Abstract
The UvrD helicase has been implicated in the disassembly of RecA nucleoprotein filaments in vivo and in vitro. We demonstrate that UvrD utilizes an active mechanism to remove RecA from the DNA. Efficient RecA removal depends on the availability of DNA binding sites for UvrD and/or the accessibility of the RecA filament ends. The removal of RecA from DNA also requires ATP hydrolysis by the UvrD helicase but not by RecA protein. The RecA-removal activity of UvrD is slowed by RecA variants with enhanced DNA-binding properties. The ATPase rate of UvrD during RecA removal is much slower than the ATPase activity of UvrD when it is functioning either as a translocase or a helicase on DNA in the absence of RecA. Thus, in this context UvrD may operate in a specialized disassembly mode.
INTRODUCTION
The Escherichia coli UvrD protein is a superfamily 1 (SF1) DNA helicase/translocase that functions in methyl-directed mismatch repair (MMR) (1,2), nucleotide excision repair (NER) (3–5) and more broadly in genome integrity maintenance. A uvrD− phenotype is characterized by an increased rate of recombination and by a constitutive induction of the SOS response (6–8), which controls expression of a number of DNA repair genes under the control of the LexA transcriptional regulator (9). Strains containing an overexpressed or hyperactive UvrD variant exhibit the opposite phenotype and are hypo-recombinant and display low mutagenicity (7,10). However, both the hyperactive and the inactive UvrD confer a DNA-damage susceptibility phenotype (7).
The helicase/translocase activity of UvrD has a 3′ to 5′ directional bias (11–13). The preference of the protein for unwinding DNA duplex substrates with a 3′ tail (12) is utilized directly in several DNA repair pathways. However, UvrD will also unwind DNA at nicks and at blunt ends (14). In NER, UvrD interacts directly with UvrB to unwind a short region of DNA containing a misincorporated deoxyribonucleotide (15,16). It also moves RNA polymerase backward to expose lesions requiring repair (4) and helps to mediate collisions between transcription and replication (17). In MMR, UvrD is recruited and positioned by MutL to displace a significant region of DNA (1–2 kbp) containing an incorrectly incorporated base (reviewed in (18)). Both NER and MMR are dependent on the classic activity of the UvrD helicase to unwind DNA. In addition to its helicase activity, UvrD can displace proteins from ssDNA. UvrD frees the ter sites of the bacterial chromosome from the Tus protein, and the translocase and/or helicase activities of UvrD may be necessary for this function (19).
Another major target of UvrD for protein displacement from DNA is the RecA protein (6,8,10,20). RecA catalyzes homologous recombination and is involved in non-mutagenic and mutagenic DNA repair (21–23). RecA is a DNA-dependent ATPase functioning in the form of a nucleoprotein filament assembled on DNA (24,25). A recA− E. coli strain displays susceptibility to DNA damage (26). RecA recombination activity is necessary for repair of DNA damage, especially the double-strand breaks that can accompany replication fork collapse.
RecA catalyzes replication fork regression in vitro (27), and UvrD may remove RecA from replication forks after repair (6,28). The lethality of a ΔpolAΔuvrD strain (15) provides additional evidence that UvrD-mediated RecA filament removal is important for replication fork maintenance. RecA filaments may be toxic under certain conditions when impaired forks are present (28). This hypothesis is supported by the fact that the lethal phenotype of a ΔuvrDΔrep strain is rescued by a knockout of any of the recFORJQ genes (28,29). The proteins RecF, RecO and RecR have been implicated in loading RecA on gapped DNA structures, such as collapsed replication forks (reviewed in (30)). However, a uvrD knockout strain has an increased recombination phenotype, while overexpressed UvrD results in reduced Hfr recombination and mutability (10). These observations suggest that UvrD may interact with RecA filaments throughout the cell. More studies are needed to understand how the two proteins augment each other's functions.
The active form of RecA protein is a nucleoprotein filament (31,32). Forming most readily on ssDNA, the filament aligns the bound single strand with homologous sequences in a duplex DNA, and promotes a reaction called DNA strand exchange. ATP is hydrolyzed during strand exchange, needed both to promote filament dissociation and the extensive branch migration associated with strand exchange (24,33–39). RecA filaments are nucleated and grow primarily on the 3′-proximal end. Dissociation occurs primarily on the 5′-proximal end. Due to the polarity of its movements on DNA, UvrD will normally encounter a RecA filament at its 3′-proximal end. The current report explores what happens next.
RecA function is regulated at many levels (40). In addition to transcriptional regulation as part of the SOS response, RecA is subject to autoregulation and to regulation by other proteins. The autoregulation is brought on by a C-terminal regulatory flap, encompassing the final 17 amino acid residues of the protein (41–43). This segment is highly charged, with seven of the 17 residues featuring negatively charged side chains. Removal of this C-terminal segment enhances a wide range of RecA functions (41–44). The regulatory proteins include the RecA loaders RecBCD and RecFOR, as well as the positive regulator DinI (45,46) and negative modulators such as the RecX protein (44,47–51) and the UvrD helicase considered here.
It is not clear how UvrD mediates the displacement of RecA filaments. Based on other UvrD functions, there are arguments for and against a requirement for a direct interaction between the two proteins. UvrD participates in a number of chromosomal maintenance processes, so targeted recruitment may require direct interactions. For example, during MMR UvrD interacts with and is stimulated by MutL to unwind a long region of DNA duplex (reviewed in (18)). A reaction lacking MutL would be highly inefficient due to the low unwinding processivity of UvrD. In fact, the stimulation by MutL is so strong that UvrD252, a mutant almost completely ATPase deficient, is still able to participate in MMR (28,52). Similarly, in NER, UvrD interacts with UvrB via the UvrD C-terminus (16). However, a C-terminal truncation of UvrD is proficient in NER repair in vitro, possibly because other areas of UvrD interact with UvrB. An interesting aspect of the UvrD ΔC phenotype is that the cells are more UV irradiation sensitive, which suggests that the interaction between UvrB and UvrD may be physiologically relevant. In yeast, the homolog of UvrD, Srs2, interacts directly with the RecA homolog Rad51 in order to disassemble Rad51 filaments on DNA (53).
The UvrD homolog PcrA, an essential helicase in gram positive bacteria, facilitates RecA filament removal from DNA by a mechanism that depends upon the RecA ATPase (54). In brief, when the PcrA helicase encounters the 3′-proximal end of a RecA filament, it halts translocation and prevents further extension of the RecA filament. The filament disassembly could then proceed by dissociation of RecA subunits at the nearby 3′-proximal end when that subunit hydrolyzes ATP. This dissociation may be facilitated in some way by the PcrA. In principle, the displacement mechanism could also be passive, with net disassembly of the RecA filament proceeding as normal from the 5′-proximal end while filament extension was stalled by the arrested PcrA. If a RecA protein mutant that cannot hydrolyze ATP is bound to the DNA, it arrests PcrA translocation, but filament disassembly does not occur (54). A single-molecule study of PcrA-mediated displacement of RecA filaments suggests a more active process for PcrA-mediated RecA displacement that involves the ATPase activity of PcrA (55). Potential active and passive RecA displacement mechanisms are contrasted in Figure 1.
Figure 1.
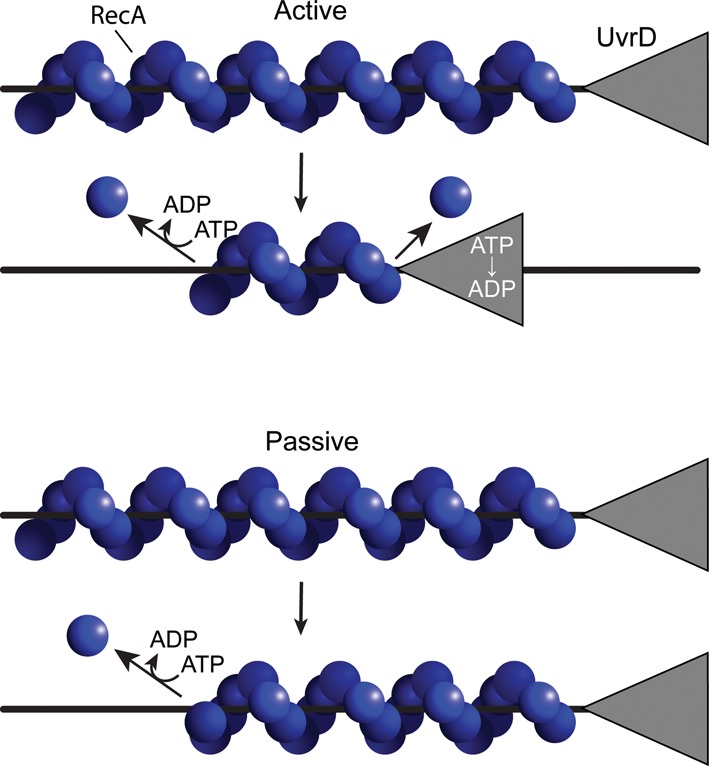
Active versus passive mechanisms for UvrD-mediated RecA displacement. Two of the more straightforward mechanisms are presented. The active mechanism involves any process in which UvrD actively removes RecA protein from the 3′-proximal end, requiring the UvrD ATPase function. This may or may not be complemented by RecA filament dissociation from the opposite filament end. In the passive mechanism, UvrD simply encounters the 3′-proximal end and prevents further filament growth there. Filament dissociation occurs entirely from the opposite end and requires the ATPase function of RecA protein. Other possible mechanisms exist. The work in this study lends support for a mechanism approximating that shown for the active scheme at top.
In contrast, the mechanism of RecA displacement by UvrD is little explored. PcrA can substitute for UvrD in Ter protein removal, suggesting that a specificity of interaction with its target is not necessary for UvrD, unless the PcrA homolog conserves an interaction site with Ter that has not been identified (19). An active mechanism has been proposed based on the observation that substoichiometric levels of UvrD result in disassembly of RecA filaments in vitro (10). However, a similar effect has been observed with the RecA inhibitor RecX, which inhibits RecA largely through passive capping rather than active displacement (48,49). Finally, the eukaryotic Srs2 homolog appears to induce a higher ATPase activity in a Rad51 filament (53). UvrD was unable to produce the same effect on Rad51 (53), although it remains possible that RecA-mediated ATP hydrolysis is involved in the UvrD-mediated displacement process as it is for PcrA. Here, we demonstrate that UvrD removes RecA protein via an active mechanism that requires UvrD-mediated ATP hydrolysis, but does not require ATP hydrolysis by RecA protein.
