Abstract
Nucleoporins are evolutionary conserved proteins mainly involved in the constitution of the nuclear pores and trafficking between the nucleus and cytoplasm, but are also increasingly viewed as main actors in chromatin dynamics and intra-nuclear mitotic events. Here, we determined the cellular localization of the nucleoporin Mlp2 in the ‘divergent’ eukaryotes Leishmania major and Trypanosoma brucei. In both protozoa, Mlp2 displayed an atypical localization for a nucleoporin, essentially intranuclear, and preferentially in the periphery of the nucleolus during interphase; moreover, it relocated at the mitotic spindle poles during mitosis. In T. brucei, where most centromeres have been identified, TbMlp2 was found adjacent to the centromeric sequences, as well as to a recently described unconventional kinetochore protein, in the periphery of the nucleolus, during interphase and from the end of anaphase onwards. TbMlp2 and the centromeres/kinetochores exhibited a differential migration towards the poles during mitosis. RNAi knockdown of TbMlp2 disrupted the mitotic distribution of chromosomes, leading to a surprisingly well-tolerated aneuploidy. In addition, diploidy was restored in a complementation assay where LmMlp2, the orthologue of TbMlp2 in Leishmania, was expressed in TbMlp2-RNAi-knockdown parasites. Taken together, our results demonstrate that Mlp2 is involved in the distribution of chromosomes during mitosis in trypanosomatids.
INTRODUCTION
In eukaryotes, nuclear pore complexes (NPCs) are macromolecular structures of 8-fold rotational symmetry which perforate the nuclear envelope and serve as gateways for the transit of macromolecules between the cytoplasm and the nucleus. NPCs are made up of nucleoporins (NUPs) which, in association with transport proteins (karyoproteins and importins) (1–7) and with a gradient of Ran-GTP/GDP (8), allow the import and export of cargos. The composition and structure of NUPs have largely been explored in Saccharomyces cerevisiae (5) and in mammals (9,10), and more recently in the divergent unicellular eukaryote Trypanosoma brucei (11). Fibrillar appendices are attached to the NPC's nucleoplasmic side and interdigitate with each other laterally, forming a structure called the nuclear basket. The nuclear basket is composed by a small variety of proteins that are structurally distinct from those of the core scaffold of the NPC, or phenylalanine-glycine (FG)-repeat containing NUPs, and which are the Translocated promoter region protein (Tpr) in mammals, Myosin-like proteins (Mlp) 1 and 2 in yeast, Megator in Drosophila and the Nuclear Pore Anchor protein (NUA) in plants. The organization and composition of NPCs are highly conserved through evolution, suggesting a common origin established early in eukaryotes (11). If this conservation reflects the classical nucleocytoplasmic transport in eukaryotes, there is more and more evidence that NPCs are implicated in other numerous biological processes (12), some of them established only in specific organisms (13). For example, several NUPs are involved in the mechanisms of chromosomal segregation, mitotic spindle formation and cytokinesis (14–22). Moreover, heterochromatin is associated with the nuclear envelope, and electron microscopy showed that the occurrence of heterochromatin exclusion zones is dependent on the large coiled coil-forming domain of Tpr (23). In addition to this direct role as a determinant of perinuclear organization and of the formation of a morphologically distinct nuclear sub-compartment, NUPs have been associated with the regulation of transcription (24–33). In particular, Megator may stimulate transcription by promoting the formation of an open chromatin environment and define transcriptionally active regions in the Drosophila genome (34). In yeast, ScMlp1 and ScMlp2 have also been identified as having a major role in nuclear architecture and in spindle pole body assembly (27,35–37). In addition, the NPC-tethered gene loops may modulate gene expression, through the so-called transcriptional memory, in reference to a gene ‘remembering’ its previous transcriptionally active state and having the ability of a faster rate of transcription initiation when reinduced following a short period of repression (38–40). ScMlp1 could play a key role in this process (39,40). Finally, ScMlp1 and ScMlp2 are thought to be involved in preservation of genome integrity, like telomere length maintenance (41,42).
In T. brucei, as well as in Leishmania major, both parasitic protozoa of the family of trypanosomatids, 22 NUPs have been identified, of which 20 in T. brucei (11) and 16 in L. major (our unpublished data) have been localized to the nuclear envelope (11). Like in S. cerevisiae, and as opposed to higher eukaryotes, two Mlp paralogues are present in their genomes. Here, we show that, in these two trypanosomatids, Mlp2 is intranuclear and located preferentially at the periphery of the nucleolus. Centromeric sequences and unconventional kinetochore proteins have been characterized (43,44) in T. brucei, although not yet in Leishmania. TbMlp2 was found adjacent or in close proximity to the centromeric sequences and the kinetochore protein TbKKT4 in the periphery of the nucleolus. The depletion of TbMlp2 disrupted chromosomal allotment during mitosis, leading to aneuploidy, which surprisingly had no effect on cell growth.
MATERIALS AND METHODS
Parasites and in vitro cultivation
Leishmania major ‘Friedlin’ promastigotes (MHOM/IL/81/Friedlin) were grown at 26°C in RPMI 1640 (Gibco BRL) supplemented with 10% fetal bovine serum (FBS) (45). Procyclic forms of the Lister 427 wild-type strain and Lister 427 29–13 cell line of T. brucei were grown at 27°C in SDM-79 (PAA Laboratories) supplemented with 10% FBS and 7-μg/ml hemin, and with 30 μg/ml of hygromycin and 10 μg/ml of geneticin for the 29–13 line (46).
Construction of the Leishmania GFP- and Ruby-fused protein expression vectors
The genes LmjF26.2660 coding for LmMlp2 and LmjF36.2510 coding for LmNup93 (orthologous of TbMlp2 and TbNup93, respectively) were selected in GeneDB. Open reading frames were analysed and characterized by pairwise sequence alignments BLAST (47) and FASTA (48) with the National Center for Biotechnology Information (NCBI) non-redundant database. The genes were polymerase chain reaction (PCR)-amplified from genomic DNA. Oligonucleotide primers used were 5′GGGCAATTGTGGCTCCAGAAGAGCATAACC3′ and 5′GGGGTTAACCTGAATCTTAAGCGGGTAGATTA3′ for LmMlp2, and 5′GGGAGATCTCATATGTTTAGCTCGACTTCCGATCTTG3′ and 5′GGGGGTACCGCACAGATACGTCCGTTCGAA3′ for LmNup93. All the constructions contained MfeI and HpaI restriction sites except for LmNup93 that contained NdeI and KpnI. The PCR products were cloned into pGEM-Teasy® (Promega) and then into both vectors pTH6cGFPn and pTH6nGFPc (49). In the RFP-fused protein expression vector, the GFP gene was replaced by the RFP gene. The conservation of the reading frame of fused proteins was systematically confirmed by nucleotide sequence analysis. For LmMlp2 coiled coil regions were also predicted by COILS (50).
