Abstract
Objectives
To evaluate whether micrRNAs associated with endometriosis are detectable in the circulation and could serve as potential noninvasive biomarkers for endometriosis.
Design
Case-control study.
Setting
University hospital.
Patient(s)
Twenty-four women with endometriosis and 24 women without the disease (controls).
Interventions
Serum samples were collected from women undergoing laparoscopy for endometriosis and other benign gynecologic disease.
Main Outcome Measure(s)
Total RNA was extracted from serum and qRT-PCR was used to determine levels of miRNA let-7a–f and miR-135a,b.
Result(s)
Levels of circulating let-7b and miR-135a were significantly decreased in women with endometriosis compared with controls, while let-7d and 7f showed a trend toward down-regulation. Let-7b expression strongly correlated with serum CA-125 levels and showed the highest AUC curve of 0.691. When the subjects were analyzed according to the phase of the menstrual cycle, expression of let-7b, 7c, 7d, and 7e were significantly lower in women with endometriosis during the proliferative phase. Using a logistic regression model, the diagnostic power of differently expressed miRNAs were evaluated, and the combination of let-7b, let-7d and let-7f during the proliferative phase yielded the highest ACU curve value of 0.929 in discriminating endometriosis from controls.
Conclusions
Several circulating miRNAs are differentially expressed in sera of patients with endometriosis compared to controls. Combination of serum let-7b, 7d and 7f levels during the proliferative phase may serve as a diagnostic marker for endometriosis.
Keywords: Biomarker, circulating miRNAs, diagnosis, endometriosis, let-7
Introduction
Endometriosis is defined as the proliferation of endometrial tissue outside the uterine cavity and is one of the most common gynecologic disorders. This disease is present in approximately 10% of all reproductive-aged women, and its prevalence increases to 20–50% in infertile women (1). Despite the high prevalence of the disease, the diagnosis of endometriosis is often delayed due to the complexity of the pathogenesis and diversity of symptoms (2, 3). Although multivariate analysis of several biomarkers validated in independent population seems promising (4), there is no definite diagnostic biomarker yet available. Imaging techniques, such as ultrasound and magnetic resonance imaging, do offer some benefit in the diagnosis of ovarian endometriomas, however, these modalities have been shown to be unreliable in the diagnosis or staging of this more common peritoneal disease (3). Therefore, the direct visualization of lesions and histologic confirmation through surgical procedures are currently employed for the definitive diagnosis of endometriosis. Although laparoscopy is a minimally invasive procedure, it requires general anesthesia, developed surgical skills, and has a high procedural cost. Additionally, laparoscopy is associated with the risk of potential intraoperative or postoperative complications.
MicroRNAs (miRNAs) are a family of endogenous, small (approximately 22 nucleotides in length), noncoding, functional RNAs and these sequences control gene expression either by translational repression or degradation of messenger RNA transcripts after targeting the 3´UTR. Increasing evidence suggests that miRNAs are pivotal regulators of development and cellular homeostasis through their control of diverse biological processes. Numerous studies have shown that aberrant miRNA expression is associated with several human diseases such as cancer, cardiovascular disorders, inflammatory diseases as well as with gynecological pathology (5–11). Recently, the presence of circulating miRNAs was demonstrated in the blood (12). Since their first description as potential diagnostic biomarkers for diffuse large B cell lymphoma (13), it has been demonstrated that circulating miRNAs may be used as noninvasive biomarkers for other conditions including autoimmune disease, sepsis, and myocardial infarction (14–16). Although the exact origin of the changes in circulating miRNA profiles is not fully understood, it has been shown that there are strong correlations between the miRNA profiles of serum/plasma and cancer tissues, suggesting that miRNAs may be released from tissues and shed into the circulation (17, 18).
MiRNAs and their target mRNAs are expressed in endometrium and differentially expressed in endometriosis (19–23). Differential expression of miRNAs between eutopic endometrium of women with and without endometriosis, and between eutopic and ectopic endometrial samples from endometriosis patients have been identified using microarray profiling (20, 21,23, 24). MiRNAs likely act as potent regulators of gene expression in the pathogenesis of endometriosis and endometriosis associated infertility (25) and with the hypothesis that plasma miRNAs might be differentially expressed in women with endometriosis, circulating miRNAs have been identified in sera and plasma of patients with endometriosis using miRNA microarray expression profiling and shown to have relatively high diagnostic power (26, 27).
We have previously shown that in women with severe endometriosis, a polymorphism of a let-7 miRNA binding site in the KRAS 3’ UTR leads to abnormal endometrial cell KRAS expression as well as increased proliferation and invasion(19). Further, in women with endometriosis, endometrial let-7 family members were expressed at lower levels. Similarly, homeobox A10 (HOXA10), an essential regulator of endometrial receptivity, has been shown to have decreased expression in women with endometriosis; the aberrant regulation in the endometrium of women with endometriosis is regulated by miR-135a and miR-135b (22). Considering aberrant expressions in endometrium and the important roles of these miRNAs in the pathogenesis of endometriosis, we hypothesized that these miRNAs would be differentially expressed in circulating blood in women with and without endometriosis. Here we evaluated the expressions of circulating let-7 and miR-135 in patients with and without endometriosis and determined the diagnostic utility of serum miRNAs as potential noninvasive biomarkers for endometriosis.
