Soil ecologists have debated the relative importance of dispersal limitation ("everything" is not "everywhere") and ecological factors in determining the structure of soil microbial communities. The relative explanatory power of spatial and ecological factors, including plant species identity and even plant relatedness, for different fractions of the soil microbial community (i.e. bacterial and fungal communities) are poorly understood. We sampled field soils in a northern California field site, and find that soil microbial community variation correlates most strongly with plant species identity, followed by soil chemistry, spatial location and plant genus.
Keywords: Coastal grassland community, niche, soil bacterial community, soil fungal community, terminal-restriction fragment length polymorphism
Abstract
Soil ecologists have debated the relative importance of dispersal limitation and ecological factors in determining the structure of soil microbial communities. Recent evidence suggests that ‘everything is not everywhere’, and that microbial communities are influenced by both dispersal limitation and ecological factors. However, we still do not understand the relative explanatory power of spatial and ecological factors, including plant species identity and even plant relatedness, for different fractions of the soil microbial community (i.e. bacterial and fungal communities). To ask whether factors such as plant species, soil chemistry, spatial location and plant relatedness influence rhizosphere community composition, we examined field-collected rhizosphere soil of seven congener pairs that occur at Bodega Bay Marine Reserve, CA, USA. We characterized differences in bacterial and fungal communities using terminal-restriction fragment length polymorphism. Plant species identity was the single best statistical predictor of both bacterial and fungal community composition in the root zone. Soil microbial community structure was also correlated with soil chemistry. The third best predictor of bacterial and fungal communities was spatial location, confirming that everything is not everywhere. Variation in microbial community composition was also related to combinations of spatial location, soil chemistry and plant relatedness, suggesting that these factors do not act independently. Plant relatedness explained less of the variation than plant species, soil chemistry, or spatial location. Despite some congeners occupying different habitats and being spatially distant, rhizosphere fungal communities of plant congeners were more similar than expected by chance. Bacterial communities from the same samples were only weakly similar between plant congeners. Thus, plant relatedness might influence soil fungal, more than soil bacterial, community composition.
Introduction
The study of microbial community structure has long centred around the debate between the hypothesis that ‘everything is everywhere’ (Bass Becking 1934), and the alternative that dispersal limitation influences microbial community structure (O'Malley 2007). Because the soil microbial community is diverse, heterogeneous and difficult to characterize (Singh et al. 2004), understanding the environmental correlates of soil microbial communities has lagged behind community ecology studies in other systems (Fierer and Ladau 2012; Fierer et al. 2012). Thus the relative explanatory powers of ecological factors (e.g. plant species identity, soil chemistry and plant relatedness) and dispersal limitation (i.e. spatial location) for determining soil microbial communities are not well characterized (reviewed in Berg and Smalla 2009). By comparing bacterial and fungal community datasets, we explore whether ecological and spatial factors structure soil microbial communities, and, if so, how bacterial and fungal communities differ.
Plant–microbial relationships are often plant species-specific (reviews in Ehrenfeld et al. 2005; Hardoim et al. 2008; Berg and Smalla 2009). For example, legume species are often associated with particular strains or species of rhizobia bacteria (Long 2001). Plants exude chemicals from their roots that can foster beneficial microbes in the rhizosphere (reviewed in Ehrenfeld et al. 2005; Bais et al. 2006; Compant et al. 2010). For example, Arabidopsis thaliana exudes malic acid in the presence of a pathogen, and malic acid attracts a beneficial bacterium, Bacillus subtilis, protecting the roots from the pathogen (Rudrappa et al. 2008). However, plant species-specific relationships are not universally found for soil microbial communities (Ehrenfeld et al. 2005), and data sets comparing bacterial and fungal communities for plant species-specificity are rare (but see, e.g. Ushio et al. 2008).
Soil chemistry also influences soil microbial community composition, diversity and activity (Ehrenfeld et al. 2005). For example, differences in pH explained 70 % of the variance in bacterial community diversity across soils sampled from North and South America (Fierer and Jackson 2006). Greater soil fertility (NPK) increases bacterial biomass and activity in grassland mesocosms and these effects interact with plant species identity in some systems (Innes et al. 2004) and not in others (Bardgett et al. 1999), thus effects of soil chemistry are often system-dependent. Whether or how these soil chemistry effects may interact with plant species identity or plant relatedness is not generally known.
Spatial location could also influence soil microbial composition, especially if microbial taxa are dispersal limited, resulting in spatial autocorrelation of microbial communities (e.g. Ettema and Wardle 2002; Fierer and Ladau 2012). Early workers argued that microbes do not experience significant dispersal limitation, and thus ‘everything is everywhere: but the environment selects’ (Bass Becking 1934, reviewed in O'Malley 2007). However, more recent work has found strong spatial autocorrelation in many microbial communities, such that spatially close communities are more similar than expected by chance, and this similarity decays with distance (Ettema and Wardle 2002). Such a pattern of spatial autocorrelation suggests that dispersal limitation does influence patterns of distribution for microbes (O'Malley 2007; Rout and Callaway 2012). The relative importance of dispersal limitation and ecological factors is still an area of ongoing research (Berg and Smalla 2009).
