Abstract
RecG is a potent, atypical, monomeric DNA helicase. It simultaneously couples ATP hydrolysis to duplex unwinding and rewinding, and to the displacement of proteins bound to the DNA. A model is presented for the localization of the enzyme to the inner membrane via its binding to SSB. Upon fork stalling, SSB targets the enzyme to the fork where it can act. RecG displays a strong preference for processing the fork in the regression direction, that is, away from the site of damage that initially led to fork arrest. Regression is mediated by strong binding of the wedge domain to the fork arms as well as to parental duplex DNA by the helicase domains. Once RecG has regressed the fork, it will dissociate leaving the now relaxed, Holliday junction-like DNA, available for further processing by enzymes such as RuvAB.
1. Introduction
Genome duplication is inherently accurate, highly processive and relies on the close interplay between the genetic recombination and DNA repair machinery (Kogoma, 1997; Kowalczykowski, 2000; Kuzminov, 1999). The need for this interplay arises due to the replication machinery frequently encountering roadblocks that have the potential to stall or collapse a replication fork (Cox, 2001; Cox et al., 2000; Seigneur et al., 1998). The types of lesions that could disrupt replication include proteins bound to the DNA ahead of the replication fork such as repair enzymes or RNA polymerase, non-coding lesions in the template DNA and either single- or double strand breaks (Kowalczykowski, 2000; Marians, 2004; McGlynn and Lloyd, 2002b). Each of the different blocks could lead to a different type of damage to the DNA and this is highlighted by the varied recombination and repair gene requirements for dealing with exposure to different types of DNA damaging agents (Marians, 2000; Marians, 2004; McGlynn and Lloyd, 2002a; Michel et al., 2004). Whatever its source, the impediment to replication fork progression has to be removed or bypassed and replication must be restarted.
In bacteria, stalled replication forks can directly restarted or regressed (Lusetti and Cox, 2002; Marians, 2000; Marians, 2004; Michel et al., 2004). That is, moved in the direction opposite to that of replisome movement (Figure 1). Although replication fork regression can in principle be spontaneous as demonstrated by Cozzarelli (Postow et al., 2001), it can also be catalyzed by a number of proteins (McGlynn and Lloyd, 2001; Robu et al., 2001; Robu et al., 2004; Seigneur et al., 1998). Over the past several years two branched DNA-specific molecular motors known as RecG and RuvAB emerged as potential key players in the regression of stalled replication forks (McGlynn et al., 2000; Seigneur et al., 1998). Studies favouring RuvAB as the dominant player came from genetic studies showing that mutations in ruvAB had disastrous effects on survival following UV-irradiation, whereas mutations in recG had only a small effect (Baharoglu et al., 2006; Michel et al., 2007; Seigneur et al., 1998). In addition, biochemical studies demonstrate that there is significant overlap in DNA substrate specificity for these two enzymes suggesting that they may act on a similar range of substrates in vivo. However, a series of recent biochemical and genetic studies strongly point to RecG being the initial, dominant player in fork regression (Abd Wahab et al., 2013; Buss et al., 2008; Manosas et al., 2014). It is only once RecG has acted, and processed the fork into a Holliday junction-like structure and which has little or no superhelical tension, can RuvAB act. This suggests a temporal sequence of events with RecG acting first, followed by RuvAB (Abd Wahab et al., 2013; Buss et al., 2008; Manosas et al., 2014). As formation of the Holliday junction-like structure is thought to be a key intermediate in many fork rescue pathways, and it is possible for it to arise via a number of processing mechanisms, this explains the strong dependence on RuvAB (Seigneur et al., 1998). That is, processing, regardless of pathway, leads to a RuvAB substrate.
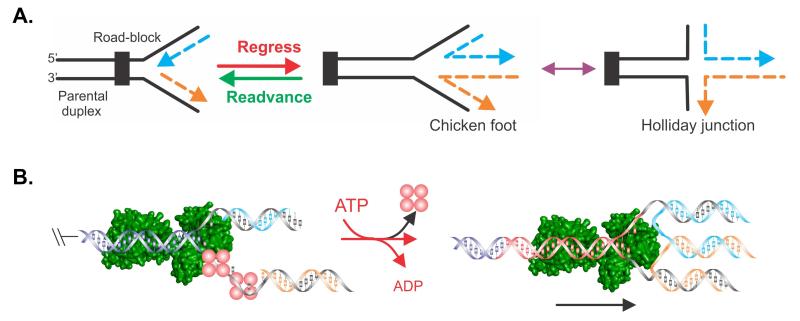
Figure 1. Schematic of events that could transpire at a stalled DNA replication fork.
(A). The fork is shown impeded and the nascent leading (blue) and lagging (orange) strands indicated. Fork regression (red arrow) would result in rightward movement of the fork, away from the site of the replication road-block, concomitant with the extrusion of a duplex region resulting from the annealing of the nascent leading and lagging strands. The resulting DNA structure has been termed the “chicken foot” and is structurally similar to a Holliday junction. Fork readvancement (not driven by RecG) would result in leftward movement of the fork in a reaction that is likely catalyzed after regression and impediment removal or repair. (B). Model of a possible mechanism of fork regression by RecG (green). Unreplicated DNA ahead of the previously advancing fork is shown in dark blue. The nascent leading and lagging strands are coloured to match those in panel A. Red spheres, SSB. Black arrow in the right panel indicates the direction of RecG translocation and consequently, of fork regression. The nascent, reannealed parental duplex is coloured black.
But how can a single, monomeric protein such as RecG, present at 7 molecules per cell, gain access to a stalled replication fork, in the milieu of 4.7Mb of DNA and numerous DNA binding proteins? The least of which is RuvAB, present at several hundred copies per cell. A number of possibilities exist. RecG could be associated with one or more protein components of the replisome so that when stalling occurs, it is already in proximity so that it can readily act. Alternatively, and as RecG binds to a variety of fork structures with affinity in the low nanomolar range, a constitutively active and omniscient presence could have disastrous effects on genome stability. Thus regulation, perhaps by keeping the enzyme in a storage form and rapidly delivering it to sites of DNA damage only when needed, may be key to enzyme function. Critically, data for both possibilities exist, and these may not be mutually exclusive.
