Climate change can have widespread and devastating impacts, but little is known about the effects of this change on the genetics of natural populations. We examined the genetics of two populations of an annual plant (Brassica rapa) before and after an extended drought in California. We found limited genetic differences between pre-drought ancestors and post-drought descendants, indicating that the two populations did not become more genetically similar, despite a greater overlap in reproductive timing after the drought. The findings of this study contribute to an understanding of the genetic impacts of climate change, which is critical in optimizing conservation efforts.
Keywords: Brassica rapa, climate change, drought, gene flow, phenological isolation, population structure, resurrection study
Abstract
The impact of environmental change on population structure is not well understood. This study aimed to examine the effect of a climate change event on gene flow over space and time in two populations of Brassica rapa that evolved more synchronous flowering times over 5 years of drought in southern California. Using plants grown from seeds collected before and after the drought, we estimated genetic parameters within and between populations and across generations. We expected that with greater temporal opportunity to cross-pollinate, due to reduced phenological isolation, these populations would exhibit an increase in gene flow following the drought. We found low but significant FST, but no change in FST or Nm across the drought, in contrast to predictions. Bayesian analysis of these data indicates minor differentiation between the two populations but no noticeable change in structure before and after the shift in flowering times. However, we found high and significant levels of FIS, indicating that inbreeding likely occurred in these populations despite self-incompatibility in B. rapa. In this system, we did not find an impact of climate change on gene flow or population structuring. The contribution of gene flow to adaptive evolution may vary by system, however, and is thus an important parameter to consider in further studies of natural responses to environmental change.
Introduction
Climate change is a major environmental concern and one of the greatest threats to biodiversity (Williams et al. 2008; Bellard et al. 2012). Climate can influence natural systems from a genetic to an ecosystem level, affecting individuals' fitness and species' survival, and is a major factor in determining community composition and ecosystem function (Bellard et al. 2012). The complex relationships between biotic and abiotic factors, combined with the intricate responses of species to a changing climate and other threats to biodiversity, make understanding the current and future impacts of global climate change difficult as well as of great importance (Parmesan and Yohe 2003; Pearson and Dawson 2003; Walther 2010).
Climate change can act as a strong selective pressure in natural populations (Jump and Peñuelas 2005; Gienapp et al. 2008; Hoffmann and Sgró 2011). Several recent studies have illustrated the potential for rapid evolution in response to changes in species' surroundings (Huey et al. 2000; Stockwell et al. 2003; Franks et al. 2007). Adaptive responses in both plants and animals frequently involve the evolution of phenological traits (Fitter and Fitter 2002; Peñuelas et al. 2002; Menzel et al. 2006; Hoffmann et al. 2010), which often depend on seasonal cues, such as temperature and/or precipitation conditions (Franks et al. 2007; Jarrad et al. 2008). Consequently, changes in climate patterns can have major impacts on phenology, and evolutionary changes in phenology may further affect adaptive ability by altering patterns of gene flow between populations (Garant et al. 2007; Franks and Weis 2009). For example, in populations that are differentially affected by changes in environmental conditions, phenology (i.e. timing of reproduction) may become more or less synchronized, impacting the opportunity for reproduction across populations.
Generally, gene flow is expected to reduce local adaptation by homogenizing populations found in differing environments, or by spreading detrimental alleles across populations (Ellstrand et al. 1989; Holt and Gomulkiewicz 1997). However, gene flow may also serve to introduce potentially adaptive alleles to populations, and increase genetic diversity, which natural selection can act upon to provide an evolutionary response (Ellstrand et al. 1989; Räsänen and Hendry 2008; North et al. 2011; Sexton et al. 2011). An improved understanding of both the effects of climate on evolutionary changes in phenology, and the consequences of these phenological changes for gene flow and local adaptation, will be valuable in predicting how species may respond to changes in climate (Garant et al. 2007).
Adaptation can provide a response to changes in climate in some natural populations (Franks et al. 2014). Franks et al. (2007) found that an extended drought in southern California led to an adaptive response in the flowering times of two populations of Brassica rapa (field mustard). The drought, lasting from 2000 to 2004, corresponded to a shift to earlier mean flowering time in both populations, causing an abbreviated growing season and resulting in more synchronous flowering times between the populations following the drought (Franks et al. 2007; Franks and Weis 2009).
Seeds were collected from both sites in May 1997 (prior to the drought) and June 2004 (immediately following the drought), at the end of the growing season for both populations in these years. A parent-offspring analysis from a greenhouse study demonstrated an additive genetic basis for this evolutionary change in flowering time (Franks et al. 2007). Additionally, further analysis has shown a decrease in phenological isolation of the two sites between 1997 (Isolation Index = 33 %) and 2004 (Isolation Index = 14 %) as a result of the change in flowering time (Franks and Weis 2009).
The current study focuses on the impacts of the change in flowering times on the genetics of these natural populations. We analysed the population genetics of this system before and after the drought to examine genetic structuring and gene flow across this phenological change. While previous studies have used population genetic data to infer historic levels of gene flow and genetic structure, this study is unique in using the ‘resurrection approach’ (Franks et al. 2008) of comparing population genetic patterns in ancestors and descendants using stored seeds.
Based on previous analyses of this system (Franks et al. 2007; Franks and Weis 2009), we expected to find an increase in gene flow between the populations following the drought, corresponding to decreased phenological, and potentially reproductive, isolation. Given the long-distance (>5 km) pollinator-mediated pollen dispersal found in related species (Devaux et al. 2005, 2007), we expected that our sites (3 km apart) would be connected by some degree of gene flow. Regardless, our approach allowed us to determine genetic changes that occurred both within and between populations over time. Changes in population structure would not only elucidate how an adaptive evolutionary change influenced genetic patterns, but may also have implications for the ability of these populations to further evolve in response to continued changes in climate. The results of this study can contribute to the growing understanding of natural responses to climate change events, which will be necessary in aiding in the conservation of species as the environment continues to change.
Methods
Study system
Brassica rapa is a weedy plant, native to Eurasia but currently distributed worldwide with cultivars and naturalized populations (Suwabe et al. 2002). The populations of interest in this study are found in southern California, where B. rapa was introduced ∼300 years ago and has become naturalized (Franks et al. 2007). In these populations, B. rapa is an annual plant, produces perfect flowers and is self-incompatible. Once an individual plant has begun to flower, many new flowers may open each day, each flower lasting ∼3 days, for the remainder of the growing season. In southern California, B. rapa germinates in the winter, flowers in the spring and continues to produce flowers until the end of the rainy season (late spring to summer). Flowers are pollinated by many species of vagile insects, such as bees and flies, allowing for the dispersal of pollen across these insects' foraging range (Davis et al. 1994; Warwick et al. 2003).