Materials and Methods
Study population
Forty eight subjects aged 18 to 48 years participated in this study after giving written informed consent. The study was approved by the Institutional Review Board of Gangnam Severance Hospital and Yale University School of Medicine. Volunteers were recruited among patients who underwent laparoscopy for various indications including pelvic masses, pelvic pain, endometriosis, infertility, and diagnostic evaluation of benign gynecologic disease between June 2010 and March 2013. Women were recruited when the following criteria were met: aged 20–50 year, no hormone therapy for at least 3 months, nonsmoker, without history or signs of other inflammatory disease undergoing surgical treatment/exploration. Exclusion criteria included post-menopausal status; previous hormone or gonadotropin-releasing hormone (GnRH) agonist use; adenomyosis; endometrial cancer, hyperplasia, or endometrial polyps; infectious diseases; chronic or acute inflammatory diseases; malignancy; autoimmune disease; and cardiovascular disease. Pre-treatment serum CA-125 levels in all patients were measured using CA-125 II electro-chemiluminescence immunoassay (ECLIA) with the Roche/Hitachi Modular Analytics E170 system (Roche Diagnostics, Tokyo, Japan). Before surgery, clinical data from each individual was collected including alcohol usage and comorbidities. Concerning classifying alcohol and non-alcohol users, the group was divided into non-drinking or drinking individuals and no attempts were made about the frequency of alcohol use. At the time of surgery, all possible endometriotic lesions were excised and sent to pathology for confirmation of diagnosis. Patients were assigned to the endometriosis group only after pathologic confirmation of excised tissue. The extent of endometriosis was determined using the American Society of Reproductive Medicine (ASRM) revised classification (28). Twenty-four patients had histologically confirmed peritoneal and/or ovarian endometriosis, with moderate-to-severe disease (Stages III and IV). Twenty-four patients participated as controls, which included 10 cases of dermoid cysts (n=10), serous cystadenoma (n=5), mucinous cystadenoma (n=3), simple ovarian cysts (n=3) and paratubal cysts (n=1). All patients had regular menstrual cycle and menstrual cycle stage was recorded for each patient as either proliferative phase, (from the beginning of menses until 14 days before the next menses), or secretory phase, (1–13 days before the next menses).
Sample Collection and RNA extraction
Blood samples were collected into 10 mL sterile tubes containing no additives before anesthesia. All subjects abstained from food for at least 8 hours prior to surgery. Samples were immediately centrifuged at 1000×g for 10 min and sediment-free serum samples were obtained. Serum aliquots were frozen at −80 °C until further analysis and thereafter were thawed only once. Total RNA was extracted from 400 µℓ of serum using the miRVana™ RNA Isolation Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s specifications, and eluted with 50 µℓ of nuclease-free water. The yield of RNA was determined using a NanoDrop ND-2000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA).
Quantitative real-time PCR for miRNAs
We employed the Poly (A) RT-PCR method using Invitrogen NCode miRNA First-Strand cDNA Synthesis MIRC-50 kit (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s instructions. Total RNA (25 ng) from each sample was reverse transcribed and miRNAs were quantified using the iQ SYBR Green supermix kit (Bio-Rad Laboratories) with the specific forward primers to let-7a–f, miR-135a and miR-135b, and the universal reverse primer complementary to the anchor primer. Reaction mixture included 2.5 µℓ of cDNA, 12.5 µℓ of iQSYBR Green Supermix, 1.0 µℓ of forward primer, 1 µℓ of Universal qPCR Primer, and 8 µℓ RNase-free water for a final reaction volume of 25 µℓ. The thermal cycling conditions were initiated by uracil-N glycosylase activation at 50 °C for 2 min and initial denaturation at 95 °C for 10 min, and then 40 cycles at 95°C for 15 sec, and annealing at 60 °C for 20 sec. Threshold cycle (Ct) and melting curves were acquired by using the quantitation and melting curve program of the Bio-Rad iCycleriQsystem (Bio-Rad Laboratories). Anchor RT primer was used as the template for negative control and U6 small nuclear RNA was used as a control to determine relative miRNA expression (29, 30). Relative mRNA level was determined using the formula 2−ΔCT. Primers for let-7a–f, miR-135a, miR-135b and the U6 genes were obtained from the W. M. Keck Oligonucleotide Synthesis Facility (Yale University): let-7a forward, TGAGGTAGTAGGTTGTATAGTT; let-7b forward, TGAGGTAGTAGGTTGTGTGGTT; let-7c forward, TGAGGTAGTAGGTTGTATGGTT; let-7d forward, AGAGGTAGTAGGTTGCATAGTT; let-7e forward, TGAGGTAGGAGGTTGTATAGTT; let-7f forward, TGAGGTAGTAGATTGTATAGTT; miR-135a forward, TATGGCTTTTTATTCCTATGTGA; mi-135b reverse, TATGGCTTTTCATTCCTATGTGA; U6 forward, CTCGCTTCGGCAGCACA, reverse, AACGCTTCACGAATTTGCGT.
Statistical Analysis
Data were expressed as mean ± standard deviation (SD) or median (interquartile range) where appropriate. Student’s t-test was used to determine the significance of difference in clinical characteristics between endometriosis and control group. The expression levels of serum miRNAs between the groups were compared using the Mann–Whitney U-test. To determine the significance of differences in correlations, Pearson’s correlation coefficient or Spearman rank correlation coefficient were calculated, where appropriate. The diagnostic performance of miRNA expression levels were assessed using receiver operating characteristic (ROC) curves to plot the test sensitivity versus its false-positive rate and determine the usefulness of a diagnostic test over a range of possible clinical results (31). The diagnostic utility of the test can be expressed as the area under the ROC curve (AUC), which was calculated as a measure of the ability of each potential biomarker to discriminate between endometriosis and control cases. An AUC of 0.5 indicates classifications assigned by chance. Based on ROC analysis, the best statistical cut-off value of miRNA expression level was calculated, which corresponds to the point at which the sum of false-positives and false-negatives is less than any other point. Sensitivity and specificity for selected cut-off points were then assessed. To assess the diagnostic power of multiple combinations of miRNAs expression levels, logistic regression model was applied. SPSS 16.0 (SPSS Inc, Chicago, IL, USA) was used for statistical analysis and a P<0.05 was considered statistically significant.