Construction of the T. brucei GFP-fused protein expression vectors and in situ tagging vectors
The gene Tb927.9.1340 coding for TbMlp2 was PCR-amplified from genomic DNA using the oligonucleotide primers 5′GGGCTCGAGATGAGCATCAGTGAGTCGGACAGC3′ and 5′GGGCATATGCTGTGGCTGCTTTACTTCCTCTGCC3′ for the GFPc construct, and 5′GGGCTCGAGAGCATCAGTGAGTCGGACAGCCTTT3′ and 5′GGGCTCGAGTTACTGTGGCTGCTTTACTTCCTCTGCC3′ for the GFPn construct. All constructs contained XhoI and XbaI restriction sites. The PCR products were cloned into pGEMTeasy® (Promega) and then into both vectors pLew79GFPc and pLew79GFPn (kindly provided by Frédéric Bringaud, Université Victor Segalen, Bordeaux 2, France). Protein expression was induced by adding tetracycline. For in situ GFP insertion at the N-terminus, the home-made vector pIS-B/GFPn (GenBank KJ417663) was used; pIS-B/GFPn-TbMlp2 was amplified with the following two primers:
5′TGTGAATGCATGCTATCCTAACTGCTTTCTCTTTCCTTCTCTATGGCAACGCATGTGTTTAAATGTAAGTGCATTGATTCAACTAGTATGGCCAAGTTGACCAG3′ and 5′CCCTCCTCCAGTGGCTTCCGGTATGGGATCGGCGAGGTGAATACTCTTTTGCAGCCAAAGGCTGTCCGACTCACTGATGCTGTTAACGGGCAATTGCTTGTACA3′. In the transfectants, the PCR product integrated in the 5′ end of the targeting gene, i.e. the native TbMlp2 gene with deletion of the start codon of the native gene. Of note, the native 3′ end was preserved. For in situ insertion of GFP at the C-terminus of TbMlp2, the PCR-mediated C-terminal in situ tagging vector pMOTag4G (51) and the following two primers were used: 5′ACTCGGGCAGTAACTTGGGATAAGTTGGTTGAGCTCATTTACCCCGTTAAAATCGGTGTCGTGGCAGAGGAAGTAAAGCAGCCACAGGGTACCGGGCCCCCCCTCGAG3′ and 5′GGTATTTCATCTCATCCTTTAGAGCCTCTCTCCCCCTCTTTCCTCTCCCCCCCCCCAATTGCAGAGGGAGCATAATATATTCTACTATGGCGGCCGCTCTAGAACTAGTGGAT3′. An N-terminus Ruby-fused TbKKT4 (Tb927.8.3680) was obtained using a pIS-B/Ruby-n vector and the following two primers: 5′CGAATATTTTCTTTTTTTTTTGCTTACGCTTTCTATTTTAGTTACACAACAGCAAAAGGGGGCTAGATCAGAGGAATAAGGATGGCCAAGCCTTTGTCTCAAG3′ and 5′AAGTGCTAAAATTCCCTGCATCTGAGTAATCAGCTGGGGATTAGATGCGAGTTGCTGTAGCACAGACAGTGGATTGTCTCCCCCTCCGCCCAGGCCGG3′. In all cases, the conservation of the reading frame of the fusion proteins was systematically confirmed by nucleotide sequence analysis and insertions were checked by PCR.
RNA interference in T. brucei
The targeted sequences were identified using TrypanoFAN (http://trypanofan.path.cam.ac.uk/software/RNAit.html) and PCR-amplified from genomic DNA. For TbMlp2 three pairs of probes were used: (i) 5′GGGCCGCGGGCAACGTGAAGAGTCGTGAA3′ and 5′GGGAAGCTTGGCATAAAGCAAGGTGGTGT3′, (ii) 5′GGGCCGCGGAGCGAGAATTAGAAATTGTGC3′ and 5′GGGAAGCTTTTCACGACTCTTCACGTTGC3′ and (iii) 5′GGGCCGCGGTGACGAACTCCAACGCTCC3′ and 5′GGGAAGCTTGGCATAAAGCAAGGTGGTGT3′. The different PCR products were cloned into pGEM-Teasy® (Promega) and then into p2T7tiB/GFP (all probes contained HindIII and SacII restriction sites). For transfection and RNA interference (RNAi) induction, 10 µg of linearized plasmid DNAs were transfected into 3.107 cells of the 29–13 cell line. An exponential protocol was used with 1500 V and 25 μF as parameters. Transfectants were grown under selective pressure with 5 μg/ml of phleomycin (Sigma®) during 15–20 days before induction by addition of 1 μg/ml of tetracycline.
Construction of the vector for the expression of LmMlp2 in the complementation assay in T. brucei
Two constructs were performed, in order (i) to produce the native form of LmMlp2 and (ii) a LmMlp2-GFP-c fused protein. LmMlp2 was PCR-amplified from genomic DNA. Oligonucleotide primers used were 5′gggcaattgTGGCTCCAGAAGAGCATAACC and 5′ggggttaacCTACTGAATCTTAAGCGGGTAG3′. The PCR product was cloned into pGEMTeasy® (Promega); insert was digested by Mfe1 and Hpa1 and then cloned into the vector pLew79cmycC. A stop codon was inserted at the end of the LmMlp2, in order to eliminate the cmyc tag at the C term of the fusion protein and to express the native form. For LmMlp2-GFP-c-fused protein, oligonucleotide primers used were 5′ggggttaacATGTGGCTCCAGAAGAGCATAAC3′ and 5′gggtctagaCTGAATCTTAAGCGGGTAGATTA3′. The PCR product was cloned into pGEMTeasy® (Promega); insert was digested by Hpa1 and Xba1 and then cloned into the vector pLew79GFPC. Conservation of the reading frame of the fusion protein was confirmed by nucleotide sequence analysis. The constructs were transfected in the first RNAi line.
Transfection of L. major
A total of 5.107 cells grown to mid-log phase were resuspended with 80 μg of plasmid DNA. Electroporation was performed in a Bio-Rad Gene pulser 2 electroporator using the following conditions: square wave protocol, 1500 V, 25μF, 2 pulses of 0.5 ms and 10 s between the pulses. The day after, the selective antibiotic hygromycin (Sigma) was added at 30 μg/ml, and stable transfectants were obtained between 1 and 2 weeks after transfection.
Transfection of T. brucei
A total of 3.107 cells grown to mid-log phase were resuspended with 10 μg of DNA. Electroporation was performed in a Bio-Rad Gene pulser 2 electroporator using the following conditions: square wave protocol, 1500 V, 25 μF, 1 pulse of 0.5 ms. The selective antibiotic was added on the following day: phleomycin (Invitrogen) at 5 μg/ml for vectors pLew79GFPc and pLew79GFPn and for vector pIS-B/GFPn, blasticidin (Euromedex) for vector pIS-B/Ruby and hygromycin (Sigma) for vector pMOT-GFP. Stable transfectants were obtained between 1 and 2 weeks after transfection. For the complementation assay, TbMlp2 RNAi knockdown parasites were transfected and the selective antibiotic was blasticidin (Euromedex, at 10 μg/ml).
Microscopy and fluorescence imaging
For intracellular localization analysis of GFP- and Ruby-fused proteins, transfected cells were grown to mid-log phase, fixed in 4% paraformaldehyde and air-dried on microscope immunofluorescence slides. Slides were finally mounted with Mowiol® (Calbiochem) and 4,6-diamino-2-phenylindole (DAPI). Cells were viewed by phase contrast, and fluorescence was visualized using appropriate filters on a Zeiss Axioplan2 microscope with an X100 objective. Digital images were captured using a Photometrics CoolSnap CDD camera (Roper Scientifics) and processed with MetaView® (Universal Imaging). Three-dimensional representations of the complete nucleus were generated from Z-stack acquisitions (25 planes of 0.25 μm). Images were deconvolved using Metamorph® (Universal Imaging). Movies were made from stack overlays obtained from Metamorph using Image J 1.37v (National Institute of Health, USA, http://rsb.info.nih.gov/ij/). For statistical significance of the localizations of TbMlp2 and the centromeres, the Chi-square test was used with Yates’ correction (with 1 degree of freedom) and P value (calculated with two tails) and calculated online using QuickCalcs from GraphPad software (http://graphpad.com/quickcalcs/contingency1.cfm).
Northern blot
A total of 1.2 × 108 cells were centrifuged. RNAs were extracted by using RNeasy® kit (Qiagen). Then, RNAs were separated on a 1.2% agarose gel added with 6% formaldehyde and MOPS 1X, and transferred onto a nylon transfer membrane. The membrane was hybridized overnight at 65°C with specific probes 32P-labelled into Denhardt buffer 50X, and then washed in increasingly stringent dilutions of saline sodium citrate (SSC).
Phosphatidylserine exposure
Exposed phosphatidylserine (PS) was detected on the outer membrane of cells using the Annexin-V-FLUOS® staining kit (Roche). Cells were washed in PBS1X and incubated for 10–15 min at 4°C with the incubation buffer of the kit. Fluorescence was measured and analysed using an FACS Calibur (Becton Dickinson, San Jose, CA, USA) and the BD CellQuest Pro software.
DNA contents
To determinate the cell DNA contents, a propidium iodide (PI) staining method was used. For this purpose, cells were washed with PBS, resuspended in 500 μl of ice-cold 70% ethanol, vortexed 1 min and incubated at 4°C. After centrifugation, cells were resuspended in PBS1X and 0.1-mg/ml RNAse A, placed 20 min at 37°C, centrifuged, incubated 10–30 min on ice with 2.5% PI and immediately analysed on a NAVIOS Flow cytometer (Beckman Coulter) using Kaluza software (Beckman Coulter).