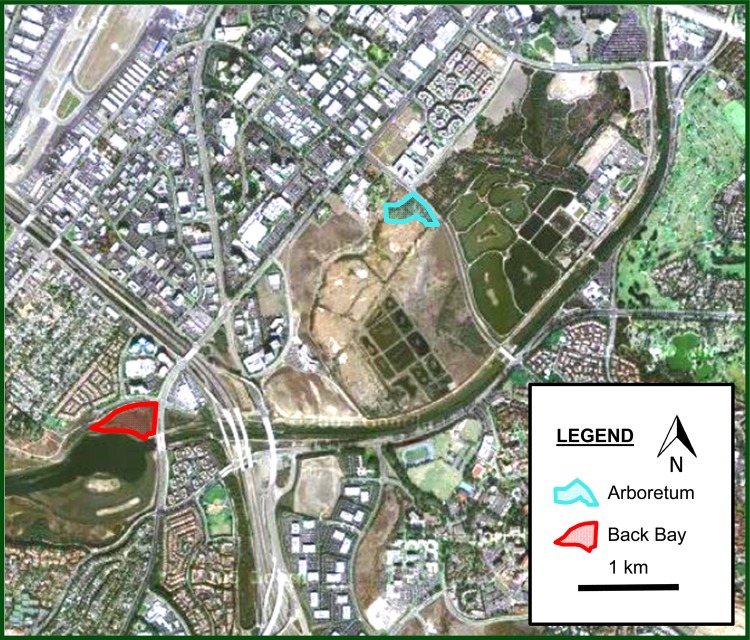
The populations were located in upper Newport Harbor Back Bay (BB) and near the San Joaquin Marsh Arboretum (Arb), in Irvine, CA, USA (Fig. 1). The two locations are ∼3 km apart and are characterized by differing soil conditions. Back Bay soil is sandy and dry in comparison to Arb, where the soil has a higher water table and clay content, aiding in moisture retention (Franke et al. 2006). These two populations also differed in their flowering time ranges, with BB flowering earlier than Arb prior to the drought (Franks et al. 2007). Both populations comprised hundreds to many thousands of individuals both before and after the drought.
Figure 1.
Map of Irvine, California showing the location of the two populations: Arboretum and Back Bay.
Samples
Seeds collected from individuals in the Arb and BB populations in 1997 and 2004 were stored at 4 °C. Over 10 000 seeds were collected from individuals across both populations to obtain representative samples of the gene pools. Seeds from this stock were grown for tissue collection on light carts in the Franks laboratory at Fordham University. Plants were watered daily and fertilized weekly yielding 259 total samples (56 Arb 1997, 39 BB 1997, 89 Arb 2004 and 75 BB 2004) (Table 1).
Table 1.
Genetic diversity characteristics by population and year: population size (N), per cent polymorphic loci (P), number of PA, number of alleles per locus (A), expected heterozygosity (He), observed heterozygosity (Ho) and inbreeding coefficient (FIS). Standard error is provided in parentheses. An asterisk indicates significance from zero at P < 0.05.
| Population | Year | N | P (%) | PA | A | He | Ho | FIS |
|---|---|---|---|---|---|---|---|---|
| BB | 1997 | 39 | 90 | 0 | 3.700 (0.633) | 0.517 (0.071) | 0.332 (0.057) | 0.369* |
| BB | 2004 | 75 | 100 | 8 | 5.000 (0.715) | 0.503 (0.066) | 0.349 (0.065) | 0.309* |
| Arb | 1997 | 56 | 100 | 4 | 4.200 (0.629) | 0.490 (0.065) | 0.332 (0.065) | 0.330* |
| Arb | 2004 | 89 | 90 | 3 | 4.100 (0.737) | 0.453 (0.068) | 0.301 (0.066) | 0.340* |
DNA extraction
Leaf tissue was collected from individuals following the development of true leaves. Approximately 50 mg of tissue per individual was removed, dried and stored in silica gel until extraction. Dried tissue was homogenized using Qiagen's TissueLyser system (Qiagen, Valencia, CA, USA). DNA was extracted using a Qiagen DNeasy plant kit following manufacturer's DNeasy 96 Protocol. This extraction process yielded 200 µL of eluted DNA, which was stored at −20 °C.
Microsatellite analysis
Ten microsatellite loci were chosen based on repeatable amplification success and high degree of polymorphism: BN12A (Szewc-McFadden et al. 1996), Na10-A08, Na10-D09, Na10-G10, Ni4-A03, Ol10-D08, Ra2-E04, Ra2-E12 (Lowe et al. 2002, 2004), BRMS-040 and BRMS-037 (Suwabe et al. 2002). PCR was carried out in 12.5 µL reactions using TC-5000 thermal cyclers (Techne, Bibby Scientific Ltd, Staffordshire, UK) to isolate and amplify microsatellite loci. These reactions included 0.5 µL of unlabelled forward and reverse primers, 0.5 µL of template DNA, RNase-free water and AmpliTaq Gold 360 Master Mix (Life Technologies Inc., Grand Island, NY, USA). Cycling protocol began with an initial denaturation at 95 °C for 5 min, followed by 30 cycles of amplification at 95 °C for 30 s, optimized annealing temperatures [see Supporting Information] for 30 s, and 72 °C for 30 s. This concluded with a final extension at 72 °C for 7 min.
A second PCR was run to incorporate a fluorescent label (WellRED Dye D2, D3 or D4) (Sigma-Aldrich Co., St. Louis, MO, USA) using 0.5 µL reverse primer and 0.5 µL fluorescently labelled primer (M13F(-20) [D2], M13F(-40) [D3] or M13R [D4]) corresponding to the tag sequence added to the 5′ end of the initial forward primer. This was included in a reaction with 0.5 µL of 1 : 100 diluted PCR product from the first PCR, along with RNase-free water and AmpliTaq Gold 360 Master Mix. This second PCR followed the same cycling protocol as the first. PCR products were run on 1 % agarose gel to confirm the presence of target microsatellite fragments. Fragment length was then analysed with a Beckman Coulter CEQ 8800 using Genetic Analysis System v.9.0 (Beckman Coulter, Fullerton, CA, USA). Samples were run with 0.5 µL of Beckman Coulter 400 bp Size Standard and PCR product at volumes according to their fluorescent label: 2.5 µL of M13F(-20), 2.5 µL of M13F(-40) and 1.5 µL of M13R. Alleles were scored using GENEMARKER v.1.90 (SoftGenetics, State College, PA, USA).