Results
Clinical Characteristics
The clinical characteristics of subjects are shown in Table 1. The mean age (± SD) of patients with endometriosis and controls was 33.08 ± 6.63 years and 32.16 ± 9.46 years, respectively. There were no significant differences in parameters such as mean age, gravidity, parity, alcohol usage and comorbidities between the two groups. Comorbidities included one case with hyperthyroidism and one case of duodenal ulcer in control group and one case of hyperthyroidism and one case of hypothyroidism in the endometriosis group. In contrast, visual analogue scale (VAS) score for pelvic pain intensity and serum CA-125 levels were significantly higher in patients with endometriosis than in controls (5.40 ± 3.37 vs. 2.08 ± 2.39, and 103.25 ± 121.75 vs.17.48 ± 6.28 IU/ml, respectively; P=0.003) (32). In this referral population all endometriosis patients had moderate-to-severe disease with mean rAFS scores of 53.93. All 24 patients had ovarian endometriosis. Among these patients, 22 patients had co-existing peritoneal disease and 8 individuals were also considered to have lesions of deep infiltrating endometriosis (Table 1).
Table 1.
Clinical characteristics of women with and without endometriosis (controls) recruited for this study.
| Endometriosis (N=24) |
Control (N=24) |
P-value | |
|---|---|---|---|
| Age (yrs) | 33.08±6.63 | 32.16±9.46 | 0.700 |
| Gravidity | 1.41 ± 1.58 | 1.29 ± 1.39 | 0.773 |
| Parity | 0.71 ± 0.85 | 0.79 ± 0.93 | 0.749 |
| Alcohol usage | 0 (0.0%) | 3 (12.5%) | 0.234 |
| Cormobidities | 2 (8.3%) | 2 (8.3%) | 1.000 |
| Pain Intensity (VAS) | 5.40 ± 3.37 | 2.08 ± 2.39 | <0.001 |
| CA-125(IU/ml) | 103.25 ± 121.75 | 17.48 ± 6.28 | 0.003 |
| r-AFS Stage | |||
| III (n) | 11 (45.8%) | NA | |
| VI (n) | 13 (54.2%) | NA | |
| r-AFS scores | 53.93 ± 6.02 | NA | |
| Distribution of endometriosis | |||
| Ovarian endometrioma | 24 (100.0%) | NA | |
| Peritoneal lesion | 22 (91.6%) | NA | |
| DIE status | |||
| With DIE lesions | 8 (33.3%) | NA | |
| Without DIE lesions | 16 (66.7%) | NA |
Data are expressed as mean ± S.D. or n (%); VAS, visual analogue scale; NA, not applicable; r-AFS, revised American Fertility Society
Expression of circulating miRNAs in sera of patients with and without endometriosis
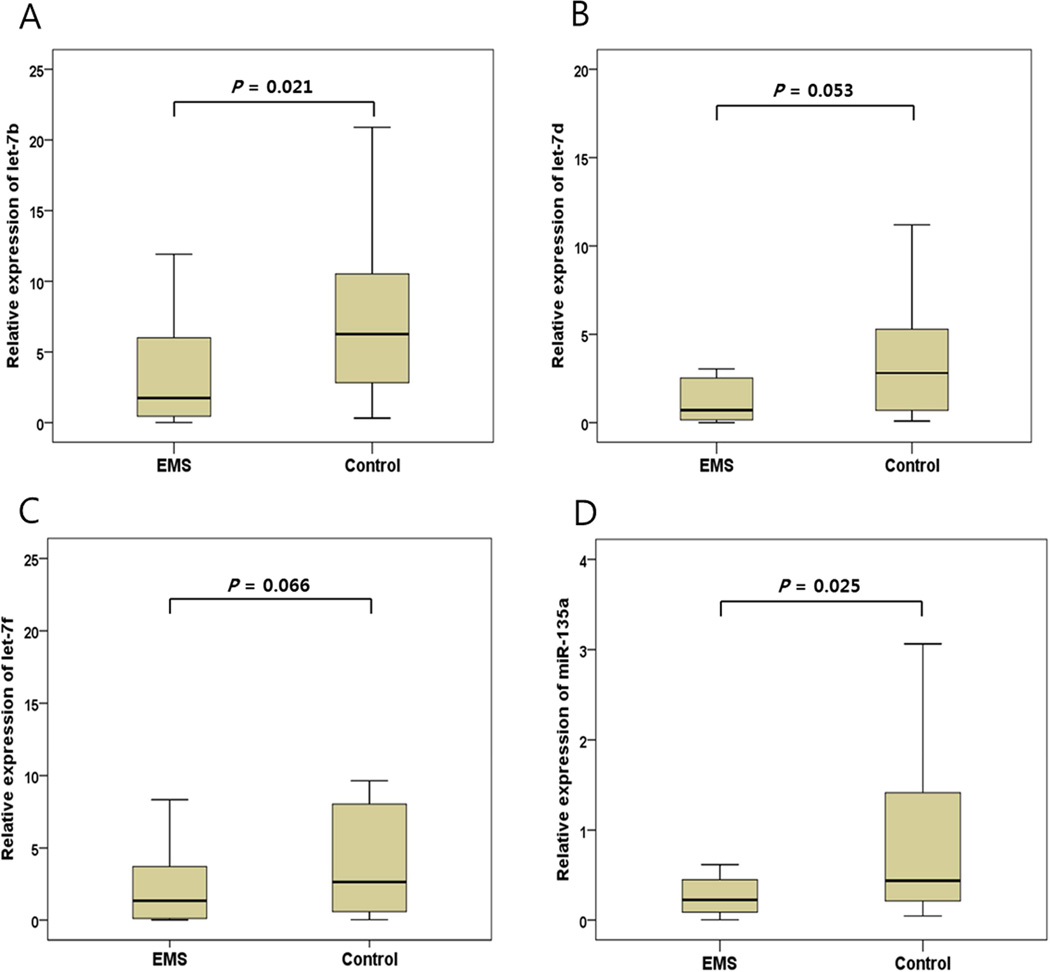
The relative expressions of let-7a-f, miR-135a and miR-135b in the serum of patients with and without endometriosis were assessed using quantitative real-time PCR. Among those miRNAs, the expression levels of let-7b and miR-135a were significantly lower in patients with endometriosis than controls (3.2-fold differences, P=0.021 and 2.0-fold differences, P=0.025, respectively), while the expression of let-7d and lef-7f showed a trend toward down-regulation (Figure 1). When correlations between miRNA expressions and clinical parameters were evaluated, there was significant negative correlations between let-7b expression levels and serum CA-125 levels (P=0.022, r=−0.333) (Supplemental Figure 1), but no other clinical parameters showed significant correlations with miRNA expression levels. Notably, when the correlations between expression levels of let-7 subtypes and miR-135 were evaluated, all let-7 subtypes including let-7a, let-7b, let-7c, let-7d, let-7e, and let-7f displayed strong positive correlations with miR-135a expression levels (r=0.512, r=0.576, r=0.413, r=0.449, r=0.490 and r=0.643, respectively) with statistical significance (P<0.001, P<0.001, P=0.004, P=0.002, P=0.001, and P<0.001, respectively).
