Abstract
Developing αβ T cells choose between the helper and cytotoxic lineages, depending upon the specificity of their T cell receptors for MHC molecules. The expression of the CD4 co-receptor on helper cells and the CD8 co-receptor on cytotoxic cells is intimately linked to this decision, and their regulation at the transcriptional level has been the subject of intense study to better understand lineage choice. Indeed, as the fate of developing T cells is decided, the expression status of these genes is accordingly locked. Genetic models have revealed important transcriptional elements and the ability to manipulate these elements in the framework of development has added a new perspective on the temporal nature of their function and the epigenetic maintenance of gene expression. We examine here the novel insights into epigenetic mechanisms that have arisen through the study of these genes.
Introduction
The T cell helper vs. cytotoxic lineage choice as a model for bi-potential fate decisions
Development of even the most complex organisms can be broken down into a series of bi-potential fate decisions: apoptosis vs. survival, proliferation vs. quiescence differentiation vs. renewal, etc. During differentiation, these choices yield cells that are increasingly restricted in lineage potential until a terminal, functional fate is reached (i.e. a neuron, an epithelial cell, a helper T cell, etc). While some stages of differentiation are plastic, bifurcation points are reached at which a cell cannot reverse course and take on alternative fates. Mechanistically, this involves the activation of a lineage specific transcriptional program (specification) and repression of the programs of alternative lineages (commitment). In many cases, these transcriptional programs must stably endure many rounds of mitosis. This is in part achieved through the binding of sequence specific transcription factors, but is also thought to be regulated by heritable epigenetic marks, which overlay important lineage information on primary DNA sequence. As we discuss below, TCRαβ T cell development, and specifically the choice between the CD4+ helper and the CD8+ cytotoxic T cell fates, is an ideal model for studying the epigenetic mechanisms of bi-potential decisions and their maintenance.
The majority of T cells in the body express TCRαβ and develop in the thymus from bone marrow derived precursors (reviewed in (Rothenberg et al., 2008)). At the earliest developmental stages, these T cell progenitors are referred to as double negatives (DN), owing to a lack of CD4 and CD8 co-receptor expression on the cell surface. DN cells proceed through multiple stages of lineage restriction and differentiation, including commitment to the TCRαβ, rather than the TCRγδ, lineage (reviewed in (Ciofani and Zuniga-Pflucker, 2010)). At the DN stage, cells rearrange the gene encoding the TCR β chain, and in-frame productive VDJ rearrangement of one allele allows cells to pass the beta selection checkpoint and proceed through multiple rounds of division. β-selected cells then up-regulate CD4 and CD8, becoming CD4+CD8+ double positive (DP) cells. DPs commence TCR α chain gene rearrangements and eventually follow one of three fates tied to the TCR: 1) unsuccessful Tcra rearrangement results in death by neglect; 2) rearrangements that result in TCRs with high avidity for peptide-MHC result in negative selection; and 3) rearrangements that result in TCRs of intermediate avidity result in positive selection (reviewed in (Starr et al., 2003)).
Following positive selection, T cells commit to either the CD4+CD8− helper lineage or the CD8+CD4− cytotoxic lineage, depending on MHC specificity. CD4 and CD8 are co-receptors for MHCII and MHCI, respectively, and during lineage choice their expression is matched to TCR specificity for either class of major histocompatibility molecules. How TCR specificity for MHC translates to co-receptor expression and lineage choice is still a matter of debate and multiple models have been proposed (reviewed in (Singer et al., 2008)). Early models suggested that the decision was either stochastic, or instructive through quantitatively or qualitatively different signals. It is now thought more likely that lineage choice is instructed by the duration of co-receptor facilitated TCR-MHC signaling. Following positive selection, all T cells downregulate Cd8 transcription, becoming CD4+CD8lo cells and attenuating potential signaling through MHCI-specific TCRs (Sarafova et al., 2005). Importantly, placing Cd4 expression under the control of Cd8 regulatory elements results in MHCII-specific cytotoxic T cells (Sarafova et al., 2005). This result is consistent with a shorter signal, following positive selection of MHCI specific TCRs, leading to the cytotoxic T cell fate, and a longer-lasting signal, upon recognition of MHCII, resulting in the helper T cell fate.
Significant progress has been made in recent years in understanding the transcriptional network that underlies CD4+ vs. CD8+ lineage choice and commitment (reviewed in (Naito and Taniuchi, 2010)). Three transcription factors have emerged as being especially important. Gata3 appears to specify the helper lineage, which is subsequently sealed by the action of the BTB-POZ transcription factor ThPOK. Gata3 up-regulates ThPOK, but that activity is not sufficient, as it is additionally required for helper cell differentiation independently of ThPOK (Hernandez-Hoyos et al., 2003; Pai et al., 2003; Wang et al., 2008b). ThPOK is not only required for CD4 T cell development, but constitutive ThPOK expression can redirect MHCI specific T cells to the helper lineage in a GATA3-dependent manner (He et al., 2005; Sun et al., 2005; Wang et al., 2008b). It is thought that ThPOK functions as a commitment factor, antagonizing the CD8-specific transcriptional program (Egawa and Littman, 2008; Muroi et al., 2008; Wang et al., 2008a). One of the factors that ThPOK antagonizes is the runt domain transcription factor Runx3, which is critical for cytotoxic lineage development (Egawa et al., 2007; Taniuchi et al., 2002a). Runx3, analogous to ThPOK, is thought to act as a commitment factor that suppresses the helper transcriptional program (Egawa and Littman, 2008). In addition, Runx3 is required for reactivating Cd8 expression and silencing Cd4 expression in MHCI selected cells, as will be discussed in more detail later (Sato et al., 2005; Taniuchi et al., 2002a; Taniuchi et al., 2002b). In addition to these critical regulators, a number of other transcription factors have been reported to play a role in lineage choice, including Myb and Tox in helper cell development, and MAZR and STAT5 (downstream of IL-7 signaling) in cytotoxic T cell development (reviewed in (Naito and Taniuchi, 2010)).
Considering the exquisite correlation between CD4 and CD8 expression and helper and cytotoxic lineage commitment, respectively, studying the transcriptional regulation of Cd4 and Cd8 has long been a strategy to uncover the factors that define lineage choice. Indeed the importance of Runx and MAZR proteins to lineage choice was first identified through examination of the Cd4 and Cd8 loci, respectively (Bilic et al., 2006; Taniuchi et al., 2002a). The fact that the loci are transiently co-expressed in DP cells and stably expressed or repressed in helper and cytotoxic cells, also makes them an excellent model to study transcriptional regulation and the mechanisms that control temporary versus permanent gene expression states. Below, we will review recent advances in the epigenetic regulation of the Cd4 and Cd8 loci; these provide insights into general mechanisms of transcriptional regulation during differentiation.
Molecular mechanism of transcriptional regulation and epigenetic propagation
To fit the few meters of DNA that exist in every eukaryotic nucleus, cells must tightly pack their genetic material. The nucleosome, consisting of 147bp of DNA wrapped around a histone octomer, is the basic packaging unit and forms the classic “beads on a string” structure observed in electron microscopy images (Olins and Olins, 1974). Histone proteins H2A, H2B, H3 and H4 (two each in a histone octamer), are positively charged and have intrinsic DNA binding affinity, independent of nucleotide sequence. Histone H1 binds between adjacent nucleosomes, creating a higher order structure called the 30nm chromatin fiber (reviewed in (Woodcock and Ghosh, 2010)).
Nucleosomal packaging can physically impede transcription by RNA polymerase and thus different degrees of packing can modulate transcriptional outcomes (reviewed in (Orphanides and Reinberg, 2000)). Looser packed, transcriptionally permissive chromatin is termed “euchromatin”, while more tightly packed, repressive chromatin is called “heterochromatin”. These chromatin states are dynamic during development and in response to extracellular signals. Covalent modifications of histone proteins (especially their protruding amino-terminal tails) and DNA are thought to be the biochemical basis for the distinction between hetero- and euchromatin. Initially, histones were found to be acetylated and this correlated with looser wrapping of the DNA (Allfrey, 1966). Additional posttranslational modifications have been identified, including methylation, phosphorylation, ubiquitination and sumoylation. Genome wide study of such modifications has allowed the correlation of some specific modifications with gene activity (Barski et al., 2007; Heintzman et al., 2007; Ji et al., 2010; Roh et al., 2007). For example, tri-methylation of Histone H3 lysine 9 (H3K9), H3K27, H4K20 and H3K79 are associated with repressed chromatin states and heterochromatin. In contrast, H3K4me1-3, histone acetylation (H3Ac and H4Ac), and mono-methylation of H3K9 and H3K27, are associated with gene activation and euchromatic regions. Thus, different modifications on different residues, the same modification on different residues, and the abundance of a single modified residue may all have unique effects on transcription. In addition to histones, DNA itself can be modified by methylation on cytosine residues, a mark that has generally been correlated with gene silencing (Bachman et al., 2003; Fuks et al., 2000), but may also facilitate transcription in certain contexts (Wu et al., 2010).