Statistical analysis
GENALEX v.6.41 (Peakall and Smouse 2006) was used to determine measures of microsatellite diversity, including number of alleles (A), expected heterozygosity (He), observed heterozygosity (Ho) and inbreeding coefficient (FIS) for each locus. ARLEQUIN v.3.5.1.3 (Excoffier and Lischer 2010) was then used to calculate Hardy–Weinberg Equilibrium (HWE) and to test for linkage disequilibrium (LD) across all loci. Departure from HWE was determined for each locus using the Markov chain method with a length of 1.0 × 105 steps (Guo and Thompson 1992). Linkage disequilibrium was determined using an Expectation Maximization algorithm with 1.0 × 104 permutations (Lewontin and Kojima 1960; Slatkin 1994; Slatkin and Excoffier 1996). The frequency of null alleles for a locus was estimated using an Expectation Maximization algorithm (Dempster et al. 1977) in GENEPOP v 4.0.10 (Raymond and Rousset 1995; Rousset 2008). These parameters describe the loci and can indicate the influence that each has in defining measures of structure in these populations.
GENALEX was also used to determine measures of allelic diversity within these populations including polymorphism (P) across all loci, the number of private alleles (PA), A, He and Ho. FIS was calculated using ARLEQUIN for each population. F-statistics, such as FIS (inbreeding) and FST (fixation), are used to describe the fixation of alleles for individuals in a population, and for populations in a metapopulation, respectively (Wright 1951; Weir and Cockerham 1984). Several analogues of FST have been developed to account for the mutational characteristics of microsatellites (Slatkin 1995; Michalakis and Excoffier 1996; Hedrick 2005; Jost 2008). In addition to FST, we also calculated ϕST (Michalakis and Excoffier 1996), which takes into account the step-wise mutation model believed to be characteristic of many microsatellites. Because FST and gene flow are inversely related (Wright 1951), we used FST to test the hypothesis that gene flow increased following the evolutionary shift to greater flowering synchrony between the populations.
Pairwise comparisons of FST and ϕST, as well as AMOVA were performed in GENALEX. To determine whether levels of fixation and gene flow changed across this period, we compared FST values across these years. Significance was established using 95 % confidence intervals (CI) as determined through 1000 bootstrapping replicates to estimate FST using GDA v.1.1 (Lewis and Zaykin 2002).
Bayesian estimates of genetic clustering probabilistically assigns individuals to populations defined by allele frequencies at multiple loci, and were determined using STRUCTURE v.2.3.2.1 (Pritchard et al. 2000) for 5.0 × 104 burn-in repetitions and 1.0 × 106 MCMC simulations at four iterations. This allowed for an estimate of the number of genetic units, K, following Evanno et al. (2005) using STRUCTURE HARVESTER web v.0.6.92 (Earl and vonHoldt 2012). Outputs of four iterations at K = 3 were run in CLUMPP v.1.1.2 (Jakobsson and Rosenberg 2007) to align these clustering results, and were then visualized using DISTRUCT v.1.1 (Rosenberg 2004).
Data were reanalysed using only loci in HWE in each population [see Supporting Information], as the analyses assume that these microsatellites are acting as neutral markers and a deviation from HWE may be an indication of an effect of selection. The same procedures were used to estimate FIS within each population, FST and ϕST between Arb and BB in 1997 and 2004 and to compare FST between the 1997 and 2004 generations. Results for these analyses are noted along with the results of analyses run with all 10 loci.
Results
Allelic characteristics
Across the populations, the loci analysed showed a mean polymorphism of 95 % per population (Table 1). Between two and nine alleles were found per locus (mean = 5.8 alleles per locus) [see Supporting Information] and between zero and eight PA were found in each population (Table 1). Of the 38 polymorphic loci (for 10 loci across four populations), 26 exhibited departure from HWE (P < 0.05). Out of 180 total locus pairs, 22 pairs across all populations showed LD. However, these locus pairs were not linked in each population. The estimated frequency of null alleles ranged from <1.0 × 10−4 to 0.3558 for all loci. The loci in HWE were found to have a lower frequency of null alleles. Therefore, supplementary analyses performed using only loci in HWE, also minimized the influence of allelic dropout.
Intra-population characteristics
We found high genetic diversity, and some changes in the number of PA, but no significant change in overall genetic diversity over time [see Supporting Information]. In the BB population, the number of PA increased from zero to eight between the sampling years (Table 1). The mean number of alleles per locus estimated for these samples did not change significantly (3.7 ± 0.633 (±SE) in 1997 to 5.0 ± 0.715 in 2004) (Table 1) [see Supporting Information]. However, in Arb, the number of PA decreased from four to three, with no significant change in the number of alleles per locus (4.2 ± 0.629 in 1997 and 4.1 ± 0.737 in 2004) (Table 1) [see Supporting Information].
All populations showed fewer than expected heterozygotes and high levels of inbreeding (FIS) (Table 1). FIS was significantly greater than zero for all populations and generations (Table 1). Levels of FIS did not change significantly over time [see Supporting Information]. Measures of FIS using only loci in HWE showed lower levels of inbreeding (between 0.065 for Arb97 and 0.085 for BB04) [see Supporting Information] than the analyses performed using all 10 loci (Table 1), which is expected as random mating is an assumption of HWE.
Genetic structure and gene flow
We estimated fixation between all pairs of populations (BB 97, BB 04, Arb 97 and Arb 04) as FST and ϕST. Fixation between populations was low but significant for all pairs except BB 97 and BB 04. FST ranged from 0.027 (between Arb 97 and Arb 04) to 0.039 (BB 97 and Arb 04) (Table 2). Pairwise comparisons of ϕST ranged from 0.067 (Arb 97 and Arb 04) to 0.108 (Arb 04 and BB 04).
Table 2.