If closely related plants are similar in root morphology (e.g. Comas and Eissenstat 2009; Valverde-Barrantes et al. 2014), root exudates, or other drivers of microbial associations, plant relatedness might also be expected to correlate with soil microbial community structure. Consistent with this hypothesis, plant clades, in some cases, have similar responses to soil biota (e.g. Brandt et al. 2009; Liu et al. 2012; Reinhart et al. 2012; Anacker et al. 2014). Closely related plants also often share characteristics of root morphology, such as the absence of many fine roots (Valverde-Barrantes et al. 2014), a trait which is correlated with higher mycorrhizal colonization (Manjunath and Habte 1991; Brundrett 2002). In addition, there is evidence, especially in Orchidaceae, of closely related plants sharing similar soil fungal associates (Jacquemyn et al. 2011; Martos et al. 2012) and close relatives sometimes respond similarly to arbuscular mycorrhizal fungi (e.g. Reinhart et al. 2012; Lugo et al. 2015). However, such relationships are by no means universal, and there are cases were plant relatedness does not correlate with effects of soil biota (e.g. Reinhart and Anacker 2014). Previous studies have often focused on whole-soil effects (i.e. plant-soil feedbacks, e.g. Burns and Strauss 2011) or have focused on a narrow fraction of the soil community (e.g. mycorrhizae, Reinhart et al. 2012; Reinhart and Anacker 2014); therefore, there is little comparative evidence about the relative influence of plant relatedness on different components of the soil microbiome (e.g. bacterial and fungal communities).
We quantified the relative contributions of plant species, soil chemistry, spatial location and plant relatedness on soil microbial communities at Bodega Bay Marine Reserve, CA, USA. (i) We predicted that plant species identity may influence soil microbial community composition, if species differ in factors, such as root exudates, which influence microbial community composition. (ii) Soil chemistry (e.g. N, pH) may correlate with microbial communities, for example, if soil microbe species differ in their nutrient preferences or soil chemistry niche axes. (iii) Spatial separation may influence microbial community composition across samples, if microbes are dispersal limited. (iv) Finally, plant congeners may have similar soil microbial communities, if close relatives have similar traits or habitat preferences, which influence soil microbial community composition.
Methods
To explore microbial community composition, we sampled field soils and used DNA fragment analysis to characterize differences among soil samples. For each of the 14 angiosperm species, we sampled soils from 6 unique collection locations in the field, at least 100 m apart, at Bodega Bay Marine Reserve, CA, USA [see Supporting Information]. These species are Cirsium occidentale, C. quercetorum, Fragaria chiloensis, F. vesca, Gilia capitata ssp. chamissonis, G. millefoliata, Plantago erecta, P. subnuda, Rumex crassus, R. occidentalis, Sanicula arctopoides, S. crassicaulis, Trifolium fucatum and T. gracilentum. Soil samples were collected from the root zone of each focal plant species to 15 cm depth, roots were removed and soils were mixed within a core. Soils therefore represent rhizosphere and non-rhizosphere soil within the active root zone of the selected plants species, which we expect to be under the influence of the plant and therefore represent microbial communities cultivated by the influence of the representative plant species. Soil samples were stored on dry ice in the field and moved to −80 °C within <12 h. To quantify the role of spatial proximity, we mapped collection locations using GPS coordinates.
Soil bacterial and fungal communities
Terminal-restriction fragment length polymorphism (TRFLP) was used to characterize the soil bacterial and fungal communities following established methods (as in, for example, Burke et al. 2008). Terminal-restriction fragment length polymorphism is a DNA fragmentation technique that quantifies differences in the soil microbial community among samples but which cannot identify microbes to specific taxa. Terminal-restriction fragment length polymorphism estimates of microbial community structure have been found to be generally robust and concur with alternative approaches such as 454 pyrosequencing in documenting microbial community differences (Pilloni et al. 2012). See details of the TRFLP methodology in the Supporting Information. In brief, for each soil sample, DNA was extracted from two 500-mg subsamples of collected soil, amplified using PCR to target bacterial or fungal-specific target regions and restriction enzymes were used to digest the PCR product. We then averaged the subsample extractions within a soil sample for analysis. Terminal-restriction fragment length polymorphism results in size fragments that represent operational taxonomic units (OTUs) in our analyses.
Soil chemistry
To characterize soil chemical properties, soil samples taken from each soil collection (n = 84) were analysed for NO3 (ppm), Olsen P (ppm), K (ppm), K (meq/100 g), Na (ppm), Ca (meq/100 g), Mg (meq/100 g), cation-exchange capacity, organic matter and pH by the Division of Agriculture and Natural Resources Analytical Laboratory at the University of California, Davis.
Phylogeny estimation
We sampled seven congeneric pairs and six soil replicates per species, and two subsamples per soil sample, to assess heterogeneity in soil microbial communities. We tested for effects of plant phylogeny, as well as tested just for effects of plant genus on microbial community composition (see below).
To estimate the phylogeny among our sampled plant species, we downloaded all available DNA regions for matK, ITS and trnL-trnF from genBank for the 14 sampled plant species [see Supporting Information]. Each species was represented in the data set by at least 1 DNA region, matK was present for 7 species, ITS for 13 and trnL-trnF for 11 [see Supporting Information]. We aligned the sequences separately for each DNA region with MEGA version 5.2.2 (Tamura et al. 2011) using MUSCLE (Edgar 2004a, b) with the default gap open penalty of −400 and then concatenated the alignments. We conducted a maximum likelihood search using Garli version 0.951 (Zwickl 2006) using a GTR + Γ + I model of evolution. To incorporate prior knowledge about angiosperm phylogeny, we enforced a constraint tree with the known relationships among genera within the Asterids (Stevens 2009). Bootstrap analysis with 100 replicates was conducted without constraints. Thus bootstrap values reflect repeatability of clades, based on the raw DNA data. We calibrated the branch lengths using r8s with the NPRS method (Sanderson 2003) and dates fixed at the Fabaceae, Rosaceae, Asteraceae, Apiaceae, Polygonaceae, Lamiales, Caryophyllales, coreeudicots, eurosid1 and easterid2 based on published fossil calibration dates (Wikström et al. 2001).
