Abstract
Recent next-generation sequencing studies have generated a comprehensive overview of the genomic landscape of Human Papillomavirus (HPV)-associated cancers. This review summarizes these findings to provide insight into the tumor biology of these cancers and potential therapeutic opportunities for HPV-driven malignancies. In addition to the tumorigenic properties of the HPV oncoproteins, integration of HPV DNA into the host genome is suggested to be a driver of the neoplastic process. Integration may confer a growth and survival advantage via enhanced expression of viral oncoproteins, alteration of critical cellular genes, and changes in global promoter methylation and transcription. Alteration of cellular genes may lead to loss of function of tumor suppressor genes, enhanced oncogene expression, loss of function of DNA repair genes, or other vital cellular functions. Recurrent integrations in RAD51B, NR4A2, and TP63, leading to aberrant forms of these proteins, are observed in both HPV-positive head and neck squamous cell carcinoma (HNSCC) and cervical carcinoma. Additional genomic alterations, independent of integration events, include recurrent PIK3CA mutations (and aberrations in other members of the PI3K pathway), alterations in receptor tyrosine kinases (primarily FGFR2 and FGFR3 in HPV-positive HNSCC, and ERBB2 in cervical squamous cell carcinoma), and genes in pathways related to squamous cell differentiation and immune responses. A number of the alterations identified are potentially targetable, which may lead to advances in the treatment of HPV-associated cancers.
Introduction
Late in 1979 Lutz Gissmann and Ethel-Michele de Villiers, working in the lab of Harald zur Hausen, successfully isolated and cloned the first Human Papillomavirus (HPV) DNA from genital warts: HPV-6. HPV-11 was cloned shortly thereafter from a laryngeal papilloma. The German research group hypothesized that HPV was the causative agent in cervical cancer. By using HPV-11 as a probe, one out of 24 cervical cancer biopsies was found to be positive. In addition, several of the other biopsies yielded faint bands, allowing speculation that these might represent the presence of related HPV types. Only a few years later, in 1983, the group isolated HPV-16 DNA, and in 1984, HPV-18 DNA, which they noted were present in about 50% and 20% of cervical cancer biopsies, respectively, as well as in several cervical cancer cell lines. Harald zur Hausen received the Nobel Prize in Physiology or Medicine in 2008 for his group’s groundbreaking discovery.
Now, over 30 years later, HPV is known to be the etiologic agent in cervical cancer, as well as in a significant proportion of anogenital cancers and head and neck squamous cell carcinoma (HNSCC) cases (in particular tonsillar and base of tongue carcinomas) (1, 2). HPV is further responsible for a variety of benign neoplasms, such as genital warts, oral papillomas, and recurrent respiratory papillomatosis. Over 150 HPV types have been identified and classified into low-risk and high-risk based on their malignant potential. The predominant high-risk type identified in cervical, anogenital, and head and neck carcinomas is HPV16.
HPV infects epithelial tissue and depends on epithelial differentiation for completion of its lifecycle (3, 4). The molecular biology of HPV during its normal life cycle and in carcinogenesis is described in several recent reviews (3, 5–9). HPV is known to drive tumorigenesis in particular through the actions of the oncoproteins E6 and E7 (10–12). These target numerous cellular pathways, such as p53 and pRB, to promote cellular immortalization, thus providing an environment amenable to viral replication. Furthermore, the virus has adapted multiple mechanisms to evade the host immune response. These include expression of viral proteins at high levels only in the upper epithelial layers where immune surveillance is limited and non-lytic release of virions without significant viraemia, through the natural epithelial shedding process. HPV further hampers the immune system by hindering Langerhans cell migration (13, 14) and activation (15), by suppressing the interferon (IFN) response (16–18), and by interfering with HLA class 1-mediated antigen presentation (19). Persistent infection with HPV leads to an environment of genomic instability and local immune suppression, which can lead to both the accumulation of genomic alterations in the host cell, as well as to the integration of the viral genome into the host genome. When these additional alterations provide a selective growth advantage to the cell, carcinogenesis may ensue.
Recent genome-wide studies (20–24) using next-generation sequencing techniques (whole genome/exome sequencing, RNA-Seq, miRNA-Seq) and methylation analyses, have described the genomic and epigenomic alterations of HPV-associated cancers. These comprehensive studies have generated novel information about how HPV integration may drive genomic instability and the progression from viral infection to cancer, as well as highlighted genomic aberrations that may be targetable in the treatment of HPV-associated cancers. This review summarizes the recent literature concerning the genomic landscape of HPV-associated cancers, and the interactions between HPV and the host-genome in cancer.
Characterization of HPV Integrations
During an infection HPV genomes are found in the nucleus as episomes (circular, extrachromosomal DNA). Integration of the viral genome, or fragments thereof, into the host genome has been noted in the majority of high-grade cervical lesions and cancers (25–28). Thus it is believed that integration occurs relatively late in the progression to high-grade cervical dysplasia. Integration has also been noted in a significant proportion of HPV-positive HNSCCs (22, 29). It has been suggested that integration disrupts the E2 open reading frame causing upregulated expression of the E6 and E7 oncoproteins (30) (E2 normally suppresses their expression (31)). Furthermore, the integrated viral transcripts confer stronger transforming capacity than those derived from episomes, due to longer half-life of transcripts (32). This promotes immortalization and transformation of these cells, and provides a selective growth advantage (3, 8, 30, 33). Integration may, however, confer a selective growth advantage to the host cells not only through its effects on the viral genome (i.e., enhanced/deregulated expression of viral oncoproteins), but also through its effects on the host genome (i.e., by affecting key cellular genes).
Akagi and colleagues (23) undertook an analysis of a panel of ten cervical and head and neck cancer cell lines (five HPV16-positive and three HPV-negative HNSCC lines, and two HPV-positive cervical lines), as well as two HPV-positive primary HNSCC samples. The HNSCC HPV-positive lines were from the following anatomic sites: oral cavity (n=1), hypopharynx (n=1), tongue (n=2), and tonsillar fossa (n=1). The two primary samples were from an oral cavity carcinoma and a tonsillar carcinoma. Samples were analyzed by whole-genome sequencing, RNA-seq, spectral karyotyping, fluorescence in situ hybridization (FISH), and other molecular assays. The majority of the lines and the two primary samples had less than 10 breakpoints in the host genome. Two of the cancer cell lines, however, had a high number of breakpoints (CaSki, a cervical carcinoma line, had 47, and UPCI:SCC090, a tongue SCC line, had 33). Akagi et al. found that breakpoints occurred throughout the viral genome, frequently fragmenting the viral genome and leading to loss of viral genes. In four out of nine cell lines E2 was missing. E6 and E7 were, however, retained and expressed in all cases. Of note, E6 and E7 were amplified within the viral-host concatemers in most samples.