Pairwise estimates of fixation (FST, ϕST) and gene flow (Nm) between populations and generations. F statistics (FST in (a) and ϕST in (b)) given below the diagonal and Nm given above. F statistics significantly different from zero (P < 0.05) are indicated with an asterisk.
| (a) FST | Arb 04 | Arb 97 | BB 04 | BB 97 | (b) ϕST | Arb 04 | Arb 97 | BB 04 | BB 97 |
|---|---|---|---|---|---|---|---|---|---|
| Arb 04 | 9.173 | 6.245 | 6.201 | Arb 04 | 3.488 | 2.057 | 2.404 | ||
| Arb 97 | 0.027* | 7.400 | 8.292 | Arb 97 | 0.067* | 2.265 | 3.001 | ||
| BB 04 | 0.038* | 0.033* | 28.961 | BB 04 | 0.108* | 0.099* | 38.347 | ||
| BB 97 | 0.039* | 0.029* | 0.009 | BB 97 | 0.094* | 0.077* | 0.006 |
In addition to pairwise comparisons, we used AMOVA to measure FST and ϕST in both 1997 and 2004. We found that estimates of FST and ϕST were low but significant and increased slightly from 1997 to 2004 (Table 3) [see Supporting Information]. AMOVA calculations of loci in HWE found low estimates of FST and ϕST between populations in both years with no change in variance within populations across this time (92 % of allelic variance was found within populations for FST estimates in 1997 and 2004, and 86 % for ϕST estimates in 1997 and 2004).
Table 3.
Hierarchical AMOVA tables. The proportion of genetic variation is partitioned among populations, within populations and within individuals. Overall fixation, FST, and corresponding Nm provided for both years (1997 in (a); 2004 in (b)). FST significantly different from zero (P < 0.05) are indicated with an asterisk.
| Source | df | SS | MS | Est. var. | % | |
|---|---|---|---|---|---|---|
| (a) 1997 | ||||||
| Among populations | 1 | 16.602 | 16.602 | 0.143 | 5 | |
| Among individuals | 93 | 319.635 | 3.437 | 0.890 | 33 | |
| Within individuals | 95 | 157.500 | 1.658 | 1.658 | 62 | |
| Total | 189 | 493.737 | 2.691 | 100 | ||
| FST | 0.053* | |||||
| Nm | 4.448 | |||||
| (b) 2004 | ||||||
| Among populations | 1 | 34.977 | 34.977 | 0.195 | 7 | |
| Among individuals | 162 | 520.151 | 3.211 | 0.801 | 31 | |
| Within individuals | 164 | 264.000 | 1.610 | 1.610 | 62 | |
| Total | 327 | 819.128 | 2.605 | 100 | ||
| FST | 0.075* | |||||
| Nm | 3.088 |
To compare fixation between the two populations before and after the drought, we permuted FST (Fig. 2) using 1000 bootstrap replications. We estimated FST to be 0.053 (0.019–0.082; 95 % CI) between BB and Arb in 1997 and 0.076 (0.024–0.136) in 2004. As these estimates of FST do not overlap with zero, this suggests significant levels of differentiation between the populations in both years. However, the estimates of FST were not significantly different between these 2 years (Fig. 2), indicating a lack of change in fixation or gene flow between the generations. Analysis using loci in HWE similarly shows no significant change in FST between Arb and BB during this time.
Figure 2.
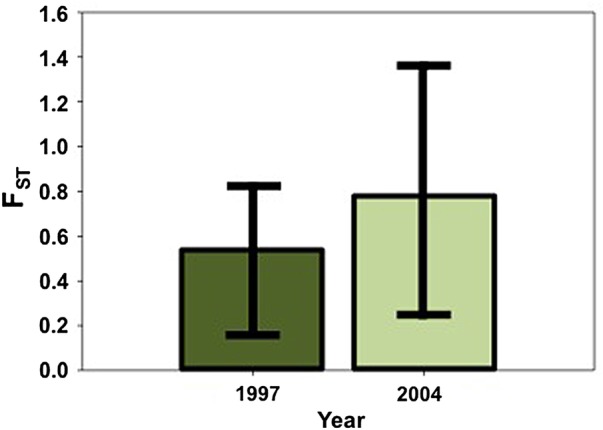
Population fixation (FST) between 1997 and 2004. FST estimated with 1000 bootstrap replications. Error bars display 95 % CI.
Bayesian analysis of this data suggests the existence of three distinct clusters (K = 3) in this system [see Supporting Information]. The greater proportion of individuals assigned to the genetic cluster represented by yellow in BB as compared with Arb indicates some genetic distinctiveness between these locations in both years. However, every individual sampled is estimated, in some proportion, to be assigned to each of these three clusters (yellow, orange and blue) (Fig. 3) suggesting low levels of structure, and consequently high levels of gene flow or genetic similarity in 1997 and 2004.
Figure 3.
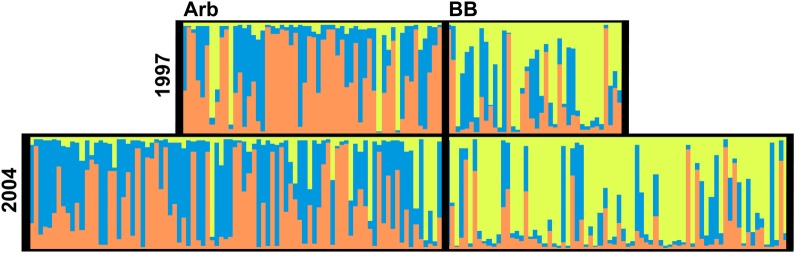
Genetic clustering assignment. Bayesian cluster analysis using allelic data from 10 microsatellite loci calculates three (K = 3) distinct genetic units (yellow, blue and orange). Assignment of these units is applied to each individual sampled in all four populations.
Discussion
The results of this study show relatively high levels of inbreeding within the Arb and BB populations, and low fixation between these locations in 1997 and 2004. Despite a previous finding of reduced phenological, and potentially reproductive isolation between these populations following a 5-year drought, we did not find a significant change in gene flow or in genetic structuring across this time.
Genetic diversity
Microsatellite data from these populations revealed high allelic diversity. Across all loci, we found 58 alleles, compared with 47 described in the literature [see Supporting Information] (Szewc-McFadden et al. 1996; Lowe et al. 2002, 2004; Suwabe et al. 2002). We found no significant shift in within population genetic parameters between 1997 and 2004 [see Supporting Information]. The presence of eight new PA in BB in 2004, as compared with 1997, may indicate an introduction of new alleles by mutation (Selkoe and Toonen 2006), as no nearby populations are known to exist to introduce new alleles via gene flow. Mutation rate is dependent on the effective population size and can vary due to environmental stresses, allowing for different trends in allelic diversity at each site (Hoffmann and Hercus 2000). However, the increase in PA in BB may also be due, fully or in part, to the relatively small sample size of BB 97 compared with BB 04 and a failure to detect low frequency alleles with our sampling effort. Thus, these populations appear to have high diversity at these loci, but there is little evidence to suggest changes in diversity over time.
