Abstract
Genomic imprinting is an epigenetic mechanism resulting in parental allele-specific gene expression. Defects in normal imprinting are found in cancer, assisted reproductive technologies, and several human syndromes. In mouse models, germline-derived DNA methylation is shown to regulate imprinting. Though imprinting is largely conserved between mammals, species- and tissue-specific domains of imprinted expression exist. Using the cynomolgus macaque (Macaca fascicularis) to assess primate-specific imprinting, we present a comprehensive view of tissue-specific imprinted expression and DNA methylation at established imprinted gene clusters. For example, like mouse and unlike human, macaque IGF2R is consistently imprinted, and the PLAGL1, INPP5F transcript variant 2, and PEG3 imprinting control regions are not methylated in the macaque germline but acquire this post-fertilization. Methylome data from human early embryos appear to support this finding. These suggest fundamental differences in imprinting control mechanisms between primate species and rodents at some imprinted domains, with implications for our understanding of the epigenetic programming process in humans and its influence on disease.
Genomic imprinting is an epigenetically regulated process resulting in gene expression from specific parental alleles. Many imprinted genes are clustered and feature both protein-coding and noncoding RNA genes (Edwards and Ferguson-Smith 2007). In mouse, differential DNA methylation at CpG-rich imprinting control regions (ICRs) is first established in gametogenesis, along with other methylation marks, and depends on the presence of DNA methyltransferases (DNMTs) (Li et al. 1993; Okano et al. 1999; Li and Sasaki 2011). During preimplantation development, protection from demethylation is essential at imprints (Li et al. 2008; Hanna and Kelsey 2014), and subsequently, additional differentially methylated regions (DMRs) can become established in response to the germline DMR (Kafri et al. 1992; Brandeis et al. 1993a,b).
Imprinted genes are involved in both pre- and post-natal growth, and metabolic and cognitive processes (Ferguson-Smith 2011; Cleaton et al. 2014). In humans, aberrant imprinting is responsible for certain developmental disorders with parental origin effects (Weksberg et al. 2003; Gicquel et al. 2005), while perturbed imprinting is regularly reported in cancers (Uribe-Lewis et al. 2011). More recently, the increased incidence of imprinting defects in infants conceived through assisted reproduction techniques emphasizes the importance of imprinting epigenetics from a very early developmental time point (Grace and Sinclair 2009).
Comparative analysis of imprinting between eu-, meta- and prototherian mammals suggests that imprinting arose relatively recently at most loci—only a few imprinted genes in Eutherians are also imprinted in marsupials, while no imprinting has been reported in the egg-laying monotreme mammals to date (Killian et al. 2000; Edwards et al. 2008; Smits et al. 2008; Renfree et al. 2009a,b). While the mouse is an informative proxy for human imprinted gene regulation, not all loci show conserved imprinting, notably in the placenta (Tycko and Morison 2002; Morison et al. 2005). Distinct differences in placental evolution, physiology, and reproductive biology of the primate and murine groups may be responsible. In contrast to an evolutionary distance of 75 million years between mouse and human, the macaque diverged 25 million years ago from human and shares many physiological similarities with humans. The added availability of the macaque genome has made this nonhuman primate a useful model for understanding recent genomic evolutionary changes (Waterston et al. 2002; Rhesus Macaque Genome Sequencing and Analysis Consortium et al. 2007; Yan et al. 2011), with further potential for understanding the evolution of epigenetic mechanisms.
In order to explore the evolution of imprinting in the primate, we surveyed established imprinted gene clusters for the conservation of imprinted gene expression and DNA methylation in the nonhuman primate, cynomolgus macaque (Macaca fascicularis). The closely related cynomolgus and rhesus (Macaca mulatta) macaques are 99.6% similar, estimated to diverge by only ∼2 million years, and share much genomic structure and similarity, both with each other and with the human genome (Hayasaka et al. 1996; Osada et al. 2008). As such, both are widely used in preclinical studies and are useful models for accessing tissues otherwise limited in human research (Bourne 1975).
Here, we provide the most comprehensive survey of imprinting in the nonhuman primate to date and investigate the conservation of primary and secondary DMRs between primates and rodents. Our findings suggest that aspects of imprinting control may differ between rodent and macaque, with important implications for epigenetic programming in normal development and disease.
Results
Conservation of allelic expression at imprinted loci in macaque
We examined somatic and extraembryonic tissues for allelic expression at a total of 32 genes known to be imprinted in either human or mouse. Macaque genomic regions analyzed were identified by orthology to known human imprinted genes, since the sequence identity between human and macaque is ∼93% (Rhesus Macaque Genome Sequencing and Analysis Consortium et al. 2007), enabling reference gene mapping and the discovery of novel polymorphisms required for allele-specific expression analysis.
Paternally imprinted genes
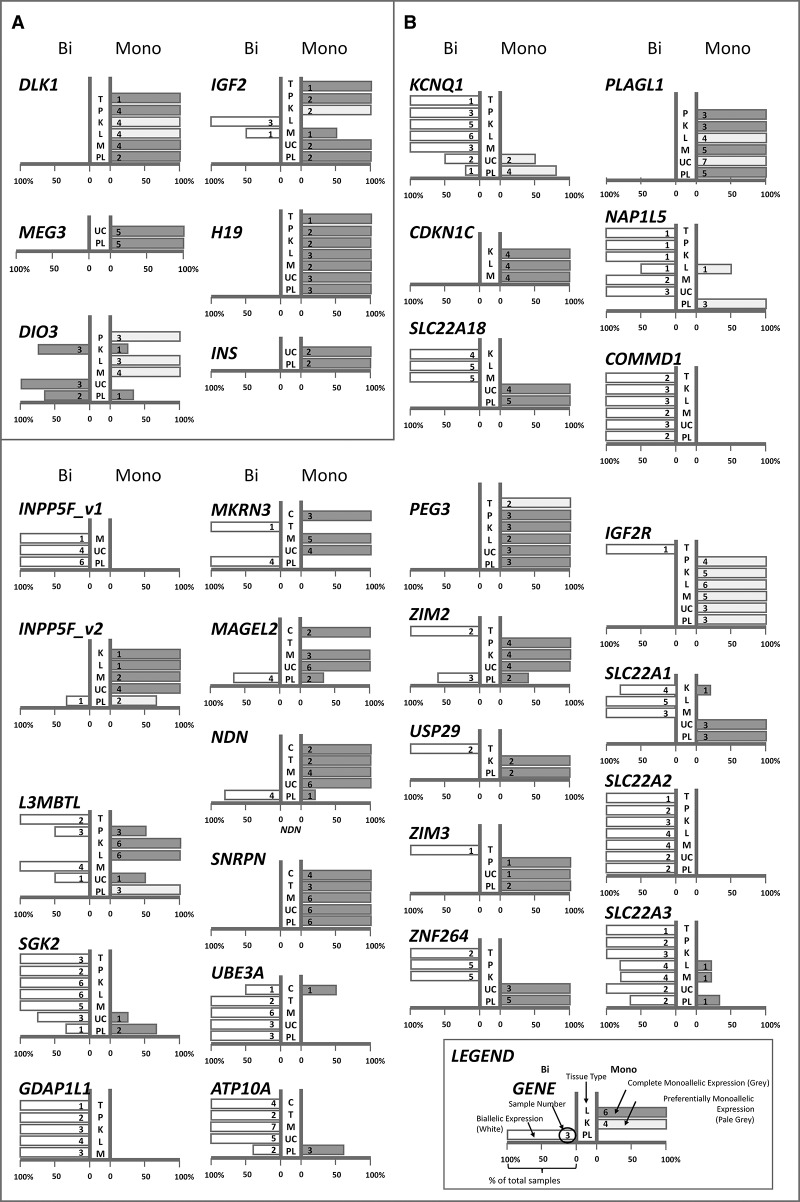
In mouse, the Igf2-H19 and Dlk1-Dio3 domains are controlled by paternal-specific germline methylation imprints at their intergenic ICRs (Kobayashi et al. 2000; de la Puente et al. 2002; Takada et al. 2002; Gabory et al. 2006; Cai and Cullen 2007). Consistent with neonatal rhesus tissues and ES cells, IGF2 and H19 are monoallelically expressed in all cynomolgus extraembryonic tissues analyzed (Fujimoto et al. 2005, 2006). H19 expression is also consistently monoallelic in all somatic tissues tested, although the corresponding tissue imprinting of IGF2 is somewhat relaxed, particularly in liver and skeletal muscle (Fig. 1A; Supplemental Table 1). Given the significance of IGF2 in fetal growth and placental development (Han and Carter 2000; Constancia et al. 2002), it is perhaps expected that imprinted expression is most robust in placenta. Post early development, the functional role of IGF2 is less clear, with relaxed biallelic imprinting reported in some somatic tissues of the macaque (Fig. 1A) and human (Davies et al. 2007; Frost et al. 2010). In addition, INS, a more distal gene, also shows consistent allele-specific expression in extraembryonic tissues (Fig. 1A).
Figure 1.
Imprinted expression profile in tissues. Individual charts show allelic expression for each gene across the main tissue types examined. (A) Paternally imprinted gene clusters. (B) Maternally imprinted gene clusters. The length of each bar represents the percentage of total samples adhering to either expression pattern—biallelic (left) or monoallelic (right). A further distinction is made between complete and preferential monoallelic expression at each gene: complete monoallelic expression (gray) and preferential monoallelic expression (pale gray). See Supplemental Figure 2 for quantitative pyrosequencing and capillary sequencing data. The number of samples analyzed is shown in each bar, and tissue type is listed in the central column of each chart. (C) Cerebellum, (T) Testes, (P) pancreas, (K) kidney, (L) liver, (M) muscle, (UC) umbilical cord, (PL) placenta. These data are also tabulated in Supplemental Table 1, with additional data available for select genes/tissues.
