Abstract
The Mre11-Rad50-Nbs1 (MRN) complex is a dynamic macromolecular machine that acts in the first steps of DNA double strand break repair, and each of its components has intrinsic dynamics and flexibility properties that are directly linked with their functions. As a result, deciphering the functional structural biology of the MRN complex is driving novel and integrated technologies to define the dynamic structural biology of protein machinery interacting with DNA. Rad50 promotes dramatic long-range allostery through its coiled-coil and zinc-hook domains. Its ATPase activity drives dynamic transitions between monomeric and dimeric forms that can be modulated with mutants modifying the ATPase rate to control end joining versus resection activities. The biological functions of Mre11’s dual endo- and exonuclease activities in repair pathway choice were enigmatic until recently, when they were unveiled by the development of specific nuclease inhibitors. Mre11 dimer flexibility, which may be regulated in cells to control MRN function, suggests new inhibitor design strategies for cancer intervention. Nbs1 has FHA and BRCT domains to bind multiple interaction partners that further regulate MRN. One of them, CtIP, modulates the Mre11 excision activity for homologous recombination repair. Overall, these combined properties suggest novel therapeutic strategies. Furthermore, they collectively help to explain how MRN regulates DNA repair pathway choice with implications for improving the design and analysis of cancer clinical trials that employ DNA damaging agents or target the DNA damage response.
Keywords: Mre11-Rad50-Nbs1, CtIP, Allostery, Double strand break repair, Dynamics, Conformational change
1. Introduction
DNA double strand break repair (DSBR) is a fascinating process that involves a plethora of proteins in a complex choreography of dynamic events in which the Mre11-Rad50-Nbs1 complex (MRN) plays critical roles. Double strand breaks (DSB) are created by exposure to ionizing radiation or chemicals, and by endogenous cellular events, including DNA replication and V(D)J recombination. For more than 40 years, scientists have strived to determine how DSBR works. By combining results piece by piece and by developing new methods, they are revealing the integrated mechanisms of this crucial phenomenon. We know there are many other interacting components for homologous recombination such as those that promote homologous DNA pairing in meiotic recombination (Zhao et al., 2014); however here we focus on the MRN complex, which acts in the first stages of DSBR. MRN has emerged as one of the main protein complexes at multiple stages in the process, impacting pathways for homologous recombination repair (HRR) and both classical and alternative non-homologous end-joining (NHEJ) (Hammel et al., 2011; Hopfner et al., 2002b; Shin et al., 2004; G. J. Williams et al., 2014; 2010; Zha et al., 2009). It detects DSBs, tethers the ends of the broken chromosomes, activates DNA damage response pathways and nucleolytically processes the DNA ends. A fascinating characteristic of MRN is that these diverse functions are possible due to its flexibility and dynamic properties that are finally being unveiled.
In 2002 an article in Nature revealed multiple conformations of the MR complex by electron microscopy (EM) (Hopfner et al., 2002a) and in 2005 a second Nature article presented a video captured by atomic force microscopy (AFM) of the MRN complex interacting with DNA (Moreno-Herrero et al., 2005; R. S. Williams and Tainer, 2005), allowing the implications of the dynamics and flexibility to be examined in the context of Mre11 and Rad50 crystal structures. These papers opened the door for the scientific community to visualize MRN in its dynamic splendor. In the solution AFM video, the long coiled-coils of Rad50 are moving between two different states upon DNA binding. In their DNA-bound form, the two 500 Å-long coiled-coils of a MRN dimer seem to be straight and rigid. When the complex leaves the DNA, the coiled-coils adopt a relaxed, bent position, similar to what is seen by EM (de Jager et al., 2001). These observations revealed that the complex is highly dynamic over a large spatial area.
In the last 10 years, tremendous efforts have been done to better understand the dynamic nature of MRN, its structure and its cellular functions. Indeed, MRN has diverse and critical functions: DSB detection, DNA end tethering, ATM activation, DNA end processing and replication fork protection or resection (reviewed in (Paull and Deshpande, 2014; Schiller et al., 2014; Symington, 2014; G. J. Williams et al., 2010)). The emerging picture is that MRN uses flexible and dynamic structures to integrate its ATPase and nuclease enzymatic activities with protein-protein and protein-DNA interactions to control biological outcomes at a DSB. To study dynamic Mre11-Rad50 complexes, we and other researchers in the field have used the power of optimal model systems including hyperthermophiles (reviewed in Shin et al., 2014). Furthermore, multiple and combined technologies have been employed, which prompted the design of our synchrotron beamline to combine crystallography and X-ray scattering (Classen et al., 2013). Indeed, the MRN complex has provided problem-driven development of multiple biophysical metrics and methods, which is noted along with the biological results.
In this review we cover the most recent conceptual advances about MRN in the context of foundational results. We first explore the current models on its roles in different pathways. Then we examine how the flexibility and dynamics of MRN’s components and partners are linked with their function. Rad50 is capable of long-range allostery and its activities are controlled by its ATPase functions, which impacts the nucleolytic activities of Mre11. Mre11 sculpts dsDNA ends and ssDNA differently and inhibitors were therefore able to separately block either endonuclease or exonuclease activities by blocking Mre11-mediated DNA conformational changes. Also, Nbs1 and CtIP are responsible for important dynamic protein-protein interactions with the Nbs1 C-terminus helping to connect MRN to ATM activation and signaling. Finally, there is growing evidence that MRN can be targeted for cancer therapeutics and knowing its dynamics and molecular mechanism is thus crucial for developing new therapeutic strategies. Overall, we aim to help the reader appreciate the dynamic and structural aspects of MRN functions. This knowledge includes growing evidence that structural dynamics can regulate repair pathway selectivity and control biological outcomes. This appreciation of the MRN complex has general biological implications, especially for the Rad50 ABC ATPase superfamily (Hopfner and Tainer, 2003), for structure-specific nucleases (Tsutakawa et al., 2014), and for phosphorylated protein binding by FHA and BRCT domains (Reinhardt and Yaffe, 2013).
2. Functional architecture of the MRN complex
The structure of the whole MRN complex or of its individual components has been examined by multiple techniques: X-ray crystallography, small-angle X-ray scattering (SAXS), analytical ultracentrifugation, inductively coupled plasma mass spectrometry (ICP-MS), dynamic light scattering (DLS), atomic force microscopy (AFM) and electron microscopy (EM). Initially, proteins from archaea and bacteria were used, since they are more stable in their purified form, but recently eukaryotic Mre11 and Nbs1 have also been studied, strengthening the previous structural findings. Before going into further details with the MRN complex, it is useful to consider a structural overview of each of its components (Fig. 1).
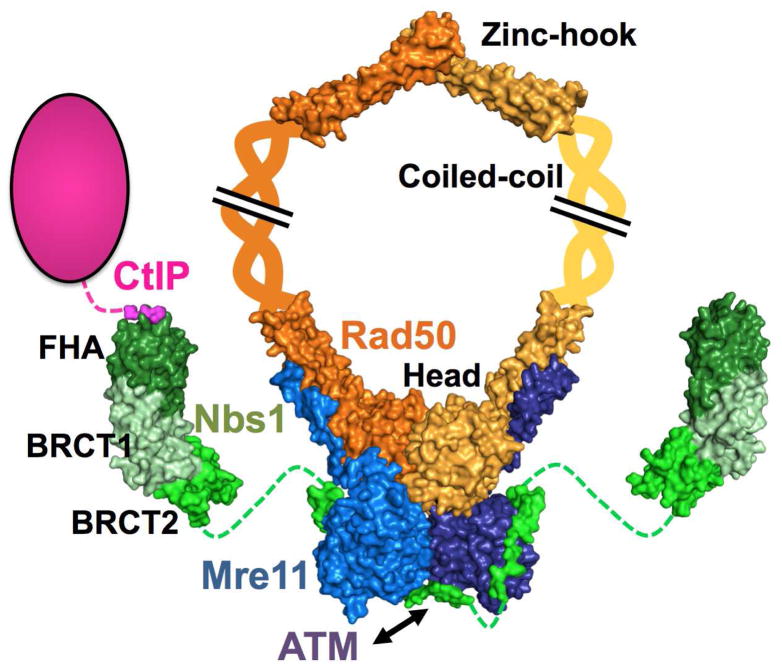
Figure 1. Architecture of the Mre11-Rad50-Nbs1 complex with its partner CtIP.
The Rad50 dimer is in orange and yellow (for each monomer) and the coiled-coils are represented as helices extending towards the zinc-hook. The Mre11 dimer is shown in blue and dark blue and interacts with Nbs1, shown in green (different shades represent the different domains). Nbs1 interacts with CtIP through its FHA domain (dark green). Interaction with ATM is shown by a black arrow. The dotted lines represent disordered protein linkers.
Rad50 is a member of the ABC ATPase superfamily and has provided insights of broad interest across the superfamily (Hopfner and Tainer, 2003). It is the largest protein of the MRN complex, with 1312 amino acids in the human enzyme (Fig. 3). Both the N- and C-terminal ends of the protein form the “head” ABC-ATPase domain, which contains the Walker A/B motifs and signature motif characteristic of this family of ATPases (Moncalian et al., 2004). It has both ATPase and adenylate kinase activities in vitro (Bhaskara et al., 2007; Paull and Gellert, 1999), but its major biological activity seems to be as an ATPase. Between the two termini of the protein, anti-parallel coiled-coil domains extend the length of the protein up to ~500 Å (Hopfner et al., 2000). Opposite to the head and at the tip of the coiled-coil, a CXXC motif creates a zinc-hook that facilitates dimerization of Rad50 (Cahill and Carney, 2007; Hopfner et al., 2002a). Indeed, two zinc-hooks (a total of four cysteines) can be attached together by coordinating a Zn2+ atom. But this is not the only part of Rad50 that interacts with another subunit. The head also dimerizes upon ATP binding: the N-terminus of one Rad50 interacts with the C-terminus of the other (and vice-versa), to form two complete active sites (Hopfner et al., 2000).