Chromatin modifications do not act in a vacuum; their effect on transcription depends not only on the specific residue modified, but also on the surrounding residues and their modifications (reviewed in (Campos and Reinberg, 2009)). Further, combinations of multiple modifications result in a context-dependent outcome on transcription (reviewed in (Lee et al., 2010). For example, H3K9me3 generally recruits HP1 to repress transcription, but in combination with phosphorylation of the adjacent H3S10, HP1 binding is abrogated and repression may be relieved (Fischle et al., 2005; Mateescu et al., 2008). Thus histone and DNA modifications superimpose a rich layer of information on the underlying DNA sequence.
Several enzyme classes, usually in the context of multi-factor complexes, catalyze chromatin modifications. Generally these complexes do not recognize specific DNA sequences, but are recruited by sequence-specific transcription factors or recognition of specific histone modifications. Further, complexes exist to write and erase most chromatin modifications. Histone acetyltransferases (HAT) add acetyl groups, while histone deacetylase (HDAC) enzymes eliminate them. Enzymes also exist to methylate and demethylate lysines and arginines. Even DNA methylation, which is considered the most stable modification, may be erased by the recently characterized Tet proteins which can modify 5-methylcytosine, resulting in a 5-hydroxymethylcytosine that may be replaced by an unmethylated cytosine residue through a base excision repair pathway (Tahiliani et al., 2009); alternatively, 5-hydroxymethylcytosine could have a stand-alone function (Ficz et al., 2011). Taken together, this means that chromatin modifications can be dynamically written and erased from the genome in the process of regulating transcription.
The functional outcome of DNA and histone modifications is at least in part brought about through their interaction with specific protein motifs. Acetylated lysines are recognized by bromodomains, while chromodomains, PHD fingers and WD40 repeats bind methylated lysines. Methylated cytosines meanwhile are read by methyl binding domains. Engagement of these modifications by individual protein subunits may recruit or modulate the activity of multi-factorial chromatin modifying complexes. Importantly, several proteins can read different modifications simultaneously to regulate their binding to nucleosomes (Bartke et al., 2010). Thus the “histone language” can be read with the aid of multiple adaptors, linking histone and DNA marks to functional outcome.
Transcriptional activity, in addition to being correlated with specific histone modifications, has been correlated with nucleosome depletion at promoter regions (Lee et al., 2004; Ozsolak et al., 2007) prior to transcriptional initiation (Petesch and Lis, 2008). It has long been known that functional DNA elements are hypersensitive to endonucleases, and such DNase I hypersensitive sites (DHS) (Wu et al., 1979) occur upon nucleosome depletion. Studies of IFNβ gene activation during viral infection revealed that a nucleosome masking the transcription start site (TSS) and TATA box was remodeled by the histone acetylation-recruited SWI/SNF chromatin remodeling complex, allowing for TFIID recruitment (Agalioti et al., 2000). Binding of the TFIID subunit TBP to the TATA box induced the nucleosome to slide further downstream, exposing the transcription start site and allowing for transcription (Lomvardas and Thanos, 2001). Thus there is a complex interplay between chromatin marks, their readers, writers and erasers, and sequence specific transcription factors in the regulation of transcription.
Views of transcriptional regulation have changed dramatically in recent years. Transcription is no longer thought to occur only in a linear fashion with regulatory elements controlling the expression of their downstream genes. Rather, it is thought to be a dynamic process involving the movement of chromatin fibers to allow co-regulated genes to come together in the subnuclear space. Osborne et al. (2004) found that active genes co-localize with reservoirs of active RNA polymerase II in what are termed “transcription factories”. Subsequently, Spilianakis et al. showed that genes on different chromosomes (the Il4 locus on chromosome 11 and Ifng on chromosome 10) could interact in a developmentally regulated manner and this interaction had a functional role in the expression of both loci, since deletion of a regulatory element on one chromosome could affect the expression of a locus on the other (2005). Such long-range movements can affect not only gene activity, but can also have an impact on the transcriptional competence of loci. For example, transient IFNγ signaling induces persistent association of the MHCII locus with promyelocytic leukemia (PML) nuclear bodies, which perpetuates histone marks through mitoses. Thus the locus is maintained in a poised state, sensitizing it to respond to lower doses of IFNγ, with faster kinetics (Gialitakis et al., 2010).
Where does epigenetic regulation fit into all of this? Differentiation is a dynamic process that relies on the inheritance of gene expression patterns. Although almost all cells within an organism have the same genetic material (antigen receptor loci excluded), the transcriptional outcomes, and thus cell type/lineage, can differ dramatically. An extra layer of stably inherited information, unique to each cell type, is provided by epigenetic modifications. Strictly speaking, epigenetic marks are not contained in the primary genetic sequence, but are stable and heritable even in the absence of their sequence-specific initiating events. For example, H3K27 trimethylation, is both catalyzed and recognized by the PRC2 methyltransferase complex (Hansen et al., 2008). The PRC2 subunit, EED, binds H3K27me3 both to recruit the PRC2 complex and to allosterically activate its methyltransferase activity, setting up a positive feedback loop to propagate the epigenetic mark through cell division without need for sequence specific factors (Margueron et al., 2009). In this context, many chromatin modifications are not in fact epigenetic. They may be by-products of the current transcriptional state of a locus, or they may require the continued action of sequence specific factors or events that initiated them. These are interesting to study in the context of how a gene is acutely transcribed or repressed, but may not help to explain how the transcriptional program of a helper or cytotoxic T cell can be stably maintained through many mitoses. In what follows, we discuss what the Cd4 and Cd8 loci have to teach us about heritable epigenetic regulation of gene expression.
Epigenetic regulation of the Cd4 locus
Cd4 expression is controlled by multiple regulatory elements (Figure 1A). DHS mapping revealed several putative regulatory elements in the locus (Adlam and Siu, 2003; Sands and Nikolic-Zugic, 1992; Sawada and Littman, 1991), and eventually led to the identification of three enhancers: the distal (E4D) and proximal enhancers (E4P) at ~24kb and ~13kb upstream of the Cd4 TSS, respectively, and the thymocyte enhancer (E4T) at ~36 kb downstream (Adlam and Siu, 2003; Sawada and Littman, 1991; Wurster et al., 1994) (Reviewed in detail by (Taniuchi et al., 2004)). In combination with the Cd4 promoter, E4P drives T cell specific transgene expression in multiple mouse lines (Blum et al., 1993; Hanna et al., 1994; Killeen et al., 1993). The developmental window during which E4P shows activity, however, was not entirely clear: while some studies found it to be active in DN and pre-selection DP cells through to mature helper and cytotoxic T cells (Blum et al., 1993; Hanna et al., 1994; Killeen et al., 1993; Manjunath et al., 1999; Sawada et al., 1994; Siu et al., 1994), others indicated that E4P activity begins post-positive selection (Adlam et al., 1997). In addition, it appears that, in the context of transgenes, E4P may lose activity in mature T cells following TCR stimulation (Manjunath et al., 1999). E4D exhibits T cell line specific activity in transient transfection assays (Wurster et al., 1994), but its in vivo relevance for Cd4 regulation has not been demonstrated and, moreover, the homologous human sequence has been implicated in control of the adjacent LAG-3 gene (Bruniquel et al., 1998) and the combination of E4D and E4P in reporter transgenes drives expression in B cells and macrophages (Siu et al., 1994). Finally E4T has been suggested to drive expression in DPs, but only in combination with E4P (Adlam and Siu, 2003). Taken together, these sometimes-conflicting studies have identified three possible Cd4 enhancers and ascribed them independent, overlapping, and cooperative functions at different developmental stages.