The DLK1 locus is another cluster with known paternal inheritance of methylation imprints and is expressed at similar developmental stages as the IGF2-H19 locus (Takada et al. 2000, 2002). DLK1 expression in the cynomolgus is fully monoallelic in placenta and some adult tissue (skeletal muscle, pancreas, testes). In adult kidney and liver, DLK1 is preferentially expressed monoallelically, suggestive of incomplete or cell-specific imprinting. Placenta and umbilical cord samples at the adjacent MEG3 (also known as GTL2) also show monoallelic expression; no somatic tissue samples were informative at this locus. Mouse Dio3 is known to be preferentially expressed from the paternal allele in embryonic tissue but biallelically expressed in the placenta (Hernandez et al. 2002; Lin et al. 2007), and maternal deletion of the Dlk1 ICR (intergenic DMR; IG-DMR) influences Dio3 imprinting despite its physical distance from the ICR (Lin et al. 2007). In our macaque samples tested, imprinted expression at DIO3 appears to be preferentially monoallelic in most somatic tissues and preferentially biallelic in placenta (Fig. 1A).
Maternally imprinted genes
Adjacent to the IGF2-H19 cluster, the KCNQ1 locus also retains its gene order and chromosomal syntenic homology between human, macaque, and mouse (Supplemental Fig. 3). Macaque KCNQ1 and SLC22A18 are largely monoallelically expressed in placenta, with a number of individuals showing preferential but incomplete monoallelic expression at KCNQ1. KCNQ1 expression was, however, biallelic in adult tissues (Fig. 1B). In mouse, imprinted expression of Kcnq1 is seen in embryos, but not in adult mice or humans. This embryonic stage-specific expression may also be true in primates and could account for the partial imprinting seen in term extraembryonic tissues (Lee et al. 1997; Caspary et al. 1998; Gould and Pfeifer 1998). Published findings on human SLC22A18 suggest that polymorphic imprinting is evident in adult liver and kidney (Dao et al. 1998; Gallagher et al. 2006), though the population frequency of this occurrence is unknown. Our analysis of SLC22A18 in macaques in these same tissues showed consistent biallelic expression (Fig. 1B).
The KCNQ1 locus further highlights that gene-ICR proximity alone does not determine imprinted expression. CDKN1C, a cyclin-dependent kinase inhibitor, is positioned downstream from the intronic KvDMR and showed monoallelic expression in cynomolgus adult tissues. No informative extraembryonic tissues were available. In mouse, Cdkn1c has a somatic promoter DMR which may contribute to stable monoallelic expression of this gene (Nowak et al. 2011), though we did not find evidence for this DMR in the primate (data not shown). Further examination of cynomolgus CDKN1C transcripts also revealed a novel transcript variant with no precedent in human or mouse CDKN1C (Supplemental Fig. 4; Nielsen et al. 2005). The genomic sequence of CDKN1C is conserved between rhesus, cynomolgus, and human, but alternative splicing results in a cynomolgus-specific transcript that, if translated, is only conserved at the cyclin-dependent kinase inhibitor domain (CDI, Pfam 02234).
The IGF2R cluster is well conserved across species and contains IGF2R, a receptor for the oppositely imprinted IGF2, and three related solute carriers, SLC22A1, SLC22A2, and SLC22A3 (Smrzka et al. 1995). In mouse, the noncoding Airn RNA transcript antisense to Igf2r is necessary for imprinting. In mouse, despite its position between the imprinted Igf2r, Slc22a2, and Slc22a3 genes, Slc22a1 is not imprinted (Lyle et al. 2000; Sleutels et al. 2002, 2003). The imprinting of human IGF2R has been questioned—various sources have suggested either polymorphic imprinting at relatively high frequency or, conversely, no imprinting (Xu et al. 1993; Wutz et al. 1998; Killian et al. 2001). In a survey of IGF2R imprinting across phylogenetic orders, it was also suggested that IGF2R imprinting was absent from an ancestor of the Euarchonta order, a mammalian order that includes humans and other primates (Killian et al. 2001). Our analysis of IGF2R allelic expression in various macaque tissues suggests that preferential monoallelic expression occurs consistently across all tissues except testes, where IGF2R expression was wholly biallelic (Fig. 1B).
Like IGF2R, SLC22A2 and SLC22A3 are also polymorphically imprinted in human term placenta, although their mouse counterparts are imprinted (Ogawa et al. 1993; Xu et al. 1993; Riesewijk et al. 1996; Monk et al. 2006). We observe biallelic expression of macaque SLC22A2 in placenta and other somatic tissues in all samples analyzed. However, though we do not see polymorphic imprinting, our sample size is small. Macaque SLC22A3 is polymorphically imprinted, and we anticipate that these genes behave similarly in macaque and human. Intriguingly, macaque SLC22A1 is imprinted in the placenta and polymorphically imprinted elsewhere, though there is no precedent for this in the human or mouse, where the gene has not previously been shown to be imprinted. Plausibly, SLC22A1 may also be polymorphically regulated in the human population, albeit restricted to late gestational extraembryonic tissues, which have not previously been examined at this locus.
Given the species variation in imprinting at this cluster, we next considered if an orthologous macaque AIRN transcript was present. Though a human homolog of mouse Airn has been reported, its in vivo expression and imprinting role is unclear (Oudejans et al. 2001; Yotova et al. 2008). Our attempts to isolate a similar transcript in macaque by 5′/3′ RACE were unsuccessful. However, we have amplified a monoallelic transcript that overlaps IGF2R intron 2 and may be indicative of macaque AIRN (Supplemental Table 1).
Other maternally imprinted loci examined in the cynomolgus macaque showed expression profiles more closely conserved with their human and mouse counterparts. The SNRPN cluster includes genes heavily involved in neural development, as exemplified by cognitive impairments in patients with imprinting defects in this region (Knoll et al. 1989). Like in human and mouse, known paternally expressed genes within the SNRPN cluster (MKRN3, MAGEL2, NDN and SNRPN) are imprinted in macaque somatic tissues including cerebellum (Nicholls and Knepper 2001), whereas the known maternally expressed counterparts (UBE3A and ATP10A) are imprinted in a tissue-specific and individual-dependent manner (imprinted UBE3A expression: cerebellum only, imprinted expression of ATP10A: placenta only) (Fig. 1B).
The product of PLAGL1, a paternally expressed imprinted gene, is proposed to be a master regulator of multiple imprinted genes (Varrault et al. 2006) and is imprinted in all tested mouse and human tissues (Arima et al. 2000; Kamiya et al. 2000; Piras et al. 2000). Macaque PLAGL1 was imprinted in most tissues examined, though imprinted expression in liver shows parental bias (Fig. 1B). In mouse liver, a biallelic alternative transcript of Plagl1 originates >50 kb upstream of the imprinted Plagl1 transcript and shares overlapping exons, confounding analysis of Plagl1 expression in mouse. A similar biallelic transcript, if present in macaques, would potentially contribute to the incomplete imprinting observed (Piras et al. 2000; Valleley et al. 2007).
The L3MBTL1 locus represents a recently characterized human imprinted gene that is not imprinted in mouse, despite their apparent sequence homology (Li et al. 2005). Imprinting at this locus was acquired after the divergence of primate from rodent, and the status of this gene in macaque helps to further refine this point of divergence. Though L3MBTL1 imprinted expression is conserved in the macaque, its adjacent genes are generally not imprinted (Fig. 1). SGK2, located immediately downstream from L3MBTL1, is polymorphically imprinted in macaque, suggesting that it could be imprinted as a bystander to the regulation at L3MBTL1 (Aziz et al. 2013). Intriguingly, we also noted that imprinted expression of L3MBTL1 and SGK2 in adult macaque peripheral blood monocytes also associated with gender, which may warrant future consideration with larger sample sizes (Supplemental Table 1).
A number of imprinted genes in the mouse are thought to originate from a retrotransposition of the X chromosome into an autosomal host gene (McCole and Oakey 2008). Of four such genes (Nap1l5, Inpp5f transcript variant 2, Mcts2, and Zrsr1), three were shown to have corresponding human orthologs (NAP1L5, INPP5F transcript variant 2, and MCTS2). The fourth, antisense transcript Zrsr1 in mouse Commd1 (also known as Murr1) arose independently in rodents after a primate-rodent divergence and resulted in Commd1 imprinting by transcriptional interference (Wang et al. 2004; Zhang et al. 2006; Wood et al. 2007b). With no primate ortholog of Zrsr1, it is not surprising that macaque COMMD1, as in human, was biallelic in all tissues analyzed (Fig. 1B). At the mouse Inpp5f locus, three transcript variants, two of which are imprinted, have been identified (Wood et al. 2007a,b; McCole and Oakey 2008). Conserved in human, imprinted expression of INPP5F transcript variant 2 but not INPP5F transcript variant 1 (hereafter abbreviated as INPP5F_v1 and INPP5F_v2, respectively) has also been reported in human fetal spinal cord, brain, heart, and tongue (Wood et al. 2007b). Likewise, in macaques, INPP5F_v1 shows no evidence of imprinted expression, while INPP5F_v2 is monoallelically expressed in all extraembryonic and somatic tissues tested (Fig. 1B).