MATERIALS AND METHODS
All chemicals were purchased from Sigma and Fisher. ATPγS was purchased from Roche.
DNA substrates
M13mp18 circular ssDNA was purified as previously described (56). M13mp18 linear ssDNA was generated by annealing a primer (ACTCTAGAGGATCCCCGGGTAC) to the virion DNA and incubating with BamHI restriction enzyme (New England Biolabs, R0136). The DNA was purified by phenol-chloroform extraction and ethanol precipitation. Poly(dT) DNA was ordered from Amersham, lot GG0076. Poly deoxythymidylate (poly(dT)) was purchased from Midland; the analysis provided indicated nearly all polymers had a length greater than 250 nucleotides. All DNA concentrations are reported in terms of total nucleotides rather than total molecules, to facilitate comparison with RecA protein concentrations.
Proteins
Wild-type (WT) RecA, RecA E38K K72R, RecA E38K and single stranded DNA binding protein (SSB) were purified as previously described (43,57–59). Their concentrations were determined utilizing the extinction coefficients: RecA: ε280 = 2.23×104 M−1cm−1 and SSB: 2.38×104 M−1cm−1. UvrD and UvrD K35I were purified as described, and concentrations were determined using the extinction coefficient 1.06 x105 M−1 (monomer) cm−1 for both proteins (60). Pyruvate kinase and lactate dehydrogenase were ordered from Sigma (P7768 and L1006, respectively).
ATPase assays
The reaction was set up by allowing first 2 μM of a RecA variant (except for the experiments shown in Figures 4 and 9, where RecA concentrations were varied as indicated on the figure) to nucleate on 3 μM M13mp18 circular ssDNA or lss M13mp18 DNA or lss poly(dT) in 1xRecA buffer (25 mM Tris-OAc (80% cation), pH 7.58, 1 mM DTT, 3 mM potassium glutamate, 10 mM magnesium acetate, 5% glycerol) for 10 min at 37ºC. The reaction included 10 U pyruvate kinase and 3 mM phosphoenolpyruvate as a regeneration system, and a coupling system consisting of 10 U of lactate dehydrogenase and 1.5 mM NADH. The reaction was started by the addition of SSB to 0.3 μM and ATP to 3 mM. When poly(dT) DNA was used, no SSB was included in the reaction. The generation of ADP was followed by measuring the consumption of NADH at 380 nm in Cary 300 (Varian) or Lambda 650 (Perkin-Elmer) as previously described (61). UvrD was added to the reactions 15 min after the start of the reaction. Total reaction volume was 130 μl final.
Figure 4.
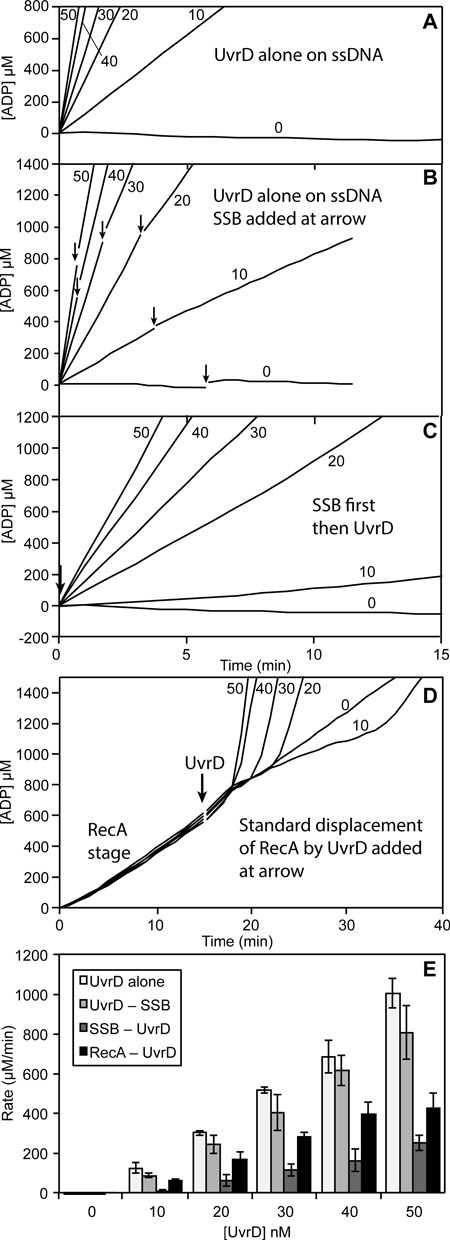
UvrD ATPase activity on circular ssDNA in the absence of RecA protein. UvrD ATPase rates were determined in the presence or absence of SSB and RecA. Values on each graph indicate the concentration of UvrD in nM. (A) UvrD alone (at the concentrations indicated, in nM) on circular M13mp18 ssDNA. (B) UvrD alone (at the concentrations indicated, in nM) with SSB (0.3 μM) added at the point indicated by an arrow. (C) UvrD (at the concentrations indicated, in nM) added to an SSB (0.3 μM)/circular ssDNA mixture as a substrate. (D) RecA/SSB/circular M13mp18 ssDNA reaction with UvrD (at the concentrations indicated, in nM) added at the point indicated by an arrow. (E) Quantification of the rates of UvrD in graphs A–D. For graphs B and D, the maximum post-UvrD addition rates observed, obtained at a time when the reaction progress has reached a new apparent steady state as indicated by approximate progress linearity, are used.
Figure 9.
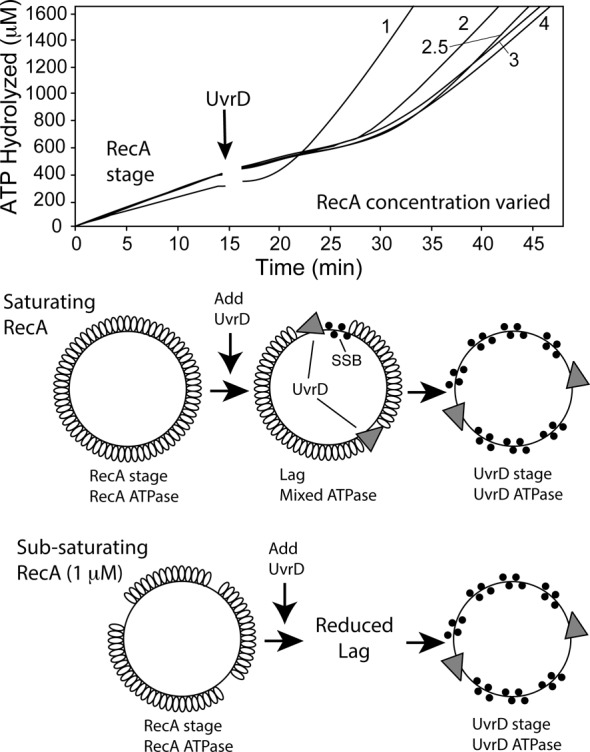
The effect of RecA concentration on UvrD capacity to dismantle RecA filaments. The indicated concentrations of RecA protein (in μM) were allowed to form active filaments on M13mp18 circular ssDNA as described in Materials and Methods. After 15 min (arrow), the reactions were challenged with 50 nM of UvrD.
Observation of the three reaction stages requires rapid mixing of components after addition of the UvrD (done in this study by hand within 5 s of addition). Aggregation of concentrated UvrD on DNA or RecA filaments or some related effect may otherwise mask observation of the effects of UvrD on the total ATPase activity as reported here.
Electron microscopy
A modified Alcian method was used to visualize RecA filaments. Activated grids were prepared as described previously (43). Samples for electron microscopy analysis were prepared as follows. The reaction was set up as the ATPase assays described above, except no DTT was used. The concentration of UvrD used in the experiments was 40 nM, calculated based on total volume. Ten microliter of the reaction volume was withdrawn and incubated with ATPγS at final concentration of 3 mM for 1–2 min at 37°C. The time points were taken as described below.
For experiments involving WT RecA and ATP, the UvrD addition occurred 15 min after the start of the reaction, and time points were taken as follows. The first time point was taken 10 min after the start of the reaction by the addition of ATP to 3 mM and SSB to 0.3 μM; the second—6–8 min after the addition of UvrD as the ATPase rate of the monitored reaction was visibly decreasing and the third—13–50 min after UvrD addition, either when the ATPase rate clearly increased or 50 min after UvrD addition the latest.
For experiments involving RecA E38K K72R, the UvrD protein was added 16 min after the start of the reaction, and time points were taken as follows. The first aliquot was taken 10 min after the start of the reaction by the addition of ATP to 3 mM and SSB to 0.3 μM; the second—6 min after the addition of UvrD and the third—50 min after UvrD addition.
For experiments involving RecA ΔC17, the UvrD protein was added 15 min after the start of the reaction, and time points were taken as follows. The first aliquot was taken 8 min after the start of the reaction by the addition of ATP to 3 mM and SSB to 0.3 μM; the second—5 min after the addition of UvrD.
The electron microscopy (EM) experiments that were done only in the presence of ATPγS and not ATP were conducted as follows. RecA (2 μM) was incubated with 3 μM M13mp18 circular ssDNA, 3 mM PEP and 10 U PK and 1xRecA buffer (see the ATPase experiments) for 10 min at 37ºC. Then, ATPγS (3 mM) and SSB (0.3 μM) were added to the mixture, and that was considered the start of the reaction. After 16 min, UvrD was added to 40 nM. Time points were taken 10 min after the start of the reaction and 6 and 50 min after the addition of UvrD.
The samples were processed as follows. An 8 μl sample of the reaction mixture described above was diluted to a final DNA concentration of 0.0004 μg/μl with 200 mM ammonium acetate, 10 mM Hepes and 10% glycerol (pH adjusted to 7.5) and adsorbed to an activated Alcian grid for 3 min. The grid was then touched to a drop of the above buffer followed by floating on a drop of the same buffer for 1 min. The sample was then stained by touching to a drop of 5% uranyl acetate followed by floating on a fresh drop of the same solution for 30 s. Finally, the grid was washed by touching to a drop of double distilled water followed by immersion in two 10-ml beakers of double distilled water. After the sample was dried, it was rotary-shadowed with platinum. This protocol is designed for visualization of complete reaction mixtures, and no attempt was made to remove unreacted material. Although this approach should yield results that give a true insight into reaction components, it does lead to samples with a high background of unreacted proteins.