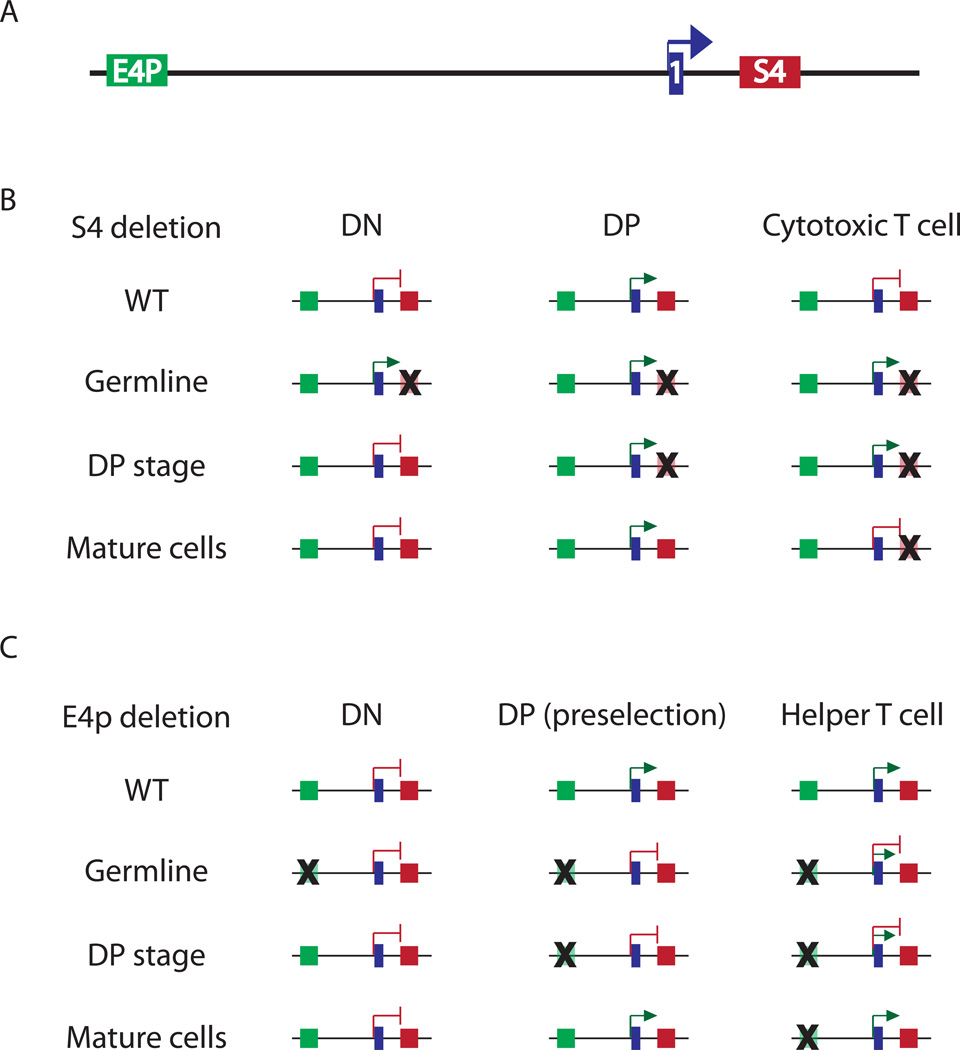
Figure 1. Epigenetic regulation of Cd4 transcription and silencing.
A) Genomic organization of the Cd4 locus: proximal enhancer (E4p, green box), exon 1 (1, blue box) and silencer (S4, red box). B) S4 deletion at different developmental stages reveals epigenetic silencing of Cd4. In WT cells, Cd4 is silenced in immature DN and mature cytotoxic T cells, but is expressed in DP cells. Germline deletion of S4 leads to inappropriate expression of Cd4 in DN and cytotoxic T cells. Inducible S4 deletion at the transition to the DP stage results in expression in cytotoxic T cells. However, inducible S4 deletion in cytotoxic T cells does not lead to ectopic expression, indicating that Cd4 is heritably silenced in mature cells (i.e. independently of S4). C) E4p deletion at different developmental stages reveals epigenetic maintenance of Cd4 expression. In WT mice, Cd4 is expressed in DP cells and in mature helper T cells. E4p deletion in the germline, or at the transition between the DN and DP stages, results in DP cells that fail to express Cd4 prior to positive selection, and in unstable Cd4 expression in helper T cells. Inducible E4p deletion in helper cells does not affect Cd4 expression, indicating epigenetic maintenance of expression.
Germline and conditional targeting of two of these enhancers, E4P and E4T, has helped to determine their relevant functions (Chong et al., 2010) (Figure 1C). E4T was found to be dispensable for Cd4 expression in TCRαβ T cells, but required for expression on a subset of lymphoid tissue inducer (LTi) cells, now commonly referred to as innate lymphoid cells, in the small intestine lamina propria. In contrast, E4P was required for Cd4 expression in pre-selection DP cells. However, CD4 was expressed following positive selection in E4P−/− thymocytes, suggesting the existence of a yet unidentified “maturation enhancer”. Importantly, E4P activity was dispensable in the periphery, as its Cre-mediated deletion in mature CD4+ T cells did not affect Cd4 expression. Moreover, E4P is required at the DP stage for stable, high level CD4 expression in mature cells, as will be discussed later. Thus this study indicated that Cd4 transcription is potentiated at the DP stage by E4P, and suggested that, after positive selection, it is regulated by an unidentified maturation enhancer whose heritable activity would be initiated in concert with E4P.
Restriction of Cd4 expression to DPs and mature helper T cells is conferred by the activity of Runt domain-containing transcription factors that bind to sites in a silencer element (S4). S4 was initially identified as a 434 bp element, in the first intron of Cd4, that suppressed transgene expression in DN and CD8+ T cells (Sawada et al., 1994). Germline deletion of S4 released the expression of CD4 in all T cells starting at the DN stage, indicating that this element was responsible for suppressing developmental stage-inappropriate Cd4 expression (Figure 1B) (Leung et al., 2001; Zou et al., 2001). Subsequently, silencer activity was found to be mediated by Runx1 and 3, in DN and cytotoxic T cells, respectively (Taniuchi et al., 2002a; Taniuchi et al., 2002b). Germline mutations in Runx binding motifs in S4 led to CD4 expression in DN and mature CD8 thymocytes. Further, deletion of Runx1 revealed that it is indispensable for Cd4 silencing in DN cells. In contrast, Runx3 deletion led to variegated Cd4 expression in mature CD8+ cells, indicating a role for Runx3-mediated silencing later in development. Thus Runx1 and 3 have stage-specific roles in mediating Cd4 repression through S4.
In collaboration with other factors, Runx1 and 3 mediate two developmental stage specific modes of Cd4 silencing: reversible and permanent. In DN cells, silencing is reversible, as Cd4 transcription must be activated upon transition to the DP stage. What is the mechanism? One model that can be constructed from recent data would involve active antagonism of E4P function by Runx1. The HEB and E2A bHLH transcription factors, which are crucial for E4P-mediated Cd4 activation between the DN and DP stages (Jones and Zhuang, 2007; Sawada and Littman, 1993), are preloaded onto E4P at the DN stage (Yu et al., 2008). In the presence of S4, however, p300 recruitment to E4P, and thus transcriptional activation, is impaired (Yu et al., 2008). This antagonism of p300 recruitment could be mediated by long-range interactions between S4, bound by Runx1, and E4P, as was recently reported (Jiang and Peterlin, 2008). In accord with this model, the bHLH-ZIP transcription factor AP4 binds E4P, interacts with Runx1, and is required for efficient silencing of Cd4 in DN cells (Egawa and Littman, 2011). Thus, a physical interaction between Runx1 and AP4 may bring E4P and S4 into close proximity, so that Runx1 or other silencer-associated factors can antagonize co-activator recruitment by HEB, E2A and other positively acting factors. Additionally Runx1-AP4 mediated interaction between S4 and E4P could prevent recruitment of E4P-bound transcriptional co-factors like P-TEFb to the promoter, precluding elongation by RNA PolII until the DP stage (Jiang et al., 2005) (Figure 2).
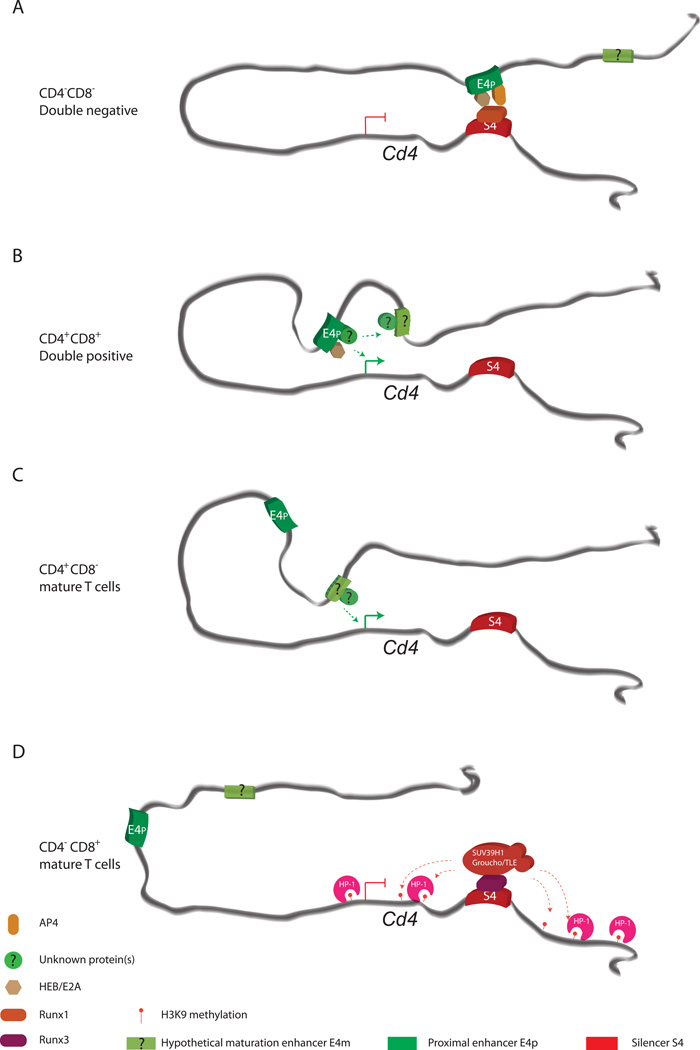
Figure 2. A model for Cd4 regulation during development.