The region from PEG3-ZNF264 has undergone a number of changes in primates. Compared to its rodent counterpart, the gene distance between PEG3 and ZIM2 has become increasingly abbreviated with primate evolution, resulting in the gradual loss of an intervening Zim1 transcript and the overlap of ZIM2 and PEG3 transcripts (Huang and Kim 2009). In macaque placenta, ZIM2 is polymorphically imprinted, while ZNF264, the most distal of these genes, is biallelically expressed in somatic tissues, in agreement with bovine data (Kim et al. 2007). ZNF264 is imprinted in macaque placenta. It is possible that the increased gene distances between ZNF264, ZIM3, and the PEG3 DMR in primates may now include elements that restrict imprinting of ZNF264 to the extraembryonic lineage (Fig. 1B). While it may be possible that the DMR we have identified may not be the critical control DMR for the PEG3 cluster, we were unable to find other somatic DMRs in the region (data not shown).
Conservation of differential methylation at orthologous imprinting control regions (ICRs) in the macaque
We next assessed putative ICRs of imprinted gene clusters by bisulfite sequencing and assumed parental origin of methylation at the ICR based on published rodent and human findings (Figs. 2, 3; Supplemental Fig. 5), since no parental macaque samples were available for confirmation. Putative ICR regions were identified using BLAT and VISTA tools against known human and/or mouse ICR regions, with coordinates shown in Supplemental Figure 3 (Kent 2002; Frazer et al. 2004). All genes and DMRs identified in the macaque retained high sequence and syntenic homology, both within individual genes and across the cluster, and were not duplicated elsewhere in the macaque genome.
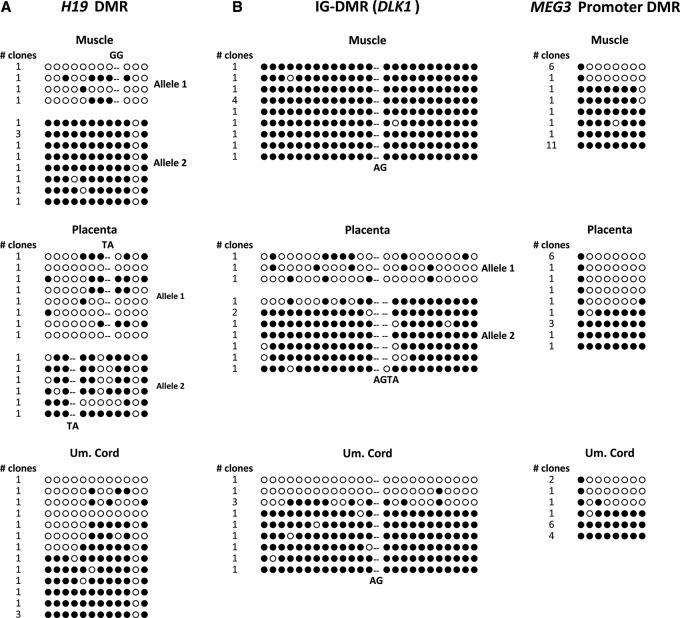
Figure 2.
Methylation at paternally imprinted gene clusters. Methylation of DMRs at the H19 (A) and DLK1 (B) clusters in extraembryonic and somatic tissues was obtained by bisulfite sequencing. Although parental/offspring samples were not available, allelic distinction between methylated/unmethylated alleles is consistent between tissues from the same individual and supports imprinting, not random allelic inactivation, at these loci. SNPs around each DMR were used to make the allelic distinctions shown.SNPs around each DMR were used to make the allelic distinctions shown.
Figure 3.
Methylation at known maternally imprinted gene clusters. Differentially methylated ICRs are evident for all maternally imprinted clusters in both somatic and extraembryonic tissues. Individual panels represent DMRs of each cluster as titled. (A) IGF2R intronic DMR. (B) INPP5F_v1 is biallelically expressed with an unmethylated promoter and is shown alongside the imprinted INPP5F_v2 for comparison. (C) KCNQ1 DMR. (D) NAP1L5 DMR. (E) PEG3 DMR. (F) SNRPN DMR (PWS-IC). (G) PLAGL1 DMR. SNPs around each DMR were used to make the allelic distinctions shown.
The macaque H19 DMR is located ∼2 kb upstream of H19, as in human and mouse. Informative cynomolgus samples show distinct differences in the degree of methylation on each parental allele, with the presumed paternal allele more heavily methylated, although sporadic CpGs interspersed within the putative DMR show an opposite pattern within each clone (Fig. 2A). Fujimoto et al. (2005) previously showed that the H19 DMR in juvenile rhesus muscle showed variable methylation, although this was determined by methylation-specific PCR and did not show base resolution methylation (Fujimoto et al. 2005; Mitalipov et al. 2007). In human, base resolution bisulfite sequencing of the H19 DMR in lymphocytes also showed variable methylation between clones, although the averaged methylation across the region approximates 50% (Kerjean et al. 2000; Borghol et al. 2006). Our results demonstrate that this complexity at the human H19 DMR is also seen in the macaque.
The DLK1 IG-DMR and MEG3 DMR were located by homology to their known human DMRs (Kagami et al. 2008, 2010). In the human, long-range interactions between these DMRs may establish the MEG3 DMR as a secondary ICR in somatic tissues, while the IG-DMR alone is responsible for placental imprinting (Kagami et al. 2010). Consistent with a more prominent role for the MEG3 DMR in somatic tissues, the macaque IG-DMR is differentially methylated in term placenta and umbilical cord; yet in adult somatic tissue (Fig. 2B; Supplemental Fig. 5), this region no longer retains allele-specific methylation. This suggests a diminished role for IG-DMR maintenance in the adult macaque following establishment of the MEG3 DMR, which is differentially methylated in extraembryonic and analyzed somatic tissues. This differs markedly from the human IG-DMR, where differential methylation appears to be retained in adult somatic tissues (Kagami et al. 2008, 2010), although hypermethylation has been observed in the human placenta (Murphy et al. 2012). In mouse, the more distal section of the IG-DMR gradually loses complete parental distinction of methylation, with somatic tissues of mid-gestation embryos showing a marked increase in maternal methylation (Sato et al. 2011). Interestingly, there is little sequence conservation between primates and rodents at the respective established IG-DMRs, with homology largely limited to the 3′ end of the DMR (Paulsen et al. 2001). Comparisons of the homologous macaque and human IG-DMR sequences also suggest that despite high overall sequence conservation of >96%, CpG site homology is far lower at ∼50% (data not shown), suggesting that additional mechanisms and sequence features may be required to maintain differential methylation at the IG-DMR and MEG3 DMR in each species.
The SNRPN DMR is known to be maternally methylated in juvenile rhesus macaque muscle and ES cell lines (Fujimoto et al. 2005; Mitalipov et al. 2007). Our results in cynomolgus macaque tissues are in agreement, and we further demonstrate that differential methylation is also seen in placenta and umbilical cord (Fig. 3). We also show that the ICRs of KCNQ1 and IGF2R are differentially methylated in somatic tissues, although some hypermethylation is evident in placenta at the IGF2R DMR, perhaps reflective of a reduced requirement for DMR maintenance in the extraembryonic lineage. This was also observed at the PEG3 DMR (Fig. 3; Supplemental Fig. 5). Imprinted genes PLAGL1, NAP1L5, and INPP5F_v2 were also associated with differential methylation at their promoter regions (Fig. 3). In comparison, the nonimprinted INPP5F_v1 promoter was unmethylated, even though this variant shares exons with the imprinted INPP5F_v2 gene.
Determination of germline imprinting in the primate
Germline imprints are DMRs acquired during gametogenesis (Ferguson-Smith 2011) and depend on DNA methyltransferases to establish and maintain these modifications (Tucker et al. 1996; Kaneda et al. 2004b; Weaver et al. 2009). DNMT mutant mice exhibit loss of methylation and perturbed imprinting (Biniszkiewicz et al. 2002; Kaneda et al. 2004a; Kato et al. 2007), and mice knockouts for specific germline DMRs have demonstrated that these serve as imprinting control regions for the locus (Thorvaldsen et al. 1998; Bielinska et al. 2000; Fitzpatrick et al. 2002; Lopes et al. 2003; da Rocha et al. 2008). The mouse has been the primary species for determining DMR germline origin (Supplemental Table 6). A small number of human germline DMR analyses have been assessed at the DLK1, H19, KCNQ1, and SNRPN loci at orthologs to the murine DMRs, though this may be subject to inherent methylation defects within the accessible patient samples (Geuns et al. 2003, 2007a,b). More recently, whole-genome bisulfite sequencing of human oocytes suggested hypermethylation across all known maternally derived ICRs (Okae et al. 2014), and we sought to establish if there were common features of these regions between human and nonhuman primates.