Intra-population genetic structuring
We found evidence for high levels of inbreeding in our populations despite the genetic self-incompatibility of B. rapa (Sobotka et al. 2000). This high level of inbreeding could be due to local seed and pollen dispersal causing mating between relatives, or to assortative mating. Some level of assortative mating was likely in this system due to the range of flowering times, as individuals may only cross with other individuals contemporaneously in flower (Weis and Kossler 2004; Franks and Weis 2009), and individuals with similar flowering times may be more likely related to one another than expected by chance. Levels of inbreeding did not change significantly between these years despite changes in the flowering phenology of each population (Franks and Weis 2009). As the drought resulted in an abbreviated growing season in 2004, this could allow for a greater proportion of individuals to be in flower at the same time thereby reducing the likelihood of potential crosses between closely related individuals. However, genetic similarity among all individuals may have allowed these populations to maintain high FIS across these generations. Ultimately, this shift in phenology did not appear to influence the genetic structuring within each population.
Fixation and gene flow over time
We found low levels of fixation, and correspondingly high levels of gene flow or shared common ancestry, between the two populations both before and after the drought. The lowest values of fixation were found between years within the same population, which is likely due to the 2004 generation maintaining much of the genetic character of its ancestral 1997 populations. However, we found very little deviation from these FST values between all other population comparisons, indicating maintenance of, and no large change in, population connectivity over these generations.
We calculated FST and ϕST in both of these generations to investigate whether a change in gene flow occurred with the decreased phenological isolation between the populations. Although these parameters have been critiqued as estimates of genetic differentiation, particularly for microsatellite data (Whitlock and McCauley 1999; Jost 2008), the focus of our analysis is to determine if the populations became more genetically similar over time, as would occur if gene flow increased. Thus, examining these parameters and their change over time is appropriate for our goals. Both FST and ϕST indicate similar patterns of fixation between the BB and Arb populations with lower, significant fixation occurring prior to the drought in 1997 and higher, significant fixation following the drought in 2004 (Table 3) [see Supporting Information]. These values of fixation are very low and represent minimal structure between the populations. The statistical significance of these measures suggests a deviation from no fixation, however the low estimates of FST may not represent biologically meaningful levels of structure. The 95 % bootstrapped CI indicate no significant change in FST or gene flow over time (Fig. 2). This result was not expected given the increased phenological opportunity for gene flow. However, in some systems, a decrease in phenological isolation may not cause changes in gene flow, particularly when minimal structure is present prior to the phenological change.
The data were also analysed using Bayesian statistics, which showed little genetic distinctiveness between these populations and generations. Three genetic clusters were recovered for this dataset, and all individuals in each population were assigned to these clusters to varying degrees (Fig. 3). This indicates a high degree of genetic relatedness of the BB and Arb sites across these years. Despite some variation across the two sites in terms of the relative proportions of each cluster, the assignment of all individuals sampled to each of the three clusters suggests that these populations had a recent shared ancestry. Furthermore, no major shift in proportion of these genetic units is evident across time (Fig. 3). Overall, this assessment of the data supports the results of FST analyses. The minimal observed change in structure and gene flow may have been due to an insignificant change in migration rate over time, or these results may have instead been impacted by the populations' genetic similarity or large effective population sizes (Neigel 2002).
Evolutionary, environmental and community considerations
The lack of an expected change in gene flow over time could be due to several factors including the evolutionary history of these populations, seed banks, a low initial degree of genetic structure between the populations or a direct effect of the drought on the movement of pollen between populations. While research has been done to examine the phenological traits of these populations, little is known about their evolutionary history preceding the initial Franks et al. (2007) study, and pollination dynamics are not well understood. The low levels of fixation maintained between the two populations across seven generations (Table 3) [see Supporting Information] could have resulted if they had been very genetically similar across the data collection period. For example, if both populations had been established by individuals from the same source population, one population had been established from the other, or they were effectively a single population in the recent past, they would exhibit low levels of fixation. In any of these cases, fixation could have increased over time, given the 3 km separating these populations, allowing for divergence through local effects of genetic drift and mutation at neutral loci. However, it is possible that neither of these processes had an effect on fixation in this time, or that high levels of gene flow diminished a trend toward fixation.
Seed banks present in these populations might influence phenology and genetics. Both populations likely have persistent seed banks, as seed viability remains high over time, especially under the dry conditions that these populations experience outside of the growing season, and as persistent seed banks were found in the related species, Brassica napus (Simard et al. 2002). The presence of a seed bank would serve to hinder the evolutionary response to selection across this climatic shift, as genotypes adapted to previous conditions could emerge after conditions changed. However, evolutionary change in phenology was previously documented in these populations across these years (Franks et al. 2007) despite the likely presence of a seed bank. Nevertheless, a seed bank could have been a contributing factor to the lack of change in gene flow seen in these populations.
Additionally, given that little information exists regarding the pollinators of this system in these years, it is unknown whether they were able to easily traverse the 3 km distance separating the populations. Some sources define 3 km as the maximum distance that insects will forage (Rieger et al. 2002), while others believe that flowering plant populations within a range of 10–20 km can still exhibit substantial levels of gene flow, depending on the type of insect pollinator (dePamphilis and Wyatt 1989; Ellstrand 1992; Cresswell et al. 2000). Therefore, understanding whether the distance between these populations is a reasonable foraging range for this system's pollinators is necessary to properly interpret these results. The observed levels of fixation suggest very high levels of gene flow but this may instead be a consequence of historic patterns of population connectivity (Heywood 1991; Slatkin 1995; Whitlock and McCauley 1999).
The landscape surrounding these two populations is highly urbanized and developed (Fig. 1), but contains managed conservation areas surrounding the populations, including the UCI Arboretum grounds and the Upper Newport Bay Nature Preserve and Ecological Reserve. Although weedy populations of other wild and landscaped flowering plants are present in the area and could potentially serve as stepping stones in pollinators moving between the two study populations, the extent to which this occurs, and the effects of urbanization in this area on pollinator movement, is not known.