Figure 1.
miRNAs are differentially expressed in sera of endometriosis patients and control. The expression levels of let-7b and mir-135a were significantly lower in patients with endometriosis than controls, whereas let-7d and 7f showed a trend toward decreased expressions (A–D). Data are represented by box-and-whisker plots. Boxes indicate the 25th and 75th percentiles, with a solid line within the box showing the median value.
Evaluation of target gene prediction for let-7b and mir-135a
Using the StarBase database (33), target genes for differently expressed miRNAs between endometriosis and controls were predicted. For let-7b, 321 genes were predicted as target sites including several genes that are known to be dysregulated in cancerous conditions and other diseases such as CCND1, HOXA9, TGFBR1 and HMGA2. Interestingly, functional annotation analysis indicated that 15 of these genes are involved in p53 signaling pathway (CASP3, CCND1, CCND2, CDKN1A, FAS, RRM2, and THBS1) and cell cycle (CCND1, CCND2, CDC25A, CDKN1A, E2F5, ESPL1, SMC1A, and YWHAZ), suggesting that let-7b may play a role in endometriosis by influencing the function of p53 proteins and cell cycle control. For mir-135a, 62 genes were predicted as targets sites including HOXA10, VLDLR and CSF1. Association between mir-135a and HOXA10 in endometriosis and ovarian cancer has been documented previously (22, 34). It has been shown that mir-135a functions as a tumor suppressor in epithelial ovarian cancer by regulation of HOXA10 expression (34) and acts as a putative tumor suppressor by directly targeting VLDLR in human gallbladder cancer (35). Also, several targeted genes are known to be involved in positive regulation of apoptotic process (DDX3X, NET1, NF1, TCF7L2, and TXNIP) and Wnt signaling pathways (FZD1, NFAT5, TBL1XR1, TCF7L2).
Expression of circulating miRNAs according to the menstrual cycle
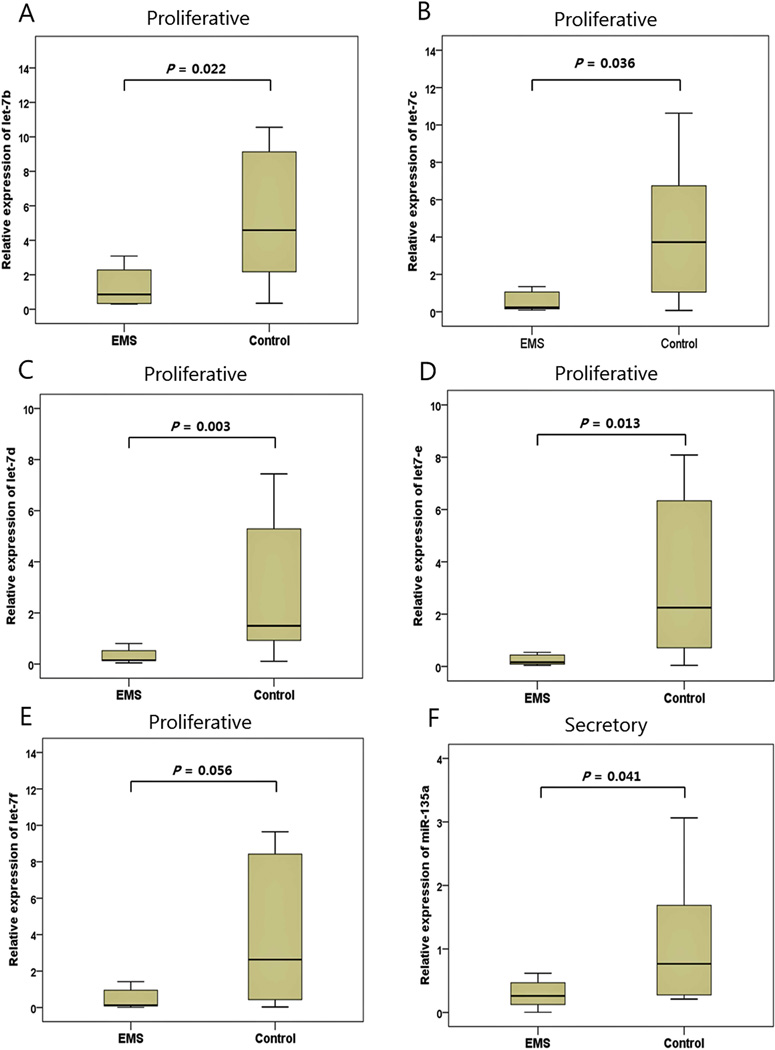
When the relative expressions of miRNAs in patients with and without endometriosis were evaluated according to the menstrual cycle, several miRNAs were found to be expressed differently in endometriosis patients. The expression of let-7a, let-7d, let-7e and let-7f were significantly higher during the secretory phase (P=0.018, P=0.033, P=0.039, and P=0.039, respectively) whereas, no cyclic differences in miRNA expression were noted in control group. When miRNA expression were compared between the endometriosis group vs controls according to the menstrual cycle, let-7b, let-7c, let-7d and let-7e were down-regulated in endometriosis patients compared to control (5.3-fold,16.9-fold, 9.9-fold and 14.0-fold, respectively) during the proliferative phase with statistical significance (P=0.022, P=0.036, P=0.003 and P=0.013, respectively), whereas the expressions of let-7f showed a trend toward down-regulation (Figure 3). During secretory phase, the expression of miR-135a was significantly lower in patients with endometriosis than controls (2.9-fold, P=0.041) (Figure 2).