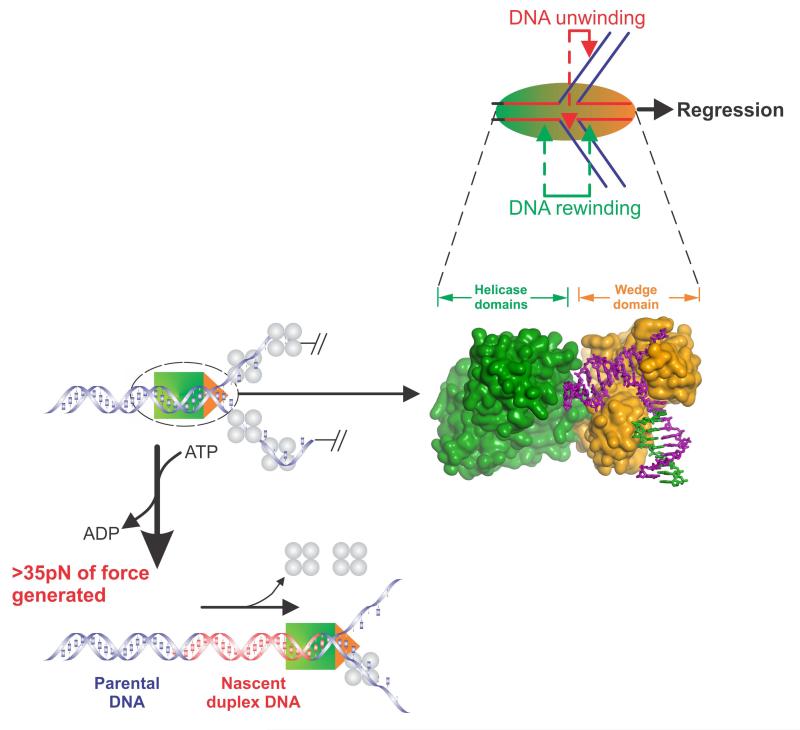
Finally, once RecG is loaded onto a stalled fork, it must possess several key activities that operate simultaneously. First, it must operate as an atypical DNA helicase. That is, it must be capable of both unwinding nascent duplex regions while simultaneously rewinding DNA both ahead of the advancing enzyme, as well as in its wake (Figure 1). Second, during the process of translocation and duplex DNA remodeling, it must generate sufficient force so as to displace proteins which may be bound to either single- or double-stranded DNA regions. A recent single molecule study demonstrates that RecG has these abilities which act in unison to efficiently catalyze fork regression generating a Holliday junction-like structure in the process (Manosas et al., 2013).
In this review, a model is proposed for RecG acting at a fork. The model is consistent with the current state of the literature and highlights the remarkable characteristics present in a single, monomeric, SuperFamily II DNA helicase.
2. Identification of RecG
The recG locus was identified as a mutation (recG162; Ala428 to Val in helicase motif III) that made cells moderately recombination deficient and sensitive to UV irradiation (Kalman et al., 1992; Lloyd, 1991; Lloyd and Buckman, 1991). The most interesting phenotypic effect of recG mutations is that RecG, like RecA, operates in all three pathways of recombination. Although the effects of recG are not nearly as severe as those of recA, noticeable recombination deficiencies are observed in rec+, recBC sbcA, and recBC sbcBC strains. In otherwise wild-type cells, these effects include sensitivity to UV, ionizing radiation, and mitomycin C, as well as decreased levels of conjugal and transductional recombination (Lloyd, 1991). These results suggested that recG could be involved in recombinational repair of both dsDNA breaks and ssDNA gaps.
Mutants lacking RecG function allow constitutive stable DNA replication (cSDR or SDR), that is, DNA replication in the absence of protein synthesis (Hong et al., 1995). They also exhibit an absolute requirement for Pol I DNA polymerase activity. They are lethal in combination with mutants in rnhA, another gene whose absence allows cSDR (Hong et al., 1995; Kogoma et al., 1993). recG mutants are somewhat more susceptible to DNA damage as alluded to above (Courcelle and Hanawalt, 2003; Whitby et al., 1994). In the specific case of UV irradiation, recG mutants lead to a PriA dependent pathological DNA replication cascade, most likely through repeated generation of new replication forks (Rudolph et al., 2010a; Rudolph et al., 2009). recG deletion mutants have a high incidence of extra replication initiation in the terminus area. This replication is the result of PriA-PriB mediated loading of DnaB at branched DNA structures that form in the absence of RecG (Rudolph et al., 2010a). Collectively, these studies have led to the proposal that RecG may be a guardian of the genome, limiting replication initiation to oriC and replication restart to resurrected forks (Rudolph et al., 2010b).
But what exactly could RecG be doing? Further clarification the role of RecG in recombination came from studies in which a recG mutation was combined with mutations in other genes involved in recombination. Modest effects were seen in combination with recB and recJ, but the most significant and insightful were those observed in ruv-recG double mutants, where ruv (resistance to UV) is mutation of either ruvA, ruvB, or ruvC (Lloyd, 1991; Lloyd and Buckman, 1991). Mutants with single mutations in either ruvA, ruvB, or ruvC have approximately similar phenotypes and are fairly recombination proficient. When the ruv mutation is present in combination with recG however, both conjugational and transductional recombination and UV resistance are dramatically reduced (30- to 500-fold greater than in the mutants with single ruv mutations), arguing that the ruv and recG genes define components of alternative resolution recombination pathways (Bennett et al., 1993). As RuvABC is known to act on recombination intermediates, this suggested that RecG could do so as well, but it would take the biochemistry and biophysics to make this exquisitely clear.