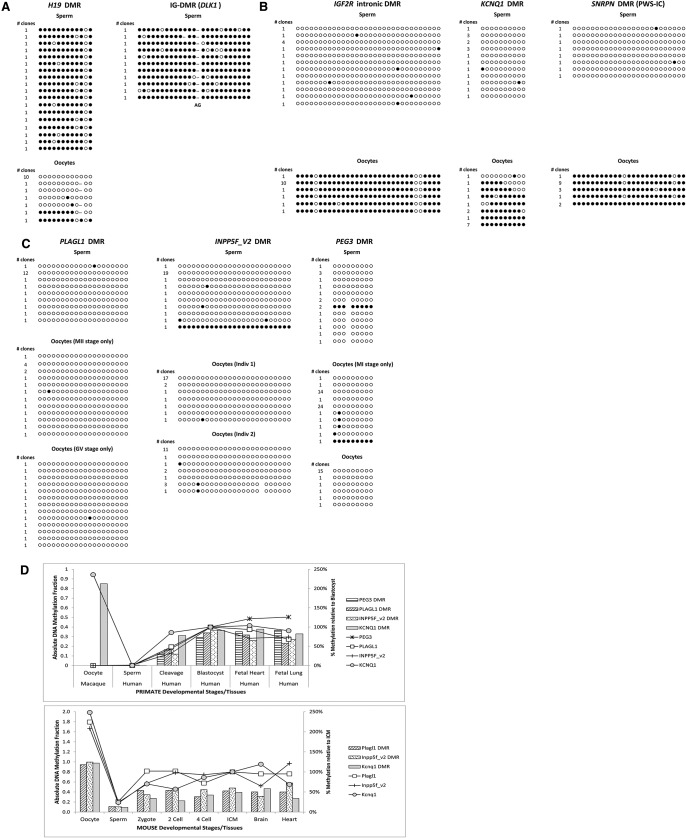
Having analyzed macaque tissues for differential methylation at putative DMRs, we next used primate gametes to assess the germline origin of these DMRs. Previously, we determined that the primate-specific imprinted gene, L3MBTL1, associates with a maternal germline imprint (Aziz et al. 2013). In this study, all eight DMRs showed germline methylation consistent with the expected parental origin of the locus (Fig. 4A–C). We also analyzed macaque oocytes and found reciprocal methylation at four of the eight loci examined, in line with their expected germline origin. Insufficient oocyte material was available to assess the DLK1 IG-DMR. Oocytes were collected from ovarian stimulated and normal cycling females, cleaned of cumulus cells and debris following enzymatic incubation and mechanical pipetting, then pooled by individual for subsequent analysis. In assurance of appropriate technical care, oocytes were largely unmethylated at the paternally imprinted H19 DMR, consistent with the human germline DMR status (Borghol et al. 2006).
Figure 4.
Germline imprints in the cynomolgus macaque. DNA methylation in macaque gametes fall into three distinct groups of paternally (A) and maternally (B) acquired germline imprints, and a third group (C) with delayed maternal imprint acquisition. Except for PLAGL1 (lower oocyte image: GV only; upper oocyte image: MII only), all other oocyte samples comprised of pooled stages (GV, MI, and MII). In D, macaque oocyte methylation is shown alongside meta-analysis of data from Smith et al. (2014), showing that the lack of oocyte methylation at PLAGL1, PEG3, and INPP5F_v2 (patterned bars) is consistent with human, where methylation is gradually acquired across the primate embryonic cleavages, and only completed by the blastocyst stage. In contrast, mouse homologs show that imprinted methylation is present in oocytes and already complete in the early zygote. The intronic KCNQ1/Kcnq1 DMR (gray bar) is shown for comparison. No data were available for mouse Peg3. (Left axis, bar chart) Absolute DNA methylation, (right axis, line graph) percent DNA methylation relative to level in ICM/blastocyst.
Similarly, at SNRPN, IGF2R, and KCNQ1, known maternal germline inherited DMRs in rodents were also fully methylated in macaque oocytes (Fig. 4B). At the human SNRPN locus, the timing of maternal imprint acquisition is debated and centers on the presence or absence of a complete imprint by the meiosis II (MII) stage in oocytes (El-Maarri et al. 2001; Geuns et al. 2003). Our sample at SNRPN comprised three oocyte stages (GV, MI, MII) and was fully methylated, in agreement with the analysis of Geuns et al. (2003) on human oocytes.
Intriguingly, we expected maternal germline methylation at three other loci (PLAGL1, PEG3, and INPP5F_v2), but instead found these largely unmethylated. To confirm that this was not due to sampling limitations, we repeated this on an independent sample, with similar outcomes (Fig. 4C). The lack of methylation in macaque oocytes at these loci suggests that the imprinting mechanisms conferring DMR identity at a germline stage may not be fully conserved between species. In the mouse, DMRs of Plagl1, Peg3, and Inpp5f_v2 are maternally inherited in the germline (Supplemental Table 6). Concordant with this expectation, macaque sperm at these DMRs are unmethylated (Fig. 4C). Yet in macaque oocytes, we found no evidence of maternal methylation, indicating that this is acquired post-fertilization, since allele-specific differential methylation was seen in somatic tissues (Fig. 3). Recently available data on the methylomes of human gametes and early embryos suggests that there are subtle but distinct differences between human and rodent development, with human genome-wide remethylation complete only in the post-implantation embryo (Guo et al. 2014; Smith et al. 2014). Meta-analysis of data from Smith et al. (2014) indicated that the three imprinted loci which, in macaque, were unmethylated in the female germline, acquired methylation during the early cleavage to blastocyst stages in human (Fig. 4D). In contrast, the intronic DMR at KCNQ1, which we show is germline in the macaque, maintains methylation from the cleavage stages through to later development. These findings are consistent with the lack of methylation we observe in the macaque oocyte at PLAGL1, PEG3, and INPP5F_v2 and suggest that maternal-specific methylation may indeed be acquired during these early primate post-fertilization divisions. No overlapping DMR coverage was found in the oocyte data sets from Guo et al. (2014). It remains to be determined if alternative maternal-specific marks, such as histone modifications, may already be in place in gametes.
Discussion
From a broad survey of known human/mouse imprinted genes in the nonhuman primate, we have found that at most clusters, imprinted expression and differential methylation is concordant with both the widespread and tissue-specific patterns observed in human and mouse. Our analysis also revealed previously unrecorded tissue-specific imprinting in healthy adult primate tissues such as kidney, muscle, pancreas, and testes.
This study also uncovered novel primate-specific imprinting features:
At the expression level, a novel transcript of CDKN1C appears to be specific to the cynomolgus macaque. The CDKN1C transcript variant results from alternative splicing between exon 1 and 2, though the expected splice junction retains the classic GT-AC donor-acceptor sequence (data not shown). In the human genome, its corresponding sequence is characterized by a highly repetitive region. Between primates, segmental duplications are associated with repetitive sequences, regions of high instability, and recombination proposed to have contributed to primate evolution (Kehrer-Sawatzki and Cooper 2008). Such alterations may promote transcriptional or regulatory changes. Furthermore, macaque-human synteny maps localize the CDKN1C-containing KCNQ1 cluster <500 kb from a chromosomal inversion in macaques, consistent with a possible shared role of direct repeats as well as longer range enhancer elements in directing transcriptional splicing (Ventura et al. 2007).
Polymorphic imprinting has been described for IGF2R in humans. At this cluster, we demonstrate that macaque imprinting appears to have features that are intermediate between human and mouse. It is clear that the IGF2R intron 2 DMR found in human is also present in the macaque. However, the polymorphic imprinting of IGF2R that has been described for human was not evident in the macaque tissues analyzed in our study. It therefore appears that imprinted IGF2R is evident in at least this old world primate species (Fig. 1; Supplemental Table 1). The species-specific difference may be genetically conferred; however, we note that a single ZFP57 binding site, required for the maintenance of imprints in early development in rodents (Li et al. 2008), is shared between the human and macaque in intron 2 of IGF2R, hence this is unlikely to be the cause of the species-specific difference.
At the IGF2R locus, a syntenic disruption occurs between human and macaque. Macaque IGF2R is located more proximal to a neocentromere feature than its human equivalent—this association might partially account for the differences observed in this cluster (Ventura et al. 2007; Rocchi et al. 2009). Further comparative genomic mapping may highlight features that contribute to imprinting regulation control.
We also observed some potential differences between human and macaque in the retention of the DMR status at the IG-DMR in somatic tissues, suggesting that the more conserved methylation status of the nearby MEG3 DMR might function as the primary ICR in somatic tissues.
In addition, we analyzed L3MBTL1 and its downstream neighbors for evidence of imprinting. Our macaque data show conserved L3MBTL1 imprinting between primate species and also expand the present understanding of tissue-specific imprinting at this locus, which is not imprinted in mouse or marsupial (Aziz et al. 2013).
Our understanding of genomic imprinting mechanisms is based primarily on genetic and epigenetic studies in mouse, with conservation of all DMRs found between mouse and human somatic tissues. Analysis of patients with parental origin effects such as Beckwith-Wiedemann and Prader-Willi/Angelman syndromes indicates that these murine germline-derived DMRs are also functionally conserved ICRs in human. However, the developmental origins of primate ICRs are not well established, and we were keen to verify if previously ascertained or assumed human germline DMRs were indeed germline in origin.
The limited availability of human gametes has precluded analysis from all but two maternal inherited loci DMRs (KCNQ1, SNRPN) and two paternally inherited loci (DLK1, H19) (Kerjean et al. 2000; El-Maarri et al. 2001; Geuns et al. 2003; Borghol et al. 2006). While reduced representative bisulfite sequencing (RRBS) data on human gametes and early embryos have recently become available, these techniques do not guarantee coverage at all loci of interest but have provided a global perspective on DNA methylation at each developmental stage (Guo et al. 2014; Smith et al. 2014). Whole-genome bisulfite sequencing of human oocytes suggests that maternally derived ICRs are hypermethylated in the human (Okae et al. 2014), including the orthologs of the three loci that we show with delayed maternal imprinting in macaque oocytes. Although averaged rather than specific locus detail is presented, this may point to species-specific differences in the acquisition of imprints between humans and nonhuman primates.
