Abstract
Eukaryotic DNA ligases seal DNA breaks in the final step of DNA replication and repair transactions via a three-step reaction mechanism that can abort if DNA ligases encounter modified DNA termini, such as the products and repair intermediates of DNA oxidation, alkylation, or the aberrant incorporation of ribonucleotides into genomic DNA. Such abortive DNA ligation reactions create 5’–adenylated nucleic acid termini in the context of DNA and RNA-DNA substrates in DNA base excision repair (BER), double strand break repair (DSBR) and ribonucleotide excision repair (RER). Aprataxin (APTX), a protein altered in the heritable neurological disorder Ataxia with Oculomotor Apraxia 1 (AOA1), acts as a DNA ligase “proofreader” to directly reverse AMP-modified nucleic acid termini in DNA- and RNA-DNA damage responses. Herein, we survey APTX function and the emerging cell biological, structural and biochemical data that has established a molecular foundation for understanding the APTX mediated deadenylation reaction, and is providing insights into the molecular bases of APTX deficiency in AOA1.
1. Introduction
In the late 1980's, Ataxia-ocular motor apraxia (AOA, now referred to as Ataxia with Oculomotor Apraxia) was identified as a novel autosomal recessive neurological syndrome. AOA is similar to the DNA repair deficiency disorder, ataxia-telangiectasia (A-T) in that it is typified by ataxia (uncoordinated movement), choreoathetosis (involuntary movement), ocular apraxia (limited eye movement) and cerebellar atrophy, but distinct in that it does not share extra-neurological features of A-T (Aicardi et al., 1988). AOA is separated into three subgroups, AOA1, AOA2, and AOA3 that are linked to mutations in either the human APTX (AOA1) gene encoding Aprataxin (APTX) (Date et al., 2001; Le Ber et al., 2003; Moreira et al., 2001) or SETX (AOA2) encoding Senataxin (SETX) (Moreira et al., 2004). The causative genetic defects in AOA3 are yet to be identified (Gueven et al., 2007; Murad et al., 2013). Missense, frameshift and nonesense mutations in APTX further produce clinically heterogeneous outcomes. APTX deficiency has also been implicated in Ataxia with coenzyme Q10 (CoQ10) deficiency (Quinzii et al., 2005) and a syndrome clinically related to a Parkinson's-like multiple system atrophy (MSA; also see Caldecott, 2008 and references therein for review).
Analysis of the APTX gene product identified cardinal features of APTX orthologs including two highly conserved regions corresponding to a histidine triad (HIT) domain closely linked to a putative C-terminal C2H2 Zn-finger (Znf) domain (Date et al., 2001; Moreira et al., 2001). The HIT superfamily of proteins is a diverse group of nucleoside hydrolases and transferases that include the fragile histidine triad protein (FHIT) and the DCPS mRNA decapping enzyme (Brenner, 2002; Gu et al., 2004; Lima et al., 1997a, 1997b). Consistent with known functions for other HIT proteins, early work established that APTX catalyzes hydrolysis of adenylate nucleotides and dinucleotides such as ATP and AP4A (di-adenosine tetraphosphate), as well as AMP-lysine, albeit with substantially lower activity compared to other HIT proteins (Kijas et al., 2006; Seidle et al., 2005).
Additional findings hinted at a role for APTX in the DNA damage response (DDR). First, APTX possesses an N-terminal forkhead associated (FHA) domain akin to that found in the DNA end damage repair factor polynucleotide kinase phosphatase (PNKP) (Andres et al., 2014; Bernstein et al., 2005; Caldecott 2003; Clements et al., 2004; Durocher et al., 2000; Koch et al., 2004; Moreira et al., 2001). Second, recombinant APTX binds DNA and RNA (Kijas et al., 2006). In connecting the dots to conceptually link the DNA/RNA binding, putative DNA repair functions, and nucleoside hydrolase activity of APTX, West and colleagues reported the seminal finding that APTX harbors a robust hydrolase activity against 5’-adenylated DNA (Ahel et al., 2006). In this capacity, APTX functions as a DNA ligase “proofreader” to directly reverse damaged 5’-adenylated termini of DNA strand breaks that have been subjected to DNA damage-induced “abortive” processing by DNA ligases (Ahel et al., 2006; Caglayan et al., 2014; Harris et al., 2009; Rass et al., 2007, 2008b; Reynolds et al., 2009; Tumbale et al., 2014) (see Fig. 1, section 1.2).
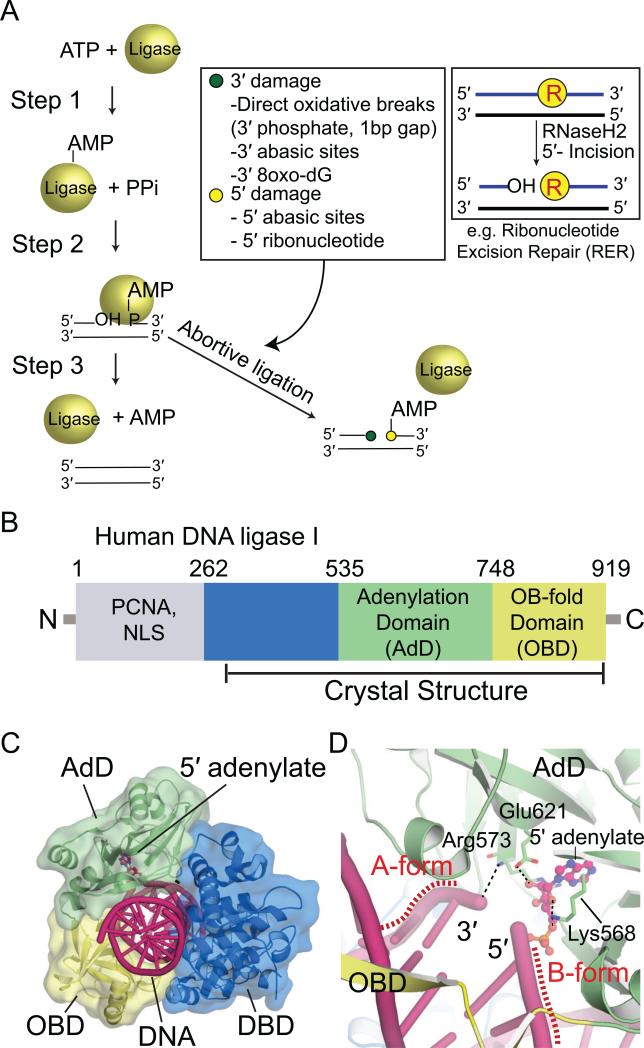
Fig. 1.
DNA ligation and sources of abortive ligation. (A) Three-step ligation reaction utilized by ATP-dependent DNA ligases. Repair of the DNA backbone is coupled to ATP hydrolysis. Inset (left): Lesions found at the 3' (green circle) and 5' (yellow circle) terminal ends of the nicked strands that can trigger abortive ligation are listed in the box. Inset (right): an example of generation of non-canonical DNA termini during ribonucleotide excision repair that triggers abortive ligation is diagrammed. (B) Human DNA ligase I (LigI) domain architecture. The N-terminal domain (gray) mediates nuclear localization and protein interactions with PCNA. The DBD (blue), AdD (green), and OBD (yellow) contain key catalytic residues that comprise the catalytic core of eukaryotic DNA ligases. (C) X-ray structure of LigI/DNA complex (PDB 1X9N) showing three domains (DBD, AdD, OBD) encircle the 5’-adenylated DNA intermediate. DNA is shown in pink and 5’-adenylate in magenta. (D) Structure of the active site of LigI showing key catalytic residues from AdD and OBD that align the nicked strands for the final strand–sealing reaction.
