Abstract
Botulinum neurotoxins (BoNTs) are among the most deadly toxins known. They act rapidly in a highly specific manner to block neurotransmitter release by cleaving the soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) complex at neuromuscular junctions. The extreme toxicity of BoNTs relies predominantly on their neurotropism that is accomplished by recognition of two host receptors, a polysialo-ganglioside and in the majority of cases a synaptic vesicle protein, through their receptor-binding domains. Two proteins, synaptotagmin and synaptic vesicle glycoprotein 2, have been identified as the receptors for various serotypes of BoNTs. Here, we review recent breakthroughs in the structural studies of BoNT–protein receptor recognitions that highlight a range of diverse mechanisms by which BoNTs manipulate host neuronal proteins for highly specific uptake at neuromuscular junctions.
Keywords: botulinum neurotoxin, botulism, synaptotagmin, synaptic vesicle glycoprotein 2, protein complex, host-pathogen recognition
Introduction
Botulinum neurotoxins (BoNTs) are designated as Tier 1 select agents by the Centers for Disease Control and Prevention (CDC). With an estimated lethal dose for human at ~ 1 ng per kg of body weight (Gill, 1982), BoNTs are among the most life threatening natural substances that raise serious concern of a possible biowarfare use (Arnon et al., 2001; Bigalke and Rummel, 2005; Binz et al., 1990). There are seven major serotypically distinct BoNTs, termed BoNT/A to BoNT/G, which comprise at least 40 different subtypes. They are also structurally and functionally related to tetanus toxin (for a recent review see (Rossetto et al., 2014)). The eighth serotype, BoNT/H, has recently been proposed but remains to be verified experimentally (Barash and Arnon, 2014; Dover et al., 2014). Interestingly, the double-faced BoNTs, especially BoNT/A, are among the top-selling drugs as prescription medicines in clinic and facial rejuvenation agents in cosmetic industries (Bigalke, 2013).
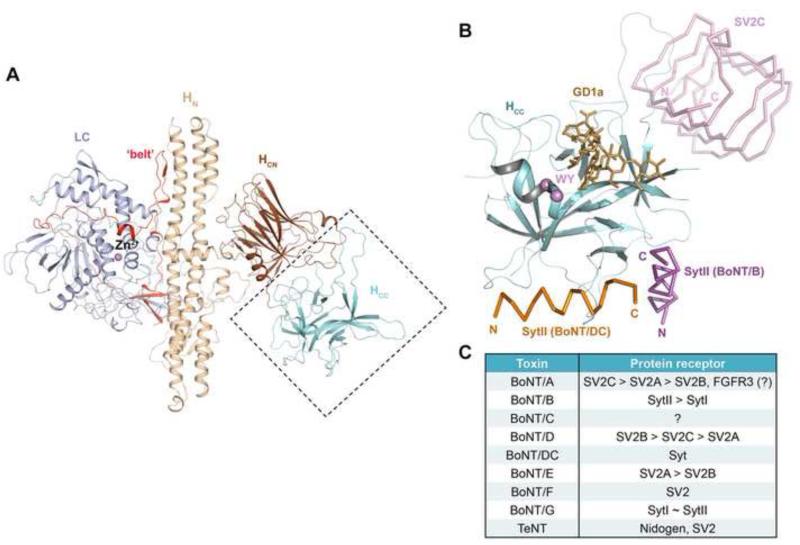
BoNT is synthesized as a ~150 kDa protein and then proteolytically cleaved into two chains, an N-terminal ~50 kDa light chain (LC) and a C-terminal ~100 kDa heavy chain (HC), which are linked by an essential disulfide bridge. Crystal structures of BoNT/A1, BoNT/B1, and BoNT/E1 all exhibit a similar tri-modular architecture (Kumaran et al., 2009; Lacy et al., 1998; Swaminathan and Eswaramoorthy, 2000). LC is a Zn2+-metalloprotease, whereas HC comprises an N-terminal ~50 kDa translocation domain (HN) and a C-terminal ~50 kDa receptor-binding domain (HC). HC is further divided into an N-terminal (HCN) and a C-terminal (HCC) subdomains (Figure 1A). Other BoNT serotypes are expected to adopt a similar architecture.
Figure 1.
The conserved structure of BoNTs and their diverse receptor-binding modes. (A) The conserved architecture of BoNTs: LC (light blue), HN (wheat), the belt region connecting LC and HN (red), HCN (brown), and HCC (cyan) (BoNT/B, PDB ID: 2NP0). (B) The HCC is a versatile receptor-binding domain. The model is built based on superposition of the structures of HCB–SytII (PDB ID: 4KBB), HCDC–SytII (PDB ID: 4ISR), and HCA–SV2C (PDB ID: 4JRA). HCCB is represented as cartoon (cyan) and the view direction is similar to that shown in panel (A). The BoNT/B-bound SytII (magenta), BoNT/DC-bound SytII (orange), and BoNT/A-bound SV2C (pink) are drawn in ribbon. GD1a, representing the polysialo-ganglioside receptor, is represented as sticks (gold). The highly conserved ganglioside-binding residues (WY) are highlighted in purple. (C) A summary of the protein receptors of various BoNT serotypes and tetanus toxin (TeNT). The preferred receptors of BoNT/A, /B, /D and /E are listed in the order of descending affinities. SytI and SytII show similar binding affinity towards BoNT/G.