Figure 3. Long-range allostery is a crucial feature of Rad50.
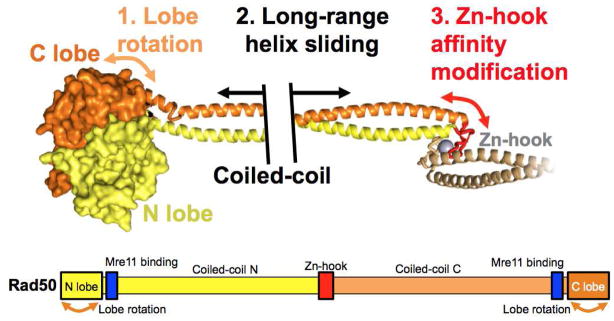
Top: ATP binding induces rotation of the C-lobe relative to the N-lobe of Rad50’s head domain. This is communicated to the zinc-hook domain through the coiled-coils, possibly by helix sliding. Bottom: schematic of Rad50’s sequence with arrows depicting segments of flexibility and dynamics.
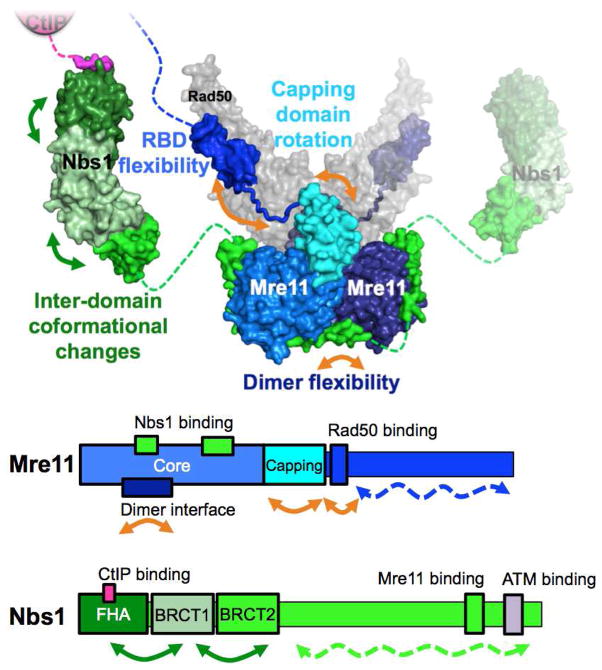
Like Rad50, Mre11 (708 residues in the human enzyme) is a dimer and it dimerizes through its core domain, located at the N-terminus of the sequence (Fig. 5). This core phosphodiesterase domain has single stranded DNA (ssDNA) endonuclease and 3′-5′ double stranded DNA (dsDNA) exonuclease activities (Hopfner et al., 2001). In the active site, five histidines, two aspartic acids and an asparagine coordinate two Mn2+ ions and are important for catalysis (Hopfner et al., 2001). Attached to the core domain, a “capping” domain is thought to help discriminate between ssDNA and dsDNA substrates and to be involved in orientating the DNA substrate within the active site (Das et al., 2010; R. S. Williams et al., 2008). The C-terminal domain of Mre11 is predicted to be disordered or flexible, except a small region forming α-helices that interact with the base of Rad50’s coiled-coils (Lammens et al., 2011; G. J. Williams et al., 2011). The rest of the C-terminal domain is responsible for protein-protein and protein-DNA interactions and is subject to post-translational modifications, including arginine methylation (Boisvert et al., 2005; Déry et al., 2008). The functional complex formed by the Mre11 and Rad50 dimers is conserved throughout all the three domains of life (named MR or M2R2) (Lim et al., 2011; Möckel et al., 2012), revealing this as a critical complex for cell biology.
Figure 5. Flexibility and dynamics are part of Mre11 and Nbs1’s structures.
Top: Structure of MRN highlighting flexible segments (arrows). Color-coding is the same as Figure 1, except Rad50 is gray and the capping domain of Mre11 is cyan. Bottom: schematic of Mre11 and Nbs1’s sequences with arrows depicting segments of flexibility and dynamics. Dashed lines and arrows represent intrinsically disordered segments.
While absent in bacteria and archaea, eukaryotes have Nbs1 (Xrs2 in Saccharomyces cerevisae), a phosphoprotein-binding and adapter subunit that plays an important role in connecting M2R2 to other DSB repair and signaling molecules. Although the stoichiometry is not fully known, recent structures support a model in which two Nbs1 interact with M2R2, as opposed to only one (Schiller et al., 2012). Thus it is a “M2R2N2” hetero-hexameric complex that is often implied when referring to the MRN complex. Starting from the N-terminus, Nbs1 possesses a fork head-associated (FHA) domain and two breast cancer-associated 1 C terminus (BRCT) domains known to bind phosphoproteins (Fig. 5). These three domains are responsible for interactions with several phosphorylated proteins (see section 7), like CtIP (R. S. Williams et al., 2009). On the other hand, the C-terminal half of Nbs1 is flexible, with the C-terminus containing adjacent motifs that interact with Mre11 and ATM.
3. A minimal model integrating diverse MRN functions
MRN is one of the first proteins to detect the DNA ends created at a DSB. Yet, how the whole complex precisely interacts with DNA in vivo is not known. After this initial DNA binding event, MRN interacts and activates ATM kinase to initiate DSB signaling and participates in DSB repair processes. In this section we present an overview of MRN’s mechanism in the two major pathways, HRR and NHEJ.
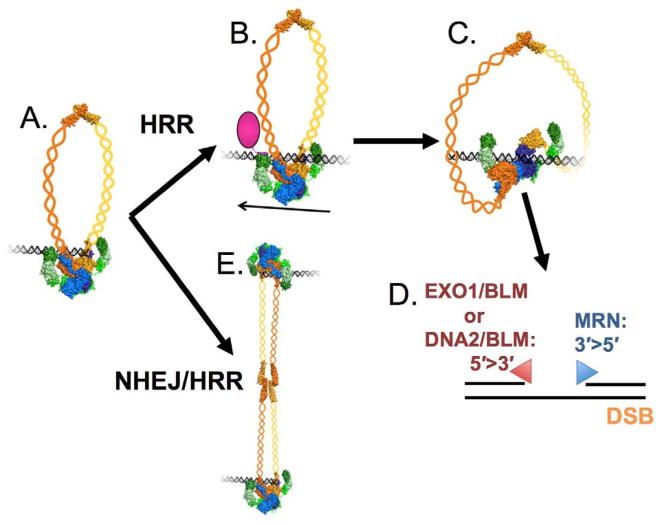
In HRR, the MRN complex has to move 100–200 base pairs away from the end of the DNA (Fig. 2A–D) (Shibata et al., 2014). Then, the protein CtIP (Sae2 in yeast) stimulates the endonuclease activity of Mre11, which makes a cut in one strand of the DNA (Cannavo and Cejka, 2014). It is currently debated whether or not CtIP has an intrinsic endonuclease activity (Makharashvili et al., 2014; H. Wang et al., 2014). From that point, Mre11 3′–5′ exonuclease activity resects the DNA strand back towards the break (Cannavo and Cejka, 2014; Garcia et al., 2011; Shibata et al., 2014; Zakharyevich et al., 2010). Simultaneously, the helicase/nuclease resection machinery (BLM in complex with either Exo1 and/or DNA2) carries out long-range 5′-3′ resection from the endonuclease cut in the opposite direction (Gravel et al., 2008; Mimitou and Symington, 2008; Zhu et al., 2008). These combined activities create the >1000 bp ssDNA overhang necessary for HRR. We know that the Rad50 coiled-coils are necessary in this process, but how they act precisely is an important question that remains to be solved. One model for the function of the Rad50 coiled-coils in both NHEJ and HRR is to tether the two DNA ends together. Instead of interacting within the dimer, the zinc-hooks interact with another MRN-DNA complex to attach them together (Fig. 2E). In yeast and in human cells, Rad50 and its hook keeps a DNA double-strand break from becoming a chromosome break (Hohl et al., 2011; Lobachev et al., 2004; Wiltzius et al., 2005). We are starting to understand how all these processes need to be controlled and finely tuned. This is what we explore in greater details in the next sections.
Figure 2. Model of MRN’s functions in DSB repair.
(A) MRN first detects and interacts with the DNA end. (B) If the process goes into HRR, MRN moves away from the break (shown with an arrow) and recruits CtIP. (C) CtIP induces changes in MRN leading to an open complex capable of nuclease activities. (D) After an initial endonuclease cut, MRN resects from 3′ to 5′ towards the DSB, with its exonuclease activity, while EXO1/BLM or DNA2/BLM resects from 5′ to 3′. (E) In NHEJ or HRR, the DNA tethering functions of MRN can be used. Color-coding is the same as in Figure 1.
4. Long-range allostery: the Rad50 coiled-coil and zinc hook
The 500 Å-long coiled-coil of Rad50, joining at one end the ABC-ATPase head and at the other end the zinc-hook, is its most striking feature. The length of the coiled-coil seems to be important for Rad50’s function, since shortening it has dramatic effects in vivo, impairing telomere maintenance and meiotic DSB formation (Hohl et al., 2011). Furthermore, NHEJ is severely affected by shortening the coiled-coils. On the other hand, homologous recombination functions are stable with a shorter coiled-coil (~300 amino acids removed), but not a very short coiled-coil (~500 amino acids removed) (Hohl et al., 2011). This could be an effect of a loss of flexibility in the coiled-coils (Hohl et al., 2011).