Together, emerging discoveries from biochemical, structural and functional studies in yeast and mammalian systems are illuminating the molecular basis for APTX functions in resolving the products of abortive DNA ligase reactions. This work forms a foundation for understanding the APTX direct DNA damage reversal deadenylation reaction, as well as insights into how human APTX mutations variably impact APTX functions in AOA1. Herein we provide an integrated overview of APTX structure, function and biology.
2. Abortive DNA ligation
Faithful repair of DNA damage typically involves a cascade of enzymatic steps including DNA damage detection, excision of damaged DNA structures (DNA bases, damaged termini) and template-directed replication of lost information (Andres et al., 2014; Ciccia and Elledge, 2010). All DNA repair (and replication) pathways share a common final step, DNA ligation (Ellenberger and Tomkinson, 2008; Lehman, 1974; Pascal et al., 2004; Tomkinson et al., 2006). Eukaryotic DNA ligases use the energy of ATP hydrolysis to catalyze phosphodiester bond formation between a 3’–OH and 5’–phosphate at a DNA strand break, thereby restoring continuity of the sugar-phosphate backbone of the DNA double helix (Lindahl and Edelman, 1968). The DNA ligation reaction proceeds through three steps (Fig. 1A). In step 1, DNA ligase hydrolyzes an ATP molecule to form an AMP–lysyl intermediate, where an active-site lysine in DNA ligase is adenylated. In step 2, the AMP is transferred to the DNA 5’–phosphate by formation of a phosphoanhydride bond, producing a 5’–adenylated DNA intermediate. In step 3, the 3’–OH attacks the phosphoanhydride, AMP is released and the nick is sealed (Lehman, 1974). The energy of ATP hydrolysis ensures the ligation reaction is thermodynamically favorable and drives it to completion.
The three mammalian DNA ligases (I, III, and IV) (Tomkinson et al., 2006) share a common 3-domain structural core: a DNA-binding domain (DBD), an adenylation domain (AdD), and an OB-fold domain (OBD) (Fig. 1B). The core is dynamic, and structural rearrangements occur when DNA ligase wraps around a nicked DNA duplex (Cotner-Gohara et al., 2010; Pascal et al., 2006; Tomkinson et al., 2006). DNA encirclement is a common mode of substrate engagement observed in DNA ligases for which high-resolution structural information is available (Fig. 1C) (Cotner-Gohara et al., 2010; Nair et al., 2007; Nandakumar et al., 2007; Pascal et al., 2004). The transition from an opened apo state to a DNA encircled active state likely functions as a molecular checkpoint for DNA ligases to interrogate their substrate DNA, and ensure the fidelity of substrate recognition during alignment of the DNA ends for catalysis.
While precise in their action on undamaged DNA strands, DNA ligases are sensitive to a variety of endogenous DNA repair intermediates, mismatches and DNA damage that can trigger “abortive ligation” (Ahel et al., 2006) (Fig. 1A). In this process, DNA ligase fails to complete the final nick-sealing step (Step 3), and an AMP adduct is left attached to the 5’–DNA terminus. Thus, rather than sealing the DNA nick to protect genomic integrity, DNA ligase abortive processing of DNA repair intermediates compounds damage by adding bulky adenylate groups to the 5’–terminus. Examples of lesions inducing abortive ligation are oxidative DNA strand breaks with gaps and 3’ phosphate termini (Ahel et al., 2006; Carroll et al. 2014) and incised 5’ abasic sites (5’-dRP) that arise during base excision repair (Caglayan et al., 2014; Rass et al., 2007).
Recently, abortive ligation was also demonstrated to occur on intermediates that arise during ribonucleotide excision repair (RER) (Tumbale et al., 2014; Williams and Kunkel, 2014). DNA polymerases erroneously incorporate ribonucleotides into DNA at high frequency relative to other common forms of DNA damage (Nick McElhinny et al., 2010a, Williams and Kunkel, 2014 ; Williams et al., 2013). The removal of ribonucleotides in RER is initiated by RNase H2 cleavage on the 5’-side of the ribonucleotide, followed by FEN1 incision (Nick McElhinny 2010b; Rydberg and Game, 2002; Sparks et al., 2012). RER produces abundant, albeit transient 5’-RNA-DNA-3’ junctions (see inset, Fig. 1A) that if engaged by DNA ligase can trigger abortive ligation and the production of “RNA-DNA damage” in the form of adenylated RNA-DNA junctions (Tumbale et al., 2014). Inspection of the DNA ligase I nicked DNA complex reveals enzyme-induced distortions of the DNA duplex through interaction of the OBD and AdD domains. In this closed state, ligase widens the major and minor groove, and binds the DNA upstream of the nick (the 3’–hydroxyl side) in A-form geometry to position the 3’– hydroxyl optimally for nucleophilic attack (Lu et al., 2000; Pascal et al., 2004) (Fig. 1D). The downstream DNA (the 5’–phosphate side) is held in a B-form conformation. This A-form to B-form transition across the DNA junction has been proposed to act as a DNA ligase fidelity mechanism that guards against ligation of substrates containing 5’ ribonucleotides, such as unprocessed Okazaki fragments (Pascal et al., 2004). Likewise, engagement of DNA substrate using the A-form to B-form transition may underlie DNA ligase I abortive ligation at 5’ ribonucleotide–containing intermediates generated during RER. However, the molecular basis for abortive DNA ligation reactions remains unclear. Both localized distortions of damaged or mismatched DNA termini engaged by DNA ligases, as well as global impacts on DNA ligase encirclement of its substrate may underlie abortive RNA/DNA ligation and the production of AMP-adducted 5’-terminal RNA or DNA ends.
3. APTX proofreads abortive ligation
Once DNA ligase has dissociated from a DNA substrate, the high concentration of cellular ATP (relative to DNA strand–breaks) promotes rapid re-adenylation of the DNA ligase active site lysine to begin the next round of catalysis (Taylor et al., 2011). For this reason, it is hypothesized that DNA ligases are unable to re-engage adenylated DNAs to complete a failed ligation reaction. Without chemical reversal of adenylation, DNA single strand break repair can stall at the final stage of ligation due to a lack of non-adenylated DNA ligase (Reynolds et al., 2009a, 2009b). Therefore, cells require enzymatic activities to reverse adenylation, to provide a second chance to process damaged termini that induced the abortive ligation, and to faithfully complete nick-sealing during ligation.
Direct DNA and RNA-DNA 5’-deadenylation is catalyzed by APTX, and has now been documented by multiple groups (Fig. 2A) (Ahel et al., 2006; Caglayan et al., 2014; Harris et al., 2009; Rass et al., 2007, 2008; Reynolds et al., 2009b; Tumbale et al., 2014). Consistent with adenylated nucleic acids being cognate substrates for APTX homologs, APTX has a >30,000-fold higher catalytic efficiency on adenylated polynucleotide substrates (Tumbale et al., 2014), compared to adenosine nucleotide substrates such as Ap4A (Kijas et al., 2006) or AMP-lysine (Kijas et al., 2006; Seidle et al., 2005). Additional backup pathways for the APTX direct reversal deadenylation repair reaction include nucleolytic cleavage of adenylated substrates by FEN1 (Rad27 in S. cerevisiae) (Caglayan et al., 2014; Daley et al. 2010), and in the case of adenylated abasic sites (AMP-5’-dRP), the AP lyase activity of DNA polymerase β can remove AMP-5’-dRP en masse (Caglayan et al., 2014).