The modular structures of BoNTs are highly adapted for their potent neuron-specific toxicity. In foodborne or intestinal botulism, BoNTs are secreted in the form of large progenitor toxin complexes composed of non-toxic non-hemagglutinin protein and other auxiliary proteins, which are essential for the absorption of BoNTs in the intestine to enter the general circulation (Fujinaga et al., 2013; Gu and Jin, 2013; Gu et al., 2012; Lee et al., 2013; Lee et al., 2014). BoNTs then travel to the neuromuscular junction (NMJ), where the HC domain specifically targets presynaptic motoneurons and toxins are endocytosed through synaptic vesicle recycling. Upon vesicular acidification, the HN domain undergoes a conformational change to form a protein channel that allows translocation of LC to the cytoplasm. LC specifically cleaves the soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) complex that forms a crucial vesicle fusion machinery. The cleavage terminates neurotransmitter release and paralyzes the affected muscle (Blasi et al., 1993; Schiavo et al., 1992; Schiavo et al., 2000).
Dual receptor model
The molecular mechanisms by which BoNTs specifically target motoneurons have attracted great attention in recent decades. It is believed that most BoNTs possess two independent binding regions in HCC for polysialo-gangliosides and neuronal protein receptors to achieve high binding affinity and specificity (Montecucco, 1986). HCC adopts a β-trefoil fold containing 12 core β- strands. Structure-based sequence alignment of HCC from different BoNT serotypes displays a low sequence conservation of ~31% on average. The conserved residues are mainly in the core of HCC that are important for maintaining the protein fold, while the surface loops connecting the β-strands are highly variable (Ginalski et al., 2000).
Polysialo-gangliosides, such as GD1a and GT1b, are a large family of glycosphingolipids that are present abundantly on the outer leaflet of the presynaptic membrane and are organized in microdomains together with some glycoproteins. BoNT/A, /B, /E, /F, and /G have a conserved ganglioside-binding site in HCC composed of a “E(Q)...H(K)...SXWY...G” motif (Figure 1B), whereas BoNT/C, /D, and /DC display two independent ganglioside-binding sites (for a more detailed review of the BoNT–ganglioside interaction, see (Rummel, 2013)). BoNT– ganglioside interaction ensures an effective initial capture and enrichment of the scarcely distributed BoNTs to the presynaptic nerve terminus, preceding the engagement of the protein receptor.
In contrast to the conserved ganglioside-binding mode, BoNTs recognize their protein receptors in a serotype-specific manner, even though only two protein receptors, synaptotagmin (Syt) and synaptic vesicle glycoprotein 2 (SV2), have been identified (Figure 1C). Syt has two major isoforms (SytI and SytII), which are calcium sensors that regulate the fusion of synaptic vesicle exocytosis (Fernandez-Chacon et al., 2001). They contain an unstructured N-terminal luminal segment and two C-terminal cytosolic calcium sensing (C2) domains, which are connected by a single transmembrane region. BoNT/B, /G and /DC have been shown to recognize the luminal domain of SytI and SytII. SV2 proteins, including three isoforms SV2A, SV2B, and SV2C, are 12-transmembrane domain glycoproteins (Bajjalieh et al., 1992; Feany et al., 1992; Janz and Sudhof, 1999). They have been shown to be the protein receptors for BoNT/A (Dong et al., 2006; Mahrhold et al., 2006), BoNT/E (Dong et al., 2008), BoNT/F (Fu et al., 2009; Rummel et al., 2009), and BoNT/D (Peng et al., 2011). Fibroblast growth factor receptor 3 is proposed to be another potential receptor of BoNT/A, but the physiological relevance of this observation awaits further study (Jacky et al., 2013). Interestingly, nidogen, an extracellular matrix protein, was recently identified as the protein receptor of tetanus toxin (Figure 1C) (Bercsenyi et al., 2014).
The two receptors are believed to function independently (Berntsson et al., 2013a) and likely play distinctive roles in the course of BoNT intoxication due to their different localizations in presynaptic neurons. Gangliosides are abundantly exposed on the membrane and Syt and SV2 are confined within the dynamic recycling synaptic vesicles, and their luminal domains are accessible to BoNTs only after synaptic vesicle fusion. Therefore, both receptors are required to ensure high binding affinity and specificity that are necessary for BoNT's extreme toxicity (Pierce et al., 1986). Remarkably, BoNTs seem to develop serotype- and/or subtype-specific mechanisms for protein receptor recognition as opposed to a conserved ganglioside-binding mode. The exact reason for this diversity remains elusive, though the unique protein-protein interactions maybe in part responsible for their distinct toxicological properties (Whitemarsh et al., 2013). In this review, we summarize the recent findings in a fast evolving research field, which reveal how BoNTs employ diverse mechanisms to hijack protein receptors.
BoNT/B–Synaptotagmin interaction
The first structures of the BoNT/B–SytII complex were reported simultaneously from two groups. The recombinant protein complex was prepared by covalently fusing the luminal domain (residues 8–61) of rat SytII to the C-terminus of HC domain (HCB) (2.15 Å) or by co-crystallizing the full length BoNT/B with a short mouse SytII peptide (residues 40–60) (2.6Å) (Chai et al., 2006; Jin et al., 2006). The two structures are essentially identical and this has been further confirmed by a recent structure of HCB in complex with SytII and GD1a (Berntsson et al., 2013a). The SytII luminal domain is unstructured but conforms to a short helix (residues 47–58) upon BoNT/B binding and docks into a hydrophobic saddle-shaped crevice on HCB. In comparison, the SytII-binding site on HCB is largely pre-formed (r.m.s.d. of 0.73 Å between the apo and complexed HCB). The SytII-binding site is adjacent to the GD1a-binding pocket, but there is no allosteric interaction between them (Berntsson et al., 2013a; Jin et al., 2006).