Indeed, EM and AFM images show that the coiled-coil is flexible and adopts multiple conformations (de Jager et al., 2001; 2004; Hopfner et al., 2001; Moreno-Herrero et al., 2005). These images show that as homodimers, their zinc-hooks and/or their ATPase heads attach two Rad50s. In some cases, the two coiled-coils are in proximity throughout their length to form a straight and rigid rod, as seen by AFM and EM (Anderson et al., 2001; Hopfner et al., 2002a; 2001). But most of the time, when both ends are attached, the coiled-coils have substantial flexibility (van Noort et al., 2003), and appear to create a curved, ovoid loop. Despite the structural insights, the importance and function of the various conformations of the Rad50 coiled-coils are still poorly understood.
How the structure of the zinc-hook relates to its function is also puzzling. Its crystal structure shows that it can join two coiled-coils in almost opposite directions (Hopfner et al., 2002a). At the apex of the coiled-coils, a loop of twelve amino acids curves back and forms the hook. In the middle, the cysteines of a CXXC motif coordinate a Zn2+ ion. The two pairs of cysteines form a tetragonal geometry around the Zn2+ ion. This structural model agrees with EM images showing arch-shaped Rad50 coils from single dimers (Hopfner et al., 2002a). Furthermore the configuration seen in the crystal structure also agrees with a model where a Rad50 dimer would attach to another Rad50 dimer, to bridge two DNA ends (Fig. 2E).
Mutational analysis of this motif and surrounding residues provided insight into its function. If the cysteines are mutated in yeast, survival after ionizing radiation is compromised and the Mre11-Rad50 interaction is impaired (Hopfner et al., 2002a), but the Rad50 dimerization through the head, the Mre11-Rad50 interaction or the interaction with DNA are not compromised (Cahill and Carney, 2007). Furthermore, removing the zinc-hook in yeast also removes the DSB repair functions of MRN and impairs telomere maintenance, but does not affect Rad50’s capability to associate with chromatin (Wiltzius et al., 2005). On the other hand, mutating the zinc-hook cysteines in mammalian cells does affect the recruitment and stability of MRN to chromosomal DSBs. This mutant fails to activate ATM, is defective in end resection and ATR activation, and thus is defective in initiating HRR-mediated DSB repair (He et al., 2012). This is further confirmed by hook mutants in mouse (Roset et al., 2014). Since Rad50 cysteine mutants are embryonic lethal, as is the Rad50 knock-out, it was necessary to mutate residues around the cysteines in order to obtain a milder phenotype. Thus two mutants were created in mouse (Rad5046: S679R+P682E and Rad5047: V683R). While homozygotes are not viable, heterozygotes can survive and are relatively proficient in DNA repair. Yet, they have problems in DNA damage response signaling through the ATM pathway, which greatly enhances tumorigenesis. At the molecular level, those Rad50-hook mutants still have the ability to dimerize and bind Zn2+, but are engaging in heterotypic dimerization.
These pieces of evidence inform us that Rad50 engages in long-range allostery through the coiled-coils: from the zinc-hook to the ATPase domain and vice versa (Fig. 3). The coiled-coils are not just a spacer or a rod to improve the reach of Rad50’s interactions, but there is a mechanical link between both ends of Rad50. The zinc hook functions are associated with NHEJ, which is consistent with its function in bridging DNA molecules that are interacting with the head. In that pathway, the zinc-hook interacts in an intermolecular fashion. On the other hand, a long coiled-coil is not necessary in homologous recombination, but the zinc hook is still important for intramolecular interactions. We emit the idea that the hook can undergo a structural transition to orient the coiled-coils of a MRN dimer in different orientations relative to one another. This transition would be communicated from the ATPase domain, through the coiled-coils. Interestingly, a similar mechanism is seen in dyneins, which are protein-motors involved in intracellular transport and other processes. Briefly, the dynein ATPase head is connected to a microtubule binding domain (MBD) by a 130 Å-long anti-parallel coiled-coil. The current “helix sliding” model suggests that conformational changes driven by ATP hydrolysis cause the two helices of the coiled-coil to slide, which changes the orientation and affinity of the MBD towards tubulin (Carter et al., 2008; Kon et al., 2009; Redwine et al., 2012). This releases the dynein from tubulin and allows movement. For Rad50, changes in the ATPase domain could also cause the coiled-coils to slide and transmit information to the zinc hook via long-range allostery. To support this model, it was seen with AFM that a transition in the orientation of the coiled-coils is dependent on DNA binding by the MRN head (Moreno-Herrero et al., 2005). Furthermore, upon nucleotide binding there are important conformational changes at the base of the coiled-coils near the ATPase head, as seen in different structures of Rad50 (Lammens et al., 2011; Lim et al., 2011; Möckel et al., 2012; G. J. Williams et al., 2011). Supporting this idea, the affinity of Rad50 to ATP and its hydrolysis are different when coiled-coils are truncated (Deshpande et al., 2014). In the next section, we explore how these changes in the ATPase head of Rad50 occur.
5. Functional dynamics of the Mre11-Rad50 ATPase head complex
Dynamic, ATP-dependent conformational changes in the Rad50 ATPase domain controls Mre11 nuclease activity and ATM signaling. In the absence of ATP, Mre11-Rad50 is in an open state, allowing access to the Mre11 active site (Fig. 4A). A crystal structure of a bacterial Mre11-Rad50 homolog (SbcD-SbcC) revealed a striking, extended conformation exists in the crystal, and the relatively close fit of the theoretical SAXS scattering curve to experimental data suggests that the bacterial complex is indeed in this rigid conformation in solution (Lammens et al., 2011). In contrast, SAXS data with archaeal Mre11-Rad50 revealed that the ATP-free complex is flexible in solution, conformationally sampling open and closed conformations (Rambo and Tainer, 2011; G. J. Williams et al., 2011). Such differences may be due to differing cellular roles of bacterial versus archaeal Mre11-Rad50 complexes. Flexibility within Mre11-Rad50 likely stems from a linker that connects the Mre11 phosphodiesterase and capping domains to a C-terminal Rad50 binding domain, which binds to the base of the Rad50 coiled coils allowing Rad50 subunits to move with respect to the Mre11 dimer (G. J. Williams et al., 2011).
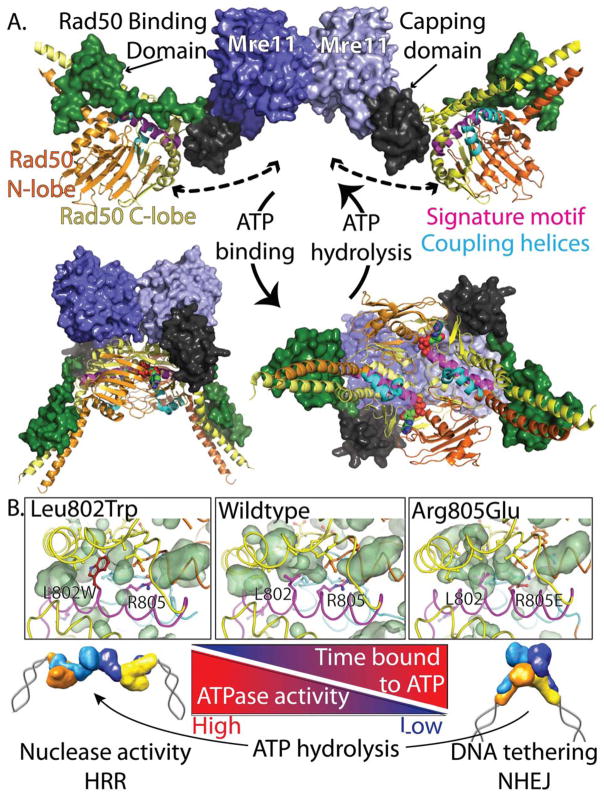
Figure 4. Rad50 ATP binding and hydrolysis controls its conformational state and pathway choice.
(A) Crystal structures showing the Mre11-Rad50 head without nucleotide (top, PDB 3QG5) and bound to nucleotide (bottom, PDB 3AV0). Mre11 (surface representation) and Rad50 (cartoon representation) are colored by subdomains as indicated (the color of the font matches the color of the domain). In the nucleotide-free state, Rad50 monomers are flexible and conformationally sample the closed state, as indicated by dashed-arrows. (B) Top: Crystal structures of P. furiosus Rad50 without nucleotide (PDB 3QKS) show a core cavity (green surface) adjacent to the signature motif (magenta) that remodels upon ATP-binding. The L802W mutation (PDB 4NCH) partially fills the core cavity, destabilizing the ATP-bound closed state. In contrast, the R805E variant (PDB 4NCI) destabilizes the Rad50 monomer resulting in a stabilization of the ATP-bound closed state. Bottom: The effects of the mutations compared to wildtype Rad50 on Mre11-Rad50 conformational states and activities are depicted.
Upon nucleotide binding, Rad50 head domains dimerize and block the Mre11 active site, as revealed by crystal structures of bacterial Mre11-Rad50 (Möckel et al., 2012) and archaeal Mre11-Rad50 (Lim et al., 2011) (Fig. 4A). SAXS results reveal that this ATP-bound Mre11-Rad50 complex is in a compact, rigid conformation (Rambo and Tainer, 2011; G. J. Williams et al., 2011), with theoretical scattering curves fitting experimental data remarkably well (Deshpande et al., 2014). In addition to relatively small conformational changes in Mre11 (Lim et al., 2011; Schiller et al., 2012), ATP-binding by Rad50 is coupled to a large conformational change within each Rad50 monomer, with the N-lobe rotating ~35° relative to the C-lobe. This conformational change allows the Walker A box of one subunit and the signature motif plus the Walker B motif of the other subunit to come together to form the active sites that sandwich two ATP molecules at the dimer interface (Hopfner et al., 2000). Furthermore, concerted changes in two coupling helices are involved in repositioning the base of the Rad50 coiled-coils and attached Mre11 Rad50 binding domain (RBD), which may be involved in regulating Mre11 activities (G. J. Williams et al., 2011).