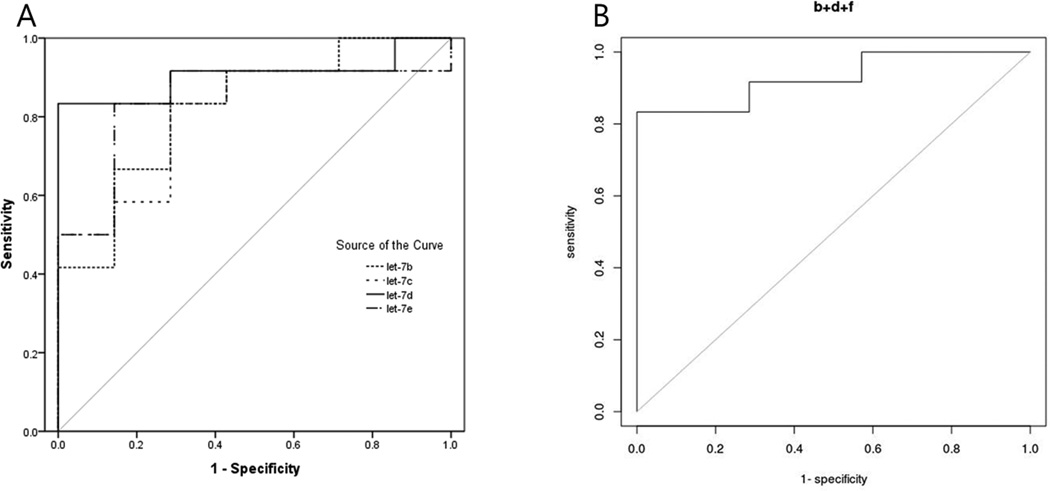
Figure 3.
Receiver operating characteristic (ROC) curves of different miRNAs during the proliferative phase. Let-7d showed the highest AUC of 0.905 (A). When the combination of the serum miRNA expression levels during the proliferative phase were utilized using the logistic regression model, the combination of serum let-7b, let-7d and let-7f showed the highest AUC value of 0.929 (B).
Figure 2.
Expression levels of miRNA in serum from endometriosis patients and controls according to the menstrual cycle. During the proliferative phase, let-7b (A), let-7c (B), let-7d (C) and let-7e (D) were significantly down-regulated in endometriosis patients compared with controls. The expression levels of let-7f showed a trend toward down-regulation (E). During the secretory phase, mir-135a was significantly down-regulated in patients with endometriosis compared with controls (F). Data are represented by box-and-whisker plots. Boxes indicate the 25th and 75th percentiles, with a solid line within the box showing the median value.
Assessment of the diagnostic value of circulating miRNAs in endometriosis
To assess the utility of circulating miRNAs in diagnosing endometriosis, we examined the ROC curve of serum miRNA that were differentially expressed between endometriosis patients and controls. The AUC of let-7b and miR-135a were 0.694 [95% confidence interval (CI): 0.543–0.844] and 0.690 (95% CI: 0.540–0.841), respectively. Because several miRNAs were found to be differentially expressed between the endometriosis patients and controls according to the menstrual cycle, we examined the ROC curve of let-7b, let-7c, let-7d, and let-7e during the proliferative phase and that of miR-135a during the secretory phase. The AUC of let-7b, let-7c, let-7d and let-7e during the proliferative phase were 0.821 (95% CI: 0.599–1.000), 0.798 (95% CI: 0.584–1.000), 0.905 (95% CI: 0.000–1.000) and 0.845 (95% CI: 0.000–1.000), respectively. The AUC of miR-135a during the secretory phase showed 0.741 (95% CI: 0.549–0.933). Because let-7d expression levels yielded the highest AUC among others, we calculated the sensitivity and specificity for let-7d expression levels, and it displayed the sensitivity and specificity of 83.3% and 100.0% at the cut-off value of 0.823 in discriminating endometriosis from controls.
To improve the diagnostic power of miRNA expressions during the proliferative phase, we applied the logistic regression model to the data for disease vs control samples. Different combinations of predictors were fed to the model and the maximum AUC score of 0.929 was achieved when using the predictors let-7b, let-7d and let-7f (Figure 3). The fitted logistic regression model is as follows:
Where P(Y=1) is the probability of a proliferative stage sample being a disease sample.