Data analysis
Soil community diversity
We ordinated the community data using non-metric multidimensional scaling. We used the ‘metaMDS’ function in the ‘vegan’ package (Oksanen et al. 2010), using 20 random starts and a Bray–Curtis distance matrix (Faith et al. 1987). We conducted ordinations with and without data transformation, and with and without removing rare taxa (taxa present in ≤7 % of subsamples) prior to ordination. These strategies did not reduce ordination stress, and we therefore present ordinations on untransformed whole community data. In addition, we conducted visual inspection of a graph of stress versus the number of dimensions in the ordination, and chose the number of axes that reduced stress below 0.20, where the graph appeared to asymptote (k = 4). NMDS ordination resulted in a reduced number of dimensions that describe differences in soil communities, and with which we could conduct further analyses.
To describe the amount of community variance potentially represented by the ordination, we conducted a post hoc partial Mantel correlation between a Euclidean distance matrix from the ordination and the total community data matrix using Bray–Curtis distances following recommendations in McCune and Grace (2002). We used multi.mantel in the phytools package with 1000 permutations (Revell 2012) to conduct this analysis and present R2 values converted to percentages.
Do plant species identity, soil chemistry, spatial location and phylogenetic relatedness predict similarity in rhizosphere microbial communities?
We used two statistical tools to explore the correlations in this data set: phylogenetic eigenvector regression (PVR) and a variance partitioning analysis. The PVR approach allows us to test for statistical significance of effects of our predictors on soil microbial community structure. Because many predictors may covary in this observational data set, the variance partitioning approach allows us to ask about the relative explanatory power of combinations of predictors and is purely descriptive.
First, we fit four PVR models, each with one of the ordination axes as the response variable; predictors in each model included the eigenvectors that represented plant species, soil chemistry, spatial location and plant phylogeny, as described below. To acquire phylogenetic eigenvectors, we first decomposed the phylogeny with 14 plant taxa using PVRdecomp in the PVR library (Diniz-Filho et al. 1998, 2012). We examined a graph of the eigenvalues against the vector number for the decomposition and used a visual break in the data to determine which eigenvectors to consider for the model selection, which included the first seven. Next, we decomposed spatial distance into a set of two eigenvectors (Bellier et al. 2007). To select a minimal model for each ordination axis, we fit a full linear model with the first seven phylogenetic eigenvectors and two spatial eigenvectors using the lm function. We then choose the subset of phylogenetic eigenvectors that were statistically significant (P < 0.05) for the reduced model (Diniz-Filho et al. 2012). This approach never resulted in more than three phylogenetic predictors. Because identical values for eigenvectors were associated with each species, species identity could not simultaneously be included in this model. We compared this full model (e.g. fungal community axis 1 ∼ plant phylogeny + spatial location) to models dropping effects from the model using a likelihood ratio test to determine the statistical significance of each predictor. Because ‘phylogeny’ and ‘spatial location’ consisted of multiple vectors, this approach tested the overall significance of these predictors of microbial community structure. To select minimal soil chemical models, we used stepAIC on a model with all soil chemical vectors and included only statistically significant predictors in the final model. This reduced variance inflation factors to below 2.5 for all predictors. Because phylogeny co-varied with soil chemistry (Spearman rank correlation range: −0.44 to 0.59) and models with both phylogeny and soil chemistry resulted in high multicollinearity (VIF > 100), phylogeny and soil chemistry effects were tested separately. Finally, we report the adjusted R2 for models with each of four classes of predictors (i.e. plant species, soil chemistry, spatial location and plant phylogeny) individually, to estimate the maximum relative explanatory power of each, adjusted for the number of predictors in the model. Because our sampling was concentrated on congeners, we also conducted a PVR analysis with plant genus as a predictor, rather than plant phylogeny, to ask whether plant genus alone could predict differences in microbial community composition.
Standard diagnostic plots were examined and residuals were normally distributed for all PVR models. We used adjusted R2 value comparisons among models to ensure that models were not over-fit. Variance inflation factors were <2.5 for all predictors in all final models (median VIF = 1.03), which is generally considered very low multicollinearity (Fox and Monette 1992; O'Brien 2007).
Finally, some of these predictors may covary, such as if closely related species prefer the same habitats and are thus spatially close together, and multiple predictors combined could influence microbial community composition. To describe the combined effects of plant phylogeny, soil chemistry and spatial location on soil community structure, we used variance partitioning methods (Desdevises et al. 2003; Peres-Neto et al. 2006; Gonçalves-Souza et al. 2014). See the Supporting Information for more details about this analysis. All analyses were conducted in the R statistical package (version 3.0.2) (R Development Core Team 2008). Data are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.9bf7f.
Results
Soil chemistry
Soils from the Bodega Bay site were primarily loamy sand with a somewhat acidic average pH of 6.07 ± 0.05 SE (range = 5.01–8.03). Organic matter was generally low with a mean 4.2 % ± 0.2 SE (range = 0.6–15.5 %). Salinity was highly variable and ranged from 12 to 390 ppm with a mean of 91.8 ± 6.7 SE at this coastal site.
