Abstract
The classification of neurons into types has been much debated since the inception of modern neuroscience. Recent experimental advances are accelerating the pace of data collection. The resulting information growth of morphological, physiological, and molecular properties encourages efforts to automate neuronal classification by powerful machine learning techniques. We review state-of-the-art analysis approaches and availability of suitable data and resources, highlighting prominent challenges and opportunities. The effective solution of the neuronal classification problem will require continuous development of computational methods, high-throughput data production, and systematic metadata organization to enable cross-lab integration.
Keywords: Neural classification, machine learning, standardization, metadata, big data
1. Data descriptors to classify neuron types
Neuron type classification is an increasingly hot topic, yet its history began with neuroscience itself [1]. Researchers routinely refer to pyramidal, stellate, granule, bipolar, or basket cells, but these names are often insufficient to describe neuronal diversity even within limited brain areas. Realizing this issue, both the European Human Brain Project and the American BRAIN initiatives identified cell-type classification among their first priorities [2,3]: “to complete a comprehensive cell census of the human brain”. The ultimate endeavor is to link neuronal types with behavior, computation, and eventually cognition. Prominent international efforts proposed initial guidelines to help organize the growing body of knowledge [4]. However, manual classification attempts are ill-equipped to deal with big data. The magnitude and complexity of neuron classification demands high-throughput technologies.
Neuroscience and computer science are mature to tackle neuronal classification by powerful mathematical approaches. Several recent studies leveraged modern computational methodologies to considerably advance the state-of-the-art [5-26]. Increasing integration of machine learning techniques with microscopic, chemical, and functional methods has already pushed bioinformatics to new heights [27]. While neuroscience is rapidly transitioning to digital data [28,29], the principles behind automatic classification algorithms remain often inaccessible to neuroscientists, limiting the potential for breakthroughs.
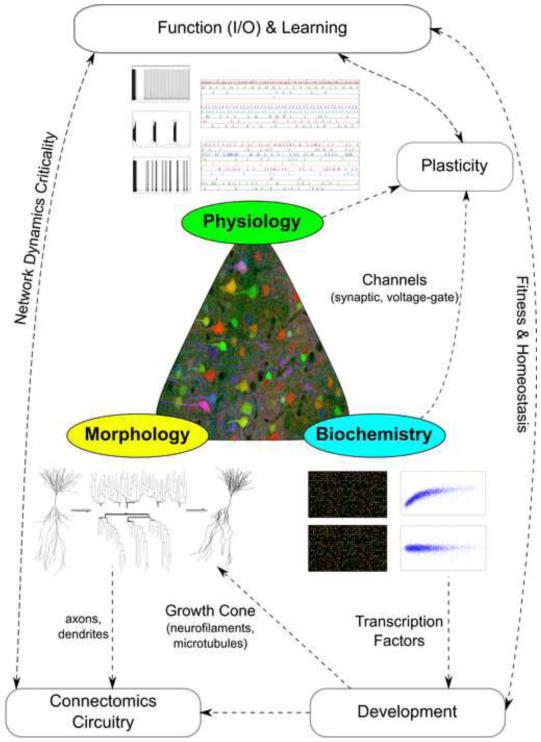
Neurons are typically characterized by their morphology, physiology, and biochemistry (Figure 1). These principal experimental approaches reflect the most prominent available techniques, namely microscopic imaging, electrical recording, and molecular analysis. These investigation domains also constitute proxies for key attributes of neuronal identity: axonal and dendritic structures establish the means for network connectivity; neuronal expression profiles provide a window onto developmental origins; and electrophysiological properties underlie signal processing. Furthermore, these features are intimately intertwined. The macromolecular machinery sculpts both neuronal excitability and circuitry, and these together define computational functions. The difficulty of the problem increases even further when considering systematic differences between species, across brain areas, and throughout development. Yet even for the most common animal models, well-defined regions of the nervous systems, and confined age ranges or developmental stages, available information on neuronal identity has so far failed to yield a broadly agreed-upon approach to neuronal classification.
Figure 1.
Basic dimensions of neuronal characterization: morphology (yellow), physiology (green), and biochemistry (blue). These feature domains are tightly interrelated with other fundamental aspects of neural identity, such as connectivity, development, and plasticity.
Much like “parts lists” preceding “exploded diagrams” in assembly kit manuals, the objective identification of neuron types is essential to understand their functional interactions [30,31,32]. After formally introducing automatic neuronal classification, we review exemplary progress, from foundational breakthroughs to recent trends, providing useful pointers to available informatics tools. We then highlight current opportunities and challenges in neuronal classification before discussing the transformative prospects of forthcoming big data.
2. Automatic neuronal classification
The term classification is often used with two related yet distinct meanings when referring to neuron types. In the narrower sense, neuronal classification is the process of dividing a group of neurons into known classes, as exemplified by the task to distinguish excitatory from inhibitory cells. The second usage of the term encompasses the above classification proper as well as the identification of the classes themselves, a step sometime referred to as categorization. This broader connotation implies the definition of distinct neuron types and the simultaneous assignment of neurons to each type.
This work reviews the automatic classification of neurons by quantitative measurements. The emphasis on minimized human intervention complements qualitative descriptions of neuron types based on expert knowledge (e.g. [33]) as well as computational models of the biophysical mechanisms differentiating neuron types (e.g. [34]). Automatic classification is primarily data-driven and hence largely blind to the researcher.
Formally, a neuronal classification dataset D (see Box 1 for a glossary of machine learning terms) consists of a set of k observed neurons, each described by (n+1) variables. The first n, known as predictive variables, are measurements on the neurons. The last variable, referred to as the class variable, specifies the neuron type. A classifier is a function γ assigning labels to observations,
Box 1. Glossary of common machine learning terms.
- Accuracy
Probability of classifying a new instance correctly. Conversely, the error is the complementary probability of classifying a new instance incorrectly. Accuracy and/or error quantify classifier performance.
- Attribute
Function (random variable) associating a value to every outcome of a (random) experiment. A discrete variable takes a numerable number of values; discrete variables are ordinal if a possible ordering exists among the values or nominal otherwise. In contrast, the domain of a continuous (or real) variable is not numerable.
- Classifier
Mathematical function that labels dataset instances from a set of possible classes.
- Clustering
see Unsupervised learning
- Data mining
Identification of previously undetected relationships and patterns in large datasets. Common data mining approaches include stages of selection, processing, transformation, learning, interpretation, and evaluation (for the last two stages, see also Knowledge discovery).
- Dataset
Collection of instances and the schema describing their structure. The most common machine learning dataset format is a matrix with columns as attributes and rows as instances. Training and test sets can be subsets of the initial dataset.
- Error
see Accuracy
- Feature
see Attribute
- Induction
Specification of a model from a training dataset. An induction algorithm is a computer program that takes a training dataset as input and outputs a model.
- Instance
Set of values (measurements) for a single observation (neuron) described by a number of variables. The terms instance, case, record or sample are indistinctly used in machine learning as referring to one and the same concept.
- Knowledge discovery
Human inspection, interpretation, validation, and refinement of patterns extracted from a data mining process.
- Model
Mathematical function that assigns labels to instances. In regression models the labels are continuous, whereas in classification models (or classifiers) the labels are discrete.
- Performance estimation
Statistical approach for predicting model correctness over future unseen samples. The most widely known methods include hold-out, cross-validation, and bootstrapping.
- Supervised classification
(also commonly referred to as Supervised learning): Techniques for model induction from a given training dataset. The task is to use instances with known classes or group structure in order to correctly predict the class (or group label) of unseen data samples.
- Semi-supervised classification/learning
Similar goal as Supervised classification, but the data contains only a (typically small) fraction of labeled instances together with a large number of unlabeled cases.
- Unsupervised classification/learning
Classes (labels) for the data are not available (or not used): the goal is to find the inherent categories, or optimal partition, of the dataset in order to maximize the proximity of instances within (relative to between) each category.
where the n-dimensional vector x = x1, …, xn contains the values for all measurements of a particular neuron, and {1, 2, …, m} are the possible neuronal classes. The real class of the given neuron, usually denoted c, is a value in that range.