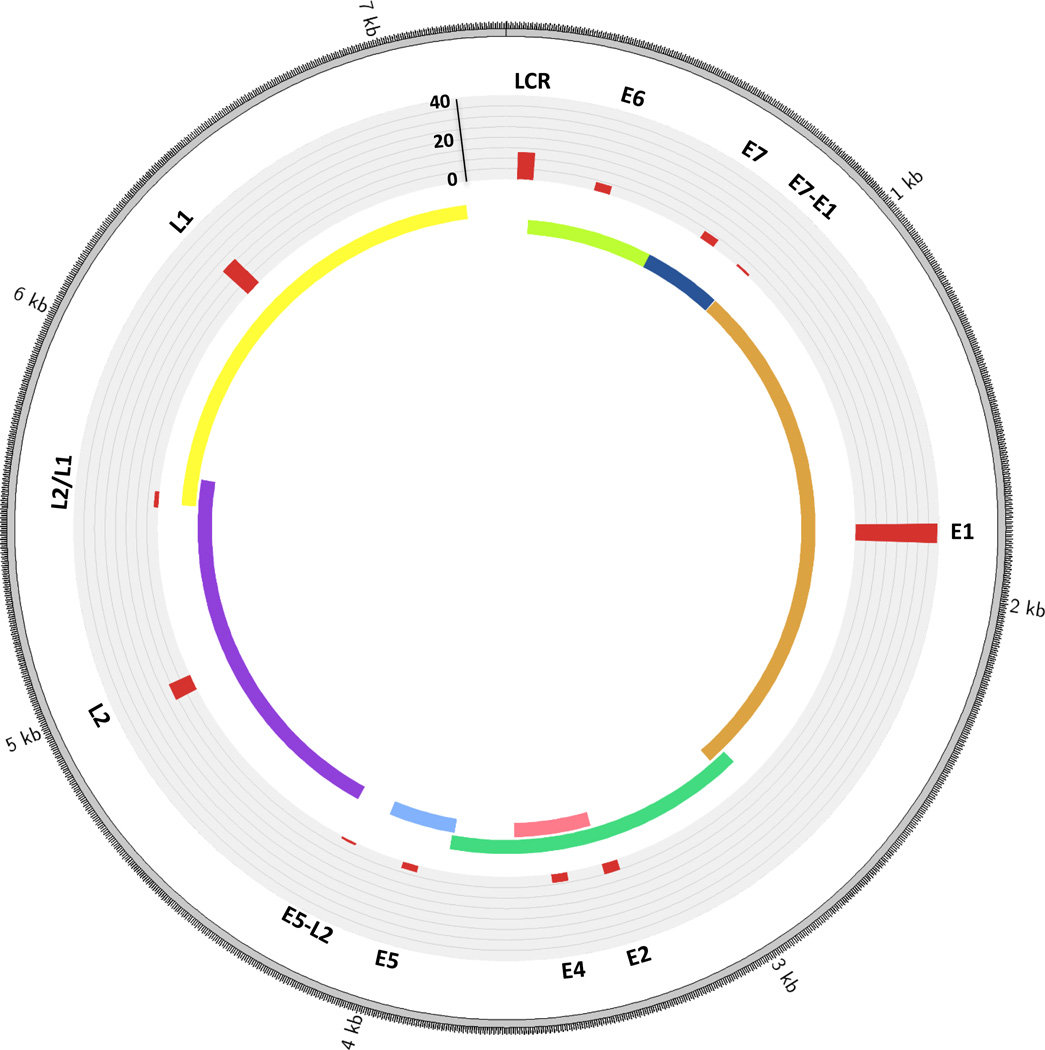
Parfenov and colleagues performed a comprehensive, genome-wide analysis of 35 primary HPV-positive HNSCCs, considering the effect of integrations on the structure of the host genome, RNA expression, and the epigenome (22). Twenty-nine of the 35 tumors (83%) had HPV16, and the remainder had HPV33 or HPV35. Twenty-five of the 35 cases (71%) had integrated HPV DNA into anywhere from one to 16 regions of the human genome (21 cases had HPV16, three HPV33 and one HPV35). Thirteen cases were from the oropharynx, ten from the oral cavity, and two from the larynx. In line with the findings of Akagi et al. (23) as well as studies in cervical cancer (34), the breakpoints mapped broadly across the viral genome, however occurred with higher observed frequency in E1 (Fig. 1). The observed breakpoints in the viral genome were nonrandom, as they were higher than expected by chance in E1, and in all but one case E6 and E7 were intact. Tumors with an integration event were associated with lower levels of HPV E2, E4 and E5, and higher levels of HPV E6 and E7 expression, compared to integration-negative tumors. However, these results, along with those of others (35), suggest that E2 ORF disruption is not mandatory for enhanced viral oncogene expression, and that E2 may be downregulated by additional mechanisms. It is important to note that not all tumors with integrated HPV showed enhanced expression of viral E6 and E7 oncoproteins, indicating that elevated levels may not be essential for the development of cancer. This suggests that HPV integration, or additional genetic alterations, can drive carcinogenesis independently of enhanced E6 and E7 expression, and that in certain cases, constitutive rather than enhanced expression of E6/E7 is sufficient for cancer development.
Figure 1.
Distribution of breakpoints across the HPV genome. The histogram (in red) indicates the number of tumors with a breakpoint in that particular gene. L2/L1 indicates a region of overlap between L2 and L1. E7-E1 refers to the area between the E7 and E1 genes, and likewise for E5-E2. Counts are based on data from the 25 HPV-positive HNSCC primary tumors with integrations analyzed by Parfenov et al. (22). Please note the HPV16 genome is depicted here, however three of the tumors had HPV33 and one had HPV35 (the structure of these is highly similar to HPV16).
Interestingly, breakpoints in a specific HPV gene did not correlate with that gene’s expression level in the samples studied by Parfenov et al. (22). This may be due to expression from intact HPV copies in the sample. Expression of viral genes post-integration may also be influenced by nearby cellular regulatory sequences (36). Conversely, in cervical cancer samples, Ojesina et al. (24) observed elevated host gene expression levels at sites of integration compared to expression levels of the same genes in tumors without integration. This was associated, in a proportion of the cases, with copy-number gains, but not at all sites, indicating that expression may be partly driven by the integrated viral promoter(s) at some sites (24).
Integrations occur throughout the human genome in both HNSCC (22, 23) and cervical cancer (23, 24, 34) (Fig. 2). Parfenov and colleagues noted, however, that this is often in regions of microhomology (1–10 bp) among the viral and host genome, and most frequently into genic regions and miRNA regions. In 54% of cases the virus integrated into a known gene and in 17% within 20 kb of a gene. Similarly, Akagi and colleagues observed enrichment of HPV integrants within 50 kb of RefSeq genes. In addition, several studies have found that HPV integrates within, or close to, fragile sites (23, 24, 34).
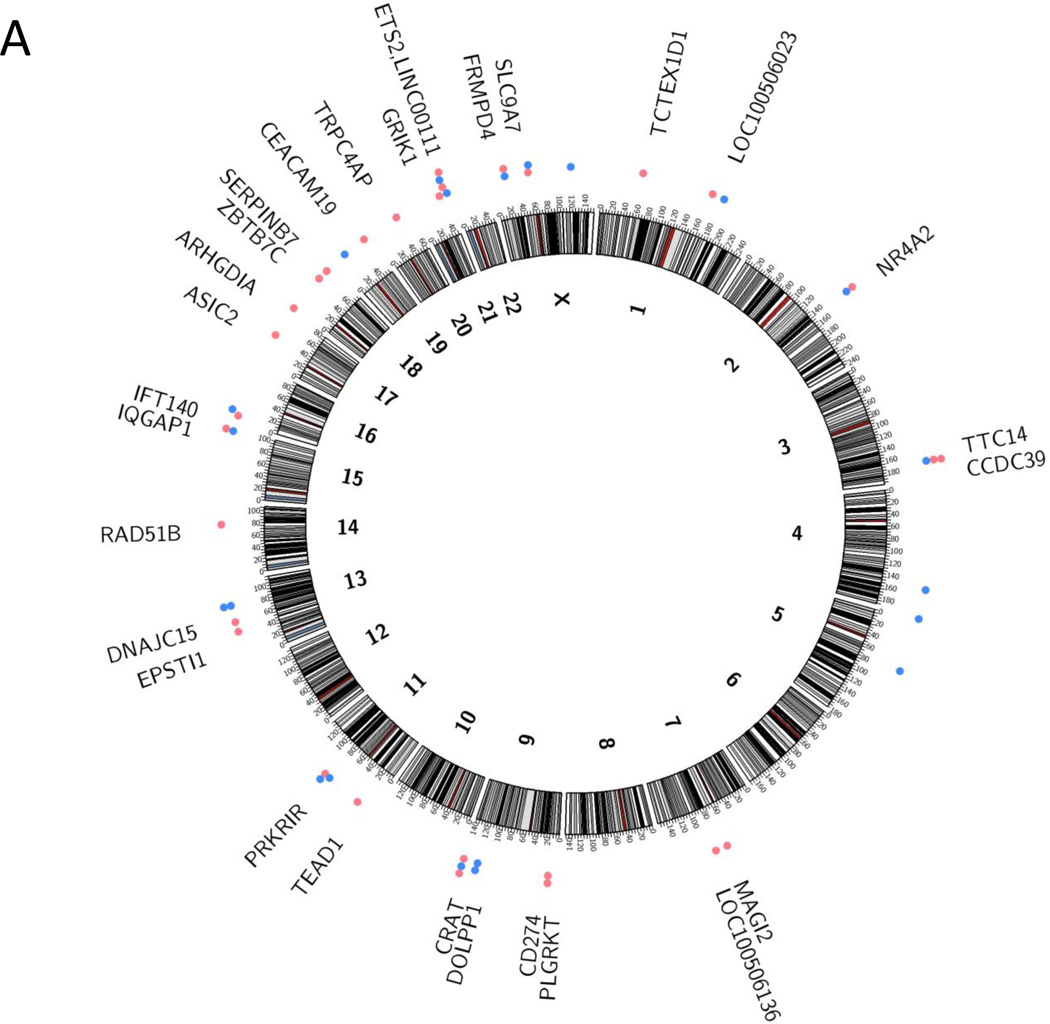
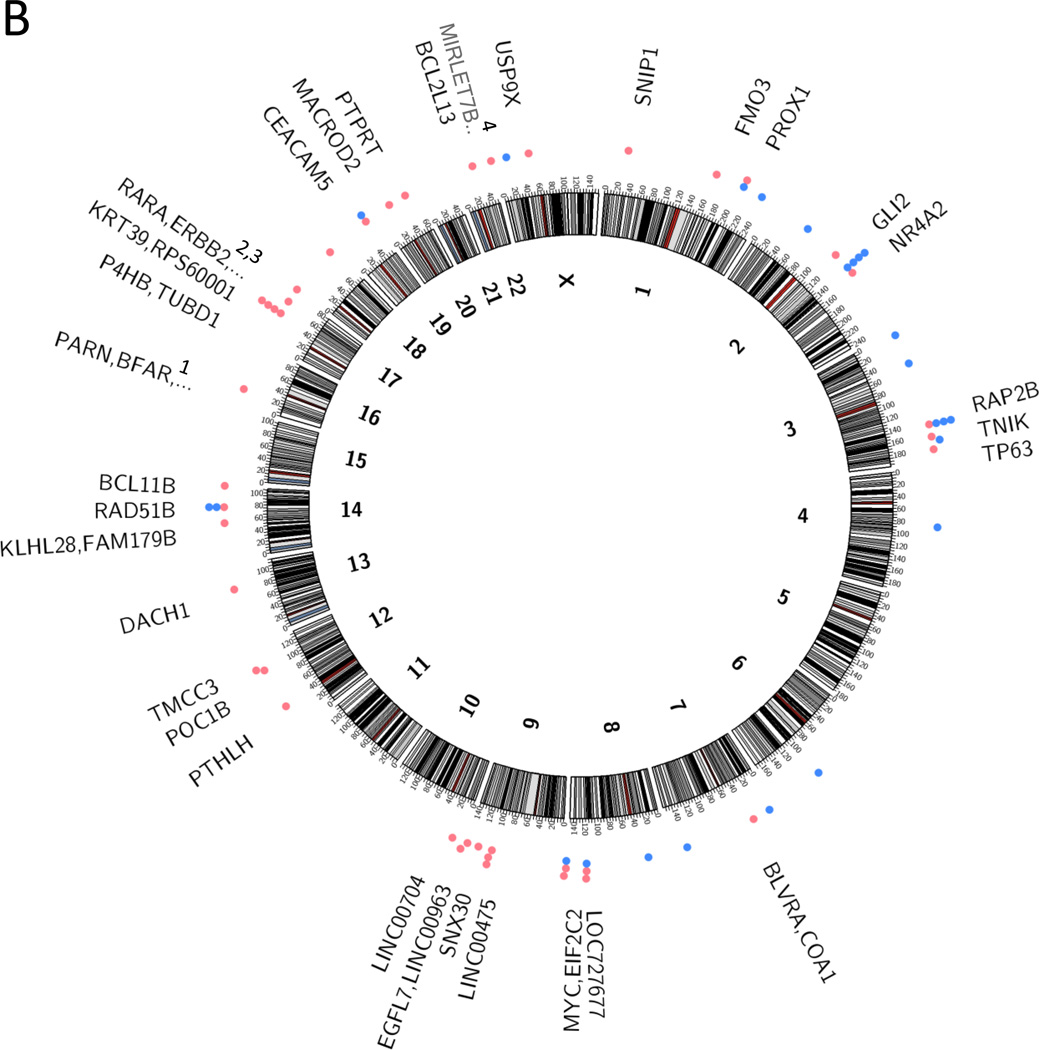
Figure 2.