Discussion
In the present study we evaluated expression of circulating let-7 and miR-135 family members as potential biomarkers for endometriosis. Our results showed that the expressions of let-7b and miR-135a were significantly down-regulated in patients with endometriosis compared to those without the disease. We also evaluated the cyclic differences of these miRNAs and discovered that the expressions of several miRNAs were different between endometriosis and controls when compared according to stage of the menstrual cycle. Although the study population included relatively high prevalence of endometriosis in this study, when the diagnostic power of differentially expressed miRNAs were evaluated, let-7d expression during the proliferative phase showed a sensitivity and specificity of 83.3% and 100.0%, respectively, at a cut-off value of 0.823. In addition, the combination of let-7b, let-7d and let-7f during the proliferative phase yielded even higher diagnostic potentials using the logistic regression model with an AUC of 0.929.
Since its first discovery in 1993, miRNAs have been shown to play critical roles in various physiological and pathological conditions. Moreover, much attention has been given to circulating miRNAs in search of new potential biomarkers for human diseases due to the fact that circulating miRNAs offer several advantages as biomarker source, such as easy accessibility, consistent reproducibility across individuals of the same species (36), and relative stability against enzymatic degradation, freezing, and thawing, or intense pH conditions (12, 37). Numerous studies indicated that circulating miRNAs may serve as reliable biomarkers for various human diseases (12, 14–16,38, 39). However, despite the fact that miRNAs have been shown to be involved in the pathogenesis of endometriosis by regulation of various transcripts associated with hypoxia, inflammation, apoptosis, tissue repair, cellular proliferation, extracellular matrix remodeling and angiogenesis (25), data concerning circulating miRNAs and endometriosis has been limited until recently (26, 27).
Let-7 was the first human miRNA discovered and one of its major functions is to promote differentiation of cells (40). However, reduced expression of multiple let-7 members have been found to be associated with human malignancies leading to the strong notion that the let-7 miRNA family also function as tumor suppressors (41), and their restoration to normal levels has been reported to suppress cancer growth (42). Interestingly, our results showed that let-7b expression levels were significantly lower in patients with endometriosis than controls, and significant negative correlations were found between the expression levels of let-7b and serum CA-125 levels. Although there is no data concerning the expression of circulating members of the let-7-family in endometriosis patients, associations between tissue let-7b expression and endometriosis has been noted previously (19). In accordance with our results, let-7b expression was significantly decreased in cultured human endometrial stromal cells of patients with endometriosis compared with those without the disease (19). Also, let-7b has been shown to be associated with cyclin D1, an estrogen-regulated gene involved in cellular proliferation and endometriosis (43). The inverse relationship between let-7b and cyclin D1 expression has been documented (44), in addition to elevated KRAS are possible mechanism regulating cell proliferation in endometriosis.
Circulating miR-135a expressions was significantly lower in patients with endometriosis and the expression levels were positively correlated with let-7 family expression. Interestingly both miR-135a and miR-135b were overexpressed in the endometrium of patients with endometriosis than controls and increased expression of these miRNAs in endometriosis led to suppression of genes required for implantation (22). There is no clear explanation for the discrepancy between the tissue and serum levels. Although the correlation of miRNA expression between tissues and the circulation have been noted in various conditions, discordance in miRNA expression patterns also has been reported (45). Moreover miR-135a functions differently in some tumor lineages, acting as oncomiR in colorectal cancers through inactivation of the adenomatous polyposis coli, however as a tumor suppressor in ovarian cancer by regulation of HOXA10 expressions (34, 46). These miRNAs may be express differently and have distinct functions depending on the environment and specific tissue types.
Interestingly, the functional annotation analysis revealed several genes that are predicted target sites for both let-7b and mir-135a and some of these genes were identified as BACH1 and IGF2BP1. BACH1 is a transcriptional repressor of the Hemeoxygenase 1 (HMOX1), which is known as a key cytoprotective enzyme involved in catalyzing heme degradation and acting as antioxidants (47), and it has been shown that let-7b miRNA directly acts on the 3’-UTR of BACH1 and negatively regulates expression of this protein, thereby up-regulating HMOX1 gene expression and increasing resistance against oxidant injury (48). IGF2BP1 belong to a conserved family of RNA-binding, oncofetal proteins involved in cell polarization, migration, morphology, metabolism, proliferation and differentiation in tumor-derived cells (49) and its association with other classical oncogenes, in particular MYC and KRAS has been documented (50). These findings suggest that these miRNAs may be involved in the pathogenesis of endometriosis via regulation of their target genes, and that their protein products may serve as new potential targets for biomarker discovery.
In the present study, we observed significantly lower expression of several miRNAs in sera from women with endometriosis compared with controls. However, the exact origin of these changes in circulating is not clear. Although evidence suggests that miRNAs may be released from tissues and shed into the circulation (17, 18), discrepancies between paired tissue–plasma miRNA expression profiles has been noted indicating that disease tissue or malignant tumor cells are not the sole source of circulating miRNAs (51). The concentration of miRNAs in the circulation is not simply a reflection of the tissue level, rather it more likely depends on the complex regulatory relationships between miRNAs and the possible regulatory relationship between tissues. While still poorly understood, circulating miRNAs may function to signal between tissues; circulating levels reflect the net production from multiple sources in a complex regulatory network.