To determine the proportion of the molecules observed that were either fully or partially coated by RecA protein or bound only by the SSB protein, at least two separate regions of two to three independent experiments were counted at an identical magnification for each sample. ‘Full’ filaments completely encompassed the circular DNA molecule or had small discontinuities in the regular striated pattern of the filament. A molecule was considered gapped if it had a detectable region of SSB-coated DNA of any size. Imaging and photography were carried out with a TECNAI G2 12 Twin Electron Microscope (FEI Co.) equipped with a GATAN 890 CCD camera. Digital images of the nucleoprotein filaments were taken at ×15 000 and ×26 000 magnification as is evident from the scale bar.
The observed lengths of the RecA filaments and the length of SSB-coated DNA were used to assign counted molecules to five categories: full filaments, medium filaments, small filaments, very small filaments or SSB/DNA molecules. Linearized DNA molecules, likely originating from shearing force during pipetting, were also counted. A RecA filament was considered a full filament if it does not have a detectable region of SSB-coated DNA or a region that appeared to reduce the filament length by <10%. Medium filaments were smaller in length than full filaments, but still had substantial regions of nucleoprotein filament. Small filaments were generally less than half the length of full filaments, and often had regions of obvious SSB binding. Very small filamented molecules are those with just detectable segments of RecA filamented regions, with the rest of the molecule coated with SSB. With the total number of molecules counted as 100%, the percentage of each type of nucleoprotein filament was calculated. At least four separate regions of the grids encompassing at least 500 DNA molecules for each time point were counted at the identical magnification for each sample.
Using this same classification scheme, filament measurements have been recently completed for between 10 and 30 molecules from each of these classes, bound to the same ssDNA substrate (62). Full filaments averaged 3.23 +/– 0.29 μm in length, with a range of 3.03–3.6 μm. Medium filaments averaged 2.32 μm in length, with a range of 1.73–2.72 μm. Small filaments averaged 1.28 μm, with a range of 0.97–1.8 μm. Very small filaments averaged 0.39 μm in length, with a range of 0.12–0.71.
RESULTS
UvrD inhibits RecA-catalyzed ATP hydrolysis
In this work, we use several assays to follow displacement of RecA by UvrD. A coupled ATPase assay is used. Although indirect, the ATPase provides a real time view of reaction progress. Both RecA and UvrD promote ATP hydrolysis, but this complication is ameliorated by the very different kcat values of the RecA and UvrD ATPase activities. UvrD catalyzes ATP hydrolysis at rates that are orders of magnitude greater than RecA, such that nM levels of UvrD will hydrolyze much more ATP than μM levels of RecA. To aid interpretation of the results, the work is complemented throughout by electron microscopy.
When RecA protein was added to circular single-stranded DNA at concentrations approximately twice that required to saturate the available DNA binding sites, the bound RecA protein hydrolyzed ATP at rates consistent with its reported kcat of ∼30 min−1 (32,63,64) (Figures 2 and 3). Based on the much higher values of ATP hydrolysis by UvrD acting as a helicase (∼70 s−1 (65,66)) or translocase (∼190 s−1 (67)), we expected a large increase in ATP consumption and ADP generation to indicate that UvrD has been successful in displacing RecA bound to DNA, and this was the case. However, upon addition of UvrD at nM levels to established RecA nucleoprotein filaments, a pronounced lag phase appeared prior to the rapid ATP hydrolysis typical of the unencumbered UvrD reaction. UvrD initially inhibited the ATP hydrolysis observed in the RecA reaction, in a manner dependent upon the concentration of UvrD (Figure 3). Under these conditions, the reaction profile thus has three pronounced stages, depending on the amount of UvrD included in the reaction. To simplify further description of the reaction, we refer to the steady state of ATP hydrolysis due only to RecA bound to ssDNA as the RecA stage (Figure 2). The period immediately after UvrD addition, characterized by a noticeable decline in the observed ATPase rate, is the lag stage. The UvrD stage encompassed the final increased rate of ATP hydrolysis that we attribute to UvrD translocation on DNA that has been freed (or largely freed) from bound RecA protein. The attribution of this final stage to UvrD is supported by the observation that the rates of ATP hydrolysis in this stage generally surpass the capacity of the available RecA alone to hydrolyze ATP under these conditions. This assignment is also supported by electron microscopy observations presented below, demonstrating that the RecA protein has indeed been displaced in this final stage of the reaction. In these experiments, RecA (2 μM) and ssDNA (M13mp18 or poly(dT) at 3 μM total nucleotide) are preincubated at 37°C for 10 min. E. coli SSB (0.3 μM monomer) and ATP (3 mM) were added to initiate ATP hydrolysis, followed by UvrD after the ATPase reaction is at steady state (usually 15 min after the RecA reaction is initiated).
Figure 2.
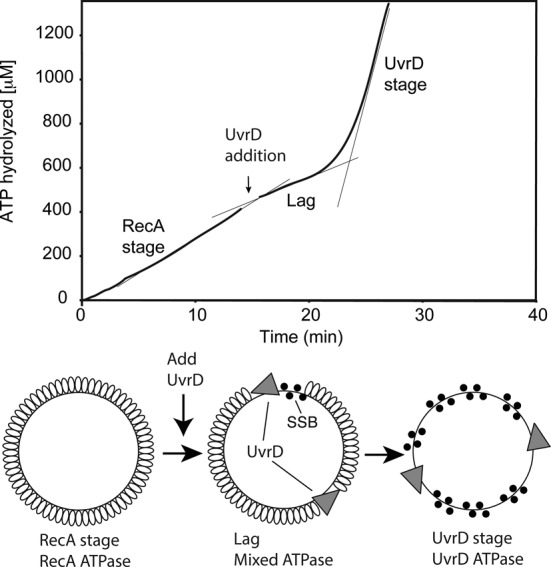
UvrD inhibits RecA-catalyzed ATPase activity. Reactions were carried out as described in Materials and Methods, and contained 3 μM circular M13mp18 ssDNA and 2 μM RecA protein. The reaction was initiated by addition of ATP (3 mM) and SSB (0.3 μM) as a mixture at t = 0. In the reaction shown, 50 nM UvrD (30 nM) was added at the point indicated by the arrow. The data following this addition have been corrected for a slight decline in absorption caused by a dilution effect. The ATP consumption profile can be divided into three stages. The first stage (prior to UvrD addition) reflects the constant rate of ATPase activity by RecA in the presence of ATP and DNA. A lag stage immediately follows the addition of UvrD and is defined by a decline in ATPase rate. The final stage is the UvrD stage, characterized by a large increase of ATP consumption (greater than the highest level of ATPase possible due to the RecA protein present) attributed to UvrD translocation on the DNA after RecA removal. Confirmation of RecA removal is presented in subsequent figures.
Figure 3.
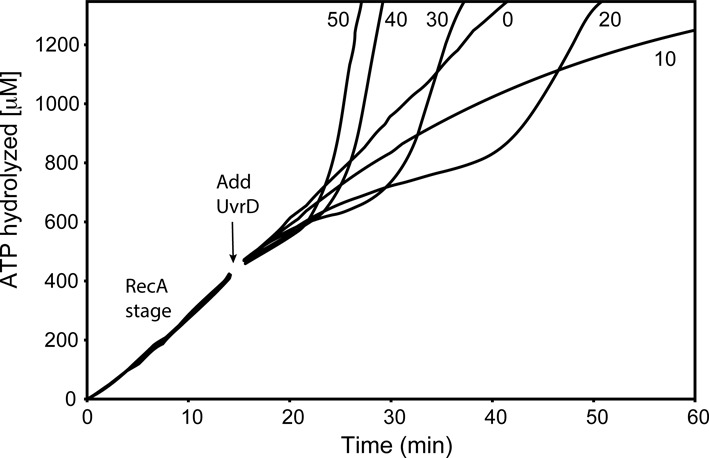
UvrD effects on RecA filaments are UvrD concentration-dependent. RecA (2 μM) filaments were allowed to form on 3 μM circular M13mp18 ssDNA in the presence of ATP and SSB (0.3 μM) as described in Materials and Methods. After 15 min (arrow), UvrD was added at the concentrations shown (in nM). The data following this addition have been corrected for a small decline in absorption caused by a dilution effect. The effects of UvrD are detected through changes in the consumption of ATP. The average rate of ATP hydrolysis in the early RecA stage (prior to UvrD addition) is 30 μM/min (yielding an apparent kcat (assuming all potential DNA binding sites are occupied by RecA) of 30 min−1), reflecting the near saturation of the 1 μM available RecA binding sites on the ssDNA.
As shown in Figure 3, the pattern observed depended upon the concentration of the added UvrD, although the three stages were evident under many conditions. The 3 μM circular M13mp18 ssDNA (concentration given in total nucleotides) translates into ∼0.4 nM M13mp18 ssDNA molecules. Thus, at 10 nM UvrD, there are ∼25 UvrD monomers per ssDNA molecule coated with RecA protein. At this concentration, the rate of ATP hydrolysis never increases above that seen with RecA alone. This amount of UvrD thus appears to be insufficient to completely displace the bound RecA protein in the 60 minute time span of the experiment, possibly due to RecA re-binding. As UvrD concentration increases, the three stages become evident at 20 nM UvrD and above. The lag becomes shorter as UvrD concentration increases, and the effects appear to saturate with the addition of 40–50 nM UvrD protein, where there are 100–125 UvrD monomers per DNA molecule.
As controls, the rates of ATP hydrolysis promoted by UvrD alone under several different conditions are provided in Figure 4. Panels A, B and C are scaled to facilitate direct comparison from one panel to the next. When UvrD is added to M13mp18 ssDNA under the conditions of the experiments of Figures 2 and 3, the rates of ATP hydrolysis are generally about twice the rates seen in the UvrD stage of the RecA displacement reaction (Figure 4A; summary in Figure 4E). If SSB (0.3 μM) is added to ongoing reactions, the rates appear to trend slightly downward (Figure 4B), but the changes are generally not statistically significant. Once bound to ssDNA, UvrD appears to be little affected by SSB. Adding SSB prior to UvrD results in a substantial decline in the observed ATPase rates, albeit with no evident lag (Figure 4C). This might reflect some form of SSB-mediated inhibition in DNA binding by UvrD. The rates seen in the final UvrD stage are again shown in Figure 4D, and the ATPase rates are summarized in Figure 4E. The rates seen in the final UvrD stage of the RecA displacement assays are approximately half of the rates seen when SSB is added after UvrD, but greater than the rates seen when SSB is added prior to UvrD. Our interpretation of this, expanded upon below, is that the lag stage represents active RecA displacement that is limited by the capacity of UvrD to access free DNA and/or free RecA filament ends. In the final UvrD stage, UvrD has accessed a DNA-bound state which allows it to translocate with minimal inhibition by SSB, perhaps slowed by intermittent encounters with re-bound RecA filaments. The rates seen in the final UvrD stage would then be a hybrid situation in which many UvrD molecules were actively translocating and a few were displacing re-bound RecA.