Bacterial communities
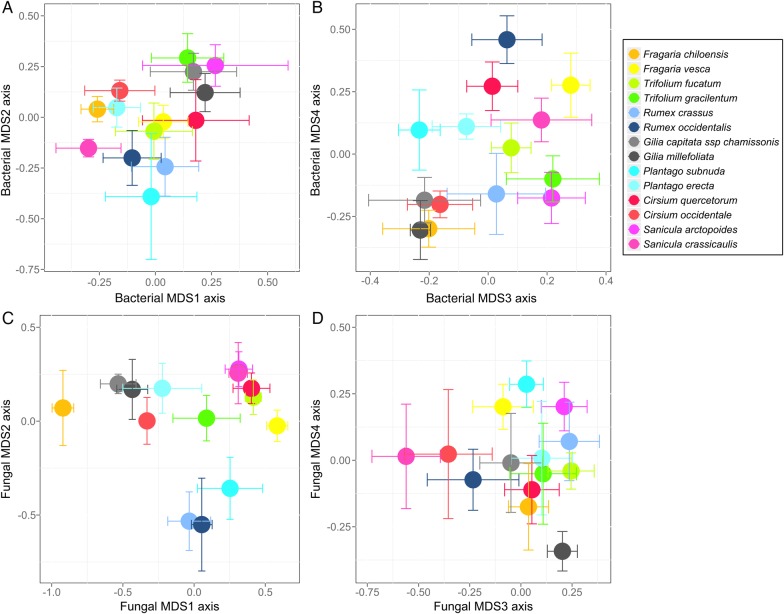
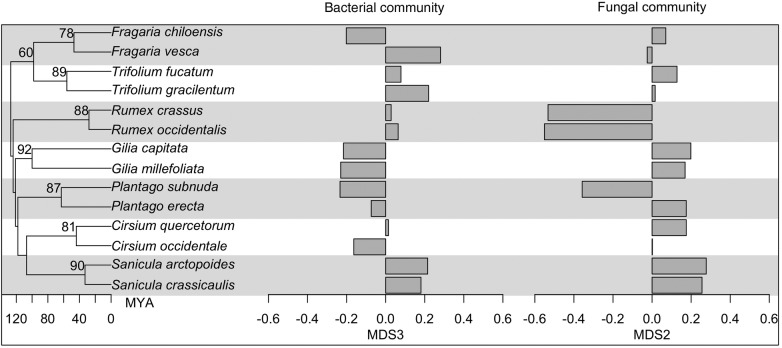
NMDS ordination resulted in a stress of 0.12 with four ordination axes (Fig. 1A and B), and the total ordination (axes 1–4 combined) explained ∼82 % of the bacterial community composition in a partial Mantel test. Bacterial communities in the rhizosphere exhibited plant species-specific patterns, and plant species explained 15, 15 and 38 % of the variance on ordination axes 2, 3 and 4, respectively (Table 1). Variation in pH explained 19 % of the variance in bacterial community composition on axis 2 (Table 1). Variation in Mg and Ca across samples explained 16 % of the variation on axis 3 (Table 1). Variation in K, Na and Ca explained 11 % of the variance on axis 4 (Table 1). Spatial location explained 19 and 16 % of the variation on ordination axes 3 and 4 (Table 1). Plant phylogeny was a significant but poor predictor of bacterial community composition, explaining 1, 5, 9 and 8 % of the variation on ordination axes 1–4 (Table 1). An alternative analysis approach with plant genus instead of plant phylogeny explained 12 % of the variance on ordination axis 3 [see Supporting Information—Table S2], and this pattern was primarily driven by the Gillia and Sanicula congeners, which were very similar in their bacterial community composition on axis 3 (Fig. 2). In the variance partitioning analysis, soil chemistry alone explained more of the variance in bacterial community composition than other factors, and combinations of predictors (e.g. spatial location, soil chemistry and plant phylogeny combined), explained additional variation [see Supporting Information—Fig. S2].
Figure 1.
NMDS ordination with species means ± 1 SE for soil samples collected in the field at Bodega Bay, CA from the rhizospheres of 14 plant species. (A and B) The bacterial community ordination with a stress of 0.12. (C and D) The fungal community ordination with a stress of 0.15.
Table 1.
Linear PVR models were used to test the effects of plant species, soil chemistry, spatial location and plant relatedness on soil microbial community composition. Model selection was conducted by retaining statistically significant phylogenetic eigenvectors (P < 0.05) for the combined data set (see text for details). Significant results (P < 0.05) are highlighted in bold.
| Predictor | Microbial community structure ordination axes |
|||||||
|---|---|---|---|---|---|---|---|---|
| MDS1 |
MDS2 |
MDS3 |
MDS4 |
|||||
| Adj. R2 | P-value | Adj. R2 | P-value | Adj. R2 | P-value | Adj. R2 | P-value | |
| Bacterial | ||||||||
| Plant species | <0.01 | 0.42 | 0.15 | 0.02 | 0.15 | 0.02 | 0.38 | <0.001 |
| Soil chemistry | <0.01 | 0.77 | 0.19 | <0.001 | 0.16 | <0.01 | 0.11 | 0.05 |
| Spatial location | <0.01 | 0.10 | <0.01 | 0.50 | 0.19 | <0.001 | 0.16 | <0.01 |
| Phylogeny | 0.01 | 0.02 | 0.05 | 0.02 | 0.09 | 0.02 | 0.08 | 0.02 |
| Fungi | ||||||||
| Plant species | 0.56 | <0.001 | 0.31 | <0.001 | 0.20 | <0.01 | 0.03 | 0.31 |
| Soil chemistry | 0.45 | <0.001 | 0.27 | <0.001 | <0.01 | 0.17 | 0.03 | 0.06 |
| Spatial location | 0.43 | <0.001 | <0.01 | 0.45 | 0.09 | <0.001 | 0.10 | 0.05 |
| Phylogeny | <0.01 | 0.07 | 0.29 | <0.001 | <0.01 | <0.01 | 0.02 | 0.66 |
Figure 2.
Soil bacterial communities were significantly, but weakly, correlated with plant phylogeny on ordination axis 3 (Table 1). Plant phylogeny explained 29 % of the variance in soil fungal communities on ordination axis 2 (Table 1).
