Abstract
Growth arrest and DNA-damage-inducible protein 45 (Gadd45) family members have been implicated in DNA demethylation in vertebrates. However, it remained unclear how they contribute to the demethylation process. Here, we demonstrate that Gadd45a promotes active DNA demethylation through thymine DNA glycosylase (TDG) which has recently been shown to excise 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) generated in Ten-eleven-translocation (Tet)—initiated oxidative demethylation. The connection of Gadd45a with oxidative demethylation is evidenced by the enhanced activation of a methylated reporter gene in HEK293T cells expressing Gadd45a in combination with catalytically active TDG and Tet. Gadd45a interacts with TDG physically and increases the removal of 5fC and 5caC from genomic and transfected plasmid DNA by TDG. Knockout of both Gadd45a and Gadd45b from mouse ES cells leads to hypermethylation of specific genomic loci most of which are also targets of TDG and show 5fC enrichment in TDG-deficient cells. These observations indicate that the demethylation effect of Gadd45a is mediated by TDG activity. This finding thus unites Gadd45a with the recently defined Tet-initiated demethylation pathway.
INTRODUCTION
Methylation at position 5 of cytosine (5-methylcytosine, 5mC) in DNA is a major epigenetic modification that regulates gene transcription and other functions of the genome (1,2). Cytosine methylation in CpG-rich regulatory gene promoters and enhancers inversely correlates with transcriptional activity of associated genes as it causes chromatin condensation and thus gene silencing. Since patterns of 5mC are subject to mitotic inheritance through maintenance methylation during DNA replication, active demethylation is required for a rapid and efficient erasure of 5mC (3). Both locus-specific and genome-wide demethylation have been documented (4). For instance, the promoter of the estrogen receptor target gene pS2 undergoes active demethylation in the cyclic activation of transcription (5). Genome-wide demethylation in primordial germ cells is believed to be important for erasing the parental methylation patterns (6). Demethylation of the zygotic genome is associated with remodeling of the parental epigenomes, presumably to establish developmental competence for the early embryo (7–10). Multiple mechanisms have been proposed to achieve active demethylation, which include direct removal of the exocyclic methyl group from the cytosine via C–C bond cleavage, replacement of the methylated cytosine base and nucleotide respectively through DNA base excision repair (BER) and nucleotide excision repair pathways (11). However, most of the proposed mechanisms have not been validated biochemically and genetically (12,13).
Compelling biochemical and genetic evidence has suggested that members of the Ten-eleven-translocation (Tet) family of DNA dioxygenases function to reverse DNA methylation (4,14). Tet enzymes catalyze the oxidation of 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (15–17). Thymine DNA glycosylase (TDG), originally identified as a DNA glycosylase for the excision of thymine and uracil mispaired with guanine, is able to recognize and excise the Tet-generated oxidation products 5fC and 5caC, leading to the incorporation of unmethylated cytosine via the BER pathway (15,18–20). The functional relevance of TDG in the regulation of DNA methylation is well-established by gene inactivation experiments at both animal (21,22) and ES cell levels (15,23,24). Given the importance of DNA methylation in stem cell biology and cancer, the study of Tet/TDG-mediated demethylation has become a major focus over the recent years. However, it is still unclear how the oxidative demethylation process is regulated.
Gadd45 (growth arrest and DNA-damage-inducible protein 45) family proteins are multi-faceted nuclear factors implicated in active DNA demethylation, apart from maintenance of genomic stability, DNA repair and suppression of cell growth (25,26). Overexpression of Gadd45a activates methylation-silenced reporter genes and promotes global DNA demethylation (27). However, despite the connection of Gadd45 proteins with DNA demethylation in several contexts, including neuronal activity-induced demethylation in the mouse brain (28) and deaminase-related demethylation in Xenopus laevis embryos (29), if and how they exactly promote DNA demethylation has remained unresolved and controversial (26,30).
In this study, we investigate the role of Gadd45a in active demethylation and activation of silenced genes. We find that Gadd45a interacts physically and functionally with TDG and contributes to DNA demethylation and gene activation in a TDG-dependent manner. In mouse ES cells, inactivation of Gadd45a/b leads to hypermethylation at loci most of which overlap with those depending on TDG for demethylation. These findings connect Gadd45 proteins with the Tet-TDG axis, functionally integrating the seemingly diverse demethylation pathways.
MATERIALS AND METHODS
Materials
Primary antibodies used for western blotting assays were as follows: anti-Flag (Sigma, F7425), anti-HA (Sigma, H6908), anti-GAPDH (Sigma, 9545). Anti-Tet2 and anti-TDG antibodies were as described previously (31). The Tdg knockout ES cell line was described (32).
Luciferase reporter assay
The reporter plasmid pCpGL-CMV-firefly luciferase was generated by subcloning the Cytomegalovirus (CMV) promoter from pcDNA3.1 (Invitrogen) into the CpG-free pCpGL-basic vector (33). By replacing the firefly luciferase gene with the renilla luciferase gene, an analogous control reporter plasmid pCpGL-CMV-renilla luciferase was constructed. The firefly plasmid was in vitro methylated with CpG methylase M.SssI (NEB) and the complete methylation was verified by digestion with methylation sensitive enzymes TaiI (Fermentas). HEK293T cells were transiently transfected in 12-well plate with 500 ng expression constructs (Tet2/TDG/Gadd45a) each, 20 ng methylated firefly luciferase reporter and 0.2 ng unmethylated renilla luciferase reporter as an internal control for normalization. Forty-six hours after transfection, luciferase activities were measured using the dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. Each experiment was repeated at least three times.
HPLC analysis of nucleosides
The nucleosides were analyzed by HPLC according to He et al. (15). Briefly, 200 μg of genomic DNA extracted from HEK293T cells were heat-denatured and hydrolyzed into mononucleotides with 0.5 U of nuclease P1 (Sigma) at 37°C overnight (the reaction buffer containing 20 mM NaAc, pH 5.3, 0.2 mM ZnSO4). The nucleotides were then dephosphorized by incubation with calf intestinal alkaline phosphatase (CIAP, TaKaRa) for at least 2 h at 37°C. The reactions were then concentrated into 35 μl and analyzed on an Agilent 1260 HPLC machine with an AQ-C18 column of 5-μm particle size, 25 cm × 4.6 mm. The mobile phase was 10 mM KH2PO4, pH 3.7, running at 0.6 ml per min and the detector was set at 280 nm. 5hmC and 5caC nucleoside standards were prepared by dephosphorylation of 5-Hydroxymethyl-dCTP and 5-Carboxy-dCTP (TriLink), and 2′-deoxycytidine (C) and 5-methyl 2′-deoxycytidine (5mC) were bought from Sigma.