Fluorescence in situ hybridization (52–54)
Trypanosoma brucei procyclics were fixed in 4% paraformaldehyde and 4% acetic acid, air-dried on microscope immunofluorescence slides and dehydrated in serial ethanol baths (50–100%). Probes were labelled with tetramethyl-rhodamine-5-dUTP® (Roche) by using the Nick Translation Mix® (Roche). Slides were then hybridized with a heat-denatured DNA probe under a sealed rubber frame at 94°C for 2 min and then overnight at 37°C. The hybridization solution contained 50% formamide, 10% dextran sulfate, 2X SSPE, 250-mg/ml salmon sperm DNA and 100 ng of labelled double strand DNA probe. After hybridization, parasites were sequentially washed in 50% formamide-2 X SSC at 37°C for 30 min, 2X SSC at 50°C for 10 min, 2X SSC at 60°C for 10 min and 4X SSC at room temperature. Slides were finally mounted in Vectashield (Vector Laboratories) with DAPI and microscopically examined; more than 200 cells per transfected strain were counted. For statistical analysis of centromere localization, data for 200 cells were compared using the chi-square test. After inhibition of TbMlp2 by RNAi, the number of copies of chromosome 1 was determined by using DNA probes targeting the alpha- and beta-tubulin genes (55).
Immunolabeling and Immuno-fluorescence in situ hybridization in T. brucei
GFP-fused TbMlp2 was expressed by adding tetracycline in transfected cells. Cells were fixed and air-dried on a slide as described above for fluorescence in situ hybridization (FISH). For immunolabeling, slides were treated with 0.2% triton in PBS1X and saturated with 20% FBS. Anti-tubulin KMX antibody (kindly provided by Keith Gull, University of Oxford, UK) (56) diluted to 1/100th was added during 1 h. After four washes with PBS1X/20% FBS, goat anti-mouse AF594 secondary antibodies (Molecular Probes, Ref. A-11005) diluted to 1/500th were added. For in situ hybridization combined with immunofluorescence, so-called immuno-FISH, slides were treated with NP40 0.1% solution 5 min and saturated with 20% FBS. Rabbit anti-GFP polyclonal antibodies (Santa Cruz Biotech. Ref. Sc-8334) diluted to 1/5000th were then added, followed by goat anti-rabbit AF488 secondary antibodies (Molecular Probes, Ref. A-11008) diluted to 1/500th. The FISH assay using a centromeric probe was achieved as described above. The centromeric probe of chromosome 3 was a 120-bp oligonucleotide obtained by PCR-amplification of genomic DNA using the oligonucleotide primers 5′GCATAATGGCGTGTTATCGC3′ and 5′GGTTATCGCCCCTACGGC3′. The centromeric probe of chromosome 2 was a synthetic oligonucleotide Alexa Fluor 488 modified at the 5′ end: 5′AF488-CGTGTTTTATGTGCAAAAGCATGTCATTA3′ (Eurofins MWG-Biotech AG).
RESULTS
Subcellular localization of Mlp2 in L. major and T. brucei
The orthologous gene of TbMlp2 identified in T. brucei (11) was also found in L. major by sequence homology in the genome database GeneDB (http://www.genedb.org). LmMlp2 is the gene product of LmjF26.2660 and displays 35% identity and 57% similarity with the protein of T. brucei. As described for T. brucei (11), in silico prediction using the COILS tool (50) found extended coiled coil domains in LmMlp2 (LmMlp2: cc105–183, 212–382, 392–530, 588–615); and no FG repeats were found. Using C- and N-terminal GFP-fused recombinant proteins, we studied the subcellular localization of this protein in L. major and in T. brucei throughout the cell cycle.
Trypanosomatids are divergent eukaryotes characterized by markedly original molecular features, in particular two independent though coordinated (nuclear and mitochondrial) cell cycles. The cell cycle course is easy to follow by DAPI staining in both L. major and in T. brucei, as the nuclear envelope and therefore the DAPI staining persist during the whole cycle; the intracellular distribution of two DNA-containing organelles, the nucleus and the kinetoplast (a dense and complex network of circular DNA molecules at the apex of the single mitochondrion), is widely used for this purpose. Thus, the correct cell cycle progress consists (i) in the duplication of the kinetoplast (2K), (ii) followed synchronously but independently by mitosis and karyokinesis (2N) and (iii) eventually by cytokinesis that depends on the ‘kinetoplast-associated’ cycle. Therefore, the end of mitosis and cytokinesis follows the kinetoplast segregation (57–60). During interphase, LmMlp2-GFP, whether N- or C-terminally tagged, displayed a punctuate localization within the nucleus, as a limited number of irregular dots (Figure 1A and Supplementary Figure S1). During mitosis, LmMlp2 relocated to the spindle poles, identified by the labelling of the mitotic spindle using the anti-tubulin monoclonal antibody KMX (Figure 2A). As TbMlp2 had been found at the nuclear envelope in interphasic cells of T. brucei (11), we checked its localization in this organism. We first classically inserted the TbMlp2 gene into the ribosomal DNA cluster: in interphasic cells, TbMlp2 was found intranuclear and preferentially at the periphery of the nucleolus (Figure 1B and C). During mitosis, like LmMlp2, TbMlp2 relocated to the spindle poles (Figure 2). These data were somehow different from those reported by DeGrasse et al. (11). Indeed, these authors, using in situ GFP-tagging at the C-terminus, reported a localization at the nuclear envelope, and not at the periphery of the nucleolus, in interphasic cells. In order to reproduce their data, we expressed both TbMlp2-GFPn and TbMlp2-GFPc from an in situ integration of the GFP (hereafter termed ‘isTbMlp2-GFPn/c’) using a home-made vector and the pMOTag4G construct, respectively. These transfections confirmed our initial data: in the large majority of interphasic cells, isTbMlp2-GFPn/c localized within the nucleus, preferentially at the periphery of the nucleolus (Figure 1C and Supplementary Figure S2). A colocalization experiment using the Ruby-tagged NUP Nup93, which clearly localized at the nuclear envelope, showed that the nuclear localization of both proteins differed, both in L. major and in T. brucei (Supplementary Figure S3). It is noteworthy, however, that if 14% of the interphasic (1N1K) cells displayed an intranuclear localization as described above, a minor proportion (<1%) of them also displayed a localization more typical of NUPs, i.e. as a ‘bead collar’ at the nuclear envelope (Supplementary Figure S2E and F). By contrast, all mitotic cells (1N2K and 2N2K) expressed the recombinant protein, showing that the expression of this protein is cell-cycle regulated. In the mitotic cells of T. brucei, as described by DeGrasse et al. (11), isTbMlp2-GFPn/c also relocated at the spindle poles (Figure 2B and C and Supplementary Figure S2C and D), and they were also present along the mitotic spindle at anaphase and telophase (Figure 2D and Supplementary Figure S2D).
Figure 1.
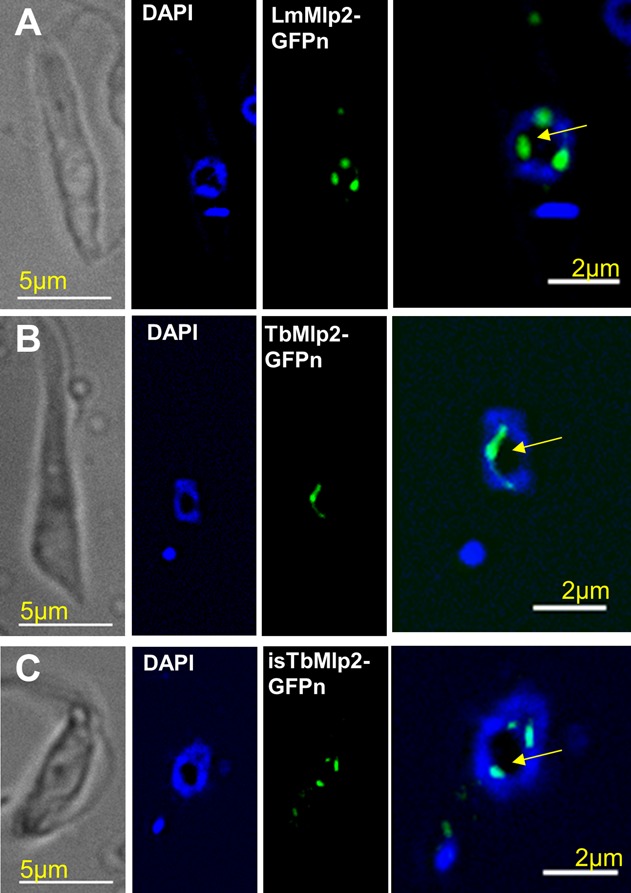
Subcellular localization of the nucleoporin Mlp2 in L. major and in T. brucei during interphase. (A) In L. major, LmMlp2 N-terminally fused to the GFP (LmMlp2-GFPn, green) was essentially found in the nucleus, as a limited number of more or less extended dots that localized at the periphery of the nucleolus (intranuclear area weakly labelled by DAPI, yellow arrow) (see also Supplementary Figure S1). The nuclear localization was similar for TbMlp2-GFPn in T. brucei, following either integration in the ribosomal DNA cluster (B) or in situ tagging (C). Of note, using C-terminal in situ tagging in T. brucei, a localization typical of nucleoporins could be seen in a very small proportion of cells (see Supplementary Figure S2E and F).