The assumed (but unknown) joint probability distribution p(x1, …,xn, c) underlying the observations can be estimated from the sample {x1, c1, … , (xk, ck)}, where superscripts refer to neurons, and subscripts to measurements of those neurons.
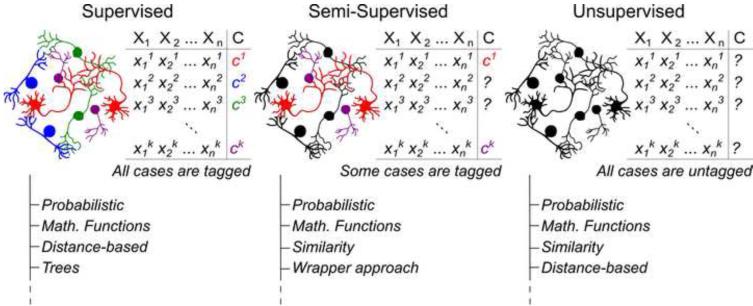
Extensive mathematical groundwork defined a wide variety of classifiers with distinct theoretical assumptions, whose effectiveness cannot be ranked a priori [35]. Automatic classification can be broadly conceptualized on the basis of available knowledge and scientific goal (Figure 2). In supervised classification, both the n measurement and the class assignment c are known for all k neurons in the dataset. The goal of supervised classification is to formulate a predictive or explanatory mapping between the measurements and the classes. For example, the values for spike amplitude, frequency, and duration from a known sample of glutamatergic and GABAergic neurons can be used to associate the spiking characteristics with their post-synaptic effects. This knowledge can be leveraged to infer network function or to deduce the neurotransmitter released by neurons in a different dataset from their spiking records. In supervised classification the number of neuron types is predetermined.
Figure 2.
Major classification approaches with representative families of algorithms. Examples of implementations with references and links to available resources are provided as Supplementary Material.
Conversely, in unsupervised classification or clustering only the set of n measurements is available, but the particular values of the class variable are unknown for all k neurons. In this case, the aim is to find the classes that best explain the measurements by grouping the neurons into identifiable clusters. Unsupervised classification also determines the number of cells types. For example, the observed variety of protein expression profiles in a set of neurons might be reducible to a restricted number of distinct, but internally consistent, expression patterns, each controlled by specific transcription factors.
In the intermediate case of semi-supervised classification, only some, but not all, neurons are labeled with known classes (Figure 2). For example, a researcher might record spiking latency, input resistance, and adaptation ratio for a set of neurons, but could only establish the morphological identity from biocytin injection in a minority of the cells. The Supplementary Material includes an expanded version of Figure 2 with extensive examples of algorithms corresponding to supervised, semi-supervised, and unsupervised classification along with references and pointers to available implementations.
3. Advances in automatic neuronal classification
This section surveys a representative selection of recent publications on automatic neuronal classification, briefly assessing biological data, computational techniques, and software implementation with particular attention on resource availability for further applications (Table 1).
Table 1.
Current machine learning techniques, software platforms, and data resources used for automatic neuronal classification.
| Refs | ML-technique | Software platform | Sample size | Data domains | Availability |
|---|---|---|---|---|---|
| [5] | ANOVA, hierarchical clustering, t-test, Gene Ontology |
MAS, dChip, R, Python, IgorPro |
36 microarrays of mouse forebrain cells (12 types) |
13,232 mRNA transcripts | 1-C* |
| [6] | Pearson correlation, ANOVA, t-test | SPSS, Prism | 520 crab stomatogastric ganglion cells (6 types) |
6 genes qPCR expression | 2-C |
| [7] | Intensity levels, SAM method, hierarchical clustering (complete linkage) |
MAS, dChip, SAM, Bioconductor, IPA, David |
29 mice forebrain astrocytes, neurons and oligodendrocytes (9 subtypes) |
45,037 mRNA probe sets | 1-C |
| [8] | Memory-guided saccade task, visuomovement index for each neuron, coefficient of variation, Wilcoxon ranksum test, Bonferroni correction |
R | 94 frontal eye field cells (3 subtypes of cells, 4 male macaques) |
1 electrophysiological (width of spikes) |
2-B |
| [9] | Hierarchical clustering (Ward’s linkage), k- means, Mann–Whitney U test, silhouette analysis |
Statistica, Matlab | 68 out of 200 cortical interneurons layers I to IV Wistar rat (6 subtypes) |
1 positional, 12 morphological, 32 electrophysiological, 10 molecular features |
2-B-C |
| [10] | Hierarchical clustering (Spearman correlation distance), t-test, fold-change, Gene Ontology |
MAS, GenMAPP, IPA |
16 microarrays of P8 mice tissues (8 subplate, 8 Layer VI cells) |
39,000 mRNA transcripts | 3-C |
| [11] | PCA, hierarchical clustering (Ward’s linkage), k-means, Mann Whitney U test, silhouette analysis |
Statistica, Clustan | 59 SOM and cortical mouse interneurons |
19 electrophysiological, 67 morphological features |
1-C |
| [12] | Hierarchical clustering, naïve Bayes, C4.5, kNN, MLP, Logistic regression, PCA, FSS |
R, WEKA | 327 mouse neocortex cells (128 pyramidal and 199 interneurons) |
65 morphological features | 1-C |
| [13] | Seven clustering algorithms: reported results from four of them (Fuzzy Gustafson-Kessel, k-means, PAM, affinity) |
Matlab | 471 mouse retinal ganglion cells (12 subtypes) |
5 electrophysiological | 3-D |
| [14] | Hierarchical clustering, fold-change, ANOVA, Gene Ontology |
Affymetrix Power Tools |
195 mouse microarrays (64 cell types) |
22,690 mRNA probe sets | 1-B |
| [15] | Coefficient of variation of interspike interval, hierarchical clustering, correlation, cross-covariance, t-tests |
Matlab, MClust | 207 ventral tegmental cells, male Long-Evans rats |
Action potential firing, dopamine receptor pharmacology |
2-B |
| [16] | Mutual information, kNN, area under the receiver operating characteristic curve (AUC) |
Matlab | 60 Drosophila melanogaster neuroblast clones |
350 whole brain images neuroblast clones |
1-A* |
| [17] | Expert crowdsourcing, k-means, Bayesian networks |
Matlab, WEKA | 320 cortical interneurons, mouse, rat, rabbit, cat, monkey and human |
6 polled axonal-derived morphological features |
1-D* |
| [18] | Unsupervised fuzzy sets clustering | Matlab | 200 cortical interneurons Wistar rat (6 subtypes) |
1 positional, 32 electrophysiological, 10 molecular features |
2-B |
| [19] | PCA, LDA, FSS, Gaussian distributions, k- means, Rand index |
Matlab | 466 P12-16 Wistar rat cortical interneurons (8 subtypes based on PING criteria [4]) |
38 electrophysiological | 3-D |
| [20] | Idem as in [9] | Statistica, Matlab | 123 interneurons of Wistar rat barrel cortex (see [9]) |
1 positional, 32 electrophysiological, 11 molecular features |
3-B-C |
| [21] | Affinity propagation clustering | Neurolucida, Matlab | 337 P13-25 mouse interneurons (4 subtypes: PV+ basket, PV+ chandelier, SOM+ Martinotti, SOM+ non- Martinotti) |
67 morphological, 20 electrophysiological features |
1-B |
| [22] | Bayesian networks, k-means, Bayesian multinets |
GeNIe, R | 320 cortical interneurons, mouse, rat, rabbit, cat, monkey and human |
Idem as in [17] | 1-C* |
| [23] | Harmonic co-clustering, wavelet smoothing | FARSIGHT Trace Editor |
728 mouse necortex pyramidal (6 subtypes), 502 axonal rat hippocampus (4 subtypes) |
130 morphological features | 1-A* |
| [24] | Hierarchical clustering | Matlab | 363 mouse retinal ganglion (15 subtypes) |
48,000 morphological voxels | 1-A |
| [25] | EM clustering, manual classification | KlustaKwik, SpikeDetekt, NeuroSuite |
6,732 Long-Evans rat hippocampus and EC cells (12 subtypes) |
31-127 electrophysiological channels |
1-A* |
| [26] | Hierarchical clustering, kNN | R, NBLAST | 16,129 Drosophila melanogaster neurons (1,052 subtypes) |
~ 1,070 spatial points | 1-A* |
| [39] | Bayesian network classifiers, FSS | R, WEKA | 237 cortical interneurons, mouse, rat, rabbit, cat, monkey and human |
214 morphological features | 1-A |
| [48] | Affinity propagation clustering, kNN | Matlab, NeuTu | 379 Drosophila melanogaster optic medulla (89 subtypes) |
10xN (user fixed) branch density similarities from morphological skeletons |
1-A |
| [59] | Hierarchical clustering (Ward’s linkage), ANOVA, Fisher test |
Statistica | 77 dorsolateral prefrontal cortex pyramidal cells, male long-tailed macaque (4 subtypes) |
16 electrophysiological parameters (membrane properties) |
3-C |