UvrD causes RecA filament disassembly
The lags seen in Figures 2 and 3 after UvrD addition could reflect some aspect of UvrD-mediated removal of the RecA protein filaments. We utilized electron microscopy to examine all stages of the reaction.
RecA filaments before UvrD addition at 10 min (RecA stage), at 21–23 min (lag) and 35 min after addition of 40 nM UvrD (UvrD stage) were visualized, categorized based on filament completeness and counted. The ATPase profile of the sampled reaction is shown in Figure 5A. Filaments prior to UvrD addition are shown in Figure 5B, and the lag and UvrD stage are shown in Figure 5C and D. Figure 5 documents a progressive decrease in RecA-DNA filament length that begins after UvrD addition. It also establishes that the higher levels of ATPase during the UvrD stage are due to UvrD, as there are virtually no RecA filaments left on the circular M13mp18 ssDNA at this reaction stage. This eliminates the possibility that the UvrD stage reflects an increase in RecA ATPase activity. The status of the filaments at each reaction stage is summarized in Figure 5E. UvrD was not able to enter a UvrD stage characterized by ATPase levels exceeding those expected for RecA alone under all conditions in these experiments, but EM revealed that RecA filaments were almost as rare during a prolonged lag stage as they were during a marked UvrD stage (data not shown). We discuss factors that affect UvrD function in the Discussion.
Figure 5.
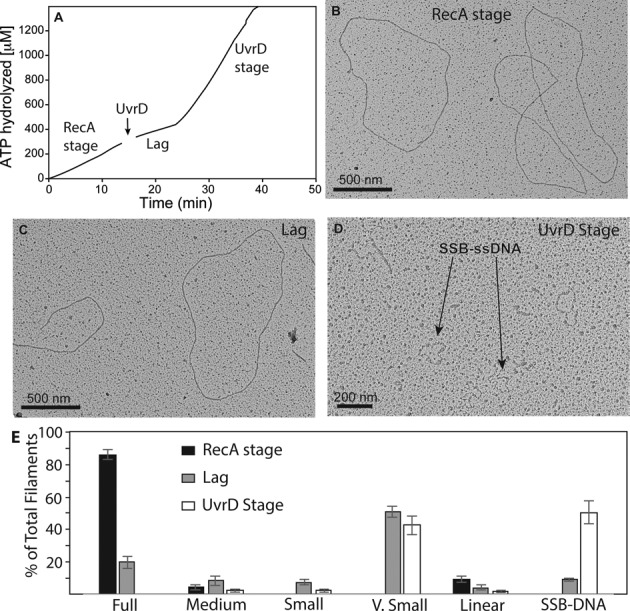
Electron microscopy of RecA filaments in the presence of UvrD. (A) An ATPase reaction from which samples were taken at the early RecA stage, during the lag and after the later UvrD stage was achieved. Samples were taken at ∼10 min, 22 min and 35 min, times that correspond to the various stages. Reaction conditions were identical to those utilized in the reaction of Figure 2, except that 40 nM UvrD helicase was utilized. The RecA stage (10 min), lag (22 min) and UvrD stage (35 min) of the UvrD-mediated displacement reaction are illustrated in panels B, C and D, respectively. A field showing SSB–ssDNA complexes by themselves can be found in Supplementary Figure S1. (E) Statistical analysis of the RecA filament distribution in the samples illustrated in panels B–D. Filament categories are explained in Materials and Methods. In general, full, medium, small and very small filaments reflect declining filament lengths. Linear molecules are the subset of filaments present on broken DNA circles. Circular ssDNA molecules coated with SSB (pointed out in panel D) do not contain visible RecA protein. The error bars are representative of a standard deviation of three independent experiments. Molecules counted (n) = 580, 675 and 870 for the RecA stage (panel B), the lag (panel C) and the UvrD stage (panel D), respectively.
UvrD-mediated ATP hydrolysis is required for efficient displacement of RecA from DNA
To further understand the dynamics of RecA-UvrD antagonism, we utilized a UvrD K35I variant that binds DNA but does not hydrolyze ATP (66). UvrD K35I inhibited the ATPase activity of RecA (Figure 6A). The pattern strongly suggests a competition by UvrD K35I with RecA protein for DNA binding sites in which UvrD K35I blocks or partially blocks RecA filament extension, but disassembly still occurs. The addition of equal amounts of the WT and mutant UvrD protein are directly compared in Figure 6B. In multiple trials, the WT UvrD protein consistently produced a somewhat greater decline in ATPase during its lag phase than did an equal addition of the K35I mutant protein. The relatively slow but continuing decline in ATPase due to addition of UvrD K35I may reflect normal RecA protein net dissociation of filaments from the 5′-proximal end. The results indicate that removal of RecA can occur via normal disassembly of RecA filaments if filament extension is blocked. The somewhat more rapid ATPase decline seen with the WT UvrD protein suggests a more active role for UvrD in RecA removal. The potential for an additional role of UvrD, besides simply blocking RecA filament extension, was further explored.
Figure 6.
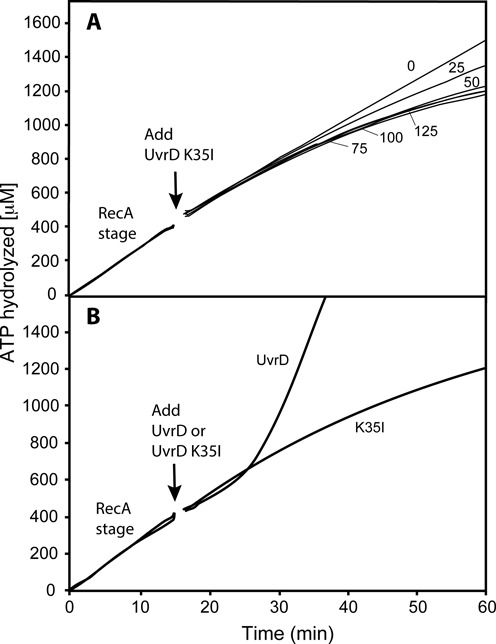
Disassembly of active RecA nucleoprotein filaments in the presence of the ATPase deficient UvrD K35I. (A) Reactions were carried out as described in Materials and Methods, but with the substitution of UvrD K35I protein for WT UvrD. The UvrD variant was added at 15 min (arrow) at the concentration (in nM) shown next to each ATPase curve. The data following this addition have been corrected for a small decline in absorption caused by a dilution effect. The average rate of ATP hydrolysis in the RecA stage is 27 μM/min (yielding an apparent kcat—assuming the 1 μM potential DNA binding sites are all occupied by RecA—of 27 min−1). (B) A comparison between the inhibition of RecA ATPase activity by WT UvrD and UvrD K35I, each added to 50 nM final concentration at 15 min (arrow).
RecA ATPase activity is not necessary for UvrD-mediated RecA filament displacement from ssDNA
To address the question of active catalysis of RecA disassembly by UvrD more closely, we resorted to the use of RecA E38K K72R. This variant forms WT-like RecA filaments on DNA. However, the mutant protein binds but does not hydrolyze ATP. The E38K mutation counters a deficiency in filament formation seen with RecA K72R (58). Once bound to ssDNA, this mutant protein does not undergo ATPase-mediated disassembly at any significant rate, and it hydrolyzes ATP with a kcat that is <1% of that of the WT protein (58). When UvrD is added to pre-formed RecA E38K K72R filaments, ATP hydrolysis is observed only after the UvrD addition (Figure 7A). The progress of the ATP hydrolysis was a function of UvrD concentration. The advantage of this experiment is that any ATPase activity observed is due only to UvrD. Initially, the ATP hydrolysis was slow—no more than 50±10 nM ATP/min/nM UvrD. When sufficient UvrD was present, this slow lag phase was followed by a rapid increase in rate. We followed up on the different stages of the reaction using electron microscopy (Figure 7B, C, D and E). Similarly to experiments containing WT RecA, full RecA nucleoprotein filaments were formed prior to UvrD addition. When viewed in the electron microscope, the filaments formed by the mutant protein (Figure 7B) exhibit somewhat more small gaps or sharp bends than the WT protein (Figure 5B), and this has been previously documented (58). Once UvrD was added, a diverse range of filament lengths was observed, while the average length declined with time (summarized in Figure 7E). Finally, the rapid increase in ATPase rate could be correlated to the near absence of RecA E38K K72R filaments on DNA (Figure 7D and E). The E38K mutation confers on this RecA protein variant a relatively rapid nucleation and enhanced filament stability on ssDNA, even though ATPase activity is generally too low to measure. The slower ATPase rates seen for UvrD protein in the UvrD stage (51.7 μM min−1 by 50 nM UvrD in the presence of RecA E38K K72R protein relative to 425.9 μM min−1 for 50 nM UvrD with WT RecA protein) may reflect an enhanced capacity of this mutant RecA protein to compete with UvrD for DNA binding sites, perhaps via rapid re-nucleation after displacement by UvrD. At higher concentrations of UvrD, RecA filaments are cleared from the DNA and any short filaments derived from new nucleation events are eliminated rapidly.
Figure 7.