Integration sites of HPV into the human genome. a) Integration sites in head and neck squamous cell carcinomas, based on data from the HPV-positive tumors analyzed by Parfenov et al. (22), and b) Integration sites in cervical carcinomas, based on data from Ojesina et al. (24). In both panels, integrations into coding regions are represented by red dots, and noncoding regions by blue dots. If a tumor had multiple insertions at the same locus it is only represented once in the diagram. Several cases had multiple genes in the region involved in the integration event:
a PARN, BFAR, PLA2G10
b ERBB2, STARD3, TCAP, PNMT, PGAP3, C17orf37, GRB7, IKZF3
c ERBB2, C17orf37, GRB7
d MIRLET7B, MIRLET7BHG, MIRLET7A3, MIR4763
Interestingly, several studies have noted that HPV integrations colocalize with somatic copy number variants, including focal amplifications, deletions, intra- and interchromosomal translocations (22–24). Akagi et al. found HPV integrants at regions of amplification (ranging from a 1.5-fold increase in HMS001 cells to a 58-fold increase in UPCI:SCC090 cells), as well as regions with deletions (spanning from 487 bp in HMS001 to 234 kb in chromosome 3 of UPCI:SCC090). They further observed that HPV insertional breakpoints frequently clustered together. Akagi et al. suggested a looping model to explain the amplifications and rearrangements noted at integration sites. In this model there is nicking of the host genome, integration of the linear HPV genome, transient formation of circular DNA containing both host and viral sequences, rolling circle amplification of this template and formation of integrated concatemers of viral-host sequences. Parfenov et al. further noted amplification events that were suggestive of excision, subsequent circularization of the integrated virus and adjacent human sequences, and maintenance of the fused viral-host genome as an episome. Both Parfenov et al. (22) and Akagi et al. (23) noted the expression of virus-host fusion transcripts.
Parfenov et al. (22) further considered whether integration was associated with clinical outcome or other clinical features (anatomic site, tumor stage, age, smoking status), but did not find any statistically significant associations. However, the sample size was quite limited. In cervical cancer patients treated with radiotherapy, Shin et al. (37) found a trend towards decreased disease-free survival in patients with only HPV integrated forms vs. patients with both integrated and episomal HPV. Further research is needed to elucidate the relationship between HPV physical status (integrated vs. episomal vs. mixed) and clinical outcome in both HNSCC and cervical cancer.
Deregulation of Key Cellular Genes by HPV Integration
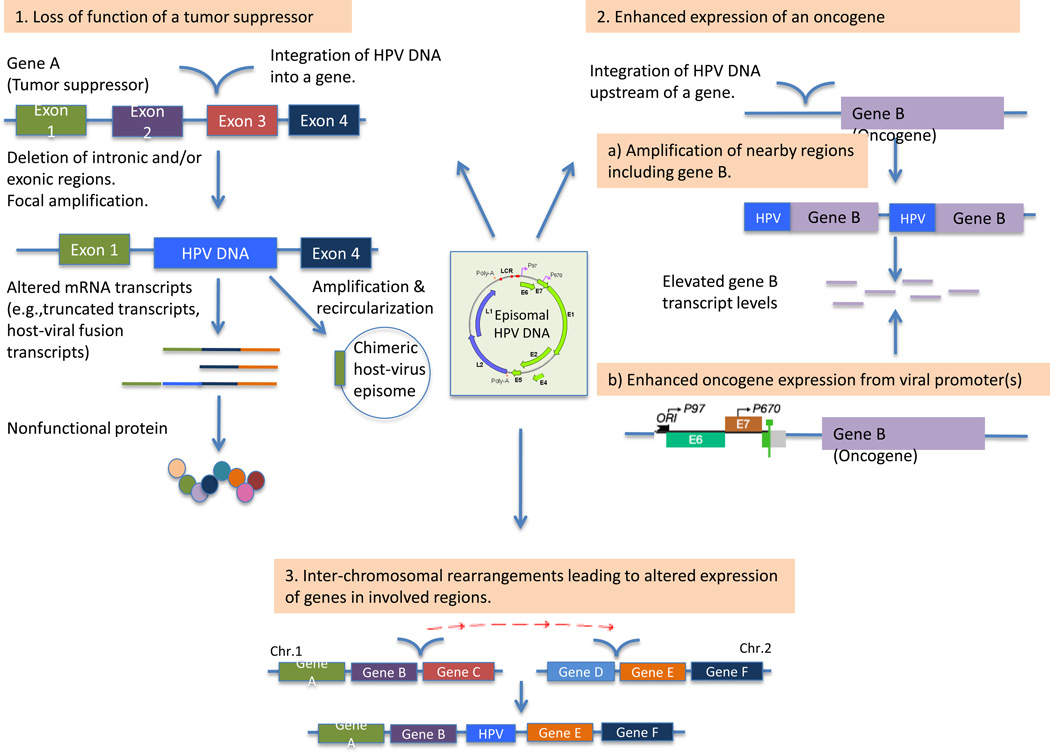
Several mechanisms by which HPV integration may confer a selective advantage have been described (Fig. 3). The first of these is loss of function by integration into a gene. Parfenov and colleagues (22), for instance, identified three integration events, in the same primary HNSCC tumor sample, into intron 8 of the RAD51B gene on chromosome 14. RAD51B is a component of the DNA double-strand break repair pathway, and loss of this gene may promote genomic instability. The integration resulted in a 28-fold amplification of a 42 kb segment of intron 8 along with parts of the viral genome. The chimeric construct was circularized, and alternative RAD51B transcripts were expressed that were likely nonfunctional. Intriguingly, integrations in this gene have also been noted in multiple cervical cancer samples by Ojesina et al. (24) related to HPV16, HPV 18 and HPV52 integrations, and by Khoury et al. in HNSCC samples (38). Similarly, integration has been described into ETS2, a tumor suppressor gene, with deletion of exon 7 and 8 at the integration site, resulting in truncated forms of the ETS2 protein (22).