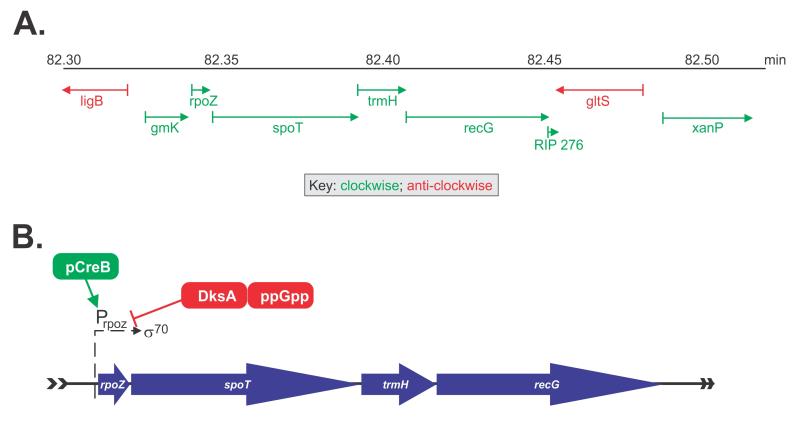
The recG gene is 2,082bp in length and encodes a monomeric, 76.43 kDa protein that is present at 7 molecules per cell (Taniguci et al., 2010). It maps to 82.4 min on the E. coli chromosome and is part of the spo operon that includes rpoZ, spoT, trmH (previously known as spoU) (262, 353) (Figure 2A)... This operon is part of the stringent response and encodes the omega subunit of RNA polymerase (rpoZ), the predominant guanosine 3′,5′-bispyrophosphate hydrolase (spoT), responsible for cellular (ppGpp) degradation and a tRNA (Guanosine-2′-O-)-methyltransferase (trmH) (Gentry and Burgess, 1986; Persson et al., 1997; Sarubbi et al., 1989). Transcription of the operon occurs in a clockwise orientation relative to the direction of replication fork progression and initiates at the rpoZ promoter which is recognized by the RNA polymerase σ70-holoenzyme (Sarubbi et al., 1989). This promoter does not contain a LexA binding site and as such, is not under the control of the SOS regulon (Lloyd and Sharples, 1991). Transcription is induced by the CreBC two component system in minimal medium growth conditions and suppressed by DksA (dnaK suppressor) bound to ppGpp (Figure 2B and (Avison et al., 2001)). (Taniguchi et al., 2010)
Figure 2. The recG gene is part of the spo operon.
(A), a schematic showing the location of the operon in the E.coli chromosome. (B), Transcription of the spo operon. Initiation occurs at the rpoZ promoter which is recognized by σ70-RNA polymerase. Positive regulation by CreB (green) and negative regulation by DksA (red) are indicated.
How can a DNA helicase function in the stringent response? The answer to this came from a study from the Lloyd group that showed a strong correlation between the ability to survive UV irradiation and the ability to synthesize ppGpp (McGlynn and Lloyd, 2000). They described how elevation of ppGpp or a subclass of stringent RNA polymerase (RNAP) mutations (rpo*) that mimic the effect of ppGpp dramatically improves survival of strains lacking RuvABC. They postulated that ppGpp and rpo* destabilize RNAP, thus reducing the incidence of stalled complexes and facilitating excision repair, thereby clearing the way for replication to continue. This follows because RNAP is modulated by the stringent response regulator ppGpp, which binds next to the active site to destabilize open complexes (Cashel et al., 1996). The interaction between RNAP and ppGpp is stabilized by DksA, which inserts a coiled-coil domain into the secondary channel of RNAP (Paul et al., 2004; Perederina et al., 2004). When these stringent response regulators are removed from the picture, there would be an increase in stalled RNAP molecules around the genome, resulting in elevated levels of fork stalling, likely requiring the actions of RecG for rescue.
3. Characterization of the RecG protein
The protein possesses ATPase and DNA helicase activities, consistent with its primary sequence containing a Walker A motif and similarity to the DEXH class of DNA and RNA helicases (Lloyd and Sharples, 1991). It has been classified as a member of the SF2 DNA helicases and nucleic acid translocases (Singleton et al., 2007). Homology to DNA helicases is limited to the C-terminal half of the protein which contains the now well recognized helicase motifs that impart the ability to bind and hydrolyze nucleoside triphosphates and couple the associated conformational changes into the ability to perform work (Singleton et al., 2001). The N-terminal half contains the wedge domain responsible for binding to forked DNA structures as explained below.
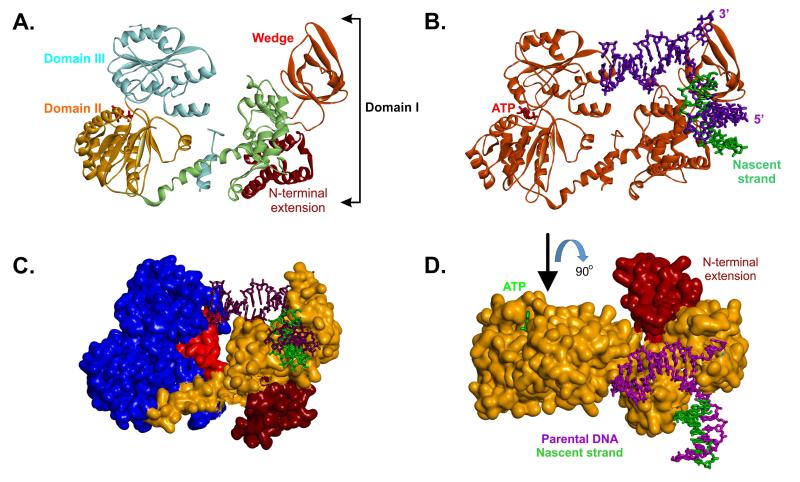
Analysis of the crystal structure of the enzyme bound to a stalled DNA replication fork substrate revealed that RecG can be divided into three structural domains (Figure 3A and (Singleton et al., 2001)). Domain 1 is the largest, comprising approximately half of the protein and contains the wedge domain which confers the ability to bind to all of the above-mentioned branched DNA structures (Figure 3A, red). Binding via the wedge is responsible for clamping the enzyme tightly onto the DNA, splitting the junction and stabilizing the unwound fork (Briggs et al., 2005). The wedge domain is coupled to domains 2 and 3, via an α-helical linker (Figure 3A, light green). In the Thermotoga structure, there is an N-terminal extension of unknown function and unique to this genus (Figure 3A and C). The remainder of the protein is split approximately equally between the two C-terminal domains (Domains 2 and 3; Figure 3A, orange and cyan regions respectively). Domains 2 and 3 contain the characteristic motifs that identify RecG as an SF2 helicase, and couple the energy released from ATP hydrolysis to drive the enzyme (Gorbalenya and Koonin, 1988; Gorbalenya et al., 1988; Singleton et al., 2001). An additional and more recently identified motif also present in this domain, is the TRG motif; (TRG = translocation by RecG; Figure 3C, red) (Mahdi et al., 2003). The TRG motif forms a helical hairpin linked to a loop projecting into the proposed dsDNA binding channel positioned between the helicase and wedge domains (Mahdi et al., 2003). Mutations in TRG disrupt unwinding of HJ and forked DNA structures in vitro Mahdi et al., 2003)
Figure 3. Domain organization of the RecG DNA helicase.