The switch between closed and open forms of Mre11-Rad50 is controlled by Rad50 ATP hydrolysis and product release, which in turn controls the nuclease activity of Mre11 (Majka et al., 2012). Cross-linking experiments verified that in the ATP-bound closed state, Mre11 nuclease activity is switched off. Furthermore, opening of the Rad50 dimer following ATP hydrolysis is required for both Mre11 endonuclease and exonuclease activities (Deshpande et al., 2014). Interestingly, Rad50 has a slow ATP hydrolysis rate (Majka et al., 2012). Moreover, elegant biochemical analysis of a T4 Rad50 homolog revealed that the chemistry of ATP hydrolysis is likely the rate-limiting step (Herdendorf et al., 2011; Herdendorf and Nelson, 2014), meaning that within the cell this ATP-bound form is likely to exist for a substantial amount of time before ATP hydrolysis and subsequent release of ADP and phosphate returns the complex to the open state for another round of the ATPase cycle. Further insights into the biological importance of open and closed states of Mre11-Rad50 and the rate of Rad50 ATPase activity emerged from investigations into point mutations designed to disrupt ATP-dependent Rad50 conformations (Deshpande et al., 2014), as discussed in more detail below.
The molecular environment surrounding the extended signature motif, which includes the α-helix following the classical ABC-ATPase signature motif, facilitates the plastic deformations necessary for ATP-dependent conformational changes (Fig. 4B). The extended signature motif is at the intersection of the N- and C-lobes of Rad50 that undergo the 35° rotation, and contains arginine residues that switch between different salt-bridge networks to coordinate subdomain rotation (G. J. Williams et al., 2011). Furthermore, adjacent to the extended signature helix is an internal protein cavity that is remodeled between ATP-free and ATP-bound states, facilitating the conformational change (Deshpande et al., 2014). To further dissect the role of different Rad50 ATP-states, mutations that either disrupted a key salt-bridge network involved in coordinating conformational changes (Arg805Glu in Pyrococcus furiosus Rad50, and equivalent conserved residues in eukaryotic Rad50) or partially filled the protein cavity (Leu802Trp in P. furiosus Rad50, and equivalent conserved residues in eukaryotic Rad50) were studied (Deshpande et al., 2014). Biochemical and biophysical results, including novel time-course SAXS experiments, revealed that these mutations result in distinct separation-of-functions that alter the rate at which Rad50 ATPase activity switches the Mre11-Rad50 complex from ATP-bound and closed to ATP-free and open conformations. The Arg805Glu mutation enhanced the closed ATP-bound conformation; resulting in reduced ATPase and nuclease activities, yet increased the binding and tethering of DNA ends. In vivo assays showed that this variant was more proficient at NHEJ than wildtype Rad50. In contrast, while the Leu802Trp mutation reduced ATP-binding, when ATP bound it was hydrolyzed more quickly than wildtype; resulting in more time spent in the open conformation and enhanced Mre11 nuclease and end-resection activities. Survival assays following clastogen exposure, as well as DSB signaling assays, revealed that both mutations were deficient in DSBR and ATM activation, showing the time Rad50 spends in the ATP-bound state is critical for MRN functions. Interestingly, this ATP-bound state has been implicated in unwinding DNA ends (Cannon et al., 2013), and is required for MRN-mediated ATM activation in vitro (J.-H. Lee et al., 2013). Recently, a crystal structure of bacterial Rad50 in complex with DNA and ATP has been solved (Rojowska et al., 2014). This revealed a biologically important Rad50-DNA binding interface that is necessary for telomere maintenance (through ATM activation), but has little impact on repair activities (Rojowska et al., 2014). However, molecular mechanisms for how ATP-bound Mre11-Rad50 unwinds DNA ends, activates ATM, and hands off DNA from Rad50 to Mre11 for nucleolytic processing remain enigmatic.
The results using Rad50 mutations imply that modulating Rad50’s ATPase activity can affect biological outcomes by favoring HRR versus NHEJ or by impacting DSB signaling. Furthermore, the long-range allostery observed for MRN suggests that alterations to the Zn-hook and/or the coiled-coils of Rad50 or of Mre11/Nbs1 subunits can impact the ABC-ATPase domain. This collective data raises the intriguing possibility that in eukaryotes post-translational modification within the MRN complex or protein partner interactions may alter Rad50 ATPase activities to control repair pathway choice and/or signaling depending on the biological context. With the progress in SAXS experiments and analysis that can provide accurate structures (Putnam et al., 2007; Schneidman-Duhovny et al., 2013), comprehensive conformations (Hura et al., 2013a; Rambo and Tainer, 2010), high throughput determination of the precise mass of assemblies in solution (Hura et al., 2009; Rambo and Tainer, 2013a), and the ability to track DNA conformations with gold nanocrystals (Hura et al., 2013b), we can expect further results on the functional flexibility and conformations of Rad50 and MRN.
6. Mre11 conformations act in Rad50’s ATP-dependent dimerization and DNA sculpting
We now have a good sampling of Mre11 catalytic domain structures from several different organisms in the Archaea (Hopfner et al., 2001; Lim et al., 2011), Bacteria (Das et al., 2010; Liu et al., 2014) and Eukarya kingdoms (Park et al., 2011; Schiller et al., 2012). They all have a common fold, with the phosphodiesterase core and capping domains, and their active site arrangement is also conserved. However, the dimer organization is different in all structures, which highlights the flexible nature of Mre11. The dimerization of Mre11 is crucial for stable DNA binding (R. S. Williams et al., 2008) and differential orientations of both dimers could be a way to control and limit the nuclease activity. Moreover, recent Mre11-DNA complex structures served as a basis to hypothesize that dimer rotation could help DNA melting by twisting the duplex (Sung et al., 2014).
The role of Mre11 in melting dsDNA is also hypothesized in a model of its enzymatic mechanism (R. S. Williams et al., 2008). In a three-step exonuclease substrate recognition mechanism, Mre11 sculpts the DNA using several flexible DNA binding motifs of the protein. More precisely, the first step is the interaction of the Mre11 dimer with the phosphates and minor groove of duplex DNA (Sung et al., 2014; R. S. Williams et al., 2008). Then, the inter-subunit orientation changes, the capping domain rotates and a helical wedge inserts into the DNA minor groove, which causes the DNA end to melt. The scissile strand is then directed towards the capping domain, a necessary step to control and identify the substrate. Finally, a conserved histidine facilitates phosphate rotation that directs the scissile strand into the Mre11 active site for 3′ to 5′ exonucleotytic cleavage, which proceeds in a processive manner. Mutating this histidine has an effect on the exonuclease activity of Mre11, but not its endonuclease activity, showing that both activities have different mechanisms. For the endonuclease mechanism, it is hypothesized that there is a ssDNA-binding pocket on the other side of the active side. Furthermore, since ssDNA is more flexible than dsDNA, the histidine critical for exonuclease activity is dispensable as there is no need for substrate rotation. These hypothesized mechanisms have proven to be useful models, yet a Mre11-DNA structure with DNA trapped in the active site would provide further molecular insights into Mre11 activities. Currently, the ability of Mre11 to sculpt DNA for specific excision is consistent with a theme for structure-specific DNA and RNA nucleases, such as the replication and repair 5′ flap nuclease FEN1 (Tsutakawa et al., 2011), the structurally non-homologous apurinic/apyrimidinic endonucleases APE1 and endonuclease IV (Tsutakawa et al., 2013), as well as multiple other structure-specific nucleases (Tsutakawa et al., 2014).
The differences in the hypothesized endonuclease versus exonuclease mechanisms were exploited for the development of inhibitors specific to each activity of Mre11 (Shibata et al., 2014). Several inhibitors were discovered using the structure of Thermotoga maritima Mre11 with the Mre11 inhibitor Mirin as a guide, and exploiting a focused chemical library. PFM01 and PFM03 inhibit the endonuclease activity of Mre11 by blocking the hypothesized ssDNA-binding groove, while PFM39 binds near the histidine involved in phosphate rotation to inhibit the exonuclease activity. Both types of inhibitors reduce DSB end resection in vivo, but create distinct phenotypes. Inhibition of the endonuclease activity directs DSBR towards NHEJ instead of HRR, whereas exonuclease inhibition causes a repair defect. This repair defect could be partially rescued by knock-out of EXO1/BLM in conjunction with exonuclease inhibition, which phenocopies endonuclease inhibition by pushing repair towards NHEJ. Collectively, these results showed that Mre11 endonuclease acts upstream of exonuclease and supports a two-step model of Mre11 activity in end resection for HRR. First, Mre11 uses its endonuclease activity to nick duplex DNA dozens of base pairs away from the break. Second, Mre11 exonuclease digests the DNA back towards the DNA break, with BLM helicase working together with either EXOI nuclease or DNA2 nuclease/helicase to digest DNA 5′-3′ (Nimonkar et al., 2011; Niu et al., 2010), in order to generate long single-strand DNA overhangs and to commit the pathway choice towards HRR (Fig. 2). These inhibitors are important tools to study pathway selection, since there is no known mutation of Mre11 impacting only the endonuclease activity.