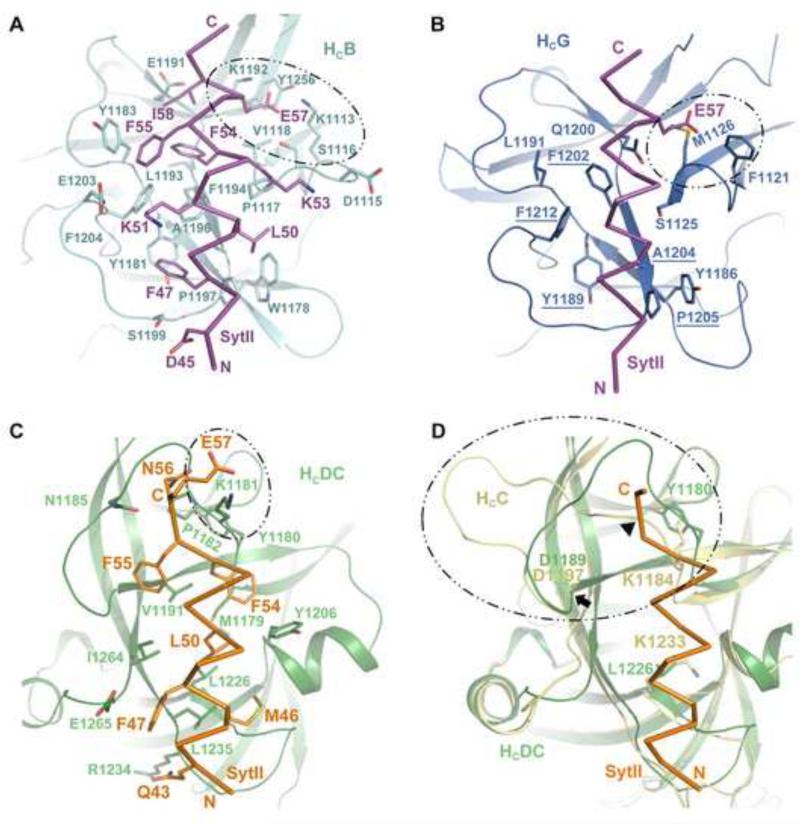
SytII binds to HCB with high affinity (dissociation constant, Kd, of ~34 nM), which is not influenced by acidic pH, a condition BoNT might encounter during vesicle acidification (Jin et al., 2006). The complex is stabilized by extensive hydrophobic contacts involving two prominent pockets on the HCB surface that are separated by a shallow ridge (Figure 2A). The N-terminal SytII residues F47 and L50 dock into a pocket surrounded by residues W1178, Y1181, A1196, and P1197 of HCB; the C-terminal SytII residues F54, F55, and I58 are embraced by V1118, Y1183, E1191 and L1193 of HCB. Surrounding the hydrophobic core, K51 of SytII forms a salt bridge with E1203 of HCB; D45 and K53 of SytII are hydrogen bonded to S1199 and D1115 of HCB, respectively. Additionally, E57 of SytII is attracted electrostatically to an electropositive pocket of HCB composed of K1192, K1113, S1116, and Y1256. The importance of the interfacial residues has been verified by in vitro pull down assay as well as toxicity assay (Chai et al., 2006; Jin et al., 2006; Willjes et al., 2013), and the SytII-binding residues are highly conserved among various BoNT/B subtypes (Chai et al., 2006). In contrast to SytII, the binding affinity of SytI to HCB is at least two orders of magnitude weaker. Although the structure of HCB–SytI remains unresolved, mutagenesis studies suggest that SytI binds to the same pocket of HCB by forming a short helix. This is strongly supported by the observation that substitution of BoNT/B-binding residues of SytI with the corresponding residues of SytII (e.g., M47F/L50I or L50I/H51N) significantly enhanced the affinity of SytI to a level similar to that of SytII (Chai et al., 2006; Jin et al., 2006).
Figure 2.
Syt is the protein receptor for BoNT/B, /G, and /DC. (A) The HCB–SytII binding interface (PDB ID: 2NM1). Residues participated in the interaction are shown as sticks. (B) Putative HCG–SytII binding pocket (PDB ID: 2VXR). Residues discussed in the text are drawn as sticks. Identical residues between HCB and HCG in the binding pocket are underlined. (C) The HCDC–SytII binding interface (PDB ID: 4ISR). The residues surrounding SytII E57 are circled. (D) Structural comparison of HCDC and HCC (PDB ID: 3R4S). Two residues that potentially differentiate HCC from HCDC (Y1180 and L1226 of HCDC and K1184 and K1233 of HCC) are shown as sticks. The arrowhead marks the position of potential steric clash between SytII and HCC. The arrow indicates the position of D1197 of HCC and D1189 of HCDC. The K1184–D1197 loop of HCC is highlighted in circle. Coloring scheme: HCB, cyan; HCB-bound SytII, magenta; HCG, dark blue; HCDC, forest; HCDC-bound SytII, orange; HCC, yellow.
It is noted that F54 of SytII, which is a key BoNT/B-binding residue and conserved across many vertebrates, is replaced by leucine in human and chimpanzee. Mutating F54 in mouse/rat SytII drastically decreased its binding to BoNT/B and also reduced the cleavage of synaptobrevin (Peng et al., 2012; Strotmeier et al., 2012). This finding thus helps clarify the observed discrepancy about the potency of BoNT/B on humans and mice, and also raises the possibility that structure-based BoNT/B modification may improve its binding to human SytII and lead to enhanced therapeutic efficacy on human.