Fungal communities
The NMDS ordination for the fungal data set had a stress of 0.15 (Fig. 1C and D), and the total ordination (axes 1–4 combined) explained ∼71 % of the fungal community composition in a partial Mantel test. Plant species identity explained 56, 31 and 20 % of the variation on ordination axes 1, 2 and 3, respectively (Table 1). Phosphorus, Ca and Mg explained 45 % of the variance on fungal axis 1 (Table 1). Variation among soil samples in pH explained 27 % of the variation in fungal community composition on axis 2 (Table 1). Spatial location also explained a significant amount of the variance in fungal community composition, explaining 43, 9 and 10 % of the variance on ordination axes 1, 3 and 4 (Table 1). Plant phylogeny explained 29 % of the variance on ordination axis 2 (Table 1). The alternative analysis with plant genus, rather than plant phylogeny, suggested that plant genus explained 16 and 30 % of the variance on ordination axes 1 and 2, respectively [see Supporting Information—Table S2]. For example, Rumex species fungal rhizosphere communities were very close in ordination space (Fig. 1C, blue points) as were Gillia congeners (Fig. 1C, grey points), and Sanicula congeners (Fig. 1C, pink points). This signal of plant genus was especially strong on ordination axis 2 (Fig. 2, Supporting Information—Table S2). Combinations of the predictors explained larger percentages of the fungal community composition than individual predictors alone [see Supporting Information—Fig. S2].
Discussion
Plant species identity was the single best statistical predictor of soil microbial community composition
Consistent with a large body of literature (reviews in Brundrett 2002; Ehrenfeld et al. 2005; Berg and Smalla 2009), we found a signal of plant species identity on the soil bacterial and fungal communities, and plant species was the single best statistical predictor of soil microbial community variation (Table 1). Variance in bacterial community composition was higher than for fungal communities, and no combination of predictors could explain a large amount of this variance [see Supporting Information—Fig. S2]. This large variance could reflect a high degree of fine-scale heterogeneity within the root zone to which bacterial communities respond. For example, the presence of fungal biomass was found to influence the community structure of important bacterial functional groups (e.g. denitrifying bacterial) at fine spatial scales within a forest system (Burke et al. 2012). This high heterogeneity of microsites for bacterial within the root zone, coupled with the high degree of species diversity within the bacterial domain, could be responsible for the weaker predictors of bacterial communities in our study. In addition, our examination of bacterial community structure using general 16S rDNA primers could miss the potential relationship between plant species and specific bacterial functional groups. Analysis of the nitrogen-fixing functional group, for example, could reveal plant effects on that functional group, because we expect some plants to have co-evolved relationships with these nitrogen-fixing bacteria (e.g. rhizobia-plant relationships; Long 2001). The weaker signal of plant species identity on bacteria could therefore be due to our focus on overall bacterial community structure and not functional group distribution, which could yield different results.
Plant species explained a significant amount of the variance in fungal community composition (Table 1). Effects of plant species identity on soil microbial communities might be the result of ‘active’ selection by plants for soil microbes (e.g. via root exudates), a ‘passive’ by-product of nutrient uptake, root structure (Singh et al. 2004) or shared habitat preferences by plants and microbes. It is increasingly accepted that soil fungi, and especially mutualistic organisms such as mycorrhizal fungi (Brundrett 2002), may form plant species-specific relationships that can affect plant growth and community composition. Plant species-specific associations with soil microbes likely contribute to the strong plant-soil feedbacks observed in many systems (Kulmatiski et al. 2008), which have important implications for coexistence and community assembly (Bever et al. 2010).
Soil chemistry was correlated with soil microbial community composition
Soil chemistry had the second strongest correlation with differences in microbial communities (Table 1). Previous studies have found strong correlations between soil chemistry and soil microbial communities at large spatial scales (e.g. Lauber et al. 2009), consistent with our results at smaller spatial scales. Variation in pH is often correlated with bacterial communities at the continent scale (Lauber et al. 2009), and we found pH to correlate with bacterial community composition. Fungal communities, including AMF and EMF communities, also frequently differ across soils with differences in pH (Kluber et al. 2012), also consistent with the correlation between fungal ordination axis 2 and pH. Further, many fungi can affect C and N availability and cycling through their saprotrophic capabilities (e.g. production of extracellular enzymes for liberation of organic nutrients) (Leake et al. 2002), though we found no strong evidence for correlations between the fungal community and N.
Spatial location was the third best predictor of microbial community composition and was correlated with soil chemistry and plant relatedness
We found significant associations between soil microbial communities and spatial location (Table 1). Thus ‘everything’ is not ‘everywhere’ in this data set, as many other workers have also found for soil microbial communities (reviews in Ettema and Wardle 2002; Rout and Callaway 2012). This is consistent with the hypothesis that dispersal limitation influences microbial distributions even at fairly restricted spatial scales [see Supporting Information—Fig. S1]. The correlations we observed between soil chemistry, plant relatedness and spatial locations on fungal community composition suggest that these factors co-varied in the field, making disentangling the relative importance of these predictors difficult [see Supporting Information—Fig. S2].
Plant congeners had similar soil fungal, but not bacterial, communities
Plant relatedness explained less of the variation in soil microbial communities than plant species, soil chemistry or spatial location (Table 1). Plant congeners had similar soil fungal (but not bacterial) communities, even when they occurred in divergent habitats. For example, congeners in Sanicula and Rumex had similar fungal communities (Fig. 2) but they are found in different habitats and are spatially distant (>1 km) from one another [see Supporting Information—Fig. S1]. Thus, spatial location and similarity in habitat preferences between close relatives are not the sole drivers of the correlation between plant relatedness and soil fungal communities. Similar root exudates or root morphology between close relatives (Comas and Eissenstat 2009; Valverde-Barrantes et al. 2014) may be other explanations for shared fungal community composition.