(A), a ribbon diagram showing the three domains of RecG (details in the text). The positions of the wedge sub-domain and N-terminal extension, present in T. maritima RecG but not E.coli, are indicated. (B), a ribbon diagram of RecG (orange) bound to a model fork substrate. The position of the ATP sandwiched between domains II and III is highlighted in red. (C), T. maritima RecG is represented as a Connelly surface. The N-terminal extension is coloured brown; the wedge and linker region are coloured orange and the helicase domains are blue. The TRG motif is coloured in red. (D), T. maritima RecG is represented as a Connelly surface and is coloured orange. The N-terminal extension is indicated. The view of the protein is rotated 90° relative to that in panel B.
In vitro analyses showed RecG to be a 3′ → 5′ polarity DNA helicase with a variety of roles in DNA repair and recombination (130,131). The 76 kDa enzyme functions as a monomer, binding specifically to stalled replication fork substrates (and a variety of structures resembling these such as R-loops, D-loops and Holliday junctions (HJ)) and subsequently processes them into structures that can be acted upon by additional members of the recombination machinery (Abd Wahab et al., 2013; Buss et al., 2008; Fukuoh et al., 1997; Lloyd and Sharples, 1993; Slocum et al., 2007; Vincent et al., 1996; Whitby et al., 1993).
In addition to being able to process a variety of branched DNA structures in vitro, RecG exhibits significant ATPase activity on (-)scDNA, ssDNA, and SSB-coated M13 ssDNA (Abd Wahab et al., 2013; Buss et al., 2008; Slocum et al., 2007). This suggests different ways for RecG to access a stalled replication fork and these are dictated by the type of DNA available. The strong preference that the enzyme exhibits for (-)scDNA in vitro, suggests that DNA must first be converted from (+) to (-)scDNA for RecG to function. Once the DNA is in this form, RecG catalyzes fork regression efficiently (McGlynn et al., 2001). Activity on SSB-coated M13 ssDNA is intriguing as it involves a species-specific, protein-protein interaction between RecG and SSB (Buss et al., 2008). This interaction is mediated through the C-terminal tail of SSB, similar to that observed for Exonuclease I, PriA, RecQ and Topoisomerase III (Shereda et al., 2008). Further, this interaction is key to RecG function at a stalled fork since the enzyme can be directly loaded onto the DNA in single-stranded regions and is consistent with the role of SSB targeting repair helicases to active forks in vivo (Abd Wahab et al., 2013; Buss et al., 2008; Slocum et al., 2007).
Once loaded at a stalled replication fork, RecG is thought to regress these away from the site(s) of DNA damage, resulting in the formation of a 4-way intermediate called a “chicken foot” that resembles a Holliday junction (Figure 1, 4 and (Manosas et al., 2013; McGlynn and Lloyd, 2000)). The rate of fork reversal is 240 bp/s with the enzyme hydrolyzing 1 ATP to track a distance of 3bp (Manosas et al., 2013; Martinez-Senac and Webb, 2005). The similarity of the resulting reversed or regressed DNA structures to Holliday Junctions suggests that following the action of RecG, further processing is carried out by RuvAB (Abd Wahab et al., 2013; Buss et al., 2008; Manosas et al., 2013; Manosas et al., 2014).
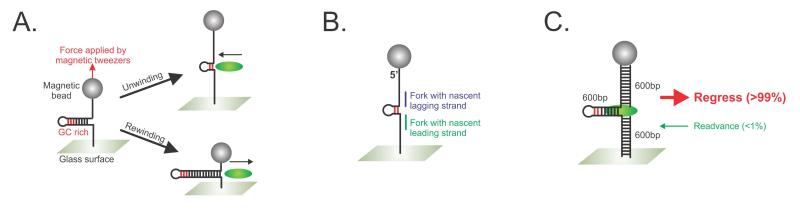
Figure 4. Magnetic tweezers have been used to determine how RecG processes model fork substrates.
(A), a schematic of the assay system. The hairpin DNA substrate consists of complementary single stranded arms that can extrude an approximately 1,200 bp hairpin that has a GC clamp (indicated in red). The substrate is tethered at one end to the surface of a glass flow cell and to a magnetic bead at the opposite end. The position of this bead can be controlled by magnetic tweezers and its position recorded with nanometer resolution. Experiments are carried out with an opposing force of 18pN (red arrow) that is applied by the magnetic tweezers. If the RecG (green sphere) catalyzes DNA unwinding, the reaction will be assisted by the force of the tweezers and the hairpin will be unwound resulting in an increase in the net length of the DNA tether. In contrast, if RecG rewinds the complementary arms, the length of the hairpin will increase as it is extruded out the back of the enzyme, concomitant with a net decrease in the length of the substrate tether. (B), DNA substrates to determine substrate specificity for RecG. To construct these, the hairpin is unwound by the application of force and in separate reactions, oligonucleotides are annealed in situ, to create forks with nascent leading (green) and lagging (blue) strands. (C), The stalled DNA replication fork substrate. The arms are approximately equal in length, and similar to reactions in panel A, the DNA tether can be shortened as the result of regression (red arrow) concomitant with extrusion of a fourth arm (the Holliday junction or chicken foot). Alternatively, if RecG readvances the fork, the resulting Holliday junction would be reversed into the fork structure shown. Adapted from (Manosas et al., 2013; Manosas et al., 2014).