| [60] | Hierarchical clustering (Ward’s linkage), k- means, PCA, t-test, silhouette analysis, bootstrap test |
IGOR Pro, R | 82 mouse interneurons (4 subtypes) |
17 electrophysiological parameters (membrane properties) |
3-A-C |
| [84] | Hierarchical clustering (Ward’s linkage), Principal Factor Analysis, Spearman correlation, Kruskal-Wallis test, Mann- Whitney U test |
Stimfit, Mathematica, Matlab, SPSS |
114 P18-25 mouse dentate gyrus GABAergic interneurons (5 subtypes) |
31 morphological, 34 electrophysiological features |
1-C |
1 – Data available online
2 – Data available upon request
3 – Data not available
A – Software code available online
B – Software code available upon request
C – Commercial software without coding
D – Software or specific code not available
– Data and/or software dedicated website
3.1. Neuronal morphology and circuit connectivity
The prominence of morphological features for identifying neuron types began with the transformative pairing of Golgi staining and optical microscopy [1]. Compared to the sensitive condition-dependence of electrophysiological and biochemical states, the shape of neurons ensures considerable robustness. To this day, axonal and dendritic structures remain pivotal in the analysis of neuronal development, pathology, computation, and circuitry [36]. Early pattern recognition applications of multi-scale wavelet energy showed shape classification promise for specific neuron types such as retinal ganglion cells [37]. Initial attempts to produce general neuron hierarchies based on morphological properties [38], however, gained limited traction. Despite recent progress, knowledge of morphological diversity and connectivity patterns is still in its infancy. The limited information about neuron types, let alone their hierarchical relations, curtails the potential of taxonomical approaches.
The applications of machine learning in automatic neuronal classification was pioneered through purely data-driven analysis of GABAergic cortical interneurons [9,12,17,18,21,22,39]. Although these studies shared the common goal to identify distinct subtypes of local-circuit inhibitory neurons, they relied on different experimental measurements and computational algorithms (Table 1). Automatic classification results were validated by expert inspection [22], previous knowledge over the classes [21], biological interpretability [12], or community consensus [17]. Classic Sholl-like analysis [40] provided surprisingly effective measurements for automatic classification (with 300 µm intervals for axons [12,17,22] and 50-100 µm intervals for dendrites [12,21]. Other relevant morphological features were dendritic convex hull area [39], volume, surface area [21], and ratio of dendritic length to surface area [12].
A recent study included several transgenic lines in an analysis of 363 mouse retinal ganglion cells [24]. Dendritic arbors three-dimensionally skeletonized from voxel coordinates were differentiated by hierarchical clustering with Euclidean distance. The clustering cut-off was chosen as the lowest level that correctly grouped the strongly defined genetic types, assessing reliability by leave-one-out validation. Fifteen classes were identified, six of which await finer genetic identification.
Incorporation of over 100 morphometric parameters from L-Measure [41] in the Farsight toolkit [42] fueled an automatic classification attempt with 1230 rodent neurons from multiple sources [23]. The widely non-uniform sample yielded neuronal groups of limited consistency, but useful morphological features were nonetheless identified that allowed successfully classification in selected cases. In a similar vein, the dendritic arbors of over 5000 neurons from NeuroMorpho.Org were classified by model-based unsupervised clustering with expectation maximization after morphometric dimensionality reduction by principal component analysis [43]. Specific combinations of measures related to branching density, overall size, tortuosity, bifurcation angles, arbor flatness, and topological asymmetry captured anatomically and functionally relevant dendritic features across a broad diversity of species and brain regions. Similar approaches enabled the automatic identification of “extreme” structural phenotypes across species and brain regions [44].
At finer microcircuit scale, automatic classification has been adopted to characterize type, densities, and geometry of cerebellar axonal boutons [45] and hippocampal dendritic spines [46]. Neuronal classification could in principle be achieved directly from network connectivity [47], but the necessary mathematical framework is still under development and will require large-scale neuron-level connectomes for empirical testing. As a proxy for connectivity, branching similarity distributions were used to classify 379 optic lobe neurons labeled from a single drosophila brain [48]. The results matched an expert curated subset of 56 categories within 91% estimated accuracy.
A new paradigm for morphological classification adapted the bioinformatics idea of sequence alignment [49]. Each of 16,129 drosophila neurons (from [50]), represented as lightweight binary arrays [16], were first embedded in a common whole-brain atlas. Then massive pairwise similarity comparisons in spatial location and local geometry yielded 1,052 differentiated phenotypic groups by affinity propagation clustering [51]. This large set of clusters was then rearranged into super-classes by hierarchical clustering with Euclidean distance. Many of these super-classes matched known drosophila neuron types and circuits, whereas others constituted as-of-yet unexplored novel families [26]. Drosophila larva is also a powerful model organism to investigate dendritic growth [52], stereotypy [53], and synaptic connectivity [54] in motor circuits.
3.2. Firing patterns and plasticity
Characterizing neurons electrophysiologically can potentially reveal their functional identity. Recent automatic identification of neuron types by spiking patterns builds on earlier descriptive analyses [55-57]. Spike width alone segregated three main types of neurons in the macaque monkey frontal eye field, namely visual, movement, and visuomovement cells [8]. The analysis simply relied on the coefficient of variation and Wilcoxon rank sum test with Bonferroni corrections for multiple comparisons.
Stepping up in complexity, twelve types of retinal ganglion cells were automatically detected using five response characteristics: on and off amplitude, latency and transience, direction selectivity, and receptive field indexes [13]. A simple firing frequency threshold with in vivo spike duration allowed separation of putative GABAergic and dopaminergic neurons in the ventral tegmentum, but clustering by response to D2-type receptor agonists and antagonists further identified several distinct subtypes [15].
Visual and somatosensory neocortices are amongst the most explored regions of the mammalian brain. With 46 quantitative electrophysiological features from 466 cortical interneurons previously identified as eight distinct types based on Petilla nomenclature [4], hierarchical classification revealed six fast-spiking and adapting subtypes [19]. Spike shape and timing were identified as key physiological features, notably firing rate, fast after-hyperpolarization depth of the first spike, as well as half-height width and latency from stimulus of the second spike. Other similar studies reported analogous distinctive electrophysiological parameters, namely rise, duration, and decay of the first two spikes, plus firing rate and spike duration at 10 Hz [21,58].
Passive and active intrinsic membrane properties proved to be key physiological properties in classifying layer 2/3 pyramidal cells in the dorsolateral prefrontal cortex of macaques [59] as well as layer 2/3 interneurons in the visual cortex of mice [60]. Both works used hierarchical clustering over 16-17 electrophysiological measurements, reporting four neuronal subtypes each. Among mouse interneurons, the firing patterns in response to moderate stimuli above rheobase and the excitatory synaptic input constitute the most informative parameter. In the case of macaque pyramidal cells, the most indicative membrane properties were spiking regularity, input resistance, minimum firing current, and the current-to-frequency relationship.