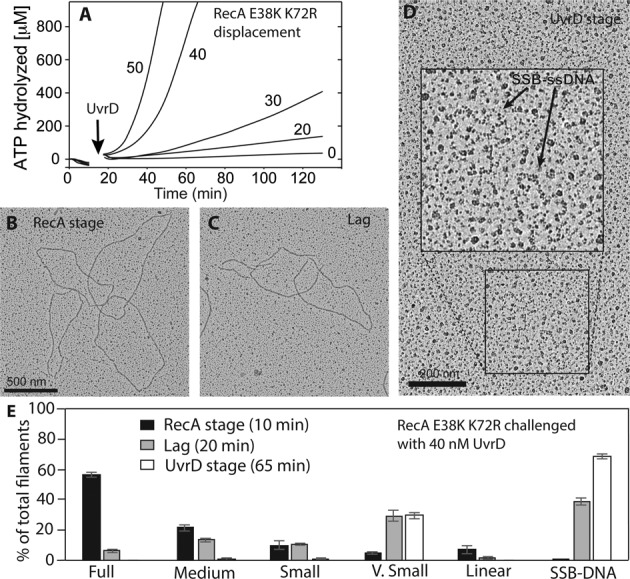
ATP hydrolysis by RecA protein is not required for UvrD-mediated disassembly of RecA filaments. (A) The RecA E38K K72R variant, which lacks detectable ATPase activity, was allowed to form filaments on DNA in the presence of ATP for 15 min. UvrD was then added to the reactions at the concentrations (in nM) shown. The data following this addition have been corrected for a small decline in absorption caused by a dilution effect. (B, C, D) ATPase reactions containing 2 μM RecA E38K K72R and 3 μM DNA, followed by a challenge with 40 nM UvrD, were set up. Samples were taken for spreading to observe RecA E38K K72R filaments under an electron microscope at 10 min (prior to UvrD addition), 20 min (5 min after UvrD addition) and at 65 min (50 min after UvrD addition, UvrD stage), respectively. (E) Statistical analysis of the types of filaments observed from the experiments illustrated in B, C and D. The results are an average of three independent experiments and the error bars represent standard deviation between the three trials. Molecules counted (n) = = 700, 1442 and 668 for the RecA stage (panel B), the lag (panel C) and the UvrD stage (panel D), respectively. Filament categories are described in Materials and Methods and in the legend to Figure 5.
Importantly, when the RecA filament-capping inhibitor RecX is added to RecA E38K K72R filaments for the same time as the longest incubation carried out here with UvrD, no disassembly of the RecA variant is observed (Gruenig,M. and Cox,M., unpublished data). E. coli RecX has been shown to inhibit RecA filament extension through binding to the 3′-proximal end of the nucleoprotein filament (68). No RecA protomers can be added to the growing end of the filament in the presence of RecX, while the 5′-proximal end continues to disassemble, resulting in eventual elimination of the filament. However, the passive mechanism of RecX requires ATP hydrolysis by RecA (49). Therefore, the results in Figure 7, which show displacement of RecA E38K K72R from DNA by UvrD, support a mechanism of active displacement of RecA by UvrD. They also demonstrate that ATP hydrolysis by RecA is not needed for this displacement.
Finally, we examined how much ATP hydrolysis is required to dismantle pre-formed RecA filaments. As mentioned earlier, the rate of ATP hydrolysis that can be measured at early times after UvrD is added to RecA E38K K72R filaments on circular ssDNA is so low that we started questioning whether it was necessary at all. To investigate further whether ATP hydrolysis by UvrD is required for RecA displacement we followed the state of RecA filaments in response to UvrD addition by EM, but we substituted ATP entirely with the non-hydrolyzable analog ATPγS. Addition of UvrD had essentially no effect on RecA filaments under these conditions (Figure 8A–D). Time points of the reaction were taken as described in the Materials and Methods section. Mostly RecA full filaments with small gaps were observed even after 50 min of UvrD exposure to RecA nucleoprotein filaments pre-formed with ATPγS (Figure 8). Addition of ATP along with the UvrD resulted in significant displacement of RecA (Figure 8D). Decreasing the concentration of ATPγS in the initial incubation and increasing the concentration of ATP added subsequently enhanced the RecA displacement effect. The overall results are consistent with a requirement for UvrD-mediated ATP hydrolysis to bring about RecA displacement from DNA.
Figure 8.
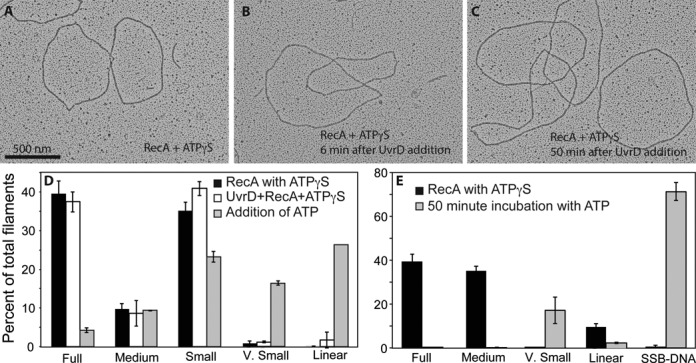
ATP hydrolysis by UvrD is required for dismantling RecA filaments. (A) RecA filaments formed on circular ssDNA after a 10 min incubation with 1 mM ATPγS. Two full filaments and a very small filament, as well as SSB-coated DNA are present in this image. (B) RecA filaments formed as in A), after a six-minute incubation with UvrD. (C) RecA filaments formed as in (A) after 50 min incubation with UvrD. No decrease in filamentation can be observed. (D) If 3 mM ATP is added with the UvrD helicase, significant disassembly of the RecA filaments is seen after 6 min. Molecules are quantified with RecA and ATPγS alone (n = 1027 molecules counted and scored), after addition of UvrD (n = 1187 molecules counted and scored) and after further addition of ATP (n = 1060 molecules counted and scored). (E) Experiment as in panel D, but with less ATPγS (30 μM) added to form the RecA filaments, addition of UvrD, and filaments examined 50 min after further addition of 3 mM ATP. Molecules counted and scored (n) = 573 prior to and (n) = 581 at 50 min after addition of ATP.
Availability of RecA-ssDNA filament ends and/or ssDNA binding sites controls the duration of the lag phase
Next, we examined what factors affect the efficiency of UvrD activity on RecA. We varied the concentration of WT RecA in the ATPase reactions to determine how effective UvrD was at competing for DNA with the RecA filament. The results suggest that when the DNA is not fully coated by RecA, the lag phase exhibited a substantial decline in duration (Figure 9, RecA = 1 μM curve versus the rest). The duration of the lag increased at higher RecA concentrations, possibly due to reduced availability of RecA filament ends and/or DNA binding sites where UvrD could act. The results suggest that the slow step in removal of RecA filaments from circular ssDNA (when RecA protein is saturating) is the ability of UvrD to gain access to those filament ends and/or DNA binding sites.
Next, we assayed for UvrD-catalyzed displacement of RecA from linear ssDNA. RecA cycles on and off short linear ssDNA molecules, as ATP hydrolysis mediates end-dependent dissociation of filaments and they are replaced with new filament nucleation events. SSB is not needed to form contiguous RecA filaments on ssDNA when secondary structure is not present, and is not added in the experiments with poly(dT). On short poly(dT) DNA (average length greater than 250 nucleotides), the lag preceding a pronounced UvrD stage completely disappeared (Figure 10A). Moreover, UvrD was able to enter the UvrD stage at lower concentrations when RecA filaments were formed on poly(dT) as compared to reactions in which RecA filaments were pre-formed on circular ssDNA (compare Figures 3–10A). This piece of evidence elucidates two important points about the UvrD mechanism. First, it supports the active displacement model. The passive mode of inhibition by RecX is partially built on the evidence that it requires greater amounts of RecX to inhibit RecA on poly(dT) than on circular ssDNA M13mp18 DNA because there are more filaments to cap on short linear ssDNA than on large circular ssDNA for the same concentration of nucleotides (48). Exactly the opposite effect is seen with UvrD. Second, the ATPase function of UvrD must be required because when UvrD K35I is added to the reactions containing RecA-poly(dT) filaments, no inhibition was observed at concentrations at which WT UvrD promotes a rapid switch to a UvrD stage (Figure 10B).
Figure 10.
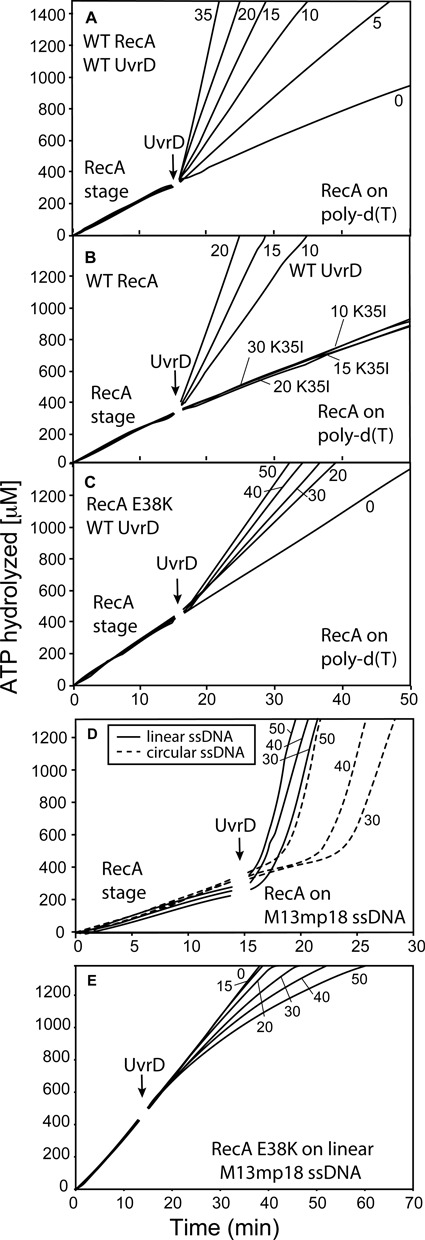
UvrD-mediated disassembly of RecA filaments formed on linear ssDNA. (A) WT RecA was allowed to form filaments on 3 μM poly(dT) DNA (average length 225 nucleotides) in the presence of ATP. All reactions in this and subsequent panels that utilize poly(dT) do not include SSB. After 15 min UvrD was titrated into the reactions at final concentrations shown on the graphs (in nM). (B) Reactions were carried out as in panel A, but UvrD K35 I was substituted for WT UvrD in the indicated reactions. (C) RecA E38K resists inhibition by UvrD. The RecA E38K variant was allowed to form filaments on poly(dT) as in panels A and B, and UvrD was titrated into the reaction at final concentrations (in nM) shown in the figure. (D) WT RecA protein was incubated in the presence of ATP either with 3 μM circular M13mp18 ssDNA (dashed lines) or linear M13mp18 ssDNA (solid lines). After 15 min UvrD was titrated into the reactions at the indicated concentrations (in nM). The average rate of RecA-mediated ATP hydrolysis on circular M13mp18 ssDNA is 24 μM/min and on lssM13 DNA is 21 μM/min. These reactions, and those in panel E, included SSB added with the ATP during the initial formation of RecA filaments. (E) The RecA E38K mutant was used. The experiment was set up as in C, but linear M13mp18 ssDNA was used in place of poly (dT).