4. Mechanism of fork access
Analysis of the variety of DNA structures on which RecG can act leads one to believe that this is a promiscuous enzyme. If left unregulated, it could lead to disastrous consequences for the cell. One way to regulate the activity of the enzyme is to tightly control the levels of expression. Consistent, only 7 RecG molecules are present per cell (Taniguchi et al., 2010). A second method, in addition to the first, would actively control the enzyme itself. Recent work by the Bianco laboratory points to the regulator as being the single stranded DNA binding protein or SSB (Yu et al., 2014).
In vitro studies show that RecG binds to SSB both in the presence and absence of DNA (Buss et al., 2008). Furthermore, binding requires the highly conserved SSB C-terminus and is species-specific. In addition, SSB also stabilizes RecG on the DNA as evidenced by 2- to 5-fold increases in the salt-titration midpoint. Once loaded, RecG binds to model fork substrates with affinities in the low nanomolar range, versus high nanomolar for RuvAB (Abd Wahab et al., 2013; Buss et al., 2008; Slocum et al., 2007). Furthermore, SSB inhibits RuvAB activity on forks while simultaneously enhancing that of RecG. Thus, SSB plays a key role in dictating the access of enzymes to stalled replication forks. In addition, RecG out-competes a 1,200-fold excess of RuvAB in the presence of model fork substrates (Abd Wahab et al., 2013). This result indicates that if both RecG and RuvAB were present at a fork concurrently, RecG would preferentially bind and process the fork.
The above-mentioned in vitro studies demonstrated SSB-RecG binding, but this was done in the presence of 27% ammonium sulfate (~1.3M). This is potentially a problem as binding is not detectable in the absence of this salt. To address this issue, Yu et. al studied binding in vivo (Yu et al., 2014). They co-expressed his-SSB and RecG, and separately, SSB+hisRecG. The resulting fractions eluted from nickel columns demonstrated the presence of both the helicase and SSB. Co-elution is observed in both 0.6 and 0.1M NaCl. These results were confirmed by double-tagging experiments. Surprisingly, stoichiometric amounts of DNA were not detected in the eluted fractions.
This led to the proposal that SSB was bound to RecG in the absence of DNA and would then target the helicase to the stalled replication fork when needed. If this were the case, then SSB, typically present at ~2,000 tetramers per cell could be the storage form of the helicase (versus 7 monomers for RecG). This was demonstrated using fluorescent-tagged enzymes and fluorescence microscopy. This study showed that when SSB was present in excess over mcherry-RecG in the absence of DNA damage, RecG (and PriA) was localized to the inner membrane (Yu et al., 2014). This study further showed that in approximately 12% of cells, WtSSB-dependent foci containing both RecG and PriA could be detected, consistent with the number of stalled forks that might be present in an undamaged, exponentially growing culture (Cox et al., 2000). Thus from these studies, it was concluded that in the absence of DNA damage SSB maintains RecG at the inner membrane, stabilizing the enzyme. RecG is delivered to the fork when stalling occurs. As SSB/RecG is associated with the membrane, fork targeting is rapid due to the alteration of the search from 3- to 1-dimensional as the replisome is likely associated with the inner membrane possibly mediated by DnaA binding (Castuma et al., 1993; Yung et al., 1990).
In a separate study, the Lloyd group demonstrated that RecG co-localizes with SeqA and the replisome (Upton et al., 2014). Furthermore, conserved arginine and tryptophan residues near the C-terminus of RecG were required for this localization. Taken on face value, these data suggest that RecG moves with the replisome, although it is unclear which protein(s) it could be associated with. The Yu study suggests it is likely SSB. It was also unclear from this study if RecG is always associated with the replisome or whether colocalization was observed in a fraction of cells as in the Yu study.
5. Modes of action at a fork
Once RecG is bound to a fork substrate, what takes place? If RecG is loaded onto the fork and the DNA is still superhelical in character, it binds with high affinity and is able to catalyze an efficient regression reaction (McGlynn et al., 2001; Slocum et al., 2007). If single-stranded DNA regions are available then targeting by SSB is expected: either the SSB bound to the ssDNA regions could direct loading of RecG or, alternatively, SSB-RecG complexes could be targeted to the fork.
The reaction catalyzed by RecG is known as fork regression (Figure 1). The purpose of regression is required to move the fork away from the site of DNA damage, in a direction opposite to that of replisome movement, while simultaneously producing DNA structure(s) which, upon further processing would result in reloading of the replisome. Regression requires a specialized DNA helicase. In this unique reaction, the enzyme must bind specifically to the branch point and translocate on the parental duplex DNA immediately ahead of the fork (Figures 1, 4 and 5). The wedge domain of RecG is required for fork binding, while the remainder of the protein is required for ATP hydrolysis-coupled dsDNA translocation (Singleton et al., 2001). Next, the enzyme must both unwind the nascent duplex regions present in the arms of the fork, while simultaneously rewinding duplex DNA both behind and ahead of the advancing motor protein (Figure 5). The effect of this rewinding is to move the fork away from the site of duplex while “reforming” parental duplex DNA and extruding a duplex arm consisting of nascent strands of DNA.
Figure 5. Components of the fork regression reaction catalyzed by RecG.
Top panel, RecG (sphere) is shown bound to a fork in the process of catalyzing regression in the direction of the black arrow. During this reaction, the enzyme couples the hydrolysis of ATP to the simultaneous DNA transactions of unwinding of the nascent heteroduplex regions (red arrows) and DNA rewinding that occurs ahead of the translocating enzyme as well as in its wake (green arrows). Bottom left panels, RecG is shown in the process of fork regression on a DNA substrate bound by the single stranded DNA binding protein. During regression, RecG generates greater than 35pN of force that is sufficient to displace the bound SSB protein. Middle panel, Connolly surface of RecG bound to a model fork. The helicase and wedge domains are coloured to match the schematics in the other panels. When viewed in this way, it is clear that the wedge domain is bound at the fork while the helicase domains translocate along duplex DNA.