Automatic classification of somatostatin-expressing interneurons, using 19 physiological features from 36 cells and in parallel 67 morphological features from 39 cells, independently identified three neuron types: Martinotti cells and two other groups with short asymmetric axons targeting layers 2/3 and bending medially [11]. Although morphological reconstructions and electrophysiological recordings were jointly available for only 16 cells, classification results were consistent across domains.
Three glutamatergic and four GABAergic archetypes were identified from 200 somatosensory cells [9]. Reanalysis of the same data with unsupervised fuzzy sets assuming a diversity continuum quantitatively confirmed the quasi-optimality of the previous separation, with the single addition of a previously unidentified subtype [18]. Key continuous features included voltage sag at hyperpolarized potentials, bursting phenotypes, input resistance, electrical excitability, and expression of GADs, calbindin, and NPY. The 123 of the same original 200 neurons that expressed reelin were reanalyzed again, corroborating the different intensity of reelin immunoreactivity among specific interneuron subtypes [20].
The above triad of studies illustrates the potential of data reuse in this field. Classification endeavors can thus benefit from the recent public release of a noteworthy dataset of 7,736 cells recorded in vivo from hippocampal regions CA1, CA3, and DG, as well as from entorhinal layers 2, 3, and 5 in rats performing multiple behavioral tasks [25]. As a preprocess stage, spiking patterns were automatically sorted by refined expectation maximization clustering. Human curation of the resulting groups with the NeuroSuite software labeled 6,732 cells. Following and extending already published criteria [61], principal cells and interneurons were distinguished based on waveforms, short-term monosynaptic interactions, and (for hippocampal neurons) bursting propensity.
3.3. Molecular markers and developmental origin
The third pillar of neuronal characterization is biochemical analysis at single-cell or whole-tissue level. Most biophysical properties of neurons are dictated by their transcriptional state, hence the emphasis on gene profiling [62-64]. For example, absolute mRNA levels of six different ion channels were measured by real-time PCR within and across six identifiable types of crab stomatogastric ganglion neurons [6]. Pearson coefficient correlation, ANOVA, and t-tests empirically demonstrated that correlated expressions of particular channel subsets determine the specific electrophysiological output.
Another seminal study used multiple DNA microarray replicas from murine forebrain [5]. Hierarchical clustering (with Euclidean distance) of the most significantly expressed genes (ANOVA, p<10−5) yielded a quantitative taxonomy of twelve major recognized populations of excitatory projection neurons and inhibitory interneurons. Despite expression heterogeneity within single regions, homologous cell types were recognized across neocortex and hippocampus, e.g. between somatosensory layers 5/6 and CA1 or between cingulate layers 2/4 and CA3. The same dataset was later reused to investigate the relationships between microRNA and mRNA patterns in each neuronal population [65] by means of weighted gene co-expression network analysis [66].
Developmental studies increasingly adopt automatic molecular classifiers to identify spatially or temporally segregated neuronal groups and precursors [67]. Among many reports of neuronal phenotyping by DNA microarray technology [7,14,68], several linked regulation of class-specific morphological development to individual proteins [69,70]. Comparative microarray mining of subplate and layer 6 of mouse cerebral cortex, confirmed by in situ hybridization and known mutant lines, revealed specific cell sub-populations identifiable by differential expression of several newly reported subplate markers [10]. Combining statistical testing, fold-change differences, and gene ontology sorting from 39,000 mRNA transcripts, feature selection yielded 383 significantly expressed genes.
Massive efforts, such as the Allen Institute’s BrainSpan, focus on brain-wide whole-genome developmental profiles [71]. Despite promising progress in drosophila models [16], large-scale fingerprinting of specific neuron types by expression patterns remains an outstanding goal in mammalian brains.
4. Challenges and opportunities
The published literature offers many more attempts and several success stories in neuronal classification. The above selection, however, is sufficient to appreciate both the scientific potential and technical struggles of this field. This section offers a critical perspective on open research issues that constitute at the same time low-hanging fruits and paradigm-shifting opportunities.
4.1. Software: seeking user-friendly general-purpose neuron classifier
Until recently, neuroscience labs typically paid little attention to their custom-developed analysis software. Countless fast-coded, scantly-commented scripts were hastily forgotten and practically lost buried in hard drives shortly after paper acceptance. Today’s growing emphasis on reproducibility, however, is strengthening the original neuroinformatics sharing plea for data and algorithms alike [72-74]. Early successes in life science software programs included ImageJ (http://imagej.nih.gov) and its expanded distribution Fiji (http://fiji.sc, originally conceived as neuroscience-specific). These popular tools’ intuitive graphic interfaces, clear documentation, platform-independence, and open-source distribution enable continuous development of new user-designed functionality. Funding agencies, philanthropic foundations, and international consortia have since fostered blooming resources such as the Allen Brain Atlas (http://brain-map.org), the Neuroimaging Informatics Tools and Resources Clearinghouse (http://nitrc.org), the International Neuroinformatics Coordinating Facility (http://incf.org), and the Neuroscience Information Framework (http://neuinfo.org).
Automatic neuron classification, however, has yet to capitalize on such momentum. While revealing an encouraging trend in code and data sharing, the brief overview of the previous section highlights an almost complete disconnect between papers (Table 1). With few notable exceptions (Farsight [23], NeuroSuite [25], and NBlast [26]), scripts are seldom re-purposed even when based on flexible analysis environments like R or Matlab. At the same time, available general-purpose machine learning software, such as Weka [75], Knime [76], or Shogun [77] are too technically demanding for widespread adoption in neuroscience (see also Supplementary Material). A promising academic market niche awaits an open-source, plugin-extensible, cross-platform package integrating multiple supervised and unsupervised machine learning techniques for automatic neuron classification by morphological, physiological, and molecular measurements alike.
4.2. The more the merrier? Data set size vs. joining realms
A common question in neuronal classification regards the minimum sample size to reliably reveal distinct types. For two neuronal populations, the estimated total number of (evenly split) neurons required to ensure a 1-α confidence in the comparison of the mean of a given measurement is
where are a priori estimates of measurement variance in the two populations, Zα is the number of standard deviations around the mean encompassing 1-α of the data, and d is the maximum allowed error (similar formulations apply to proportion calculations [78] and statistical hypothesis testing [79]). For instance, the total neuritic length per neuron for drosophila glutamatergic and GABAergic cells in NeuroMorpho.Org 6.0 is 2735±2546 and 2032±1579 µm (mean ± standard deviation), respectively. Assuming the difference between the averages (703 µm) as the limit for the error and a value of 1.96 for (corresponding to α=0.05 for normal distributions), the necessary sample size would be 70 (35 neurons from each group). The outcome of this computation is often prohibitively incompatible with experimental constraints [80]. In machine learning practice, a sample size of 30 is often the lower limit for estimating each classification parameter. This lower bound assures a minimum robustness in the estimation given a Gaussian assumption through the central limit theorem [81]. It is essential, however, to verify the assumed normal distribution of the data, e.g. with the Saphiro-Wilk test [82].
Yet determining the optimal sample for neuronal classification is not just a matter of size. Given the strong (if still largely unknown) inter-dependence of neuronal morphology, physiology, and biochemistry (e.g. [83,84]), characterizing a smaller sample of neurons across different domains may be more conducive to discovery than separately collecting the same measurements from each of three larger neuron samples. Nevertheless, 21 of 27 classification studies summarized in Table 1 used single experimental domains, and only one dataset combined all three major dimensions [9,18]. The few successes in automatically inducing neuron prototypes in one domain [12,19] have yet to be extended to global classification algorithms combining structural, electrical, and molecular information [85].