DNA fragmentation tools, like TRFLP, do not identify taxa without additional work. Because our soil sampling procedure may have broken some plant roots, endophytic fungi could be contributing to the pattern we observed. Our general ITS fungal primers preferentially amplify non-arbuscular mycorrhizal fungi (D. J. Burke, pers. obs.; Taylor et al. 2014), suggesting that arbuscular mycorrhizal fungi are not driving the correlation that we observed between plant phylogeny and fungal rhizosphere community. Given the species-specific nature of some mycorrhizal associations (Brundrett 2002), we may be underestimating the importance of host plant relatedness in influencing fungal rhizosphere communities.
Meta-analyses suggest that plant relatedness does not explain much of the overall variance in plant-soil feedbacks, where plant performance is compared between conspecific soils and heterospecific soils (Mehrabi and Tuck 2015); however, phylogenetic relatedness in plant-soil feedbacks did improve predictions of plant abundance for 57 species within an old field community (Anacker et al. 2014). Some fractions of the soil community may be similar between closely related plants. For example, arbuscular mycorrhizal effects are similar between closely related plants (Reinhart et al. 2012; Lugo et al. 2015) and some mycorrhizal associates are specialized to particular plant clades (Brundrett 2002). Our results suggest that plant relatedness correlates with the fungal fraction of the soil community, but only weakly with the bacterial community. Thus the relative importance of fungal and bacterial components of the soil microbiota to plant performance may influence whether close relatives have similar plant–soil feedback effects on one another (Burns and Strauss 2011).
Conclusions
Plant species identity was the best predictor of bacterial and fungal community composition in the soil, followed by soil chemistry, spatial location and plant relatedness. Because ‘everything’ is not ‘everywhere’ in microbial communities, spatial location can confound our attempts to predict microbial community structure. Future work should use experimental manipulations to disentangle covarying predictors, such as soil chemistry, spatial location and plant relatedness in the field. Understanding the feedbacks between soil microbial community structure and plant communities may benefit from considering the evolutionary relationships among plants.
Sources of Funding
Our work was funded by the National Science Foundation (United States) DEB (11-20387) and by the Holden Arboretum Trust and the Corning Institution for Education and Research.
Contributions by the Authors
J.H.B. contributed to data collection and analysis and wrote the manuscript. B.L.A. collected soil samples in the field, sent soil samples for nutrient analysis and contributed to data analysis. S.Y.S. co-conceived the project and data collection design, discussed data analysis and commented on drafts. D.J.B. trained J.H.B. and undergraduate researchers in TRFLP techniques in the laboratory, mentored undergraduates in the laboratory and contributed to data analysis and manuscript writing. All authors contributed to manuscript revision.
Conflict of Interest Statement
None declared.
Supporting Information
The following additional information is available in the online version of this article –
Table S1. Plant species sampled to examine rhizosphere soil microbial communities at Bodega Bay, CA, USA.
Figure S1. Map of the spatial sampling locations for the 14 species and 6 replicate soil collections per species at Bodega Bay, CA, USA.
Figure S2. Variance partitioning analysis on the bacterial and fungal data sets.
Table S2. Linear PVR models were used to test the effects of plant species, spatial location and plant genus on soil microbial community composition.
Acknowledgements
Special thanks to J. Sones and P. Conners for assistance with sampling locations. Thanks also to A. Flanigan, M. MacBeth and X. Zhao for assistance in the laboratory and P. Peres-Neto for analysis advice. We thank K. Abbott, M. Benard, C. Cope, K. Dananay, S. Diamond, R. Martin, J. Murphy, B. Nolting, A. Osvaldsson, H. Rollins and R. Snyder, for constructive feedback on the manuscript.
Literature Cited
- Anacker BL, Klironomos JN, Maherali H, Reinhart KO, Strauss SY. 2014. Phylogenetic conservatism in plant-soil feedback and its implications for plant abundance. Ecology Letters 17:1613–1621. 10.1111/ele.12378 [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annual Review of Plant Biology 57:233–266. 10.1146/annurev.arplant.57.032905.105159 [DOI] [PubMed] [Google Scholar]
- Bardgett RD, Mawdsley JL, Edwards S, Hobbs PJ, Rodwell JS, Davies WJ. 1999. Plant species and nitrogen effects on soil biological properties of temperate upland grasslands. Functional Ecology 13:650–660. 10.1046/j.1365-2435.1999.00362.x [DOI] [Google Scholar]
- Bass Becking L. 1934. Geobiologie of Inleiding Tot de Milieukunde. The Hague: Van Stockum & Zoon. [Google Scholar]
- Bellier E, Monestiez P, Durbec J-P, Candau J-N. 2007. Identifying spatial relationships at multiple scales: principal coordinates of neighbour matrices (PCNM) and geostatistical approaches. Ecography 30:385–399. 10.1111/j.0906-7590.2007.04911.x [DOI] [Google Scholar]
- Berg G, Smalla K. 2009. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiology Ecology 68:1–13. 10.1111/j.1574-6941.2009.00654.x [DOI] [PubMed] [Google Scholar]