We obtained oocytes from both normal and ovarian stimulated female macaques and manually removed surrounding cumulus cells by multiple washes and pipetting with increasingly narrower bores. The presence of two clones with partial methylation at the H19 DMR in oocytes suggests the presence of maternal germline epimutation, or that despite our best efforts, our oocyte sample may have rare somatic cell contamination (no contamination was evident at other loci using the same 46-cell sample). Nonetheless, >87% of the reads represented unmethylated clones, and we interpret this to mean that, as in the mouse, the maternal allele at the H19 DMR is predominantly unmethylated in the female germline.
In mice, the exact acquisition timing of maternal germline imprints differs between loci but is completed by the MII stage (Obata and Kono 2002; Hiura et al. 2006). While superovulation in mice can also contribute to a dose-dependent effect on imprinting, this is evident only with oocytes derived from overstimulation, with the Snrpn, Peg3, and Kcnq1 DMRs showing loss of methylation in oocytes derived from high dose ovulation protocols (Market-Velker et al. 2009). In macaque, these potentially sensitive loci appear unaffected by our stimulation protocol, since the majority of imprinted loci examined show the expected methylation patterns. At SNRPN and KCNQ1, normal maternal imprints were evident in superovulated macaque oocytes. Results for DMRs at H19, KCNQ1, IGF2R, and SNRPN also confirmed canonical germline imprinting, in agreement with known imprints from nonprimate species.
Unexpectedly, three DMRs of established germline origin in rodents did not acquire their DMR status in the macaque germline. At the PLAGL1, INPP5F_v2, and PEG3 DMRs, no methylation was found in either the sperm or oocytes examined. These findings question the central dogma that DNA methylation at all ICRs is established during gametogenesis and is subsequently resistant to the genome-wide demethylation events that occur post-fertilization. While genomic rearrangements between species, particularly at the PEG3 cluster, may not conclusively point toward the regions analyzed as the sole or critical DMR for the cluster, we did not find other somatic DMRs at the USP29 nor ZNF264 promoters within the cluster, suggesting we have not missed other regions with the potential to be germline, unless they are transitory.
Additionally, we do not anticipate that the germline absence of methylation observed at these loci is an artifact associated with oocyte maturation or superovulation, since PLAGL1 methylation was absent in an exclusively MII oocyte sample from one individual, yet SNRPN methylation was complete in a separate oocyte sample that included less mature GV oocytes. Neither do we suspect conversion failure at rare non-CG methylated primer targets. Instead, we postulate that post-oocyte acquisition of methylation at PLAGL1, INPP5F_v2, and PEG3 is a primate phenomenon. It is notable that in the human methylome, global DNA hypomethylation was seen in the early human cleavage stages (Guo et al. 2014; Smith et al. 2014). More specifically, we looked at imprinted genes with coverage in these methylome data sets. Consistent with our findings, DNA methylation at PLAGL1, PEG3, and INPP5F_v2 was only partially acquired by the cleavage stage, whereas the KCNQ1 DMR was methylated at levels similar to somatic tissues by this early developmental time point and corresponding full maternal imprints already present in oocytes (Smith et al. 2014).
When compared to other imprinted loci with parental-specific DNA methylation already acquired in the germline, it is possible that parental histone marks may be different at PLAGL1, INPP5F_v2, and PEG3, leading to the post-fertilization differential recruitment of DNMTs. Notably, the DNMT profile in primates differs from mouse, with a marked relative reduction of DNMT3 de novo methylases in primates at the oocyte stage, during which imprints are established in rodents (Huntriss et al. 2004; Vassena et al. 2005). However, within this study, we were unable to assess allele-specific histone modifications. In primates, DNMT3A levels rise ∼10-fold as oocytes progress to fertilized pronuclei-stage zygotes. Plausibly, this surge of DNMT enzymes may assist in completion of germline DMR methylation while the parental chromosomes are still separated (Vassena et al. 2005). Alternatively, there may be species-specific genetic differences in the occurrence of oocyte-specific upstream promoters, thought to drive transcription required for oocyte-specific acquisition of methylation at imprinted ICRs (Smallwood and Kelsey 2012).
What might be the implications of deferred maternal methylation at these genes in primates? Prior to the maternal-zygotic transition, the emphasis of regulatory mechanisms depends more on post-transcriptional activities, not zygotic transcription. In primates, the zygotic genome is activated later than the very early two-cell activation found in mice. To this end, we speculate that the selection for pre-fertilization imprint acquisition via maternal germline methylation is relaxed in the primate, with little negative impact on the early embryo. It is noteworthy that the early germline-derived imprinted X inactivation described in mice is not found in human (Okamoto et al. 2011). The dosage of imprinted genes such as PLAGL1 and PEG3 is important for postnatal metabolism. Maternal modulation of their epigenetic status after, rather than before fertilization, may be permissible in species such as primates with a later onset of zygotic transcription. This might provide an adaptive programming mechanism sensitive to environmental resources that provides a selective advantage to offspring.
With our findings, it is evident that though imprinted expression is largely conserved between Eutherians, the timing and exact mechanisms employed might involve subtle yet profound differences between species. We suggest that these findings in a nonhuman primate emphasize the importance of post-fertilization events in imprinting control (Hanna and Kelsey 2014).
Methods
Nonhuman primate use
Adult cynomolgus macaques were housed at the A*STAR Non-human Primate Facility. Animals 5–10 yr of age and surplus to the breeding colony requirements at the center were euthanized, and tissues collected for further analysis. Cynomolgus birth tissues were collected from a breeding facility in Vietnam. All animal protocols and experiments were approved and conducted in accordance with requirements of the SingHealth Institutional Animal Care and Use Committee (IACUC #2009/SHS/509). No primate parent-offspring pairs were available for parent-specific allelic expression analysis.
Ovarian stimulation and oocyte retrieval
Regular monthly cycling female macaques (n = 6) between 5 and 8 yr of age were administered with follicle stimulating hormone (rhFSH, 5.5–7 units/kg, twice daily, GONAL-f, Merck-Serono) for 7–10 d from the onset of menstruation. From Day 7, luteinizing hormone (rhLH, 20 units/kg, twice daily, Luveris, Merck-Serono) and gonadotropin releasing hormone antagonist (GnRH antagonist, 5 μg/kg, once daily, Cetrotide, Merck-Serono) were also delivered intra-muscularly. Animals were monitored by ultrasound from Day 8 and administered with a single chorionic gonadotropin-α dose when more than five follicles (>3 mm) were visible (rhCG-a, 1000 units, Ovidrel, Merck-Serono). Twenty-four to twenty-seven hours later, animals were subjected to an ovariectomy. Cumulus-oocyte complexes (COCs) were aspirated from follicles and placed in equilibrated KSOM medium (Sigma-Aldrich) overnight at 37°C, 5% CO2 to allow for maturation of any immature oocytes. The following morning, healthy COCs were placed in M2 medium (Sigma) containing 0.025% trypsin and 1 mg/mL hyaluronidase, and cumulus cells were removed by mechanical pipetting. Denuded oocytes were washed in consecutive rounds of M2 medium and classified by nuclear maturity. Oocytes from the same individual were pooled for DNA analysis, numbers as follows: H19, INPP5F (Indiv. 1), and SNRPN, n = 46 (GV-1, MI-25, MII-20); INPP5F (Indiv. 2), n = 17; PEG3, n = 14; PLAGL1, GV-6, MII-12; KCNQ1, n = 36 (MI-28, MII-8).
Sperm retrieval
Testes were retrieved from adult male macaques euthanized for nonfertility-related studies at the animal facility. Surface blood vessels were bled to reduce blood contamination, washed, and then placed in a dish with fresh PBS. To release sperm, the epididymis was cut repeatedly with scissors and gently agitated for 10 min at room temperature to allow motile sperm to swim out. This PBS suspension was then spun down at 1500g, 15 min, 4°C to remove excess volume and the pellet used for DNA analysis.
DNA isolation, bisulfite conversion, and PCR
DNA isolated from sperm was based on a two-step lysis procedure as previously described (Alcivar et al. 1989; Carracedo 2005). Contaminating somatic cells were first lysed by placing the sperm pellet in buffer containing 0.8 mg/mL Proteinase K (Roche Applied Sciences), 30 min at 37°C. Subsequently, sperm were lysed by addition of fresh buffer with 0.8 mg/mL Proteinase K and 40 mM DTT, incubated overnight at 55°C, and used for standard DNA extraction. Oocytes were subject to multiple freeze-thaw cycles, then digested for 90 min in buffer containing 0.25 mg/mL Proteinase K and 2.5 μM SDS at 37°C—protocol modified from Zuccotti and Monk (1995). Proteinase K was inactivated at 98°C for 15 min. The lysed cell extract was used directly for bisulfite conversion. Prior to DNA extraction, whole-blood samples were subject to lysis of red blood cells in a hypotonic buffer (20 mM Tris-Cl, pH 8.0, 0.1 M NaCl, 25 mM EDTA, 0.5% SDS). The white cell pellet was lysed with proteinase K and used for standard DNA extraction. All other solid tissues were first homogenized in gentleMACS M tubes (Miltenyi Biotec) before standard DNA extraction. Bisulfite conversions were done according to manufacturers’ protocols (Zymo EZ DNA Methylation Gold or Qiagen EpiTect Bisulfite Kit). The SequalPrep Long Polymerase PCR kit (Invitrogen) was used for all bisulfite-PCRs, using the recommended PCR program and 40 cycles. Picked clones for each sample were capillary-sequenced and analyzed using a BiQ Analyzer (Bock et al. 2005). Bisulfite conversion efficiencies for all samples considered ranged from 95% to 100%, with an overall average of 99%, comparable to the reported efficiencies of the bisulfite conversion kits. Capillary sequences for a single sample were mapped to the in silico bisulfite converted reference genome, and conversion efficiencies were calculated by counting the number of unconverted, non-CpG cytosine residues in the amplicon, against the total number of non-CpG cytosines in the original unconverted sequence. Occurrences of these nonconverted sequences were rare, and we note that they cannot formally be distinguished from C/T polymorphisms that may exist on the parental alleles. Animals used are not inbred, and any allelic differences shown on the bisulfite lollipop diagrams were determined at a non-C/T SNP present within the amplicon used for bisulfite sequencing. If any, identical capillary sequences of separate clones were taken to be nonindependent clones resulting from PCR amplification—only a single representation of these clones is shown in each bisulfite lollipop diagram, with the number of clones clearly shown by the side.