The potential of circulating miRNAs as diagnostic biomarkers for endometriosis has only recently been described, (26, 27). Similar to these two studies, our analysis also showed higher AUC when combination of two or more circulating miRNAs were used, and the diagnostic power of our combined marker was higher than those of commonly used diagnostic indexes of endometriosis, such as CA-125, IL-8, IL-6, CA125, TNF-α, high sensitivity C-reactive protein, and CA19-9, (32, 52). However, in contrast to our study which focused on miRNAs that have been shown to be associated with endometriosis, previous studies selected target miRNAs for validation based on microarray-based miRNA expression profiling of sera or plasma of patients with and without the disease (26, 27). Since the associations between the majority of the selected miRNAs and the role in the endometriosis lesions have not been described previously, it will be interesting to determine the roles of those miRNAs in the regulation of endometrium and endometriosis. Also, to the best of our knowledge, this is the first study to evaluate the cyclic differences of circulating miRNAs in endometriosis, and the best AUC was achieved in the proliferative phase after logistic regression. It is interesting, however, that cyclic variations were only noted in sera of patients with endometriosis, but not in those without the disease. These findings are in agreement with a study showing that circulating miRNAs levels do not fluctuate in healthy women through the menstrual cycle (53). Cyclic differences in circulating miRNAs were associated with endometriosis, and are further evidence of the role of aberrant miRNA regulation as an essential component of endometriosis (22).
There are several limitations to this study. Although all patients underwent laparoscopic surgery and were proven to be free of endometriosis, our control group had various non-endometriotic cysts, which may have altered the serum microRNA profiles. However, the presence of pathology other than endometriosis may have more closely mimicked application in clinical practice where the differential diagnosis often includes other gynecologic disease. Also, because only patients with advanced stage of the disease were included in this study, further research involving a large number of patients is warranted to investigate the diagnostic accuracy in minimal-to-mild disease. Lastly, this study lacks validation of the data in a separate sample set. Therefore, validations of these potential markers in independent populations should be considered.
In conclusion, we have shown that let-7b and miR-135a, miRNAs, that we have described previously to be associated with endometriosis (19, 22), are detectable in serum samples and are differentially expressed between patients with and without endometriosis. We also showed that the expression of these miRNAs have cyclic differences. The combinations of several circulating miRNAs are a potential diagnostic biomarker for endometriosis. Despite several limitations such as small sample size, inclusion of only moderate-to-severe cases, and use of non-endometriosis patients with benign gynecologic disease as controls, the relatively high diagnostic value during proliferative phase make these circulating miRNAs worthy of further consideration as a novel diagnostic marker for endometriosis.
Supplementary Material
Acknowledgments
Funding: This study was supported by National Institute of Health Grant U54 HD052668.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author’s roles:
SiHyun Cho- substantial contributions to conception and design analysis, interpretation of data, drafting the article and revising, final approval of the version to be published
LeventMutlu- analysis and interpretation of data, revising the article, final approval of the version to be published
Olga Grechukhina- substantial contributions to conception and design, revising the article, final approval of the version to be published
Hugh S. Taylor- substantial contributions to conception and design, drafting the article and revising, reviewing the article, final approval of the version to be published
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Taylor RN, Lebovic DI, Mueller MD. Angiogenic factors in endometriosis. Ann N Y Acad Sci. 2002;955:89–100. doi: 10.1111/j.1749-6632.2002.tb02769.x. discussion 18, 396–406. [DOI] [PubMed] [Google Scholar]
- 2.Husby GK, Haugen RS, Moen MH. Diagnostic delay in women with pain and endometriosis. Acta Obstet Gynecol Scand. 2003;82:649–653. doi: 10.1034/j.1600-0412.2003.00168.x. [DOI] [PubMed] [Google Scholar]
- 3.Dunselman GA, Vermeulen N, Becker C, Calhaz-Jorge C, D'Hooghe T, De Bie B, et al. ESHRE guideline: management of women with endometriosis. Hum Reprod. 2014;29:400–412. doi: 10.1093/humrep/det457. [DOI] [PubMed] [Google Scholar]
- 4.Vodolazkaia A, El-Aalamat Y, Popovic D, Mihalyi A, Bossuyt X, Kyama CM, et al. Evaluation of a panel of 28 biomarkers for the non-invasive diagnosis of endometriosis. Hum Reprod. 2012;27:2698–2711. doi: 10.1093/humrep/des234. [DOI] [PubMed] [Google Scholar]
- 5.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 6.Akao Y, Nakagawa Y, Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26:311–320. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 7.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 8.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 10.Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K, et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68:10307–10314. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsh EE, Lin Z, Yin P, Milad M, Chakravarti D, Bulun SE. Differential expression of microRNA species in human uterine leiomyoma versus normal myometrium. Fertil Steril. 2008;89:1771–1776. doi: 10.1016/j.fertnstert.2007.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 14.Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58:1284–1292. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, Wan XJ, et al. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem Biophys Res Commun. 2010;394:184–188. doi: 10.1016/j.bbrc.2010.02.145. [DOI] [PubMed] [Google Scholar]