Figure 2.
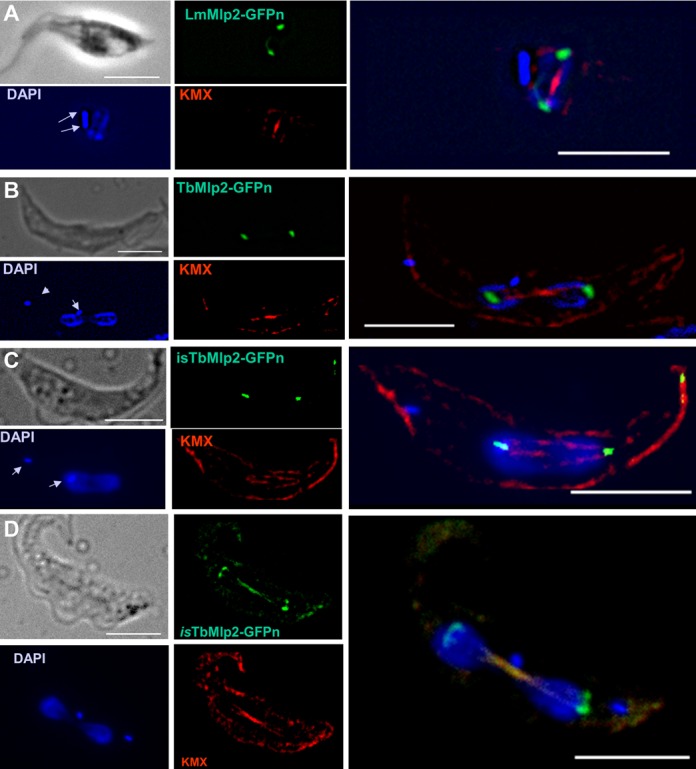
Subcellular localization of the nucleoporin Mlp2 in L. major and in T. brucei during mitosis. (A) During mitosis, LmMlp2 repositioned at the poles of the mitotic spindle, here visualized using a specific anti-tubulin antibody, KMX (red). A similar relocation was observed when the TbMlp2-GFPn recombinant gene was inserted in the ribosomal DNA cluster (B) or in situ (C, D). DAPI staining shows the duplicated (A) and then segregated kinetoplasts (B–D) (arrows) as well as dividing nuclei. Scale bar 5 μm.
Subcellular localization of the centromeres of chromosomes 2 and 3 in T. brucei and colocalization assay with TbMpl2
Whereas centromeric sequences are not characterized in Leishmania, they have been so in T. brucei where they are classically made of tandem repeat motifs. These repeats are more or less degenerated and shared by several large chromosomes of this parasite (61). We localized the centromeres in T. brucei cells by FISH, using two specific DNA probes, one targeting the centromeric sequences of chromosome 2 and the other one those of chromosome 3. In interphasic cells, the centromeres of both chromosomes 2 and 3 were located in the periphery of the nucleolus (Figure 3). As said above, TbMlp2 was also partially located at the nucleolar periphery during interphase. We then used immuno-FISH, combining the same DNA probes and a specific anti-GFP antibody, and found that TbMlp2-GFP was located adjacent or in close proximity to centromeres in 89% of the interphasic cells expressing the protein (Figures 4 and 5 and Supplementary Movie 1). These data were confirmed by the localization of one of the recently described unconventional kinetoplastid kinetochore proteins, TbKKT4 (Figure 6). In addition, the difference between this specific distribution of TbMlp2 and a random distribution of centromeres in the nucleus was highly significant (P < 10−4).
Figure 3.
Nuclear localization of the centromeric sequences of chromosomes 2 and 3 in T. brucei. DNA probes specific for the centromeres of chromosome 2 (green) and chromosome 3 (red) were used for FISH. These centromeric sequences were found at the periphery of the nucleolus (yellow arrow). Scale bar 2 μm.
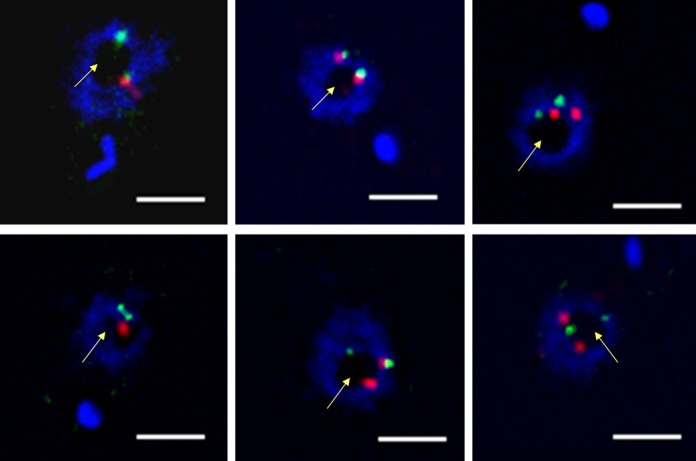
Figure 4.
Localization of TbMlp2 and centromeres of chromosomes 2 and 3 in T. brucei at interphase. In interphasic cells, the in situ tagged recombinant protein isTbMlp2-GFPn (green) and the centromeres of chromosomes 2 and 3 ((A) and (B), respectively, red) were all located at the periphery of the nucleolus (intranuclear area weakly labelled by DAPI, yellow arrow) and the centromeres found adjacent or in close proximity to TbMlp2. Bar 5 μm.
Figure 5.
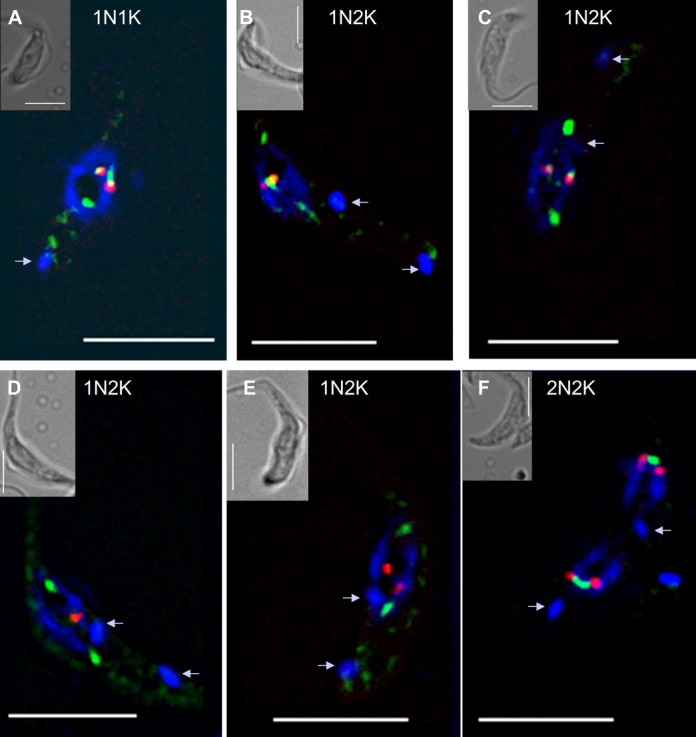
Dynamics of the relocation of TbMlp2 and of the centromeres during the cell cycle progress. (A) In interphasic cells (1N1K), both isTbMlp2-GFPn (green) and the centromeres of chromosome 3 (red) were found at the periphery of the nucleolus (see Figure 4). (B–D) During mitosis, which begins after the duplication of the kinetoplasts (arrow in 1N2K cells), a progressive migration of isTbMlp2-GFPn towards the mitotic spindle poles was observed. (E, F) The migration of the centromeres took place after isTb-Mlp2-GFPn had reached the pole. During their migration, centromeres were identified as two dots, but at the end of karyokinesis, the two dots were visible in each sister nucleus, materializing the two chromosome homologues. For chromosome 2, see Supplementary Figure S4. Scale bar 5 μm.
