Abstract
Transcription in eukaryotes is associated with two major changes in chromatin organization. Firstly, nucleosomal histones are continuously replaced by new histones, an event that in yeast occurs predominantly at transcriptionally active promoters. Secondly, histones become modified post-translationally at specific lysine residues. Some modifications, including histone H3 trimethylation at lysine 4 (H3K4me3) and acetylation at lysines 9 (H3K9ac) and 14 (H3K14ac), are specifically enriched at active promoters where histones exchange, suggesting a possible causal relationship. Other modifications accumulate within transcribed regions and one of them, H3K36me3, is thought to prevent histone exchange. Here we explored the relationship between these four H3 modifications and histone turnover at a few selected genes. Using lysine-to-arginine mutants and a histone exchange assay, we found that none of these modifications plays a major role in either promoting or preventing histone turnover. Unexpectedly, mutation of H3K56, whose acetylation occurs prior to chromatin incorporation, had an effect only when introduced into the nucleosomal histone. Furthermore, we used various genetic approaches to show that histone turnover can be experimentally altered with no major consequence on the H3 modifications tested. Together, these results suggest that transcription-associated histone turnover and H3 modification are two correlating but largely independent events.
INTRODUCTION
A central aspect of gene transcription in eukaryotes is that the DNA template is packaged into a highly compact nucleoprotein structure called chromatin. The basic repeating unit of chromatin is the nucleosome, in which the DNA wraps around an octamer of the histone proteins H3, H4, H2A and H2B (1). Nucleosomes represent a major obstacle to transcription factor binding at gene promoters and subsequent transcription elongation by the RNA polymerase. Accordingly, major changes in the nucleosomal structure and stability must take place, either as a requirement for gene induction or as a consequence of transcription.
One important and widely studied nucleosome alteration is the reversible post-translational modification of histones, among which the best known are the acetylation and methylation of lysine residues (2). Most of these modifications are evolutionarily conserved from yeast to human and many occur in the N-terminal histone tails protruding from the nucleosome core. Genome-wide chromatin immunoprecipitation (ChIP) studies in Saccharomyces cerevisiae (3,4), Drosophila (5,6) and human cells (7,8) revealed that actively transcribed genes are typically enriched for specific histone acetyl and methyl marks, with some mapping in the promoter and others in the transcribed region of genes. For example, acetylation of lysines 9 and 14 on histone H3 (H3K9/14ac) and trimethylation of H3K4 (H3K4me3) occur almost universally at the promoters and 5′ ends of genes (9), whereas H3K36 trimethylation (H3K36me3) accumulates preferentially within gene bodies (10,11). These modifications are brought about or removed by specific histone modifying activities, which are locally recruited to transcriptionally active genes by activators or the elongating RNA polymerase II (Pol II) and/or function in a global, untargeted fashion (12,13). Histone modifications are thought to facilitate transcription initiation, either by directly loosening the chromatin structure at promoters or by providing docking sites for chromatin remodeling and transcription factors, and to contribute to transcription elongation and maintenance of a proper chromatin structure over gene bodies (11,14,15).
Another, less well understood and more drastic transcription-coupled chromatin event is the turnover of histones, that is, the replacement of ‘old’ histones by ‘new’ histones in the chromatin. This process is regulated by various histone chaperones that either promote incorporation of new histones to replace those evicted by chromatin remodeling and transcription factors, or that prevent incorporation of new histones by favoring retention of the original histones (16). Histone turnover has been proposed to have a role in the kinetics of gene induction and repression, in adding or erasing histone modifications associated with transcription and in preventing spreading of histone marks across chromatin (17). Interestingly, genome-wide studies in yeast (18,19) and in mammalian cells (20) revealed that the profile of histone exchange tightly correlates, either positively or negatively, with that of specific histone modifications that typically mark transcriptionally active genes. Thus, histone exchange is generally highest at active promoters, where H3K4me3 and H3K9/14ac accumulate, and either absent or less apparent within transcribed regions, which are typically enriched for H3K36me3 (4,18,20).
A strong correlation between histone exchange and histone H3 modifications suggests a causal relationship. For example, some modifications may trigger the exchange of histones thus making the chromatin structure more dynamic. Consistent with such a possibility, acetylation of lysines 9 and 14 on histone H3 has been reported to promote nucleosome eviction both in vitro (21) and in vivo (22,23). Alternatively, the exchange of histones may introduce new modifications that could destabilize the nucleosome or serve as epigenetic marks to facilitate transcription. One well described example is the acetylation of lysine 56 on histone H3 (H3K56ac), which occurs prior to and is thought to be required for incorporation of the histone into chromatin (19,24). Here we addressed this question for four histone H3 N-terminal modification marks typically associated with active transcription. Our results suggest that despite correlating, these marks neither have a major impact on nor accumulate as a consequence of histone turnover in yeast. We also show that, in G1-arrested cells, all four modifications persist long after TATA-binding protein (TBP) dissociation from promoters and hence transcriptional shutdown.
MATERIALS AND METHODS
Yeast strains, expression constructs and growth conditions
The yeast strains used in Figures 1–3 are derived from DY8862 (relevant genotype MATa, hht1-hhf1::LEU2, hht2-hhf2::kanMX3 + YCp-TRP1(HHT2-HHF2), ade2, trp1-1, ura3-1; kindly provided by David Stillman, University of Utah) that contains a disruption of both the HHT1-HHF1 and HHT2-HHF2 loci and expresses wild-type HHT2 and HHF2 from a TRP1 plasmid. The strains expressing mutant alleles of HHT2 as the sole source of histone H3 in Figure 2 were generated as follows. The HHT2-HHF2 locus encoding histones H3 and H4 was PCR-amplified from yeast genomic DNA using primers that introduced convenient restriction sites and directly cloned into plasmids pRS314 and pRS316 carrying, respectively, the TRP1 and URA3 selectable markers. The histone H3 coding region in pRS314 was then mutated by polymerase chain reaction (PCR) amplification using mutagenic primers and subcloning of the resulting fragments to replace the wild-type H3 coding sequence in the original plasmid. The resulting constructs were introduced by plasmid shuffle into a derivative of strain DY8862 expressing wild-type HHT2 and HHF2 from the pRS316 construct described above. The anchor-away strains containing SPT6-FRB (Figures 4 and 6), HPC2-FRB (Figure 5), or both SPT6-FRB and HPC2-FRB alleles (Figure 6) are all derived from the parental strain HHY221 (relevant genotype MATa, tor1-1, fpr1::loxP-LEU2-loxP, RPL13A-2×FKBP12::loxP, ade2-1, trp1-1, his3-11, ura3) (25). The SPT6-FRB allele was generated by PCR amplification of the FRB-KanMX6 cassette from plasmid pFA6a-FRB-KanMX6 (Euroscarf No. P30578) and insertion at the carboxyl terminus of SPT6 by one step integration. HPC2-FRB was constructed in the same way using plasmid pFA6a-FRB-HIS3MX6 (Euroscarf No. P30579). The TBP-FRB strain used in Figure 7 has been described (25,26).
Figure 1.