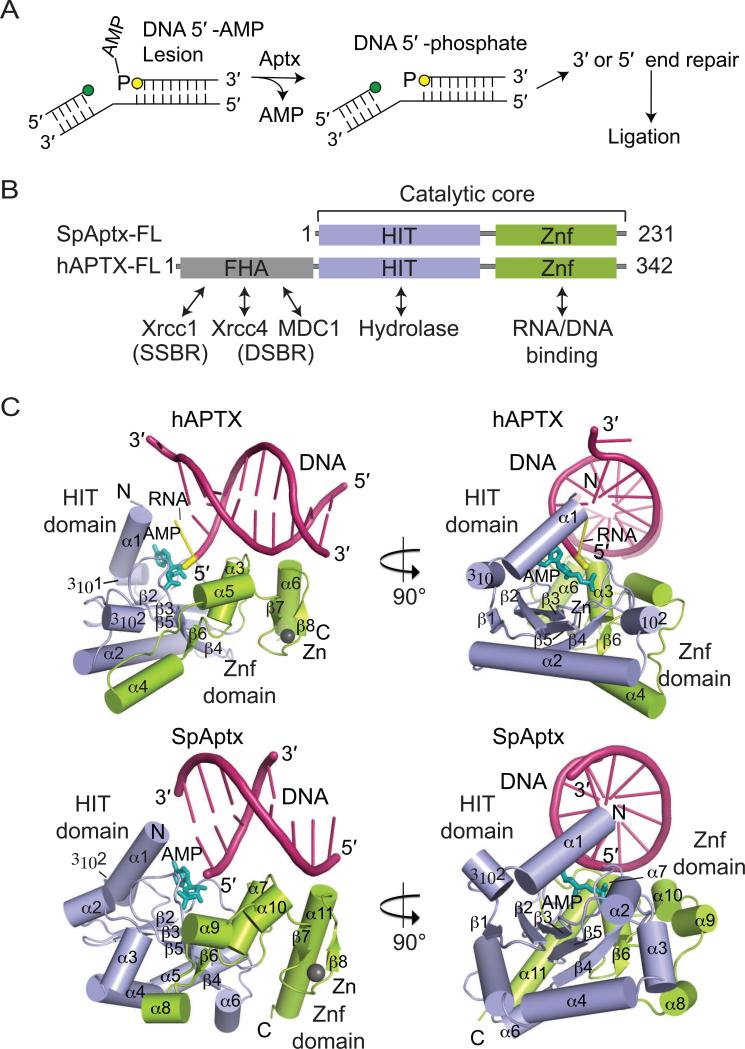
Fig. 2.
APTX and deadenylation of 5’-adenylated intermediates. (A) Lesions at the 3’ (green circle) or 5’ (yellow circle) terminal ends of the nicked strands can trigger abortive ligation. APTX hydrolyses 5’-adenylates to restore the 5’-phosphate of the nicked strand. Other DNA repair pathways can remove the lesions before DNA ligase reseals the nick. (B) Domain architectures of hAPTX and SpAptx. The N-terminal FHA domain (gray) mediates protein interactions and cellular localization in humans. The Histidine Triad domain (HIT) (blue) and the Zn finger motif-containing C-term domain (Znf, orange) comprise the catalytic core of APTX. (C) Orthogonal views of the hAPTX (PBD 4NDF) and SpAptx(PDB 3SZQ) X–ray structures. The HIT (blue) and Znf (green) domains are represented as cylindrical α–helices and β-strands. DNA is colored in pink, 5'-ribonucleotide in yellow, AMP in teal, and Zn in gray.
APTX's role in DNA damage responses are further supported by observations that APTX deficient AOA1 cells display increased chromosome aberrations following treatment with the anticancer topoisomerase inhibitor camptothecin (Mosesso et al., 2005) and elevated levels of oxidative DNA damage (Harris et al., 2009; Hirano et al., 2007). The Schizosaccharomyces pombe Aptx homolog was identified in a genome wide screen for genes conferring sensitivity to the DNA damaging agent 4-nitroquinoline-1-oxide. Genetic evidence in S. cerevisiae further implicate the budding yeast Aptx ortholog (Hnt3p) in oxidative and alkylation DNA damage repair (Daley et al. 2010). Deletion of HNT3 in a genetic background that promotes elevated levels of ribonucleotides in genomic DNA confers severe cellular proliferation defects and activation of the S-phase DNA damage checkpoint (Tumbale et al., 2014). In this role, Aptx is hypothesized to promote repair of adenylated RNA-DNA junctions arising during RER, which is a model consistent with efficient processing of adenylated RNA-DNA junctions in vitro.
4. APTX molecular architecture
Two highly conserved amino acid sequence motifs typify the HIT-Znf region of APTX. The first is the histidine triad motif HΦHΦHΦΦ (Φ=hydrophobic residue) of the HIT domain, and the second is a C-terminal Zn-binding (Znf) domain with a sequence motif C(x2)C(x11-12)H(x3)H/E (x=any amino acid), (Date et al., 2001; Moreira et al., 2001; Tumbale et al., 2011). This core HIT-Znf architecture is conserved in APTX orthologs with bona-fide polynucleotide adenylate hydrolase activity including plant, yeast and vertebrate homologs, suggesting that the Znf domain imparts critical substrate specificity to the Aprataxins (Rass et al., 2008) (Fig. 2B). The core HIT-Znf domain is augmented with a forkhead associated (FHA) domain in vertebrate Aprataxins. Although the FHA is dispensable for catalytic activity (Kijas et al., 2006; Tumbale et al., 2014), it confers phosphorylation-regulated binding of APTX to multiple protein partners. These interactions aid in integrating APTX deadenylation activity with multi-protein complexes where DNA ligases act (see Andres et al, 2014 for recent review). The APTX–FHA domain interacts with proteins involved in single-strand break repair (Xrcc1) (Clements et al., 2004; Date et al., 2004; Luo et al., 2004), as well as double-strand break repair (DSBR) proteins including MDC1 (Becherel et al., 2010) and Xrcc4 (Clements et al., 2004). However, the significance of Aptx association with DSBR factors remains unclear. Arabidposis thaliana (At) APTX also contains an additional kinase domain, as well as a macrodomain (Rass et al., 2008). This kinase domain is related to mammalian PNKP, and macro domains are associated with poly(ADP)-ribose PAR binding and metabolism in DNA base excision repair (Sharifi et al., 2013; Feijs et al., 2013). So, in addition to deadenylation repair, the plant Aprataxins may have added multi-functional roles in DNA strand break repair.
5. The APTX HIT-Znf catalytic core
Overall, the human and yeast APTX HIT domains resemble other HIT superfamily nucleotide hydrolases including FHIT, PKCI (Lima et al., 1997a; Lima et al., 1997b), Hint1 (Dolot et al., 2012) and Hint2 (Maize et al., 2013). This canonical HIT architecture is typified by a central five-stranded anti-parallel β-sheet encircled by a number of helical elements on three sides of the sheet (Fig. 2C). HIT dimerization and domain swapping in FHIT, PKCI, Hint1, and Hint2 completes assembly of the nucleotide hydrolase active site in trans across a dimeric pseudo 2-fold interface (for review see Brenner, 2002). In this respect, APTX is distinct from many members of the HIT superfamily. In APTX, the HIT-Znf interface replaces the domain swapping to mold the active site cleft, and create an extended, electropositive nucleic acid substrate interaction interface (Tumbale et al., 2011; Tumbale et al., 2014). It is this unique fusion of the asymmetric HIT-Znf interface that imparts the DNA and RNA-DNA binding interaction specificities to APTX, differentiating it as a hydrolase of asymmetric polynucleotideadenylates. The DCSP mRNA decapping enzyme is another HIT domain protein that processes polynucleotide substrates. Like APTX, the core HIT-fold chemistry of DCPS is embedded within a larger protein scaffold which confers important substrate and regulatory scaffolding (Gu et al., 2004).