A) At the DN stage, S4 interacts with E4p enhancer, preventing it from interacting with, and activating, the Cd4 promoter. This interaction could be mediated through association between E4p-bound AP-4 and S4-bound Runx1. B) In DP cells, S4 is inactivated, allowing E4p to interact with the Cd4 promoter to drive transcription. Deletion of E4p suggests that it may activate a yet unidentified maturation enhancer (E4m), or collaborate with this enhancer to activate an epigenetic state required for subsequent high-level, stable Cd4 expression in mature helper T cells. C) In mature helper cells, Cd4 is expressed independently of E4p, possibly due to epigenetic mechanisms or E4p-mediated activation of the putative E4m enhancer. D) In cytotoxic T cells, Cd4 is silenced by S4 in a Runx3 and HP-1 dependent manner. Runx3 could recruit transcriptional co-repressors such as SUV39H1 and Groucho/TLE to the locus to deposit repressive histone marks, such as Histone H3K9 methylation. These marks could in turn recruit HP-1 to epigenetically suppress transcription. The role of DNA methylation remains to be further investigated and thus is not shown here. TCF-1α/LEF-1 binding on E4p is not shown.
It should be noted that reversible Cd4 silencing in DNs is likely more complicated than this model suggests. Genetic studies have demonstrated that BAF57 and BRG subunits of the SWI/SNF-like chromatin remodeling complexes are critical for Cd4 silencing in DN cells (Chi et al., 2003; Chi et al., 2002). Interestingly, dominant negative BAF57 expression or T cell specific Brg deletion results in decreased chromatin accessibility and Runx1 binding at S4 accompanied by CD4 de-repression in DN cells (Wan et al., 2009), indicating that SWI/SNF contributes to reversible silencing by remodeling chromatin to allow for Runx1 recruitment. In contrast to SWI/SNF, the NuRD chromatin remodeling complex has been implicated in reversing Cd4 silencing. Deletion of the Mi2b NuRD subunit allows Cd4 silencing to continue past the DN stage (Naito et al., 2007). Taking into account that NuRD is generally considered a repressive chromatin remodeling complex, it is tempting to speculate that NuRD could remodel S4 chromatin to a state inaccessible to Runx1, eliminating silencer function during the transition from DN to DP. Taken together, it appears that reversible silencing requires chromatin remodeling by SWI/SNF, which allows Runx1 to bind to S4 and interact with E4P-bound AP4, thus actively repressing transcriptional elongation at Cd4.
In contrast to transient Cd4 silencing in DN cells, silencing in mature cytotoxic T cells appears to be permanent and mediated epigenetically (Figure 1B). Mutation of individual critical transcription factor binding sites in S4 (including one Runx binding motif) led to partial, but uniform, CD4 de-repression in DN cells, but variegated CD4 expression on CD8+ T cells (Taniuchi et al., 2002a; Taniuchi et al., 2002b). This variegated pattern is reminiscent of position effect variegation (PEV) of transgene expression, which is mediated by heterochromatin spreading from adjacent loci and its stable propagation through multiple mitoses (Fodor et al., 2010). The epigenetic nature of Cd4 silencing was confirmed by the finding of continued stable silencing of CD4 expression through multiple rounds of cell division following Cre-mediated deletion of S4 in mature CD8+ T cells (Zou et al., 2001). Thus S4 initiates Cd4 silencing during development, and this silenced state may be epigenetically propagated in the absence of S4.
These data indicate that epigenetic silencing of Cd4 involves two distinct mechanisms: 1) a silenced state is first initiated after positive selection by factors associated with S4, and 2) silencing is maintained in mature cells independently of S4..Runx3 binding to S4 is clearly required for the initiation of silencing, but not maintenance (Taniuchi et al., 2002a; Taniuchi et al., 2002b). To understand maintenance, we initially assessed DNA methylation, as it is required to maintain heterochromatin-dependent X chromosome inactivation (Sado et al., 2000). We found that pharmacological inhibition of DNA methyltransferase activity with 5-azacytidine did not induce CD4 expression in proliferating CD8+ cells, and argued that DNA methylation does not have a key role in maintenance (Zou et al., 2001). However, the time frame of the experiment, with a relatively small number of mitoses, precluded reaching a definitive conclusion, and this issue needs to be further explored with mice mutant for DNA methyltransferase genes. Intriguingly, overexpression of the heterochromatin protein HP-1β partially rescued silencing in mice in which silencer function was compromised by deletion of the binding domain for a critical, but yet unidentified, transcription factor, indicating that HP-1 proteins and the H3K9 methylation marks that they recognize may contribute to silencing (Taniuchi et al., 2002b) (Figure 2D). In accord with this finding, Runx transcription factors can associate with the Groucho/TLE corepressor complex and the SUV39H1 H3K9 methyltransferase to mediate repression (Levanon et al., 1998; Reed-Inderbitzin et al., 2006). The Runx3 VWRPY motif is required for interactions with Groucho/TLE (its importance for interactions with SUV39H1 is unknown) and for epigenetic Cd4 silencing in mature CD8+ cells (Yarmus et al., 2006), indicating that the Groucho/TLE co-repressor or other factors that interact with Runx3 through this domain are required for silencing. Importantly, the Runx1 VWRPY motif is dispensable for its silencing function in DNs (Telfer et al., 2004), highlighting the two modes of Cd4 silencing: transient in DNs and permanent/epigenetic in CD8+ T cells.
What other mechanisms could be involved in the initiation and maintenance of epigenetic Cd4 silencing? X chromosome inactivation (XCI) is reminiscent of this process as an X chromosome inactivation center (Xic) is crucial for XCI in the inner cell mass (in mice), but is not necessary to maintain the inactive X in a silenced state in more differentiated cells (Brown and Willard, 1994; Wutz and Jaenisch, 2000). Interestingly, the non-coding XIST (nc)RNA encoded within the Xic is required for initiation but not maintenance of XCI (Csankovszki et al., 1999). Similarly, imprinted genes are silenced on one parental allele through the action of ncRNAs (Reviewed in O'Neill, 2005). Thus it will be interesting to determine if ncRNAs play a role in either the initiation or maintenance of the silenced state of Cd4. An interesting possibility is that Runx3 or another silencer binding factor functions to tether a ncRNA to the Cd4 locus to initiate silencing, similarly to the recently described role of YY1 in tethering XIST to the X chromosome undergoing inactivation (Jeon and Lee, 2011).
As illustrated in this discussion, Cd4 silencing deserves vigorous study in the future, as it can provide potentially novel insight into the mechanisms of establishment and inheritance of gene repression. Importantly, this is a unique system to study these events as in contrast to XCI, which occurs stochastically on either of the two X chromosomes, Cd4 silencing occurs in a developmentally regulated manner dependent on extracellular signals (i.e. TCR specificity for MHCI). This dependency on extracellular cues may provide unique insights into the initiation of epigenetic silencing, that may not be revealed by XCI initiation due to its stochastic nature. Finally, the Cd4 system is less difficult to work with: knowledge of the signals that induce Cd4 silencing, as well as the fact that T cell differentiation occurs in post-natal mice, render this system more easily manipulated than the embryonic inner cell mass where XCI is initiated.
In addition to silencing, recent work from our laboratory has shown that the active transcription state of Cd4 in helper T cells is also propagated epigenetically, i.e. independently of the genetic element that initially activates Cd4 transcription (Figure 1C). As mentioned earlier, E4P is required for CD4 expression in DP thymocytes (Chong et al., 2010). However, positive selection of E4P−/− thymocytes activates CD4 expression in T helper lineage cells, possibly through a putative “maturation enhancer”, leading to a reduced population of CD4+ helper T cells. These cells had a broader distribution and lower amount of CD4 expression than wild type cells, and lost expression upon TCR stimulation and proliferation. However, conditional deletion of E4P in mature peripheral CD4+ cells by retroviral transduction of Cre had no effect on the stability or level of CD4 expression, even after many rounds of division. Thus, like the mirror opposite of the silencer, E4P sets an active epigenetic state of the Cd4 locus in helper T cells, which can then be propagated in its absence.