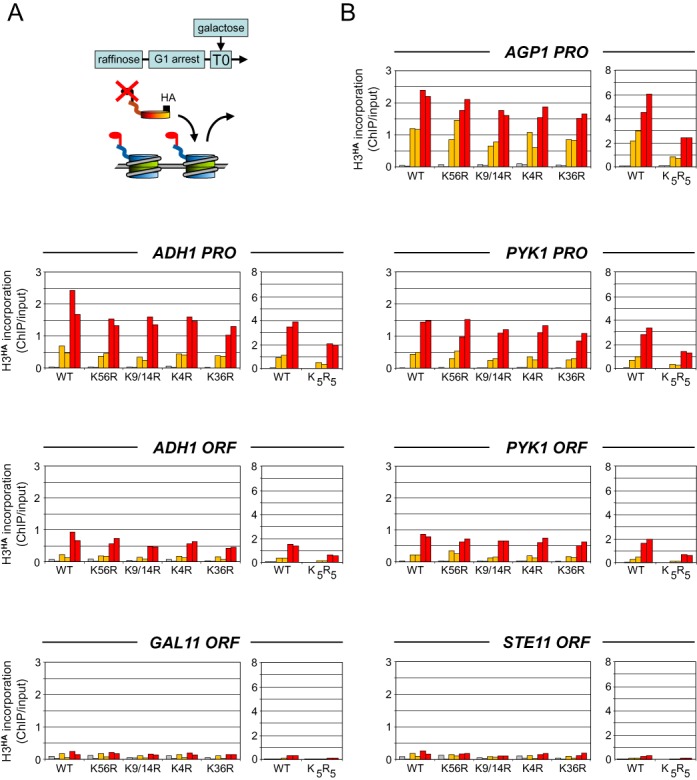
Transcription-dependent incorporation of histone H3 modification mutants in a wild-type chromatin background. (A) Schematic diagram illustrating the experimental approach. Yeast strains expressing a galactose-inducible HA-tagged histone H3 or derivatives with specific N-terminal lysine-to-arginine mutations to prevent their modification were grown overnight in raffinose and arrested in G1 by alpha factor. Expression of H3HA was induced by galactose at T0 (min). Its incorporation into chromatin, which indirectly measures histone exchange (31), was monitored at various time points by quantitative ChIP analysis using antibodies against the HA tag. (B) Time course of incorporation of wild-type H3HA and the indicated modification mutants at the promoter (PRO) and in the coding region (ORF) of the highly transcribed AGP1, ADH1 and PYK1 genes, the weakly expressed GAL11 gene and the inactive STE11 gene. K5R5 carries a K9/14/18/23/27R quintuple mutation. Shown are the amounts of tagged histones detected just prior to (gray bars) and at 30 min (orange bars) and 60 min (red bars) after galactose addition. Data are expressed as the percentage of input DNA recovered. A control western blot for galactose activation of the H3HA proteins is shown in Supplementary Figure S1. Shown are the results of two independent biological experiments.
Figure 3.
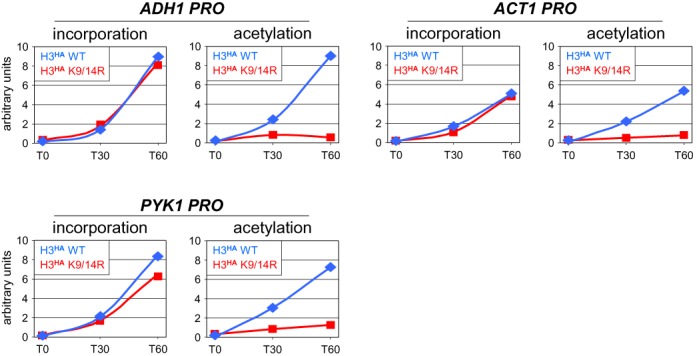
Histone H3 newly incorporated at promoters is acetylated at K9. The K9 acetylation status of newly incorporated H3HA was determined in a time course experiment by sequential Chromatin Immunoprecipitation (SeqChIP) (27). Incorporation of newly synthesized wild-type H3HA (blue lines) or H3HA(K9/14R) as a control (red lines) were monitored in G1-arrested cells at the indicated active promoters by quantitative ChIP as before (left panels). The purified H3HA–DNA complexes were subsequently subjected to a second round of immunoprecipitation using antibodies against H3K9ac (right panels). The ChIP signals for H3HA (incorporation) and H3K9ac (acetylation) are expressed in arbitrary units to facilitate comparison. The results for the three promoters are directly comparable. An independent biological experiment is shown in Supplementary Figure S6.
Figure 2.
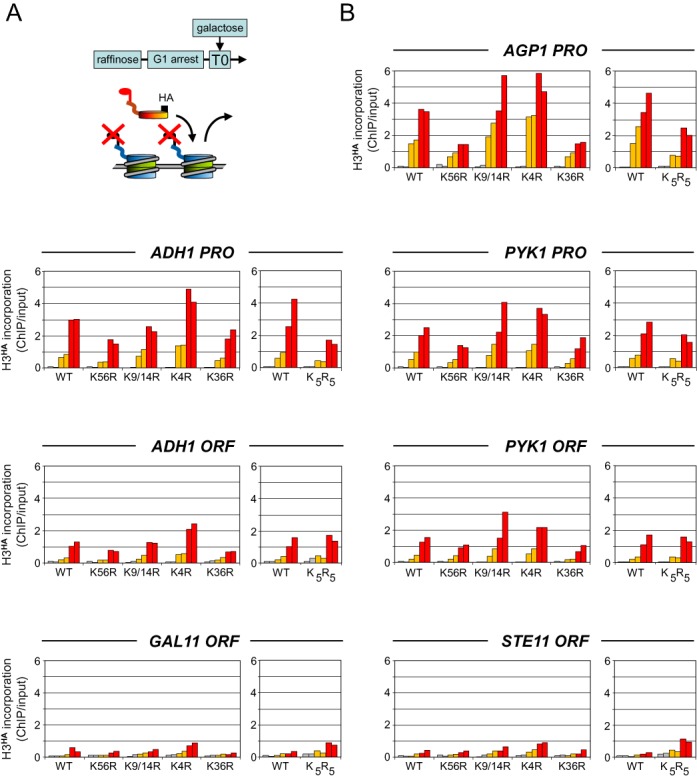
Transcription-dependent incorporation of wild-type histone H3 in a mutant chromatin background. (A) Similar experiment as in Figure 1 but measuring the incorporation of wild-type H3HA in a chromatin containing histone H3 modification mutants. Yeast strains deleted for both chromosomal H3 and H4 loci and expressing plasmid-encoded histone H4 and either wild-type or mutant histone H3 variants from their native promoters were grown in raffinose and arrested in G1 by alpha factor. Expression of wild-type H3HA was then induced at T0 (min) by adding galactose to the medium. (B) The time course of H3HA incorporation at the indicated loci and histone mutant strains was determined by quantitative ChIP analysis as in Figure 1. ChIP values are expressed as the percentage of input DNA recovered as in Figure 1. A western blot to assess for galactose activation of the H3HA proteins is shown in Supplementary Figure S1.
Figure 4.