- 16.D'Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–2773. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 18.Wang F, Zheng Z, Guo J, Ding X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010;119:586–593. doi: 10.1016/j.ygyno.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 19.Grechukhina O, Petracco R, Popkhadze S, Massasa E, Paranjape T, Chan E, et al. A polymorphism in a let-7 microRNA binding site of KRAS in women with endometriosis. EMBO Mol Med. 2012;4:206–217. doi: 10.1002/emmm.201100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohlsson Teague EM, Van der Hoek KH, Van der Hoek MB, Perry N, Wagaarachchi P, Robertson SA, et al. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. 2009;23:265–275. doi: 10.1210/me.2008-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;13:797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- 22.Petracco R, Grechukhina O, Popkhadze S, Massasa E, Zhou Y, Taylor HS. MicroRNA 135 regulates HOXA10 expression in endometriosis. J Clin Endocrinol Metab. 2011;96:E1925–E1933. doi: 10.1210/jc.2011-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramon LA, Braza-Boils A, Gilabert-Estelles J, Gilabert J, Espana F, Chirivella M, et al. microRNAs expression in endometriosis and their relation to angiogenic factors. Hum Reprod. 2011;26:1082–1090. doi: 10.1093/humrep/der025. [DOI] [PubMed] [Google Scholar]
- 24.Dai L, Gu L, Di W. MiR-199a attenuates endometrial stromal cell invasiveness through suppression of the IKKbeta/NF-kappaB pathway and reduced interleukin-8 expression. Mol Hum Reprod. 2012;18:136–145. doi: 10.1093/molehr/gar066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teague EM, Print CG, Hull ML. The role of microRNAs in endometriosis and associated reproductive conditions. Hum Reprod Update. 2010;16:142–165. doi: 10.1093/humupd/dmp034. [DOI] [PubMed] [Google Scholar]
- 26.Wang WT, Zhao YN, Han BW, Hong SJ, Chen YQ. Circulating microRNAs identified in a genome-wide serum microRNA expression analysis as noninvasive biomarkers for endometriosis. J Clin Endocrinol Metab. 2013;98:281–289. doi: 10.1210/jc.2012-2415. [DOI] [PubMed] [Google Scholar]
- 27.Jia SZ, Yang Y, Lang J, Sun P, Leng J. Plasma miR-17-5p, miR-20a and miR-22 are down-regulated in women with endometriosis. Hum Reprod. 2013;28:322–330. doi: 10.1093/humrep/des413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.American Society for Reproductive Medicine. Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil Steril. 1997;67:817–821. doi: 10.1016/s0015-0282(97)81391-x. [DOI] [PubMed] [Google Scholar]
- 29.Kuwabara Y, Ono K, Horie T, Nishi H, Nagao K, Kinoshita M, et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet. 2011;4:446–454. doi: 10.1161/CIRCGENETICS.110.958975. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, et al. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56:1830–1838. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- 31.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 32.Mol BW, Bayram N, Lijmer JG, Wiegerinck MA, Bongers MY, van der Veen F, et al. The performance of CA-125 measurement in the detection of endometriosis: a meta-analysis. Fertil Steril. 1998;70:1101–1108. doi: 10.1016/s0015-0282(98)00355-0. [DOI] [PubMed] [Google Scholar]
- 33.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang W, Jiang Y, Mu X, Xu L, Cheng W, Wang X. MiR-135a functions as a tumor suppressor in epithelial ovarian cancer and regulates HOXA10 expression. Cell Signal. 2014;26:1420–1426. doi: 10.1016/j.cellsig.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Zhou H, Guo W, Zhao Y, Wang Y, Zha R, Ding J, et al. MicroRNA-135a Acts as a Putative Tumor Suppressor by Directly Targeting VLDLR in Human Gallbladder Cancer. Cancer Sci. 2014 doi: 10.1111/cas.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 37.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka M, Oikawa K, Takanashi M, Kudo M, Ohyashiki J, Ohyashiki K, et al. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One. 2009;4:e5532. doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Jerome T, Laurie P, Louis B, Pierre C. Enjoy the Silence: The Story of let-7 MicroRNA and Cancer. Curr Genomics. 2007;8:229–233. doi: 10.2174/138920207781386933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pellegrini C, Gori I, Achtari C, Hornung D, Chardonnens E, Wunder D, et al. The expression of estrogen receptors as well as GREB1, c-MYC, and cyclin D1, estrogen-regulated genes implicated in proliferation, is increased in peritoneal endometriosis. Fertil Steril. 2012;98:1200–1208. doi: 10.1016/j.fertnstert.2012.06.056. [DOI] [PubMed] [Google Scholar]
- 44.Guo Y, Yan K, Fang J, Qu Q, Zhou M, Chen F. Let-7b expression determines response to chemotherapy through the regulation of cyclin D1 in glioblastoma. J Exp Clin Cancer Res. 2013;32:41. doi: 10.1186/1756-9966-32-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jarry J, Schadendorf D, Greenwood C, Spatz A, van Kempen LC. The validity of circulating microRNAs in oncology: Five years of challenges and contradictions. Mol Oncol. 2014 doi: 10.1016/j.molonc.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 47.Zhu Z, Wilson AT, Mathahs MM, Wen F, Brown KE, Luxon BA, et al. Heme oxygenase-1 suppresses hepatitis C virus replication and increases resistance of hepatocytes to oxidant injury. Hepatology. 2008;48:1430–1439. doi: 10.1002/hep.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou W, Tian Q, Steuerwald NM, Schrum LW, Bonkovsky HL. The let-7 microRNA enhances heme oxygenase-1 by suppressing Bach1 and attenuates oxidant injury in human hepatocytes. Biochim Biophys Acta. 2012;1819:1113–1122. doi: 10.1016/j.bbagrm.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bell JL, Wachter K, Muhleck B, Pazaitis N, Kohn M, Lederer M, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70:2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mongroo PS, Noubissi FK, Cuatrecasas M, Kalabis J, King CE, Johnstone CN, et al. IMP-1 displays cross-talk with K-Ras and modulates colon cancer cell survival through the novel proapoptotic protein CYFIP2. Cancer Res. 2011;71:2172–2182. doi: 10.1158/0008-5472.CAN-10-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suryawanshi S, Vlad AM, Lin HM, Mantia-Smaldone G, Laskey R, Lee M, et al. Plasma microRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer. Clin Cancer Res. 2013;19:1213–1224. doi: 10.1158/1078-0432.CCR-12-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.May KE, Conduit-Hulbert SA, Villar J, Kirtley S, Kennedy SH, Becker CM. Peripheral biomarkers of endometriosis: a systematic review. Hum Reprod Update. 2010;16:651–674. doi: 10.1093/humupd/dmq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rekker K, Saare M, Roost AM, Salumets A, Peters M. Circulating microRNA Profile throughout the menstrual cycle. PLoS One. 2013;8:e81166. doi: 10.1371/journal.pone.0081166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.