To explore the issue further, we used a RecA E38K variant that has reduced disassembly rates and more rapid filament nucleation rates, perhaps due to enhanced cooperativity between RecA protomers (69). This protein lacks the K72R mutation and hydrolyzes ATP at rates similar to the WT protein. When RecA E38K is preincubated with poly(dT), and UvrD is added to the reaction, UvrD proceeds to an evident UvrD stage (with elevated ATPase levels) immediately (Figure 10C). Again, the abundance of easily accessible RecA-DNA filament ends appears to enhance UvrD efficiency. However, the rates of ATP hydrolysis in the UvrD stage decline. For example, with 20 nM UvrD, the ATPase rate is 125 μM min−1 when UvrD challenges the WT RecA protein, but only 41 μM min−1 when UvrD challenges the RecA E38K protein. The greater efficiency with which RecA E38K nucleates onto ssDNA may require more frequent removal by UvrD and thus lower net rates of UvrD-mediated ATP hydrolysis than are seen with the WT RecA protein in Figure 10A.
The disappearance of the lag stage could be due to the much shorter RecA filament lengths in this experiment, the greater accessibility of the RecA-DNA filament ends, and/or the absence of SSB from the reactions that contained poly(dT). We repeated the same experiment using linearized ssM13mp18 DNA. With this much longer and complex ssDNA, RecA filaments were again formed with the aid of added SSB, to eliminate the secondary structure in this DNA substrate and form contiguous filaments. The use of that DNA substrate resulted in restoration of a lag stage, but its duration was greatly reduced when compared to the lag observed when circular ssM13 was used (Figure 10D). The great reduction in the lag, even with SSB present, suggests that an absence of SSB does not give rise to the reduction in the lag seen in Figure 10A–C. This suggests again that the lack of a lag in Figure 10A reflects a greater accessibility to RecA filament ends and/or DNA binding sites. The substitution of RecA E38K for the WT protein again reduced the effect of UvrD, even at UvrD levels that would result in a prominent transition to a UvrD stage when WT RecA is used (Figure 10E). Overall, the results suggest that the longer lags seen when RecA is bound at saturating levels to circular ssDNA are due to a slow step in gaining access to viable sites where UvrD can carry out its RecA displacement activity, either RecA filament ends or DNA binding sites that transiently appear in the RecA filament. Once UvrD is bound at the appropriate RecA filament end, RecA displacement occurs quite rapidly, but at rates that can be slowed significantly by a RecA mutant with enhanced DNA-binding properties such as RecA E38K. When free RecA filament ends are present, or when RecA concentration declines so that filament gaps appear, the lags in the UvrD-mediated ATPase are greatly reduced.
The displacement of RecA by UvrD is blocked by deletion of the RecA C-terminus
As detailed in the Introduction, the C-terminal 17 amino acid residues of RecA represent a highly charged regulatory flap that affects many RecA functions. Removal of this flap enhances RecA protein binding to ssDNA in a manner similar to the RecA E38K mutant protein. We carried out experiments to determine if this flap affected the RecA displacement reaction catalyzed by UvrD. The experimental design is similar to that in Figure 3, with 30 nM UvrD added to filaments of WT RecA or RecAΔC17 at the arrow in Figure 11A. The results of the ATPase trials, repeated three times, indicate that nucleoprotein filaments of the RecAΔC17 protein are minimally affected by UvrD. This interpretation is supported by the electron microscopy experiments in Figure 11B. The resistance of RecAΔC17 to UvrD is not complete, but it is clearly substantial.
Figure 11.
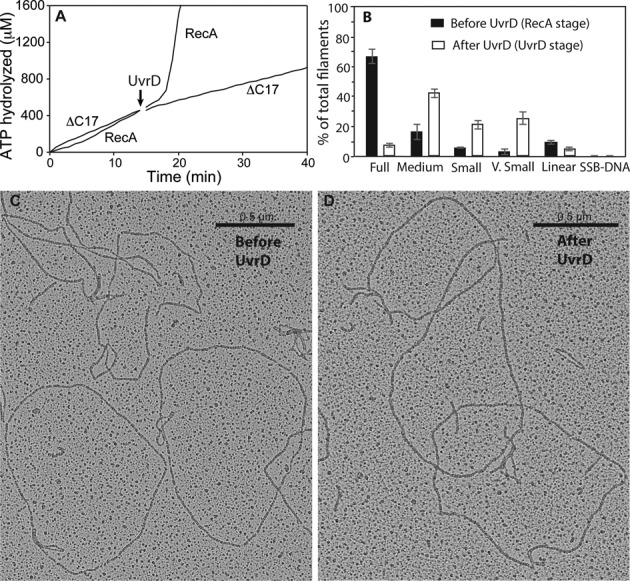
UvrD-mediated displacement of RecA protein requires the RecA C-terminus. WT RecA or RecA ΔC17 were added to 2 μM under standard reaction conditions. After 15 min, 30 nM UvrD was added to each. (A) While the WT RecA is removed as expected, leading to normal acquisition of the UvrD stage, the RecA ΔC17 variant is not removed and no change in ATPase rates is observed. (B) Electron microscopy. Molecules counted and scored prior to UvrD addition (n) = 876, n = 1355 molecules counted and scored 5 min after UvrD addition.
It is possible that this is due to a role for the RecA C-terminus in the RecA displacement mechanism—a direct interaction with UvrD. Alternatively, it could simply reflect the enhanced binding of RecAΔC17 to ssDNA relative to the WT protein. As may be the case for RecA E38K, a more persistent RecAΔC17 filament may simply resist displacement by UvrD.
DISCUSSION
Our primary conclusion is that UvrD displaces RecA filaments using an active mechanism approximating the active mode illustrated in Figure 1. The ATPase activity of UvrD is required for displacement; the ATPase activity of RecA protein is not. The results largely eliminate three possible displacement mechanisms. These are (i) a completely passive mechanism in which UvrD might encounter and block the 3′-proximal end of a RecA filament and thus block elongation at that end, while RecA protein dissociates from the opposite filament end (the passive mode in Figure 1); (ii) a simple competition for DNA binding sites (normal RecA dissociation from the 3′-proximal end, especially when RecA is not hydrolyzing ATP, is insufficient to account for the rapid rate of UvrD-mediated displacement) or (iii) a UvrD-activated higher rate of ATP hydrolysis in RecA (that would accelerate normal RecA filament disassembly; instead, RecA-mediated ATP hydrolysis is not required). Instead, UvrD is actively displacing RecA subunits, whether RecA is hydrolyzing ATP or not. We also establish that the efficiency of the displacement reaction can be limited by the availability of DNA binding sites and/or the accessibility of the 3′-proximal end of the growing RecA filament. UvrD function appears to be distinct from that of its PcrA homolog, at least with respect to its lack of a requirement for the RecA ATPase activity (54).
RecA is not the first protein for which an active displacement mechanism from DNA by UvrD has been demonstrated. UvrD has been shown to actively remove the Tus protein from the ter sites on the E. coli chromosome (19). It would be interesting to directly compare the dynamics of UvrD catalyzed removal of RecA and Tus. The slow ATPase rates of UvrD during RecA removal suggest that UvrD does not translocate on the ssDNA at its normal rate during this process. In fact, the UvrD stage (Figure 2) is most likely the translocase mode of UvrD, to which the protein switches after most RecA has been removed from the DNA. Based on low ATPase rates observed during the lag stage, especially during the lags accompanying removal of RecA E38K K72R mutant filaments that contribute no background ATP hydrolysis, we estimate that the kcat for UvrD-mediated ATP hydrolysis during RecA removal is no more than 70 s−1. Intriguingly, this is close to the rate of ATP hydrolysis during the unwinding of duplex DNA by a UvrD dimer.
There are several possible avenues for an active UvrD displacement mechanism. In principle, UvrD could interact with any part of the RecA filament, perhaps triggering a cooperative local filament collapse. However, given the known properties of UvrD and RecA, as well as the lack of a requirement for the RecA ATPase, we propose that the simplest and most likely disassembly mechanism would involve UvrD displacing RecA filaments from 3′-proximal end. Simple translocation of UvrD could displace the RecA protein without need for a specific interaction between the two proteins. UvrD hydrolyzes 1 ATP per nucleotide translocated even at low concentrations of ATP (70). This argues that there is little to no slippage of UvrD even when not bound to ATP. If RecA filaments slow the translocation as expected, then the ATPase activity will slow as well. RecA mutants with enhanced DNA-binding properties could further slow or prevent the progress of UvrD in a displacement mode, such as seen here with the RecA E38K and RecAΔC17 mutants. A mechanism featuring direct and specific interactions between UvrD and RecA is also possible, although the current work provides no evidence for such an interaction. The ATPase activity of UvrD must be intact for Tus removal (19). The need for the UvrD ATPase activity for RecA displacement also suggests that the active translocase activity of UvrD plays a role in the protein displacement process.
A UvrD monomer is able to translocate along ssDNA (67,70,71). The UvrD helicase function in vitro involves an oligomeric protein (at least a dimer) (66,71–73). Since there is no duplex DNA present in our experiments, we hypothesize that the active form of UvrD in RecA displacement is the monomeric species, although our experiments do not directly bear on this issue. In principle, multiple subunits of UvrD could cooperate in the displacement reaction much like the cooperative inchworm mechanism proposed for displacement of DNA binding proteins by the bacteriophage T4 Dda helicase (74).
Helicases without ATPase function can displace RecA protein in principle. A PcrA mutant devoid of ATPase and helicase activity possesses RecA displacement activity (75). However, the result can be easily explained by a competition by PcrA and RecA for DNA binding sites. Evidence has been provided more recently that PcrA acts by a mechanism that requires ATP hydrolysis by RecA (54). As shown in Figure 6A, UvrD K35I can eventually inhibit RecA ATPase activity, brought about by a net RecA filament disassembly accompanying RecA-mediated ATP hydrolysis. The properties of the normal RecA filament disassembly process, in particular its reliance on ATP hydrolysis, explain why the RecA filament needs to hydrolyze ATP when the K35I mutant of UvrD is used for RecA filament displacement. The same property of RecA may explain some published observations with PcrA. Without RecA-mediated ATP hydrolysis, the pre-formed RecA nucleoprotein filaments will not disassemble in the time span of the current experiments.