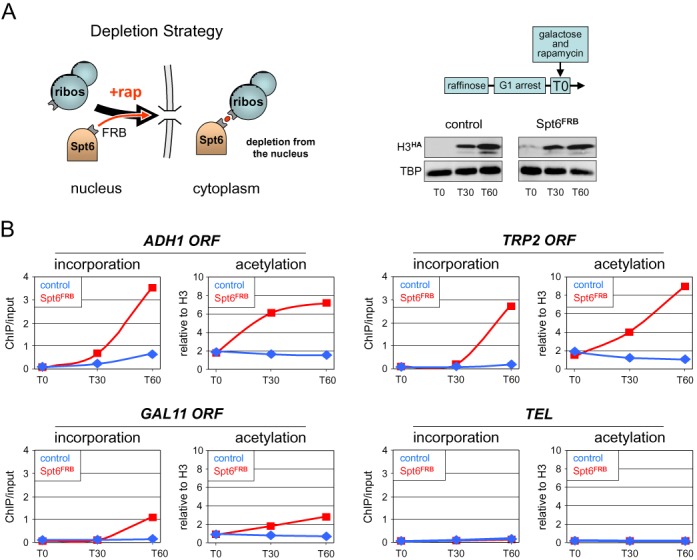
Spt6 inactivation leads to increased histone incorporation and H3K9 acetylation within transcribed regions. (A) Schematic illustration of the anchor-away strategy. Upon addition of rapamycin (+ rap, red dot), Spt6 fused to the rapamycin-binding domain FRB is rapidly exported out of the nucleus through its interaction with a ribosomal protein carrying the complementary FKBP rapamycin-binding domain (for details, see (25)). Right panel: experimental diagram. Cells from a Spt6-anchor-away strain (Spt6FRB) containing the galactose-inducible H3HA construct and from an isogenic control strain (control) lacking the Spt6FRB fusion were arrested in G1 by alpha factor. At T0, galactose and rapamycin were added to both cultures to, respectively, induce H3HA expression and deplete Spt6FRB from the nucleus. Shown on the lower right is a western blot analysis of H3HA expression and TBP as a control in the two strains. (B) The amounts of newly incorporated H3HA (left panels) and the levels of H3K9ac (right panels) at the indicated ORFs were monitored in the Spt6-anchor-away (Spt6FRB, red lines) and control (control, blue lines) strains by quantitative ChIP using antibodies against the HA tag or against H3K9ac. Data for H3HA incorporation are expressed as the percentage of input DNA recovered. The ChIP signals for H3K9ac were normalized to those obtained using a core histone H3 antibody for each genomic region to account for variations in nucleosome occupancy during the experiment and between different loci. TEL is a subtelomeric heterochromatin region on chromosome VI where Spt6 depletion is without consequence. An independent biological experiment is shown in Supplementary Figure S7.
Figure 6.
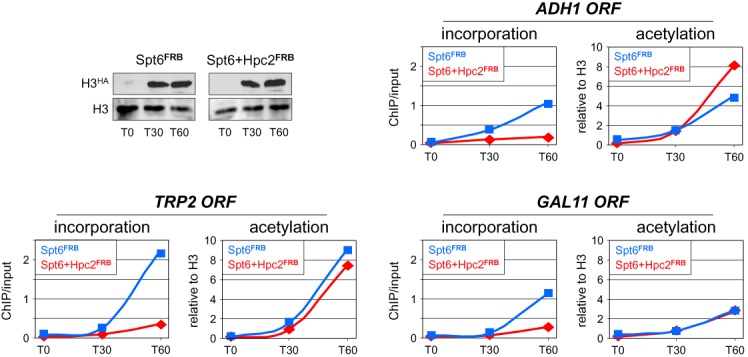
Inactivation of Hpc2 blocks the increase in histone exchange but not in H3K9ac induced by Spt6 inactivation. Same time course experiment for H3HA incorporation into chromatin (left panels) and total H3K9ac levels (right panels) as in Figure 4 but under conditions where either Spt6 alone (Spt6FRB, blue lines) or Spt6 and Hpc2 (Spt6+Hpc2FRB, red lines) are depleted from the nucleus by addition of rapamycin together with galactose at T0. The ChIP values are expressed as in Figure 4. Shown on the top left is a western blot analysis using antibodies against HA or the core histone H3 to assess for galactose activation of H3HA and constant expression levels of endogenous H3 in the two strains. Total H3K9ac levels remained largely unchanged under these experimental conditions (Supplementary Figure S5D). An independent biological experiment is shown in Supplementary Figure S9.
Figure 5.
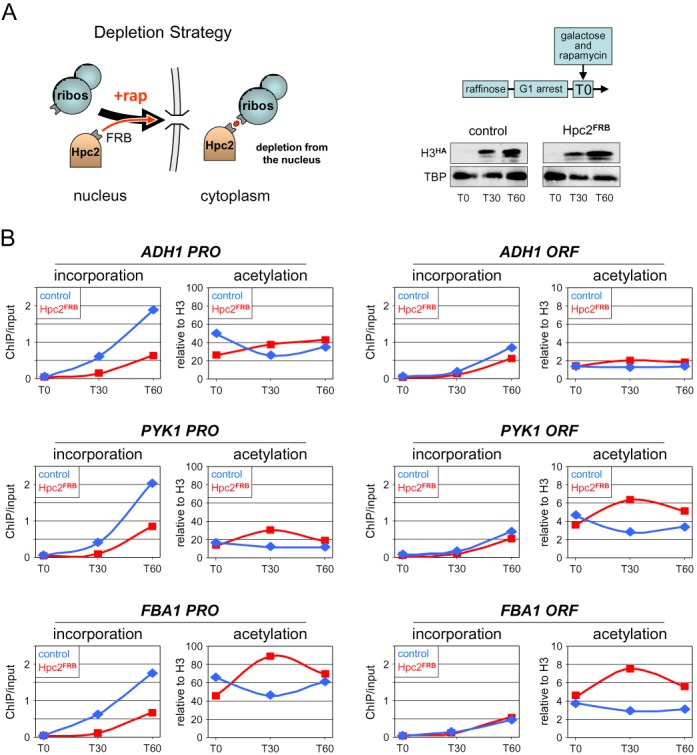
Inactivation of histone chaperone Hpc2 leads to decreased histone turnover at active promoters without concomitant decrease in H3K9ac. (A) Experimental diagram. Cells from an Hpc2-anchor-away strain (Hpc2FRB) containing the galactose-inducible H3HA construct and from an isogenic control strain (control) lacking the Hpc2FRB fusion were arrested in G1 by alpha factor. At T0, galactose and rapamycin were added to both cultures to, respectively, induce H3HA expression and deplete Hpc2FRB from the nucleus. Shown on the right is a western blot analysis of H3HA expression in the two strains. (B) The amounts of newly incorporated H3HA (left panels) and the levels of H3K9ac (right panels) at the promoters (PRO) and coding regions (ORF) of the indicated highly expressed genes were monitored by ChIP in the control (blue lines) and the Hpc2FRB strains (red lines) as in Figure 4. Note the different scales used for H3K9ac at PRO and ORF. An independent biological experiment is shown in Supplementary Figure S8.
Figure 7.