SNP analysis
To identify informative individuals for subsequent analysis of allele-specific expression, genomic DNA surrounding exon regions were amplified by PCR and sequenced. Novel SNPs were identified by alignment, using the rhesus macaque genomic sequence as reference (Rhesus Genome, version rheMac2). These SNPs are available through NCBI dbSNP (accession numbers in Supplemental Table 10).
RNA isolation, reverse transcription and pyrosequencing for allele-specific expression
Tissues for RNA extraction were homogenized in TRIzol (Invitrogen) and processed according to the manufacturer's protocol. Following phase separation, the aqueous phase was further treated with DNase I and purified on RNeasy mini columns according to the manufacturer's protocol (Qiagen). Reverse transcription was primed with random hexamers (Hi Capacity cDNA Reverse Transcription Kit, Applied Biosystems) and cDNA used for subsequent PCR with AccuPrime Taq (Invitrogen) for 30–35 cycles. RT-PCR products were subjected to standard capillary sequencing and the allele specificity of expression determined by the presence or absence of multiple nucleotide peaks at a predetermined polymorphic location. Pyrosequencing primers were designed to cover the same polymorphism but included a 5′ biotin primer on one end for capture on sepharose beads. Briefly, cDNA was amplified with HotStarTaq Master Mix (Qiagen) at 55°C for 40 cycles. The RT-PCR product was then bound to sepharose beads via the biotin tag and denatured to generate single-stranded DNA to allow annealing of an internal sequencing primer. Pyrosequencing was performed in AQ mode on a Qiagen PyroMark Q24 machine using 10 μL of amplified cDNA product and 0.3 μM of sequencing primer.
Data access
M. fascicularis SNPs used in this study have been uploaded to NCBI dbSNP (www.ncbi.nlm.nih.gov/SNP), with reference to assembly GCF_000364345.1 (Macaca_fascicularis_5.0), using the following accession numbers: ss1414417769, ss1414417873, ss1536213772. These SNPs are also reported in Supplemental Table 10. Cynomolgus-specific CDKN1C transcripts from adult tissues have been submitted to NCBI GenBank (www.ncbi.nlm.nih.gov/genbank/) with accession numbers KP238484 (liver), KP238485 (muscle), KP238486 (kidney), KP238487 (lung). Capillary sequence data used in this study are accessible through NCBI Trace Archive (www.ncbi.nlm.nih.gov/Traces) under the TI accession numbers 2340903719–2340904081. Qiagen PyroMark data is available online in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank the following individuals for their technical support in the preparation of this study: Bhuvaneshwari Shunmuganathan, Samantha Juanita Liew, David Sin, Yong-Chee Tan, Trisse S.K. Goh, Rico Tan, Vilma Templonuevo, and Carnette Custodio Pulma. We also thank Emilia E.C. Tng, Aileen A.L. Lim, Pan Hong, and Peter Gluckman for discussions, support, and advice. This study was conducted by all authors while at the Singapore Institute for Clinical Research and was fully supported by funding from the Agency for Science, Technology and Research, Singapore.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.183301.114.
Freely available online through the Genome Research Open Access option
References
- Alcivar AA, Hake LE, Millette CF, Trasler JM, Hecht NB. 1989. Mitochondrial gene expression in male germ cells of the mouse. Dev Biol 135: 263–271. [DOI] [PubMed] [Google Scholar]
- Arima T, Drewell RA, Oshimura M, Wake N, Surani MA. 2000. A novel imprinted gene, HYMAI, is located within an imprinted domain on human chromosome 6 containing ZAC. Genomics 67: 248–255. [DOI] [PubMed] [Google Scholar]
- Aziz A, Baxter EJ, Edwards C, Cheong CY, Ito M, Bench A, Kelley R, Silber Y, Beer PA, Chng K, et al. 2013. Cooperativity of imprinted genes inactivated by acquired chromosome 20q deletions. J Clin Invest 123: 2169–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinska B, Blaydes SM, Buiting K, Yang T, Krajewska-Walasek M, Horsthemke B, Brannan CI. 2000. De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat Genet 25: 74–78. [DOI] [PubMed] [Google Scholar]
- Biniszkiewicz D, Gribnau J, Ramsahoye B, Gaudet F, Eggan K, Humpherys D, Mastrangelo MA, Jun Z, Walter J, Jaenisch R. 2002. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol Cell Biol 22: 2124–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. 2005. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 21: 4067–4068. [DOI] [PubMed] [Google Scholar]
- Borghol N, Lornage J, Blachere T, Sophie Garret A, Lefevre A. 2006. Epigenetic status of the H19 locus in human oocytes following in vitro maturation. Genomics 87: 417–426. [DOI] [PubMed] [Google Scholar]
- Bourne GH. 1975. The rhesus monkey. Academic Press, New York. [Google Scholar]
- Brandeis M, Ariel M, Cedar H. 1993a. Dynamics of DNA methylation during development. Bioessays 15: 709–713. [DOI] [PubMed] [Google Scholar]
- Brandeis M, Kafri T, Ariel M, Chaillet JR, McCarrey J, Razin A, Cedar H. 1993b. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J 12: 3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Cullen BR. 2007. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13: 313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo Á. 2005. Forensic DNA typing protocols. Humana Press, Totowa, NJ. [Google Scholar]
- Caspary T, Cleary MA, Baker CC, Guan XJ, Tilghman SM. 1998. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol Cell Biol 18: 3466–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaton MA, Edwards CA, Ferguson-Smith AC. 2014. Phenotypic outcomes of imprinted gene models in mice: elucidation of pre- and postnatal functions of imprinted genes. Annu Rev Genomics Hum Genet 15: 93–126. [DOI] [PubMed] [Google Scholar]
- Constancia M, Hemberger M, Hughes J, Dean W, Ferguson-Smith A, Fundele R, Stewart F, Kelsey G, Fowden A, Sibley C, et al. 2002. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature 417: 945–948. [DOI] [PubMed] [Google Scholar]
- Dao D, Frank D, Qian N, O'Keefe D, Vosatka RJ, Walsh CP, Tycko B. 1998. IMPT1, an imprinted gene similar to polyspecific transporter and multi-drug resistance genes. Hum Mol Genet 7: 597–608. [DOI] [PubMed] [Google Scholar]
- da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. 2008. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet 24: 306–316. [DOI] [PubMed] [Google Scholar]
- Davies W, Isles AR, Humby T, Wilkinson LS. 2007. What are imprinted genes doing in the brain? Epigenetics 2: 201–206. [DOI] [PubMed] [Google Scholar]
- de la Puente A, Hall J, Wu YZ, Leone G, Peters J, Yoon BJ, Soloway P, Plass C. 2002. Structural characterization of Rasgrf1 and a novel linked imprinted locus. Gene 291: 287–297. [DOI] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC. 2007. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol 19: 281–289. [DOI] [PubMed] [Google Scholar]
- Edwards CA, Mungall AJ, Matthews L, Ryder E, Gray DJ, Pask AJ, Shaw G, Graves JA, Rogers J; SAVOIR Consortium, et al. 2008. The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol 6: e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarri O, Buiting K, Peery EG, Kroisel PM, Balaban B, Wagner K, Urman B, Heyd J, Lich C, Brannan CI, et al. 2001. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet 27: 341–344. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC. 2011. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet 12: 565–575. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins MJ. 2002. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet 32: 426–431. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res 32: W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost JM, Monk D, Stojilkovic-Mikic T, Woodfine K, Chitty LS, Murrell A, Stanier P, Moore GE. 2010. Evaluation of allelic expression of imprinted genes in adult human blood. PLoS One 5: e13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto A, Mitalipov SM, Clepper LL, Wolf DP. 2005. Development of a monkey model for the study of primate genomic imprinting. Mol Hum Reprod 11: 413–422. [DOI] [PubMed] [Google Scholar]
- Fujimoto A, Mitalipov SM, Kuo HC, Wolf DP. 2006. Aberrant genomic imprinting in rhesus monkey embryonic stem cells. Stem Cells 24: 595–603. [DOI] [PubMed] [Google Scholar]
- Gabory A, Ripoche MA, Yoshimizu T, Dandolo L. 2006. The H19 gene: regulation and function of a non-coding RNA. Cytogenet Genome Res 113: 188–193. [DOI] [PubMed] [Google Scholar]
- Gallagher E, McGoldrick A, Chung WY, McCormack O, Harrison M, Kerin M, Dervan PA, McCann A. 2006. Gain of imprinting of SLC22A18 sense and antisense transcripts in human breast cancer. Genomics 88: 12–17. [DOI] [PubMed] [Google Scholar]
- Geuns E, De Rycke M, Van Steirteghem A, Liebaers I. 2003. Methylation imprints of the imprint control region of the SNRPN-gene in human gametes and preimplantation embryos. Hum Mol Genet 12: 2873–2879. [DOI] [PubMed] [Google Scholar]