UHPLC- MS/MS analysis of mononucleosides
UHPLC-MS/MS analysis of modified mononucleosides was carried out according to Yin et al. (34). It was performed on an Agilent 1290 UHPLC system coupled with a G6410B triple quadrupole mass spectrometer (Agilent Technologies, Palo Alto, CA, USA). An isocratic elution with 5.0% methanol, 95% water and 0.1% formic acid running at 0.25 ml/min was used for UHPLC separation of mononucleosides. The eluate from the column was injected into electrospray ionization-triple quadrupole mass spectrometry. Positive multiple reaction monitoring modes were used: m/z 242→126 for 5mC (collision energy, 5eV); m/z 258→142 for 5hmC (5eV); m/z 256→140 for 5fC (5eV); m/z 272→156 for 5caC (5eV). 2′-deoxyguanosine (dG) in all samples was also measured to normalize the amount of DNA on column. Each sample was analyzed at least three times. The frequency of modified mononucleosides was calculated by corresponding standard curves.
Establishment of TDG KO HEK293T cell lines
Targeting of TDG in HEK293T cells was carried out by using TALEN mediated homologous recombination. TALENs against TDG were designed by online software TAL Effector Nucleotide Targeter 2.0 (35) to target the exon 2 of TDG. The TALENs were composed of domains targeting to the left arm sequence: 5′-TCAGCTATTCCCTTCAGCA-3′ and the right arm sequence: 5′-TCAGTTGTTGAAATGGAAA-3′. TALEN constructs were assembled using FastTALE TALEN Assembly Kit (SiDanSai). The TALENs plasmids and targeting vectors (PGK-hygromycin and PGK-puromycin) were transfected into HEK293T cells using Lipofectamine 2000. Selective medium with 200 μg/ml hygromycin and 1.5 μg/ml puromycin was applied 48 h after transfection and replaced every two days. Individual positive colonies were picked and expanded, which were further characterized by genotyping polymerase chain reaction (PCR) and western blotting using antibody against TDG.
Generation of Gadd45a/Gadd45b double-knockout mouse ES cell lines
Gene targeting of Gadd45a/b was accomplished by using CRISPR/Cas9 system according to Ran et al. (36). The target sequences were designed on the website (http://crispr.genome-engineering.org/) to disrupt the first exon containing the start codon of the Gadd45a or Gadd45b genes. Expression vector px330 containing the Cas9 and mCherry genes was digested with BbsI and the linearized plasmid purified using QIAquick PCR Purification Kit (Qiagen). Paired oligonucleotides for sgRNA at each targeted site (Supplementary Table S1) were annealed and ligated to the linearized vector. The plasmids were confirmed by sequencing.
V6.5 ES cells were cultured on feeder cells with standard ES cell culture conditions. Cells were transfected with px330-sgRNA plasmids using Lipofectamine 2000. After 48 h, mCherry-positive cells were sorted by flow cytometry using BD FACS Aria II (BD) and cultured on feeder layers. After recovering for 4–6 days, individual colonies were picked and genotyped by PCR genotyping and sequencing.
Reduced representation bisulfite sequencing (RRBS)
Reduced representation bisulfite sequencing (RRBS) was performed as described in Guo et al. (32) with slight modification. Briefly, genomic DNA was digested by MspI (Fermentas) and the 200–500 bp DNA fragments were selected to undergo bisulfite conversion. Analysis of RRBS data was performed according to Guo et al. (32), the adaptor-trimmed reads were mapped to the mouse genome (mm9) using Bismark (v. 0.76, http://www.bioinformatics.babraham.ac.uk/projects/bismark/). The methylation level of each single CpG site was calculated using the number of RRBS-measured C (methylated) divided by the sum of measured C (methylated) and T (unmethylated). CpG sites with at least five uniquely mapped reads were chosen for further analysis.
The differentially methylated CpG sites were identified if the absolute methylation level difference was >25% between wild-type (WT) and Gadd45 DKO ES cells (one-tailed Fisher's exact test with P < 0.05, with Benjamini–Hochberg false discovery rate <0.05). The neighboring hypermethylated (or hypomethylated) CpG sites were merged to form a differentially methylated region if there was at most one CpG site between these two hypermethylated (or hypomethylated) CpG sites and the distance between them was <100 bp.
Preparation of the 5fC/5caC-containing plasmid (oxi-5mC plasmid)
For the preparation of oxi-5mC plasmid DNA, 100 ng of M.SssI-methylated pCpGL-CMV-firefly plasmids were oxidized by using 2 μg of the TET2 protein (37) in the reaction buffer (50 mM HEPES, pH 8.0, 50 mM NaCl, 2 mM ascorbic acid, 1mM 2-oxoglutarate, 100 μM Fe(NH4)2(SO4)2, 1 mM adenosine triphosphate (ATP) and 1 mM Dithiothreitol (DTT)) for 1 h at 37°C. The reaction was then added with 1 μg of the TET2 protein for further incubation of 1 h. The plasmid DNA was recovered by phenol–chloroform extraction and the oxidative level of the DNA was tested by M.SssI-assisted bisulfite sequencing (MAB-seq) (32).
Isolation of transfected plasmid
Forty-eight hours after transfection, nuclei were extracted from the transfected HEK293T cells using the Wizard Genomic DNA Purification Kit (Promega) according to the manufacturer's instructions. The plasmid DNA was then isolated using phenol–chloroform extraction.
Bisulfite sequencing
A total of 200 ng of genomic DNA were treated with the EZ DNA Methylation-Direct Kit (Zymo Research). Specific genomic regions were PCR-amplified using Taq HS enzyme (Takara). The PCR products were gel-purified by using Gel Extraction Kit (Qiagen) and cloned into pMD 19-T (Takara) for sequencing. Data were analyzed by an online called BISMA (38) (http://services.ibc.uni-stuttgart.de/BDPC/BISMA/) and duplicated clones were deleted by this software.
M.SssI-assisted bisulfite sequencing (MAB-seq)
MAB-seq was performed to map sequence-specific 5fC and 5caC distribution as described (31,32). A total of 100 ng of genomic DNA were methylated by M.SssI (NEB) following the vendor's instructions. The methylated DNA sample was then purified by phenol–chloroform extraction. Bisulfite conversion, sequencing and data analysis were performed as described above.
Recombinant protein expression and purification
For purification of Flag-tagged proteins, coding sequences were cloned into a modified pcDNA4 (Invitrogen) vector. The expression plasmids were transfected into HEK293T cells using polyethylenimine (PEI, Sigma). After culturing for 48 h, the cells were harvested and lysed in lysis buffer (50 mM Tris, pH 7.5, 500 mM NaCl, 1% NP-40, 1× protease inhibitors without ethylenediaminetetraacetic acid (EDTA)). The recombinant proteins were purified by using the FLAG M2 affinity gel (Sigma) according to the manufacturer's instructions. The eluted proteins were dialyzed against storage buffer (20 mM HEPES, pH 7.4, 50 mM NaCl and 50% glycerol) for 4 h at 4°C and then stored at −80°C.