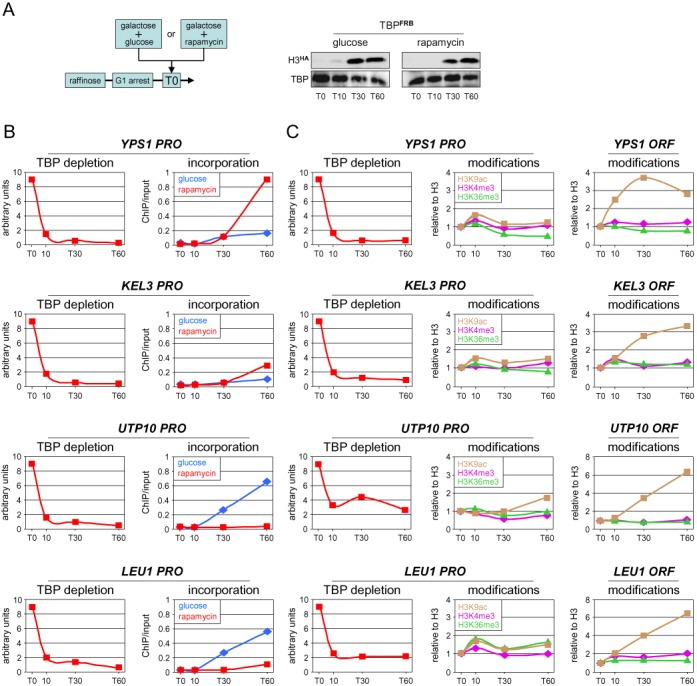
Loss of TBP from active promoters impacts differently on histone turnover and H3 modifications. (A) Cells from a TBP-anchor-away strain (26) carrying the galactose inducible H3HA construct were arrested in G1 as before. Galactose was then added at T0 to induce expression of H3HA together with rapamycin to deplete TBPFRB from the nucleus (rapamycin). As a control, H3HA expression was induced without TBP depletion by replacing rapamycin with glucose (glucose) to limit galactose induction of the tagged histone when TBP remains nuclear. Shown on the right is a Western blot analysis for H3HA expression under both conditions. (B) TBP promoter occupancy and H3HA incorporation under TBP-depletion (rapamycin, red lines) or control (glucose, blue lines) conditions were monitored by quantitative ChIP using antibodies against TBP and the HA tag. The TBP ChIP values are relative to those measured at T0 for each gene, which were arbitrarily set to 9.0. Results for H3HA incorporation are expressed as the percentage of input DNA recovered. Note that TBP occupancy drops very rapidly, reaching close to background levels already at T10 (min) and that the delay in H3HA incorporation is likely due to the slow kinetics of galactose induction (A). (C) TBP occupancy and the absolute levels of the indicated histone H3 modifications were monitored under TBP depletion conditions using antibodies against TBP and the relevant histone H3 modifications. The ChIP signals (normalized to H3) for H3K4me3 (right panels, violet), H3K9ac (brown) and H3K36me3 (green) are relative to those measured at T0, which were set to 1 to facilitate comparison. (B) and (C) are from two separate experiments. An independent biological experiment is shown in Supplementary Figure S10.
To allow efficient G1 arrest by alpha factor, all strains were MATa and disrupted for the BAR1 gene that encodes a secreted protease that cleaves and inactivates alpha factor. This was done by one-step replacement with—depending on marker availability—the loxP-KlURA3-loxP cassette or the hygromycin B resistance gene hphMX4 amplified by PCR from, respectively, plasmids pUG72 (Euroscarf No. P30117) and pAG32 (Euroscarf No. P30106). Disruption was confirmed by PCR analysis. Cells were arrested in G1 by treatment with 200 ng/ml (600 ng/ml for the anchor-away strains) alpha factor (Primm srl, Italy) for 3 h, at which time >95% of cells displayed the elongated ‘shmoo’ phenotype associated with G1 arrest. For ChIP analysis of histone exchange, cells were transformed with a C-terminal triple-HA-tagged histone H3 variant (H3HA) expressed under control of a galactose-inducible promoter from an ADE2-marked single copy plasmid in Figures 1 and 7 and from pRS314 in all other Figures (26). Mutants thereof were generated by PCR amplification as above. All constructs were confirmed by sequencing of the open reading frame (ORF).
Cells were grown overnight at 30°C to a density at OD600 nm of about 0.4–0.5 in Casamino acid medium lacking adenine or tryptophan supplemented with 2% raffinose and 0.1% glucose. Cells were then arrested in G1 and expression of H3HA was induced by directly adding 2% galactose. Where indicated, rapamycin (LC laboratories) was added at the same time to a final concentration of 4 μg/ml to deplete the FRB-tagged proteins from the nucleus. In Figure 7, glucose (0.5% final) instead of rapamycin was added as a control to limit galactose induction of H3HA. Culture aliquots were removed just prior to (T0) and at the indicated time points after galactose addition and immediately processed for ChIP and western blot analyses.
Western blotting
Whole-cell extracts were prepared by boiling the pelleted cells for 10 min in RIPA Buffer (50 mM HEPES [pH 7.5], 2 mM ethylenediaminetetraacetic acid (EDTA), 0.25 M NaCl, 0.1% sodium dodecyl sulphate (SDS), 0.1% DOC, 1% Triton X-100) mixed 3:1 with 4× Loading Sample Buffer (125 mM Tris–HCl [pH 6.8], 50% Glycerol, 4% SDS, 700 mM dithiothreitol (DTT), 0.02% Bromophenol Blue). The membranes were probed with 1:10 000 anti-HA monoclonal antibody (#16B12, Covance), 1:10 000 rabbit polyclonal anti-TBP antibodies (a kind gift from Laurie Stargell, Colorado State University), 1:10 000 rabbit polyclonal anti-acetyl-Histone H3 (Lys9) antibody (#07-352, Millipore) or 1:10 000 rabbit polyclonal anti-H3 antibodies (#1791, Abcam). Horseradish peroxidase–conjugated goat anti-mouse (Biorad, 1:10 000) or anti-rabbit (Biorad, 1:10 000) IgG were used as secondary antibodies. Detection was carried out with Immobilon Western Chemiluminescent HRP substrate (Millipore).
Chromatin immunoprecipitation (ChIP) and quantitative PCR
A detailed protocol for ChIP and quantitative PCR analysis can be found at http://www.mimo.unige.ch/STRUBIN_LABb.htm. Briefly, whole-cell extracts equivalent to about 2 × 108 yeast cells were immunoprecipitated with anti-HA antibodies (2 μl, #16B12, Covance), anti-H3 antibodies (1 μl, #1791, Abcam), anti-acetyl-H3K9 (2 μl, #07-352, Millipore), anti-trimethyl-H3K4 (2 μl, #07-473, Millipore) and anti-trimethyl-H3K36 (2 μl, #9050, Abcam). The recovered DNA and at least two standard dilutions of the input DNA were quantified in duplicate by real-time PCR using the KAPA SYBR FAST qPCR Kit Master Mix (2×) Universal (Kapa Biosystems) and the Biorad CFX96 Real-time PCR System. Sequences of the oligonucleotide primers are available upon request. The immunoprecipitation (IP) value for a given region was calculated as the ratio between the IP signal and the respective input DNA signal to correct for variation between different samples and primer pairs. Quantitative sequential ChIP was performed as previously described (27). All data are representative of at least two completely independent experiments. Independent biological replicates can be found in Supplementary Material.