Besides its importance in break repair, Mre11 acts along with BRCA1, BRCA2/FANCD1 and the ATPase SMARCAL1 to protect or degrade replication forks (Mason et al., 2014; Schlacher et al., 2011; 2012), consistent with its crystal structures with two DNA ends or a Y-DNA mimic for a replication fork (R. S. Williams et al., 2008). Mre11 furthermore provides a backup removal of covalently attached proteins at the replication fork (K. C. Lee et al., 2012), which is also a function of TDP2 (Shi et al., 2012). While the mechanistic basis for Mre11 function at the replication fork is poorly understood, characterizing distinct roles of Mre11 versus other nucleases in this context will be important keys to understanding replication fidelity.
We further hypothesize that flexibility of Mre11 has a role in regulation. Indeed, the flexibility of the Mre11 dimer appears to be higher in eukaryotes, as highlighted by stabilization of Schizosaccharomyces pombe Mre11 dimers by Nbs1 interaction (Schiller et al., 2012). Indeed, in structures of Mre11 with Nbs1, two Nbs1 molecules interact with the Mre11 dimer (Schiller et al., 2012). Only 14 residues of Nbs1 are involved in this interaction (Fig. 5). Interestingly, a 7-residue segment from one Nbs1 molecule is making contacts with both Mre11 subunits simultaneously, stabilizing the Mre11 dimer. The interaction site is on the opposite surface of Mre11 to its active site and has flexible loops unique to eukaryotes. The binding of Nbs1 on this site induces an ordering of the loops and a 30° rotation of the Mre 11 dimer, which could have an impact on bound Rad50 or interactions with other proteins such as ATM or CtIP. Such flexibility may provide a target for the development of inhibitors by finding ligands that bind and stabilize an inactive conformation of Mre11 dimer. In the next section we explore how flexibility of MRN protein-protein interactions regulates pathway choices.
7. Nbs1 and CtIP regulate pathway choice
Nbs1 can be viewed as a sensor as well as an effector. Through its interaction across the Mre11 dimer interface, it senses the conformation of Mre11, which in turn is impacted by Rad50 ATP-state. The biological importance of this is still unclear, but we know that mutations in Nbs1 can increase the nuclease activity of Mre11 during replication and thus increases the number of DSBs (Crown et al., 2013). Another biological role of the Nbs1-Mre11 interaction could be linked with ATM signaling (J.-H. Lee and Paull, 2005; 2004). Indeed, the Mre11-interacting segment of Nbs1 is adjacent to the ATM-interaction motif, located at the C-terminus of the protein (You et al., 2005). To support that model, Nbs1 and its connection of MRN to ATM are able to regulate the Mre11 nuclease based upon an examination of Mre11 activity in Ataxia-telangiectasia cells (Rahal et al., 2010). Furthermore, mutations of Mre11 can dissect DSBR and ATM activation functions revealing key roles for Nbs1 partner Mre11 in both processes (Limbo et al., 2012). Modulation of Mre11-Rad50 conformation could be sensed by Nbs1, which in turn recruits ATM for its activation.
A second sensor interface of Nbs1 is its ordered N-terminal domain, which interacts with different proteins, and thus senses the damage status of the cell (R. S. Williams et al., 2009). More specifically it interacts with disordered segments of proteins, and senses their phosphorylated form. Its two phosphoprotein-binding sites (one on the FHA domain and one on the BRCT-repeat domain) confers several binding modes (R. S. Williams et al., 2009). CtIP (Lloyd et al., 2009; R. S. Williams et al., 2009) MDC1 (Hari et al., 2010; Lloyd et al., 2009; Spycher et al., 2008), ATR (Olson et al., 2007), and WRN (Cheng et al., 2004; Kobayashi et al., 2010) have been shown to interact directly with Nbs1. Notably, the structure of Nbs1 with a CTP1-based phosphopeptide (ortholog of CtIP in S. pombe) has been solved and informs how Nbs1 acts as a sensor (R. S. Williams et al., 2009). CTP1 binding induces a long-range conformational change in the N-terminal domain of Nbs1, which alters the interface between BRCT1 and BRCT2. This suggests a cross-talk between the two phosphoprotein-binding sites, which could modulate the interaction with the other protein partners. This mechanism could be a way for Nbs1 to coordinate the repair pathway choice.
Beyond its roles as a sensor, Nbs1 also recruits proteins to the DNA damage site, for them to achieve their function. This includes CtIP, which has been at the center of a lot of attention and debate recently, with its precise role and function being unclear. Unfortunately, no structures are currently available that could help us understand its mechanism. Some groups have suggested that CtIP has an intrinsic endonuclease activity (Makharashvili et al., 2014; H. Wang et al., 2014), which agrees with the bidirectional resection model presented in section 3. Also agreeing with that model it was shown that Sae2 (CtIP ortholog in S. cerevisiae) stimulates the endonuclease activity of MRN, but has no intrinsic endonuclease activity (Cannavo and Cejka, 2014). A possible model is that CtIP is physically modulating the structure of MRN in order to stimulate Mre11’s endonuclease activity.
8. Clinical and therapeutic opportunities
Recently, cancer centers and the US National Cancer Institute (NCI) have initiated research programs around “failed” clinical trials of anti-cancer drugs (Mullard, 2014). These trials were rejected, since the drug was not reducing the cancer’s progression in most patients. But some patients responded very well to the drug and were cured, thus they are named “exceptional responders”. By studying them, researchers hypothesize that they can tailor drug regimes specifically for individual patients, the idea behind personalized medicine. If we know why those people are exceptional responders using genomics, whole-genome sequencing or biomarkers, we will be able to apply the same remedy to patients bearing the same genotype.
A great example out of this approach is the discovery of an exceptional responder in the clinical trial of a CHK1 kinase inhibitor combined with the DNA-damaging agent irinotecan (Al-Ahmadie et al., 2014). CHK1 is a substrate of ATR in the DNA damage response. Briefly, whole-genome sequencing of the tumor before chemotherapy identified the RAD50 mutant Leu1237Phe as the mutation responsible for the exceptional response. Leu-1237 is part of the D-loop of RAD50, which is close to the Walker B box, involved in ATP hydrolysis. It is also located at the dimer interface and the side chain of the conserved residue in archaeal Rad50 switches between a buried and solvent-exposed position in the dimer and monomer crystal structures, respectively. The Leu1237Phe mutant could thus be deficient in dimerization or in its rate of ATP hydrolysis, processes intrinsically linked. Further biochemical characterization is necessary to conclude the precise impact of this mutation, but Rad50-Leu1237Phe is deficient in ATM signaling (Al-Ahmadie et al., 2014).
In considering the biophysical data, we herein suggest another approach that uses the knowledge derived from exceptional responders. The precise molecular understanding of the deficient mutated protein could be used to design drugs that mimic the mutation. Such drugs, when used in combination with other therapies, would work for a wide range of patients. For example, if we could design a drug against Rad50 that mimics the effect of the Leu1237Phe mutation, combined with irinotecan and the CHK1 inhibitor, this drug would be successful for most patients having the same cancer, not to only a subset of them.
9. Combining biophysics and molecular biology to understand MRN’s structural flexibility
Crystal structures of the individual MRN domains and multiple complexes have provided detailed knowledge for mechanistic understanding. Crystallography provides precise structures but less information about flexibility, although temperature factors can identify functional flexible regions when corrected for crystal contacts (Tainer et al., 1984). Unveiling the roles of flexibility in DNA damage responses is an ongoing process. Flexibility for interaction and functional access to DNA ends makes sense geometrically, and is seen in systems such as the Ligase-PCNA interaction (Pascal et al., 2006). Yet, intra-molecular interactions of flexible regions can contribute to the stability of the folded region and can act in intermolecular interaction, making interpretations of functions challenging by single methods (Hegde et al., 2013). Thus, combining biophysical techniques together with molecular biology will be crucial to understand flexibility of macromolecular machines.
SAXS is an ideal biophysical technique to be combined with other structural biology techniques. Its development helped study Mre11, Rad50 (along with other members of the ABC ATPases family) and Nbs1. For example, gold nanocrystal SAXS was created to provide a means to examine DNA conformations at high sensitivity along a complex reaction pathway in the context of multiple protein components (Hura et al., 2013b). Similarly, the ABC-ATPase MutSβ was used to develop a SAXS-based method to objectively and comprehensively compare conformational states in solution (Hura et al., 2013a). Also, we know that water molecules are part of the active site and other concave surface features and that residue composition impact bound waters (Kuhn et al., 1992; 1995). An exciting recent observation is that our refined macromolecular crystal structure models are in general not limited by data noise or error but by our ability to properly model the water structure and model flexibility (Holton et al., 2014). We can expect that further advances in biophysical methods that define flexibility and bound water structures may allow more accurate and complete models of dynamic complexes, including MRN.
Besides the dynamics of defined domains and complexes, it will be important to look at the impacts of post-translational modifications on MRN. Dynamic protein-protein interactions and complex assemblies mediating DNA damage response are often initiated or regulated by protein post-translational modification, such as phosphorylation, methylation, ubiquitination, and sumoylation. SAXS has proven useful to define the activity and the structural impacts of these modifications (Datta et al., 2009; Duda et al., 2008; Konarev et al., 2014). Indeed, SAXS and molecular biology showed that the ubiquitin ligase RNF4 selectively recognizes and ubiquitinates of SUMO-modified histones, therefore it integrates signaling by SUMO and ubiquitin (Groocock et al., 2014). We know from SAXS analyses of NHEJ that phosphorylation of DNA-PKcs causes a functionally important conformational change to promote opening and release from DNA (Hammel et al., 2010). We can expect to see analogous conformational regulation for MRN conformations and flexibility (Deshpande et al., 2014). Indeed, independent modes of MRN phosphorylation have been reported, with CDK1 orthologs phosphorylating MRN during mitosis and, independently, ATM orthologs phosphorylating MRN after DNA damage (Martin et al., 2014; Simoneau et al., 2014). This suggests that MRN integrates signals from an intricate network of phosphoregulatory mechanisms to modulate DSB responses. Furthermore, Rad17 phosphorylation by ATM at a novel Thr622 site results in its interaction with NBS1, facilitating recruitment of the MRN complex and ATM to the DSB, thereby enhancing ATM signaling (Q. Wang et al., 2014).