- Geuns E, De Temmerman N, Hilven P, Van Steirteghem A, Liebaers I, De Rycke M. 2007a. Methylation analysis of the intergenic differentially methylated region of DLK1-GTL2 in human. Eur J Hum Genet 15: 352–361. [DOI] [PubMed] [Google Scholar]
- Geuns E, Hilven P, Van Steirteghem A, Liebaers I, De Rycke M. 2007b. Methylation analysis of KvDMR1 in human oocytes. J Med Genet 44: 144–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gicquel C, Rossignol S, Cabrol S, Houang M, Steunou V, Barbu V, Danton F, Thibaud N, Le Merrer M, Burglen L, et al. 2005. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat Genet 37: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Gould TD, Pfeifer K. 1998. Imprinting of mouse Kvlqt1 is developmentally regulated. Hum Mol Genet 7: 483–487. [DOI] [PubMed] [Google Scholar]
- Grace KS, Sinclair KD. 2009. Assisted reproductive technology, epigenetics, and long-term health: a developmental time bomb still ticking. Semin Reprod Med 27: 409–416. [DOI] [PubMed] [Google Scholar]
- Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J, et al. 2014. The DNA methylation landscape of human early embryos. Nature 511: 606–610. [DOI] [PubMed] [Google Scholar]
- Han VK, Carter AM. 2000. Spatial and temporal patterns of expression of messenger RNA for insulin-like growth factors and their binding proteins in the placenta of man and laboratory animals. Placenta 21: 289–305. [DOI] [PubMed] [Google Scholar]
- Hanna CW, Kelsey G. 2014. The specification of imprints in mammals. Heredity 113: 176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka K, Fujii K, Horai S. 1996. Molecular phylogeny of macaques: implications of nucleotide sequences from an 896-base pair region of mitochondrial DNA. Mol Biol Evol 13: 1044–1053. [DOI] [PubMed] [Google Scholar]
- Hernandez A, Fiering S, Martinez E, Galton VA, St. Germain D. 2002. The gene locus encoding iodothyronine deiodinase type 3 (Dio3) is imprinted in the fetus and expresses antisense transcripts. Endocrinology 143: 4483–4486. [DOI] [PubMed] [Google Scholar]
- Hiura H, Obata Y, Komiyama J, Shirai M, Kono T. 2006. Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells 11: 353–361. [DOI] [PubMed] [Google Scholar]
- Huang JM, Kim J. 2009. DNA methylation analysis of the mammalian PEG3 imprinted domain. Gene 442: 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntriss J, Hinkins M, Oliver B, Harris SE, Beazley JC, Rutherford AJ, Gosden RG, Lanzendorf SE, Picton HM. 2004. Expression of mRNAs for DNA methyltransferases and methyl-CpG-binding proteins in the human female germ line, preimplantation embryos, and embryonic stem cells. Mol Reprod Dev 67: 323–336. [DOI] [PubMed] [Google Scholar]
- Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. 1992. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev 6: 705–714. [DOI] [PubMed] [Google Scholar]
- Kagami M, Sekita Y, Nishimura G, Irie M, Kato F, Okada M, Yamamori S, Kishimoto H, Nakayama M, Tanaka Y, et al. 2008. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet 40: 237–242. [DOI] [PubMed] [Google Scholar]
- Kagami M, O'Sullivan MJ, Green AJ, Watabe Y, Arisaka O, Masawa N, Matsuoka K, Fukami M, Matsubara K, Kato F, et al. 2010. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet 6: e1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya M, Judson H, Okazaki Y, Kusakabe M, Muramatsu M, Takada S, Takagi N, Arima T, Wake N, Kamimura K, et al. 2000. The cell cycle control gene ZAC/PLAGL1 is imprinted—a strong candidate gene for transient neonatal diabetes. Hum Mol Genet 9: 453–460. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. 2004a. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429: 900–903. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Sado T, Hata K, Okano M, Tsujimoto N, Li E, Sasaki H. 2004b. Role of de novo DNA methyltransferases in initiation of genomic imprinting and X-chromosome inactivation. Cold Spring Harb Symp Quant Biol 69: 125–129. [DOI] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. 2007. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16: 2272–2280. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Cooper DN. 2008. Molecular mechanisms of chromosomal rearrangement during primate evolution. Chromosome Res 16: 41–56. [DOI] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjean A, Dupont JM, Vasseur C, Le Tessier D, Cuisset L, Paldi A, Jouannet P, Jeanpierre M. 2000. Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum Mol Genet 9: 2183–2187. [DOI] [PubMed] [Google Scholar]
- Killian JK, Byrd JC, Jirtle JV, Munday BL, Stoskopf MK, MacDonald RG, Jirtle RL. 2000. M6P/IGF2R imprinting evolution in mammals. Mol Cell 5: 707–716. [DOI] [PubMed] [Google Scholar]
- Killian JK, Nolan CM, Wylie AA, Li T, Vu TH, Hoffman AR, Jirtle RL. 2001. Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum Mol Genet 10: 1721–1728. [DOI] [PubMed] [Google Scholar]
- Kim J, Bergmann A, Choo JH, Stubbs L. 2007. Genomic organization and imprinting of the Peg3 domain in bovine. Genomics 90: 85–92. [DOI] [PubMed] [Google Scholar]
- Knoll JH, Nicholls RD, Magenis RE, Graham JM Jr, Lalande M, Latt SA. 1989. Angelman and Prader-Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am J Med Genet 32: 285–290. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Wagatsuma H, Ono R, Ichikawa H, Yamazaki M, Tashiro H, Aisaka K, Miyoshi N, Kohda T, Ogura A, et al. 2000. Mouse Peg9/Dlk1 and human PEG9/DLK1 are paternally expressed imprinted genes closely located to the maternally expressed imprinted genes: mouse Meg3/Gtl2 and human MEG3. Genes Cells 5: 1029–1037. [DOI] [PubMed] [Google Scholar]
- Lee MP, Hu RJ, Johnson LA, Feinberg AP. 1997. Human KVLQT1 gene shows tissue-specific imprinting and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nat Genet 15: 181–185. [DOI] [PubMed] [Google Scholar]
- Li Y, Sasaki H. 2011. Genomic imprinting in mammals: its life cycle, molecular mechanisms and reprogramming. Cell Res 21: 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. 1993. Role for DNA methylation in genomic imprinting. Nature 366: 362–365. [DOI] [PubMed] [Google Scholar]
- Li J, Bench AJ, Piltz S, Vassiliou G, Baxter EJ, Ferguson-Smith AC, Green AR. 2005. L3mbtl, the mouse orthologue of the imprinted L3MBTL, displays a complex pattern of alternative splicing and escapes genomic imprinting. Genomics 86: 489–494. [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. 2008. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell 15: 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SP, Coan P, da Rocha ST, Seitz H, Cavaille J, Teng PW, Takada S, Ferguson-Smith AC. 2007. Differential regulation of imprinting in the murine embryo and placenta by the Dlk1-Dio3 imprinting control region. Development 134: 417–426. [DOI] [PubMed] [Google Scholar]
- Lopes S, Lewis A, Hajkova P, Dean W, Oswald J, Forne T, Murrell A, Constancia M, Bartolomei M, Walter J, et al. 2003. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum Mol Genet 12: 295–305. [DOI] [PubMed] [Google Scholar]
- Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davies C, Barlow DP. 2000. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet 25: 19–21. [DOI] [PubMed] [Google Scholar]
- Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR. 2009. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet 19: 36–51. [DOI] [PubMed] [Google Scholar]
- McCole RB, Oakey RJ. 2008. Unwitting hosts fall victim to imprinting. Epigenetics 3: 258–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitalipov S, Clepper L, Sritanaudomchai H, Fujimoto A, Wolf D. 2007. Methylation status of imprinting centers for H19/IGF2 and SNURF/SNRPN in primate embryonic stem cells. Stem Cells 25: 581–588. [DOI] [PubMed] [Google Scholar]
- Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, Feil R, Moore GE. 2006. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci 103: 6623–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morison IM, Ramsay JP, Spencer HG. 2005. A census of mammalian imprinting. Trends Genet 21: 457–465. [DOI] [PubMed] [Google Scholar]
- Murphy SK, Huang Z, Hoyo C. 2012. Differentially methylated regions of imprinted genes in prenatal, perinatal and postnatal human tissues. PLoS One 7: e40924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD, Knepper JL. 2001. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet 2: 153–175. [DOI] [PubMed] [Google Scholar]
- Nielsen EM, Hansen L, Stissing T, Yanagisawa K, Borch-Johnsen K, Poulsen P, Vaag A, Hansen T, Pedersen O. 2005. Studies of variations of the cyclin-dependent kinase inhibitor 1C and the cyclin-dependent kinase 4 genes in relation to type 2 diabetes mellitus and related quantitative traits. J Mol Med 83: 353–361. [DOI] [PubMed] [Google Scholar]