For purification of Glutathione S-transferase (GST) fusion proteins, GST-tagged proteins were overexpressed in Escherichia coli strain BL21 (DE3) Codon-Plus-RIL (Stratagene). Cells were grown at 37°C in LB medium to an optical density of 0.6–0.8 at 600 nm and then induced with 0.2 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at 20°C overnight. Purification was performed using Glutathione Sepharose 4B (GE Healthcare) according to the manufacturer's instructions.
Purification of TDG used in the in vitro glycosylase assay was described before (39).
GST-pull down assay
To confirm the interaction of TDG and Gadd45a, GST pull-down assay was performed essentially as described (40). A total of 4 μg of the GST–TDG fusion protein purified from E. coli were incubated with 20 μl Glutathione Sepharose 4B beads in the binding buffer [20 mM Tris–HCl (pH 7.9), 0.1 M NaCl, 1 mM EDTA, 5 mM MgCl2, 0.1% NP-40, 1 mM DTT, 0.2 mM phenylmethanesulfonyl fluoride (PMSF) ] in a total volume of 200 μl at 4°C for 2 h. Then 1 μg of Flag-Gadd45a was added to the slurry and incubated at 4°C for another 2 h. The Sepharose beads were washed three times with the washing buffer [20 mM Tris–HCl (pH 7.9), 0.15 M NaCl, 1 mM EDTA, 5 mM MgCl2, 0.1% NP-40, 1 mM DTT, 0.2 mM PMSF] and bound proteins were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and visualized by western blotting.
The interaction of Flag-TDG and GST-Gadd45a was determined using the same protocol.
Yeast two-hybrid assay
To confirm the interaction between Gadd45a and TDG, Gadd45a was cloned in-frame with GAL4 DNA binding domain (GAL4DBD) in the bait vector pGBKT7 (Clontech) and TDG fragments were fused with the GAL4 activation domain (GAL4AD) in the prey vector pGADT7. Yeast was co-transformed with pGBKT7-Gadd45a and pGADT7-TDG. Colonies were selected on the SD medium lacking His, Leu, Trp and adenine according to the recommended protocol (Clontech).
Base release assay
The 60-mer double-stranded oligonucleotide substrates containing different modifications were prepared by annealing of an unlabeled upper strand oligonucleotide 5′-TAGACATTGCCCTCGAGGTACCATGGATCCGATGTCGACCTCAAACCTAGACGAATTCCG-3′ to a 5′-fluorescein-labeled lower oligonucleotide strand 5′-F-CGGAATTCGTCTAGGTTTGAGGTXGACATCGGATCCATGGTACCTCGAGGGCAATGTCTA-3′, where X = T or 5caC.
Multiple turnover base release assays were carried out in a total volume of 200 μl containing 25 nM of the labeled DNA substrate (G•T), 100 nM of unlabeled DNA substrate, 25 nM of recombinant TDG and 1 μM Gadd45a (or BSA) in 1× reaction buffer [50 mM Tris–HCl (pH 8.0), 1 mM EDTA, 1 mM DTT] at 37°C and 20 μl samples were taken at indicated time points. Generated AP-sites were cleaved by the addition of NaOH to a final concentration of 100 mM and heating to 99°C for 10 min. Subsequently, DNA was ethanol precipitated overnight at −20°C in 0.3 M Na-acetate (pH 5.2) and in the presence of 0.4 mg/ml carrier tRNA. The DNA was collected by centrifugation (20 min, 20 000 g, 4°C) and washed in 70% ethanol. Air-dried pellets were resuspended in loading buffer (1× Tris-Borate-EDTA (TBE) buffer, 90% formamide), heated at 99°C for 5 min and then immediately chilled on ice. Reaction products were separated on 15% denaturing polyacrylamide gels in 1× TBE. The fluorescein-labeled DNA was visualized with a Typhoon 9400 (GE Healthcare) and quantified using the ImageQuant TL software (GE Healthcare).
Single turnover base release assays were carried out as described above in a total volume of 200 μl containing 25 nM of the labeled DNA substrate (G•5caC), 250 nM recombinant TDG and 1 μM Gadd45a in 1× reaction buffer [50 mM Tris–HCl (pH 8.0), 1 mM EDTA, 1 mM DTT] at 30°C and 20 μl samples were taken at indicated time points.
RESULTS
Gadd45a activates the expression of a methylated reporter gene in cooperation with Tet and TDG
Active DNA demethylation can be achieved by a concerted action of Tet and TDG (41). We reasoned that other factors previously proposed to contribute to demethylation might exert their effects by modulating the Tet-TDG axis. To test this possibility, we used a cell-based firefly luciferase reporter assay to test anti-gene silencing effects of factors of interest (Supplementary Table S2). The luciferase-reporter plasmids were free of CpGs except for the 0.6-kb CMV promoter (33) regions, which contained 37 CpGs that we fully methylated in vitro by reaction with the CpG-specific bacterial methyltransferase M.SssI prior to transfection. Methylation of the CMV promoter conferred 100-fold repression of the firefly luciferase activity but co-transfection of the methylated reporter plasmid with Tet2 catalytic domain (Tet2CD) together with TDG, increased the activity by 13-fold, thus partially alleviating the repression (Figure 1A). Gadd45a, which we tested as a candidate for a modulatory factor, was able to stimulate the silenced reporter expression by 35-fold, but only when co-transfected with Tet2CD and TDG (Figure 1A). Expression of Gadd45a did not affect the protein levels of Tet2CD and TDG in transfected cells (Supplementary Figure S1). Moreover, its effect was dependent on the enzymatic activities of Tet and TDG because no stimulation effect was observed in co-transfection with the catalytically inactive mutants. By contrast, Gadd45a in combination with Tet and TDG had only a two-fold stimulatory effect on expression of the reporter gene on an unmethylated plasmid (Figure 1B). These observations suggest that Gadd45a cooperates with Tet and TDG in the activation of a methylation-silenced reporter gene.
Figure 1.
