a;16:4013–4023. doi: 10.1091/mbc.E05-02-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler H, Mueller AK, Matuschewski K. Unusual properties of Plasmodium falciparum actin: new insights into microfilament dynamics of apicomplexan parasites. FEBS letters. 2005b;579:655–660. doi: 10.1016/j.febslet.2004.12.037. [DOI] [PubMed] [Google Scholar]
- Sheiner L, Santos JM, Klages N, Parussini F, Jemmely N, Friedrich N, Ward GE, Soldati-Favre D. Toxoplasma gondii transmembrane microneme proteins and their modular design. Molecular microbiology. 2010 doi: 10.1111/j.1365-2958.2010.07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Sibley LD. Toxoplasma aldolase is required for metabolism but dispensable for host-cell invasion. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3567–3572. doi: 10.1073/pnas.1315156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK, Sattler JM, Chatterjee M, Huttu J, Schuler H, Kursula I. Crystal structures explain functional differences in the two actin depolymerization factors of the malaria parasite. The Journal of biological chemistry. 2011;286:28256–28264. doi: 10.1074/jbc.M111.211730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hora R, Belrhali H, Chitnis CE, Sharma A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature. 2006;439:741–744. doi: 10.1038/nature04443. [DOI] [PubMed] [Google Scholar]
- Skillman KM, Diraviyam K, Khan A, Tang K, Sept D, Sibley LD. Evolutionarily divergent, unstable filamentous actin is essential for gliding motility in apicomplexan parasites. PLoS pathogens. 2011;7:e1002280. doi: 10.1371/journal.ppat.1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Springer TA. Structures of the Toxoplasma gliding motility adhesin. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4862–4867. doi: 10.1073/pnas.1403059111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Koksal AC, Lu C, Springer TA. Shape change in the receptor for gliding motility in Plasmodium sporozoites. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21420–21425. doi: 10.1073/pnas.1218581109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano F, Putignani L, Guida S, Crisanti A. Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (thrombospondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Experimental parasitology. 1998a;90:195–198. doi: 10.1006/expr.1998.4324. [DOI] [PubMed] [Google Scholar]
- Spano F, Putignani L, Naitza S, Puri C, Wright S, Crisanti A. Molecular cloning and expression analysis of a Cryptosporidium parvum gene encoding a new member of the thrombospondin family. Molecular and biochemical parasitology. 1998b;92:147–162. doi: 10.1016/s0166-6851(97)00243-0. [DOI] [PubMed] [Google Scholar]
- Srinivasan P, Beatty WL, Diouf A, Herrera R, Ambroggio X, Moch JK, Tyler JS, Narum DL, Pierce SK, Boothroyd JC, Haynes JD, Miller LH. Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13275–13280. doi: 10.1073/pnas.1110303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan P, Yasgar A, Luci DK, Beatty WL, Hu X, Andersen J, Narum DL, Moch JK, Sun H, Haynes JD, Maloney DJ, Jadhav A, Simeonov A, Miller LH. Disrupting malaria parasite AMA1-RON2 interaction with a small molecule prevents erythrocyte invasion. Nature communications. 2013;4:2261. doi: 10.1038/ncomms3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SSGCID. Staker BL, Edwards TE, Sankaran B. Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Toxoplasma gondii. Protein Data Bank. 2011 [Google Scholar]
- Sultan AA, Thathy V, Frevert U, Robson KJ, Crisanti A, Nussenzweig V, Nussenzweig RS, Menard R. TRAP is necessary for gliding motility and infectivity of plasmodium sporozoites. Cell. 1997;90:511–522. doi: 10.1016/s0092-8674(00)80511-5. [DOI] [PubMed] [Google Scholar]
- Tardieux I, Baines I, Mossakowska M, Ward GE. Actin-binding proteins of invasive malaria parasites and the regulation of actin polymerization by a complex of 32/34-kDa proteins associated with heat shock protein 70kDa. Molecular and biochemical parasitology. 1998;93:295–308. doi: 10.1016/s0166-6851(98)00044-9. [DOI] [PubMed] [Google Scholar]
- Taylor DW, Evans CB, Aley SB, Barta JR, Danforth HD. Identification of an apically-located antigen that is conserved in sporozoan parasites. The Journal of protozoology. 1990;37:540–545. doi: 10.1111/j.1550-7408.1990.tb01262.x. [DOI] [PubMed] [Google Scholar]