6. Recognition of DNA and RNA-DNA by APTX
A two-point DNA binding mechanism of APTX is well suited for engaging adenylated DNA or RNADNA ends and nicks. The most distinctive aspect of the APTX-RNA/DNA interaction interfaces is an APTX helical “wedge” formed by HIT helix α1 (Fig. 3). This conserved DNA binding element caps the adenylated strand with a highly conserved aromatic side chain (SpAptx Phe34 or hAPTX Trp167) that ring-stacks with the terminal exposed nucleobase of the adenylated nucleotide. Similarly positioned helical elements form the walls of the adenylate-binding pocket of other HIT proteins. In APTX, however, elongation of the wedge helix enables dynamic interactions with both the AMP lesion and the exposed base stack on the 5’-end of the damaged DNA strand (Fig. 3A-C; discussed below in section 2.4)
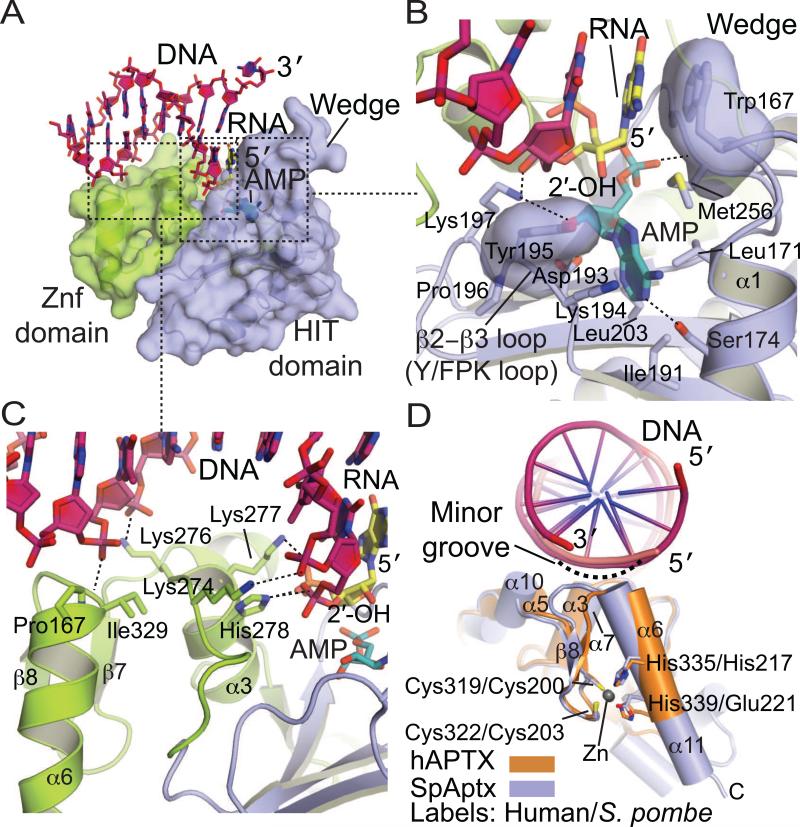
Fig. 3.
APTX–DNA interactions. (A) Molecular details of the APTX Znf-DNA interactions. The HIT (blue) and Znf (green) domains are represented as cylindrical α–helices and β-strands. DNA is colored in pink, 5’-ribonucleotide in yellow, and AMP in teal. (B) Molecular details of the APTX HIT-DNA interactions, colored as in panel A (C) Surface representation of the APTX HIT-Znf DNA interaction interface. An extended DNA binding surface comprised of the HIT (blue) and Znf (green) mediates DNA (pink) contacts. (D) Superposition of the Znf domains of APTX (orange, PDB 4NDF) and SpAptx (blue, PDB 3SZQ). DNA is colored in pink and Zn in gray. C2H2 (hAPTX) and C2HE (SpAptx) Znf motifs adopt a similar fold and the Zn is coordinated by four zinc binding residues (Cys319, Cys322, His335 and His339 of hAPTX and Cys200, Cys203, His217, Glu221 of SpAptx).
The second major DNA binding interface involves undamaged DNA strand binding by the Znf domain. The APTX Zn2+–binding ββα core is structurally related to the ubiquitous family of DNA binding C2H2 transcription factors including the prototypical Zif268 (Klug 2010; Pavletich and Pabo, 1991). Interestingly, APTX Zn2+ ligands can be of either the C2H2 (Cys2-His2 in vertebrate APTX) or C2HE (Cys2-His-Glu in fungal Aptx), which both fold into very similar DNA damage recognition elements (Fig. 3D). To our knowledge, a C2HE Zn binding ββα fold had not been observed previous to the determination of SpAptx structure. This observation is significant, as C2H2 zinc fingers are widespread in nature, and the size of this protein class may be even larger given that a comparable fold is assembled using C2HE as Zn2+ ligands. APTX employs this fold to recognize the minor groove of DNA in a sequence-independent binding mode. The helical dipole of the ββα element is directed precisely at the sugar phosphate backbone. This contrasts the sequence–specific DNA–binding C2H2 Zn-finger domains that are typically arrayed in tandem to bind nucleic acids of varied lengths using major groove sequence specific interactions.
A central role of the ββα-core in substrate interactions is fully consistent with biochemical studies demonstrating that a C-terminal 22-amino acid deletion that removes the ββα core of APTX deletion ablates ssDNA and dsDNA binding (Kijas et al., 2006). Likewise, mutation of the Zn2+-coordinating cysteine residues blocks APTX binding to nicked and nicked-adenylate substrates, and has a severe impact on deadenylation activity (Rass et al., 2007). The HIT domain of Aprataxin also contributes conserved helical elements that augment the ββα Znf core. In particular, helix α3 in hAPTX or α7 in S. pombe makes extensive critical interactions with the 5’-strand proximal to the active site (Fig. 3C,D). Charge reversal mutations of S. pombe α7 Aptx basic DNA binding residues ablate DNA binding and deadenylation activity (Tumbale et al., 2011), underscoring the importance of the fused HIT-Znf DNA binding surface in directing SpAptx activity.
7. RNA-DNA substrate distortions
Together the Znf and α1 helical wedge extensively engage the adenylated strand, orienting it for catalysis (Fig. 3B). The asymmetry of the HIT–ZnF assembly provides polarity for the interaction with the nucleic acid substrate, facilitating specificity for processing at 5’–adenylated DNA ends. Overall the nucleic acid bound substrates of both human and S. pombe complexes display similar substrate distortions from canonical B-DNA. Most noteworthy is the bound A-form conformations of the 5’ terminal 2–3 nucleotides of the damaged strand. A-form binding is observed for both DNA and RNA-DNA substrates, and is consistent with functional roles for Aprataxin processing adenylated substrates from abortive ligation arising during base excision repair of oxidative DNA damage, and also adenylated RNA-DNA substrates in RER (Section 1.2) using a common substrate– binding mode.
Given that the wedge helix almost completely occludes the duplex DNA base stack, wedging is presumably critical for APTX recognition and substrate engagement of cognate nicked DNA substrates, although the precise nature of this interaction awaits structural elucidation of APTX bound to such substrates. Coincident with A-form binding and wedging is the distortion (SpAptx) or complete unpairing (hAPTX) of the 5’ terminal base pair. The mechanism of DNA end interaction explains the ability of APTX to unwind the terminal adenylated base pair during DNA binding (Rass et al., 2008). We speculate the observed substrate distortions and implied DNA nick bending are critical for accessing and positioning the scissile phosphoanhydride linkage for catalysis in the context of variable DNA end structures (damaged dirty ends) on the 5’ and 3’ sides of the nick. This implied nick distortion mechanism by APTX may also be relevant to the coordinated processing of toxic strand break intermediates during repair (Tumbale et al., 2011). For example, nick distortion may be important for relaying substrates to downstream DNA repair enzymes, such as PNKP or POLβ.