Techniques such as feature creation [86] and feature subset selection [87,88] could alleviate the technical obstacles of cross-modal data acquisition or the substantial labor of massive human curation to integrate experimental evidence (e.g. Hippocampome.Org [89]). Although multimodal classification can in principle be organized in hierarchical stages, probabilistic clustering and biclustering are better-suited alternatives to overcome the limitation of classic k-means clustering. Advanced probabilistic graphical models [90] such as Bayesian or Markov networks and factor or chain graphs are designed to combine different domains allowing uncertainty, partial knowledge, and expert intervention. These approaches can account for cross-interactions of distinct neuronal features, such as molecular expression, dendritic morphology, and neuronal excitability [91], while enabling exploration of new inter-relationships from experimental data [92] and digital atlases [93].
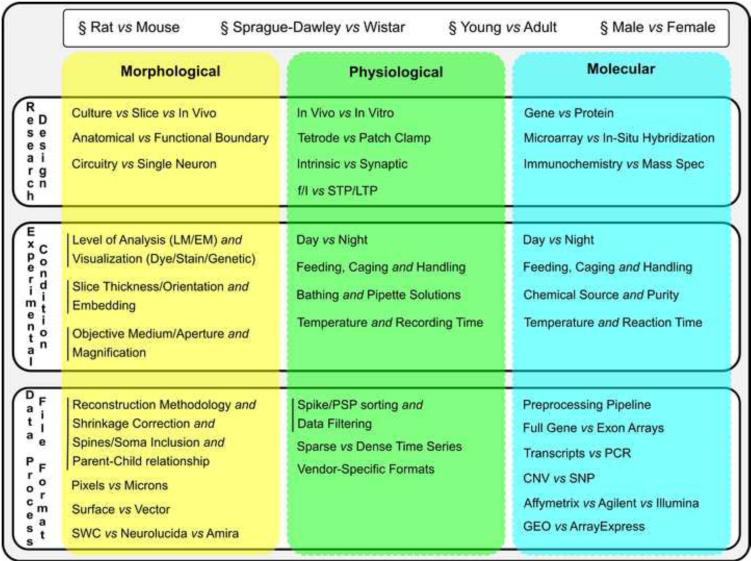
4.3. perimental design: diversity, annotation, and integration
Neuronal characterization involves a range of animal models, research designs, experimental conditions, and data formats (Box 2). Such variety allows addressing a correspondingly broad set of open questions, but it also introduces additional issues, exemplified by the non-trivial comparison of findings from drosophila to humans or even from cat and monkey [94]. In practice, successful integration of data from multiple labs for neuronal classification, although possible [14], remains extremely rare even within single research domains. One of the main difficulties consists of explicitly accounting for all potential experimental differences.
Box 2.
Research heterogeneity in neuronal classification: general metadata, research design, experimental condition, and data format across morphological, physiological, and molecular dimensions.
Certain details, such as those specifying the animal subject, transcend experimental domains; others are specific to morphological, physiological or molecular approaches (e.g. those related to labeling, recording, and sequencing, respectively). Some conditions are mutually exclusive (e.g. in vivo versus in vitro), whereas others provide complementary information (slice thickness and orientation). Thorough knowledge of all these metadata is as essential as the data themselves, not only to integrate datasets in subsequent research, but also to maximize reproducibility [95].
The diversity of neuroscience research makes it unfeasible to standardize the experimental conditions for neuronal classification. Standardizing the reporting of experimental conditions is far more viable and just as effective. Despite ongoing metadata standardization efforts, community consensus is still lacking [96]. Open information exchange formats such as the MIAME protocol [97] have gained traction in genomics [98]. Adoption of common data standards enables validation approaches to attenuate the impact of data variability across labs [99].
More generally, all known metadata may be encoded as confounding variables in statistical models. These variables typically correlate with both the dependent and independent variables (the neuron type and measurements, respectively), but those interactions are often difficult to recognize. Undetected conditional dependences may lead to spurious classification results. In contrast, confounding variables with identified bivariate dependence are (independently predictive) covariates, and their inclusion improves the classification outcome.
Neuronal classification could benefit tremendously from the growing machine learning toolset for metadata integration, including generalized linear models [100] such as logistic regression. Matrix eQTL [101] provides a user-friendly R package to test some of these linear models. Multitask lasso models [102] and sparse regularized graphical models [103] also allow analysis of dependencies between metadata variables as well as of their interactions with the predictive variables. Other relevant techniques, including support vector machines with covariate support [104] and random forests [105], are well-established in machine learning but largely unknown in neuroscience. Pervasive cross-pollination between these disciplines could benefit neuronal classification tremendously. A more extensive list of classification approaches, algorithms, and references is provided as Supplementary Material.
5. Future prospects: expected impact of big data
The number of publications on automatic neuron classification from the past two years exceeded those in the previous decade. This progression might accelerate even further through integration of domains, development of neuroscientist-friendly software, collection of exhaustive metadata, inclusion of corrective covariates, and overall standardization of analyses, formats, and reporting [106]. Such exciting new endeavors require cross-disciplinary collaboration within and between research teams and institutions. Equally noteworthy is the increasing dataset size from dozen neurons [5,7,8] to hundreds [13,19], to thousands [25,26] in just eight years. This “big data” trend, also just beginning and likely to continue, is paralleled by paradigmatic changes in the required infrastructure to process data and in the complexity of models to describe data.
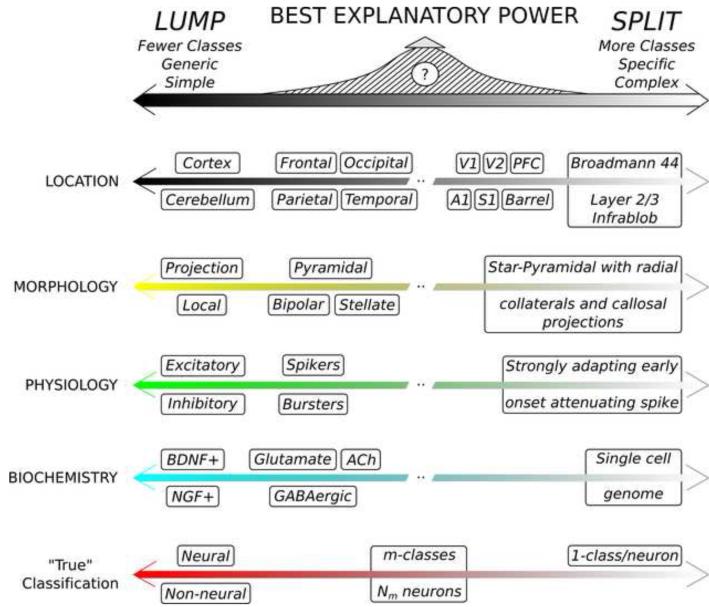
Neuronal classification evolved from its descriptive origin to modern quantitative analysis, but the field remains fraught with the classic dilemma between lumpers and splitters [107]. On the one hand, all neurons share common properties that distinguish them from non-neural cells, and can thus be aptly considered a single class. On the other, no two neurons are ever identical; therefore each neuron could be viewed as its own class. Although these extremes shed no light on neuronal identity, they illustrate the complementary advantages and disadvantages of simpler but generic vs. specific but complex classifications [108]. For example, somatic location can be defined at coarser anatomical level (cortical, retinal, spinal, cerebellar, etc.) or with finer-grained distinction of sub-regions and layers. Similar choices apply to morphological, physiological, and biochemical characterization (Figure 3). Even approaches that assume structured continuous distributions of neuronal characteristics need to set the granularity which eventually defines neuronal archetypes [18]. Are there a “right number” of neuron types?
Figure 3.
Optimal classification balance of neuronal lumping and splitting. Neuron types can be defined at higher or lower resolution in each data domain. The best explanatory power maximizes the trade-off between description complexity and captured generalization.