Figure 3.
Mechanisms by which integration may lead to the deregulation of key cellular genes. The figure highlights mechanisms by which integration of HPV DNA into the host genome may lead to alteration of critical cellular genes. These include: (1) disruption of a tumor suppressor gene, (2a) by amplification of an oncogene, or (2b) by enhanced expression of an oncogene from a viral promoter. Integration may also cause (3) more extensive intra- or inter-chromosomal rearrangements, resulting in altered expression of multiple genes in the involved regions.
A second mechanism by which integration may lead to deregulation of key cellular genes is by amplification and subsequent over-expression of these genes. In one example HPV integrated upstream of the NR4A2 oncogene, resulting in a 284-fold amplification of a 75 kb genomic region encompassing the NR4A2 gene, and overexpression of NR4A2. Interestingly, this tumor exhibited low levels of E6 and E7, suggesting that other factors were important for tumorigenesis in this case. Integration near NR4A2 was also noted in one cervical sample by Ojesina et al (24). Additional examples include amplification of the oncogenes FOXE1 and PIM1 in UPCI:SCC090 cells (tongue SCC), and the solute carrier, SLC47A2, in UM-SCC-104 cells (oral cavity SCC) (23). In cervical cancer, several cases of integration near or within the MYC gene locus have also been described (24, 39–41).
Lastly, HPV insertion is associated with intra- and interchromosomal rearrangements. Parfenov et al. described one HNSCC case where there was a rearrangement between chromosomes 3 and 13 near the site of integration. The integration was in a nongenic region, however the translocation involved a region of chromosome 3 containing tumor protein p63 regulated 1 (TPRG1) and TP63, and on chromosome 13 the Krüppel-like factor 5 (KLF5) gene. The regions involved in the rearrangement were amplified and led to increased expression of KLF5, TP63, and TPRG1. Of note, KLF5 is a transcription factor known to regulate proliferation and has been implicated in a number of cancer types (42). TP63 is a transcription factor, with an important role in epithelial development, and which has been implicated as an important oncogene in squamous cell cancers (43, 44). The function of TPRG1 is not well characterized. Aberrant expression of TP63 secondary to HPV integration was also noted in UM-SCC-47 cells (23) and in cervical cancer (24). Fig. 2 provides an overview of the integration sites into the human genome identified in recent studies of HNSCC and cervical cancer.
Interestingly, Parfenov et al. (22) also showed that DNA methylation profiles are distinct for HPV-positive tumors with integration compared to those without integration. Some of the differentially methylated genes were the tumor suppressors BARX2 and IRX4, and the oncogenes SIM2 and CTSE. The mechanism by which integration alters the methylation profile remains to be elucidated.
The co-localization of HPV integrations with alterations that may lead to loss or gain of function in key cancer genes, in particular the presence of recurrent integration in specific genes, highly suggests that integration contributes to tumorigenesis. Further work is, however, needed to more fully characterize and validate the impact of HPV integration on these cellular genes to gain a deeper understanding of the cancer biology in these cases.
Additional Genomic Alterations in HPV-Associated Cancers
Additional genomic alterations, not associated with HPV integration events, have been described in HPV-driven cancers. These are believed to contribute to tumor development.
HNSCC
Several genome-wide studies of HNSCC have suggested that HPV-driven cases display less genomic complexity compared to HPV-negative cases, which is associated with excessive smoking and alcohol consumption (45). A comprehensive study of 279 HNSCC tumors by the Cancer Genome Atlas group did not confirm this finding (36 of the tumors were HPV-positive, composed of 21 oropharyngeal, 12 oral cavity, one laryngeal, and one hypopharyngeal carcinoma). However, the majority of HPV-positive patients in the study were also smokers, and displayed CpG transversions, a mutation class typically associated with smoking (along with the expected virus-associated Tp*Cp (A/C/T) substitution mutations). Seiwert et al. have shown that smokers (both HPV-negative and HPV-positive) display a higher mutational burden (21). Although HPV-positive status confers a favorable prognosis, patients with >10 pack-year smoking history have a poorer prognosis (46). The increased genomic complexity, the presence of unfavorable alterations (i.e. p53 mutations), as well as changes to the immune environment, in tumors associated with smoking may contribute to the poorer prognosis.
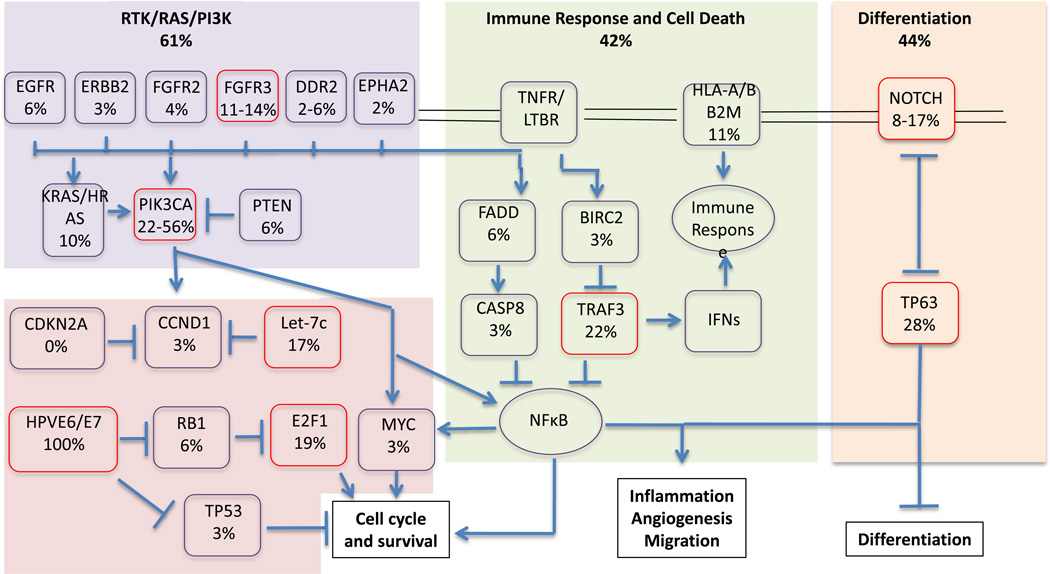
Both HPV-positive and negative HNSCC tumors have been shown to have recurrent focal amplifications of 3q26/28, which includes factors involved in squamous lineage transcription, such as TP63 and SOX2, as well as the oncogene, PIK3CA (20, 21) (Fig. 4). In addition to amplification of PIK3CA, mutations in PIK3CA have been also been found to be enriched in HPV-positive HNSCC in a number of studies (20, 21, 47, 48). Importantly, PIK3CA alterations have been reported to be potential therapeutic biomarkers in this patient population (48). In addition, KRAS, NRAS and HRAS alterations have been reported in about 10% of cases (21). These alterations converge upon NF-kB transcription factors that promote cell survival, migration, inflammation and angiogenesis. Of note, RAS mutations have been associated with poor outcome in other cancer types (49), as well as resistance to anti-EGFR therapies in non-small cell lung cancer and colorectal cancer (50). The clinical implications of KRAS mutations in HPV-positive tumors are currently unknown.