- Nowak K, Stein G, Powell E, He LM, Naik S, Morris J, Marlow S, Davis TL. 2011. Establishment of paternal allele-specific DNA methylation at the imprinted mouse Gtl2 locus. Epigenetics 6: 1012–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata Y, Kono T. 2002. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J Biol Chem 277: 5285–5289. [DOI] [PubMed] [Google Scholar]
- Ogawa O, McNoe LA, Eccles MR, Morison IM, Reeve AE. 1993. Human insulin-like growth factor type I and type II receptors are not imprinted. Hum Mol Genet 2: 2163–2165. [DOI] [PubMed] [Google Scholar]
- Okae H, Chiba H, Hiura H, Hamada H, Sato A, Utsunomiya T, Kikuchi H, Yoshida H, Tanaka A, Suyama M, et al. 2014. Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet 10: e1004868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf JP, Renard JP, Duranthon V, et al. 2011. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472: 370–374. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. [DOI] [PubMed] [Google Scholar]
- Osada N, Hashimoto K, Kameoka Y, Hirata M, Tanuma R, Uno Y, Inoue I, Hida M, Suzuki Y, Sugano S, et al. 2008. Large-scale analysis of Macaca fascicularis transcripts and inference of genetic divergence between M. fascicularis and M. mulatta. BMC Genomics 9: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudejans CB, Westerman B, Wouters D, Gooyer S, Leegwater PA, van Wijk IJ, Sleutels F. 2001. Allelic IGF2R repression does not correlate with expression of antisense RNA in human extraembryonic tissues. Genomics 73: 331–337. [DOI] [PubMed] [Google Scholar]
- Paulsen M, Takada S, Youngson NA, Benchaib M, Charlier C, Segers K, Georges M, Ferguson-Smith AC. 2001. Comparative sequence analysis of the imprinted Dlk1-Gtl2 locus in three mammalian species reveals highly conserved genomic elements and refines comparison with the Igf2-H19 region. Genome Res 11: 2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras G, El Kharroubi A, Kozlov S, Escalante-Alcalde D, Hernandez L, Copeland NG, Gilbert DJ, Jenkins NA, Stewart CL. 2000. Zac1 (Lot1), a potential tumor suppressor gene, and the gene for ε-sarcoglycan are maternally imprinted genes: identification by a subtractive screen of novel uniparental fibroblast lines. Mol Cell Biol 20: 3308–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree MB, Hore TA, Shaw G, Graves JA, Pask AJ. 2009a. Evolution of genomic imprinting: insights from marsupials and monotremes. Annu Rev Genomics Hum Genet 10: 241–262. [DOI] [PubMed] [Google Scholar]
- Renfree MB, Papenfuss AT, Shaw G, Pask AJ. 2009b. Eggs, embryos and the evolution of imprinting: insights from the platypus genome. Reprod Fertil Dev 21: 935–942. [DOI] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium, Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, et al. 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316: 222–234. [DOI] [PubMed] [Google Scholar]
- Riesewijk AM, Schepens MT, Welch TR, van den Berg-Loonen EM, Mariman EM, Ropers HH, Kalscheuer VM. 1996. Maternal-specific methylation of the human IGF2R gene is not accompanied by allele-specific transcription. Genomics 31: 158–166. [DOI] [PubMed] [Google Scholar]
- Rocchi M, Stanyon R, Archidiacono N. 2009. Evolutionary new centromeres in primates. Prog Mol Subcell Biol 48: 103–152. [DOI] [PubMed] [Google Scholar]
- Sato S, Yoshida W, Soejima H, Nakabayashi K, Hata K. 2011. Methylation dynamics of IG-DMR and Gtl2-DMR during murine embryonic and placental development. Genomics 98: 120–127. [DOI] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810–813. [DOI] [PubMed] [Google Scholar]
- Sleutels F, Tjon G, Ludwig T, Barlow DP. 2003. Imprinted silencing of Slc22a2 and Slc22a3 does not need transcriptional overlap between Igf2r and Air. EMBO J 22: 3696–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Kelsey G. 2012. De novo DNA methylation: a germ cell perspective. Trends Genet 28: 33–42. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A. 2014. DNA methylation dynamics of the human preimplantation embryo. Nature 511: 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits G, Mungall AJ, Griffiths-Jones S, Smith P, Beury D, Matthews L, Rogers J, Pask AJ, Shaw G, VandeBerg JL, et al. 2008. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet 40: 971–976. [DOI] [PubMed] [Google Scholar]
- Smrzka OW, Fae I, Stoger R, Kurzbauer R, Fischer GF, Henn T, Weith A, Barlow DP. 1995. Conservation of a maternal-specific methylation signal at the human IGF2R locus. Hum Mol Genet 4: 1945–1952. [DOI] [PubMed] [Google Scholar]
- Takada S, Tevendale M, Baker J, Georgiades P, Campbell E, Freeman T, Johnson MH, Paulsen M, Ferguson-Smith AC. 2000. Δ-like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr Biol 10: 1135–1138. [DOI] [PubMed] [Google Scholar]
- Takada S, Paulsen M, Tevendale M, Tsai CE, Kelsey G, Cattanach BM, Ferguson-Smith AC. 2002. Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2-H19. Hum Mol Genet 11: 77–86. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev 12: 3693–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KL, Beard C, Dausmann J, Jackson-Grusby L, Laird PW, Lei H, Li E, Jaenisch R. 1996. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev 10: 1008–1020. [DOI] [PubMed] [Google Scholar]
- Tycko B, Morison IM. 2002. Physiological functions of imprinted genes. J Cell Physiol 192: 245–258. [DOI] [PubMed] [Google Scholar]
- Uribe-Lewis S, Woodfine K, Stojic L, Murrell A. 2011. Molecular mechanisms of genomic imprinting and clinical implications for cancer. Expert Rev Mol Med 13: e2. [DOI] [PubMed] [Google Scholar]
- Valleley EM, Cordery SF, Bonthron DT. 2007. Tissue-specific imprinting of the ZAC/PLAGL1 tumour suppressor gene results from variable utilization of monoallelic and biallelic promoters. Hum Mol Genet 16: 972–981. [DOI] [PubMed] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, et al. 2006. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell 11: 711–722. [DOI] [PubMed] [Google Scholar]
- Vassena R, Dee Schramm R, Latham KE. 2005. Species-dependent expression patterns of DNA methyltransferase genes in mammalian oocytes and preimplantation embryos. Mol Reprod Dev 72: 430–436. [DOI] [PubMed] [Google Scholar]
- Ventura M, Antonacci F, Cardone MF, Stanyon R, D'Addabbo P, Cellamare A, Sprague LJ, Eichler EE, Archidiacono N, Rocchi M. 2007. Evolutionary formation of new centromeres in macaque. Science 316: 243–246. [DOI] [PubMed] [Google Scholar]
- Wang Y, Joh K, Masuko S, Yatsuki H, Soejima H, Nabetani A, Beechey CV, Okinami S, Mukai T. 2004. The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented 1-rs1 gene. Mol Cell Biol 24: 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Weaver JR, Susiarjo M, Bartolomei MS. 2009. Imprinting and epigenetic changes in the early embryo. Mamm Genome 20: 532–543. [DOI] [PubMed] [Google Scholar]
- Weksberg R, Smith AC, Squire J, Sadowski P. 2003. Beckwith-Wiedemann syndrome demonstrates a role for epigenetic control of normal development. Hum Mol Genet 12: R61–R68. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Bourc'his D, Bestor TH, Oakey RJ. 2007a. Allele-specific demethylation at an imprinted mammalian promoter. Nucleic Acids Res 35: 7031–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Roberts RG, Monk D, Moore GE, Schulz R, Oakey RJ. 2007b. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet 3: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Smrzka OW, Barlow DP. 1998. Making sense of imprinting the mouse and human IGF2R loci. Novartis Found Symp 214: 251–259; discussion 260–253. [DOI] [PubMed] [Google Scholar]
- Xu Y, Goodyer CG, Deal C, Polychronakos C. 1993. Functional polymorphism in the parental imprinting of the human IGF2R gene. Biochem Biophys Res Commun 197: 747–754. [DOI] [PubMed] [Google Scholar]
- Yan G, Zhang G, Fang X, Zhang Y, Li C, Ling F, Cooper DN, Li Q, Li Y, van Gool AJ, et al. 2011. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat Biotechnol 29: 1019–1023. [DOI] [PubMed] [Google Scholar]
- Yotova IY, Vlatkovic IM, Pauler FM, Warczok KE, Ambros PF, Oshimura M, Theussl HC, Gessler M, Wagner EF, Barlow DP. 2008. Identification of the human homolog of the imprinted mouse Air non-coding RNA. Genomics 92: 464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Joh K, Yatsuki H, Wang Y, Arai Y, Soejima H, Higashimoto K, Iwasaka T, Mukai T. 2006. Comparative analyses of genomic imprinting and CpG island-methylation in mouse Murr1 and human MURR1 loci revealed a putative imprinting control region in mice. Gene 366: 77–86. [DOI] [PubMed] [Google Scholar]
- Zuccotti M, Monk M. 1995. Methylation of the mouse Xist gene in sperm and eggs correlates with imprinted Xist expression and paternal X-inactivation. Nat Genet 9: 316–320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.