This and several other outstanding questions that emerged in this review are summarized in Box 3. Coevolving high-throughput data production and large-scale analysis tools may soon begin to offer objective answers. Specifically, once the available data are sufficiently large to be statistically representative, theory can define the most powerful explanatory model. For instance, the Minimum Description Length (MDL) criterion [109] selects models with least data requirements. A practical example is variable binning from numerical to categorical by entropy discretization [110]. Similarly, the Bayesian Information Criterion (BIC) [111] optimizes the tradeoff between model complexity and accuracy by penalizing the inclusion of excessive explanatory variables. We find the purely data-driven determination of the number of neuron classes by BIC particularly appealing, because it jointly minimizes the costs of lumping (regressing different neurons to fewer prototypes) and splitting (accounting for different prototypes), both measured in information units.
Box 3. Outstanding questions in automatic neuronal classification.
How many types of neurons are there? Quantitative criteria, such as the Bayesian Information Criterion, may provide an objective answer by identifying the granularity with the best explanatory power for the available data.
What is the nature of the neuronal morphology-physiology-biochemistry relationship? Interactions reported to date range from quasi-independence to tight correlations and include both direct influences and homeostatic compensation, often resulting in complex phenotypic combinations.
Do neuron types constitute sharply segregated categories or a continuum? Although evidence abounds for clearly identifiable neuronal classes, more data are needed to ascertain the overlap extent of quantitative measurements between categories. The answer is likely to vary by species and brain region.
Can neuron types be described by a consistent taxonomical hierarchy? A hierarchical organization implies ranking the classification relevance of available measurements based on underlying functions and mechanisms (connectivity, signal transmission, information processing, plasticity etc.).
How do developmental programs and functional adaptation interact to produce observed phenotypes? Neuron types are the combined result of ontogeny and experience. What are the key genetic and computational variables guiding the emergence of distinct neuronal classes?
Existing neuroscience knowledge is still far from adequate to satisfactorily attempt closure on neuronal classification. Ideally the morphological, physiological, and molecular observable would be explained in terms of both developmental origins [112] and behavioral relevance [113], ultimately connecting the two [114], but more data are undoubtedly required. Continuous technological advances and the prospects of industrial-scale societal efforts in data collection may soon close this gap. This expected progress will initially increase the complexity of neuronal classification by revealing a much larger number of types than currently known. Upon accumulating a critical amount of evidence, however, automatic classification will begin to simplify the explanatory model by defining increasingly more effective descriptors. The resulting clearer view of neuronal organization should reveal major findings on computation and cognition.
Supplementary Material
Highlights.
The automation of neuron type classification is advancing ever more rapidly.
Accelerating data collection makes machine learning necessary for neuronal classification.
We review analysis approaches, algorithm classes, and available resources.
Opportunities include software development, data standardization and integration.
Acknowledgements
This work was supported in part by grants R01 NS39600 and NS86082 from the National Institutes of Health (NINDS), ONR MURI 14101-0198 from the Office of Naval Research, and Keck NAKFI to GAA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Cajal SR. El nuevo concepto de la histología de los centros nerviosos. Rev. Cienc. Méd. 1892;18:457–476. [Google Scholar]
- [2].Insel TR, et al. The NIH BRAIN Initiative. Science. 2013;340:687–688. doi: 10.1126/science.1239276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Markram H. The Human Brain Project. Sci. Am. 2012;306:50–55. doi: 10.1038/scientificamerican0612-50. [DOI] [PubMed] [Google Scholar]
- [4].Ascoli GA, et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat. Rev. Neurosci. 2008;9:557–568. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sugino K, et al. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat. Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- [6].Schulz DJ, et al. Quantitative expression profiling of identified neurons reveals cell-specific constraints on highly variable levels of gene expression. Proc. Natl. Acad. Sci. USA. 2007;104:13187–13191. doi: 10.1073/pnas.0705827104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cahoy JD, et al. A Transcriptome Database for Astrocytes, Neurons, and Oligodendrocytes: A New Resource for Understanding Brain Development and Function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cohen JY, et al. Biophysical Support for Functionally Distinct Cell Types in the Frontal Eye Field. J. Neurophysiol. 2009;101:912–916. doi: 10.1152/jn.90272.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Karagiannis A, et al. Classification of NPY-Expressing Neocortical Interneurons. J. Neurosci. 2009;29:3642–3659. doi: 10.1523/JNEUROSCI.0058-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hoerder-Suabedissen A, et al. Novel markers reveal subpopulations of subplate neurons in the murine cerebral cortex. Cereb. Cortex. 2009;19:1738–1750. doi: 10.1093/cercor/bhn195. [DOI] [PubMed] [Google Scholar]
- [11].McGarry LM, et al. Quantitative classification of somatostatin-positive neocortical interneurons identifies three interneuron subtypes. Front. Neural Circuits. 2010 doi: 10.3389/fncir.2010.00012. DOI: 10.3389/fncir.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Guerra L, et al. Comparison between supervised and unsupervised classifications of neuronal cell types: A case study. Dev. Neurobiol. 2011;71:71–82. doi: 10.1002/dneu.20809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Farrow K, Masland RH. Physiological clustering of visual channels in the mouse retina. J. Neurophysiol. 2011;105:1516–1530. doi: 10.1152/jn.00331.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Okaty BW, et al. A quantitative comparison of cell-type-specific microarray gene expression profiling methods in the mouse brain. PLoS One. 2011 doi: 10.1371/journal.pone.0016493. DOI: 10.1371/journal.pone.0016493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li W, et al. Quantitative unit classification of ventral tegmental area neurons in vivo. J. Neurophysiol. 2012;107:2808–2820. doi: 10.1152/jn.00575.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Masse NY, et al. A mutual information approach to automate identification of neuronal clusters in Drosophila brain images. Front. Neuroinform. 2012 doi: 10.3389/fninf.2012.00021. DOI: 10.3389/fninf.2012.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].DeFelipe J, et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat. Rev. Neurosci. 2013;14:202–216. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Battaglia D, et al. Beyond the frontiers of neuronal types. Front. Neural Circuits. 2013 doi: 10.3389/fncir.2013.00013. DOI: 10.3389/fncir.2013.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Druckmann S, et al. A hierarchical structure of cortical interneuron electrical diversity revealed by automated statistical analysis. Cereb. Cortex. 2013;23:2994–3006. doi: 10.1093/cercor/bhs290. [DOI] [PubMed] [Google Scholar]
- [20].Pohlkamp T, et al. Characterization and Distribution of Reelin-Positive Interneuron Subtypes in the Rat Barrel Cortex. Cereb. Cortex. 2013;24:3046–3058. doi: 10.1093/cercor/bht161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Santana R, et al. Classification of neocortical interneurons using affinity propagation. Front. Neural Circuits. 2013 doi: 10.3389/fncir.2013.00185. DOI: 10.3389/fncir.2013.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].López-Cruz PL, et al. Bayesian network modeling of the consensus between experts: An application to neuron classification. Int. J. Approx. Reason. 2014;55:3–22. [Google Scholar]