Crystal structures have also revealed important results pertaining to flexibility and activity by defining the metal binding sites in Mre11 and Rad50 domains. Frequently, metal ions are an over-looked issue in characterizing macromolecular complexes. Mre11 and Rad50 bind multiple metal ions; so far zinc, magnesium, and manganese have been defined crystallographically. Identifying and characterizing the roles of metal ions is often challenging, in part because they may not be correctly incorporated into the protein in recombinant expression hosts, and may thus require purification from native biomass (Cvetkovic et al., 2010; Yannone et al., 2012). Even partial loss of metal ions impacts proper function and increases flexibility and misassembly, as seen for the stress response enzyme superoxide dismutase (Pratt et al., 2014) and the DNA repair and transcription helicase XPD (Fan et al., 2008). Biophysical techniques allowing the accurate identification and measurement of metal ions will be important in future research.
One thing that can guide flexible enzyme-substrate recognition and allows flexibility of the enzyme’s binding site is the negatively charged DNA and an appropriate positive patch on the repair enzyme surface (Huffman et al., 2005; Mol et al., 2000; Slupphaug et al., 1996). Such positive patches are seen on both Mre11 and Rad50 (Hopfner et al., 2001) and are consistent with subsequent DNA-complex crystal structures (Rojowska et al., 2014; R. S. Williams et al., 2008). Indeed electrostatics are part of DNA mimicry that can regulate DNA repair processes (Mol et al., 1995; Putnam et al., 1999; Putnam and Tainer, 2005). In general, electrostatic potential gradients can increase the rate of interactions (Getzoff et al., 1983) as well as properly orient two macromolecules for productive collisions (Roberts et al., 1991). In the future it will be interesting to understand how the distinct Mre11 and Rad50 DNA binding sites cooperate to bind and then process DNA substrates.
10. Perspective and conclusion
In eukaryotic cells, the initiation of end resection has emerged as a critical regulatory step to differentiate between homology-dependent and end-joining repair of DSBs and to impact replication fork protection versus replication fork excision. MRN is the key complex in these two essential components of cell biology. Therefore, a bottom up mechanistic understanding is worthwhile for its biological and medical impacts.
Since the first human DNA repair enzymes revealed DNA bending and nucleotide flipping in damage recognition (Slupphaug et al., 1996), structural biology has revealed conformational changes and flexibility as a theme in recognition specificity. With MRN we have come to appreciate how the dynamics and kinetics of the system can modulate distinct outcomes. Mre11 and Rad50 both sculpt the DNA to achieve specificity as seen for base repair enzymes (Slupphaug et al., 1996), nucleotide excision repair enzymes (Fuss and Tainer, 2011) and proteins that provide connections between DNA repair pathways (Dalhus et al., 2013). Going forward we expect that hybrid methods, such as SAXS combined with crystallography or NMR, will play key roles in examining functionally important structural flexibility in solution under near physiological conditions (Putnam et al., 2007; Rambo and Tainer, 2013b).
To conclude, MRN is a highly dynamic protein complex, with different parts being flexible in both their structure and mechanism. Understanding the dynamics of the MRN complex is helping drive novel and combined biophysical methods and is furthermore essential for the detailed knowledge necessary to both understand and predict biological responses to DSBs, and for the development of novel medical approaches to cancer.
Acknowledgments
Work for this review and the authors’ efforts on the MRN complex were funded by NIH R01CA117638 and P01CA092584. J.L.V. is recipient of a fellowship from the Canadian Research Institutes of Health Research (CIHR).
Abbreviations
- AFM
atomic force microscopy
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related protein
- BRCT
BRCA1 C-terminus domain
- DSB
DNA double strand break
- DSBR
double strand break repair
- EM
electron microscopy
- FHA
forkhead-associated domain
- HRR
homologous recombination repair
- MRN
Mre11-Rad50-Nbs1
- NHEJ
non-homologous end-joining
- SAXS
small-angle X-ray scattering
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Ahmadie H, Iyer G, Hohl M, Asthana S, Inagaki A, Schultz N, Hanrahan AJ, Scott SN, Brannon AR, McDermott GC, Pirun M, Ostrovnaya I, Kim P, Socci ND, Viale A, Schwartz GK, Reuter V, Bochner BH, Rosenberg JE, Bajorin DF, Berger MF, Petrini JHJ, Solit DB, Taylor BS. Synthetic Lethality in ATM-Deficient RAD50-Mutant Tumors Underlies Outlier Response to Cancer Therapy. Cancer Discov. 2014;4:1014–1021. doi: 10.1158/2159-8290.CD-14-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DE, Trujillo KM, Sung P, Erickson HP. Structure of the Rad50 x Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J Biol Chem. 2001;276:37027–37033. doi: 10.1074/jbc.M106179200. [DOI] [PubMed] [Google Scholar]
- Bhaskara V, Dupré A, Lengsfeld B, Hopkins BB, Chan A, Lee JH, Zhang X, Gautier J, Zakian V, Paull TT. Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes. Mol Cell. 2007;25:647–661. doi: 10.1016/j.molcel.2007.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F-M, Déry U, Masson JY, Richard S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005;19:671–676. doi: 10.1101/gad.1279805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill D, Carney JP. Dimerization of the Rad50 protein is independent of the conserved hook domain. Mutagenesis. 2007;22:269–274. doi: 10.1093/mutage/gem011. [DOI] [PubMed] [Google Scholar]
- Cannavo E, Cejka P. Sae2 promotes dsDNA endonuclease activity within Mre11–Rad50–Xrs2 to resect DNA breaks. Nature. 2014;514:122–125. doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- Cannon B, Kuhnlein J, Yang S-H, Cheng A, Schindler D, Stark JM, Russell R, Paull TT. Visualization of local DNA unwinding by Mre11/Rad50/Nbs1 using single-molecule FRET. Proc Natl Acad Sci USA. 2013;110:18868–18873. doi: 10.1073/pnas.1309816110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Garbarino JE, Wilson-Kubalek EM, Shipley WE, Cho C, Milligan RA, Vale RD, Gibbons IR. Structure and functional role of dynein’s microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W-H, Kobbe von C, Opresko PL, Arthur LM, Komatsu K, Seidman MM, Carney JP, Bohr VA. Linkage between Werner syndrome protein and the Mre11 complex via Nbs1. J Biol Chem. 2004;279:21169–21176. doi: 10.1074/jbc.M312770200. [DOI] [PubMed] [Google Scholar]
- Classen S, Hura GL, Holton JM, Rambo RP, Rodic I, McGuire PJ, Dyer K, Hammel M, Meigs G, Frankel KA, Tainer JA. Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J Appl Crystallogr. 2013;46:1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown KN, Savytskyy OP, Malik SB, Logsdon J, Williams RS, Tainer JA, Zolan ME. A mutation in the FHA domain of Coprinus cinereus Nbs1 Leads to Spo11-independent meiotic recombination and chromosome segregation. G3 (Bethesda) 2013;3:1927–1943. doi: 10.1534/g3.113.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovic A, Menon AL, Thorgersen MP, Scott JW, Poole FL, Jenney FE, Lancaster WA, Praissman JL, Shanmukh S, Vaccaro BJ, Trauger SA, Kalisiak E, Apon JV, Siuzdak G, Yannone SM, Tainer JA, Adams MWW. Microbial metalloproteomes are largely uncharacterized. Nature. 2010;466:779–782. doi: 10.1038/nature09265. [DOI] [PubMed] [Google Scholar]
- Dalhus B, Nilsen L, Korvald H, Huffman J, Forstrøm RJ, McMurray CT, Alseth I, Tainer JA, Bjørås M. Sculpting of DNA at abasic sites by DNA glycosylase homolog mag2. Structure. 2013;21:154–166. doi: 10.1016/j.str.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, Moiani D, Axelrod HL, Miller MD, McMullan D, Jin KK, Abdubek P, Astakhova T, Burra P, Carlton D, Chiu H-J, Clayton T, Deller MC, Duan L, Ernst D, Feuerhelm J, Grant JC, Grzechnik A, Grzechnik SK, Han GW, Jaroszewski L, Klock HE, Knuth MW, Kozbial P, Krishna SS, Kumar A, Marciano D, Morse AT, Nigoghossian E, Okach L, Paulsen J, Reyes R, Rife CL, Sefcovic N, Tien HJ, Trame CB, van den Bedem H, Weekes D, Xu Q, Hodgson KO, Wooley J, Elsliger M-A, Deacon AM, Godzik A, Lesley SA, Tainer JA, Wilson IA. Crystal structure of the first eubacterial Mre11 nuclease reveals novel features that may discriminate substrates during DNA repair. J Mol Biol. 2010;397:647–663. doi: 10.1016/j.jmb.2010.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta AB, Hura GL, Wolberger C. The structure and conformation of Lys63-linked tetraubiquitin. J Mol Biol. 2009;392:1117–1124. doi: 10.1016/j.jmb.2009.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager M, Trujillo KM, Sung P, Hopfner K-P, Carney JP, Tainer JA, Connelly JC, Leach DRF, Kanaar R, Wyman C. Differential arrangements of conserved building blocks among homologs of the Rad50/Mre11 DNA repair protein complex. J Mol Biol. 2004;339:937–949. doi: 10.1016/j.jmb.2004.04.014. [DOI] [PubMed] [Google Scholar]