Figure 4.
Signaling pathways deregulated in HPV-associated HNSCC. Red boxes highlight the most frequently altered components. Pathway alterations include homozygous deletions, focal amplifications and somatic mutations. Data is based on results from TCGA (20) and Seiwert and colleagues (21).
FGFR2 and FGFR3 mutations have been identified among 17.6% of HPV-positive tumors. FGFR2 mutations include N569D and N569K mutations, and FGFR3 has primarily S249C mutations. Both FGFR2 N569K and FGFR3 S249C have been described in several cancer types, and are sensitive to FGFR inhibitors (51, 52). FGFR-TACC3 fusions, previously identified in glioblastoma (53) and bladder cancer (54), have also been identified in HPV-positive HNSCC.
In addition, TNF-receptor associated factor 3 gene (TRAF3) deletions and truncating mutations have been described in HPV-associated HNSCCs (20). TRAF3 is involved in innate and adaptive antiviral responses. Loss of TRAF3 promotes aberrant NF-kB signaling, and has been associated with hematologic malignancies and nasopharyngeal carcinomas (55, 56). Other immune response genes (i.e. HLA-A, HLA-B) were also altered in HNSCC (21).
Furthermore, genes involved in DNA-repair (BRCA1, BRCA2, ATM, FANCG, FANCA, FANCD2, RAD51B) are altered in HPV-positive HNSCC (21). Of note, RAD51B has also been reported as an integration target by multiple studies (22, 24). These alterations, including RAD51B, have also been described in patients with Fanconi anemia. These are patients who are at extreme risk of developing squamous cell carcinomas (57, 58). Alterations in DNA-repair genes have been suggested to contribute to the chemo- and/or radiosensitivity of HPV-positive tumors (21).
HPV-positive HNSCC shares many common altered genes and pathways with HPV-negative HNSCC (eg. NOTCH, MLLs, RAS, TP63) (20, 21). Many of these alterations have also been observed in HPV-positive HNSCC cell lines (23). However, unlike in HPV-negative HNSCC, TP53 and CDKN2A are intact in the majority of HPV-positive HNSCC (20–23). Table 1 provides an overview of frequently altered genes in HPV-positive HNSCCs.
Table 1. Genes with recurrent somatic mutations in HPV-associated HNSCC and cervical carcinomas.
The approximate frequency with which the genes are mutated in HNSCC is based on data from Seiwert et al. (21) and TCGA (20), and for the cervical carcinomas on data from Ojesina et al. (24).
| Gene | Description | Approximate frequency (%) |
|---|---|---|
| HPV-positive HNSCC | ||
| PIK3CA | Phosphatidylinosital-4,5-bisphosphate 3-kinase, catalytic subunit alpha | 22–56 |
| TRAF3 | TNF receptor-associated factor 3 | 22 |
| TP63 | Tumor protein p63 | 28 |
| FGFR3 | Fibroblast growth factor receptor 3 | 11–14 |
| MLL3 | Lysine (K)-specific methyltransferase 2C | 10 |
| MLL2 | Lysine (K)-specific methyltransferase 2B | 10 |
| FLG | Filaggrin | 12 |
| NOTCH1 | Notch 1 | 8–17 |
| DDX3X | DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked | 8 |
| KRAS | Kirsten rat sarcoma viral oncogene homolog | 6 |
| CYLD | Cylindromatosis (turban tumor syndrome) | 6 |
| EGFR | Epidermal growth factor receptor | 6 |
| PTEN | Phosphatase and tensin homolog | 6 |
| DDR2 | Discoidin Domain Receptor 2 | 2–6 |
| Cervical Squamous Cell Carcinoma | ||
| EP300 | E1A binding protein p300 | 16 |
| FBXW7 | F-box and WD repeat domain containing 7 | 15 |
| PIK3CA | Phosphatidylinosital-4,5-bisphosphate 3-kinase, catalytic subunit alpha | 14 |
| HLA-B | Major histocompatibility complex, class I, B | 9 |
| TP53 | Tumor protein p53 | 9 |
| MAPK1 | Mitogen-activated protein kinase 1 | 8 |
| PTEN | Phosphatase and tensin homologue | 6 |
| ERBB2 | V-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 | 5 |
| STK11 | Serine/threonine kinase 11 | 4 |
| NFE2L2 | Nuclear factor, erythroid 2-like 2 | 4 |
| Cervical Adenocarcinoma | ||
| ELF3 | E74-like factor 3 | 13 |
| CBFB | Core binding factor, beta subunit | 8 |
Cervical cancer
A recent study employing whole-exome sequencing and transcriptome sequencing in cervical carcinomas found a different pattern of recurrent mutations in cervical squamous cell carcinomas compared to adenocarcinomas (24) (Table 1). Squamous cell carcinomas were found to have a higher rate of nonsilent mutations than adenocarcinomas (4.2 mutations vs. 1.6 mutations per Mb). Similarly to HPV-associated HNSCC, PIK3CA was found to be frequently mutated in cervical squamous cell carcinomas. TP53 and PTEN mutations, which are frequently observed in HPV-negative HNSCC, were also identified in cervical squamous cell carcinomas. In addition, recurrent mutations were found in EP300, FBXW7, HLA-B, MAPK1, ERBB2, STK11 (also known as LKB1), and NFE2L2 in squamous cell carcinomas, and ELF3 and CBFB in adenocarcinomas (Table 1). The majority of these have been implicated in other cancers as well, for instance EP300 and FBXW7 mutations have been identified in both endometrial and head and neck cancers (59, 60), STK11 in lung cancers (61, 62), and ERBB2 in breast (63), gastric, esophageal (64), and lung cancers (65). Interestingly, as in HNSCC, there were mutations in genes involved in antigen presentation, such as HLA-A and beta2-microglobulin, and other immune response genes such IFNγ and JAK2, suggesting that these alterations may synergize with HPV infection in the pathogenesis of squamous cell carcinomas (24).
Clinical Perspectives
Next-generation sequencing techniques for determination of HPV status
HPV-positive oropharyngeal squamous cell carcinoma has a significantly better prognosis independent of stage at diagnosis compared to HPV-negative oropharyngeal carcinoma (46, 66–71). The latter is primarily associated with heavy tobacco and alcohol exposure. HPV status in non-oropharyngeal HNSCC, including cancers of the oral cavity, hypopharynx and larynx, has not been clearly associated with a similar improved prognosis, suggesting that either HPV may not be playing a major role in pathogenesis of HNSCC outside of the oropharynx or that other features are more important for prognosis at these anatomic sites (72–75). Given the relation to prognosis it is essential to correctly identify HPV-positive cases.
Multiple methods for determining HPV status are available. Immunohistochemistry for p16 expression is used by many centres as a surrogate marker of HPV infection and a prognostic biomarker, as it is a simple and inexpensive assay. The gold standard is detection of E6/E7 mRNA, however this may be less sensitive depending on the quality of the clinical sample. The use of p16 alone may misclassify a small subset of tumors in which HPV is present and p16 expression has been lost by an independent mechanism. This is of particular relevance in patients that have both HPV and a positive smoking or alcohol abuse history, as p16 may be mutated in these patients. The prognosis of this subgroup of patients requires further examination. Similarly multiple HPV-positive patients (with and without HPV integration) identified in Parfenov et al. (22) had low expression of or absence of E6/E7 expression, and these may be misclassified by E6/E7 mRNA detection.
Next-generation techniques, as described herein, can also detect HPV with high sensitivity (20–23, 76), and has further been suggested to be useful for studying HPV-variant epidemiology (76). As these next-generation sequencing methods become increasingly applied in the clinic it will be important to further define the sensitivity and specificity of these methods for HPV detection, and to define which are optimal for clinical use. A discussion of the different methods and their advantages and limitations, as relating to their clinical use, is provided in a number of recent reviews (77, 78). Restricted gene expression and mutation profiling for alterations with well-described clinical significance is most clinically feasible at present. These methods have recently been shown to be applicable also to paraffin embedded tissue (79) widening the applicability and affordability of these methods.