The mechanisms underlying this positive epigenetic state are not entirely clear. Mature E4P−/− CD4+ T cells expressed lower levels of CD4 than WT despite normal levels of histone acetylation across the Cd4 locus. In contrast, decreased Cd4 transcription in these cells correlated with reduced H3K4me3 at the promoter. Upon TCR-induced proliferation, both histone modifications were lost in those E4P−/− cells that lost Cd4 expression. Considering that there was no concomitant increase in repressive histone marks such as H3K9me3 and H3K27me3 across the locus when compared to WT CD4+ cells, it appears that instability of Cd4 expression is due to the loss of activating marks rather than active silencing. Could differences in H3K4 methylation account for stable (WT) vs. unstable (E4P−/−) memory? In yeast, H3K4me3 has been shown to persist after transcription has ceased, and thus has been postulated to serve as memory of previous transcriptional activity, though admittedly not through a cell cycle (Ng et al., 2003). Intriguingly, studies in mammalian cells have suggested longer memory through H3K4me3. Memory in the murine inflammatory response has been linked to persistent H3K4 trimethylation and H4 acetylation, and competence to recruit the SWI/SNF subunit Brg1 (Foster et al., 2007). In studies of IFNγ priming of MHCII gene transcription, H3K4me2 is stably transmitted through cell cycles, correlating with increased responsiveness to secondary stimulation (Gialitakis et al., 2010). Thus, one simple model would be that H3K4 methylation might allow transmission through the cell cycle, though the exact mechanism is still unclear.
What are other possible mechanisms for the propagation of active Cd4 transcription? Two obvious suspects come to mind: DNA hypomethylation and deposition of histone variants. DNA demethylation is involved in activating epigenetic memory in the Il2 and FoxP3 loci. Activation of naïve T cells leads to demethylation of the Il2 promoter, which in turn allows for faster and stronger transcriptional response following secondary stimulation (Bruniquel and Schwartz, 2003; Murayama et al., 2006). At the FoxP3 locus, DNA demethylation allows binding of positively acting factors including Runx1 and FoxP3 itself, completing a feed-forward loop, which stabilizes FoxP3 expression (Bruno et al., 2009; Lal et al., 2009; Polansky et al., 2008; Williams and Rudensky, 2007; Zheng et al., 2010). In the case of Cd4 expression, E4P-dependent hypomethylation of specific motifs in the locus may allow a transcription factor(s) to bind and maintain Cd4 expression. Another, non-mutually exclusive, possibility is that deposition of specific histone variants potentiates memory. The variant H2A.Z has been shown to be required for memory (faster reactivation) of genes in yeast (Brickner et al., 2007). While no difference in H2A.Z occupancy was found at the Cd4 promoter between WT and E4P−/− CD4+ cells (Chong et al., 2010), it remains possible that H2A.Z underlies stable Cd4 expression as it is also thought to play critical roles in enhancer accessibility (He et al., 2010). Thus E4P-dependent H2A.Z deposition at an unidentified enhancer element could promote stable Cd4 expression. Another candidate is the histone variant H3.3, which acts at the MyoD promoter to allow persistent (and inappropriate) MyoD expression in Xenopus muscle cell nuclei that have been reprogrammed by two rounds of nuclear transfer into embryos (Ng and Gurdon, 2008). Finer examination of DNA methylation and histone variant occupancy in WT and E4P−/− cells will be required to evaluate these possibilities.
The mechanisms underpinning epigenetic memory of Cd4 transcription are still unclear. Nevertheless, this system clearly deserves to be studied extensively, since, despite the widespread notion that self-propagating epigenetic mechanisms are critical to the developmental regulation of many genes, the activation and silencing of Cd4 remain rare examples of bona fide heritability in mammalian gene expression.
Epigenetic regulation of the Cd8 locus
The Cd8 locus consists of the Cd8a and Cd8b genes, separated by ~35kb in mice, and ~25kb in humans. CD8 is expressed either as a homo-dimer of CD8αα molecules, for example on intraepithelial lymphocytes (IEL) and CD8+ DCs, or as a hetero-dimer of CD8αβ molecules on DP thymocytes and TCRαβ cytotoxic T cells. Thus Cd8a and Cd8b genes can be both co- and independently regulated (reviewed in (Taniuchi et al., 2004)). Here we focus mainly on Cd8 locus regulation in the TCRαβ lineage, and what hints this gives us into epigenetic mechanisms of gene regulation.
Cd8 locus expression is controlled by multiple enhancers. While the Cd8a promoter was not sufficient to drive lineage specific expression, an 80kb fragment of the mouse locus stretching from 2kb upstream of Cd8b to 25kb downstream of Cd8a could drive tissue specific expression (Hostert et al., 1997a). Thus all cis-regulatory elements critical for appropriate Cd8a/b expression were present in this interval. This study also identified four DHS clusters (CI-IV), as putative regulatory regions. Further transgenic studies based on these DHS clusters identified specific regions that regulate Cd8 expression (Ellmeier et al., 1997; Ellmeier et al., 1998; Hostert et al., 1998; Hostert et al., 1997a; Hostert et al., 1997b; Kieffer et al., 1996; Kieffer et al., 1997; Zhang et al., 1998; Zhang et al., 2001). The E8I (CIII-1,2) enhancer drove transgene expression in mature CD8+ cells and in IEL (Ellmeier et al., 1997; Ellmeier et al., 1998; Hostert et al., 1997b). The E8III (CIV-3) enhancer drove expression in DP thymocytes, and E8II (CIV-4,5) in DPs and mature CD8+ cells. The E8IV (CIV-1,2) element was more promiscuous than the above enhancers, driving low level expression in CD4+ T cells as well as DP and CD8+ T cells (Ellmeier et al., 1998; Feik et al., 2005). Finally, while E8V (CII) exhibited no enhancer function by itself, a combination of E8V and E8I drove expression in DP cells in addition to mature CD8+ cells (Hostert et al., 1998; Hostert et al., 1997b). These studies indicated that regulation of Cd8 locus expression in a cell type- and stage-specific manner is achieved through complex interactions between multiple and sometimes apparently redundant regulatory elements.
To determine the in vivo function of the individual Cd8 locus enhancers in controlling Cd8a and Cd8b expression, knockout studies were undertaken. Deletion of E8I, which drives transgene activity in IELs and mature CD8+ cells, reduced CD8αα and CD8αβ expression on IEL by 40–80%, but left CD8 expression on TCRαβ DPs and mature CD8+ cells largely unaffected; there was only a minor decrease (10–20%) in CD8 expression at the CD8SP stage (Ellmeier et al., 1998; Hostert et al., 1998). There was no detectable phenotype in E8II−/− animals, suggesting that loss of E8II activity may be compensated for by other enhancers (Ellmeier et al., 2002). Indeed, combined deletion of E8II and E8I, or E8II and E8III, resulted in variegated CD8 expression in DP cells (Ellmeier et al., 2002; Feik et al., 2005), indicating incomplete relief of heterochromatin-mediated Cd8 repression. Combined E8II/E8I deletion also reduced CD8 expression by 30% on mature CD8+ T cells in the periphery. Similarly to the combined deletions above, elimination of E8V resulted in variegated CD8 expression on DPs, as well as reduced CD8 expression on mature CD8+ cells (down 20%) (Garefalaki et al., 2002). These results indicate that while E8I is specifically required for CD8αα expression on IEL and DCs, the E8I, E8II, E8III and E8V elements all contribute to relieving heterochromatin mediated Cd8 locus repression at the DN to DPs stage, and to maintaining high-level CD8αβ expression in mature CD8+ cells (Figure 3).
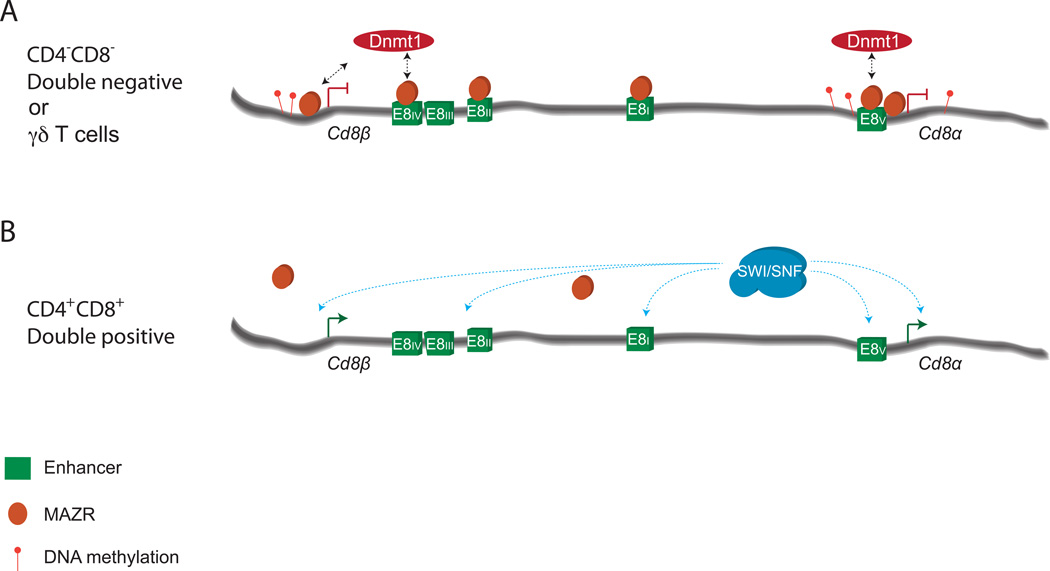
Figure 3. A model for the epigenetic repression of Cd8 locus transcription.