8. Detection of 5’-Adenylated (5’–AMP) lesions
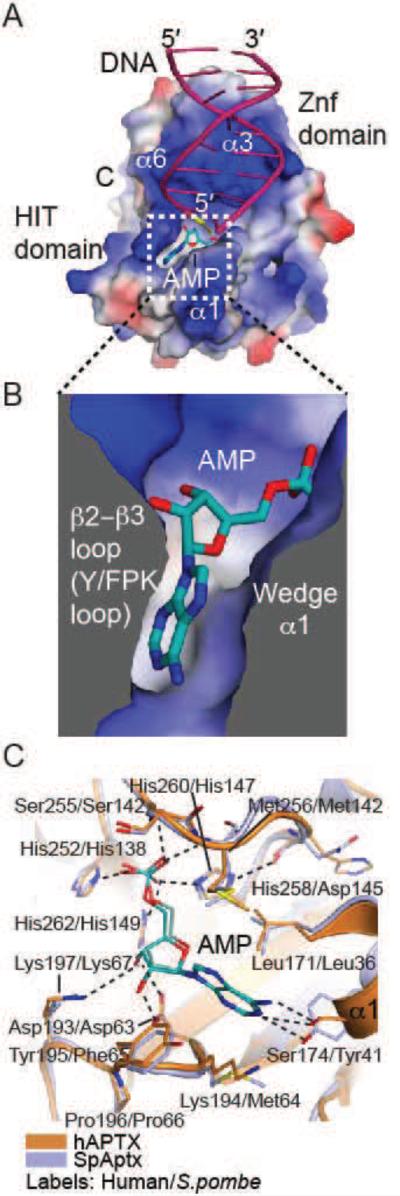
The 5’–AMP moiety of abortive ligation products is the unique feature that distinguishes them amongst the pool of undamaged DNA substrates produced during DNA replication and repair. DNA binding studies demonstrated that APTX has enhanced affinity for adenylated substrates over a non-adenylated equivalents indicating that adenylate recognition is a key feature of substrate–recognition by APTX (Rass et al., 2007). Accordingly, both S. pombe and human APTX make extensive interactions with the 5’–AMP that is bound in a deep pocket formed at the interface between the HIT and Znf domains (Fig. 4A-C). A comparison of S. pombe and human APTX structures reveals that the DNA binding region, active site, and 5’–AMP binding–pocket are highly conserved. This is not the case with regions distal to the substrate-binding site, which are more divergent at the primary sequence and structural level. The 5’–AMP is sandwiched between elements of the (Y/F)PK-loop (aka the β2-β3 loop) and the α1 helical wedge (Fig. 4B). Several hydrogen bonds from active site residues and backbones amides orient the AMP phosphate for catalysis (Fig. 4C). The ribose moiety is recognized by three hydrogen–bonds to the 2’– and 3’–hydroxyls, and the nucleobase is bound between four hydrophobic side–chains (hAPTX/SpAptx: Tyr195/Phe65, Lys194/Met64, Met256/142, and Leu171/36). Interestingly, the Watson–Crick face of the adenine base of the lesion is recognized by hydrogen bonds to Ser174/Tyr41. Thus the molecular features of the APTX lesion-binding pocket explain APTX specificity for processing of adenylated nucleic acid termini. Intriguingly, reversibility of the serine hydrogen bond donor/acceptor would also enable the recognition of guanine in the lesion-binding pocket. Consistent with this, SpAptx has recently been reported to catalyze removal of GMP from 3’–GMP–capped DNA ends (Das et al., 2014). We speculate that GMP binds in an analogous manner to AMP during a deguanylation reaction.
Fig. 4.
APTX AMP interactions. (A) Surface potential charge representation (red – negatively charged, blue – positively charged, gray – neutral/hydrophobic) of APTX with the bound AMP (teal), 5’ terminal RNA (yellow), and DNA (pink). The AMP binding pocket is outlined in a dotted-line box. (B) Helix α1 (Wedge) and β2-β3 loop (Y/FPK loop) elements comprise the AMP binding pocket. (C) Structure of the AMP binding pockets of APTX (orange, PDB 4NDF) and SpAptx (blue, PDB 3SZQ), showing conserved active–site architecture. AMP is colored in teal.
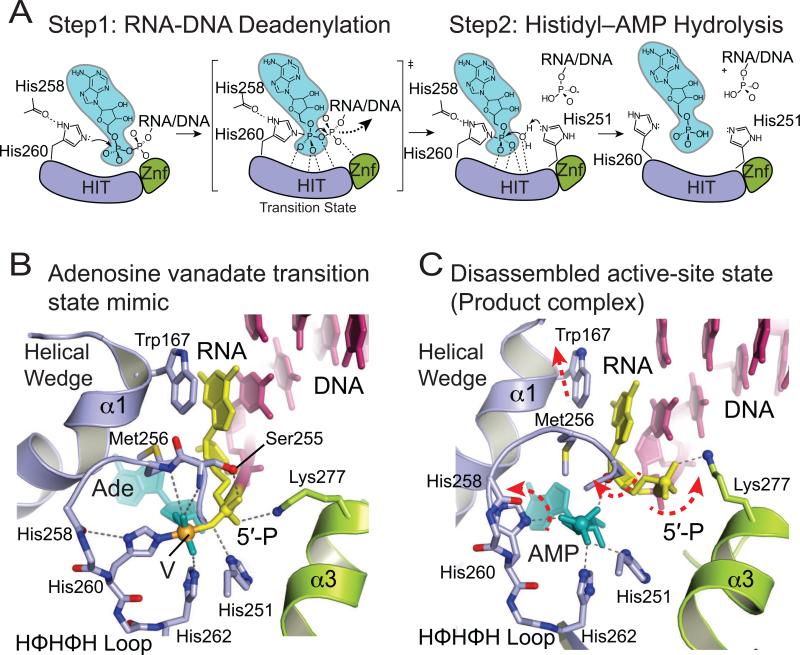
9. APTX catalytic mechanism
Together, integrated analysis from structural and biochemical reports support a two-step mechanism for hydrolysis of 5’–AMP from RNA-DNA and DNA (Fig. 5A) (Ahel et al., 2006; Rass et al., 2008; Tumbale et al., 2011; Tumbale et al., 2014). In the first step, the active site nucleophile (His260 of the HIT loop in APTX) attacks the 5’–adenylate phosphoanhydride forming a transient enzyme-AMP intermediate and releasing a DNA 5’– phosphate product. Several hydrogen bonds with active–site residues and peptide backbone amides stabilize the negative charge on the transition–state of step 1, which has been characterized in a vanadate transition state mimic of RNA-DNA bound APTX (Tumbale et al. 2014) (Fig. 5B). The nucleophile (His260) is activated by a hydrogen bond to the carbonyl of His258, which is an interaction conserved in other HIT proteins (Brenner, 2002), while His251 is proposed to protonate the 5’–phosphate leaving group. The second step involves hydrolysis of the His260-AMP-RNA/DNA intermediate, using chemistry that is essentially the reverse of the first step with water replacing the 5’–phosphate, and with His251 proposed to act as a general base to deprotonate a water nucleophile. While APTX homologs bind their DNA 5’ phosphate product with moderate affinity in the nanomolar range (Rass et al, 2007; Tumbale et al., 2011), AMP can compete for binding of 5’-phosphorylated DNA nicked substrate in SpAptx (Tumbale et al., 2011). Thus, while both AMP and 5’-phosphorylated products can be accommodated in the active site of APTX, charge-charge repulsion of juxtaposed phosphates (Fig. 5C) may ultimately promote product RNA-DNA substrate release.
Fig. 5.