- Bever JD, Dickie IA, Facelli E, Facelli JM, Klironomos J, Moora M, Rillig MC, Stock WD, Tibbett M, Zobel M. 2010. Rooting theories of plant community ecology in microbial interactions. Trends in Ecology and Evolution 25:468–478. 10.1016/j.tree.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt AJ, Seabloom EW, Hosseini PR. 2009. Phylogeny and provenance affect plant–soil feedbacks in invaded California grasslands. Ecology 90:1063–1072. 10.1890/08-0054.1 [DOI] [PubMed] [Google Scholar]
- Brundrett MC. 2002. Coevolution of roots and mycorrhizas of land plants. New Phytologist 154:275–304. 10.1046/j.1469-8137.2002.00397.x [DOI] [PubMed] [Google Scholar]
- Burke DJ, Dunham SM, Kretzer AM. 2008. Molecular analysis of bacterial communities associated with the roots of Douglas fir (Pseudotsuga menziesii) colonized by different ectomycorrhizal fungi. FEMS Microbiology Ecology 65:299–309. 10.1111/j.1574-6941.2008.00491.x [DOI] [PubMed] [Google Scholar]
- Burke DJ, Smemo KA, López-Gutiérrez JC, DeForest JL. 2012. Soil fungi influence the distribution of microbial functional groups that mediate forest greenhouse gas emissions. Soil Biology and Biochemistry 53:112–119. 10.1016/j.soilbio.2012.05.008 [DOI] [Google Scholar]
- Burns JH, Strauss SY. 2011. More closely related species are more ecologically similar in an experimental test. Proceedings of the National Academy of Sciences of the USA 108:5302–5307. 10.1073/pnas.1013003108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas LH, Eissenstat DM. 2009. Patterns in root trait variation among 25 co-existing North American forest species. New Phytologist 182:919–928. 10.1111/j.1469-8137.2009.02799.x [DOI] [PubMed] [Google Scholar]
- Compant S, Clément C, Sessitsch A. 2010. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biology and Biochemistry 42:669–678. 10.1016/j.soilbio.2009.11.024 [DOI] [Google Scholar]
- Desdevises Y, Legendre P, Azouzi L, Morand S. 2003. Quantifying phylogenetically structured environmental variation. Evolution 57:2647–2652. 10.1111/j.0014-3820.2003.tb01508.x [DOI] [PubMed] [Google Scholar]
- Diniz-Filho JAF, de Sant’Ana CER, Bini LM. 1998. An eigenvector method for estimating phylogenetic inertia. Evolution 52:1247–1262. 10.2307/2411294 [DOI] [PubMed] [Google Scholar]
- Diniz-Filho JAF, Bini LM, Rangel TF, Morales-Castilla I, Olalla-Tárraga MA, Rodríguez MA, Hawkins BA. 2012. On the selection of phylogenetic eigenvectors for ecological analyses. Ecography 35:239–249. 10.1111/j.1600-0587.2011.06949.x [DOI] [Google Scholar]
- Edgar RC. 2004a. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:1–19. 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004b. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenfeld JG, Ravit B, Elgersma K. 2005. Feedback in the plant-soil system. Annual Review of Environment and Resources 30:75–115. 10.1146/annurev.energy.30.050504.144212 [DOI] [Google Scholar]
- Ettema CH, Wardle DA. 2002. Spatial soil ecology. Trends in Ecology and Evolution 17:177–183. 10.1016/S0169-5347(02)02496-5 [DOI] [Google Scholar]
- Faith DP, Minchin PR, Belbin L. 1987. Compositional dissimilarity as a robust measure of ecological distance. Vegetatio 69:57–68. 10.1007/BF00038687 [DOI] [Google Scholar]
- Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proceedings of the National Academy of Sciences of the USA 103:626–631. 10.1073/pnas.0507535103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Ladau J. 2012. Predicting microbial distributions in space and time. Nature Methods 9:549–551. 10.1038/nmeth.2041 [DOI] [PubMed] [Google Scholar]
- Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG. 2012. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proceedings of the National Academy of Sciences of the USA 109:21390–21395. 10.1073/pnas.1215210110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J, Monette G. 1992. Generalized collinearity diagnostics. Journal of the American Statistical Association 87:178–183. 10.1080/01621459.1992.10475190 [DOI] [Google Scholar]
- Gonçalves-Souza T, Diniz-Filho JAF, Romero GQ. 2014. Disentangling the phylogenetic and ecological components of spider phenotypic variation. PLoS ONE 9:e89314 10.1371/journal.pone.0089314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardoim PR, van Overbeek LS, van Elsas JD. 2008. Properties of bacterial endophytes and their proposed role in plant growth. Trends in Microbiology 16:463–471. 10.1016/j.tim.2008.07.008 [DOI] [PubMed] [Google Scholar]
- Innes L, Hobbs PJ, Bardgett RD. 2004. The impacts of individual plant species on rhizosphere microbial communities in soils of different fertility. Biology and Fertility of Soils 40:7–13. 10.1007/s00374-004-0748-0 [DOI] [Google Scholar]
- Jacquemyn H, Merckx V, Brys R, Tyteca D, Cammue BPA, Honnay O, Lievens B. 2011. Analysis of network architecture reveals phylogenetic constraints on mycorrhizal specificity in the genus Orchis (Orchidaceae). New Phytologist 192:518–528. 10.1111/j.1469-8137.2011.03796.x [DOI] [PubMed] [Google Scholar]
- Kluber LA, Carrino-Kyker SR, Coyle KP, DeForest JL, Hewins CR, Shaw AN, Smemo KA, Burke DJ. 2012. Mycorrhizal response to experimental pH and P manipulation in acidic hardwood forests. PLoS ONE 7:e48946 10.1371/journal.pone.0048946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulmatiski A, Beard KH, Stevens JR, Cobbold SM. 2008. Plant-soil feedbacks: a meta-analytical review. Ecology Letters 11:980–992. 10.1111/j.1461-0248.2008.01209.x [DOI] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Applied and Environmental Microbiology 75:5111–5120. 10.1128/AEM.00335-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake JR, Donnelly DP, Boddy L. 2002. Interactions between ecto-mycorrhizal and saprotrophic fungi. In: Heijden MGA, Sanders IR, eds. Mycorrhizal ecology. Berlin: Springer, 345–371. [Google Scholar]