Real-time RT-PCR analysis
The mRNA levels were quantified by real-time RT-PCR analysis using total RNA prepared by the hot-phenol extraction method from yeast strains grown to an OD600 of about 0.4–0.5. Reverse transcription was performed on 1 μg of RNA treated with RNAse-free DNAse using M-MLV reverse transcriptase (Promega) and an oligo(dT)15 primer. Real-time PCR quantification was carried out as described above using primer pairs specific for the indicated genes. The values were normalized to those obtained for the 18S rRNA to account for differences in total RNA across samples.
RESULTS
Histone turnover in the absence of transcription-associated H3 modifications
As a first step toward understanding the relationship between histone modifications and histone turnover associated with active transcription, we explored whether specific post-translational modifications of histone H3 may have a causal role in histone dynamics. We concentrated on H3K4me3 and H3K9/14ac, which are enriched within the promoter regions of actively transcribed genes where histones dynamically exchange (4,18), and H3K36me3 that accumulates over gene bodies where it has been implicated in suppressing histone exchange (11,28). We also considered H3K56ac, a modification mark added on free histone H3 prior to its incorporation into chromatin and reported to stimulate histone turnover (19,24,29). We mutated the relevant lysine residues to arginine, either individually or in combination, to prevent their modification. Since redundant roles for N-terminal histone acetylation in nucleosome assembly have been reported (30), we also simultaneously mutated all five potential H3 N-terminal acetylation sites, K9/14/18/23/27. To test the consequence of the mutations on histone turnover, we used hemagglutinin (HA) epitope-tagged versions expressed from a galactose-inducible promoter. The proteins were tagged at the C-terminus to avoid interference with the modification of the N-terminal lysines. Expression of the tagged histones was induced by galactose and their incorporation into chromatin was monitored in a time course by ChIP analysis using antibodies against the HA tag (Figure 1A). Newly synthesized H3HA is expected to replace endogenous H3 at regions where nucleosomes are dynamic and therefore its incorporation indirectly measures histone exchange (18,19,31). To exclude any replication associated events, cells were arrested at the G1 phase of the cell cycle by treatment with alpha factor (31). Wild-type H3HA rapidly accumulated at the promoters of the highly transcribed AGP1, ADH1 and PYK1 genes, which are among those showing the highest histone turnover rates in genome-wide studies (18,19), less so in the corresponding ORFs, and almost not at all at the lowly expressed GAL11 and inactive STE11 genes, as reported (Figure 1 and Supplementary Figure S1) (18,19,31). Strikingly, all single and double H3HA mutants exhibited essentially the same overall profile of incorporation (Figure 1). It is only when all five potential N-terminal acetylation sites were mutated that H3HA incorporation was affected, decreasing by about two-fold at all sites tested (K5R5 in Figure 1). Thus, none of these lysine modifications is essential for transcription-dependent incorporation of newly synthesized histone H3.
In a reciprocal experiment, we monitored the incorporation of wild-type H3HA into a mutated chromatin background using yeast strains expressing the histone mutants from the natural promoter as the sole source of histone H3 (Figure 2A). Among the five strains only K56R grew more slowly (Supplementary Figure S2A), in agreement with previous studies (32–34), whereas galactose induction of H3HA occurred with similar kinetics in all strains (Supplementary Figure S1). This is consistent with these lysine modifications playing no essential role. We then measured H3HA incorporation in these strains in a time course ChIP experiment as before. Compared to wild-type, a slight increase, at least at some loci, occurred in a K4R chromatin background (Figure 2B). By contrast, incorporation remained mostly unaffected in the K9/14R mutant strain, while in the K56R strain it was slightly but reproducibly decreased at all tested loci, consistent with K56ac facilitating histone eviction (Figure 2B) (29,35). The same was observed when the K9/14R and K56R mutations were present on both the endogenous and the tagged histone (Supplementary Figures S3 and S4). A modest decrease in H3HA incorporation was also detected in the K5R5 quintuple acetylation mutant, but this occurred only at promoter regions. Unexpectedly, if anything H3HA incorporation was generally reduced in the K36R strain (Figure 2B), although it increased by about two-fold across the silent STE11 gene, as reported (28) (Supplementary Figure S2B). However, the levels at STE11 remained far below those observed at highly dynamic regions (Supplementary Figure S2B). Thus, post-translational modification of these H3 lysine residues may fine-tune but are not essential for either eviction of the original histone or incorporation of new histones.
Newly incorporated histone H3 is acetylated at Lysine 9
We then addressed the opposite possibility, namely, that some H3 modifications may arise as a consequence of histone turnover. We concentrated mainly on H3K9ac, since there is evidence that this mark is present on soluble histone H3 (36). We therefore examined whether newly incorporated H3HA is acetylated at K9 by sequential Chromatin Immunoprecipitation (SeqChIP) (27) (Figure 3). Specifically, H3HA was induced by galactose and after cross-linking to DNA immunoprecipitated using anti-HA antibodies. A fraction of the purified H3HA–DNA complexes was then subjected to a second round of immunoprecipitation using antibodies specific for H3K9ac, to selectively isolate DNA bound to acetylated H3HA. To test for specificity, the same experiment was performed with a strain expressing histone mutant H3HA(K9/14R). As shown in Figure 3, wild-type H3HA (blue lines) and the mutant (red lines) were incorporated at similar levels at the three highly dynamic promoters tested, ADH1, ACT1 and PYK1 (Figure 3, left panels). This confirms that modification at these lysine residues is not causally implicated in histone deposition (Figure 1). A similar kinetics of increase in ChIP signals was observed for wild-type H3HA in the SeqChIP with anti-H3K9ac antibodies (right panels in Figure 3, blue lines). This reflects specific acetylation at K9, as no major changes were observed with the H3HA(K9/14R) mutant (right panels in Figure 3, red lines). Thus, K9 acetylation closely parallels H3HA incorporation into chromatin, suggesting that this mark is added either prior to or rapidly after incorporation.
Increased histone exchange within transcribed regions leads to increased H3K9ac
If H3K9ac is a consequence of histone exchange, then experimentally increasing or decreasing histone exchange should impact on the H3K9 steady-state acetylation levels. Previous work has shown that inactivation of the histone chaperone Spt6 causes a transcription-dependent reduction in nucleosome occupancy over coding regions (37). We found here that this correlates with a concomitant increase in incorporation of new histones (see below). We therefore investigated whether inactivation of Spt6 would lead to an increase in H3K9ac. For this, we used the anchor away method (25), which allows for rapid inactivation of a nuclear protein fused to the rapamycin binding domain FRB via its rapamycin-mediated export to the cytoplasm (Figure 4A). An anchor-away strain carrying an FRB-tagged SPT6 allele at the native locus and the parental strain were arrested at G1 with alpha factor. Galactose was then added to induce the expression of H3HA together with rapamycin to trigger nuclear depletion of Spt6 in the anchor-away strain. Incorporation of H3HA and H3K9ac were monitored at different time points by ChIP. Galactose activation of H3HA was comparable in both strains (Figure 4A). However, while H3HA incorporation in the parental strain (left panels in Figure 4B, blue curves) was low within the ORF of the highly transcribed ADH1 gene and essentially absent within the moderately expressed TRP2 gene and the lowly transcribed GAL11 gene, it dramatically increased at all sites in the Spt6 depletion strain (red curves). Strikingly, increased H3HA incorporation was associated with a marked increase in H3K9ac within these regions, but not at TEL were Spt6 depletion is without consequence (Figure 4B, right panels). These effects were not attributable to changes in transcription or in total histone acetylation levels due to varying growth conditions or Spt6 depletion; there were no major differences in steady-state mRNA and H3K9ac levels in the course of the experiment (Supplementary Figure S5A–C). Thus, the finding that newly incorporated H3 is acetylated at K9 and that increasing histone exchange leads to increased H3K9ac steady-state levels suggests that H3K9ac may occur as a consequence of histone turnover. However, the following experiments argue that this is not the case.