- de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- Déry U, Coulombe Y, Rodrigue A, Stasiak A, Richard S, Masson JY. A glycine-arginine domain in control of the human MRE11 DNA repair protein. Mol Cell Biol. 2008;28:3058–3069. doi: 10.1128/MCB.02025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande RA, Williams GJ, Limbo O, Williams RS, Kuhnlein J, Lee J-H, Classen S, Guenther G, Russell P, Tainer JA, Paull TT. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 2014;33:482–500. doi: 10.1002/embj.201386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss JO, Tainer JA. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair (Amst) 2011;10:697–713. doi: 10.1016/j.dnarep.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Phelps SEL, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getzoff ED, Tainer JA, Weiner PK, Kollman PA, Richardson JS, Richardson DC. Electrostatic recognition between superoxide and copper, zinc superoxide dismutase. Nature. 1983;306:287–290. doi: 10.1038/306287a0. [DOI] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groocock LM, Nie M, Prudden J, Moiani D, Wang T, Cheltsov A, Rambo RP, Arvai AS, Hitomi C, Tainer JA, Luger K, Perry JJP, Lazzerini-Denchi E, Boddy MN. RNF4 interacts with both SUMO and nucleosomes to promote the DNA damage response. EMBO Rep. 2014;15:601–608. doi: 10.1002/embr.201338369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, Pique ME, Fang S, Mahaney BL, Weinfeld M, Schriemer DC, Lees-Miller SP, Tainer JA. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. Journal of Biological Chemistry. 2011;286:32638–32650. doi: 10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, Abolfath RM, Chen DJ, Lees-Miller SP, Tainer JA. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. Journal of Biological Chemistry. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari FJ, Spycher C, Jungmichel S, Pavic L, Stucki M. A divalent FHA/BRCT-binding mechanism couples the MRE11-RAD50-NBS1 complex to damaged chromatin. EMBO Rep. 2010;11:387–392. doi: 10.1038/embor.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Shi LZ, Truong LN, Lu CS, Razavian N, Li Y, Negrete A, Shiloach J, Berns MW, Wu X. Rad50 zinc hook is important for the Mre11 complex to bind chromosomal DNA double-stranded breaks and initiate various DNA damage responses. Journal of Biological Chemistry. 2012;287:31747–31756. doi: 10.1074/jbc.M112.384750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Tsutakawa SE, Hegde PM, Holthauzen LMF, Li J, Oezguen N, Hilser VJ, Tainer JA, Mitra S. The disordered C-terminal domain of human DNA glycosylase NEIL1 contributes to its stability via intramolecular interactions. J Mol Biol. 2013;425:2359–2371. doi: 10.1016/j.jmb.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdendorf TJ, Albrecht DW, Benkovic SJ, Nelson SW. Biochemical Characterization of Bacteriophage T4 Mre11-Rad50 Complex. Journal of Biological Chemistry. 2011;286:2382–2392. doi: 10.1074/jbc.M110.178871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdendorf TJ, Nelson SW. Catalytic Mechanism of Bacteriophage T4 Rad50 ATP Hydrolysis. Biochemistry. 2014;53:5647–5660. doi: 10.1021/bi500558d. [DOI] [PubMed] [Google Scholar]
- Hohl M, Kwon Y, Galván SM, Xue X, Tous C, Aguilera A, Sung P, Petrini JHJ. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat Struct Mol Biol. 2011;18:1124–1131. doi: 10.1038/nsmb.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton JM, Classen S, Frankel KA, Tainer JA. The R-factor gap in macromolecular crystallography: an untapped potential for insights on accurate structures. FEBS J. 2014;281:4046–4060. doi: 10.1111/febs.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BAL, Karcher A, Henderson B, Bodmer JL, McMurray CT, Carney JP, Petrini JHJ, Tainer JA. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002a;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Hopfner K-P, Putnam CD, Tainer JA. DNA double-strand break repair from head to tail. Curr Opin Struct Biol. 2002b;12:115–122. doi: 10.1016/s0959-440x(02)00297-x. [DOI] [PubMed] [Google Scholar]
- Hopfner K-P, Tainer JA. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr Opin Struct Biol. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hura GL, Budworth H, Dyer KN, Rambo RP, Hammel M, McMurray CT, Tainer JA. Comprehensive macromolecular conformations mapped by quantitative SAXS analyses. Nat Methods. 2013a;10:453–454. doi: 10.1038/nmeth.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, Tsutakawa SE, Jenney FE, Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MWW, Tainer JA. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, Tsai C-L, Claridge SA, Mendillo ML, Smith JM, Williams GJ, Mastroianni AJ, Alivisatos AP, Putnam CD, Kolodner RD, Tainer JA. DNA conformations in mismatch repair probed in solution by X-ray scattering from gold nanocrystals. Proc Natl Acad Sci USA. 2013b;110:17308–17313. doi: 10.1073/pnas.1308595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi J, Okui M, Asaithamby A, Burma S, Chen BPC, Tanimoto K, Matsuura S, Komatsu K, Chen DJ. WRN participates in translesion synthesis pathway through interaction with NBS1. Mech Ageing Dev. 2010;131:436–444. doi: 10.1016/j.mad.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Imamula K, Roberts AJ, Ohkura R, Knight PJ, Gibbons IR, Burgess SA, Sutoh K. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat Struct Mol Biol. 2009;16:325–333. doi: 10.1038/nsmb.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev PV, Kachalova GS, Ryazanova AY, Kubareva EA, Karyagina AS, Bartunik HD, Svergun DI. Flexibility of the linker between the domains of DNA methyltransferase SsoII revealed by small-angle X-ray scattering: implications for transcription regulation in SsoII restriction-modification system. PLoS ONE. 2014;9:e93453. doi: 10.1371/journal.pone.0093453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn LA, Siani MA, Pique ME, Fisher CL, Getzoff ED, Tainer JA. The interdependence of protein surface topography and bound water molecules revealed by surface accessibility and fractal density measures. J Mol Biol. 1992;228:13–22. doi: 10.1016/0022-2836(92)90487-5. [DOI] [PubMed] [Google Scholar]
- Kuhn LA, Swanson CA, Pique ME, Tainer JA, Getzoff ED. Atomic and residue hydrophilicity in the context of folded protein structures. Proteins. 1995;23:536–547. doi: 10.1002/prot.340230408. [DOI] [PubMed] [Google Scholar]
- Lammens K, Bemeleit DJ, Möckel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermüller C, Söding J, Strässer K, Hopfner KP. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Mand MR, Deshpande RA, Kinoshita E, Yang SH, Wyman C, Paull TT. Ataxia telangiectasia-mutated (ATM) kinase activity is regulated by ATP-driven conformational changes in the Mre11/Rad50/Nbs1 (MRN) complex. Journal of Biological Chemistry. 2013;288:12840–12851. doi: 10.1074/jbc.M113.460378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lee KC, Padget K, Curtis H, Cowell IG, Moiani D, Sondka Z, Morris NJ, Jackson GH, Cockell SJ, Tainer JA, Austin CA. MRE11 facilitates the removal of human topoisomerase II complexes from genomic DNA. Biol Open. 2012;1:863–873. doi: 10.1242/bio.20121834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HS, Kim JS, Park YB, Gwon GH, Cho Y. Crystal structure of the Mre11-Rad50-ATPγS complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 2011;25:1091–1104. doi: 10.1101/gad.2037811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Moiani D, Kertokalio A, Wyman C, Tainer JA, Russell P. Mre11 ATLD17/18 mutation retains Tel1/ATM activity but blocks DNA double-strand break repair. Nucleic Acids Res. 2012;40:11435–11449. doi: 10.1093/nar/gks954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Tian LF, Liu YP, An XM, Tang Q, Yan XX, Liang DC. Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair. Acta Crystallogr D Biol Crystallogr. 2014;70:299–309. doi: 10.1107/S139900471302693X. [DOI] [PubMed] [Google Scholar]
- Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol. 2004;14:2107–2112. doi: 10.1016/j.cub.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Majka J, Alford B, Ausio J, Finn RM, McMurray CT. ATP hydrolysis by RAD50 protein switches MRE11 enzyme from endonuclease to exonuclease. Journal of Biological Chemistry. 2012;287:2328–2341. doi: 10.1074/jbc.M111.307041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makharashvili N, Tubbs AT, Yang SH, Wang H, Barton O, Zhou Y, Deshpande RA, Lee JH, Lobrich M, Sleckman BP, Wu X, Paull TT. Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol Cell. 2014;54:1022–1033. doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RM, Kerr M, Teo MTW, Jevons SJ, Koritzinsky M, Wouters BG, Bhattarai S, Kiltie AE. Post-transcriptional regulation of MRE11 expression in muscle-invasive bladder tumours. Oncotarget. 2014;5:993–1003. doi: 10.18632/oncotarget.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason AC, Rambo RP, Greer B, Pritchett M, Tainer JA, Cortez D, Eichman BF. A structure-specific nucleic acid-binding domain conserved among DNA repair proteins. Proc Natl Acad Sci USA. 2014;111:7618–7623. doi: 10.1073/pnas.1324143111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, Mosbaugh DW, Tainer JA. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell. 1995;82:701–708. doi: 10.1016/0092-8674(95)90467-0. [DOI] [PubMed] [Google Scholar]
- Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- Moncalian G, Lengsfeld B, Bhaskara V, Hopfner K-P, Karcher A, Alden E, Tainer JA, Paull TT. The rad50 signature motif: essential to ATP binding and biological function. J Mol Biol. 2004;335:937–951. doi: 10.1016/j.jmb.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- Möckel C, Lammens K, Schele A, Hopfner K-P. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 2012;40:914–927. doi: 10.1093/nar/gkr749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. Learning from exceptional drug responders. Nat Rev Drug Discov. 2014;13:401–402. doi: 10.1038/nrd4338. [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, Ira G, Sung P. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E, Nievera CJ, Lee AY-L, Chen L, Wu X. The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J Biol Chem. 2007;282:22939–22952. doi: 10.1074/jbc.M702162200. [DOI] [PubMed] [Google Scholar]
- Park YB, Chae J, Kim YC, Cho Y. Crystal Structure of Human Mre11: Understanding Tumorigenic Mutations. Structure/Folding and Design. 2011;19:1591–1602. doi: 10.1016/j.str.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, Classen S, Tomkinson AE, Tainer JA, Ellenberger T. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol Cell. 2006;24:279–291. doi: 10.1016/j.molcel.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Deshpande RA. The Mre11/Rad50/Nbs1 complex: Recent insights into catalytic activities and ATP-driven conformational changes. Exp Cell Res. 2014;329:139–147. doi: 10.1016/j.yexcr.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt AJ, Shin DS, Merz GE, Rambo RP, Lancaster WA, Dyer KN, Borbat PP, Poole FL, Adams MWW, Freed JH, Crane BR, Tainer JA, Getzoff ED. Aggregation propensities of superoxide dismutase G93 hotspot mutants mirror ALS clinical phenotypes. Proc Natl Acad Sci USA. 2014;111:E4568–76. doi: 10.1073/pnas.1308531111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J Mol Biol. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Tainer JA. Protein mimicry of DNA and pathway regulation. DNA Repair (Amst) 2005;4:1410–1420. doi: 10.1016/j.dnarep.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Rahal EA, Henricksen LA, Li Y, Williams RS, Tainer JA, Dixon K. ATM regulates Mre11-dependent DNA end-degradation and microhomology-mediated end joining. Cell Cycle. 2010;9:2866–2877. doi: 10.4161/cc.9.14.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr Opin Struct Biol. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013a;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annu Rev Biophys. 2013b;42:415–441. doi: 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- Redwine WB, Hernández-López R, Zou S, Huang J, Reck-Peterson SL, Leschziner AE. Structural basis for microtubule binding and release by dynein. Science. 2012;337:1532–1536. doi: 10.1126/science.1224151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt HC, Yaffe MB. Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat Rev Mol Cell Biol. 2013;14:563–580. doi: 10.1038/nrm3640. [DOI] [PubMed] [Google Scholar]
- Roberts VA, Freeman HC, Olson AJ, Tainer JA, Getzoff ED. Electrostatic orientation of the electron-transfer complex between plastocyanin and cytochrome c. J Biol Chem. 1991;266:13431–13441. [PubMed] [Google Scholar]
- Rojowska A, Lammens K, Seifert FU, Direnberger C, Feldmann H, Hopfner K-P. Structure of the Rad50 DNA double-strand break repair protein in complex with DNA. EMBO J. 2014;33:2847–2859. doi: 10.15252/embj.201488889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roset R, Inagaki A, Hohl M, Brenet F, Lafrance-Vanasse J, Lange J, Scandura JM, Tainer JA, Keeney S, Petrini JHJ. The Rad50 hook domain regulates DNA damage signaling and tumorigenesis. Genes Dev. 2014;28:451–462. doi: 10.1101/gad.236745.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller CB, Lammens K, Guerini I, Coordes B, Feldmann H, Schlauderer F, Möckel C, Schele A, Strässer K, Jackson SP, Hopfner K-P. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat Struct Mol Biol. 2012;19:693–700. doi: 10.1038/nsmb.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller CB, Seifert FU, Linke-Winnebeck C, Hopfner KP. Structural Studies of DNA End Detection and Resection in Homologous Recombination. Cold Spring Harbor Perspectives in Biology. 2014;6:a017962. doi: 10.1101/cshperspect.a017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys J. 2013;105:962–974. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K, Kurahashi K, Gao R, Tsutakawa SE, Tainer JA, Pommier Y, Aihara H. Structural basis for recognition of 5′-phosphotyrosine adducts by Tdp2. Nat Struct Mol Biol. 2012;19:1372–1377. doi: 10.1038/nsmb.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, Ismail A, Ismalaj E, Petricci E, Neale MJ, Bristow RG, Masson JY, Wyman C, Jeggo PA, Tainer JA. DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities. Mol Cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DS, Chahwan C, Huffman JL, Tainer JA. Structure and function of the double-strand break repair machinery. DNA Repair (Amst) 2004;3:863–873. doi: 10.1016/j.dnarep.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Shin DS, Pratt AJ, Tainer JA. Archaeal genome guardians give insights into eukaryotic DNA replication and damage response proteins. Archaea. 2014;2014:206735. doi: 10.1155/2014/206735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau A, Robellet X, Ladouceur AM, D’Amours D. Cdk1-dependent regulation of the Mre11 complex couples DNA repair pathways to cell cycle progression. Cell Cycle. 2014;13:1078–1090. doi: 10.4161/cc.27946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Li F, Park YB, Kim JS, Kim AK, Song OK, Kim J, Che J, Lee SE, Cho Y. DNA end recognition by the Mre11 nuclease dimer: insights into resection and repair of damaged DNA. EMBO J. 2014;33:2422–2435. doi: 10.15252/embj.201488299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS. End Resection at Double-Strand Breaks: Mechanism and Regulation. Cold Spring Harbor Perspectives in Biology. 2014;6:a016436. doi: 10.1101/cshperspect.a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tainer JA, Getzoff ED, Alexander H, Houghten RA, Olson AJ, Lerner RA, Hendrickson WA. The reactivity of anti-peptide antibodies is a function of the atomic mobility of sites in a protein. Nature. 1984;312:127–134. doi: 10.1038/312127a0. [DOI] [PubMed] [Google Scholar]
- Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, Cooper PK, Grasby JA, Tainer JA. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Lafrance-Vanasse J, Tainer JA. The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once. DNA Repair. 2014;19:95–107. doi: 10.1016/j.dnarep.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Shin DS, Mol CD, Izumi T, Arvai AS, Mantha AK, Szczesny B, Ivanov IN, Hosfield DJ, Maiti B, Pique ME, Frankel KA, Hitomi K, Cunningham RP, Mitra S, Tainer JA. Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes. Journal of Biological Chemistry. 2013;288:8445–8455. doi: 10.1074/jbc.M112.422774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noort J, van Der Heijden T, de Jager M, Wyman C, Kanaar R, Dekker C. The coiled-coil of the human Rad50 DNA repair protein contains specific segments of increased flexibility. Proc Natl Acad Sci USA. 2003;100:7581–7586. doi: 10.1073/pnas.1330706100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li Y, Truong LN, Shi LZ, Hwang PYH, He J, Do J, Cho MJ, Li H, Negrete A, Shiloach J, Berns MW, Shen B, Chen L, Wu X. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol Cell. 2014;54:1012–1021. doi: 10.1016/j.molcel.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Goldstein M, Alexander P, Wakeman TP, Sun T, Feng J, Lou Z, Kastan MB, Wang X-F. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. EMBO J. 2014;33:862–877. doi: 10.1002/embj.201386064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GJ, Hammel M, Radhakrishnan SK, Ramsden D, Lees-Miller SP, Tainer JA. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst) 2014;17:110–120. doi: 10.1016/j.dnarep.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 2010;9:1299–1306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GJ, Williams RS, Williams JS, Moncalian G, Arvai AS, Limbo O, Guenther G, SilDas S, Hammel M, Russell P, Tainer JA. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat Struct Mol Biol. 2011;18:423–431. doi: 10.1038/nsmb.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JNM, Iwasaki H, Russell P, Tainer JA. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Tainer JA. A nanomachine for making ends meet: MRN is a flexing scaffold for the repair of DNA double-strand breaks. Mol Cell. 2005;19:724–726. doi: 10.1016/j.molcel.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Wiltzius JJW, Hohl M, Fleming JC, Petrini JHJ. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat Struct Mol Biol. 2005;12:403–407. doi: 10.1038/nsmb928. [DOI] [PubMed] [Google Scholar]
- Yannone SM, Hartung S, Menon AL, Adams MWW, Tainer JA. Metals in biology: defining metalloproteomes. Curr Opin Biotechnol. 2012;23:89–95. doi: 10.1016/j.copbio.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Ma Y, Tang S, Hwang PYH, Boiteux S, Hunter N. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol Cell. 2010;40:1001–1015. doi: 10.1016/j.molcel.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S, Boboila C, Alt FW. Mre11: roles in DNA repair beyond homologous recombination. Nat Struct Mol Biol. 2009;16:798–800. doi: 10.1038/nsmb0809-798. [DOI] [PubMed] [Google Scholar]
- Zhao W, Saro D, Hammel M, Kwon Y, Xu Y, Rambo RP, Williams GJ, Chi P, Lu L, Pezza RJ, Camerini-Otero RD, Tainer JA, Wang H-W, Sung P. Mechanistic insights into the role of Hop2-Mnd1 in meiotic homologous DNA pairing. Nucleic Acids Res. 2014;42:906–917. doi: 10.1093/nar/gkt924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]