BoNT/G–Synaptotagmin interaction
BoNT/G binds similarly to SytI and SytII, and the binding affinities are weaker than BoNT/B–SytII but stronger than BoNT/B–SytI (Rummel et al., 2007; Stenmark et al., 2010). The structure of the apo HCG is highly similar to HCB (Schmitt et al., 2010; Stenmark et al., 2010). Structure-based mutagenesis study suggests that BoNT/B and /G may utilize a homologous binding pocket to recognize SytI and SytII. Among the fourteen amino acids that constitute the SytII-binding pocket on HCB, five residues situated at the hydrophobic core of the interface are conserved in HCG (Figure 2B, underlined) and mutating these residues significantly diminishes the binding of BoNT/G to SytII (Willjes et al., 2013). Moreover, two reciprocal mutations, Y1186W and L1191Y, significantly enhanced the binding of BoNT/G to SytII with concomitant decrease of BoNT/B binding (Willjes et al., 2013). It is suggested that SytII largely retains a helical conformation and makes hydrophobic contact to HCG. However, the SytII-binding mode on HCB and HCG may diverge at the C-terminus of SytII around E57, where Syt may adopt a conformation with reduced helix length and/or a bend in its C-terminus upon binding to BoNT/G as opposed to BoNT/B (Willjes et al., 2013). It should be noted that the Syt-HCG binding model is still not fully understood. Two independent studies reported contradictory results concerning the effect of several SytII mutations. For example, one study showed that mutating hydrophobic residues of SytII (F47, F55 and L50) to alanine disrupted BoNT/G binding, but these mutants showed no effect in another independent study (Stenmark et al., 2010; Willjes et al., 2013). High resolution structure of BoNT/G in complex with SytII or SytI is needed for clarification.
BoNT/DC–Synaptotagmin interaction
BoNT/DC is a mosaic toxin composed of the BoNT/D-like LC and HN domains, and a BoNT/C-like HC. HCDC is closely related to HCC (~64% sequence identity), but only 22 % and 24 % identical to HCB and HCG, respectively. Considering that BoNT/C may only use gangliosides for cell surface binding without the need for a protein receptor (Karalewitz et al., 2012; Strotmeier et al., 2011), it is surprising that BoNT/DC functionally diverges from BoNT/C and binds to Syt. Nevertheless, BoNT/DC exhibits ~10-fold lower binding affinity to SytII than BoNT/B, while its affinity to SytI is unknown (Peng et al., 2012).
As hinted by their low sequence identities, the Syt-binding pocket of BoNT/DC is distinct from BoNT/B and BoNT/G, and the orientation of the bound Syt differs by ~90o (Figure 1B) (Berntsson et al., 2013b; Peng et al., 2012). The HCDC-binding regions of SytII and SytI include residues 42–57 and 36–50, respectively. Similar to the HCB–SytII complex, the HCDC–SytII interaction is mainly driven by hydrophobic contacts, while electrostatic interactions are found at the two termini of the SytII helix. The hydrogen bonding pairs include Q43SytII–R1234HcDC and N56SytII– N1185/P1182HcDC. A salt bridge is formed between E57SytII and K1181HcDC (Figure 2C). Based on structure comparison, we suspect that different affinities displayed by HCDC and HCB are likely attributed to different electrostatic interactions. For example, residues F55–E57 of SytII adopt a helical turn when binding to the HCB pocket, but they form an extended structure when binding to HCDC (Figure 2A & 2C). Accordingly, SytII E57 forms extensive hydrogen bonds and salt bridges that docks into the electrostatic positive pocket of HCB. Such extensive electrostatic interactions are lost in HCDC, where SytII E57 is exposed on the surface of HCDC, held by a lysine residue.
In spite of high sequence identity between HCC and HCDC, HCC lacks some key hydrophobic residues needed for Syt binding (Berntsson et al., 2013b). For instance, HCDC Y1180 and L1226 are replaced by lysine in HCC (K1184 and K1233), and the Y1180K and L1226K mutations abolished BoNT/DC–Syt interaction (Peng et al., 2012). Additionally, we noted that HCC contains four extra residues in a key surface loop that embraces the C-terminal part of Syt (K1184–D1197 in BoNT/C and Y1180– D1189 in BoNT/DC) that could potentially block the docking of Syt (Figure 2D) (Peng et al., 2012).
It is worth noting that the C-terminus of SytII in both HCB and HCDC complexes is located at a similar position (Figure 1B). Since the C-terminus of SytII precedes its transmembrane region, the different Syt-binding modes utilized by BoNT/B and BoNT/DC presumably would not affect the positioning of the toxin on the neuronal membrane. The two distinct binding pockets recognizing the same region of Syt suggests the possibility of a functional convergent evolution. The convergence may offer a selective advantage to ensure a proper spatial constraint on BoNTs that allows double receptor binding and facilitates subsequent internalization and translocation (Berntsson et al., 2013b).
BoNT/A1–SV2C interaction
Similar to the serotype-specific BoNT binding to Syt discussed above, various BoNTs seem to develop different mechanisms recognizing three isoforms of SV2. For example, BoNT/A has the highest affinity to SV2C followed by SV2A and 2B (Dong et al., 2006); BoNT/D preferentially binds SV2B, and to a lesser extent, SV2A and 2C (Peng et al., 2011); BoNT/E exhibits higher binding affinity to SV2A over 2B but does not bind SV2C (Dong et al., 2008; Mahrhold et al., 2013; Peng et al., 2011). The binding-sites for BoNT/A and BoNT/E have been mapped to the large luminal domain 4 (LD4) of SV2, which is necessary and sufficient to mediate the entry of both toxins (Dong et al., 2008; Dong et al., 2006). However, BoNT/D does not interact with SV2-LD4, suggesting a distinct mechanism (Peng et al., 2011). BoNT/F has also been reported to bind SV2, but the details remain unclear (Figure 1C) (Fu et al., 2009; Rummel et al., 2009).