Decreasing histone turnover does not impact on H3K9ac
To further assess the possible causal role of histone turnover in H3K9ac, we performed a reciprocal experiment in which histone incorporation was decreased, expecting that this would result in reduced H3K9ac steady state levels. The histone regulatory (HIR) complex is one of the chaperones that have been implicated in the deposition of new histones during transcription (38,39). Incorporation of newly synthesized H3HA was indeed reduced at the highly active ADH1 promoter in strains deleted for the Hpc2 subunit of the HIR complex (Supplemental Figure S5E). We therefore measured the effect of inactivating Hpc2 on H3K9ac. To minimize indirect effects, we used the anchor-away method to conditionally and rapidly export Hpc2 out of the nucleus (Figure 5A). Incorporation of newly synthesized H3HA under these conditions was partially reduced at the three active promoters tested (Figure 5B, left panels). This was neither due to inefficient galactose activation of H3HA (Figure 5A) nor to increased expression of endogenous H3, which acts as competitor and has been reported to be negatively regulated by the HIR complex in response to alpha factor arrest (Figure 6) (40). Indeed, H3HA incorporation remained largely unaffected in the ORFs (Figure 5B, right panels). Unexpectedly, the reduced incorporation of H3HA observed at promoter regions was not accompanied by a corresponding decrease in H3K9ac (Figure 5B, right panels). Thus, H3K9ac and histone turnover can be experimentally uncoupled.
We performed a similar experiment to assess whether blocking the increase in H3HA incorporation within ORFs that occurs following Spt6 inactivation would also block the associated increase in H3K9ac (Figure 4). For this, we generated an anchor-away yeast strain in which both Spt6 and Hpc2 could be depleted simultaneously from the nucleus by addition of rapamycin (Figure 6). Despite a similar induction by galactose, H3HA was incorporated much less efficiently at the three tested loci when Hpc2 was inactivated together with Spt6 (left panels in Figure 6, red curves). However, the increase in H3K9ac observed when only Spt6 is depleted was still present (Figure 6, right panels). Hence, experimentally decreasing histone turnover has no impact on H3K9ac, suggesting that this histone modification does not results as a consequence of histone turnover.
Histone turnover and modifications following PIC depletion
Previous work has shown that the binding of general transcription factor TBP, and thus likely of the entire the preinitiation complex (PIC) (41), to active RNA Pol II promoters is very dynamic and that nuclear depletion of TBP by anchor away causes a rapid drop in TBP promoter occupancy (26). It was also found that loss of TBP results in increased or decreased histone turnover at some promoters (26). We therefore examined whether any transcription-associated histone H3 modification would show a similar trend in this situation (Figure 7). For this, we selected four genes identified in a genome-wide analysis (A. Hughes et al., unpublished results) that show either a strong increase or nearly complete loss of H3HA incorporation following TBP dissociation from the promoter (Figure 7B). No corresponding changes in H3K4me3, H3K9ac and H3K36me3 were observed at any promoter under these conditions (Figure 7C). Unexpectedly, while H3K4me3 and H3K36me3 also remained unchanged in the downstream coding regions, H3K9ac increased progressively to reach levels several-fold higher than those measured under transcription conditions (Figure 7C, right panels). These results provide further evidence for a lack of causal relationship between histone turnover and transcription-associated histone H3 modifications. The persistence of these modifications following transcriptional shutdown is consistent with them having a role in transcriptional memory (28,42,43).
DISCUSSION
Uncoupling transcription-associated histone turnover and H3 modifications
Transcription in eukaryotes is associated with local changes in chromatin including post-translational modifications of histones, nucleosome remodeling and histone turnover, a process by which old histones are replaced by new histones (44). While there is overwhelming evidence that these events closely correlate with active transcription, the nature of their inter-relationship and whether they play an instructive role in the transcription process or merely occur as a consequence of transcription is still a matter for investigation. Here we explored the relationship between histone turnover and four histone H3 modifications typically associated with active transcription. These include H3K4me3 and H3K9/14ac, which accumulate at active promoters, and H3K36me3 that is enriched over the body of transcribed genes (4,18). We found that mutating the corresponding lysine residues to arginine had little consequence on histone turnover at both the promoters and coding regions of the genes we examined. Only when H3K56—whose acetylation has been implicated in histone turnover (29,45)—was mutated did we observe a decrease in new histone incorporation (Figures 1 and 2). However, the effect was modest and present only when the mutation was introduced into the chromosomal histone (Figures 1 and 2), consistent with a role for H3K56ac in nucleosome eviction (29,35). Thus, none of the transcription-associated N-terminal H3 modifications we examined is instrumental in promoting histone exchange.
In a series of reciprocal experiments, we found that increasing or decreasing histone exchange, by inactivating specific histone chaperones or depleting TBP from promoters, was mostly without consequence on the histone H3 steady-state modification levels. This was true even at the most dynamic promoters, where one would expect to see an effect if histone modifications were deposited or erased as a consequence of histones exchanging. A similar situation has been observed in mouse embryonic stem cells, where no difference in H3K4me3 enrichment was observed at promoters under conditions of reduced nucleosome turnover dynamics (46). Thus, despite correlating both tightly with active transcription, the histone H3 N-terminal modifications examined and the process of histone turnover are largely independent events.
One situation where an enrichment in H3 acetylation occurred concurrently with an increase in histone exchange was within transcribed regions following inactivation of Spt6. An increase in H3 steady-state acetylation at lysines 9 and 14 has also been observed in fission yeast lacking Spt6 (47). Inactivation of Spt6 not only induces increased incorporation of new histones, as we show here, but also results in a transcription-coupled loss of nucleosomes (37). A similar transcription-dependent loss of nucleosomes and concomitant increase in H3 acetylation was observed upon inactivation of Spt16 (48). By contrast, depleting TBP from promoters, or inactivating histone chaperone Hpc2, two experimental conditions that we found affect histone turnover at promoters without impacting on the histone H3 modification state, had little or no effect on H3 steady-state occupancy at the genes we tested (data not shown). The increase in H3 acetylation upon inactivation of Spt6 or Spt16 may therefore be a consequence of histone eviction rather than histone exchange. Perhaps the unacetylated histone H3 is preferentially evicted from transcribed regions in the absence of the histone chaperones.