Another potential explanation for the absence of a significant change in gene flow with decreased phenological isolation is the effect of the drought, or other unknown changes, on pollinators. As B. rapa is primarily pollinated by insects, gene flow would require the foraging activity of these pollinators. As climate change is negatively impacting many systems across the globe (Hannah et al. 2002; Parmesan and Yohe 2003), the effect of this drought may have altered the population sizes of insects in the area or caused some modification of their behaviour that impacted their ability to transfer pollen across this range (Ollerton 1996; Rafferty and Ives 2011). Therefore, if the potential for increased gene flow existed in these populations over this drought period, negative impacts of the drought on pollinators could have had effects throughout the community, which may have minimized the level of gene flow achieved through this change (Ollerton 1996; Williams et al. 2008; Walther 2010; Rafferty and Ives 2011). As little is known about the evolutionary history of this system and about the community at the time of this drought, any variation in gene flow or genetic structuring cannot be directly attributed to a change in climate. However, an effect of the drought on the pollinators in this area would likely have consequences for gene flow (Cresswell et al. 2000).
Conclusions
Despite the lack of a significant change in genetic structuring of these populations in response to a climate change event, this case highlights the importance of understanding the effects of environmental changes on gene flow between natural populations. Further research examining the genetic responses of natural populations to climate change across multiple systems will be useful in increasing our understanding of the evolutionary consequences of climate change (Franks and Hoffmann 2012) and establishing a baseline understanding with which to advise management decisions.
Sources of Funding
Funding was provided by a Sigma Xi Grant-in-Aid-of-Research (G20101015154375) (USA) to R.S.W., a Faculty Research Grant from Fordham and a grant (DEB-1142784) from the National Science Foundation (USA) to S.J.F. and by the New York Botanical Garden.
Contributions by the Authors
All authors took part in designing the experiment, discussing the results and composing and editing the manuscript. R.S.W. carried out the research, analysed the data and drafted the manuscript.
Conflict of Interest Statement
None declared.
Supporting Information
The following additional information is available in the online version of this article–
Table S1. ‘HWE estimates of FIS’ provides estimates of FIS, and corresponding 95 % CI, from 1000 bootstrap replicates using only loci found to be in HWE. Loci used are listed for each population and generation.
Table S2. ‘HWE estimates of FST’ provides estimates of FST, and corresponding 95 % CI (when available), from 1000 bootstrap replicates using only loci found to be in HWE. Loci used are listed for each generation.
Table S3. ‘Hierarchical AMOVA tables using loci in HWE’ (FST) provides estimates of FST and the distribution of molecular variance for both generations, using only loci in HWE.
Table S4. ‘Hierarchical AMOVA tables using loci in HWE’ (ϕST) provides estimates of ϕST and the distribution of molecular variance for both generations, using only loci in HWE.
Table S5. ‘Loci in HWE’ provides a list of the loci estimated to be in HWE for each population and generation.
Table S6. ‘Microsatellite loci characteristics’ lists allelic parameters calculated for each of the 10 loci used in this study.
Table S7. ‘Hierarchical AMOVA tables’ provides estimates of ϕST and the distribution of molecular variance for both generations, using all 10 loci.
Table S8. ‘Within population parameters’ provides estimates of standard deviation for genetic parameters of each population and generation.
Table S9. ‘STRUCTURE likelihood outputs’ provides likelihood scores used to determine the optimal number of clusters comprising these populations.
Table S10. ‘Within population parameters over time’ provides the results of Student's t-test comparing within population genetic parameters across generations.
Table S11. ‘CI for within population fixation’ provides the 95 % CI corresponding to estimates of FIS from 1000 bootstrap replicates, for each population and generation.
Table S12. ‘Estimates of FIS across populations’ provides global estimates of FIS for each generation, and across generations.
Acknowledgements
We thank Evon Hekkala, Jim Lewis, Anthony Caragiulo, Anna Nowogrodski, Vinson Doyle and Rebecca Lalchan for assistance with this research, as well as two anonymous reviewers for helpful comments which improved this manuscript. Seeds were made available by Art Weis and Emily Austen.
Literature Cited
- Bellard C, Bertelsmeier C, Leadley P, Thuiller W, Courchamp F. 2012. Impacts of climate change on the future of biodiversity. Ecology Letters 15:365–377. 10.1111/j.1461-0248.2011.01736.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell JE, Osborne JL, Goulson D. 2000. An economic model of the limits to foraging range in central place foragers with numerical solutions for bumblebees. Ecological Entomology 25:249–255. 10.1046/j.1365-2311.2000.00264.x [DOI] [Google Scholar]
- Davis AR, Sawhney VK, Fowke LC, Low NH. 1994. Floral nectar secretion and ploidy in Brassica rapa and B napus (Brassicaceae). I. Nectary size and nectar carbohydrate production and composition. Apidologie 25:602–614. 10.1051/apido:19940611 [DOI] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. 1977. Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 39:1–38. [Google Scholar]
- dePamphilis CW, Wyatt R. 1989. Hybridization and introgression in buckeyes (Aesculus: Hippocastanaceae): a review of the evidence and a hypothesis to explain long-distance gene flow. Systematic Botany 14:593–611. 10.2307/2419005 [DOI] [Google Scholar]