- [23].Lu Y, et al. Quantitative arbor analytics: Unsupervised harmonic co-clustering of populations of brain cell arbors based on L-Measure. Neuroinformatics. 2015;13:47–63. doi: 10.1007/s12021-014-9237-2. [DOI] [PubMed] [Google Scholar]
- [24].Sümbül U, et al. A genetic and computational approach to structurally classify neuronal types. Nat. Commun. 2014 doi: 10.1038/ncomms4512. DOI: 10.1038/ncomms4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mizuseki K, et al. Neurosharing: large-scale data sets (spike, LFP) recorded from the hippocampal-entorhinal system in behaving rats. F1000Res. 2014 doi: 10.12688/f1000research.3895.1. DOI: 10.12688/f1000research.3895.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Costa M, et al. NBLAST: Rapid, sensitive comparison of neuronal structure and construction of neuron family databases. BioRxiv. 2014 doi: 10.1016/j.neuron.2016.06.012. DOI: http://dx.doi.org/10.1101/006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Larrañaga P, et al. Machine learning in bioinformatics. Brief. Bioinform. 2006;7:86–112. doi: 10.1093/bib/bbk007. [DOI] [PubMed] [Google Scholar]
- [28].Akil H, et al. Challenges and opportunities in mining neuroscience data. Science. 2011;331:708–712. doi: 10.1126/science.1199305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Parekh R, Ascoli GA. Neuronal Morphology Goes Digital: A Research Hub for Cellular and System Neuroscience. Neuron. 2013;77:1017–1038. doi: 10.1016/j.neuron.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Stone J. Parallel Processing in the Visual System: The Classification of Retinal Ganglion Cells and its Impact on the Neurobiology of Vision. Springer. 1983 [Google Scholar]
- [31].Bota M, Swanson LW. The neuron classification problem. Brain. Res. Rev. 2007;56:79–88. doi: 10.1016/j.brainresrev.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sharpee TO. Toward functional classification of neuronal types. Neuron. 2014;83:1329–1334. doi: 10.1016/j.neuron.2014.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zaitsev AV. Classification and Function of GABAergic Interneurons of the Mammalian Cerebral Cortex. Biochemistry (Moscow) Supplement Series A: Membrane and Cell Biology. 2013;7:245–259. [Google Scholar]
- [34].Mensi S, et al. Parameter extraction and classification of three cortical neuron types reveals two distinct adaptation mechanisms. J. Neurophysiol. 2012;107:1756–1775. doi: 10.1152/jn.00408.2011. [DOI] [PubMed] [Google Scholar]
- [35].Wolpert D. The lack of a priori distinctions between learning algorithms. Neural Comput. 1996;8:1341–1390. [Google Scholar]
- [36].Parekh R, Ascoli GA. Quantitative Investigations of Axonal and Dendritic Arbors: Development, Structure, Function, and Pathology. Neuroscientist. 2014 doi: 10.1177/1073858414540216. DOI: 10.1177/1073858414540216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cesar Junior RM, Costa LF. Neural cell classification by wavelets and multiscale curvature. Biol. Cybern. 1998;79:347–360. doi: 10.1007/s004220050484. [DOI] [PubMed] [Google Scholar]
- [38].Bota M, Swanson LW. BAMS neuroanatomical ontology: Design and implementation. Front. Neuroinform. 2008 doi: 10.3389/neuro.11.002.2008. DOI: 10.3389/neuro.11.002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mihaljevic B, et al. Bayesian network classifiers for categorizing cortical GABAergic interneurons. Neuroinformatics. 2014 doi: 10.1007/s12021-014-9254-1. DOI: 10.1007/s12021-014-9254-1. [DOI] [PubMed] [Google Scholar]
- [40].Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- [41].Scorcioni R, et al. L-Measure: a web-accessible tool for the analysis, comparison and search of digital reconstructions of neuronal morphologies. Nat. Protoc. 2008;3:866–876. doi: 10.1038/nprot.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Luisi J, et al. The FARSIGHT Trace Editor: An Open Source Tool for 3D Inspection and Efficient Pattern Analysis Aided Editing of Automated Neuronal Reconstructions. Neuroinformatics. 2011;9:305–315. doi: 10.1007/s12021-011-9115-0. [DOI] [PubMed] [Google Scholar]
- [43].Polavaram S, et al. Statistical analysis and data mining of digital reconstructions of dendritic morphologies. Front. Neuroanat. 2014 doi: 10.3389/fnana.2014.00138. DOI: 10.3389/fnana.2014.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zawadzki K, et al. Morphological homogeneity of neurons: searching for outlier neuronal cells. Neuroinformatics. 2012;10:379–389. doi: 10.1007/s12021-012-9150-5. [DOI] [PubMed] [Google Scholar]
- [45].Brown K, et al. Digital morphometry of rat cerebellar climbing fibers reveals distinct branch and bouton types. J. Neurosci. 2012;32:14670–14684. doi: 10.1523/JNEUROSCI.2018-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sündermann F, et al. High-Resolution Imaging and Evaluation of Spines in Organotypic Hippocampal Slice Cultures. In: Skaper SD, editor. In Neurotrophic Factors. Vol. 846. Humana Press; 2012. pp. 277–293. [DOI] [PubMed] [Google Scholar]
- [47].Marchette DJ, et al. Investigation of a random graph model for neuronal connectivity. In Joint Mtg. Am. Math. Soc. 2012 1077-62-424. [Google Scholar]
- [48].Zhao T, Plaza SM. Automatic Neuron Type Identification by Neurite Localization in the Drosophila Medulla. 2014 arXiv:1409.1892. [Google Scholar]
- [49].McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chiang AS, et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr. Biol. 2011;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- [51].Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- [52].Kim MD, et al. Patterning and organization of motor neuron dendrites in the Drosophila larva. Dev. Biol. 2009;336:213–221. doi: 10.1016/j.ydbio.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tsechpenakis G, et al. Motor neuron morphology estimation for its classification in the Drosophila brain. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society; 2011. pp. 7755–7758. IEEE. [DOI] [PubMed] [Google Scholar]
- [54].Chang X, et al. Part-based motor neuron recognition in the Drosophila ventral nerve cord. Neuroimage. 2013;90:33–42. doi: 10.1016/j.neuroimage.2013.12.023. [DOI] [PubMed] [Google Scholar]
- [55].Bornstein JC, et al. Electrophysiological characterization of myenteric neurons: how do classification schemes relate? J. Auton. Nerv. Syst. 1994;48:1–15. doi: 10.1016/0165-1838(94)90155-4. [DOI] [PubMed] [Google Scholar]
- [56].Oshio K, et al. Neuron classification based on temporal firing patterns by the dynamical analysis with changing time resolution (DCT) method. Biol. Cybern. 2003;88:438–449. doi: 10.1007/s00422-003-0396-8. [DOI] [PubMed] [Google Scholar]
- [57].Salganicoff M, et al. Unsupervised waveform classification for multi-neuron recordings: a real-time, software-based system. I. Algorithms and implementation. J. Neurosci. Methods. 1988;25:181–187. doi: 10.1016/0165-0270(88)90132-x. [DOI] [PubMed] [Google Scholar]
- [58].Katai S, et al. Classification of extracellularly recorded neurons by their discharge patterns and their correlates with intracellularly identified neuronal types in the frontal cortex of behaving monkeys. Eur. J. Neurosci. 2010;31:1322–1338. doi: 10.1111/j.1460-9568.2010.07150.x. [DOI] [PubMed] [Google Scholar]
- [59].Zaitsev AV, et al. Electrophysiological classes of layer 2/3 pyramidal cells in monkey prefrontal cortex. J. Neurophysiol. 2012;108:595–609. doi: 10.1152/jn.00859.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Helm J, et al. Subgroups of parvalbumin-expressing interneurons in layers 2/3 of the visual cortex. J. Neurophysiol. 2013;109:1600–1613. doi: 10.1152/jn.00782.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mizuseki K, et al. Theta Oscillations Provide Temporal Windows for Local Circuit Computation in the Entorhinal Hippocampal Loop. Neuron. 2009;64:267–280. doi: 10.1016/j.neuron.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Nelson SB, et al. The problem of neuronal cell types: a physiological genomics approach. Trends Neurosci. 2006;29:339–345. doi: 10.1016/j.tins.2006.05.004. [DOI] [PubMed] [Google Scholar]