H3K36me3 and histone turnover
A recent genome-wide study suggests that a main function of H3K36me3 is to suppress histone exchange across the body of transcribed genes (28,49). It was shown that deletion of Set2, the enzyme that methylates H3 at residue K36, results in increased histone exchange specifically over coding regions, as well as increased H3K56ac, a modification that occurs prior to nucleosomal assembly (50) and thus accumulates at genomic regions where histones exchange (19,29,51). An increase in H3K56ac over coding regions was also observed in yeast cells expressing a K36A point mutant of H3 (28). However, histone exchange was not directly assessed in the K36 mutant background. We found that blocking H3K36 methylation by a lysine-to-arginine mutation leads to a modest increase in H3HA incorporation across the coding region of the inactive STE11 gene. This is consistent with the reported increase in H3K56ac (28). However, the incorporation levels remained far below those detected at highly dynamics sites (Supplementary Figure S2B). Furthermore, the K36R mutation had no major impact on histone exchange at the other coding regions we examined (Figures 1 and 2). It is therefore likely that other mechanisms in addition to H3K36me3 are involved in suppressing histone exchange over gene bodies.
Specific roles for histone chaperones Spt6, FACT and HIR in histone turnover
Several histone chaperones have been implicated in histone removal, histone deposition and/or histone exchange during transcription (16). Among these are Spt6, FACT and the HIR complex. Both Spt6 and FACT are essential for viability and they both travel with the elongating RNA Pol II (52), thus showing a very similar pattern of association across the genome (53). As alluded to above, inactivation of Spt6 leads to a general loss of nucleosomes (37), initiation from cryptic promoters (54) and, as we show here, increased incorporation of new histones over transcribed regions (Figure 4). Exactly the same was observed upon inactivation of the Spt16 subunit of FACT (48,55) and was interpreted as indications that a main function of FACT is to reassemble the original nucleosomes evicted by the elongating polymerase (48). Thus, although both chaperones are involved in multiple other and, sometimes, distinct processes (37), they likely function in a common pathway to maintain a proper chromatin and prevent incorporation of new histones over transcribed regions (17,52).
In contrast, inactivation of the HIR complex by anchor-away-mediated nuclear depletion of its Hpc2 subunit resulted in decreased H3 incorporation. Intriguingly, Hpc2 depletion by itself altered histone dynamics at active promoters but did not change turnover within the coding regions we examined (Figure 5). However, the increase in turnover at coding regions upon Spt6 depletion was blunted by concomitant Hpc2 depletion (Figure 6). This is fully consistent with the earlier proposal that the HIR complex has a role in compensating defects associated with spt6 or spt16 mutations (56). It further suggests that the HIR complex has an essential role in regulating turnover at promoters and largely redundant roles at transcribed regions that are revealed only when multiple factors are compromised. This is reminiscent of the reported overlapping roles of the HIR complex and the CAF-1 histone deposition complex in replication-independent histone turnover (57).
Histone exchange and histone modifications, how and to what end?
In summary, we have shown that four major transcription-coupled histone H3 N-terminal modifications are neither the cause nor the consequence of histone turnover at active genes. What, then, triggers histone modifications and, independently, histone exchange, and what might be their biological significance? Histone modifications are widely viewed as resulting from the selective recruitment of histone-modifying activities by activators or by the elongating RNA Pol II and as playing a key causal role in regulating transcription initiation and elongation (2). However, we found that, in G1-arrested cells, H3K9ac, H3K4me3 and H3K36me3 all persist long after loss of TBP from promoters and thus transcriptional shutdown (Figure 7). The same was observed in mammalian cells following treatment with the RNA Pol II inhibitor α-amanitin and dissociation of the polymerase and all histone-modifying factors examined (58). A persistence of H3K4me3 after transcriptional repression of galactose-regulated genes in yeast has also been reported (42,59). This is consistent with these histone marks also potentially serving another, longer-term purpose and with erasure of these marks occurring mainly during S phase (60). Unexpectedly, H3K9ac was not only maintained but actually increased by several fold specifically over gene bodies upon TBP depletion and thus transcriptional shutdown (Figure 7). The increase was not linked to changes in histone turnover or nucleosome occupancy and it occurred relatively slowly after TBP departure. This suggests an involvement of globally acting histone acetylases (12). It further raises the possibility that a main function of the Rpd3S histone deacetylase complex that travels with the elongating RNA Pol II is to prevent untargeted acetylation within transcribed regions (28).
Histone exchange at active promoters, on the other hand, could simply be explained by a constant competition between nucleosomes and the PIC, which is highly dynamic (26,61), for binding to promoter DNA. Earlier and more recent studies are consistent with this view (62–64). However, loss of RNA Pol II, which results in the complete disassembly of the PIC, leads to changes in the positioning but has relatively little effect on promoter nucleosome occupancy (65). Furthermore, although we provide evidence here that the PIC is indeed largely responsible for histone turnover at some promoters (Figure 7), this appears not to be the case for most RNA Pol II genes; indeed, histones keep exchanging at many promoters independently of PIC assembly and RNA Pol II transcription (26) (A. Hughes et al., unpublished results). Further investigation will tell whether trans-acting factors (66) or some intrinsic nucleosome-destabilizing property of promoter sequences (67) plays a decisive role in histone turnover and to what extent the process is important for gene regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are most grateful to David Stillman for the yeast strain DY8862, to Françoise Stutz for the strains deleted for Hpc2, to Laurie Stargell for the anti-TBP antibodies and to Yoselin Grimaldi for helpful discussions.
FUNDING
Swiss National Science Foundation [31003A-127384, 310030-149626 to M.S.]; Canton of Geneva. Funding for open access charge: Swiss National Science Foundation [31003A-127384, 310030-149626 to M.S.]; Canton of Geneva.
Conflict of interest statement. None declared.