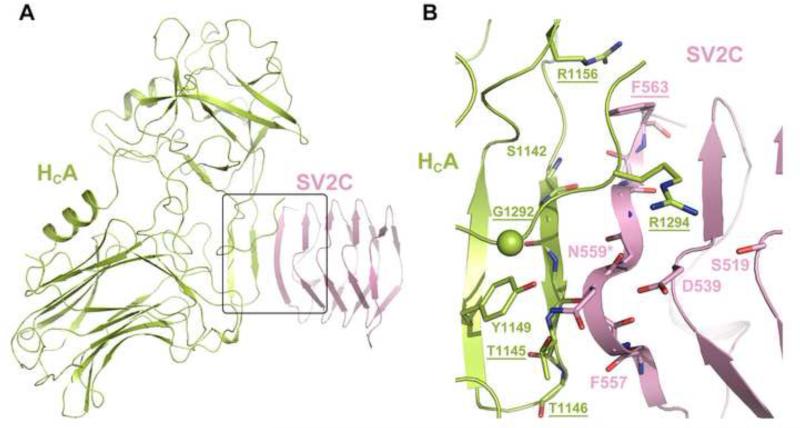
The first crystal structure of HCA in complex with the recombinant human SV2CLD4 was reported recently (Benoit et al., 2014). The structure reveals dominating backbone-backbone interactions between two short β–sheets, involving residues around E556–F563 of SV2C and R1140–N1147 of BoNT/A1 (Figure 3). In addition, the co-crystal structure suggests that a cation-π stacking interaction between F563 of human SV2C and R1156 of BoNT/A1 is crucial for binding, as mutating F563 of SV2C (F563A) or R1156 of HCA (R1156E) significantly decreased the binding (Benoit et al., 2014). Interestingly, a cation-π interaction is specific only to the pair of BoNT/A1 and human SV2C because F563 is replaced by a Leu in human SV2A and 2B as well as SV2A/B/C in rats and mice. Even though rat SV2C cannot form the cation-π interaction, mutating R1156 on HCA to a Met still led to a significant decrease of binding affinity (Strotmeier et al., 2014). At the same time, R1156 in BoNT/A1 is replaced by Met or Glu in BoNT/A2–A5. Indeed, BoNT/A2 that has a Glu at the equivalent position is even more potent than BoNT/A1 despite its lower enzymatic activity, which implies a better cell binding and/or translocation of BoNT/A2 in comparison to BoNT/A1 (Strotmeier et al., 2014; Whitemarsh et al., 2013). These findings suggest that BoNT/A subtypes may behave differently when recognizing SV2.
Figure 3.
The structure of HCA1–SV2C complex (PDB ID: 4JRA). The HCA1 and SV2C are colored in lime and pink, respectively, and the interacting residues are shown as sticks. G1292 is represented as a sphere. The conserved N-linked glycosylation site (N559) is marked with an asterisk. Mutations that disrupted the complex interactions are underlined.
Rummel et al. independently mapped the rat SV2C-binding site on BoNT/A1 using systematic mutagenesis combined with GST pull-down and mouse phrenic nerve (MPN) assay (Strotmeier et al., 2014). They found that the R1156M mutant only moderately decreased the toxicity to ~40-50% compared with the WT BoNT/A1. In contrast, mutating residue G1292, which does not directly bind to SV2C based on the crystal structure, to Arg caused a dramatic ~300-fold decrease of BoNT/A1 toxicity (Strotmeier et al., 2014). These results thus suggest that some critical BoNT/A–SV2 interactions, potentially involving the glycans of SV2, have not been resolved in the current crystal structure.
Indeed, a wealth of information suggests that N-linked glycosylation of SV2 is crucial for BoNT recognition. Among BoNT/A, /D, /E, and /F, BoNT/A is the only one that could bind to E. coli-derived unglycosylated SV2 in vitro. However, knocking out a conserved N-linked glycosylation site on SV2A (N573Q, homologous to N516 and N559 on SV2B and 2C, respectively) significantly decreased the entry of BoNT/A1 when the toxin was used at a low concentration, representing a more physiological relevant condition in disease botulism (Dong et al., 2008). Glycosylation at N573 of SV2A is critical for its interaction with BoNT/E because the single point mutation N573Q completely blocked the entry of the toxin (Dong et al., 2008). Furthermore, deglycosylation of SV2 inhibited its binding to both HCE and HCF (Fu et al., 2009). All these findings suggest that both the protein moiety and the N-linked glycans of SV2 are important for BoNT binding. However, the current crystal structure of HCA–SV2C complex provides little insight into this issue because the SV2C-LD4 was produced in E. coli and thus not glycosylated. In this structure, N559 of SV2C forms hydrogen bonds with T1145 and Y1149 of BoNT/A1; the N559A mutant did not seem to affect toxin binding (Figure 3) (Benoit et al., 2014). Future structural and functional studies using the glycosylated SV2 will help to understand the physiological role of SV2 glycosylation in BoNT recognition.
Functional implications and future perspectives
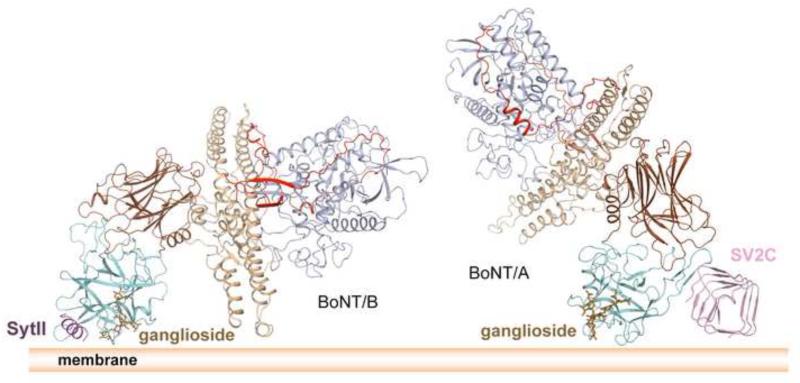
Being anchored by two receptors imposes geometric restraints on how BoNTs orientate on the cell surface, which is important for the subsequent toxin endocytosis and translocation of LC to the cytoplasm (Benoit et al., 2014; Berntsson et al., 2013b; Chai et al., 2006; Jin et al., 2006) (Figure 4). It is proposed that HC of BoNT/B is anchored within the proximity of the membrane by ganglioside and Syt, and its HN touches the membrane with the end of its long helices, which might be needed for entry of HN into the membrane during LC translocation (Berntsson et al., 2013a; Jin et al., 2006) (Figure 4, left). BoNT/B and Syt likely stay associated at luminal pH because their interaction is pH independent (Jin et al., 2006). However, HN of BoNT/A likely does not touch the membrane when anchored by GT1b and SV2, given the structure of the apo BoNT/A1 (Benoit et al., 2014; Lacy et al., 1998) (Figure 4, right). Therefore, BoNT/A may need a conformational change to “activate” the HN function by bringing it closer to the membrane. This could be triggered by the acidic luminal pH of synaptic vesicles, as HC of BoNT/A could reorient relative to HN through a flexible peptide linker at acidic pH (Gu et al., 2012; Matsui et al., 2014). Alternatively, it is noted that BoNT/A1–SV2C interaction was weakened by five-fold under acidic pH, and the complex may dissociate upon vesicle acidification (Benoit et al., 2014). Residue H564 of human SV2C was speculated to be the key pH-sensing residue. However, in human, rat and mouse SV2B, this histidine is substituted by Glu that has a much lower pKa (Ka is the acid dissociation constant). Mutagenesis studies and further functional studies are needed to appreciate the physiological relevance of this observation.