- Devaux C, Lavigne C, Falentin-Guyomarc'h H, Vautrin S, Lecomte J, Klein EK. 2005. High diversity of oilseed rape pollen clouds over an agro-ecosystem indicates long-distance dispersal. Molecular Ecology 14:2269–2280. 10.1111/j.1365-294X.2005.02554.x [DOI] [PubMed] [Google Scholar]
- Devaux C, Lavigne C, Austerlitz F, Klein EK. 2007. Modelling and estimating pollen movement in oilseed rape (Brassica napus) at the landscape scale using genetic markers. Molecular Ecology 16:487–499. 10.1111/j.1365-294X.2006.03155.x [DOI] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4:359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Ellstrand NC. 1992. Gene flow by pollen: implications for plant conservation genetics. Oikos 63:77–86. 10.2307/3545517 [DOI] [Google Scholar]
- Ellstrand NC, Devlin B, Marshall DL. 1989. Gene flow by pollen into small populations: data from experimental and natural stands of wild radish. Proceedings of the National Academy of Sciences of the USA 86:9044–9047. 10.1073/pnas.86.22.9044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14:2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10:564–567. 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- Fitter AH, Fitter RSR. 2002. Rapid changes in flowering time in British plants. Science 296:1689–1691. 10.1126/science.1071617 [DOI] [PubMed] [Google Scholar]
- Franke DM, Ellis AG, Dharjwa M, Freshwater M, Fujikawa M, Padron A, Weis AE. 2006. A steep cline in flowering time for Brassica rapa in southern California: population-level variation in the field and the greenhouse. International Journal of Plant Sciences 167:83–92. 10.1086/497648 [DOI] [Google Scholar]
- Franks SJ, Hoffmann AA. 2012. Genetics of climate change adaptation. Annual Review of Genetics 46:185–208. 10.1146/annurev-genet-110711-155511 [DOI] [PubMed] [Google Scholar]
- Franks SJ, Weis AE. 2009. Climate change alters reproductive isolation and potential gene flow in an annual plant. Evolutionary Applications 2:481–488. 10.1111/j.1752-4571.2009.00073.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ, Sim S, Weis AE. 2007. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proceedings of the National Academy of Sciences of the USA 104:1278–1282. 10.1073/pnas.0608379104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ, Avise JC, Bradshaw WE, Conner JK, Etterson JR, Mazer SJ, Shaw RG, Weis AE. 2008. The resurrection initiative: storing ancestral genotypes to capture evolution in action. BioScience 58:870–873. 10.1641/B580913 [DOI] [Google Scholar]
- Franks SJ, Weber JJ, Aitken SN. 2014. Evolutionary and plastic responses to climate change in terrestrial plant populations. Evolutionary Applications 7:123–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garant D, Forde SE, Hendry AP. 2007. The multifarious effects of dispersal and gene flow on contemporary adaptation. Functional Ecology 21:434–443. 10.1111/j.1365-2435.2006.01228.x [DOI] [Google Scholar]
- Gienapp P, Teplitsky C, Alho JS, Mills JA, Merilä J. 2008. Climate change and evolution: disentangling environmental and genetic responses. Molecular Ecology 17:167–178. 10.1111/j.1365-294X.2007.03413.x [DOI] [PubMed] [Google Scholar]
- Guo SW, Thompson EA. 1992. Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics 48:361–372. 10.2307/2532296 [DOI] [PubMed] [Google Scholar]
- Hannah L, Midgley GF, Lovejoy T, Bond WJ, Bush M, Lovett JC, Scott D, Woodward FI. 2002. Conservation of biodiversity in a changing climate. Conservation Biology 16:264–268. 10.1046/j.1523-1739.2002.00465.x [DOI] [PubMed] [Google Scholar]
- Hedrick PW. 2005. A standardized genetic differentiation measure. Evolution 59:1633–1638. 10.1111/j.0014-3820.2005.tb01814.x [DOI] [PubMed] [Google Scholar]
- Heywood JS. 1991. Spatial analysis of genetic variation in plant populations. Annual Review of Ecology, Evolution, and Systematics 22:335–355. 10.1146/annurev.es.22.110191.002003 [DOI] [Google Scholar]
- Hoffmann AA, Hercus MJ. 2000. Environmental stress as an evolutionary force. BioScience 50:217–226. 10.1641/0006-3568(2000)050[0217:ESAAEF]2.3.CO;2 [DOI] [Google Scholar]
- Hoffmann AA, Sgró CM. 2011. Climate change and evolutionary adaptation. Nature 470:479–485. 10.1038/nature09670 [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Camac JS, Williams RJ, Papst W, Jarrad FC, Wahren C-H. 2010. Phenological changes in six Australian subalpine plants in response to experimental warming and year-to-year variation. Journal of Ecology 98:927–937. 10.1111/j.1365-2745.2010.01667.x [DOI] [Google Scholar]
- Holt RD, Gomulkiewicz R. 1997. How does immigration influence local adaptation? A reexamination of a familiar paradigm. The American Naturalist 149:563–572. 10.1086/286005 [DOI] [Google Scholar]
- Huey RB, Gilchrist GW, Carlson ML, Berrigan D, Serra L. 2000. Rapid evolution of a geographic cline in size in an introduced fly. Science 287:308–309. 10.1126/science.287.5451.308 [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. 10.1093/bioinformatics/btm233 [DOI] [PubMed] [Google Scholar]
- Jarrad FC, Wahren C-H, Williams RJ, Burgman MA. 2008. Impacts of experimental warming and fire on phenology of subalpine open-heath species. Australian Journal of Botany 56:617–629. 10.1071/BT08018 [DOI] [Google Scholar]
- Jost L. 2008. GST and its relatives do not measure differentiation. Molecular Ecology 17:4015–4026. 10.1111/j.1365-294X.2008.03887.x [DOI] [PubMed] [Google Scholar]
- Jump AS, Peñuelas J. 2005. Running to stand still: adaptation and the response of plants to rapid climate change. Ecology Letters 8:1010–1020. 10.1111/j.1461-0248.2005.00796.x [DOI] [PubMed] [Google Scholar]
- Lewis PO, Zaykin D. 2002. Genetic Data Analysis (GDA): computer program for the analysis of allelic data, version 1.1 [online]. http://hydrodictyon.eeb.uconn.edu/people/plewis/software.php.