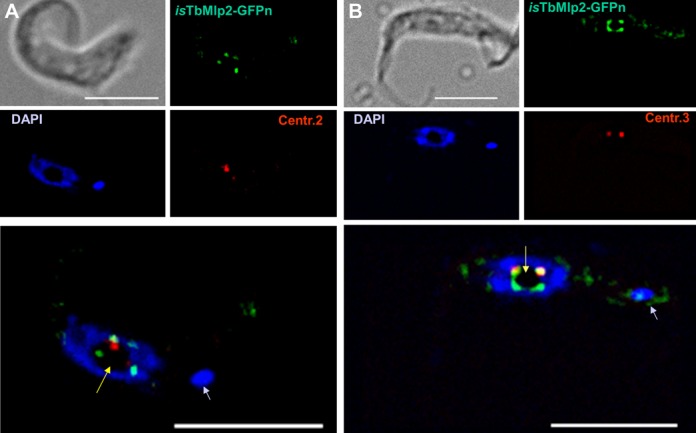
Figure 6.
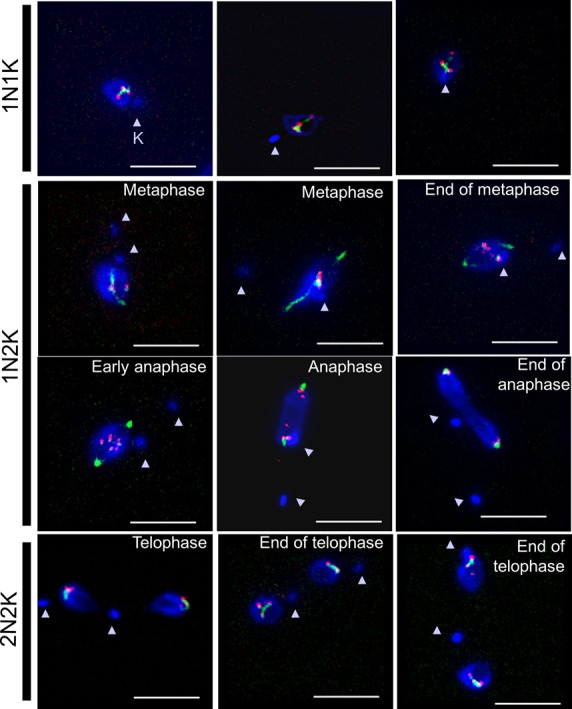
Dynamics of the relocation of TbMlp2 and of TbKKT4 during the cell cycle progress. The localization dynamics of one of the unconventional kinetochore proteins recently discovered in T. brucei (TbKKT4) were examined together with those of TbMlp2. First row: before the onset of mitosis (1N1K cells), both isTbMlp2-GFPn (green) and isTbKKT4-Ruby-n (red) clearly displayed an intranucleolar localization. Second row: at metaphase, TbMlp2 progressively migrated towards the spindle poles, with some of it still visible at the centre of the nucleus and metaphasic plate (right), where TbKKT4 remained. Third row: at anaphase, TbMlp2 concentrated at the spindle poles, while TbKKT4 in turn migrated to the poles. Fourth row: at telophase (2N2K cells), TbKKT4, like the centromeres (Figure 5F), was again concentrated adjacent or close to TbMlp2. Arrowheads: kinetoplasts (K). Scale bar 5 μm.
Differential migration of TbMlp2 and of the centromeres/TbKKT4 during mitosis
Using immuno-FISH in T. brucei, we then determined the spatiotemporal dynamics of TbMlp2 and both chromosome 2 and 3 centromeres throughout the cell cycle. As described above, in interphasic cells, both these centromeres and TbMlp2 were located at the periphery of the nucleolus (Figures 4 and 5A). During mitosis, we observed a differential migration of TbMlp2 and the centromeres (Figure 5 and Supplementary Figure S4): after the duplication of the kinetoplast, TbMlp2 was seen progressively migrating away from the centromeres towards the spindle poles (Figure 5B and C). The position of the centromeres remained unchanged until TbMlp2 had completed its migration to the spindle poles (Figure 5D); then the centromeres themselves started migrating to the poles (Figure 5E). At the end of karyokinesis, centromeres were again found in the vicinity of TbMlp2 at the spindle poles (Figure 5F and Supplementary Figure S4). This shows a differential localization dynamics of TbMlp2 and the centromeres throughout the cell cycle progress. We observed a similar differential migration between TbMlp2 and TbKKT4 (Figure 6). At the beginning of mitosis, TbMlp2 and TbKKT4 were still observed close to each other; during metaphase, TbMlp2 migrated towards the mitotic spindle poles, whereas TbKKT4 remained at the metaphasic plate. It was only during anaphase that TbKKT4 migrated towards the poles and rejoined TbMlp2 (Figure 6).
TbMlp2 is not essential but is required for correct chromosomal distribution in mitosis
In order to get an insight into the function of Mlp2, and since L. major lacks a functional RNAi pathway (62), we knocked down the protein using RNAi in T. brucei. RNAi-mediated expression inhibition of TbMlp2 did not yield any cell growth reduction (Figure 7A). In flow cytometry analysis, no significant increase of apoptotic or necrotic cells was observed (Figure 7B). TbMlp2 RNAi knockdown was essentially followed by a progressive increase in the proportion of cells with an intermediate DNA content between 2C and 4C, suggestive of either a delay/blockage in S phase or of aneuploidy (Figure 7C). However, the first hypothesis was not compatible with the absence of cell growth defect. The localization of Mlp2 to the poles of the mitotic spindle led us to investigate whether its depletion affected the distribution of chromosomes during mitosis. For this purpose, the copy number of chromosome 1 homologues was analysed in FISH experiments using a DNA probe targeting the α and β-tubulin gene cluster, spanning at least 60 kb on the megabase chromosome 1 (55). The number of homologues was determined in individual cells in the non-induced and induced population at days 1, 2, 3 and 4 post-induction (Figure 8 and Supplementary Figure S5). As expected, the non-induced cell population was disomic for chromosome 1 (52,54). In the induced population, the mean copy number of chromosome 1 varied in time: the TbMlp2-RNAi resulted in a decrease in disomic cells (already significant at day 2, and down to 50% at day 3) associated with a significant increase in trisomic cells (up to 25% at day 4) as well as in tetra-, penta- and hexasomic cells. In order to confirm the correlation between this phenotype and TbMlp2 depletion, complementation assays with LmMlp2, the orthologue of TbMlp2 in Leishmania, were performed. Either the native form of LmMlp2 or the GFP-fused protein LmMlp2-GFPc was used for this purpose; in both cases, the gene was inserted into the ribosomal DNA cluster. The expression in TbMlp2-depleted cells of both the native LmMlp2 (Figure 8B) and of LmMlp2-GFPc (Figure 8C) partially restored diploidy. FISH analysis using a chromosome 1-specific probe performed at day 4 after complementation showed that the percentage of disomic cells increased, while that of aneuploid cells decreased (Figure 8B). Similarly, flow cytometry analysis showed a reduction in the cell population between the 2C and 4C peaks (Figure 8D).
Figure 7.
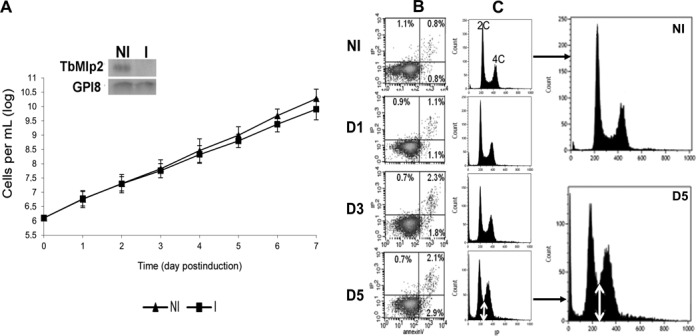
The RNAi-mediated knockdown of TbMlp2 induced a progressive increase of cells with DNA contents between 2C and 4C, but no cell growth reduction. (A) RNAi-mediated inhibition of TbMlp2 expression had no significant effect on cell growth. Insets: three independent RNAi experiments were performed, of which two targeted distinct sequences of TbMlp2. Northern blot controls of TbMlp2-RNAi; NI: non-induced cells. I: tetracyclin-induced cells at day 2. (B) Flow cytometry analysis: measure of phosphatidylserine exposure through the Annexin V assay after TbMlp2-RNAi. NI: non-induced cells; D1, D3, D5: tetracyclin-induced cells at day 1, day 3 and day 5 post-induction. Following depletion of TbMlp2, the exposure of phosphatidylserine to the outer membrane was not significantly altered. (C) Effect of the expression inhibition of TbMlp2 upon DNA contents in induced cells at D1, D3 and D5 versus non-induced cells. Depletion of TbMlp2 induced a progressive increase of cells with DNA contents between 2C and 4C (double white arrow).