Figure 4.
The structural models of BoNT/B (left) and BoNT/A (right) binding on the cell surface. The color scheme follows that of Figure 1.
In recent years, we have seen a tremendous progress on structural studies of BoNT and their protein receptors, which have unraveled diverse receptor-recognition strategies used by BoNTs to achieve neurotropism. These new findings have raised more questions than they have answered. For example, how can the dominating backbone-backbone protein interactions observed in the BoNT/A1–SV2C complex achieve the high binding specificity needed to avoid off-target binding of BoNT? Whether and how does glycosylation of the protein receptor contribute to BoNT– neuron recognition? Do the different BoNT subtypes within a serotype recognize the same protein receptor in the same manner? What is the translocation-competent conformation of BoNTs upon dual receptor binding, and how does this strategic orientation of BoNTs on membrane correlate to the largely unknown HN-mediated LC translocation? Understanding the BoNT–protein receptor recognition has profound implication in developing novel anti-BoNT strategies, such as developing peptide inhibitors, antibodies, or small molecules to target the interacting sites and specifically block BoNT binding to motoneurons. On the other hand, the structures also provide a framework for structure-based evolution of the toxin molecules to improve their potency for therapeutic applications.
Acknowledgments
This work was supported in part by National Institute of Allergy and Infectious Diseases (NIAID) grant R01AI091823 to R.J..
Abbreviations
- BoNT
botulinum neurotoxin
- LC
light chain
- HC
heavy chain
- HN
N-terminal domain of HC
- HC
C-terminal domain of HC
- HCN
N-terminal subdomain of HC
- HCC
C-terminal subdomain of HC
- Syt
synaptotagmin
- SV2
synaptic vesicle glycoprotein 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Hauer J, Layton M, Lillibridge S, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Swerdlow DL, Tonat K. Botulinum toxin as a biological weapon: medical and public health management. JAMA. 2001;285:1059–70. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- Bajjalieh SM, Peterson K, Shinghal R, Scheller RH. SV2, a brain synaptic vesicle protein homologous to bacterial transporters. Science. 1992;257:1271–3. doi: 10.1126/science.1519064. [DOI] [PubMed] [Google Scholar]
- Barash JR, Arnon SS. A novel strain of Clostridium botulinum that produces type B and type H botulinum toxins. J Infect Dis. 2014;209:183–91. doi: 10.1093/infdis/jit449. [DOI] [PubMed] [Google Scholar]
- Benoit RM, Frey D, Hilbert M, Kevenaar JT, Wieser MM, Stirnimann CU, McMillan D, Ceska T, Lebon F, Jaussi R, Steinmetz MO, Schertler GF, Hoogenraad CC, Capitani G, Kammerer RA. Structural basis for recognition of synaptic vesicle protein 2C by botulinum neurotoxin A. Nature. 2014;505:108–11. doi: 10.1038/nature12732. [DOI] [PubMed] [Google Scholar]
- Bercsenyi K, Schmieg N, Bryson JB, Wallace M, Caccin P, Golding M, Zanotti G, Greensmith L, Nischt R, Schiavo G. Tetanus toxin entry. Nidogens are therapeutic targets for the prevention of tetanus. Science. 2014;346:1118–23. doi: 10.1126/science.1258138. [DOI] [PubMed] [Google Scholar]
- Berntsson RP, Peng L, Dong M, Stenmark P. Structure of dual receptor binding to botulinum neurotoxin B. Nat Commun. 2013a;4:2058. doi: 10.1038/ncomms3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berntsson RP, Peng L, Svensson LM, Dong M, Stenmark P. Crystal structures of botulinum neurotoxin DC in complex with its protein receptors synaptotagmin I and II. Structure. 2013b;21:1602–11. doi: 10.1016/j.str.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigalke H. Botulinum toxin: application, safety, and limitations. Curr Top Microbiol Immunol. 2013;364:307–17. doi: 10.1007/978-3-642-33570-9_14. [DOI] [PubMed] [Google Scholar]
- Bigalke H, Rummel A. Medical aspects of toxin weapons. Toxicology. 2005;214:210–20. doi: 10.1016/j.tox.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Binz T, Kurazono H, Wille M, Frevert J, Wernars K, Niemann H. The complete sequence of botulinum neurotoxin type A and comparison with other clostridial neurotoxins. J Biol Chem. 1990;265:9153–8. [PubMed] [Google Scholar]
- Blasi J, Chapman ER, Link E, Binz T, Yamasaki S, De Camilli P, Sudhof TC, Niemann H, Jahn R. Botulinum neurotoxin A selectively cleaves the synaptic protein SNAP-25. Nature. 1993;365:160–3. doi: 10.1038/365160a0. [DOI] [PubMed] [Google Scholar]
- Chai Q, Arndt JW, Dong M, Tepp WH, Johnson EA, Chapman ER, Stevens RC. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature. 2006;444:1096–100. doi: 10.1038/nature05411. [DOI] [PubMed] [Google Scholar]