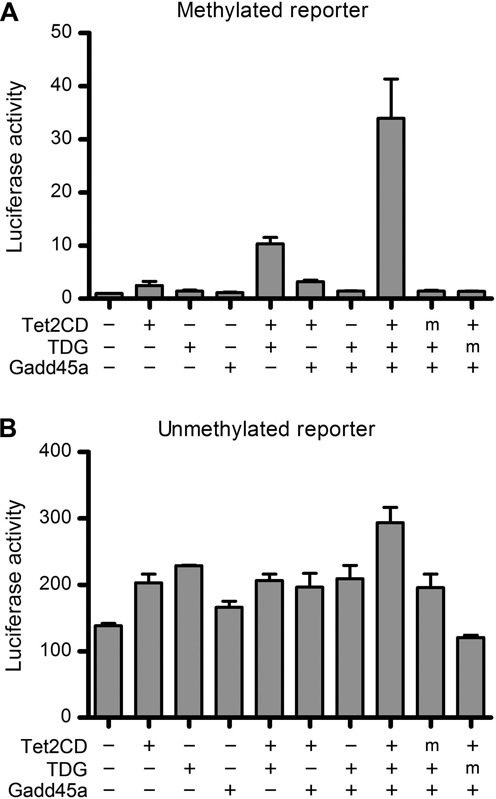
Gadd45a synergizes with Tet and TDG in activation of a methylated reporter gene in a dual luciferase reporter assay. A methylated (A) or unmethylated (B) pCpGL-CMV-firefly luciferase reporter was co-transfected into HEK293T cells with Gadd45a, Tet2CD and TDG in various combinations indicated. Shown is the firefly luciferase activity normalized to the level of control pCpGL-CMV-renilla luciferase relative to that of cells without Tet, TDG and Gadd45 expression, which is set to 1. ‘m’ denotes catalytically inactive mutants of Tet2CD and TDG. Data are represented as the mean ± SEM of three independent experiments.
Reduction of 5fC and 5caC by Gadd45a in HEK293T cells overexpressing Tet2
Since the effect of Gadd45a on methylated reporter activation depends on catalytically active Tet and TDG, we next investigated whether Gadd45a might impact on the functions of Tet and TDG in transfected cells. The occurrence of 5caC in the genomic DNA of HEK293T cells transfected with a Tet enzyme could be detected by HPLC analysis (15). We took advantage of this system to examine whether Gadd45a can regulate the level of 5caC generated in transfected cells. As reported previously (15), 5caC could be detected in Tet2-transfected cells (Figure 2A). However, the 5caC level was reduced to 30% by co-transfection of Gadd45a with Tet2, while the 5mC and 5hmC levels were unchanged. Gadd45a expression did not influence the protein expression of transfected Tet2 and endogenous TDG (Supplementary Figure S2A and B). Drastic reduction in 5fC and 5caC levels was confirmed by triple quadruple mass spectrometry quantification (Figure 2B). The selective removal of 5fC and 5caC but not 5mC and 5hmC could suggest that Gadd45a might not affect the oxidation function of Tet2 but promote the function of the endogenous TDG which is likely rate-limiting in removing genomic 5fC and 5caC from HEK293T cells.
Figure 2.
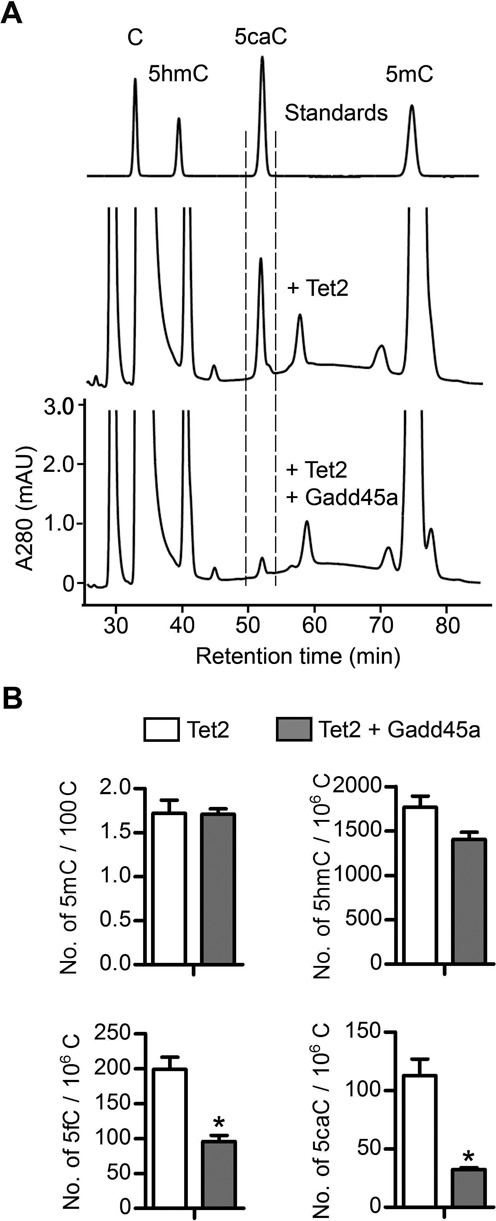
Ectopic expression of Gadd45a reduces genomic 5fC and 5caC generated by Tet2 in transfected HEK293T cells. (A) HPLC analysis of 5caC in the genomic DNA of cells overexpressing full-length Tet2 with and without Gadd45a. The y-axis indicates OD280nm of nucleoside preparations from genomic DNA. Defined nucleosides were used as standards (C, 5mC, 5hmC and 5caC). Dashed lines indicate the elution position of 5caC. (B) Mass spectrometric quantification of modified cytosines (5mC, 5hmC, 5fC and 5caC) in the genomic DNA of cells ectopically expressing Tet2 alone or with Gadd45a. Data are represented as the mean ± SEM from two independent experiments, and are analyzed by two-tailed t-test, P < 0.05 (*).
Gadd45a interacts with TDG
Having demonstrated the functional relationship between Gadd45a and TDG in reporter gene reactivation and the regulatory effect of Gadd45a on the 5caC and 5fC levels in transfected cells, we wondered whether there is a direct protein–protein interaction between them. As shown in Figure 3A, purified Flag-TDG from HEK293T cells could bind with bacterial recombinant protein GST-Gadd45a. The protein association could also be shown with co-immunoprecipitation (Co-IP) assay using extracts from HEK293T cells transfected with epitope-tagged TDG and Gadd45a (Figure 3B). To further confirm and characterize the interaction, we performed yeast two-hybrid assay. As expected, yeasts harboring both a Gadd45a bait and a TDG prey construct could grow on the selective medium lacking His, Leu, Trp and adenine because of the activation of reporter genes due to the interaction between Gadd45a and TDG (Figure 3C). To map the interacting domains, deletion analysis was performed with the bait and prey constructs. We found that the N-terminal domain of Gadd45a (amino acids1–137) mediates the interaction with TDG via a N (amino acids 1–132) and a C-terminal (amino acids 178–397) domain of TDG (Figure 3D). These data provide a confirmation of the physical interaction between Gadd45a and TDG.
Figure 3.