A) CpGs at the Cd8a and Cd8b promoters, as well as the E8V enhancer, are hypermethylated in DN and γδ T cells. Loss of this methylation due to DNMT1 deletion leads to inappropriate CD8 expression in these cells. MAZR associates with multiple regulatory elements in the Cd8 locus, and represses Cd8α/β transcription at the DN to DP transition, possibly through interactions with DNA methyltransferases. B) Downregulation of MAZR expression correlates with the relief of epigenetic Cd8α/β repression at the DP stage. Components of the SWI/SNF chromatin remodeling complex, and multiple enhancer elements (E8I, E8II, E8III, and E8V), contribute to the efficient activation of Cd8α/β expression in DP cells.
This begs the question: why does the Cd8 locus contain multiple, seemingly redundant enhancers? Recent work in Drosophila demonstrated that apparently redundant enhancers are critical to maintaining expression in response to genetic and environmental stimuli (Frankel et al., 2010). Considering the importance of precisely controlled CD8 expression in lineage choice and cytotoxic T cell development (Fung-Leung et al., 1991; Sarafova et al., 2005), it is possible that this may also be the evolutionary driving force behind the development of multiple Cd8 locus enhancers. Another possibility is that seemingly overlapping Cd8 locus enhancers are important for the expression of Cd8a or Cd8b individually (Taniuchi et al., 2004). Most of the above work relied on surface CD8αβ protein detection to infer Cd8 gene expression, but murine CD8β cannot be expressed on the surface in the absence of CD8α (Devine et al., 2000). Thus, examination of Cd8a and Cd8b transcription in various enhancer knockout mouse strains would be required to evaluate if individual enhancers are important for the expression of either gene individually.
CD8 expression appears to be regulated through multiple epigenetic mechanisms. The variegated CD8 expression phenotypes of DP thymocytes from E8I/II, E8II/III and CII knockout mice suggest that the Cd8 locus becomes activated during the DN to DP transition through reversal of potentially heritable repressive marks (Ellmeier et al., 2002; Feik et al., 2005; Garefalaki et al., 2002). Interestingly, expression of a dominant negative mutant of the Baf57 SWI/SNF complex subunit, or haploinsufficiency of the Brg subunit, resulted in diminished CD8 expression on DPs (Chi et al., 2002). Further, combining these two genetic defects in SWI/SNF members resulted in variegated CD8 expression on DPs. Thus, the SWI/SNF complex is critical for activation of CD8 expression at the transition from the DN4 to the DP stage (Figure 3).
The activation of Cd8 expression is not only controlled by positive regulators; DNA methylation and the zinc finger protein MAZR (zfp278) also play critical roles in epigenetically repressing the locus (Figure 3). MAZR is highly expressed in DN cells, and is downregulated at the DP stage as CD8 expression is activated (Bilic et al., 2006). Forced expression of MAZR results in variegated CD8 expression at the DP stage (Bilic et al., 2006). Further, variegated CD8 expression on E8I/II deficient DP cells (Ellmeier et al., 2002) was partially relieved by MAZR deletion (Sakaguchi et al., 2010), suggesting that E8I/II antagonizes MAZR-mediated Cd8 silencing during the DN to DP transition. This effect may be mediated through the direct binding of MAZR as has been observed at multiple elements in the Cd8 locus (Bilic et al., 2006). Analysis of CD8 expression in mice with combined deletion of MAZR and various Cd8 locus elements could reveal elements through which MAZR functions to repress Cd8. Interestingly, deletion of the maintenance DNA methyltransferase Dnmt1 on an E8I/II deficient background also partially rescued variegated CD8 expression (Bilic et al., 2006). While it is tempting to speculate that MAZR contributes to Cd8 repression through recruitment or maintenance of DNA methylation, no clear link has been established between MAZR and initiating or maintenance DNA methyltransferases. In keeping with the observation that Dnmt1 contributes to Cd8 locus repression, E8V sequences are differentially methylated between CD8+ and CD8- cells (e.g. WT DP cells vs. liver or CD8- E8I/II DP cells) (Bilic et al., 2006; Carbone et al., 1988; Hamerman et al., 1997; Lee et al., 2001). Further, Dnmt1 deletion results in ectopic CD8 expression on TCRγδ cells (Lee et al., 2001). Thus, DNA methylation silences Cd8a/b expression outside of the TCRαβ lineage, and, along with MAZR, maintains Cd8a/b repression until the DP stage, apparently through epigenetic mechanisms.
Long distance interactions between Cd4 and Cd8 co-receptor loci during lineage choice
In addition to cis-regulatory elements in each locus, it appears that Cd4 and Cd8 expression may be regulated by long distance interactions between loci and specific nuclear compartments. For example, the majority of Cd4 alleles associate with peri-centromeric heterochromatin (PCH) in CD8+ but not CD4+ T cells; similarly cd8 alleles preferentially associate with PCH in CD4+ but not CD8+ T cells (Collins et al., 2011; Delaire et al., 2004; Merkenschlager et al., 2004). These results indicate that lineage specific repression/silencing of these loci may result from localization to PCH, consistent with heterochromatic silencing. More intriguingly, we have found that the Cd4 and Cd8 loci dynamically associate during T cell development (Collins et al., 2011). The loci are closely associated in cis at the DP stage, separate slightly immediately after positive selection (CD4+CD8lo stage), and then are more closely associated in CD8+ than in CD4+ T cells. Association between the two loci is regulated by cis-acting elements in each locus (the E8I and E8II Cd8 enhancers and Cd4 silencer promote association in DPs and CD8+ T cells), as well as the transcription factors that govern lineage choice (Runx proteins promote associations in DPs and CD8+ T cells, while ThPOK antagonizes these associations in CD4+ T cells). Intriguingly, this phenomenon is evolutionarily conserved, as the CD4 and CD8 loci associate closely in human CD8+, but not CD4+, T cells. It is tempting to speculate that long distance association of the Cd4 and Cd8 loci in DP cells allows for co-receptor expression to be precisely and oppositely regulated during lineage commitment to the helper and cytotoxic lineages. This is indeed reminiscent of observations that the Th1 and Th2 cytokine loci interact in naïve T cells and separate upon polarization, and that the deletion of a DHS site in the Th2 locus delays activation of the Th1 locus during Th1 cell differentiation (Lee et al., 2005). Coordinated regulation of both cytokine and co-receptor loci may be facilitated by the close association of these loci in naïve helper T cells and immature DP cells, respectively.
Concluding remarks
Study of co-receptor gene expression during T cell development has yielded insights into transcriptional regulatory mechanisms in general. While Cd4 and Cd8 expression are tightly coordinated throughout development, the molecular mechanisms that govern their simultaneous repression (DN stage), simultaneous expression (DP stage) and mutually exclusive expression (single positive stage) are quite different. At the DN stage Cd4 is actively repressed by Runx1, while Cd8 appears epigenetically repressed in a heterochromatin- and DNA methylation-dependent fashion. In DP cells, silencer function is abrogated to allow Cd4 expression, while multiple enhancers are activated to overcome epigenetic silencing of Cd8. At this stage, expression of either gene is reversible. Finally, in helper T cells, transcription of Cd4 is epigenetically maintained and Cd8 expression is extinguished through unknown mechanisms, while in cytotoxic T cells Cd4 is epigenetically silenced in a Runx3 dependent manner and Cd8 transcription is presumably actively maintained through enhancer function. Thus to achieve two basic transcriptional outputs—on or off—for Cd4 and Cd8, developing T cells appear to use multiple different mechanisms, some epigenetic and some not. This is reminiscent of chromatin profiling in Drosophila, which identified different “colors” or flavors of chromatin, defined by the occupancy of unique sets of chromatin modifying complexes, chromatin readers, transcriptional regulators, and transcription factors (Filion et al., 2010). Indeed these different chromatin colors were associated with different levels of transcription, as well as genes belonging to different functional categories. For example, there were two types of active chromatin, one encompassing genes with broad expression patterns, and another enriched in genes linked to specific tissues. Detailed analysis of the protein structure and chromatin modifications at the Cd4 and Cd8 loci may reveal similarly distinct types of chromatin flavors linked to active and repressed states (i.e. the Cd4 locus in helper vs. cytotoxic cells), and instability or heritability of each of those states at different developmental stages (i.e. the Cd4 locus in DP vs. helper T cells). Clearly further characterization of these loci will yield critical insights into the epigenetic regulation of lineage specific transcriptional programs.