Our results suggest that the length of the lag stage depends largely upon UvrD access to RecA filament ends and/or DNA binding sites. Observation of substantial lags is seen only when UvrD is added to RecA filaments formed on circular ssDNA, under conditions of RecA saturation where such ends or binding sites should be limiting. When RecA concentrations are reduced to sub-saturating levels, the duration of the lag declines substantially (Figure 9). The lag is abolished at all UvrD concentrations when added to RecA filaments pre-formed on short linear poly(dT) DNA where filaments are short and ends abundant (Figure 10A–C). The lag duration declines considerably when linear ssDNA replaces circular ssDNA, where filament ends would be exposed (Figure 10D). Even when RecA is present at saturating levels on circular ssDNA, the length of the lag also declines as UvrD concentration increases (Figure 3). This might be attributed to subtle discontinuities in the RecA filament on the otherwise saturated circular ssDNA that are less accessible than an open filament end (and less apparent in the electron microscope), but that can be exploited by UvrD when its concentration is sufficient. The ssDNA binding site size for UvrD is ∼10 ± 2 nucleotides (76), potentially allowing it to exploit short RecA filament discontinuities much better than SSB (with a site size of 30+ nucleotides (59,77)). Alternatively, UvrD may possess some capacity to slowly integrate into a contiguous RecA nucleoprotein filament and create a new end or binding site.
It is interesting to speculate that UvrD may be recruited to the 3′-proximal end of the RecA nucleoprotein filament through a specific interaction. UvrD interacts with a number of proteins to perform its activities in NER and MMR (16,18), so it is possible that dismantling RecA requires a similar interaction. Also, the UvrD homolog Srs2 has been shown to interact with Rad51 through its C-terminus (53). The C-termini of UvrD, Rep and Srs2 are not conserved, so it is possible that they have adapted for species-specific interactions. The main argument against a specific interaction is the fact that PcrA from different bacteria can substitute for UvrD in E. coli. Therefore, the question of a direct interaction between UvrD and RecA remains open. If it exists, the interaction is likely weak and transient.
UvrD monomers exhibit binding specificity for a ssDNA/dsDNA junction (13). If bound near the 3′ end of the gap, UvrD might be optimally positioned both to displace any RecA filament that had nucleated in the gap and to prevent filament extension into the adjoining duplex DNA. A UvrD monomer cannot unwind DNA, so it should simply dissociate or stop when it reaches the other end of the gap (13). These features of UvrD function may play a role in its capacity to limit the lifetime of RecA filaments on the DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Dr Joshua Sokoloski for discussions and comments on the manuscript. The authors acknowledge use of instrumentation supported by the UW MRSEC (DMR-1121288) and the UW NSEC (DMR-0832760).
FUNDING
N.I.H. [GM 32335 to M.M.C. and GM045948 to T.M.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Hickson I.D., Arthur H.M., Bramhill D., Emmerson P.T. The E. coli uvrD gene product is DNA helicase II. Mol. Gen. Genet. 1983;190:265–270. doi: 10.1007/BF00330649. [DOI] [PubMed] [Google Scholar]
- 2.Modrich P. Mechanisms and biological effects of mismatch repair. Annu. Rev. Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 3.Atkinson J., Guy C.P., Cadman C.J., Moolenaar G.F., Goosen N., McGlynn P. Stimulation of UvrD helicase by UvrAB. J. Biol. Chem. 2009;284:9612–9623. doi: 10.1074/jbc.M808030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epshtein V., Kamarthapu V., McGary K., Svetlov V., Ueberheide B., Proshkin S., Mironov A., Nudler E. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505:372–377. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manelyte L., Guy C.P., Smith R.M., Dillingham M.S., McGlynn P., Savery N.J. The unstructured C-terminal extension of UvrD interacts with UvrB, but is dispensable for nucleotide excision repair. DNA Repair. 2009;8:1300–1310. doi: 10.1016/j.dnarep.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lestini R., Michel B. UvrD controls the access of recombination proteins to blocked replication forks. EMBO J. 2007;26:3804–3814. doi: 10.1038/sj.emboj.7601804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centore R.C., Leeson M.C., Sandler S.J. UvrD303, a hyperhelicase mutant that antagonizes RecA-dependent SOS expression by a mechanism that depends on its C terminus. J. Bacteriol. 2009;191:1429–1438. doi: 10.1128/JB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long J.E., Renzette N., Sandler S.J. Suppression of constitutive SOS expression by recA4162 (I298V) and recA4164 (L126V) requires UvrD and RecX in Escherichia coli K-12. Mol. Microbiol. 2009;73:226–239. doi: 10.1111/j.1365-2958.2009.06765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker G.C. The SOS response of Escherichia coli. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia Coli and SalmonellaTyphimurium: Cellular and Molecular Biology. Vol. 2. Washington, DC: American Society for Microbiology; 1987. pp. 1346–1357. [Google Scholar]
- 10.Veaute X., Delmas P., Selva M., Jeusset J., Le Cam E., Matic I., Fabre F., Petit M.A. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matson S.W. Escherichia coli helicase II (uvrD gene product) translocates unidirectionally in a 3′ to 5′ direction. J. Biol. Chem. 1986;261:10169–10175. [PubMed] [Google Scholar]
- 12.Runyon G.T., Lohman T.M. Escherichia coli helicase II (uvrD) protein can completely unwind fully duplex linear and nicked circular DNA. J. Biol. Chem. 1989;264:17502–17512. [PubMed] [Google Scholar]
- 13.Tomko E.J., Jia H.F., Park J., Maluf N.K., Ha T., Lohman T.M. 5′-Single-stranded/duplex DNA junctions are loading sites for E. coli UvrD translocase. EMBO J. 2010;29:3826–3839. doi: 10.1038/emboj.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Runyon G.T., Bear D.G., Lohman T.M. Escherichia coli helicase II (UvrD) protein initiates DNA unwinding at nicks and blunt ends. Proc. Natl Acad. Sci. U.S.A. 1990;87:6383–6387. doi: 10.1073/pnas.87.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moolenaar G.F., Moorman C., Goosen N. Role of the Escherichia coli nucleotide excision repair proteins in DNA replication. J. Bacteriol. 2000;182:5706–5714. doi: 10.1128/jb.182.20.5706-5714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manelyte L., Guy C.P., Smith R.M., Dillingham M.S., McGlynn P., Savery N.J. The unstructured C-terminal extension of UvrD interacts with UvrB, but is dispensable for nucleotide excision repair. DNA Repair (Amst) 2009;8:1300–1310. doi: 10.1016/j.dnarep.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudolph C.J., Mahdi A.A., Upton A.L., Lloyd R.G. RecG protein and single-strand DNA exonucleases avoid cell lethality associated with PriA helicase Activity in Escherichia coli. Genetics. 2010;186:473–492. doi: 10.1534/genetics.110.120691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matson S.W., Robertson A.B. The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic Acids Res. 2006;34:4089–4097. doi: 10.1093/nar/gkl450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bidnenko V., Lestini R., Michel B. The Escherichia coli UvrD helicase is essential for Tus removal during recombination-dependent replication restart from Ter sites. Mol. Microbiol. 2006;62:382–396. doi: 10.1111/j.1365-2958.2006.05382.x. [DOI] [PubMed] [Google Scholar]
- 20.Persky N.S., Lovett S.T. Mechanisms of recombination: lessons from E. coli. Crit. Rev. Biochem. Mol. Biol. 2008;43:347–370. doi: 10.1080/10409230802485358. [DOI] [PubMed] [Google Scholar]
- 21.Cox M.M. The nonmutagenic repair of broken replication forks via recombination. Mutat. Res. Fund. Mol. Mech. Mutagen. 2002;510:107–120. doi: 10.1016/s0027-5107(02)00256-7. [DOI] [PubMed] [Google Scholar]
- 22.Anderson D.G., Kowalczykowski S.C. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 23.Jiang Q., Karata K., Woodgate R., Cox M.M., Goodman M.F. The active form of DNA polymerase V is UmuD’2C•RecA•ATP. Nature. 2009;460:359–363. doi: 10.1038/nature08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arenson T.A., Tsodikov O.V., Cox M.M. Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J. Mol. Biol. 1999;288:391–401. doi: 10.1006/jmbi.1999.2705. [DOI] [PubMed] [Google Scholar]
- 25.Bell C.E. Structure and mechanism of Escherichia coli RecA ATPase. Mol. Microbiol. 2005;58:358–366. doi: 10.1111/j.1365-2958.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- 26.Ishii Y., Kondo S. Comparative analysis of deletion and base-change mutabilities of Escherichia coli B strains differing in DNA repair capacity (wild-type, uvrA-, polA-, recA-) by various mutagens. Mutat. Res. 1975;27:27–44. doi: 10.1016/0027-5107(75)90271-7. [DOI] [PubMed] [Google Scholar]
- 27.Robu M.E., Inman R.B., Cox M.M. RecA protein promotes the regression of stalled replication forks in vitro. Proc. Natl Acad. Sci. U.S.A. 2001;98:8211–8218. doi: 10.1073/pnas.131022698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lestini R., Michel B. UvrD and UvrD252 counteract RecQ, RecJ, and RecFOR in a rep mutant of Escherichia coli. J. Bacteriol. 2008;190:5995–6001. doi: 10.1128/JB.00620-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petit M.A., Ehrlich D. Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J. 2002;21:3137–3147. doi: 10.1093/emboj/cdf317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel B., Boubakri H., Baharoglu Z., LeMasson M., Lestini R. Recombination proteins and rescue of arrested replication forks. DNA Repair. 2007;6:967–980. doi: 10.1016/j.dnarep.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 31.Cox M.M. The bacterial RecA protein: structure, function, and regulation. In: Rothstein R, Aguilera A, editors. Topics in Current Genetics: Molecular Genetics of Recombination. Heidelberg: Springer-Verlag; 2007. pp. 53–94. [Google Scholar]