Figure 8.
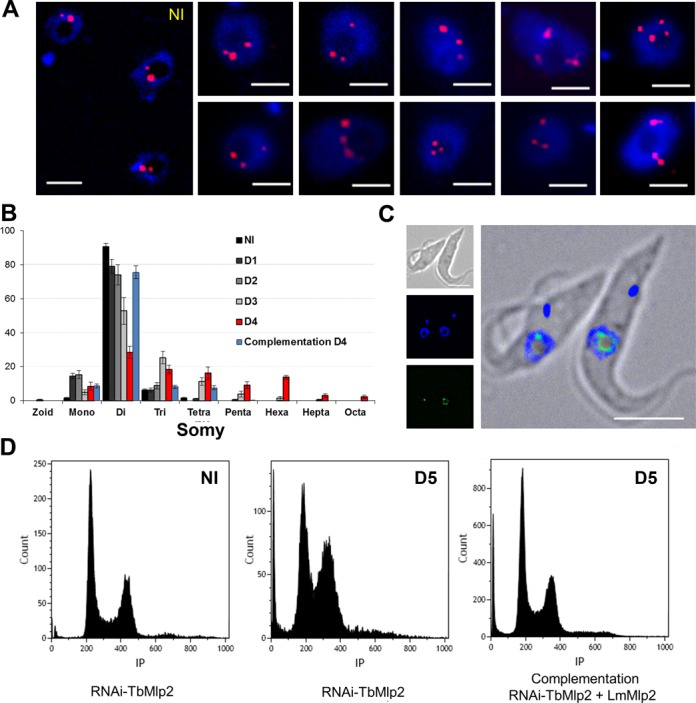
Induction of aneuploidy following RNAi-mediated knockdown of TbMlp2. (A) RNA interference targeting TbMlp2 yielded variable numbers of chromosome 1 homologues (red) per nucleus. Cells on day 4 after RNAi induction were probed for FISH with an alpha–beta tubulin probe. Disomic non-induced cells (NI) are shown on the left panel. Bar: 2 μm. (B) Time evolution of chromosome 1 somy patterns observed in FISH from day 1 to day 4 after TbMlp2-RNAi, and partial restoration of diploidy at day 4 after complementation by the non-tagged LmMlp2; y-axis = cell percentages. (C) In another complementation assay using LmMlp2-GFPc, the recombinant protein was expressed in T. brucei and found at the periphery of the nucleolus. (D) Flow cytometry analysis of DNA contents after complementation with LmMlp2: in non-induced cells (NI), at Day 5 after induction, and at Day 5 after induction but with complementation with LmMlp2. Like in Figure 7C, the TbMlp2 knockdown (centre) induced an increase in cells with DNA contents between 2C and 4C as compared with non-induced cells (left). Complementation of the RNAi-knocked down parasites with LmMlp2 (right) yielded a strong shift of this phenotype towards that of the non-induced cells.
DISCUSSION
In the present study, we first report that Mlp2 in L. major and T. brucei is essentially intranuclear and that its localization is cell cycle-dependent. The protein is located at the periphery of the nucleolus during interphase; and during mitosis, it relocates to the mitotic spindle poles in both organisms. This feature is reminiscent of several other NUPs that display a cell cycle-dependent dynamic localization within the nucleus and interact with the spindle or spindle poles during mitosis: among other examples, in yeast, ScMlp2 directly links spindle pole body components and is essential for correct spindle pole body assembly (63); in Drosophila, Megator, the Mlp orthologue, concentrates from the nuclear interior to the spindle matrix (64,65); and in mammals, the microtubule-bound Rae1 (Ribonucleic Acid Export 1) recruits NuMA (Nuclear Mitotic Apparatus protein) to the mitotic spindle and the cohesin subunit SMC1 to the spindle poles (20,66–68).
It is noteworthy that DeGrasse et al. (11) localized TbMlp2 exclusively at the inner side of the nuclear envelope (as a constituent of the basket of the NPC) during interphase, whereas we found it essentially in an intranuclear position with all the constructs we used, including the one used by DeGrasse et al. (pMOT-GFPc). Although we cannot explain this discrepancy between both data sets, we believe that our conclusions are sound as they are strongly supported by the consistency of the localization obtained (i) with the two expression systems used in T. brucei and whichever the location of the GFP tag on the protein in both systems, and (ii) with the episomal expression of both C- and N-terminally GFP-tagged proteins in L. major. Incidentally, in a more recent paper, the same groups as DeGrasse et al. localized TbMlp2 (termed TbNUP92) both intranuclear and at the nuclear envelope (69).
Using the centromeric sequences of T. brucei chromosomes 2 and 3, we then showed that, in a large majority of interphasic cells, TbMlp2 is adjacent or in close proximity to the centromeres at the periphery of the nucleolus. From the literature, the mitotic function of NUPs is often associated with their localization to kinetochores in mitosis. Tpr, the mammalian homologue of Mlp1/2, and its plant orthologue NUA (70) directly bind to Mad1 and Mad2 spindle assembly checkpoint (SAC) proteins and recruits them to the kinetochores (71–73). Mad1 and Mad2 thus localize to kinetochores in mammalian and plant, as well as yeast, prometaphase cells (70,73–75); in mammals, this is also mediated by the NUP Nup153 (19). In Aspergillus nidulans, AnMlp1, AnMad1 and AnMad2 also localize on and around kinetochores until telophase, when they transiently localize near the spindle but not at kinetochores (76). The common theme therefore appears to be that AnMlp1, like ScMlp1/2, Tpr and Megator (72–73,75,77) play a role in the spatiotemporal regulation of SAC proteins. Here, the vast majority of interphasic cells expressing Mlp2 displayed an intranuclear localization of the protein; yet, isTbMlp2 showed a localization typical of NUPs, i.e. as a ‘bead collar’ at the nuclear envelope, in a very small proportion of cells, suggesting that this localization is transient. By contrast, it is strongly expressed during mitosis, suggesting that this apparently ‘divergent’ NUP is mainly involved in the mitotic process.
The whole of our results supports the hypothesis of an interaction between TbMlp2 and the kinetochores in T. brucei. Yet, TbMlp2 has not been found among the 19 unconventional kinetochore proteins recently described in T. brucei (44). Actually, TbMlp2 localized in close proximity to the kinetochore protein TbKKT4 before the onset of mitosis. During metaphase, it partially dissociated from TbKKT4, until anaphase where it was entirely dissociated from TbKKT4 and the centromeres which remained at the centre of the dividing nucleus. From the end of anaphase onwards, the centromeres/kinetochores were found at the mitotic spindle poles, again adjacent to TbMlp2 (Figures 5 and 6). The role of TbMlp2 therefore remains unclear, but a direct or indirect involvement of this protein in chromosomal distribution during mitosis is further supported by the induction of aneuploidy after TbMlp2 depletion and the partial restoration of ploidy after complementation. It should be stressed here that we used chromosome-specific probes to affirm a chromosomal distribution defect, whereas the similar conclusion drawn by Holden et al. (of a “lowered fidelity of telomere segregation") was poorly supported by the experiments since they used telomeric (and not chromosome-specific) probes (69). In total, we hypothesized that, without integrating the protein complex constituting the kinetochore, TbMlp2 might participate for example to the assembly and/or dynamics of these complexes.