- Tham WH, Healer J, Cowman AF. Erythrocyte and reticulocyte binding-like proteins of Plasmodium falciparum. Trends in parasitology. 2012;28:23–30. doi: 10.1016/j.pt.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Tham WH, Wilson DW, Lopaticki S, Schmidt CQ, Tetteh-Quarcoo PB, Barlow PN, Richard D, Corbin JE, Beeson JG, Cowman AF. Complement receptor 1 is the host erythrocyte receptor for Plasmodium falciparum PfRh4 invasion ligand. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17327–17332. doi: 10.1073/pnas.1008151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JK, Triglia T, Reed MB, Cowman AF. A novel ligand from Plasmodium falciparum that binds to a sialic acid-containing receptor on the surface of human erythrocytes. Molecular microbiology. 2001;41:47–58. doi: 10.1046/j.1365-2958.2001.02484.x. [DOI] [PubMed] [Google Scholar]
- Tolia NH, Enemark EJ, Sim BK, Joshua-Tor L. Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell. 2005;122:183–193. doi: 10.1016/j.cell.2005.05.033. [DOI] [PubMed] [Google Scholar]
- Tomley FM, Clarke LE, Kawazoe U, Dijkema R, Kok JJ. Sequence of the gene encoding an immunodominant microneme protein of Eimeria tenella. Molecular and biochemical parasitology. 1991;49:277–288. doi: 10.1016/0166-6851(91)90071-d. [DOI] [PubMed] [Google Scholar]
- Tonkin ML, Crawford J, Lebrun ML, Boulanger MJ. Babesia divergens and Neospora caninum apical membrane antigen 1 structures reveal selectivity and plasticity in apicomplexan parasite host cell invasion. Protein science : a publication of the Protein Society. 2013;22:114–127. doi: 10.1002/pro.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ML, Beck JR, Bradley PJ, Boulanger MJ. The inner membrane complex sub-compartment proteins critical for replication of the apicomplexan parasite Toxoplasma gondii adopt a Pleckstrin homology fold. The Journal of biological chemistry. 2014a doi: 10.1074/jbc.M114.548891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ML, Grujic O, Pearce M, Crawford J, Boulanger MJ. Structure of the micronemal protein 2 A/I domain from Toxoplasma gondii. Protein science : a publication of the Protein Society. 2010;19:1985–1990. doi: 10.1002/pro.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ML, Halavaty AS, Ramaswamy R, Ruan J, Igarashi M, Ngo HM, Boulanger MJ. Structural and functional divergence of the aldolase fold in Toxoplasma gondii. Journal of molecular biology. 2014b doi: 10.1016/j.jmb.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ML, Roques M, Lamarque MH, Pugniere M, Douguet D, Crawford J, Lebrun M, Boulanger MJ. Host cell invasion by apicomplexan parasites: insights from the co-structure of AMA1 with a RON2 peptide. Science (New York, N.Y.) 2011;333:463–467. doi: 10.1126/science.1204988. [DOI] [PubMed] [Google Scholar]
- Tordai H, Banyai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS letters. 1999;461:63–67. doi: 10.1016/s0014-5793(99)01416-7. [DOI] [PubMed] [Google Scholar]
- Tossavainen H, Pihlajamaa T, Huttunen TK, Raulo E, Rauvala H, Permi P, Kilpelainen I. The layered fold of the TSR domain of P. falciparum TRAP contains a heparin binding site. Protein science : a publication of the Protein Society. 2006;15:1760–1768. doi: 10.1110/ps.052068506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treeck M, Sanders JL, Elias JE, Boothroyd JC. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites' boundaries. Cell host & microbe. 2011;10:410–419. doi: 10.1016/j.chom.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremp AZ, Dessens JT. Malaria IMC1 membrane skeleton proteins operate autonomously and participate in motility independently of cell shape. The Journal of biological chemistry. 2011;286:5383–5391. doi: 10.1074/jbc.M110.187195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T, Thompson J, Caruana SR, Delorenzi M, Speed T, Cowman AF. Identification of proteins from Plasmodium falciparum that are homologous to reticulocyte binding proteins in Plasmodium vivax. Infection and immunity. 2001;69:1084–1092. doi: 10.1128/IAI.69.2.1084-1092.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottein F, Triglia T, Cowman AF. Molecular cloning of a gene from Plasmodium falciparum that codes for a protein sharing motifs found in adhesive molecules from mammals and plasmodia. Molecular and biochemical parasitology. 1995;74:129–141. doi: 10.1016/0166-6851(95)02489-1. [DOI] [PubMed] [Google Scholar]