- Dong M, Liu H, Tepp WH, Johnson EA, Janz R, Chapman ER. Glycosylated SV2A and SV2B mediate the entry of botulinum neurotoxin E into neurons. Mol Biol Cell. 2008;19:5226–37. doi: 10.1091/mbc.E08-07-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Yeh F, Tepp WH, Dean C, Johnson EA, Janz R, Chapman ER. SV2 is the protein receptor for botulinum neurotoxin A. Science. 2006;312:592–6. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- Dover N, Barash JR, Hill KK, Xie G, Arnon SS. Molecular characterization of a novel botulinum neurotoxin type H gene. J Infect Dis. 2014;209:192–202. doi: 10.1093/infdis/jit450. [DOI] [PubMed] [Google Scholar]
- Feany MB, Lee S, Edwards RH, Buckley KM. The synaptic vesicle protein SV2 is a novel type of transmembrane transporter. Cell. 1992;70:861–7. doi: 10.1016/0092-8674(92)90319-8. [DOI] [PubMed] [Google Scholar]
- Fernandez-Chacon R, Konigstorfer A, Gerber SH, Garcia J, Matos MF, Stevens CF, Brose N, Rizo J, Rosenmund C, Sudhof TC. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–9. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- Fu Z, Chen C, Barbieri JT, Kim JJ, Baldwin MR. Glycosylated SV2 and gangliosides as dual receptors for botulinum neurotoxin serotype F. Biochemistry. 2009;48:5631–41. doi: 10.1021/bi9002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga Y, Sugawara Y, Matsumura T. Uptake of botulinum neurotoxin in the intestine. Curr Top Microbiol Immunol. 2013;364:45–59. doi: 10.1007/978-3-642-33570-9_3. [DOI] [PubMed] [Google Scholar]
- Gill DM. Bacterial toxins: a table of lethal amounts. Microbiol Rev. 1982;46:86–94. doi: 10.1128/mr.46.1.86-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginalski K, Venclovas C, Lesyng B, Fidelis K. Structure-based sequence alignment for the beta-trefoil subdomain of the clostridial neurotoxin family provides residue level information about the putative ganglioside binding site. FEBS Lett. 2000;482:119–24. doi: 10.1016/s0014-5793(00)01954-2. [DOI] [PubMed] [Google Scholar]
- Gu S, Jin R. Assembly and function of the botulinum neurotoxin progenitor complex. Curr Top Microbiol Immunol. 2013;364:21–44. doi: 10.1007/978-3-642-33570-9_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Rumpel S, Zhou J, Strotmeier J, Bigalke H, Perry K, Shoemaker CB, Rummel A, Jin R. Botulinum neurotoxin is shielded by NTNHA in an interlocked complex. Science. 2012;335:977–81. doi: 10.1126/science.1214270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacky BP, Garay PE, Dupuy J, Nelson JB, Cai B, Molina Y, Wang J, Steward LE, Broide RS, Francis J, Aoki KR, Stevens RC, Fernandez-Salas E. Identification of fibroblast growth factor receptor 3 (FGFR3) as a protein receptor for botulinum neurotoxin serotype A (BoNT/A) PLoS Pathog. 2013;9:e1003369. doi: 10.1371/journal.ppat.1003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz R, Sudhof TC. SV2C is a synaptic vesicle protein with an unusually restricted localization: anatomy of a synaptic vesicle protein family. Neuroscience. 1999;94:1279–90. doi: 10.1016/s0306-4522(99)00370-x. [DOI] [PubMed] [Google Scholar]
- Jin R, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature. 2006;444:1092–5. doi: 10.1038/nature05387. [DOI] [PubMed] [Google Scholar]
- Karalewitz AP, Fu Z, Baldwin MR, Kim JJ, Barbieri JT. Botulinum neurotoxin serotype C associates with dual ganglioside receptors to facilitate cell entry. J Biol Chem. 2012;287:40806–16. doi: 10.1074/jbc.M112.404244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran D, Eswaramoorthy S, Furey W, Navaza J, Sax M, Swaminathan S. Domain organization in Clostridium botulinum neurotoxin type E is unique: its implication in faster translocation. J Mol Biol. 2009;386:233–45. doi: 10.1016/j.jmb.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Lacy DB, Tepp W, Cohen AC, DasGupta BR, Stevens RC. Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nat Struct Biol. 1998;5:898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- Lee K, Gu S, Jin L, Le TT, Cheng LW, Strotmeier J, Kruel AM, Yao G, Perry K, Rummel A, Jin R. Structure of a bimodular botulinum neurotoxin complex provides insights into its oral toxicity. PLoS Pathog. 2013;9:e1003690. doi: 10.1371/journal.ppat.1003690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Zhong X, Gu S, Kruel AM, Dorner MB, Perry K, Rummel A, Dong M, Jin R. Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex. Science. 2014;344:1405–10. doi: 10.1126/science.1253823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrhold S, Rummel A, Bigalke H, Davletov B, Binz T. The synaptic vesicle protein 2C mediates the uptake of botulinum neurotoxin A into phrenic nerves. FEBS Lett. 2006;580:2011–4. doi: 10.1016/j.febslet.2006.02.074. [DOI] [PubMed] [Google Scholar]
- Mahrhold S, Strotmeier J, Garcia-Rodriguez C, Lou J, Marks JD, Rummel A, Binz T. Identification of the SV2 protein receptor-binding site of botulinum neurotoxin type E. Biochem J. 2013;453:37–47. doi: 10.1042/BJ20130391. [DOI] [PubMed] [Google Scholar]