- 32.Lusetti S.L., Cox M.M. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- 33.Bedale W.A., Cox M. Evidence for the coupling of ATP hydrolysis to the final (extension) phase of RecA protein-mediated DNA strand exchange. J. Biol. Chem. 1996;271:5725–5732. doi: 10.1074/jbc.271.10.5725. [DOI] [PubMed] [Google Scholar]
- 34.Cox J.M., Tsodikov O.V., Cox M.M. Organized unidirectional waves of ATP hydrolysis within a RecA filament. PLoS Biol. 2005;3:231–243. doi: 10.1371/journal.pbio.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J.I., Cox M.M., Inman R.B. On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. II. Four-strand exchanges. J. Biol. Chem. 1992;267:16444–16449. [PubMed] [Google Scholar]
- 36.Kim J.I., Cox M.M., Inman R.B. On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. I. Bypassing a short heterologous insert in one DNA substrate. J. Biol. Chem. 1992;267:16438–16443. [PubMed] [Google Scholar]
- 37.Lindsley J.E., Cox M.M. Assembly and disassembly of RecA protein filaments occurs at opposite filament ends: relationship to DNA strand exchange. J. Biol. Chem. 1990;265:9043–9054. [PubMed] [Google Scholar]
- 38.Shan Q., Cox M.M. On the mechanism of RecA-mediated repair of double-strand breaks: no role for four-strand DNA pairing intermediates. Mol. Cell. 1998;1:309–317. doi: 10.1016/s1097-2765(00)80031-3. [DOI] [PubMed] [Google Scholar]
- 39.Shan Q., Cox M.M., Inman R.B. DNA strand exchange promoted by RecA K72R. Two reaction phases with different Mg2+ requirements. J. Biol. Chem. 1996;271:5712–5724. doi: 10.1074/jbc.271.10.5712. [DOI] [PubMed] [Google Scholar]
- 40.Cox M.M. Regulation of bacterial RecA function. Crit. Rev. Biochem. Mol. Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- 41.Eggler A.L., Lusetti S.L., Cox M.M. The C terminus of the Escherichia coli RecA protein modulates the DNA binding competition with single-stranded DNA-binding protein. J. Biol. Chem. 2003;278:16389–16396. doi: 10.1074/jbc.M212920200. [DOI] [PubMed] [Google Scholar]
- 42.Lusetti S.L., Shaw J.J., Cox M.M. Magnesium ion-dependent activation of the RecA protein involves the C terminus. J. Biol. Chem. 2003;278:16381–16388. doi: 10.1074/jbc.M212916200. [DOI] [PubMed] [Google Scholar]
- 43.Lusetti S.L., Wood E.A., Fleming C.D., Modica M.J., Korth J., Abbott L., Dwyer D.W., Roca A.I., Inman R.B., Cox M.M. C-terminal deletions of the Escherichia coli RecA protein - Characterization of in vivo and in vitro effects. J. Biol. Chem. 2003;278:16372–16380. doi: 10.1074/jbc.M212917200. [DOI] [PubMed] [Google Scholar]
- 44.Drees J.C., Lusetti S.L., Cox M.M. Inhibition of RecA protein by the Escherichia coli RecX protein - Modulation by the RecA C terminus and filament functional state. J. Biol. Chem. 2004;279:52991–52997. doi: 10.1074/jbc.M409050200. [DOI] [PubMed] [Google Scholar]
- 45.Lusetti S.L., Drees J.C., Stohl E.A., Seifert H.S., Cox M.M. The DinI and RecX proteins are competing modulators of RecA function. J. Biol. Chem. 2004;279:55073–55079. doi: 10.1074/jbc.M410371200. [DOI] [PubMed] [Google Scholar]
- 46.Lusetti S.L., Voloshin O.N., Inman R.B., Camerini-Otero R.D., Cox M.M. The DinI protein stabilizes RecA protein filaments. J. Biol. Chem. 2004;279:30037–30046. doi: 10.1074/jbc.M403064200. [DOI] [PubMed] [Google Scholar]
- 47.Baitin D.M., Gruenig M.C., Cox M.M. SSB antagonizes RecX-RecA interaction. J. Biol. Chem. 2008;283:14198–14204. doi: 10.1074/jbc.M801511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drees J.C., Lusetti S.L., Chitteni-Pattu S., Inman R.B., Cox M.M. A RecA filament capping mechanism for RecX protein. Mol. Cell. 2004;15:789–798. doi: 10.1016/j.molcel.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 49.Gruenig M.C., Stohl E.A., Chitteni-Pattu S., Seifert H.S., Cox M.M. Less Is More: Neisseria gonorrhoeae RecX protein stimulates recombination by inhibiting RecA. J. Biol. Chem. 2010;285:37188–37197. doi: 10.1074/jbc.M110.171967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pages V., Koffel-Schwartz N., Fuchs R.P. recX, a new SOS gene that is co-transcribed with the recA gene in Escherichia coli. DNA Repair. 2003;2:273–284. doi: 10.1016/s1568-7864(02)00217-3. [DOI] [PubMed] [Google Scholar]
- 51.Stohl E.A., Brockman J.P., Burkle K.L., Morimatsu K., Kowalczykowski S.C., Siefert H.S. Escherichia coli RecX inhibits RecA recombinase and coprotease activities in vitro and in vivo. J. Biol. Chem. 2003;278:2278–2285. doi: 10.1074/jbc.M210496200. [DOI] [PubMed] [Google Scholar]
- 52.Washburn B.K., Kushner S.R. Characterization of DNA helicase-II from a uvrD252 mutant of Escherichia coli. J. Bacteriol. 1993;175:341–350. doi: 10.1128/jb.175.2.341-350.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antony E., Tomko E.J., Xiao Q., Krejci L., Lohman T.M., Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fagerburg M.V., Schauer G.D., Thickman K.R., Bianco P.R., Khan S.A., Leuba S.H., Anand S.P. PcrA-mediated disruption of RecA nucleoprotein filaments-essential role of the ATPase activity of RecA. Nucleic Acids Res. 2012;40:8416–8424. doi: 10.1093/nar/gks641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park J., Myong S., Niedziela-Majka A., Lee K.S., Yu J., Lohman T.M., Ha T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neuendorf S.K., Cox M.M. Exchange of RecA protein between adjacent RecA protein-single-stranded DNA complexes. J. Biol. Chem. 1986;261:8276–8282. [PubMed] [Google Scholar]
- 57.Petrova V., Chitteni-Pattu S., Drees J.C., Inman R.B., Cox M.M. An SOS inhibitor that binds to free RecA protein: the PsiB protein. Mol. Cell. 2009;36:121–130. doi: 10.1016/j.molcel.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gruenig M.C., Renzette N., Long E., Chitteni-Pattu S., Inman R.B., Cox M.M., Sandler S.J. RecA-mediated SOS induction requires an extended filament conformation but no ATP hydrolysis. Mol. Microbiol. 2008;69:1165–1179. doi: 10.1111/j.1365-2958.2008.06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lohman T.M., Overman L.B. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 60.Runyon G.T., Wong I., Lohman T.M. Overexpression, purification, DNA binding, and dimerization of the Escherichia coli uvrD gene product (helicase II) Biochemistry. 1993;32:602–612. doi: 10.1021/bi00053a028. [DOI] [PubMed] [Google Scholar]
- 61.Morrical S.W., Lee J., Cox M.M. Continuous association of Escherichia coli single-stranded DNA binding protein with stable complexes of RecA protein and single-stranded DNA. Biochemistry. 1986;25:1482–1494. doi: 10.1021/bi00355a003. [DOI] [PubMed] [Google Scholar]
- 62.Piechura J.R., Tseng T.-L., Hsu H.-F., Byrne R.T., Windgassen T.A., Chitteni-Pattu S., Battista J.R., Li H.-W., Cox M.M. Directed evolution of extreme resistance to ionizing radiation: how RecA protein adaptation contributes. DNA Repair. 2015;26:30–43. doi: 10.1016/j.dnarep.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cox M.M. Why does RecA protein hydrolyze ATP. Trends Biochem. Sci. 1994;19:217–222. doi: 10.1016/0968-0004(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 64.Cox M.M. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 65.Ali J.A., Lohman T.M. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 66.Maluf N.K., Fischer C.J., Lohman T.M. A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J. Mol. Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 67.Fischer C.J., Maluf N.K., Lohman T.M. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 68.Drees J.C., Chitteni-Pattu S., McCaslin D.R., Inman R.B., Cox M.M. Inhibition of RecA protein function by the RdgC protein from Escherichia coli. J. Biol. Chem. 2006;281:4708–4717. doi: 10.1074/jbc.M513592200. [DOI] [PubMed] [Google Scholar]
- 69.Lavery P.E., Kowalczykowski S.C. Biochemical basis of the constitutive repressor cleavage activity of recA730 protein. A comparison to recA441 and recA803 proteins. J. Biol. Chem. 1992;267:20648–20658. [PubMed] [Google Scholar]
- 70.Tomko E.J., Fischer C.J., Lohman T.M. Single-stranded DNA translocation of E. coli UvrD monomer is tightly coupled to ATP hydrolysis. J. Mol. Biol. 2012;418:32–46. doi: 10.1016/j.jmb.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee K.S., Balci H., Jia H.F., Lohman T.M., Ha T. Direct imaging of single UvrD helicase dynamics on long single-stranded DNA. Nat. Commun. 2013;4:1878. doi: 10.1038/ncomms2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maluf N.K., Lohman T.M. Self-association equilibria of Escherichia coli UvrD helicase studied by analytical ultracentrifugation. J. Mol. Biol. 2003;325:889–912. doi: 10.1016/s0022-2836(02)01276-7. [DOI] [PubMed] [Google Scholar]
- 73.Maluf N.K., Ali J.A., Lohman T.M. Kinetic mechanism for formation of the active, dimeric UvrD helicase-DNA complex. J. Biol. Chem. 2003;278:31930–31940. doi: 10.1074/jbc.M304223200. [DOI] [PubMed] [Google Scholar]
- 74.Byrd A.K., Raney K.D. Displacement of a DNA binding protein by Dda helicase. Nucleic Acids Res. 2006;34:3020–3029. doi: 10.1093/nar/gkl369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anand S.P., Zheng H., Bianco P.R., Leuba S.H., Khan S.A. DNA helicase activity of PcrA is not required for the displacement of RecA protein from DNA or inhibition of RecA-mediated strand exchange. J. Bacteriol. 2007;189:4502–4509. doi: 10.1128/JB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Runyon G.T., Lohman T.M. Kinetics of Escherichia coli helicase II-catalyzed unwinding of fully duplex and nicked circular DNA. Biochemistry. 1993;32:4128–4138. doi: 10.1021/bi00066a039. [DOI] [PubMed] [Google Scholar]
- 77.Lohman T.M., Ferrari M.E. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