References
- Adlam M, Duncan DD, Ng DK, Siu G. Positive selection induces CD4 promoter and enhancer function. Int Immunol. 1997;9:877–887. doi: 10.1093/intimm/9.6.877. [DOI] [PubMed] [Google Scholar]
- Adlam M, Siu G. Hierarchical interactions control CD4 gene expression during thymocyte development. Immunity. 2003;18:173–184. doi: 10.1016/s1074-7613(03)00021-9. [DOI] [PubMed] [Google Scholar]
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Allfrey VG. Structural modifications of histones and their possible role in the regulation of ribonucleic acid synthesis. Proc Can Cancer Conf. 1966;6:313–335. [PubMed] [Google Scholar]
- Bachman KE, Park BH, Rhee I, Rajagopalan H, Herman JG, Baylin SB, Kinzler KW, Vogelstein B. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 2003;3:89–95. doi: 10.1016/s1535-6108(02)00234-9. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilic I, Koesters C, Unger B, Sekimata M, Hertweck A, Maschek R, Wilson CB, Ellmeier W. Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR. Nat Immunol. 2006;7:392–400. doi: 10.1038/ni1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum MD, Wong GT, Higgins KM, Sunshine MJ, Lacy E. Reconstitution of the subclass-specific expression of CD4 in thymocytes and peripheral T cells of transgenic mice: identification of a human CD4 enhancer. J Exp Med. 1993;177:1343–1358. doi: 10.1084/jem.177.5.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Willard HF. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- Bruniquel D, Borie N, Hannier S, Triebel F. Regulation of expression of the human lymphocyte activation gene-3 (LAG-3) molecule, a ligand for MHC class II. Immunogenetics. 1998;48:116–124. doi: 10.1007/s002510050411. [DOI] [PubMed] [Google Scholar]
- Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- Bruno L, Mazzarella L, Hoogenkamp M, Hertweck A, Cobb BS, Sauer S, Hadjur S, Leleu M, Naoe Y, Telfer JC, et al. Runx proteins regulate Foxp3 expression. J Exp Med. 2009;206:2329–2337. doi: 10.1084/jem.20090226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Carbone AM, Marrack P, Kappler JW. Demethylated CD8 gene in CD4+ T cells suggests that CD4+ cells develop from CD8+ precursors. Science. 1988;242:1174–1176. doi: 10.1126/science.2460926. [DOI] [PubMed] [Google Scholar]
- Chi TH, Wan M, Lee PP, Akashi K, Metzger D, Chambon P, Wilson CB, Crabtree GR. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19:169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- Chi TH, Wan M, Zhao K, Taniuchi I, Chen L, Littman DR, Crabtree GR. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418:195–199. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- Chong MM, Simpson N, Ciofani M, Chen G, Collins A, Littman DR. Epigenetic propagation of CD4 expression is established by the Cd4 proximal enhancer in helper T cells. Genes Dev. 2010;24:659–669. doi: 10.1101/gad.1901610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Zuniga-Pflucker JC. Determining gammadelta versus alphass T cell development. Nat Rev Immunol. 2010;10:657–663. doi: 10.1038/nri2820. [DOI] [PubMed] [Google Scholar]
- Collins A, Hewitt SL, Chaumeil J, Sellars M, Micsinai M, Allinne J, Parisi F, Nora EP, Bolland DJ, Corcoran AE, et al. RUNX transcription factor-mediated association of Cd4 and Cd8 enables coordinate gene regulation. Immunity. 2011;34:303–314. doi: 10.1016/j.immuni.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- Delaire S, Huang YH, Chan SW, Robey EA. Dynamic repositioning of CD4 and CD8 genes during T cell development. J Exp Med. 2004;200:1427–1435. doi: 10.1084/jem.20041041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine L, Kieffer LJ, Aitken V, Kavathas PB. Human CD8 beta, but not mouse CD8 beta, can be expressed in the absence of CD8 alpha as a beta beta homodimer. J Immunol. 2000;164:833–838. doi: 10.4049/jimmunol.164.2.833. [DOI] [PubMed] [Google Scholar]
- Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellmeier W, Sunshine MJ, Losos K, Hatam F, Littman DR. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity. 1997;7:537–547. doi: 10.1016/s1074-7613(00)80375-1. [DOI] [PubMed] [Google Scholar]
- Ellmeier W, Sunshine MJ, Losos K, Littman DR. Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity. 1998;9:485–496. doi: 10.1016/s1074-7613(00)80632-9. [DOI] [PubMed] [Google Scholar]
- Ellmeier W, Sunshine MJ, Maschek R, Littman DR. Combined deletion of CD8 locus cis-regulatory elements affects initiation but not maintenance of CD8 expression. Immunity. 2002;16:623–634. doi: 10.1016/s1074-7613(02)00309-6. [DOI] [PubMed] [Google Scholar]
- Feik N, Bilic I, Tinhofer J, Unger B, Littman DR, Ellmeier W. Functional and molecular analysis of the double-positive stage-specific CD8 enhancer E8III during thymocyte development. J Immunol. 2005;174:1513–1524. doi: 10.4049/jimmunol.174.3.1513. [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Fodor BD, Shukeir N, Reuter G, Jenuwein T. Mammalian Su(var) genes in chromatin control. Annu Rev Cell Dev Biol. 2010;26:471–501. doi: 10.1146/annurev.cellbio.042308.113225. [DOI] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Fung-Leung WP, Schilham MW, Rahemtulla A, Kundig TM, Vollenweider M, Potter J, van Ewijk W, Mak TW. CD8 is needed for development of cytotoxic T cells but not helper T cells. Cell. 1991;65:443–449. doi: 10.1016/0092-8674(91)90462-8. [DOI] [PubMed] [Google Scholar]
- Garefalaki A, Coles M, Hirschberg S, Mavria G, Norton T, Hostert A, Kioussis D. Variegated expression of CD8 alpha resulting from in situ deletion of regulatory sequences. Immunity. 2002;16:635–647. doi: 10.1016/s1074-7613(02)00308-4. [DOI] [PubMed] [Google Scholar]
- Gialitakis M, Arampatzi P, Makatounakis T, Papamatheakis J. Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol Cell Biol. 2010;30:2046–2056. doi: 10.1128/MCB.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamerman JA, Page ST, Pullen AM. Distinct methylation states of the CD8 beta gene in peripheral T cells and intraepithelial lymphocytes. J Immunol. 1997;159:1240–1246. [PubMed] [Google Scholar]
- Hanna Z, Simard C, Laperriere A, Jolicoeur P. Specific expression of the human CD4 gene in mature CD4+ CD8− and immature CD4+ CD8+ T cells and in macrophages of transgenic mice. Mol Cell Biol. 1994;14:1084–1094. doi: 10.1128/mcb.14.2.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- Hostert A, Garefalaki A, Mavria G, Tolaini M, Roderick K, Norton T, Mee PJ, Tybulewicz VL, Coles M, Kioussis D. Hierarchical interactions of control elements determine CD8alpha gene expression in subsets of thymocytes and peripheral T cells. Immunity. 1998;9:497–508. doi: 10.1016/s1074-7613(00)80633-0. [DOI] [PubMed] [Google Scholar]
- Hostert A, Tolaini M, Festenstein R, McNeill L, Malissen B, Williams O, Zamoyska R, Kioussis D. A CD8 genomic fragment that directs subset-specific expression of CD8 in transgenic mice. J Immunol. 1997a;158:4270–4281. [PubMed] [Google Scholar]
- Hostert A, Tolaini M, Roderick K, Harker N, Norton T, Kioussis D. A region in the CD8 gene locus that directs expression to the mature CD8 T cell subset in transgenic mice. Immunity. 1997b;7:525–536. doi: 10.1016/s1074-7613(00)80374-x. [DOI] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. YY1 Tethers Xist RNA to the Inactive X Nucleation Center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Peterlin BM. Differential chromatin looping regulates CD4 expression in immature thymocytes. Mol Cell Biol. 2008;28:907–912. doi: 10.1128/MCB.00909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Zhang F, Kurosu T, Peterlin BM. Runx1 binds positive transcription elongation factor b and represses transcriptional elongation by RNA polymerase II: possible mechanism of CD4 silencing. Mol Cell Biol. 2005;25:10675–10683. doi: 10.1128/MCB.25.24.10675-10683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–870. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer LJ, Bennett JA, Cunningham AC, Gladue RP, McNeish J, Kavathas PB, Hanke JH. Human CD8 alpha expression in NK cells but not cytotoxic T cells of transgenic mice. Int Immunol. 1996;8:1617–1626. doi: 10.1093/intimm/8.10.1617. [DOI] [PubMed] [Google Scholar]
- Kieffer LJ, Yan L, Hanke JH, Kavathas PB. Appropriate developmental expression of human CD8 beta in transgenic mice. J Immunol. 1997;159:4907–4912. [PubMed] [Google Scholar]