Paradoxically, the aneuploidy induced by TbMlp2 depletion was not accompanied by any effect on cell growth in vitro. In higher eukaryotes, aneuploidy is known to have deleterious effects, but mosaic aneuploidy is a constitutive feature in Leishmania sp. (54,78). Here, in in vitro culture conditions, T. brucei was shown to also be able to tolerate aneuploidy. Although the chromosome duplication process does not appear as straightforward in T. brucei as it is in Leishmania, trisomy has been reported in T. brucei following genetic exchange (79). Also, polysomy of a 6-Mb chromosome was reported in a drug-resistant strain following selection by mycophenolic acid (80), showing that under particular constraints, aneuploidy can occur in this organism. This might be explained by distinctive features shared by both trypanosomatids like (i) a genomic organization in large non-functionally related polycistronic units (81), (ii) the near absence of RNA Pol II promoters and (iii) a weak regulation of gene expression at the transcription level (82). These features could explain why, constitutively in Leishmania and here in a mutant T. brucei cell line, gene dosage variations may have no or little effect on in vitro cell growth. Like in many ‘divergent’ unicellular eukaryotes the biological core processes of trypanosomatids remain ill-known, in particular many steps of mitosis, such as mitotic checkpoints and spindle assembly. For example, there is a clear deficit in the number of kinetochores, and no centrosome can be seen at the spindle poles. The data presented here show that Mlp2 is involved in chromosomal distribution during mitosis. The precise function of TbMlp2 remains to be elucidated, and we anticipate that this understanding will challenge the accepted views about mitosis in classical models, shedding light upon the setting up of ‘exotic mitotic mechanisms’ throughout evolution (83).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Frédéric Bringaud (Université Victor Segalen Bordeaux 2) for providing the T. brucei 29–13 cell line and T. brucei expression vectors; Derrick Robinson (Université Victor Segalen Bordeaux 2) for providing the T. brucei wild-type cell line; Michael Oberholzer (UCLA) for his gift of the pMOTTag4 construct; and TrypanoFAN (director Mark Field) who made the RNAi vector available to us. We also wish to thank Keith Gull (University of Oxford) for his generous gift of the monoclonal antibody KMX. We gratefully acknowledge Vicky Diakou (RIO Imaging Platform in Montpellier) for advice on and assistance in fluorescence microscopy and Cédric Mongellaz (RIO Imaging Platform in Montpellier) for help in handling the FACSCalibur (Beckton Dickinson®, San Jose, CA, USA) and the BD CellQuest Pro software.
Footnotes
Present address: Yvon Sterkers, Michel Pagès and Laurence Lachaud, Laboratoire de Parasitologie-Mycologie, CHRU de Montpellier, 39, Avenue Charles Flahault, 34295 Montpellier Cedex 5, France.
FUNDING
‘Investissements d'avenir’ Program, the Agence Nationale de la Recherche (ANR-11-LABX-0024-01 ‘PARAFRAP’); the Centre National de la Recherche Scientifique (CNRS); the French Ministry of Research; the Centre Hospitalier Universitaire of Montpellier.
Conflict of interest statement. None declared.
REFERENCES
- 1.Davis L.I. The nuclear pore complex. Annu. Rev. Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- 2.Pante N., Aebi U. Molecular dissection of the nuclear pore complex. Crit. Rev. Biochem. Mol. Biol. 1996;31:153–199. doi: 10.3109/10409239609106583. [DOI] [PubMed] [Google Scholar]
- 3.Gorlich D., Mattaj I.W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 4.Nigg E.A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 5.Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., Chait B.T. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wente S.R. Gatekeepers of the nucleus. Science. 2000;288:1374–1377. doi: 10.1126/science.288.5470.1374. [DOI] [PubMed] [Google Scholar]
- 7.Adam S.A. The nuclear pore complex. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-reviews0007. Reviews0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wente S.R., Rout M.P. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a000562. a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasu S.K., Forbes D.J. Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 2001;13:363–375. doi: 10.1016/s0955-0674(00)00221-0. [DOI] [PubMed] [Google Scholar]
- 10.Cronshaw J.M., Krutchinsky A.N., Zhang W., Chait B.T., Matunis M.J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeGrasse J.A., DuBois K.N., Devos D., Siegel T.N., Sali A., Field M.C., Rout M.P., Chait B.T. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics. 2009;8:2119–2130. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capelson M., Hetzer M.W. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 2009;10:697–705. doi: 10.1038/embor.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raices M., D'Angelo M.A. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat. Rev. Mol. Cell Biol. 2012;13:687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- 14.Bukata L., Parker S.L., D'Angelo M.A. Nuclear pore complexes in the maintenance of genome integrity. Curr. Opin. Cell Biol. 2013;25:378–386. doi: 10.1016/j.ceb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Salina D., Enarson P., Rattner J.B., Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 2003;162:991–1001. doi: 10.1083/jcb.200304080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joseph J., Dasso M. The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Lett. 2008;582:190–196. doi: 10.1016/j.febslet.2007.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawlaty M.M., Malureanu L., Jeganathan K.B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J.M. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashizume C., Nakano H., Yoshida K., Wong R.W. Characterization of the role of the tumor marker Nup88 in mitosis. Mol. Cancer. 2010;9:119. doi: 10.1186/1476-4598-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lussi Y.C., Shumaker D.K., Shimi T., Fahrenkrog B. The nucleoporin Nup153 affects spindle checkpoint activity due to an association with Mad1. Nucleus. 2010;1:71–84. doi: 10.4161/nucl.1.1.10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong R.W., Blobel G. Cohesin subunit SMC1 associates with mitotic microtubules at the spindle pole. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15441–15445. doi: 10.1073/pnas.0807660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatel G., Fahrenkrog B. Nucleoporins: leaving the nuclear pore complex for a successful mitosis. Cell Signal. 2011;23:1555–1562. doi: 10.1016/j.cellsig.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 22.De Souza C.P., Osmani S.A. Double duty for nuclear proteins–the price of more open forms of mitosis. Trends Genet. 2009;25:545–554. doi: 10.1016/j.tig.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krull S., Dorries J., Boysen B., Reidenbach S., Magnius L., Norder H., Thyberg J., Cordes V.C. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 2010;29:1659–1673. doi: 10.1038/emboj.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brickner J.H., Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Light W.H., Brickner D.G., Brand V.R., Brickner J.H. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell. 2010;40:112–125. doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabal G.G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., Buc H., Feuerbach-Fournier F., Olivo-Marin J.C., Hurt E.C., et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 27.Luthra R., Kerr S.C., Harreman M.T., Apponi L.H., Fasken M.B., Ramineni S., Chaurasia S., Valentini S.R., Corbett A.H. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J. Biol. Chem. 2007;282:3042–3049. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- 28.Dieppois G., Iglesias N., Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taddei A., Van Houwe G., Hediger F., Kalck V., Cubizolles F., Schober H., Gasser S.M. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 30.Ellisdon A.M., Jani D., Kohler A., Hurt E., Stewart M. Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1. J. Biol. Chem. 2010;285:3850–3856. doi: 10.1074/jbc.M109.070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohler A., Schneider M., Cabal G.G., Nehrbass U., Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 32.Kurshakova M.M., Krasnov A.N., Kopytova D.V., Shidlovskii Y.V., Nikolenko J.V., Nabirochkina E.N., Spehner D., Schultz P., Tora L., Georgieva S.G. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 2007;26:4956–4965. doi: 10.1038/sj.emboj.7601901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rougemaille M., Dieppois G., Kisseleva-Romanova E., Gudipati R.K., Lemoine S., Blugeon C., Boulay J., Jensen T.H., Stutz F., Devaux F., et al. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell. 2008;135:308–321. doi: 10.1016/j.cell.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 34.Vaquerizas J.M., Suyama R., Kind J., Miura K., Luscombe N.M., Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galy V., Olivo-Marin J.C., Scherthan H., Doye V., Rascalou N., Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403:108–112. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- 36.Zhao X., Wu C.Y., Blobel G. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 2004;167:605–611. doi: 10.1083/jcb.200405168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niepel M., Strambio-de-Castillia C., Fasolo J., Chait B.T., Rout M.P. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J. Cell Biol. 2005;170:225–235. doi: 10.1083/jcb.200504140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hampsey M., Singh B.N., Ansari A., Laine J.P., Krishnamurthy S. Control of eukaryotic gene expression: gene loops and transcriptional memory. Adv. Enzyme Regul. 2011;51:118–125. doi: 10.1016/j.advenzreg.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan-Wong S.M., Wijayatilake H.D., Proudfoot N.J. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laine J.P., Singh B.N., Krishnamurthy S., Hampsey M. A physiological role for gene loops in yeast. Genes Dev. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hediger F., Dubrana K., Gasser S.M. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J. Struct. Biol. 2002;140:79–91. doi: 10.1016/s1047-8477(02)00533-6. [DOI] [PubMed] [Google Scholar]