Genomic landscape of HPV-associated cancers
The studies discussed in the current review have generated considerable insight into the genomic landscape of HPV-associated cancers. Nevertheless, the sample size examined to date remains small for both HNSCC and cervical cancer, and similar studies for other HPV-associated cancers, such as anal, penile and vulvar cancers have yet to be performed. It is important to emphasize the heterogeneity of HPV-related tumors at different anatomical sites with regards to clinical behavior. The genomic landscape of HPV-associated HNSCC and cervical squamous cell carcinomas described here highlights both similarities and differences. This is consistent with prior studies on chromosomal alterations (80), gene expression patterns (81), and expression of miRNA (82) which demonstrate similarities, but also differences between HPV-positive HNSCC and cervical cancer. Additional studies are needed to further delineate the heterogeneity of these and other HPV-associated tumor types at the genomic level.
Further research is also needed to determine the frequency with which specific alterations occur in these cancers, the role of these alterations in tumorigenesis, and the clinical implications of these alterations. Several of the alterations described herein have been implicated not only in tumor formation, but also in response to therapy, and as such may serve as prognostic biomarkers. RAD51B, a protein involved in DNA repair, is disrupted by HPV integration in both HNSCC and cervical cancer. Deficiency of RAD51B has been shown to sensitize to chemotherapy and radiation therapy in in vitro models (83) and has been suggested to contribute to the favourable response to therapy of HPV-associated cancers (21). Overexpression of the oncogene NR4A2, which has also been identified as a recurrent integration site, has been shown to confer an unfavorable prognosis in colorectal cancer patients (84). PIK3CA alterations have also been reported as therapeutic biomarkers in HNSCC (48).
Several of the genomic alterations identified are therapeutically targetable, such as mutations in the PI3K pathway in both HNSCC and cervical cancer, FGFR aberrations in HNSCC and ERBB2 in cervical cancer. Importantly, this could decrease toxicity associated with chemo-radiation therapies and sequelae associated with these therapies (85). Trials with PI3K/AKT/mTOR inhibitors are underway in both HNSCC and cervical cancer (86, 87) (NCT02113878, NCT02051751, NCT01602315, NCT02145312). Interestingly PI3K/mTOR inhibition has previously been shown to sensitize cancer cells to radiation and chemotherapy (88–90). The FGFR3 mutation at position 249 and FGFR3-TACC3 fusions identified in a number of HPV-positive HNSCC cases have shown promising therapeutic response to FGFR inhibitors in pre-clinical (52) and clinical studies (91, 92). ERBB2 inhibition has well-established therapeutic efficacy in HER2 positive breast cancer and clinical trials are currently underway to consider these agents in cervical cancers with ERBB2 alterations (NCT02342587). Of note, several of the identified alterations are in tumor suppressor genes, which remain a challenge in terms of targeted therapy.
Lastly, it is important to note that prophylactic HPV vaccination is available and approved for the prevention of anogenital and cervical carcinoma. It remains to be validated for the prevention of HPV-associated HNSCC, however early studies suggest the vaccine prevents oral HPV infection (93). Public health strategies to increase vaccination coverage remain the most cost-effective and beneficial approach for reducing disease burden.
Conclusions
HPV infection and subsequent viral protein expression creates an environment suitable for viral replication, whereby keratinocytes are maintained in a proliferative state and the immune system is down-modulated. This environment is also amenable to accumulation of genetic alterations and viral integration, and subsequent tumor formation. Integration affects both the viral genome and the host genome, likely conferring additional neoplastic selective pressure, by one or more of the following mechanisms: 1) enhanced expression of viral oncoproteins, 2) alteration of critical cellular genes (leading to increased expression of oncogenic proteins, decreased expression of tumor suppressor proteins, altered DNA repair mechanisms, or modulation of the immune system), and 3) changes in global promoter methylation and transcription.
Comprehensive characterization of genomic alterations in HPV-associated cancers has highlighted multiple potential biomarkers and therapeutic targets. However, the number of HPV-positive tumor samples that have been comprehensively analyzed using genome-wide studies remains small, and larger patient cohorts will be helpful to further detail integration events and other HPV-associated genomic alterations, as well as to study the clinical implications of these aberrations. More detailed studies of the functional impact of integration on various cellular proteins will be useful in characterizing the cellular pathways that become deregulated and how this leads to tumor progression. Similarly, further research is necessary to understand how distinct methylation patterns arise in HPV-integrated compared to non-HPV-integrated cancers, and the consequences of these patterns on tumor biology and clinical outcomes. Further research regarding the clinical implications of the observed genomic alterations will be imperative for accurate stratification of patients to targeted therapies, radiation therapy and chemotherapy.
Acknowledgments
Grant Support
M. Rusan was supported by an EliteForsk Travel scholarship (Danish Council for Independent Research) and a PhD scholarship from Aarhus University. P.S. Hammerman was supported by the NCI of the NIH under award number K08CA163677.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L, et al. Global burden of human papillomavirus and related diseases. Vaccine. 2012;30(Suppl 5):F12–F23. doi: 10.1016/j.vaccine.2012.07.055. [DOI] [PubMed] [Google Scholar]
- 2.Gillison ML, Alemany L, Snijders PJ, Chaturvedi A, Steinberg BM, Schwartz S, et al. Human papillomavirus and diseases of the upper airway: head and neck cancer and respiratory papillomatosis. Vaccine. 2012;30(Suppl 5):F34–F54. doi: 10.1016/j.vaccine.2012.05.070. [DOI] [PubMed] [Google Scholar]
- 3.Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 4.Kajitani N, Satsuka A, Kawate A, Sakai H. Productive lifecycle of human papillomaviruses that depends upon squamous epithelial differentiation. Front Microbiol. 2012;3:152. doi: 10.3389/fmicb.2012.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoory T, Monie A, Gravitt P, Wu TC. Molecular epidemiology of human papillomavirus. J Formos Med Assoc. 2008 Mar;107:198–217. doi: 10.1016/S0929-6646(08)60138-2. [DOI] [PubMed] [Google Scholar]
- 6.Hebner CM, Laimins LA. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev Med Virol. 2006;16:83–97. doi: 10.1002/rmv.488. [DOI] [PubMed] [Google Scholar]
- 7.Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond) 2006 May;110:525–541. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- 8.Lehoux M, D’Abramo CM, Archambault J. Molecular mechanisms of human papillomavirus-induced carcinogenesis. Public Health Genomics. 2009;12:268–280. doi: 10.1159/000214918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stanley M. Pathology and epidemiology of HPV infection in females. Gynecol Oncol. 2010;117(2 Suppl):S5–S10. doi: 10.1016/j.ygyno.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 10.Hawley-Nelson P, Vousden KH, Hubbert NL, Lowy DR, Schiller JT. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 13.Guess JC, McCance DJ. Decreased migration of Langerhans precursor-like cells in response to human keratinocytes expressing human papillomavirus type 16 E6/E7 is related to reduced macrophage inflammatory protein-3alpha production. J Virol. 2005;79:14852–14862. doi: 10.1128/JVI.79.23.14852-14862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laurson J, Khan S, Chung R, Cross K, Raj K. Epigenetic repression of E-cadherin by human papillomavirus 16 E7 protein. Carcinogenesis. 2010;31:918–926. doi: 10.1093/carcin/bgq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fausch SC, Fahey LM, Da Silva DM, Kast WM. Human papillomavirus can escape immune recognition through Langerhans cell phosphoinositide 3-kinase activation. J Immunol. 2005;174:7172–7178. doi: 10.4049/jimmunol.174.11.7172. [DOI] [PubMed] [Google Scholar]