- [63].Bernard A, et al. Shifting the paradigm: new approaches for characterizing and classifying neurons. Curr. Opin. Neurobiol. 2009;19:530–536. doi: 10.1016/j.conb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- [64].Ng L, et al. An anatomic gene expression atlas of the adult mouse brain. Nat. Neurosci. 2009;12:356–362. doi: 10.1038/nn.2281. [DOI] [PubMed] [Google Scholar]
- [65].Tsang J, et al. MicroRNA-Mediated Feedback and Feedforward Loops Are Recurrent Network Motifs in Mammals. Mol. Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Winden KD, et al. The organization of the transcriptional network in specific neuronal classes. Mol. Syst. Biol. 2009 doi: 10.1038/msb.2009.46. DOI: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tricoire L, et al. A blueprint for the spatiotemporal origins of mouse hippocampal interneuron diversity. J. Neurosci. 2011;31:10948–10970. doi: 10.1523/JNEUROSCI.0323-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Okaty BW, et al. Cell type-specific transcriptomics in the brain. J. Neurosci. 2011;31:6939–6943. doi: 10.1523/JNEUROSCI.0626-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hawrylycz MJ, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sorensen SA, et al. Correlated Gene Expression and Target Specificity Demonstrate Excitatory Projection Neuron Diversity. Cereb. Cortex. 2013 doi: 10.1093/cercor/bht243. DOI: 10.1093/cercor/bht243. [DOI] [PubMed] [Google Scholar]
- [71].Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Ascoli GA. Mobilizing the base of neuroscience data: the case of neuronal morphologies. Nat. Rev. Neurosci. 2006;7:318–324. doi: 10.1038/nrn1885. [DOI] [PubMed] [Google Scholar]
- [73].Teeters JL, et al. Data Sharing for Computational Neuroscience. Neuroinformatics. 2008;6:47–55. doi: 10.1007/s12021-008-9009-y. [DOI] [PubMed] [Google Scholar]
- [74].Sharpe D. Why the resistance to statistical innovations? Bridging the communication gap. Psychol Methods. 2013;18:572–582. doi: 10.1037/a0034177. [DOI] [PubMed] [Google Scholar]
- [75].Hall M, et al. The WEKA Data Mining Software: An Update. SIGKDD Explorations. 2009;11:10–18. [Google Scholar]
- [76].Berthold MR, et al. KNIME: The Konstanz Information Miner. In: Preisach C, et al., editors. Data Analysis, Machine Learning and Applications. Springer; Berlin Heidelberg: 2007. pp. 319–326. [Google Scholar]
- [77].Sonnenburg S, et al. The SHOGUN Machine Learning Toolbox. J. Mach. Learn. Res. 2010;11:1799–1802. [Google Scholar]
- [78].Machin D, et al. Sample Size Tables for Clinical Studies. BMJ Books; 2008. [Google Scholar]
- [79].Cohen J. Statistical Power Analysis for the Behavioral Sciences. Routledge; 1988. [Google Scholar]
- [80].Button KS, et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013;14:365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- [81].Rice JA. Mathematical Statistics and Data Analysis. Duxbury Advanced Series; 2007. [Google Scholar]
- [82].Razali N, Wah YB. Power comparisons of Shapiro–Wilk, Kolmogorov–Smirnov, Lilliefors and Anderson–Darling tests. Journal of Statistical Modeling and Analytics. 2011;2:21–33. [Google Scholar]
- [83].Cossart R, et al. Interneurons targeting similar layers receive synaptic inputs with similar kinetics. Hippocampus. 2006;16:408–420. doi: 10.1002/hipo.20169. [DOI] [PubMed] [Google Scholar]
- [84].Hosp JA, et al. Morpho-physiological criteria divide dentate gyrus interneurons into classes. Hippocampus. 2014;24:189–203. doi: 10.1002/hipo.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Parra P, et al. How Many Subtypes of Inhibitory Cells in the Hippocampus? Neuron. 1998;20:983–993. doi: 10.1016/s0896-6273(00)80479-1. [DOI] [PubMed] [Google Scholar]
- [86].Guyon I, et al., editors. Feature Extraction: Foundations and Applications. Springer; 2006. [Google Scholar]
- [87].Liu H, Motoda H, editors. Computational Methods of Feature Selection. Chapman and Hall/CRC; 2007. [Google Scholar]
- [88].Saeys Y, et al. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- [89].Ascoli GA. The coming of age of the Hippocampome. Neuroinformatics. 2010;8:1–3. doi: 10.1007/s12021-010-9063-0. [DOI] [PubMed] [Google Scholar]
- [90].Koller D. Probabilistic Graphical Models: Principles and Techniques. The MIT Press; 2009. [Google Scholar]
- [91].Krichmar JL, et al. Effects of β-Catenin on Dendritic Morphology and Simulated Firing Patterns in Cultured Hippocampal Neurons. Biol. Bull. 2006;211:31–43. doi: 10.2307/4134575. [DOI] [PubMed] [Google Scholar]
- [92].Hashimoto Y, et al. Uncovering genes required for neuronal morphology by morphology-based gene trap screening with a revertible retrovirus vector. FASEB J. 2012 doi: 10.1096/fj.12-207530. DOI: 10.1096/fj.12-207530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Shcherbatyy V, et al. A Digital Atlas of Ion Channel Expression Patterns in the Two-Week-Old Rat Brain. Neuroinformatics. 2014 doi: 10.1007/s12021-014-9247-0. DOI: 10.1007/s12021-014-9247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Shapley R, Hugh Perry V. Cat and monkey retinal ganglion cells and their visual functional roles. Trends Neurosci. 1986;9:229–235. [Google Scholar]
- [95].Collins FS, Tabak LA. NIH plans to enhance reproducibility. Nature. 2014;505:612–613. doi: 10.1038/505612a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Hamilton DJ, et al. An ontological approach to describing neurons and their relationships. Front. Neuroinform. 2012 doi: 10.3389/fninf.2012.00015. DOI: 10.3389/fninf.2012.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Brazma A, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- [98].The 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Taylor JMG, et al. Validation of Biomarker-Based Risk Prediction Models. Clin. Cancer Res. 2008;14:5977–5983. doi: 10.1158/1078-0432.CCR-07-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Vidaurre D, et al. A Survey of L1 Regression. Int. Stat. Rev. 2013;81:361–387. [Google Scholar]
- [101].Shabalin AA. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Chen X, et al. A two-graph guided multi-task Lasso approach for eQTL mapping. In: Lawrence ND, Girolami M, editors. International Conference on Artificial Intelligence and Statistics; 2012. pp. 208–217. JMLR:W&CP 22. [Google Scholar]
- [103].Zhang X, et al. Learning Transcriptional Regulatory Relationships Using Sparse Graphical Models. PLoS ONE. 2012 doi: 10.1371/journal.pone.0035762. DOI: 10.1371/journal.pone.0035762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Li L, et al. ccSVM: correcting Support Vector Machines for confounding factors in biological data classification. Bioinformatics. 2011;27:i342–i348. doi: 10.1093/bioinformatics/btr204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Siroky DS. Navigating Random Forests and related advances in algorithmic modeling. Statist. Surv. 2009;3:147–163. [Google Scholar]
- [106].Sejnowski TJ, et al. Putting big data to good use in neuroscience. Nat. Neurosci. 2014;17:1440–1441. doi: 10.1038/nn.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Simpson GG. The Principles of Classification and a Classification of Mammals. Bull. Am. Mus. Nat. Hist. 1945;85:22–24. [Google Scholar]
- [108].Seung HS, Sümbül U. Neuronal cell types and connectivity: lessons from the retina. Neuron. 2014;83:1262–1272. doi: 10.1016/j.neuron.2014.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Rissanen J. Modeling by shortest data description. Automatica. 1978;14:465–658. [Google Scholar]
- [110].Fayyad U, Irani K. Multi-interval discretization of continuous-valued attributes for classification learning. In: Heckerman D, Mamdani EH, editors. Proceedings of the International Joint Conference on Uncertainty in Artificial Intelligence; 1993. pp. 1022–1027. Morgan Kaufmann. [Google Scholar]
- [111].Schwarz GE. Estimating the dimension of a model. Ann. Stat. 1978;6:461–464. [Google Scholar]
- [112].Jessel TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- [113].Vogelstein JT, et al. Discovery of brainwide neural-behavioral maps via multiscale unsupervised structure learning. Science. 2014;344:386–392. doi: 10.1126/science.1250298. [DOI] [PubMed] [Google Scholar]
- [114].Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.