APTX deadenylation reaction mechanism. (A) Steps of the proposed two–step APTX reaction mechanism. APTX HIT domain is colored in blue, Znf in green, AMP in teal. (B) Molecular details of active site environment during the transition state shown in the structure of APTX/AMP/RNA-DNA/Vanadate transition state mimic complex (PDB 4NDG). (C) Molecular details of the active-site disassembled state shown by the structure of APTX bound to product RNA/DNA (PDB 4NDF).
In the human APTX structures, the active site histidine triad HΦHΦHΦΦ loop is found in two dramatically different conformers — one in an “assembled active site” conformation, and another in a disassembled conformation (Fig. 5C). These conformations differ significantly in two respects. First, the disassembled conformation has the active site nucleophile rotated out of alignment for catalysis. Concomitant with this rotation are significant coordinated movements of the HIT loop and wedge helix. It is hypothesized that active site assembly is coupled to binding both the 5’–terminal end of the RNA/DNA substrate and the AMP lesion, which induces a transition to the active conformation (Tumbale et al., 2014). Such an assembly/disassembly cycle may be important for substrate induced fit active site assembly, product release, ensuring enzyme fidelity for polynucleotide substrates, or all of the above.
10. APTX disease associated mutations
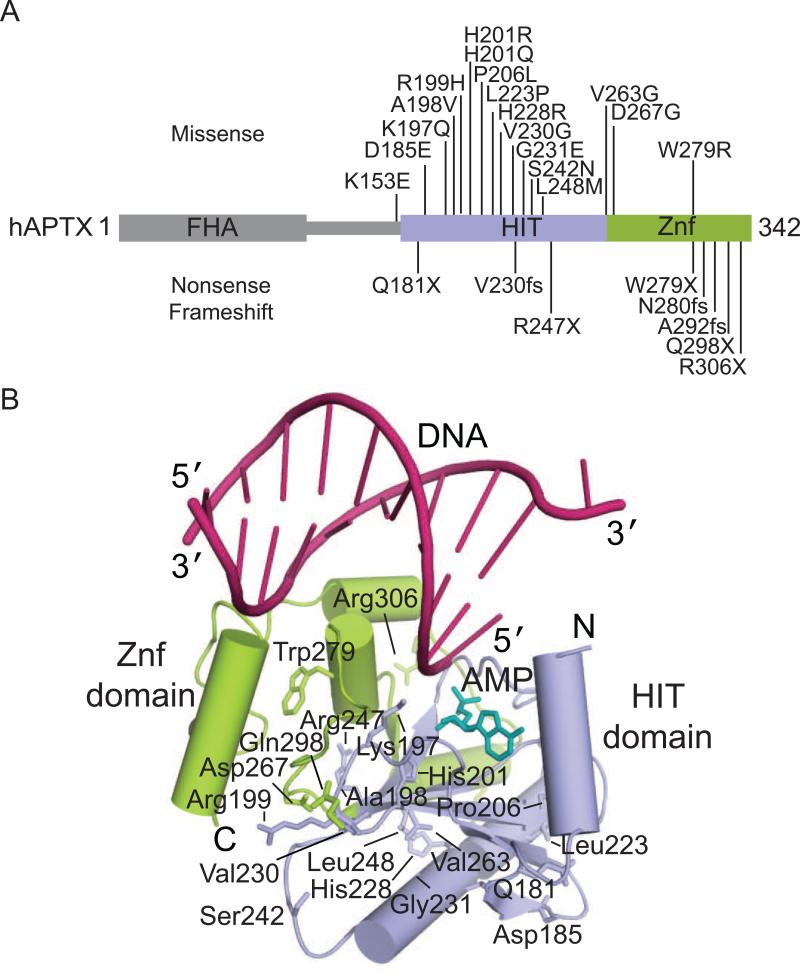
All reported AOA1 mutations occur within the HIT and Znf domains of APTX (Date et al., 2004; Le Ber et al., 2003; Moreira et al., 2001). These mutations include nonsense truncations, frameshifts, and missense mutations predicted to produce destabilized and/or inactivate mutant proteins (Tumbale et al., 2014). Significantly, APTX mutations confer heterogeneous outcomes for afflicted individuals. Some mutations are recessive, while other mutations confer mild forms of disease, yet others exhibit apparent dominant inheritance (eg. L248M) (Castellotti et al., 2011; Tranchant et al., 2003). The precise molecular basis for heterogeneous modulation of APTX function by mutagenic inactivation remains unclear. However, integrated results from biochemical and structural analyses indicate that many mutations variably affect protein folding, catalysis and RNA-DNA damage interactions.
Mapping the positions of AOA1 mutations onto the structure of APTX shows that they are distributed throughout the catalytic domain (Fig. 6A), with the majority of AOA1 mutations located in the protein core (Fig. 6B). Interestingly, although several single-nucleotide polymorphisms (SNPs) are found within the FHA domain (Sherry et al. 2001), no mutations linked to AOA1 are found in this region implying that mis-localization of APTX is not sufficient to cause AOA1. Furthermore, yeast Aptx homologs (Aptx, HNT3) lack an FHA domain entirely, suggesting that function(s) conferred by the FHA domain are not required for ancient conserved function of APTX impacted by mutation in disease.
Fig. 6.
Ataxia Oculomotor Apraxia 1 (AOA1) mutations. (A) Residues mutated in AOA1 are located in the HIT (blue) and Znf (green) domains of APTX. (B) The location of residues mutated in AOA1 mapped onto the X-ray structure of human APTX (PDB 4NDF). DNA is colored in pink, AMP in teal.
Several mutations (D185E, A198V, P206L, L223P, G231E, R247X, W279X, V263G, D267G, R306X) are predicted to affect protein stability by destabilizing or truncating the folded core of APTX (Tumbale et al., 2014). Although the impact of most of the missense variants on APTX catalytic activity on adenylated DNA or RNA-DNA has not been reported, AMP-lysine hydrolase activity was analyzed for several AOA1 variants. AOA1 mutant K197Q and W279R display impaired AMP-lysine hydrolase activity, while many other mutant proteins (A198V, P206L, V263G, D267G, W279X) are catalytically inactive (Seidle et al., 2005). Two mutations (H201R and H201Q) lie within the active site and are expected to impair catalytic activity. AOA1 mutation R199H is located on the solvent exposed side of the YPK substrate-binding loop. Interestingly, purified recombinant R199H protein retains substantial activity and stability (Seidle et al., 2005), suggesting other factors such as loss of protein-protein interactions may also contribute to AOA1. The K197Q (Tranchant et al., 2003), mutation is found as a compound heterozygous mutant with W279X, and confers a late onset AOA1. Lys 197 is located on the YPK loop where it stabilizes the adenylated strand of substrate through DNA and AMP interactions. The molecular basis for the K197Q defect is described in a high-resolution structure of this mutant bound to an RNA-DNA substrate, which unveiled substrate-binding pocket distortions that account for the significant impact on RNA-DNA deadenylation activity (Tumbale et al., 2014). Given that it is difficult to rationalize effects of some AOA1 linked mutations (e.g. S242N, L248M) it appears that phenotypic variability associated with APTX mutations is complex. More work is needed to decipher the genotype-phenotype interactions in AOA1 and other APTX linked maladies.
11. Future directions
Results from synergistic biochemical, cell biological and structural interrogation of the APTX gene product have greatly advanced our understanding of APTX functional roles in DNA and RNA-DNA damage responses. Several unanswered questions remain. For instance, how does APTX integrate with other players in the DNA and RNA-DNA damage responses to act in multistep DNA repair reactions? We lack molecular information defining APTX interactions with adenylated nicked DNA or RNA-DNA substrates, the cognate substrates for APTX in DNA single strand break repair and RER, underlining a current gap in our knowledge regarding the APTX lesion processing mechanism that needs to be addressed in future studies. Furthermore, precisely how do APTX mutations that cause AOA1 impact APTX structure and function? Of note, APTX localizes to the mitochondria (Sykora et al., 2011), as well as the nucleolus (Becherel et. al, 2006), and a better understanding of APTX functions in these sub-cellular compartments is needed. Furthermore, our understanding of the molecular and cellular contexts in which APTX deficiency contributes to tissue-specific cerebellar degeneration in disease remains limited.