- Lewontin RC, Kojima K. 1960. The evolutionary dynamics of complex polymorphisms. Evolution 14:450–472. [Google Scholar]
- Lowe AJ, Jones AE, Raybould AF, Trick M, Moule CL, Edwards KJ. 2002. Transferability and genome specificity of a new set of microsatellite primers among Brassica species of the U triangle. Molecular Ecology Notes 2:7–11. 10.1046/j.1471-8286.2002.00126.x [DOI] [Google Scholar]
- Lowe AJ, Moule C, Trick M, Edwards KJ. 2004. Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theoretical and Applied Genetics 108:1103–1112. 10.1007/s00122-003-1522-7 [DOI] [PubMed] [Google Scholar]
- Menzel A, Sparks TH, Estrella N, Koch E, Aasa A, Ahas R, Alm-Kübler K, Bissolli P, Braslavská O, Briede A, Chmielewski FM, Crepinsek Z, Curnel Y, Dahl Å, Defila C, Donnelly A, Filella Y, Jatczak K, Måge F, Mestre A, Nordli Ø, Peñuelas J, Pirinen P, Remišová V, Scheifinger H, Striz M, Susnik A, Van Vliet AJH, Wielgolaski F-E, Zach S, Zust A. 2006. European phenological response to climate change matches the warming pattern. Global Change Biology 12:1969–1976. 10.1111/j.1365-2486.2006.01193.x [DOI] [Google Scholar]
- Michalakis Y, Excoffier L. 1996. A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics 142:1061–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigel JE. 2002. Is FST obsolete? Conservation Genetics 3:167–173. 10.1023/A:1015213626922 [DOI] [Google Scholar]
- North A, Pennanen J, Ovaskainen O, Laine A-L. 2011. Local adaptation in a changing world: the roles of gene-flow, mutation, and sexual reproduction. Evolution 65:79–89. 10.1111/j.1558-5646.2010.01107.x [DOI] [PubMed] [Google Scholar]
- Ollerton J. 1996. Reconciling ecological processes with phylogenetic patterns: the apparent paradox of plant-pollinator systems. The Journal of Ecology 84:767–769. 10.2307/2261338 [DOI] [Google Scholar]
- Parmesan C, Yohe G. 2003. A globally coherent fingerprint of climate change impacts across natural systems. Nature 421:37–42. 10.1038/nature01286 [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6:288–295. 10.1111/j.1471-8286.2005.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson RG, Dawson TP. 2003. Predicting the impacts of climate change on the distribution of species: are bioclimate envelope models useful? Global Ecology and Biogeography 12:361–371. 10.1046/j.1466-822X.2003.00042.x [DOI] [Google Scholar]
- Peñuelas J, Filella I, Comas P. 2002. Changed plant and animal life cycles from 1952 to 2000 in the Mediterranean region. Global Change Biology 8:531–544. 10.1046/j.1365-2486.2002.00489.x [DOI] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafferty NE, Ives AR. 2011. Effects of experimental shifts in flowering phenology on plant-pollinator interactions. Ecology Letters 14:69–74. 10.1111/j.1461-0248.2010.01557.x [DOI] [PubMed] [Google Scholar]
- Räsänen K, Hendry AP. 2008. Disentangling interactions between adaptive divergence and gene flow when ecology drives diversification. Ecology Letters 11:624–636. 10.1111/j.1461-0248.2008.01176.x [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity 86:248–249. [Google Scholar]
- Rieger MA, Lamond M, Preston C, Powles SB, Roush RT. 2002. Pollen-mediated movement of herbicide resistance between commercial canola fields. Science 296:2386–2388. 10.1126/science.1071682 [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. 2004. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes 4:137–138. 10.1046/j.1471-8286.2003.00566.x [DOI] [Google Scholar]
- Rousset F. 2008. Genepop'007: a complete re-implementation of the Genepop software for Windows and Linux. Molecular Ecology Resources 8:103–106. 10.1111/j.1471-8286.2007.01931.x [DOI] [PubMed] [Google Scholar]
- Selkoe KA, Toonen RJ. 2006. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecology Letters 9:615–629. 10.1111/j.1461-0248.2006.00889.x [DOI] [PubMed] [Google Scholar]
- Sexton JP, Strauss SY, Rice KJ. 2011. Gene flow increases fitness at the warm edge of a species’ range. Proceedings of the National Academy of Sciences of the USA 108:11704–11709. 10.1073/pnas.1100404108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard M-J, Légère A, Pageau D, Lajeunesse J, Warwick S. 2002. The frequency and persistence of volunteer canola (Brassica napus) in Québec cropping systems. Weed Technology 16:433–439. 10.1614/0890-037X(2002)016[0433:TFAPOV]2.0.CO;2 [DOI] [Google Scholar]
- Slatkin M. 1994. Linkage disequilibrium in growing and stable populations. Genetics 137:331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Excoffier L. 1996. Testing for linkage disequilibrium in genotypic data using the Expectation-Maximization algorithm. Heredity 76:377–383. 10.1038/hdy.1996.55 [DOI] [PubMed] [Google Scholar]
- Sobotka R, Sáková L, Curn V. 2000. Molecular mechanisms of self-incompatibility in Brassica. Current Issues in Molecular Biology 2:103–112. [PubMed] [Google Scholar]
- Stockwell CA, Hendry AP, Kinnison MT. 2003. Contemporary evolution meets conservation biology. Trends in Ecology and Evolution 18:94–101. 10.1016/S0169-5347(02)00044-7 [DOI] [Google Scholar]
- Suwabe K, Iketani H, Nunome T, Kage T, Hirai M. 2002. Isolation and characterization of microsatellites in Brassica rapa L. Theoretical and Applied Genetics 104:1092–1098. 10.1007/s00122-002-0875-7 [DOI] [PubMed] [Google Scholar]
- Szewc-McFadden AK, Kresovich S, Bliek SM, Mitchell SE, McFerson JR. 1996. Identification of polymorphic, conserved simple sequence repeats (SSRs) in cultivated Brassica species. Theoretical and Applied Genetics 93:534–538. 10.1007/BF00417944 [DOI] [PubMed] [Google Scholar]
- Walther G-R. 2010. Community and ecosystem responses to recent climate change. Philosophical Transactions of the Royal Society B: Biological Sciences 365:2019–2024. 10.1098/rstb.2010.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick SI, Simard MJ, Légère A, Beckie HJ, Braun L, Zhu B, Mason P, Séguin-Swartz G, Stewart CN. 2003. Hybridization between transgenic Brassica napus L. and its wild relatives: Brassica rapa L., Raphanus raphanistrum L., Sinapis arvensis L., and Erucastrum gallicum (Willd.) O.E. Schulz. Theoretical and Applied Genetics 107:528–539. 10.1007/s00122-003-1278-0 [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. 10.2307/2408641 [DOI] [PubMed] [Google Scholar]
- Weis AE, Kossler TM. 2004. Genetic variation in flowering time induces phenological assortative mating: quantitative genetic methods applied to Brassica rapa. American Journal of Botany 91:825–836. 10.3732/ajb.91.6.825 [DOI] [PubMed] [Google Scholar]
- Whitlock MC, McCauley DE. 1999. Indirect measures of gene flow and migration: FST ≠ 1/(4Nm+ 1). Heredity 82:117–125. 10.1038/sj.hdy.6884960 [DOI] [PubMed] [Google Scholar]
- Williams SE, Shoo LP, Isaac JL, Hoffmann AA, Langham G. 2008. Towards an integrated framework for assessing the vulnerability of species to climate change. PLoS Biology 6:2621–2626. 10.1371/journal.pbio.0060325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. 1951. The genetical structure of populations. Annals of Human Genetics 15:323–354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.