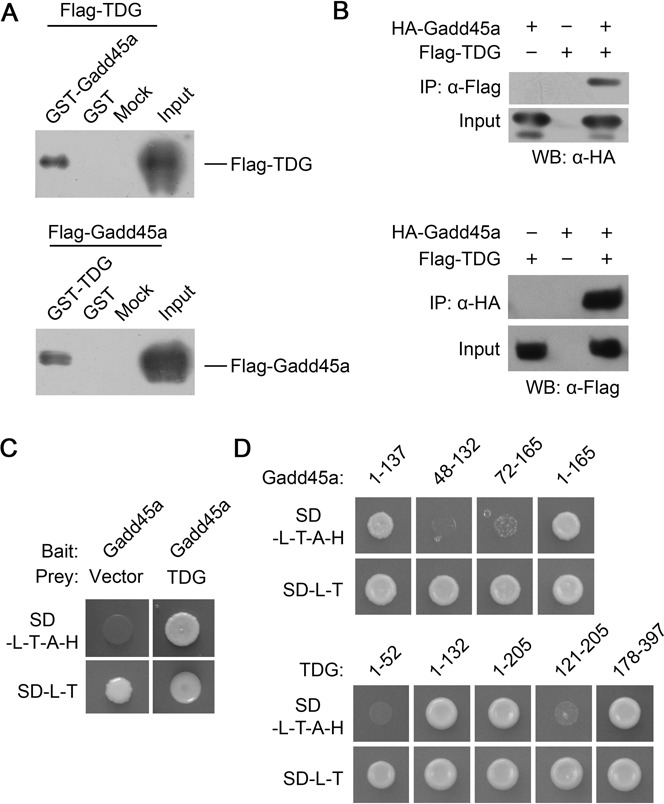
Gadd45a interacts with TDG in vitro. (A) GST and Flag pull-down assay. Western blot analysis of Flag-TDG in fractions obtained from a GST pull-down assay using GST-Gadd45a (lane 1) or GST (lane 2) (upper panel) and viceversa (lower panel). Mock is the control without GST or its fusion. Ten percent of the input was loaded. (B) Co-immunoprecipitation (Co-IP) assay. Immunoprecipitates of lysates of HEK293T cells transfected with HA-Gadd45a and Flag-TDG alone or in combination were resolved by SDS-PAGE and detected by western blotting with the indicated antibodies. (C) Yeast two-hybrid assay. Appearance of colonies in the SD-L-T-A-H selective medium indicates that the reporter genes have been activated due to the interaction between the bait (GAL4DBD-Gadd45a) and prey (TDG-GAL4AD) hybrid proteins. Yeast strains harboring the Gadd45a bait construct and an empty prey vector were used as negative controls. (D) Mapping of Gadd45a-TDG interaction domains. The protein regions tested in yeast two-hybrid assays are indicated above.
The effect of Gadd45a on 5caC removal is exclusively mediated by TDG
TDG is the only known enzyme capable of specifically excise 5fC and 5caC from DNA (15). To determine whether Gadd45a affects the reduction of 5caC in genomic DNA through the endogenous TDG, we generated HEK293T cell lines deficient in TDG. The TDG gene was disrupted by using TALEN technology (Supplementary Figure S3). Exon 2 was replaced in the two alleles, by the hygromycin-resistance gene in one gene copy and by the puromycin-resistance gene in the other. Deletion of this exon would lead to a frame-shift behind the first seven N-terminal codons. Genomic PCR confirmed that the exon 2 sequences had been deleted, with the acquisition of drug selection markers in the established knockout cell line (Figure 4A). Loss of TDG protein expression was confirmed by western blot analysis using an anti-TDG antibody (Figure 4B). As shown in the HPLC analysis of genomic DNA from transfected cells, expression of Gadd45a in TDG-deficient cells did not change the level of 5caC generated by Tet2 (Figure 4C). Mass spectrometry analyses of the genomic DNA samples confirmed that levels of 5fC and 5caC were not changed by Gadd45a expression in TDG-knockout cells (Figure 4D). The possibility that Gadd45a may increase the expression level of Tet2 thus masking its effect by generating higher level of 5caC was ruled out (Supplementary Figure S2C). Additionally, the complete removal of 5caC upon re-expression of ectopic TDG in the knockout cells validated the cellular competence for TDG function. These results indicate that the function of Gadd45a in DNA demethylation is realized exclusively through its effect on TDG.
Figure 4.
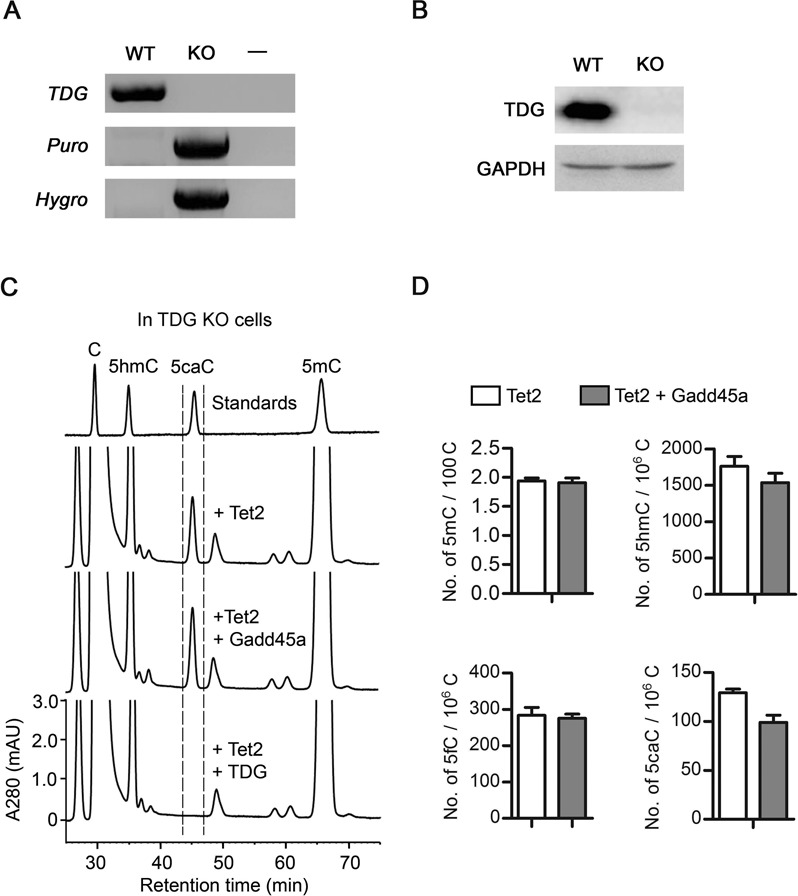
Reduction of genomic 5caC by Gadd45a depends on TDG in transfected HEK293T cells. (A) Confirmation of the TDG gene knockout (KO) in HEK293T cells by PCR genotyping of genomic DNA. The two TDG alleles present in wild-type (WT) cells were replaced by puromycin (Puro) and hygromycin (Hygro) drug selection markers in KO cells (Supplementary Figure S3). (B) Loss of the endogenous TDG protein in the established knockout cell line confirmed by western analysis using anti-TDG antibody. Detection with anti-GAPDH served as a loading control. (C) HPLC analysis of genomic 5caC in TDG-knockout cells expressing ectopic full-length Tet2 alone and together with Gadd45a. Transfection of the TDG control eliminates 5caC generated by Tet2, as previously reported (15). (D) Mass spectrometry quantification of modified cytosines in the genomic DNA of TDG knockout cells ectopically expressing Tet2 alone or Tet2 together with and Gadd45a. Data are represented as the mean ± SEM from two independent experiments.