- 42.Loeillet S., Palancade B., Cartron M., Thierry A., Richard G.F., Dujon B., Doye V., Nicolas A. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair (Amst) 2005;4:459–468. doi: 10.1016/j.dnarep.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 43.Obado S.O., Bot C., Echeverry M.C., Bayona J.C., Alvarez V.E., Taylor M.C., Kelly J.M. Centromere-associated topoisomerase activity in bloodstream form Trypanosoma brucei. Nucleic Acids Res. 2011;39:1023–1033. doi: 10.1093/nar/gkq839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akiyoshi B., Gull K. Discovery of unconventional kinetochores in kinetoplastids. Cell. 2014;156:1247–1258. doi: 10.1016/j.cell.2014.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dubessay P., Blaineau C., Bastien P., Pages M. Chromosome fragmentation in leishmania. Methods Mol. Biol. 2004;270:353–378. doi: 10.1385/1-59259-793-9:353. [DOI] [PubMed] [Google Scholar]
- 46.Wirtz E., Leal S., Ochatt C., Cross G.A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 47.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 48.Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U.S.A. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dubessay P., Blaineau C., Bastien P., Tasse L., Van Dijk J., Crobu L., Pages M. Cell cycle-dependent expression regulation by the proteasome pathway and characterization of the nuclear targeting signal of a Leishmania major Kin-13 kinesin. Mol. Microbiol. 2006;59:1162–1174. doi: 10.1111/j.1365-2958.2005.05013.x. [DOI] [PubMed] [Google Scholar]
- 50.Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 51.Oberholzer M., Morand S., Kunz S., Seebeck T. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 2006;145:117–120. doi: 10.1016/j.molbiopara.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 52.Ersfeld K., Gull K. Partitioning of large and minichromosomes in Trypanosoma brucei. Science. 1997;276:611–614. doi: 10.1126/science.276.5312.611. [DOI] [PubMed] [Google Scholar]
- 53.Freitas-Junior L.H., Bottius E., Pirrit L.A., Deitsch K.W., Scheidig C., Guinet F., Nehrbass U., Wellems T.E., Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 54.Sterkers Y., Lachaud L., Crobu L., Bastien P., Pagès M. FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in Leishmania major. Cell. Microbiol. 2011;13:274–283. doi: 10.1111/j.1462-5822.2010.01534.x. [DOI] [PubMed] [Google Scholar]
- 55.Bessat M., Ersfeld K. Functional characterization of cohesin SMC3 and segregation of large and minichromosomes in Trypanosoma brucei. Mol. Microbiol. 2009;71:1371–1385. doi: 10.1111/j.1365-2958.2009.06611.x. [DOI] [PubMed] [Google Scholar]
- 56.Sasse R., Gull K. Tubulin post-translational modifications and the construction of microtubular organelles in Trypanosoma brucei. J. Cell Sci. 1988;90:577–589. doi: 10.1242/jcs.90.4.577. [DOI] [PubMed] [Google Scholar]
- 57.Sherwin T., Gull K. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1989;323:573–588. doi: 10.1098/rstb.1989.0037. [DOI] [PubMed] [Google Scholar]
- 58.Woodward R., Gull K. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J. Cell Sci. 1990;95:49–57. doi: 10.1242/jcs.95.1.49. [DOI] [PubMed] [Google Scholar]
- 59.Siegel T.N., Hekstra D.R., Cross G.A. Analysis of the Trypanosoma brucei cell cycle by quantitative DAPI imaging. Mol. Biochem. Parasitol. 2008;160:171–174. doi: 10.1016/j.molbiopara.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ambit A., Woods K., Cull B., Coombs G., Mottram J. Morphological events during the cell cycle of Leishmania major. Eukaryot. Cell. 2011;10:1429–1438. doi: 10.1128/EC.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Echeverry M.C., Bot C., Obado S.O., Taylor M.C., Kelly J.M. Centromere-associated repeat arrays on Trypanosoma brucei chromosomes are much more extensive than predicted. BMC Genomics. 2012;13:29. doi: 10.1186/1471-2164-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolev N.G., Tschudi C., Ullu E. RNA interference in protozoan parasites: achievements and challenges. Eukaryot. Cell. 2011;10:1156–1163. doi: 10.1128/EC.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strambio-De-Castillia C., Niepel M., Rout M.P. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 64.Qi H., Rath U., Wang D., Xu Y.Z., Ding Y., Zhang W., Blacketer M.J., Paddy M.R., Girton J., Johansen J., et al. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol. Biol. Cell. 2004;15:4854–4865. doi: 10.1091/mbc.E04-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johansen K.M., Johansen J. Cell and molecular biology of the spindle matrix. Int. Rev. Cytol. 2007;263:155–206. doi: 10.1016/S0074-7696(07)63004-6. [DOI] [PubMed] [Google Scholar]
- 66.Blower M.D., Nachury M., Heald R., Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 67.Wong R.W., Blobel G., Coutavas E. Rae1 interaction with NuMA is required for bipolar spindle formation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19783–19787. doi: 10.1073/pnas.0609582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong R.W. An update on cohesin function as a ‘molecular glue’ on chromosomes and spindles. Cell Cycle. 2010;9:1754–1758. doi: 10.4161/cc.9.9.11806. [DOI] [PubMed] [Google Scholar]
- 69.Holden J.M., Koreny L., Obado S., Ratushny A.V., Chen W.M., Chiang J.H., Kelly S., Chait B.T., Aitchison J.D., Rout M.P., et al. Nuclear pore complex evolution: a trypanosome Mlp analogue functions in chromosomal segregation but lacks transcriptional barrier activity. Mol Biol Cell. 2014;25:1421–1436. doi: 10.1091/mbc.E13-12-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ding D., Muthuswamy S., Meier I. Functional interaction between the Arabidopsis orthologs of spindle assembly checkpoint proteins MAD1 and MAD2 and the nucleoporin NUA. Plant Mol. Biol. 2012;79:203–216. doi: 10.1007/s11103-012-9903-4. [DOI] [PubMed] [Google Scholar]
- 71.Xu X.M., Rose A., Meier I. NUA activities at the plant nuclear pore. Plant Signal. Behav. 2007;2:553–555. doi: 10.4161/psb.2.6.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakano H., Funasaka T., Hashizume C., Wong R.W. Nucleoporin translocated promoter region (Tpr) associates with dynein complex, preventing chromosome lagging formation during mitosis. J. Biol. Chem. 2010;285:10841–10849. doi: 10.1074/jbc.M110.105890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee S.H., Sterling H., Burlingame A., McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22:2926–2931. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iouk T., Kerscher O., Scott R.J., Basrai M.A., Wozniak R.W. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scott R.J., Lusk C.P., Dilworth D.J., Aitchison J.D., Wozniak R.W. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:4362–4374. doi: 10.1091/mbc.E05-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.De Souza C.P., Hashmi S.B., Nayak T., Oakley B., Osmani S.A. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol. Biol. Cell. 2009;20:2146–2159. doi: 10.1091/mbc.E08-08-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lince-Faria M., Maffini S., Orr B., Ding Y., Claudia F., Sunkel C.E., Tavares A., Johansen J., Johansen K.M., Maiato H. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J. Cell Biol. 2009;184:647–657. doi: 10.1083/jcb.200811012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sterkers Y., Lachaud L., Bourgeois N., Crobu L., Bastien P., Pages M. Novel insights into genome plasticity in Eukaryotes: mosaic aneuploidy in Leishmania. Mol. Microbiol. 2012;86:15–23. doi: 10.1111/j.1365-2958.2012.08185.x. [DOI] [PubMed] [Google Scholar]
- 79.Gibson W., Garside L., Bailey M. Trisomy and chromosome size changes in hybrid trypanosomes from a genetic cross between Trypanosoma brucei rhodesiense and T. b. brucei. Mol. Biochem. Parasitol. 1992;51:189–199. doi: 10.1016/0166-6851(92)90069-v. [DOI] [PubMed] [Google Scholar]
- 80.Wilson K., Berens R.L., Sifri C.D., Ullman B. Amplification of the inosinate dehydrogenase gene in Trypanosoma brucei gambiense due to an increase in chromosome copy number. J. Biol. Chem. 1994;269:28979–28987. [PubMed] [Google Scholar]
- 81.Ivens A.C., Peacock C.S., Worthey E.A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clayton C., Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 83.Drechsler H., McAinsh A.D. Exotic mitotic mechanisms. Open Biol. 2012;2:120140. doi: 10.1098/rsob.120140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