- 16.Ronco LV, Karpova AY, Vidal M, Howley PM. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnard P, Payne E, McMillan NA. The human papillomavirus E7 protein is able to inhibit the antiviral and anti-growth functions of interferon-alpha. Virology. 2000;277:411–419. doi: 10.1006/viro.2000.0584. [DOI] [PubMed] [Google Scholar]
- 18.Park JS, Kim EJ, Kwon HJ, Hwang ES, Namkoong SE, Um SJ. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. Implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J Biol Chem. 2000;275:6764–6769. doi: 10.1074/jbc.275.10.6764. [DOI] [PubMed] [Google Scholar]
- 19.Ashrafi GH, Haghshenas MR, Marchetti B, O’Brien PM, Campo MS. E5 protein of human papillomavirus type 16 selectively downregulates surface HLA class I. Int J Cancer. 2005;113:276–283. doi: 10.1002/ijc.20558. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker TP, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. 2015;21:632–641. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parfenov M, Pedamallu CS, Gehlenborg N, Freeman SS, Danilova L, Bristow CA, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci U S A. 2014;111:15544–15549. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akagi K, Li J, Broutian TR, Padilla-Nash H, Xiao W, Jiang B, et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014;24:185–199. doi: 10.1101/gr.164806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullen AP, Reid R, Campion M, Lorincz AT. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasm. J Virol. 1991;65:606–612. doi: 10.1128/jvi.65.2.606-612.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daniel B, Mukherjee G, Seshadri L, Vallikad E, Krishna S. Changes in the physical state and expression of human papillomavirus type 16 in the progression of cervical intraepithelial neoplasia lesions analysed by PCR. J Gen Virol. 1995;76:2589–2593. doi: 10.1099/0022-1317-76-10-2589. [DOI] [PubMed] [Google Scholar]
- 27.Klaes R, Woerner SM, Ridder R, Wentzensen N, Duerst M, Schneider A, et al. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 1999;59:6132–6136. [PubMed] [Google Scholar]
- 28.Pirami L, Giache V, Becciolini A. Analysis of HPV16, 18, 31, and 35 DNA in pre-invasive and invasive lesions of the uterine cervix. J Clin Pathol. 1997;50:600–604. doi: 10.1136/jcp.50.7.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Begum S, Cao D, Gillison M, Zahurak M, Westra WH. Tissue distribution of human papillomavirus 16 DNA integration in patients with tonsillar carcinoma. Clin Cancer Res. 2005;11:5694–5699. doi: 10.1158/1078-0432.CCR-05-0587. [DOI] [PubMed] [Google Scholar]
- 30.Romanczuk H, Howley PM. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc Natl Acad Sci U S A. 1992;89:3159–3163. doi: 10.1073/pnas.89.7.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thierry F, Yaniv M. The BPV1-E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory region. EMBO J. 1987;6:3391–3397. doi: 10.1002/j.1460-2075.1987.tb02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeon S, Lambert PF. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci U S A. 1995;92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeon S, Allen-Hoffmann BL, Lambert PF. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol. 1995;69:2989–2997. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wentzensen N, Vinokurova S, von Knebel Doeberitz M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 2004;64:3878–3884. doi: 10.1158/0008-5472.CAN-04-0009. [DOI] [PubMed] [Google Scholar]
- 35.Sathish N, Abraham P, Peedicayil A, Sridharan G, John S, Chandy G. Human papillomavirus 16 E6/E7 transcript and E2 gene status in patients with cervical neoplasia. Mol Diagn. 2004;8:57–64. doi: 10.1007/BF03260048. [DOI] [PubMed] [Google Scholar]
- 36.von Knebel Doeberitz M, Bauknecht T, Bartsch D, zur Hausen H. Influence of chromosomal integration on glucocorticoid-regulated transcription of growth-stimulating papillomavirus genes E6 and E7 in cervical carcinoma cells. Proc Natl Acad Sci U S A. 1991;88:1411–1415. doi: 10.1073/pnas.88.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin HJ, Joo J, Yoon JH, Yoo CW, Kim JY. Physical status of human papillomavirus integration in cervical cancer is associated with treatment outcome of the patients treated with radiotherapy. PLoS One. 2014;9:e78995. doi: 10.1371/journal.pone.0078995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khoury JD, Tannir NM, Williams MD, Chen Y, Yao H, Zhang J, et al. Landscape of DNA virus associations across human malignant cancers: analysis of 3,775 cases using RNA-Seq. J Virol. 2013;87:8916–8926. doi: 10.1128/JVI.00340-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durst M, Croce CM, Gissmann L, Schwarz E, Huebner K. Papillomavirus sequences integrate near cellular oncogenes in some cervical carcinomas. Proc Natl Acad Sci U S A. 1987;84:1070–1074. doi: 10.1073/pnas.84.4.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferber MJ, Thorland EC, Brink AA, Rapp AK, Phillips LA, McGovern R, et al. Preferential integration of human papillomavirus type 18 near the c-myc locus in cervical carcinoma. Oncogene. 2003;22:7233–7242. doi: 10.1038/sj.onc.1207006. [DOI] [PubMed] [Google Scholar]
- 41.Wentzensen N, Ridder R, Klaes R, Vinokurova S, Schaefer U, Doeberitz M. Characterization of viral-cellular fusion transcripts in a large series of HPV16 and 18 positive anogenital lesions. Oncogene. 2002;21:419–426. doi: 10.1038/sj.onc.1205104. [DOI] [PubMed] [Google Scholar]
- 42.McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays. 2007;29:549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X, Lu H, Yan B, Romano RA, Bian Y, Friedman J, et al. DeltaNp63 versatilely regulates a Broad NF-kappaB gene program and promotes squamous epithelial proliferation, migration, and inflammation. Cancer Res. 2011;71:3688–3700. doi: 10.1158/0008-5472.CAN-10-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graziano V, De Laurenzi V. Role of p63 in cancer development. Biochim Biophys Acta. 2011;1816:57–66. doi: 10.1016/j.bbcan.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011 Aug 26;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lechner M, Frampton GM, Fenton T, Feber A, Palmer G, Jay A, et al. Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV- tumors. Genome Med. 2013;5:49. doi: 10.1186/gm453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3:761–769. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohba T, Toyokawa G, Kometani T, Nosaki K, Hirai F, Yamaguchi M, et al. Mutations of the EGFR and K-ras genes in resected stage I lung adenocarcinoma and their clinical significance. Surg Today. 2014;44:478–486. doi: 10.1007/s00595-013-0589-2. [DOI] [PubMed] [Google Scholar]
- 50.Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 51.Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre-Garau X, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 52.Liao RG, Jung J, Tchaicha J, Wilkerson MD, Sivachenko A, Beauchamp EM, et al. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 2013;73:5195–5205. doi: 10.1158/0008-5472.CAN-12-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22:795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chung GT, Lou WP, Chow C, To KF, Choy KW, Leung AW, et al. Constitutive activation of distinct NF-kappaB signals in EBV-associated nasopharyngeal carcinoma. J Pathol. 2013;231:311–322. doi: 10.1002/path.4239. [DOI] [PubMed] [Google Scholar]
- 57.Romick-Rosendale LE, Lui VW, Grandis JR, Wells SI. The Fanconi anemia pathway: repairing the link between DNA damage and squamous cell carcinoma. Mutat Res. 2013;743–744:78–88. doi: 10.1016/j.mrfmmm.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le Gallo M, O’Hara AJ, Rudd ML, Urick ME, Hansen NF, O’Neil NJ, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet. 2012;44:1310–1315. doi: 10.1038/ng.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu C, Fillmore CM, Koyama S, Wu H, Zhao Y, Chen Z, et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell. 2014;25:590–604. doi: 10.1016/j.ccr.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsai LH, Chen PM, Cheng YW, Chen CY, Sheu GT, Wu TC, et al. LKB1 loss by alteration of the NKX2-1/p53 pathway promotes tumor malignancy and predicts poor survival and relapse in lung adenocarcinomas. Oncogene. 2014;33:3851–3860. doi: 10.1038/onc.2013.353. [DOI] [PubMed] [Google Scholar]