Gadd45a promotes conversion of 5fC and 5caC to unmodified cytosine in a TDG-dependent manner
As shown above, Gadd45a reduces the buildup of genomic 5fC and 5caC formed by ectopic Tet2 in transfected cells. To directly demonstrate the role of Gadd45a, we prepared in vitro a 5fC/5caC-containing reporter plasmid and transfected the plasmid with the Gadd45a expression plasmid into HEK293T cells. The recovered reporter plasmid DNA was then analyzed by M.SssI-assisted bisulfite sequencing (MAB-seq) which allows to distinguish unmodified cytosines from 5fC/5caC in a specific sequence at single-base resolution (31,32). We first confirmed the efficient methylation (5mC 99.2%) and oxidation (5fC + 5caC; 89.2%) of the CpG sites in the CMV promoter region of the prepared plasmid (oxi-5mC) using bisulfite-seq and MAB-seq (Figure 5A; Supplementary Figure S4). Interestingly, in the recovered plasmid upon co-transfection with Gadd45a, the combined level of 5fC and 5caC was reduced to 58% while the DNA without Gadd45a co-transfection retained the majority of 5fC/5caC (84.3 versus 89.2%, Figure 5B). Consistent with the idea of TDG activity mediating the Gadd45a effect, overexpression of TDG also reduced the 5fC/5caC level to 51%. Moreover, Gadd45a had no effect on the 5fC/5caC level in TDG KO cells (89.5 versus 89.2%, Figure 5B), indicating that the endogenous TDG is essential to mediate the function of Gadd45a in HEK293T cells. Taken together, Gadd45a promotes demethylation by positively impacting TDG for the conversion of 5fC and 5caC into C.
Figure 5.
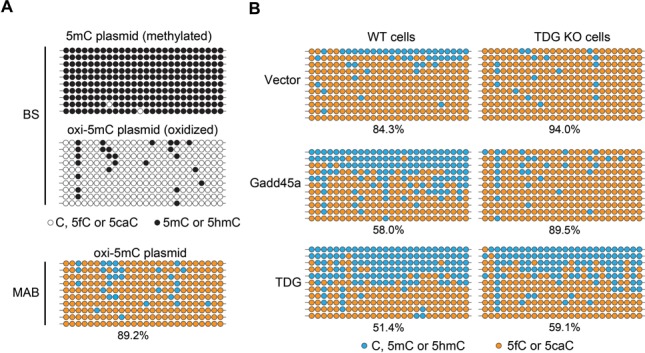
Gadd45a promotes conversion of 5fC and 5caC into unmodified cytosine in a TDG-dependent manner in transfected cells. (A) Characterization of oxidized pCpGL-CMV reporter plasmid DNA (oxi-5mC plasmid). Plasmid DNA was methylated in vitro with CpG methyltransferase M.SssI and then oxidized using human TET2 enzyme to obtain oxi-5mC DNA. BS-seq and MAB-seq profiles show occurrence of expected modifications at CpGs in the CMV promoter region. There was about 10% 5mC or 5hmC remaining in the oxi-5mC DNA due to incomplete oxidization with TET2. The white circles denote unmodified C or higher oxidized forms of 5mC (5fC and 5caC) and the black circles denote 5mC or 5hmC in BS analysis. The blue circles in the MAB-seq profile represent C, 5mC or 5hmC. The orange circles represent higher oxidized forms (5fC or 5caC). (B) MAB-seq analysis of oxidized reporter plasmid DNA recovered from transfected HEK293T cells. The oxi-5mC plasmid DNA was transfected into WT or TDG knockout cells with Gadd45a, TDG or vector control. Percentages of the oxi-5mC bases (5fC and 5caC) are indicated.
Gadd45a/b knockout causes locus-specific hypermethylation in mouse ES cells
If Gadd45a positively regulates Tet/TDG-mediated DNA demethylation in ES cells, Gadd45a deficiency would lead to hypermethylation in genomic regions known to be the target loci of active demethylation. To eliminate the effect of potential functional redundancy with Gadd45b that has 55% protein sequence identity and has a similar mRNA expression level with Gadd45a in ES cells, we generated Gadd45a/Gadd45b double-knockout (DKO) mouse ES cell lines using the CRISPR/Cas9 system. Nucleotide insertions and deletions at the targeted sites were identified which would lead to frame shift mutation or early termination, thus gene inactivation (Supplementary Figure S5). To compare genomic methylation between DKO and the WT ES cells, we performed genome-scale single-base-resolution analysis using RRBS. In the DKO cells, no obvious methylation difference was observed at the genome scale (Supplementary Figure S6). Both the mutant and WT ES cells have a CpG methylation frequency of around 33% in the RRBS-covered genome part and a similar overall gene distribution pattern. However, 68 specific regions were found hypermethylated which contained more than 4 CpG sites of significantly increased methylation (Supplementary Table S3), as exemplified by the genomic loci from the Plagl1 and Cilp2 genes (Figure 6A). In sharp contrast, no hypomethylated regions could be identified in these DKO ES cells. Primer-based conventional bisulfite sequencing confirmed significantly elevated methylation in the corresponding regions of Plagl1 and Cilp2 (Figure 6B). Interestingly, these two regions also gained hypermethylation in control ES cells deficient in TDG (Figure 6B). Since 5mC and 5hmC are not distinguishable in conventional bisulfite sequencing, we performed Tet-assisted bisulfite sequencing (TAB-seq) to determine 5hmC in the Plagl1 and Cilp2 regions. We found 5hmC very rare and no increase in both cell lines deficient in Gadd45 or TDG (Supplementary Figure S7). This indicates that the hypermethylation was actually composed of 5mC rather than 5hmC. Further inspection of the Gadd45 deficiency–caused hypermethylated regions revealed that most of them were overlapped with 5hmC and 5fC-enriched regions previously mapped in ES cells (24,42). Among the 68 hypermethylated regions, 50 are overlapped with 174 655 regions reported to be enriched in 5hmC and 48 are overlapped with 120 052 regions enriched in 5fC (Figure 6C). By comparison, much fewer regions randomly chosen from the RRBS-covered sequences are overlapped with 5hmC or 5fC regions. Overall, these results support the conclusion that Gadd45 proteins functionally cooperate with the Tet and TDG enzymes in demethylation of genomic targets in mouse ES cells.
Figure 6.