- Matsui T, Gu S, Lam KH, Carter LG, Rummel A, Mathews II, Jin R. Structural basis of the pH-dependent assembly of a botulinum neurotoxin complex. J Mol Biol. 2014;426:3773–82. doi: 10.1016/j.jmb.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco C. How do tetanus and botulinum neurotoxins bind to neuronal membranes? Trends Biochem Sci. 1986;11:314–17. [Google Scholar]
- Peng L, Berntsson RP, Tepp WH, Pitkin RM, Johnson EA, Stenmark P, Dong M. Botulinum neurotoxin D-C uses synaptotagmin I and II as receptors, and human synaptotagmin II is not an effective receptor for type B, D-C and G toxins. J Cell Sci. 2012;125:3233–42. doi: 10.1242/jcs.103564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Tepp WH, Johnson EA, Dong M. Botulinum neurotoxin D uses synaptic vesicle protein SV2 and gangliosides as receptors. PLoS Pathog. 2011;7:e1002008. doi: 10.1371/journal.ppat.1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce EJ, Davison MD, Parton RG, Habig WH, Critchley DR. Characterization of tetanus toxin binding to rat brain membranes. Evidence for a high-affinity proteinase-sensitive receptor. Biochem J. 1986;236:845–52. doi: 10.1042/bj2360845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto O, Pirazzini M, Montecucco C. Botulinum neurotoxins: genetic, structural and mechanistic insights. Nat Rev Microbiol. 2014;12:535–49. doi: 10.1038/nrmicro3295. [DOI] [PubMed] [Google Scholar]
- Rummel A. Double Receptor Anchorage of Botulinum Neurotoxins Accounts for their Exquisite Neurospecificity. Curr Top Microbiol Immunol. 2013;364:61–90. doi: 10.1007/978-3-642-33570-9_4. [DOI] [PubMed] [Google Scholar]
- Rummel A, Eichner T, Weil T, Karnath T, Gutcaits A, Mahrhold S, Sandhoff K, Proia RL, Acharya KR, Bigalke H, Binz T. Identification of the protein receptor binding site of botulinum neurotoxins B and G proves the double-receptor concept. Proc Natl Acad Sci U S A. 2007;104:359–64. doi: 10.1073/pnas.0609713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rummel A, Hafner K, Mahrhold S, Darashchonak N, Holt M, Jahn R, Beermann S, Karnath T, Bigalke H, Binz T. Botulinum neurotoxins C, E and F bind gangliosides via a conserved binding site prior to stimulation-dependent uptake with botulinum neurotoxin F utilising the three isoforms of SV2 as second receptor. J Neurochem. 2009;110:1942–54. doi: 10.1111/j.1471-4159.2009.06298.x. [DOI] [PubMed] [Google Scholar]
- Schiavo G, Benfenati F, Poulain B, Rossetto O, Polverino de Laureto P, DasGupta BR, Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–5. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- Schiavo G, Matteoli M, Montecucco C. Neurotoxins affecting neuroexocytosis. Physiol Rev. 2000;80:717–66. doi: 10.1152/physrev.2000.80.2.717. [DOI] [PubMed] [Google Scholar]
- Schmitt J, Karalewitz A, Benefield DA, Mushrush DJ, Pruitt RN, Spiller BW, Barbieri JT, Lacy DB. Structural analysis of botulinum neurotoxin type G receptor binding. Biochemistry. 2010;49:5200–5. doi: 10.1021/bi100412v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark P, Dong M, Dupuy J, Chapman ER, Stevens RC. Crystal structure of the botulinum neurotoxin type G binding domain: insight into cell surface binding. J Mol Biol. 2010;397:1287–97. doi: 10.1016/j.jmb.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotmeier J, Gu S, Jutzi S, Mahrhold S, Zhou J, Pich A, Eichner T, Bigalke H, Rummel A, Jin R, Binz T. The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites. Mol Microbiol. 2011;81:143–56. doi: 10.1111/j.1365-2958.2011.07682.x. [DOI] [PubMed] [Google Scholar]
- Strotmeier J, Mahrhold S, Krez N, Janzen C, Lou J, Marks JD, Binz T, Rummel A. Identification of the synaptic vesicle glycoprotein 2 receptor binding site in botulinum neurotoxin A. FEBS Lett. 2014;588:1087–93. doi: 10.1016/j.febslet.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotmeier J, Willjes G, Binz T, Rummel A. Human synaptotagmin-II is not a high affinity receptor for botulinum neurotoxin B and G: increased therapeutic dosage and immunogenicity. FEBS Lett. 2012;586:310–3. doi: 10.1016/j.febslet.2011.12.037. [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Eswaramoorthy S. Structural analysis of the catalytic and binding sites of Clostridium botulinum neurotoxin B. Nat Struct Biol. 2000;7:693–9. doi: 10.1038/78005. [DOI] [PubMed] [Google Scholar]
- Whitemarsh RC, Tepp WH, Bradshaw M, Lin G, Pier CL, Scherf JM, Johnson EA, Pellett S. Characterization of botulinum neurotoxin A subtypes 1 through 5 by investigation of activities in mice, in neuronal cell cultures, and in vitro. Infect Immun. 2013;81:3894–902. doi: 10.1128/IAI.00536-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willjes G, Mahrhold S, Strotmeier J, Eichner T, Rummel A, Binz T. Botulinum neurotoxin G binds synaptotagmin-II in a mode similar to that of serotype B: tyrosine 1186 and lysine 1191 cause its lower affinity. Biochemistry. 2013;52:3930–8. doi: 10.1021/bi4003502. [DOI] [PubMed] [Google Scholar]