- 63.Bose R, Kavuri SM, Searleman AC, Shen W, Shen D, Koboldt DC, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3:224–237. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dulak AM, Schumacher SE, van Lieshout J, Imamura Y, Fox C, Shim B, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012;72:4383–4393. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Greulich H, Kaplan B, Mertins P, Chen TH, Tanaka KE, Yun CH, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci U S A. 2012;109:14476–14481. doi: 10.1073/pnas.1203201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 67.Sedaghat AR, Zhang Z, Begum S, Palermo R, Best S, Ulmer KM, et al. Prognostic significance of human papillomavirus in oropharyngeal squamous cell carcinomas. Laryngoscope. 2009;119:1542–1549. doi: 10.1002/lary.20533. [DOI] [PubMed] [Google Scholar]
- 68.Ragin CC, Taioli E. Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: review and meta-analysis. Int J Cancer. 2007;121:1813–1820. doi: 10.1002/ijc.22851. [DOI] [PubMed] [Google Scholar]
- 69.Lassen P, Eriksen JG, Hamilton-Dutoit S, Tramm T, Alsner J, Overgaard J. Effect of HPV-associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. J Clin Oncol. 2009;27:1992–1998. doi: 10.1200/JCO.2008.20.2853. [DOI] [PubMed] [Google Scholar]
- 70.Lassen P, Eriksen JG, Krogdahl A, Therkildsen MH, Ulhoi BP, Overgaard M, et al. The influence of HPV-associated p16-expression on accelerated fractionated radiotherapy in head and neck cancer: evaluation of the randomised DAHANCA 6&7 trial. Radiother Oncol. 2011;100:49–55. doi: 10.1016/j.radonc.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 71.Sethi S, Ali-Fehmi R, Franceschi S, Struijk L, van Doorn LJ, Quint W, et al. Characteristics and survival of head and neck cancer by HPV status: a cancer registry-based study. Int J Cancer. 2012;131:1179–1186. doi: 10.1002/ijc.26500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chung CH, Zhang Q, Kong CS, Harris J, Fertig EJ, Harari PM, et al. p16 Protein Expression and Human Papillomavirus Status As Prognostic Biomarkers of Nonoropharyngeal Head and Neck Squamous Cell Carcinoma. J Clin Oncol. 2014;32:3930–3938. doi: 10.1200/JCO.2013.54.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lassen P, Primdahl H, Johansen J, Kristensen CA, Andersen E, Andersen LJ, et al. Impact of HPV-associated p16-expression on radiotherapy outcome in advanced oropharynx and non-oropharynx cancer. Radiother Oncol. 2014;113:310–316. doi: 10.1016/j.radonc.2014.11.032. [DOI] [PubMed] [Google Scholar]
- 74.Wilson DD, Rahimi AS, Saylor DK, Stelow EB, Jameson MJ, Shonka DC, et al. p16 not a prognostic marker for hypopharyngeal squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2012;138:556–561. doi: 10.1001/archoto.2012.950. [DOI] [PubMed] [Google Scholar]
- 75.Wilson DD, Crandley EF, Sim A, Stelow EB, Majithia N, Shonka DC, Jr, et al. Prognostic significance of p16 and its relationship with human papillomavirus in pharyngeal squamous cell carcinomas. JAMA Otolaryngol Head Neck Surg. 2014;140:647–653. doi: 10.1001/jamaoto.2014.821. [DOI] [PubMed] [Google Scholar]
- 76.Arroyo LS, Smelov V, Bzhalava D, Eklund C, Hultin E, Dillner J. Next generation sequencing for human papillomavirus genotyping. J Clin Virol. 2013;58:437–442. doi: 10.1016/j.jcv.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 77.Xuan J, Yu Y, Qing T, Guo L, Shi L. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 2013;340:284–295. doi: 10.1016/j.canlet.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loyo M, Li RJ, Bettegowda C, Pickering CR, Frederick MJ, Myers JN, et al. Lessons learned from next-generation sequencing in head and neck cancer. Head Neck. 2013;35:454–463. doi: 10.1002/hed.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saba NF, Wilson M, Doho G, DaSilva J, Benjamin Isett R, Newman S, et al. Mutation and transcriptional profiling of formalin-fixed paraffin embedded specimens as companion methods to immunohistochemistry for determining therapeutic targets in oropharyngeal squamous cell carcinoma (OPSCC): a pilot of proof of principle. Head Neck Pathol. 2014 Sep;:19. doi: 10.1007/s12105-014-0566-0. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilting SM, Smeets SJ, Snijders PJ, van Wieringen WN, van de Wiel MA, Meijer GA, et al. Genomic profiling identifies common HPV-associated chromosomal alterations in squamous cell carcinomas of cervix and head and neck. BMC Med Genomics. 2009;2:32. doi: 10.1186/1755-8794-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lajer CB, Garnaes E, Friis-Hansen L, Norrild B, Therkildsen MH, Glud M, et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. Br J Cancer. 2012;106:1526–1534. doi: 10.1038/bjc.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lio YC, Schild D, Brenneman MA, Redpath JL, Chen DJ. Human Rad51C deficiency destabilizes XRCC3, impairs recombination, and radiosensitizes S/G2-phase cells. J Biol Chem. 2004;279:42313–42320. doi: 10.1074/jbc.M405212200. [DOI] [PubMed] [Google Scholar]
- 84.Han Y, Cai H, Ma L, Ding Y, Tan X, Liu Y, et al. Nuclear orphan receptor NR4A2 confers chemoresistance and predicts unfavorable prognosis of colorectal carcinoma patients who received postoperative chemotherapy. Eur J Cancer. 2013;49:3420–3430. doi: 10.1016/j.ejca.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 85.Mirghani H, Amen F, Blanchard P, Moreau F, Guigay J, Hartl DM, et al. Treatment de-escalation in HPV-positive oropharyngeal carcinoma: ongoing trials, critical issues and perspectives. Int J Cancer. 2015;136:1494–1503. doi: 10.1002/ijc.28847. [DOI] [PubMed] [Google Scholar]
- 86.Hou MM, Liu X, Wheler J, Naing A, Hong D, Coleman RL, et al. Targeted PI3K/AKT/mTOR therapy for metastatic carcinomas of the cervix: a phase I clinical experience. Oncotarget. 2014;5:11168–11179. doi: 10.18632/oncotarget.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jimeno A, Shirai K, Choi M, Laskin J, Kochenderfer M, Spira A, et al. A randomized, phase 2 trial of cetuximab with or without PX-866, an irreversible oral phosphatidylinositol 3-kinase inhibitor, in patients with relapsed or metastatic head and neck squamous cell cancer. Ann Oncol. 2014 Dec;:18. doi: 10.1093/annonc/mdu574. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 88.Marklein D, Graab U, Naumann I, Yan T, Ridzewski R, Nitzki F, et al. PI3K inhibition enhances doxorubicin-induced apoptosis in sarcoma cells. PLoS One. 2012;7:e52898. doi: 10.1371/journal.pone.0052898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bender A, Opel D, Naumann I, Kappler R, Friedman L, von Schweinitz D, et al. PI3K inhibitors prime neuroblastoma cells for chemotherapy by shifting the balance towards pro-apoptotic Bcl-2 proteins and enhanced mitochondrial apoptosis. Oncogene. 2011;30:494–503. doi: 10.1038/onc.2010.429. [DOI] [PubMed] [Google Scholar]
- 90.Gil del Alcazar CR, Hardebeck MC, Mukherjee B, Tomimatsu N, Gao X, Yan J, et al. Inhibition of DNA double-strand break repair by the dual PI3K/mTOR inhibitor NVP-BEZ235 as a strategy for radiosensitization of glioblastoma. Clin Cancer Res. 2014;20:1235–1248. doi: 10.1158/1078-0432.CCR-13-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dienstmann R, Bahleda R, Adamo B, Rodon J, Varga A, Gazzah A, et al. First in human study of JNJ-42756493, a potent pan fibroblast growth factor receptor (FGFR) inhibitor in patients with advanced solid tumors [abstract]; Proceedings of the 105th Meeting of the American Association for Cancer Research; 2014 Apr 5–9; San Diego, CA. Philadelphia (PA): AACR. 2014. Abstract nr CT325. [Google Scholar]
- 92.Sequist LV, Cassier P, Varga A, Tabernero J, Schellens JHM, Delord J-P, et al. Phase I study of BGJ398, a selective pan-FGFR inhibitor in genetically preselected advanced solid tumors [abstract]; Proceedings of the 105th Meeting of the American Association for Cancer Research; 2014 Apr 5–9; San Diego, CA. Philadelphia (PA): AACR. 2014. Abstract nr CT326. [Google Scholar]
- 93.Herrero R, Quint W, Hildesheim A, Gonzalez P, Struijk L, Katki HA, et al. Reduced prevalence of oral human papillomavirus (HPV) 4 years after bivalent HPV vaccination in a randomized clinical trial in Costa Rica. PLoS One. 2013;8:e68329. doi: 10.1371/journal.pone.0068329. [DOI] [PMC free article] [PubMed] [Google Scholar]