A recent single molecule study demonstrated RecG is capable of driving an efficient regression reaction (Manosas et al., 2014). In this study, a combination of optical and magnetic tweezers were used. Schematics of assays done with magnetic tweezers are shown in Figure 4. The hairpin substrate was used to demonstrate that RecG catalyzes strand annealing or duplex rewinding, a key component of fork regression. To understand how RecG would process a fork with gaps in either the lagging or leading strands, two substrates were constructed in situ. Here, the hairpin in Figure 4A was unzipped by the application of force and then in separate reactions, complementary oligonucleotides were introduced and allowed to bind to the leading or lagging strand arms. The results show that although RecG can process both DNA substrates, there is a strong preference for a substrate with a nascent lagging strand, as shown by the large difference in ton (1.8 ±0.1s versus 15 ±1 for the nascent leading strand). This means that RecG binds ten-fold faster to forks with lagging nascent strand than to forks with leading nascent strands, consistent with bulk-phase studies (Buss et al., 2008; Slocum et al., 2007). Finally, a DNA substrate was constructed to study fork regression itself (Figure 4C). Surprisingly, and even though the duplex arms are equivalent, the directional preference of RecG is almost 100% in the regression direction; virtually no fork readvancing was detected. The enzyme could however be induced to switch directions only when the magnesium ion concentration was altered.
During fork regression, RecG uses the energy stored in ATP to catalyze both DNA unwinding of the nascent heteroduplex arms and rewinding of these unwound strands (Figure 5, top panel). In the process, it makes extensive contacts with both the arms of the fork and the duplex DNA ahead of the fork (to the left of the fork in Figure 5). Contact with the arms of the fork is mediated by the wedge domain while contact with the duplex DNA is mediated via the helicase domains (Manosas et al., 2013; Manosas et al., 2014; Singleton et al., 2001). Surprisingly, during regression, RecG is able to generate more than 30pN of force. When asked to act against an opposing force of 30-35pN, there is only a 40% reduction in rewinding rate. Above 35pN, the enzyme rapidly dissociates from the DNA without stalling. The ability of this monomeric enzyme to work against such a large opposing force is significant, given that the multi-subunit RNA polymerase stalls completely at 30-35 pN (Wang et al., 1998). In contrast, by extrapolation of the data in Manosas et al, RecG is predicted to stall at ~50pN (Manosas et al., 2013; Manosas et al., 2014). This is significant for a monomeric enzyme, but not surprising given the role RecG plays in DNA metabolism. It must generate sufficient force to clear the DNA of any obstacles bound to the arms of the fork. In fact it readily displaces bound SSB protein, which binds to DNA with very high affinity (Figure 1B and 5, bottom).
To facilitate an efficient fork regression reaction, RecG must unwind two DNA duplexes simultaneously. How can this be achieved? As is evident in Figure 5, the two DNA channels present in the wedge domain are wide enough to accommodate single strands of DNA. This would suggest that as the DNA arms corresponding to the leading and lagging strands of the fork are pulled into the wedge, the nascent strands would be unwound from the parental DNA, allowing them to reanneal ahead of the translocating enzyme as shown in Figure 1B.
6. Conclusion
The monomeric enzyme RecG, catalyzes stalled DNA replication fork regression. Although RecG possesses the classic DNA helicase motifs it is not simply an enzyme that separates DNA duplexes into their component strands. Instead, it does the opposite of what a DNA helicase is supposed to do: it also rewinds unwound DNA duplexes. In the process of doing so it generates sufficient force to clear the DNA of any bound proteins so that further processing and repair of damage sites can ensue.
I propose that in the cell, RecG is stabilized by binding to SSB thereby preventing its degradation and preventing it from acting promiscuously. Furthermore, binding to SSB localizes RecG to the inner membrane (Figure 6). This is the storage form of the enzyme either as a RecG-SSB complex or a RecG-SSB-PriA complex. Once DNA damage leading to fork arrest and replisome dissociation occurs, SSB would rapidly transfer RecG to the fork where regression can take place. Due to the high affinity of SSB for ssDNA combined with the high affinity of RecG for fork structures, RecG is able to outcompete other proteins such as RuvAB. This enables RecG to act first, move the fork away from the site of damage (regression), allow repair to occur to the initial site of damage as well as additional fork processing by other DNA helicases and/or nucleases.
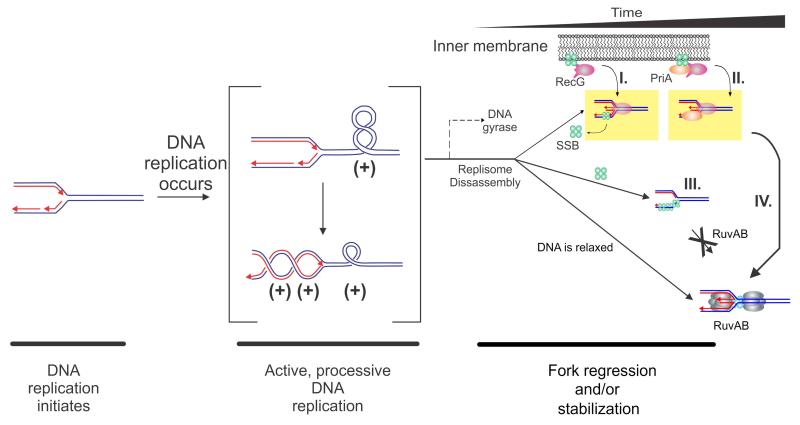
Figure 6. Mechanisms for targeting RecG to a stalled DNA replication fork.