- Turley S, Khamrui S, Bergman LW, Hol WG. The compact conformation of the Plasmodium knowlesi myosin tail interacting protein MTIP in complex with the C-terminal helix of myosin A. Molecular and biochemical parasitology. 2013;190:56–59. doi: 10.1016/j.molbiopara.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquiza M, Patarroyo MA, Mari V, Ocampo M, Suarez J, Lopez R, Puentes A, Curtidor H, Garcia J, Rodriuez LE, Vera R, Torres A, Laverde M, Robles AP, Patarroyo ME. Identification and polymorphism of Plasmodium vivax RBP-1 peptides which bind specifically to reticulocytes. Peptides. 2002;23:2265–2277. doi: 10.1016/s0196-9781(02)00267-x. [DOI] [PubMed] [Google Scholar]
- Vahokoski J, Bhargav SP, Desfosses A, Andreadaki M, Kumpula EP, Martinez SM, Ignatev A, Lepper S, Frischknecht F, Siden-Kiamos I, Sachse C, Kursula I. Structural differences explain diverse functions of Plasmodium actins. PLoS pathogens. 2014;10:e1004091. doi: 10.1371/journal.ppat.1004091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann K, Pfander C, Burstroem C, Ahras M, Goulding D, Rayner JC, Frischknecht F, Billker O, Brochet M. The alveolin IMC1h is required for normal ookinete and sporozoite motility behaviour and host colonisation in Plasmodium berghei. PloS one. 2012;7:e41409. doi: 10.1371/journal.pone.0041409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliez-Le Normand B, Tonkin ML, Lamarque MH, Langer S, Hoos S, Roques M, Saul FA, Faber BW, Bentley GA, Boulanger MJ, Lebrun M. Structural and functional insights into the malaria parasite moving junction complex. PLoS pathogens. 2012;8:e1002755. doi: 10.1371/journal.ppat.1002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan KL, Carruthers VB, Sibley LD, Ajioka JW. Molecular characterisation of an expressed sequence tag locus of Toxoplasma gondii encoding the micronemal protein MIC2. Molecular and biochemical parasitology. 1997;84:203–214. doi: 10.1016/s0166-6851(96)02796-x. [DOI] [PubMed] [Google Scholar]
- Wertheimer SP, Barnwell JW. Plasmodium vivax interaction with the human Duffy blood group glycoprotein: identification of a parasite receptor-like protein. Experimental parasitology. 1989;69:340–350. doi: 10.1016/0014-4894(89)90083-0. [DOI] [PubMed] [Google Scholar]
- Wesseling JG, Smits MA, Schoenmakers JG. Extremely diverged actin proteins in Plasmodium falciparum. Molecular and biochemical parasitology. 1988;30:143–153. doi: 10.1016/0166-6851(88)90107-7. [DOI] [PubMed] [Google Scholar]
- Withers-Martinez C, Haire LF, Hackett F, Walker PA, Howell SA, Smerdon SJ, Dodson GG, Blackman MJ. Malarial EBA-175 region VI crystallographic structure reveals a KIX-like binding interface. Journal of molecular biology. 2008;375:773–781. doi: 10.1016/j.jmb.2007.10.071. [DOI] [PubMed] [Google Scholar]
- Wong W, Skau CT, Marapana DS, Hanssen E, Taylor NL, Riglar DT, Zuccala ES, Angrisano F, Lewis H, Catimel B, Clarke OB, Kershaw NJ, Perugini MA, Kovar DR, Gulbis JM, Baum J. Minimal requirements for actin filament disassembly revealed by structural analysis of malaria parasite actin-depolymerizing factor 1. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9869–9874. doi: 10.1073/pnas.1018927108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KE, Hjerrild KA, Bartlett J, Douglas AD, Jin J, Brown RE, Illingworth JJ, Ashfield R, Clemmensen SB, de Jongh WA, Draper SJ, Higgins MK. Structure of malaria invasion protein RH5 with erythrocyte basigin and blocking antibodies. Nature. 2014a doi: 10.1038/nature13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MH, Clough B, Rackham MD, Rangachari K, Brannigan JA, Grainger M, Moss DK, Bottrill AR. Validation of N-myristoyltransferase as an antimalarial drug target using an integrated chemical biology approach. 2014b;6:112–121. doi: 10.1038/nchem.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R, Pathak PP, Shukla VK, Jain A, Srivastava S, Tripathi S, Krishna Pulavarti SV, Mehta S, Sibley LD, Arora A. Solution structure and dynamics of ADF from Toxoplasma gondii. Journal of structural biology. 2011;176:97–111. doi: 10.1016/j.jsb.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap A, Azevedo MF, Gilson PR, Weiss GE, O'Neill MT, Wilson DW, Crabb BS, Cowman AF. Conditional expression of apical membrane antigen 1 in Plasmodium falciparum shows it is required for erythrocyte invasion by merozoites. Cellular microbiology. 2014;16:642–656. doi: 10.1111/cmi.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.