We anticipate that additional integrated APTX functional and structural studies will provide testable structural paradigms for RNA-DNA damage sensing and processing by DNA-nick and -end cleansing enzymes in the DNA and RNA-DNA damage responses, and better establish a molecular platform for understanding APTX dysfunction in neurodegenerative disease. Although AOA1 is a rare disorder, outcomes of this research have implications for understanding modes of APTX mutagenic inactivation in disease and individual genetic susceptibility to environmentally induced DNA damage. In a broader sense, inhibitors that modulate APTX DNA repair activities may ultimately be useful for treatment of cancers, or possibly as a co-therapy with existing chemotherapeutics that induce “dirty” DNA breaks.
Acknowledgements
Our studies are supported by the NIH Intramural Program: National Institute of Environmental Health Sciences, 1Z01ES102765 to R.S.W. We thank J Williams, S Andres and B Wallace for comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- Aicardi J, Barbosa C, Andermann E, Andermann F, Morcos R, Ghanem Q, Fukuyama Y, Awaya Y, Moe P. Ataxia-ocular motor apraxia: a syndrome mimicking ataxia-telangiectasia. Ann. Neurol. 1988;24:497–502. doi: 10.1002/ana.410240404. [DOI] [PubMed] [Google Scholar]
- Andres SN, Schellenberg MJ, Wallace BD, Tumbale P, Williams RS. Recognition and repair of chemically heterogeneous structures at DNA ends. Environ. Mol. Mutagen. 2014 doi: 10.1002/em.21892. doi: 10.1002/em.21892. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba Y, Uitti RJ, Boylan KB, Uehara Y, Yamada T, Farrer MJ, Couchon E, Batish SD, Wszolek ZK. Aprataxin (APTX) gene mutations resembling multiple system atrophy. Parkinsonism Relat. Disord. 2007;13:139–142. doi: 10.1016/j.parkreldis.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Becherel OJ, Gueven N, Birrell GW, Schreiber V, Suraweera A, Jakob B, Taucher-Scholz G, Lavin MF. Nucleolar localization of aprataxin is dependent on interaction with nucleolin and on active ribosomal DNA transcription. Hum Mol Genet. 2006;15:2239–2249. doi: 10.1093/hmg/ddl149. [DOI] [PubMed] [Google Scholar]
- Becherel OJ, Jakob B, Cherry AL, Gueven N, Fusser M, Kijas AW, Peng C, Katyal S, McKinnon PJ, Chen J, et al. CK2 phosphorylation-dependent interaction between aprataxin and MDC1 in the DNA damage response. Nucleic Acids Res. 2010;38:1489–1503. doi: 10.1093/nar/gkp1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Karimi-Busheri F, Mani RS, Galicia S, Koch CA, Cass CE, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Brenner C. Hint, Fhit, and GalT: function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry. 2002;41:9003–9014. doi: 10.1021/bi025942q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglayan M, Batra VK, Sassa A, Prasad R, Wilson SH. Role of polymerase beta in complementing aprataxin deficiency during abasic-site base excision repair. Nat. Struct. Mol. Biol. 2014;21:497–499. doi: 10.1038/nsmb.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW. DNA single-strand break repair and spinocerebellar ataxia. Cell. 2003;112:7–10. doi: 10.1016/s0092-8674(02)01247-3. [DOI] [PubMed] [Google Scholar]
- Caldecott KW. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- Carroll J, Page TK, Chiang SC, Kalmar B, Bode D, Greensmith L, Mckinnon PJ, Thorpe JR, Hafezparast M, El-Khamisy SF. Expression of a pathogenic mutation of SOD1 sensitizes aprataxin-deficient cells and mice to oxidative stress and triggers hallmarks of premature ageing. Hum. Mol. Genet. 2014 doi: 10.1093/hmg/ddu500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellotti B, Mariotti C, Rimoldi M, Fancellu R, Plumari M, Caimi S, Uziel G, Nardocci N, Moroni I, Zorzi G, et al. Ataxia with oculomotor apraxia type1 (AOA1): novel and recurrent aprataxin mutations, coenzyme Q10 analyses, and clinical findings in Italian patients. Neurogenetics. 2011;12:193–201. doi: 10.1007/s10048-011-0281-x. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements PM, Breslin C, Deeks ED, Byrd PJ, Ju L, Bieganowski P, Brenner C, Moreira MC, Taylor AM, Caldecott KW. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst.) 2004;3:1493–1502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Cotner-Gohara E, Kim IK, Hammel M, Tainer JA, Tomkinson AE, Ellenberger T. Human DNA ligase III recognizes DNA ends by dynamic switching between two DNA-bound states. Biochemistry. 2010;49:6165–6176. doi: 10.1021/bi100503w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Wilson TE, Ramotar D. Genetic interactions between HNT3/Aprataxin and RAD27/FEN1 suggest parallel pathways for 5' end processing during base excision repair. DNA Repair (Amst.) 2010;9:690–699. doi: 10.1016/j.dnarep.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U, Chauleau M, Ordonez H, Shuman S. Impact of DNA3'pp5'G capping on repair reactions at DNA 3' ends. Proc. Natl. Acad. Sci. USA. 2014;111:11317–11322. doi: 10.1073/pnas.1409203111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date H, Igarashi S, Sano Y, Takahashi T, Takano H, Tsuji S, Nishizawa M, Onodera O. The FHA domain of aprataxin interacts with the C-terminal region of XRCC1. Biochem. Biophys. Res. Commun. 2004;325:1279–1285. doi: 10.1016/j.bbrc.2004.10.162. [DOI] [PubMed] [Google Scholar]
- Date H, Onodera O, Tanaka H, Iwabuchi K, Uekawa K, Igarashi S, Koike R, Hiroi T, Yuasa T, Awaya Y, et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat. Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- Dolot R, Ozga M, Wlodarczyk A, Krakowiak A, Nawrot B. A new crystal form of human histidine triad nucleotide-binding protein 1 (hHINT1) in complex with adenosine 5'-monophosphate at 1.38 A resolution. Acta. Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012;68:883–888. doi: 10.1107/S1744309112029491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Smerdon SJ, Yaffe MB, Jackson SP. The FHA domain in DNA repair and checkpoint signaling. Cold Spring Harb. Symp. Quant. Biol. 2000;65:423–431. doi: 10.1101/sqb.2000.65.423. [DOI] [PubMed] [Google Scholar]
- Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijs KL, Forst AH, Verheugd P, Luscher B. Macrodomain-containing proteins: regulating new intracellular functions of mono(ADP-ribosyl)ation. Nat. Rev. Mol. Cell Biol. 2013;14:443–451. doi: 10.1038/nrm3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Fabrega C, Liu SW, Liu H, Kiledjian M, Lima CD. Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity. Mol. Cell. 2004;14:67–80. doi: 10.1016/s1097-2765(04)00180-7. [DOI] [PubMed] [Google Scholar]
- Gueven N, Chen P, Nakamura J, Becherel OJ, Kijas AW, Grattan-Smith P, Lavin MF. A subgroup of spinocerebellar ataxias defective in DNA damage responses. Neuroscience. 2007;145:1418–1425. doi: 10.1016/j.neuroscience.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Harris JL, Jakob B, Taucher-Scholz G, Dianov GL, Becherel OJ, Lavin MF. Aprataxin, poly-ADP ribose polymerase 1 (PARP-1) and apurinic endonuclease 1 (APE1) function together to protect the genome against oxidative damage. Hum. Mol. Genet. 2009;18:4102–4117. doi: 10.1093/hmg/ddp359. [DOI] [PubMed] [Google Scholar]
- Hirano M, Yamamoto A, Mori T, Lan L, Iwamoto TA, Aoki M, Shimada K, Furiya Y, Kariya S, Asai H, et al. DNA single-strand break repair is impaired in aprataxin-related ataxia. Ann. Neurol. 2007;61:162–174. doi: 10.1002/ana.21078. [DOI] [PubMed] [Google Scholar]