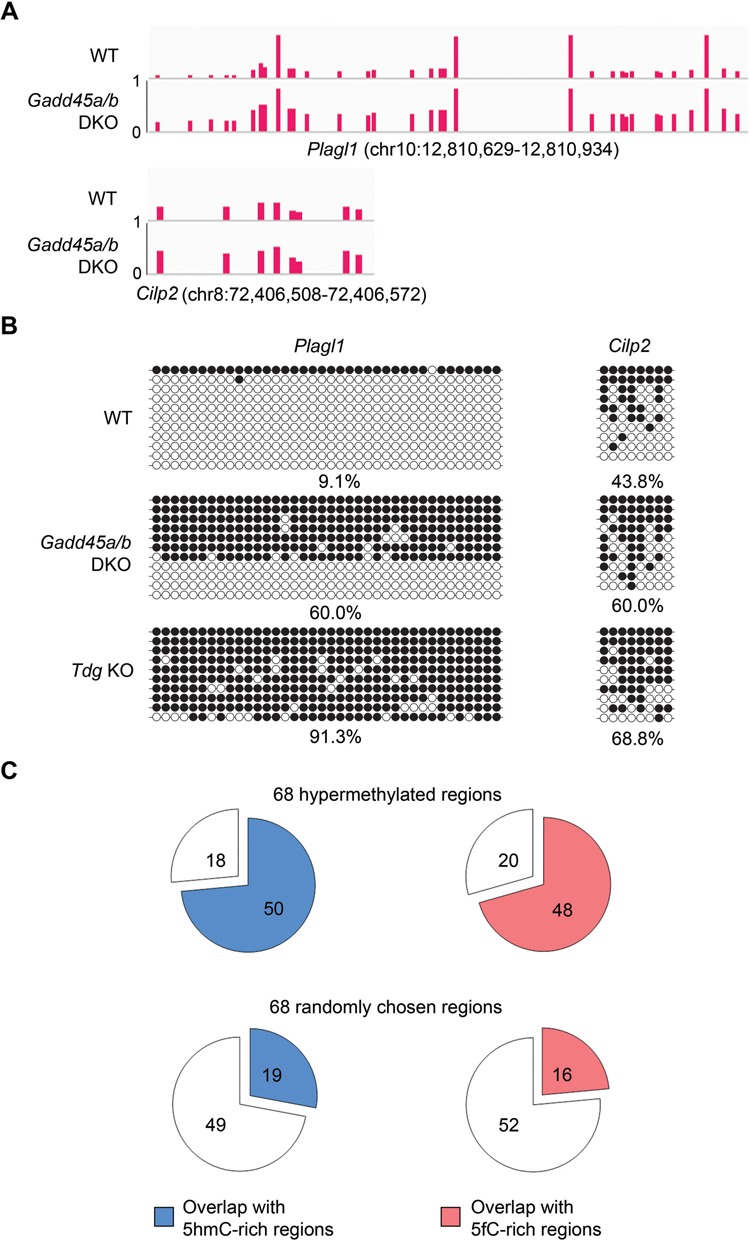
Locus-specific hypermethylation in Gadd45a/b DKO mouse ES cells. (A) Representative loci of hypermethylation in Gadd45a/b DKO ES cells mapped by RRBS analysis. Vertical bars indicate the methylation level (0–1) at individual CpGs in the genomic regions indicated. (B) Confirmation of increased methylation at the selected loci by bisulfite sequencing in WT, Gadd45a/b DKO and Tdg KO ES cells. Black and white circles denote methylated and unmethylated CpG sites respectively. Percentages of the CpGs sites resistant to bisulfite conversion are indicated below. (C) Comparison of the 68 hypermethylated regions identified in Gadd45a/b DKO ES cells (top pie charts) or 68 randomly chosen RRBS covered regions (bottom) with the pool of 5hmC-rich regions (24,42) (left) and the pool of 5fC-rich regions (24) (right) previously identified in Tdg KO ES cells.
DISCUSSION
Mechanisms of DNA demethylation have been intensively studied but remain elusive. There exist a wide variety of factors that belong to different pathways but appear to regulate DNA methylation negatively (13). As biochemical evidence revealing their participation in the demethylation process is lacking in most cases, it is plausible that the role of these factors in DNA methylation control may be indirect. In the presented work, we have examined the role of Gadd45a in the context of the recently proposed Tet-TDG mediated demethylation pathway (41). Our data indicate that Gadd45a is involved in oxidative demethylation by regulating the activity of TDG in cells.
The Tet family of proteins, or DNA dioxygenases by nature, have emerged as the most plausible enzymes for initiation of active demethylation, owing to their ability to oxidize the methyl group of 5mC (14,43). While the simple oxidation of 5mC to 5hmC will demethylate DNA passively through replicative dilution, further oxidation to 5fC and 5caC generates substrates for TDG, which will remove these bases, triggering their replacement by unmethylated cytosine, potentially via a conventional BER process (19,41). As demethylation is a highly regulated process, we reasoned that auxiliary factors including those previously implicated in demethylation may function to modulate the activity of the Tet-TDG system. Among such factors is the multifunctional Gadd45a initially identified to antagonize methylation by its activation effect on methylated reporter genes (27). Gadd45a was proposed to promote demethylation potentially through XPG-dependent DNA repair (27,44) or BER of AID-based deamination products (22,29). Our finding that Gadd45a serves as a regulator in the Tet-TDG pathway is supported by several lines of evidence. First, Gadd45a synergizes with Tet and TDG to activate methylated reporter gene in transfected cells (Figure 1), and overexpression of Gadd45a alone cannot reactive the methylated reporter gene, consistent with the previous report by Jin et al. (30). Second, Gadd45a interacts with TDG physically in vitro (Figure 3). Third, Gadd45a potentiates TDG glycosylase to remove 5fC and 5caC from genomic DNA of transfected HEK293T cells (Figure 2). Forth, endogenous TDG is required to mediate the effect of ectopic Gadd45a in decreasing Tet2-generated 5caC (Figures 4 and 5). Lastly, deletion of Gadd45a/b in mouse ES cells leads to hypermethylation at specific genomic loci which also gain increased methylation in Tdg-deficient cells and are enriched in 5fC (Figure 6). Since the molecular functions of Gadd45a appear to be diverse (26), it is noteworthy that Gadd45a promotes demethylation specifically through TDG in our studies. However, since the Gadd45 deficiency-caused hypermethylated regions are much fewer than the previously mapped 5hmC and 5fC enriched regions in ES cells (24,42) (68 versus 174 655 and 120 052), Gadd45 may only contribute to the control of a small fraction of Tet-TDG regulated genomic targets.