- Killeen N, Sawada S, Littman DR. Regulated expression of human CD4 rescues helper T cell development in mice lacking expression of endogenous CD4. EMBO J. 1993;12:1547–1553. doi: 10.1002/j.1460-2075.1993.tb05798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP, Reid SP, Levy DE, Bromberg JS. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182:259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Lee GR, Spilianakis CG, Flavell RA. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat Immunol. 2005;6:42–48. doi: 10.1038/ni1148. [DOI] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Leung RK, Thomson K, Gallimore A, Jones E, Van den Broek M, Sierro S, Alsheikhly AR, McMichael A, Rahemtulla A. Deletion of the CD4 silencer element supports a stochastic mechanism of thymocyte lineage commitment. Nat Immunol. 2001;2:1167–1173. doi: 10.1038/ni733. [DOI] [PubMed] [Google Scholar]
- Levanon D, Goldstein RE, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci U S A. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Thanos D. Nucleosome sliding via TBP DNA binding in vivo. Cell. 2001;106:685–696. doi: 10.1016/s0092-8674(01)00490-1. [DOI] [PubMed] [Google Scholar]
- Manjunath N, Shankar P, Stockton B, Dubey PD, Lieberman J, von Andrian UH. A transgenic mouse model to analyze CD8(+) effector T cell differentiation in vivo. Proc Natl Acad Sci U S A. 1999;96:13932–13937. doi: 10.1073/pnas.96.24.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B, Bourachot B, Rachez C, Ogryzko V, Muchardt C. Regulation of an inducible promoter by an HP1beta-HP1gamma switch. EMBO Rep. 2008;9:267–272. doi: 10.1038/embor.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Amoils S, Roldan E, Rahemtulla A, O'Connor E, Fisher AG, Brown KE. Centromeric repositioning of coreceptor loci predicts their stable silencing and the CD4/CD8 lineage choice. J Exp Med. 2004;200:1437–1444. doi: 10.1084/jem.20041127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A, Sakura K, Nakama M, Yasuzawa-Tanaka K, Fujita E, Tateishi Y, Wang Y, Ushijima T, Baba T, Shibuya K, et al. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J. 2006;25:1081–1092. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, Kawamoto H, Taniuchi I. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- Naito T, Gomez-Del Arco P, Williams CJ, Georgopoulos K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2beta determine silencer activity and Cd4 gene expression. Immunity. 2007;27:723–734. doi: 10.1016/j.immuni.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Naito T, Taniuchi I. The network of transcription factors that underlie the CD4 versus CD8 lineage decision. Int Immunol. 2010;22:791–796. doi: 10.1093/intimm/dxq436. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- O'Neill MJ. The influence of non-coding RNAs on allele-specific gene expression in mammals. Hum Mol Genet. 2005;141(Spec No):R113–R120. doi: 10.1093/hmg/ddi108. [DOI] [PubMed] [Google Scholar]
- Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000;407:471–475. doi: 10.1038/35035000. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, Olek S, Hamann A, von Boehmer H, Huehn J. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38:1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- Reed-Inderbitzin E, Moreno-Miralles I, Vanden-Eynden SK, Xie J, Lutterbach B, Durst-Goodwin KL, Luce KS, Irvin BJ, Cleary ML, Brandt SJ, et al. RUNX1 associates with histone deacetylases and SUV39H1 to repress transcription. Oncogene. 2006;25:5777–5786. doi: 10.1038/sj.onc.1209591. [DOI] [PubMed] [Google Scholar]
- Roh TY, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Fenner MH, Tan SS, Tam P, Shioda T, Li E. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev Biol. 2000;225:294–303. doi: 10.1006/dbio.2000.9823. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Hombauer M, Bilic I, Naoe Y, Schebesta A, Taniuchi I, Ellmeier W. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat Immunol. 2010;11:442–448. doi: 10.1038/ni.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands JF, Nikolic-Zugic J. T cell-specific protein-DNA interactions occurring at the CD4 locus: identification of possible transcriptional control elements of the murine CD4 gene. Int Immunol. 1992;4:1183–1194. doi: 10.1093/intimm/4.10.1183. [DOI] [PubMed] [Google Scholar]
- Sarafova SD, Erman B, Yu Q, Van Laethem F, Guinter T, Sharrow SO, Feigenbaum L, Wildt KF, Ellmeier W, Singer A. Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity. 2005;23:75–87. doi: 10.1016/j.immuni.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Sato T, Ohno S, Hayashi T, Sato C, Kohu K, Satake M, Habu S. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity. 2005;22:317–328. doi: 10.1016/j.immuni.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Sawada S, Littman DR. Identification and characterization of a T-cell-specific enhancer adjacent to the murine CD4 gene. Mol Cell Biol. 1991;11:5506–5515. doi: 10.1128/mcb.11.11.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S, Littman DR. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol Cell Biol. 1993;13:5620–5628. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu G, Wurster AL, Duncan DD, Soliman TM, Hedrick SM. A transcriptional silencer controls the developmental expression of the CD4 gene. EMBO J. 1994;13:3570–3579. doi: 10.1002/j.1460-2075.1994.tb06664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi I, Ellmeier W, Littman DR. The CD4/CD8 lineage choice: new insights into epigenetic regulation during T cell development. Adv Immunol. 2004;83:55–89. doi: 10.1016/S0065-2776(04)83002-5. [DOI] [PubMed] [Google Scholar]
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002a;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- Taniuchi I, Sunshine MJ, Festenstein R, Littman DR. Evidence for distinct CD4 silencer functions at different stages of thymocyte differentiation. Mol Cell. 2002b;10:1083–1096. doi: 10.1016/s1097-2765(02)00735-9. [DOI] [PubMed] [Google Scholar]
- Telfer JC, Hedblom EE, Anderson MK, Laurent MN, Rothenberg EV. Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes. J Immunol. 2004;172:4359–4370. doi: 10.4049/jimmunol.172.7.4359. [DOI] [PubMed] [Google Scholar]
- Wan M, Zhang J, Lai D, Jani A, Prestone-Hurlburt P, Zhao L, Ramachandran A, Schnitzler GR, Chi T. Molecular basis of CD4 repression by the Swi/Snf-like BAF chromatin remodeling complex. Eur J Immunol. 2009;39:580–588. doi: 10.1002/eji.200838909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wildt KF, Castro E, Xiong Y, Feigenbaum L, Tessarollo L, Bosselut R. The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity. 2008a;29:876–887. doi: 10.1016/j.immuni.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, Bosselut R. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol. 2008b;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Wong YC, Elgin SC. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979;16:807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Wu HH, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurster AL, Siu G, Leiden JM, Hedrick SM. Elf-1 binds to a critical element in a second CD4 enhancer. Mol Cell Biol. 1994;14:6452–6463. doi: 10.1128/mcb.14.10.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Yarmus M, Woolf E, Bernstein Y, Fainaru O, Negreanu V, Levanon D, Groner Y. Groucho/transducin-like Enhancer-of-split (TLE)-dependent and -independent transcriptional regulation by Runx3. Proc Natl Acad Sci U S A. 2006;103:7384–7389. doi: 10.1073/pnas.0602470103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Wan M, Zhang J, Wu J, Khatri R, Chi T. Nucleoprotein structure of the CD4 locus: implications for the mechanisms underlying CD4 regulation during T cell development. Proc Natl Acad Sci U S A. 2008;105:3873–3878. doi: 10.1073/pnas.0800810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XL, Seong R, Piracha R, Larijani M, Heeney M, Parnes JR, Chamberlain JW. Distinct stage-specific cis-active transcriptional mechanisms control expression of T cell coreceptor CD8 alpha at double- and single-positive stages of thymic development. J Immunol. 1998;161:2254–2266. [PubMed] [Google Scholar]
- Zhang XL, Zhao S, Borenstein SH, Liu Y, Jayabalasingham B, Chamberlain JW. CD8 expression up to the double-positive CD3(low/intermediate) stage of thymic differentiation is sufficient for development of peripheral functional cytotoxic T lymphocytes. J Exp Med. 2001;194:685–693. doi: 10.1084/jem.194.5.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou YR, Sunshine MJ, Taniuchi I, Hatam F, Killeen N, Littman DR. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat Genet. 2001;29:332–336. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]