- Kijas AW, Harris JL, Harris JM, Lavin MF. Aprataxin forms a discrete branch in the HIT (histidine triad) superfamily of proteins with both DNA/RNA binding and nucleotide hydrolase activities. J. Biol. Chem. 2006;281:13939–13948. doi: 10.1074/jbc.M507946200. [DOI] [PubMed] [Google Scholar]
- Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- Koch CA, Agyei R, Galicia S, Metalnikov P, O'Donnell P, Starostine A, Weinfeld M, Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Ber I, Moreira MC, Rivaud-Pechoux S, Chamayou C, Ochsner F, Kuntzer T, Tardieu M, Said G, Habert MO, Demarquay G, et al. Cerebellar ataxia with oculomotor apraxia type 1: clinical and genetic studies. Brain. 2003;126:2761–2772. doi: 10.1093/brain/awg283. [DOI] [PubMed] [Google Scholar]
- Lehman IR. DNA ligase: structure, mechanism, and function. Science. 1974;186:790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- Lima CD, D'Amico KL, Naday I, Rosenbaum G, Westbrook EM, Hendrickson WA. MAD analysis of FHIT, a putative human tumor suppressor from the HIT protein family. Structure. 1997a;5:763–774. doi: 10.1016/s0969-2126(97)00231-1. [DOI] [PubMed] [Google Scholar]
- Lima CD, Klein MG, Hendrickson WA. Structure-based analysis of catalysis and substrate definition in the HIT protein family. Science. 1997b;278:286–290. doi: 10.1126/science.278.5336.286. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Edelman GM. Polynucleotide ligase from myeloid and lymphoid tissues. Proc. Natl. Acad. Sci. USA. 1968;61:680–687. doi: 10.1073/pnas.61.2.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XJ, Shakked Z, Olson WK. A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- Luo H, Chan DW, Yang T, Rodriguez M, Chen BP, Leng M, Mu JJ, Chen D, Songyang Z, Wang Y, et al. A new XRCC1-containing complex and its role in cellular survival of methyl methanesulfonate treatment. Mol. Cell. Biol. 2004;24:8356–8365. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maize KM, Wagner CR, Finzel BC. Structural characterization of human histidine triad nucleotide-binding protein 2, a member of the histidine triad superfamily. FEBS J. 2013;280:3389–3398. doi: 10.1111/febs.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, et al. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat. Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- Moreira MC, Klur S, Watanabe M, Nemeth AH, Le Ber I, Moniz JC, Tranchant C, Aubourg P, Tazir M, Schols L, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat. Genet. 2004;36:225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- Mosesso P, Piane M, Palitti F, Pepe G, Penna S, Chessa L. The novel human gene aprataxin is directly involved in DNA single-strand-break repair. Cell. Mol. Life. Sci. 2005;62:485–491. doi: 10.1007/s00018-004-4441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad NA, Cullen JK, McKenzie M, Ryan MT, Thorburn D, Gueven N, Kobayashi J, Birrell G, Yang J, Dork T, et al. Mitochondrial dysfunction in a novel form of autosomal recessive ataxia. Mitochondrion. 2013;13:235–245. doi: 10.1016/j.mito.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Nair PA, Nandakumar J, Smith P, Odell M, Lima CD, Shuman S. Structural basis for nick recognition by a minimal pluripotent DNA ligase. Nat. Struct. Mol. Biol. 2007;14:770–778. doi: 10.1038/nsmb1266. [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Nair PA, Shuman S. Last stop on the road to repair: structure of E. coli DNA ligase bound to nicked DNA-adenylate. Mol. Cell. 2007;26:257–271. doi: 10.1016/j.molcel.2007.02.026. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA. 2010a;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 2010b;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal JM, O'Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, Classen S, Tomkinson AE, Tainer JA, Ellenberger T. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol. Cell. 2006;24:279–291. doi: 10.1016/j.molcel.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Quinzii CM, Kattah AG, Naini A, Akman HO, Mootha VK, DiMauro S, Hirano M. Coenzyme Q deficiency and cerebellar ataxia associated with an aprataxin mutation. Neurology. 2005;64:539–541. doi: 10.1212/01.WNL.0000150588.75281.58. [DOI] [PubMed] [Google Scholar]
- Rass U, Ahel I, West SC. Actions of aprataxin in multiple DNA repair pathways. J. Biol. Chem. 2007;282:9469–9474. doi: 10.1074/jbc.M611489200. [DOI] [PubMed] [Google Scholar]
- Rass U, Ahel I, West SC. Molecular mechanism of DNA deadenylation by the neurological disease protein aprataxin. J. Biol. Chem. 2008;283:33994–34001. doi: 10.1074/jbc.M807124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JJ, El-Khamisy SF, Caldecott KW. Short-patch single-strand break repair in ataxia oculomotor apraxia-1. Biochem. Soc. Trans. 2009a;37:577–581. doi: 10.1042/BST0370577. [DOI] [PubMed] [Google Scholar]
- Reynolds JJ, El-Khamisy SF, Katyal S, Clements P, McKinnon PJ, Caldecott KW. Defective DNA ligation during short-patch single-strand break repair in ataxia oculomotor apraxia 1. Mol. Cell. Biol. 2009b;29:1354–1362. doi: 10.1128/MCB.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc. Natl. Acad. Sci. USA. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidle HF, Bieganowski P, Brenner C. Disease-associated mutations inactivate AMP-lysine hydrolase activity of Aprataxin. J. Biol. Chem. 2005;280:20927–20931. doi: 10.1074/jbc.M502889200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32:1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, Burgers PM. RNase H2-initiated ribonucleotide excision repair. Mol. Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykora P, Croteau DL, Bohr VA, Wilson DM., 3rd Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc Natl Acad Sci U S A. 2011;108:7437–7442. doi: 10.1073/pnas.1100084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MR, Conrad JA, Wahl D, O'Brien PJ. Kinetic mechanism of human DNA ligase I reveals magnesium-dependent changes in the rate-limiting step that compromise ligation efficiency. J. Biol. Chem. 2011;286:23054–23062. doi: 10.1074/jbc.M111.248831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranchant C, Fleury M, Moreira MC, Koenig M, Warter JM. Phenotypic variability of aprataxin gene mutations. Neurology. 2003;60:868–870. doi: 10.1212/01.wnl.0000048562.88536.a4. [DOI] [PubMed] [Google Scholar]
- Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. DNA ligases: structure, reaction mechanism, and function. Chem. Rev. 2006;106:687–699. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- Tumbale P, Appel CD, Kraehenbuehl R, Robertson PD, Williams JS, Krahn J, Ahel I, Williams RS. Structure of an aprataxin-DNA complex with insights into AOA1 neurodegenerative disease. Nat. Struct. Mol. Biol. 2011;18:1189–1195. doi: 10.1038/nsmb.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbale P, Williams JS, Schellenberg MJ, Kunkel TA, Williams RS. Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity. Nature. 2014;506:111–115. doi: 10.1038/nature12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Kunkel TA. Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst.) 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Smith DJ, Marjavaara L, Lujan SA, Chabes A, Kunkel TA. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol. Cell. 2013;49:1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]