It is curious how Gadd45a exactly regulates TDG activity in cells. In light of the direct interaction between the two proteins and the stimulation of TDG-dependent DNA demethylation by Gadd45a in cells, we examined the possibility that Gadd45a might impact on the enzymatic activity of TDG in vitro using purified proteins. Under multiple-turnover conditions, addition of an excess of Gadd45a stimulated initial processing by as well as overall efficiency of TDG but it didn't improve its rate limiting turnover (Supplementary Figure S8A and B). The stimulation by Gadd45a, however, was in a similar range as observed when a molar equivalent of BSA was added to the reaction, indicating a non-specific effect of the protein on the structural integrity of TDG. Likewise, under single-turnover conditions, the addition of Gadd45a had no measurable effect on TDG activity, suggesting that Gadd45a does not modulate TDG substrate recognition potential (Supplementary Figure S8C and D). Taken together, Gadd45a does not appear to stimulate TDG in substrate binding, processing or turnover in a specific manner. We therefore favor a model whereby Gadd45a promotes Tet-TDG-mediated demethylation by coordinating the recruitment of TDG into demethylation complexes. Gadd45a (and Gadd45b) could act as a scaffold protein which interacts with other epigenetic regulators (26) and directs TDG to the target loci in the context of chromatin to efficiently excise 5fC and 5caC. This is consistent with the recently proposed model in which recruitment of both Gadd45a and TDG is implicated in demethylation of the tumor suppressor gene TCF21 targeted by a long non-coding RNA (45). Further investigation will be needed to elucidate the exact molecular mechanism by which Gadd45 proteins regulate the function of the TDG enzyme in specific chromatin contexts.
ACCESSION NUMBER
The Gene Expression Omnibus accession number for the RRBS data reported in this manuscript is GSE62496.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank M. Rehli for providing pCpGL-basic and L. Li for Tdg knockout ES cell line. We also thank Y. Xu and L. Hu for providing the recombinant human TET2 protein.
FUNDING
Breakthrough Project of Strategic Priority Program of the Chinese Academy of Sciences [XDB13000000]; National Science Foundation of China [31230039, 31221001]; National Science & Technology Major Project ‘Key New Drug Creation and Manufacturing Program’ of China [2014ZX09507-002 to G.-L.X.]. Funding for open access charge: Breakthrough Project of Strategic Priority Program of the Chinese Academy of Sciences [XDB13000000]; National Science Foundation of China [31230039, 31221001]; National Science & Technology Major Project ‘Key New Drug Creation and Manufacturing Program’ of China [2014ZX09507-002 to G.-L.X.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 3.Simonsson S., Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat. Cell Biol. 2004;6:984–990. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- 4.Seisenberger S., Peat J.R., Hore T.A., Santos F., Dean W., Reik W. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013;368:20110330. doi: 10.1098/rstb.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kangaspeska S., Stride B., Metivier R., Polycarpou-Schwarz M., Ibberson D., Carmouche R.P., Benes V., Gannon F., Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 6.Hajkova P., Erhardt S., Lane N., Haaf T., El-Maarri O., Reik W., Walter J., Surani M.A. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 7.Mayer W., Niveleau A., Walter J., Fundele R., Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 8.Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W., Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 9.Gu T.P., Guo F., Yang H., Wu H.P., Xu G.F., Liu W., Xie Z.G., Shi L., He X., Jin S.G., et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 10.Wossidlo M., Nakamura T., Lepikhov K., Marques C.J., Zakhartchenko V., Boiani M., Arand J., Nakano T., Reik W., Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 11.Walsh C.P., Xu G.L. Cytosine methylation and DNA repair. Curr. Top. Microbiol. Immunol. 2006;301:283–315. doi: 10.1007/3-540-31390-7_11. [DOI] [PubMed] [Google Scholar]
- 12.Ooi S.K., Bestor T.H. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Wu H., Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastor W.A., Aravind L., Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maiti A., Drohat A.C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohli R.M., Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs A.L., Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121:1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortazar D., Kunz C., Selfridge J., Lettieri T., Saito Y., MacDougall E., Wirz A., Schuermann D., Jacobs A.L., Siegrist F., et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 22.Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., Le Coz M., Devarajan K., Wessels A., Soprano D., et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen L., Wu H., Diep D., Yamaguchi S., D'Alessio A.C., Fung H.L., Zhang K., Zhang Y. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song C.X., Szulwach K.E., Dai Q., Fu Y., Mao S.Q., Lin L., Street C., Li Y., Poidevin M., Wu H., et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fornace A.J. Jr, Alamo I. Jr, Hollander M.C. DNA damage-inducible transcripts in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niehrs C., Schafer A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012;22:220–227. doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Barreto G., Schafer A., Marhold J., Stach D., Swaminathan S.K., Handa V., Doderlein G., Maltry N., Wu W., Lyko F., et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 28.Ma D.K., Jang M.H., Guo J.U., Kitabatake Y., Chang M.L., Pow-Anpongkul N., Flavell R.A., Lu B., Ming G.L., Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rai K., Huggins I.J., James S.R., Karpf A.R., Jones D.A., Cairns B.R. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin S.G., Guo C., Pfeifer G.P. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu X., Zhang L., Mao S.Q., Li Z., Chen J., Zhang R.R., Wu H.P., Gao J., Guo F., Liu W., et al. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell. 2014;14:512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Guo F., Li X., Liang D., Li T., Zhu P., Guo H., Wu X., Wen L., Gu T.P., Hu B., et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15:447–458. doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Klug M., Rehli M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics. 2006;1:127–130. doi: 10.4161/epi.1.3.3327. [DOI] [PubMed] [Google Scholar]
- 34.Yin R., Mao S.Q., Zhao B., Chong Z., Yang Y., Zhao C., Zhang D., Huang H., Gao J., Li Z., et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 2013;135:10396–10403. doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- 35.Doyle E.L., Booher N.J., Standage D.S., Voytas D.F., Brendel V.P., Vandyk J.K., Bogdanove A.J. TAL effector-nucleotide targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu L., Li Z., Cheng J., Rao Q., Gong W., Liu M., Shi Y.G., Zhu J., Wang P., Xu Y. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155:1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 38.Rohde C., Zhang Y., Reinhardt R., Jeltsch A. BISMA–fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010;11:230. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunz C., Focke F., Saito Y., Schuermann D., Lettieri T., Selfridge J., Schar P. Base excision by thymine DNA glycosylase mediates DNA-directed cytotoxicity of 5-fluorouracil. PLoS Biol. 2009;7:e91. doi: 10.1371/journal.pbio.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen T.N., Goodrich J.A. Protein-protein interaction assays: eliminating false positive interactions. Nat. Methods. 2006;3:135–139. doi: 10.1038/nmeth0206-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu G.L., Walsh C.P. Enzymatic DNA oxidation: mechanisms and biological significance. BMB Rep. 2014;47:609–618. doi: 10.5483/BMBRep.2014.47.11.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu M., Hon G.C., Szulwach K.E., Song C.X., Zhang L., Kim A., Li X., Dai Q., Shen Y., Park B., et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan L., Shi Y.G. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitz K.M., Schmitt N., Hoffmann-Rohrer U., Schafer A., Grummt I., Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol. Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Arab K., Park Y.J., Lindroth A.M., Schafer A., Oakes C., Weichenhan D., Lukanova A., Lundin E., Risch A., Meister M., et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol. Cell. 2014;55:604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