- Liu X, Liang M, Etienne RS, Wang Y, Staehelin C, Yu S. 2012. Experimental evidence for a phylogenetic Janzen-Connell effect in a subtropical forest. Ecology Letters 15:111–118. 10.1111/j.1461-0248.2011.01715.x [DOI] [PubMed] [Google Scholar]
- Long SR. 2001. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiology 125:69–72. 10.1104/pp.125.1.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugo MA, Reinhart KO, Menoyo E, Crespo EM, Urcelay C. 2015. Plant functional traits and phylogenetic relatedness explain variation in associations with root fungal endophytes in an extreme arid environment. Mycorrhiza 25:85–95. 10.1007/s00572-014-0592-5 [DOI] [PubMed] [Google Scholar]
- Manjunath A, Habte M. 1991. Root morphological-characteristics of host species having distinct mycorrhizal dependency. Canadian Journal of Botany-Revue Canadienne De Botanique 69:671–676. [Google Scholar]
- Martos F, Munoz F, Pailler T, Kottke I, Gonneau C, Selosse M-A. 2012. The role of epiphytism in architecture and evolutionary constraint within mycorrhizal networks of tropical orchids. Molecular Ecology 21:5098–5109. 10.1111/j.1365-294X.2012.05692.x [DOI] [PubMed] [Google Scholar]
- McCune B, Grace JB. 2002. Analysis of ecological communities. MjM Software Design, Gleneden Beach, OR, USA, 304 pp. [Google Scholar]
- Mehrabi Z, Tuck SL. 2015. Relatedness is a poor predictor of negative plant-soil feedbacks. New Phytologist 205:1071–1075. 10.1111/nph.13238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien RM. 2007. A caution regarding rules of thumb for variance inflation factors. Quality and Quantity 41:673–690. 10.1007/s11135-006-9018-6 [DOI] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2010. Vegan: community ecology package. R package version 1.17-2 ed. [Google Scholar]
- O'Malley MA. 2007. The nineteenth century roots of ‘everything is everywhere’. Nature Reviews Microbiology 5:647–651. 10.1038/nrmicro1711 [DOI] [PubMed] [Google Scholar]
- Peres-Neto PR, Legendre P, Dray S, Borcard D. 2006. Variation partitioning of species data matrices: estimation and comparison of fractions. Ecology 87:2614–2625. 10.1890/0012-9658(2006)87[2614:VPOSDM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Pilloni G, Granitsiotis MS, Engel M, Lueders T. 2012. Testing the limits of 454 pyrotag sequencing: reproducibility, quantitative assessment and comparison to T-RFLP fingerprinting of aquifer microbes. PLoS ONE 7:e40467 10.1371/journal.pone.0040467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2008. R: a language and environment for statistical computing, 2.8.1 edVienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Reinhart KO, Anacker BL. 2014. More closely related plants have more distinct mycorrhizal communities. AoB PLANTS 6:plu051; doi:10.1093/aobpla/plu051 10.1093/aobpla/plu051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart KO, Wilson GWT, Rinella MJ. 2012. Predicting plant responses to mycorrhizae: integrating evolutionary history and plant traits. Ecology Letters 15:689–695. 10.1111/j.1461-0248.2012.01786.x [DOI] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3:217–223. 10.1111/j.2041-210X.2011.00169.x [DOI] [Google Scholar]
- Rout ME, Callaway RM. 2012. Interactions between exotic invasive plants and soil microbes in the rhizosphere suggest that ‘everything is not everywhere’. Annals of Botany 110:213–222. 10.1093/aob/mcs061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrappa T, Czymmek KJ, Pare PW, Bais HP. 2008. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiology 148:1547–1556. 10.1104/pp.108.127613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. 2003. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19:301–302. 10.1093/bioinformatics/19.2.301 [DOI] [PubMed] [Google Scholar]
- Singh BK, Millard P, Whiteley AS, Murrell JC. 2004. Unravelling rhizosphere–microbial interactions: opportunities and limitations. Trends in Microbiology 12:386–393. 10.1016/j.tim.2004.06.008 [DOI] [PubMed] [Google Scholar]
- Stevens PF. 2009. Angiosperm phylogeny website. Version 9 ed. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DL, Hollingsworth TN, McFarland JW, Lennon NJ, Nusbaum C, Ruess RW. 2014. A first comprehensive census of fungi in soil reveals both hyperdiversity and fine-scale niche partitioning. Ecological Monographs 84:3–20. 10.1890/12-1693.1 [DOI] [Google Scholar]
- Ushio M, Wagai R, Balser TC, Kitayama K. 2008. Variations in the soil microbial community composition of a tropical montane forest ecosystem: does tree species matter? Soil Biology and Biochemistry 40:2699–2702. 10.1016/j.soilbio.2008.06.023 [DOI] [Google Scholar]
- Valverde-Barrantes OJ, Smemo KA, Blackwood CB. 2014. Fine root morphology is phylogenetically structured, but nitrogen is related to the plant economics spectrum in temperate trees. Functional Ecology; 10.1111/1365-2435.12384. [DOI] [Google Scholar]
- Wikström N, Savolainen V, Chase MW. 2001. Evolution of the angiosperms: calibrating the family tree. Proceedings of the Royal Society B: Biological Sciences 268:2211–2220. 10.1098/rspb.2001.1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD Dissertation, The University of Texas, Austin. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.