A model of the topological domains of a segment of the E. coli chromosome undergoing replication is shown. This figure is adapted from (Buss et al., 2008; Slocum et al., 2007). Parental DNA is coloured blue and nascent daughter DNA is coloured red with arrowheads indicating 3′-ends. The RecG-SSB and RecG-PriA-SSB complexes are shown associated with the inner membrane as suggested by Yu et al (Yu et al., 2014). Once the fork encounters a block, one of several temporally spaced events may occur with RecG +/-SSB (and/or PriA) acting first followed by RuvAB. (I) If DNA gyrase acts prior to the dissociation of the replication machinery (i.e., within the 5-7 minute window following fork stalling), the (+)scDNA is converted to (-)scDNA. RecG (either in complex with SSB or released from SSB) binds to the (-)scDNA and drives fork regression. If the replisome disassembles exposing gaps in either strand, the gap will be rapidly bound by SSB (green spheres) in complex with RecG (pink), which then regresses the fork. RecG may displace SSB or co-translocation may occur, but this remains an open question. (II). The replisome disassembles and is instead bound RecG-SSB-PriA, leading to fork stabilization as suggested by Tanaka (Tanaka and Masai, 2006). (III). Following replisome disassembly, regions of exposed ssDNA are bound by SSB. SSB inhibits RuvAB, but is a target for either RecG, RecG-SSB, PriA or PriA-SSB. (IV), Once fork processing has taken place, a substrate resembling a Holliday junction is formed and is then acted upon by RuvAB
Acknowledgments
Work in the Bianco laboratory is supported by NIH Grant GM100156 to PRB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abd Wahab S, Choi M, Bianco PR. Characterization of the ATPase activity of RecG and RuvAB proteins on model fork structures reveals insight into stalled DNA replication fork repair. J Biol Chem. 2013;288:26397–409. doi: 10.1074/jbc.M113.500223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avison MB, Horton RE, Walsh TR, Bennett PM. Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. J Biol Chem. 2001;276:26955–61. doi: 10.1074/jbc.M011186200. [DOI] [PubMed] [Google Scholar]
- Baharoglu Z, Petranovic M, Flores MJ, Michel B. RuvAB is essential for replication forks reversal in certain replication mutants. EMBO J. 2006;25:596–604. doi: 10.1038/sj.emboj.7600941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Dunderdale HJ, West SC. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell. 1993;74:1021–31. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- Briggs GS, Mahdi AA, Wen Q, Lloyd RG. DNA binding by the substrate specificity (wedge) domain of RecG helicase suggests a role in processivity. J Biol Chem. 2005;280:13921–7. doi: 10.1074/jbc.M412054200. [DOI] [PubMed] [Google Scholar]
- Buss JA, Kimura Y, Bianco PR. RecG interacts directly with SSB: implications for stalled replication fork regression. Nucleic Acids Res. 2008;36:7029–42. doi: 10.1093/nar/gkn795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VJ, Vinella D. The stringent response. In: Neidhardt FC, I. RC, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella Cellular and Molecular Biology. ASM Press; Washington, DC: 1996. pp. 1458–1496. [Google Scholar]
- Castuma CE, Crooke E, Kornberg A. Fluid membranes with acidic domains activate DnaA, the initiator protein of replication in Escherichia coli. J Biol Chem. 1993;268:24665–8. [PubMed] [Google Scholar]
- Courcelle J, Hanawalt PC. RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet. 2003;37:611–46. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- Cox MM. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annual Review of Genetics. 2001;35:53–82. doi: 10.1146/annurev.genet.35.102401.090016. [DOI] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Fukuoh A, Iwasaki H, Ishioka K, Shinagawa H. ATP-dependent resolution of R-loops at the ColE1 replication origin by Escherichia coli RecG protein, a Holliday junction-specific helicase. EMBO J. 1997;16:203–9. doi: 10.1093/emboj/16.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry DR, Burgess RR. The cloning and sequence of the gene encoding the omega subunit of Escherichia coli RNA polymerase. Gene. 1986;48:33–40. doi: 10.1016/0378-1119(86)90349-5. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. One more conserved sequence motif in helicases. Nucleic Acids Res. 1988;16:7734. doi: 10.1093/nar/16.15.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. A conserved NTP-motif in putative helicases. Nature. 1988;333:22. doi: 10.1038/333022a0. [DOI] [PubMed] [Google Scholar]
- Hong X, Cadwell GW, Kogoma T. Escherichia coli RecG and RecA proteins in R-loop formation. EMBO J. 1995;14:2385–92. doi: 10.1002/j.1460-2075.1995.tb07233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman M, Murphy H, Cashel M. The nucleotide sequence of recG, the distal spo operon gene in Escherichia coli K-12. Gene. 1992;110:95–9. doi: 10.1016/0378-1119(92)90449-y. [DOI] [PubMed] [Google Scholar]
- Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiology & Molecular Biology Reviews. 1997;61:212–38. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T, Hong X, Cadwell GW, Barnard KG, Asai T. Requirement of homologous recombination functions for viability of the Escherichia coli cell that lacks RNase HI and exonuclease V activities. Biochimie. 1993;75:89–99. doi: 10.1016/0300-9084(93)90029-r. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends in Biochemical Sciences. 2000;25:156–65. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiology and Molecular Biology Reviews. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. table of content. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RG. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J Bacteriol. 1991;173:5414–8. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RG, Buckman C. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J Bacteriol. 1991;173:1004–11. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RG, Sharples GJ. Molecular organization and nucleotide sequence of the recG locus of Escherichia coli K-12. J Bacteriol. 1991;173:6837–43. doi: 10.1128/jb.173.21.6837-6843.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RG, Sharples GJ. Processing of recombination intermediates by the RecG and RuvAB proteins of Escherichia coli. Nucleic Acids Res. 1993;21:1719–25. doi: 10.1093/nar/21.8.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annual Review of Biochemistry. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- Mahdi AA, Briggs GS, Sharples GJ, Wen Q, Lloyd RG. A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J. 2003;22:724–34. doi: 10.1093/emboj/cdg043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosas M, Perumal SK, Bianco P, Ritort F, Benkovic SJ, Croquette V. RecG and UvsW catalyse robust DNA rewinding critical for stalled DNA replication fork rescue. Nat Commun. 2013;4:2368. doi: 10.1038/ncomms3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosas M, Perumal SK, Bianco PR, Ritort F, Benkovic SJ, Croquette V. Corrigendum: RecG and UvsW catalyse robust DNA rewinding critical for stalled DNA replication fork rescue. Nat Commun. 2014;5:4210. doi: 10.1038/ncomms3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marians KJ. Replication and recombination intersect. Current Opinion in Genetics & Development. 2000;10:151–6. doi: 10.1016/s0959-437x(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Marians KJ. Mechanisms of replication fork restart in Escherichia coli. Philos Trans R Soc Lond B Biol Sci. 2004;359:71–7. doi: 10.1098/rstb.2003.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Senac MM, Webb MR. Mechanism of translocation and kinetics of DNA unwinding by the helicase RecG. Biochemistry. 2005;44:16967–76. doi: 10.1021/bi0512851. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd R. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd R. Action of RuvAB at replication fork structures. J Biol Chem. 2001;276:41938–44. doi: 10.1074/jbc.M107945200. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd R. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 2002a;18:413–9. doi: 10.1016/s0168-9525(02)02720-8. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd R. Replicating past lesions in DNA. Mol Cell. 2002b;10:700–1. doi: 10.1016/s1097-2765(02)00687-1. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG, Marians KJ. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc Natl Acad Sci U S A. 2001;98:8235–40. doi: 10.1073/pnas.121007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P, Mahdi A, Lloyd R. Characterisation of the catalytically active form of RecG helicase. Nucleic Acids Res. 2000;28:2324–32. doi: 10.1093/nar/28.12.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B, Boubakri H, Baharoglu Z, LeMasson M, Lestini R. Recombination proteins and rescue of arrested replication forks. DNA Repair (Amst) 2007;6:967–80. doi: 10.1016/j.dnarep.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12783–8. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–22. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel--structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Persson BC, Jager G, Gustafsson C. The spoU gene of Escherichia coli, the fourth gene of the spoT operon, is essential for tRNA (Gm18) 2′-O-methyltransferase activity. Nucleic Acids Research. 1997;25:4093–7. doi: 10.1093/nar/25.20.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Ullsperger C, Keller RW, Bustamante C, Vologodskii AV, Cozzarelli NR. Positive torsional strain causes the formation of a four-way junction at replication forks. Journal of Biological Chemistry. 2001;276:2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- Robu ME, Inman RB, Cox MM. RecA protein promotes the regression of stalled replication forks in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8211–8. doi: 10.1073/pnas.131022698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Inman RB, Cox MM. Situational repair of replication forks: roles of RecG and RecA proteins. Journal of Biological Chemistry. 2004;279:10973–81. doi: 10.1074/jbc.M312184200. [DOI] [PubMed] [Google Scholar]
- Rudolph CJ, Mahdi AA, Upton AL, Lloyd RG. RecG protein and single-strand DNA exonucleases avoid cell lethality associated with PriA helicase activity in Escherichia coli. Genetics. 2010a;186:473–92. doi: 10.1534/genetics.110.120691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Briggs GS, Lloyd RG. Is RecG a general guardian of the bacterial genome? DNA Repair (Amst) 2010b;9:210–23. doi: 10.1016/j.dnarep.2009.12.014. [DOI] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Harris L, Lloyd RG. Pathological replication in cells lacking RecG DNA translocase. Mol Microbiol. 2009;73:352–66. doi: 10.1111/j.1365-2958.2009.06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarubbi E, Rudd KE, Xiao H, Ikehara K, Kalman M, Cashel M. Characterization of the spoT gene of Escherichia coli. J Biol Chem. 1989;264:15074–82. [PubMed] [Google Scholar]
- Seigneur M, Bidnenko V, Ehrlich S, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–30. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton M, Dillingham M, Wigley D. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- Slocum SL, Buss JA, Kimura Y, Bianco PR. Characterization of the ATPase activity of the Escherichia coli RecG protein reveals that the preferred cofactor is negatively supercoiled DNA. J Mol Biol. 2007;367:647–64. doi: 10.1016/j.jmb.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Masai H. Stabilization of a stalled replication fork by concerted actions of two helicases. Journal of Biological Chemistry. 2006;281:3484–93. doi: 10.1074/jbc.M510979200. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–8. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton AL, Grove JI, Mahdi AA, Briggs GS, Milner DS, Rudolph CJ, Lloyd RG. Cellular location and activity of Escherichia coli RecG proteins shed light on the function of its structurally unresolved C-terminus. Nucleic Acids Res. 2014;42:5702–14. doi: 10.1093/nar/gku228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SD, Mahdi AA, Lloyd RG. The RecG branch migration protein of Escherichia coli dissociates R-loops. J Mol Biol. 1996;264:713–21. doi: 10.1006/jmbi.1996.0671. [DOI] [PubMed] [Google Scholar]
- Wang M, Schnitzer M, Yin H, Landick R, Gelles J, Block S. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–7. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- Whitby MC, Ryder L, Lloyd RG. Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell. 1993;75:341–50. doi: 10.1016/0092-8674(93)80075-p. [DOI] [PubMed] [Google Scholar]
- Whitby MC, Vincent SD, Lloyd RG. Branch migration of Holliday junctions: identification of RecG protein as a junction specific DNA helicase. EMBO J. 1994;13:5220–8. doi: 10.1002/j.1460-2075.1994.tb06853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Tan HY, Choi M, Stanenas A, Byrd A, Raney K, Cohan C, Bianco PR. The E.coli DNA Helicases RecG and PriA bind to SSB in vivo to facilitate rescue of stalled DNA replication forks. DNA repair. 2014 submitted. [Google Scholar]
- Yung BY, Crooke E, Kornberg A. Fate of the DnaA initiator protein in replication at the origin of the Escherichia coli chromosome in vitro. J. Biol. Chem. 1990;265:1282–5. [PubMed] [Google Scholar]