REFERENCES
- 1.Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Campos E.I., Reinberg D. Histones: annotating chromatin. Annu. Rev. Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 3.Rando O.J. Global patterns of histone modifications. Curr. Opin. Genet. Dev. 2007;17:94–99. doi: 10.1016/j.gde.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 5.Schubeler D., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kharchenko P.V., Alekseyenko A.A., Schwartz Y.B., Minoda A., Riddle N.C., Ernst J., Sabo P.J., Larschan E., Gorchakov A.A., Gu T., et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Liu C.L., Kaplan T., Kim M., Buratowski S., Schreiber S.L., Friedman N., Rando O.J. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Martelot G., Canella D., Symul L., Migliavacca E., Gilardi F., Liechti R., Martin O., Harshman K., Delorenzi M., Desvergne B., et al. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012;10:e1001442. doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smolle M., Workman J.L. Transcription-associated histone modifications and cryptic transcription. Biochim. Biophys. Acta. 2013;1829:84–97. doi: 10.1016/j.bbagrm.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katan-Khaykovich Y., Struhl K. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 2002;16:743–752. doi: 10.1101/gad.967302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imoberdorf R.M., Topalidou I., Strubin M. A role for gcn5-mediated global histone acetylation in transcriptional regulation. Mol. Cell. Biol. 2006;26:1610–1616. doi: 10.1128/MCB.26.5.1610-1616.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yun M., Wu J., Workman J.L., Li B. Readers of histone modifications. Cell Res. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musselman C.A., Lalonde M.E., Cote J., Kutateladze T.G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avvakumov N., Nourani A., Cote J. Histone chaperones: modulators of chromatin marks. Mol. Cell. 2011;41:502–514. doi: 10.1016/j.molcel.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Rando O.J., Winston F. Chromatin and transcription in yeast. Genetics. 2012;190:351–387. doi: 10.1534/genetics.111.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dion M.F., Kaplan T., Kim M., Buratowski S., Friedman N., Rando O.J. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 19.Rufiange A., Jacques P.E., Bhat W., Robert F., Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Kraushaar D.C., Jin W., Maunakea A., Abraham B., Ha M., Zhao K. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 2013;14:R121. doi: 10.1186/gb-2013-14-10-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luebben W.R., Sharma N., Nyborg J.K. Nucleosome eviction and activated transcription require p300 acetylation of histone H3 lysine 14. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19254–19259. doi: 10.1073/pnas.1009650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinke H., Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 23.Govind C.K., Zhang F., Qiu H., Hofmeyer K., Hinnebusch A.G. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 24.Li Q., Zhou H., Wurtele H., Davies B., Horazdovsky B., Verreault A., Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haruki H., Nishikawa J., Laemmli U.K. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 26.Grimaldi Y., Ferrari P., Strubin M. Independent RNA polymerase II preinitiation complex dynamics and nucleosome turnover at promoter sites in vivo. Genome Res. 2014;24:117–124. doi: 10.1101/gr.157792.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geisberg J.V., Struhl K. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res. 2004;32:e151. doi: 10.1093/nar/gnh148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkatesh S., Smolle M., Li H., Gogol M.M., Saint M., Kumar S., Natarajan K., Workman J.L. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan T., Liu C.L., Erkmann J.A., Holik J., Grunstein M., Kaufman P.D., Friedman N., Rando O.J. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 2008;4:e1000270. doi: 10.1371/journal.pgen.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shahbazian M.D., Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 31.Jamai A., Imoberdorf R.M., Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 32.Mann R.K., Grunstein M. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 1992;11:3297–3306. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Recht J., Tsubota T., Tanny J.C., Diaz R.L., Berger J.M., Zhang X., Garcia B.A., Shabanowitz J., Burlingame A.L., Hunt D.F., et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin Y., Rodriguez A.M., Stanton J.D., Kitazono A.A., Wyrick J.J. Simultaneous mutation of methylated lysine residues in histone H3 causes enhanced gene silencing, cell cycle defects, and cell lethality in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:6832–6841. doi: 10.1128/MCB.00745-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams S.K., Truong D., Tyler J.K. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo M.H., Brownell J.E., Sobel R.E., Ranalli T.A., Cook R.G., Edmondson D.G., Roth S.Y., Allis C.D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 37.Ivanovska I., Jacques P.E., Rando O.J., Robert F., Winston F. Control of chromatin structure by spt6: different consequences in coding and regulatory regions. Mol. Cell. Biol. 2011;31:531–541. doi: 10.1128/MCB.01068-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amin A.D., Vishnoi N., Prochasson P. A global requirement for the HIR complex in the assembly of chromatin. Biochim. Biophys. Acta. 2013;1819:264–276. doi: 10.1016/j.bbagrm.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 39.Kim H.J., Seol J.H., Han J.W., Youn H.D., Cho E.J. Histone chaperones regulate histone exchange during transcription. EMBO J. 2007;26:4467–4474. doi: 10.1038/sj.emboj.7601870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu H., Kim U.J., Schuster T., Grunstein M. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:5249–5259. doi: 10.1128/mcb.12.11.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuras L., Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 42.Ng H.H., Robert F., Young R.A., Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Q., Yoon Y., Yu Y., Parnell E.J., Garay J.A., Mwangi M.M., Cross F.R., Stillman D.J., Bai L. Stochastic expression and epigenetic memory at the yeast HO promoter. Proc. Natl. Acad. Sci. U.S.A. 2013;110:14012–14017. doi: 10.1073/pnas.1306113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith E., Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol. Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen C.C., Carson J.J., Feser J., Tamburini B., Zabaronick S., Linger J., Tyler J.K. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banaszynski L.A., Wen D., Dewell S., Whitcomb S.J., Lin M., Diaz N., Elsasser S.J., Chapgier A., Goldberg A.D., Canaani E., et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kato H., Okazaki K., Iida T., Nakayama J., Murakami Y., Urano T. Spt6 prevents transcription-coupled loss of posttranslationally modified histone H3. Sci. Rep. 2013;3:2186. doi: 10.1038/srep02186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jamai A., Puglisi A., Strubin M. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol. Cell. 2009;35:377–383. doi: 10.1016/j.molcel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 49.Smolle M., Workman J.L., Venkatesh S. reSETting chromatin during transcription elongation. Epigenetics. 2013;8:10–15. doi: 10.4161/epi.23333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masumoto H., Hawke D., Kobayashi R., Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 51.Tsubota T., Berndsen C.E., Erkmann J.A., Smith C.L., Yang L., Freitas M.A., Denu J.M., Kaufman P.D. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duina A.A. Histone chaperones Spt6 and FACT: similarities and differences in modes of action at transcribed genes. Genet. Res. Int. 2011:625210. doi: 10.4061/2011/625210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayer A., Lidschreiber M., Siebert M., Leike K., Soding J., Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 54.Kaplan C.D., Laprade L., Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 55.Morillo-Huesca M., Maya D., Munoz-Centeno M.C., Singh R.K., Oreal V., Reddy G.U., Liang D., Geli V., Gunjan A., Chavez S. FACT prevents the accumulation of free histones evicted from transcribed chromatin and a subsequent cell cycle delay in G1. PLoS Genet. 2010;6:e1000964. doi: 10.1371/journal.pgen.1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Formosa T., Ruone S., Adams M.D., Olsen A.E., Eriksson P., Yu Y., Rhoades A.R., Kaufman P.D., Stillman D.J. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics. 2002;162:1557–1571. doi: 10.1093/genetics/162.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopes da Rosa J., Holik J., Green E.M., Rando O.J., Kaufman P.D. Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics. 2011;187:9–19. doi: 10.1534/genetics.110.123117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kouskouti A., Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kundu S., Horn P.J., Peterson C.L. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Radman-Livaja M., Liu C.L., Friedman N., Schreiber S.L., Rando O.J. Replication and active demethylation represent partially overlapping mechanisms for erasure of H3K4me3 in budding yeast. PLoS Genet. 2010;6:e1000837. doi: 10.1371/journal.pgen.1000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Werven F.J., van Teeffelen H.A., Holstege F.C., Timmers H.T. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat. Struct. Mol. Biol. 2009;16:1043–1048. doi: 10.1038/nsmb.1674. [DOI] [PubMed] [Google Scholar]
- 62.Han M., Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 63.Gossett A.J., Lieb J.D. In vivo effects of histone H3 depletion on nucleosome occupancy and position in Saccharomyces cerevisiae. PLoS Genet. 2012;8:e1002771. doi: 10.1371/journal.pgen.1002771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zanton S.J., Pugh B.F. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 2006;20:2250–2265. doi: 10.1101/gad.1437506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiner A., Hughes A., Yassour M., Rando O.J., Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X., Bai L., Bryant G.O., Ptashne M. Nucleosomes and the accessibility problem. Trends Genet. 2011;27:487–492. doi: 10.1016/j.tig.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 67.Sekinger E.A., Moqtaderi Z., Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


