Abstract
Human catechol-O-methyltransferase (COMT; EC 2.1.1.6) catalyzes the transfer of the methyl group to a variety of endogenous and exogenous catechol substrates using S-adenosyl-L-methionine as the methyl donor. This enzymatic O-methylation plays an important role in the inactivation of biologically-active and toxic catechols. A number of studies in recent years have sought to characterize the polymorphism of human COMTs, and also to determine the catalytic activity of polymorphic enzymes. We report here the identification of a new mutant form of the human COMT gene with triplet point mutations, which encodes the D51G/S60F/K162R mutant of the soluble COMT and the D101G/S110F/K212R mutant of the membrane-bound COMT. Kinetic analysis showed that these new mutant COMTs had essentially the same kinetic characteristics and catalytic activity as the wild-type COMTs for the O-methylation of 2-hydroxyestradiol and 4-hydroxyestradiol in vitro, but the mutants have a significantly reduced thermostability at 37°C. In addition, the mutant enzymes have different binding affinities for S-adenosyl-L-methionine compared with the wild-type COMTs. In agreement with our biochemical observations, molecular modeling studies also showed that the mutant human COMT proteins shared nearly the same overall structures as the wild-type proteins. The binding energy values of the mutant COMTs in complex with catechol estrogen substrates were similar to those of the wild-type COMTs bound with the same substrates.
Introduction
Human catechol-O-methyltransferase (COMT; EC 2.1.1.6)3 catalyzes the transfer of the methyl group to a wide variety of endogenous and exogenous catechol substrates by using S-adenosyl-L-methionine (AdoMet) as the methyl donor [1–8]. In the central nervous system, COMT metabolically deactivates catecholamine neurotransmitters (dopamine and norepinephrine) through O-methylation. A decrease in the methylation of catecholamine neurotransmitters has been suggested to be a contributing factor in Parkinson's disease [3, 9] and also a number of mental disorders [10–18]. In addition to catecholamine neurotransmitters, the endogenous catechol estrogens, such as 2- and 4-hydroxyestradiol (2-OH-E2 and 4-OH-E2), which are major oxidative metabolites of 17β-estradiol (E2) formed by cytochrome P450 isoforms in humans [19, 20], are also rapidly O-methylated by COMT, in a manner analogous to the O-methylation of catecholamines [21]. A number of studies have demonstrated that metabolic O-methylation provides effective inactivation/detoxification of the procarcinogenic catechol estrogen intermediates. Besides, 2-methoxyestradiol (the major O-methylation product of 2-OH-E2) has strong apoptotic, antiangiogenic, and anticancer actions (reviewed in refs. [3, 21]). Hence, metabolic O-methylation of catechol estrogens may not only inactivate the catechol estrogen intermediates, but may also simultaneously produce 2-methoxyestradiol that has significant anticancer activity [21]. These two concurrent processes are thought to be beneficial for reducing the risk of estrogen-induced cancers [21]. In line with this suggestion, many epidemiological studies have shown that women, homozygous with a thermolabile variant (V108M) of COMT, have an increased risk of estrogen-associated cancers [22–29], thus providing support for this interesting hypothesis.
Given the potentially important biological consequences associated with a reduced COMT catalytic activity in vivo, a number of studies in recent years have sought to investigate the polymorphism of the human COMT gene, as well as its effect on the catalytic activity of the enzyme [30–33]. At present, a total of 14 polymorphisms are listed in the NCBI SNP database for human COMT gene (summarized in Table 1), and all of them are single point mutations. Among them, only three of the COMT mutants have been studied for their enzymatic activity and functional differences with the wild-type COMT. The V108M mutant of human S-COMT was found to retain nearly the same catalytic ability for the O-methylation of catechol estrogen substrates as the wild-type COMT, but it had a reduced thermostability [29–32]. In comparison, the A22S mutant of the human S-COMT had a lower catalytic activity and also reduced thermostability [33].
Table 1.
The COMT polymorphisms listed in the NCBI SNP database (http://www.ncbi.nlm.nih.gov/SNP).
| Amino acid positions (S / MB) |
Amino acid residue | Effect on the enzyme functions | |
|---|---|---|---|
| Wild type | Mutant type | ||
| – /9 | L | F | Unknown (unpublished) |
| – / 34 | C | S | Unknown (unpublished) |
| 12 / 62 | H | H* | No functional change expected |
| 22 / 72 | A | S | Lower catalytic activity and reduced thermostability [33] |
| 23 / 73 | Q | Q* | No functional change expected |
| 42 / 92 | V | M | Unknown (unpublished) |
| 52 / 102 | A | T | Reduced thermostability [33] |
| 62 / 112 | L | L* | No functional change expected |
| 84 / 134 | A | A* | No functional change expected |
| 86 / 136 | L | A | Unknown (unpublished) |
| 96 / 146 | A | V | Unknown (unpublished) |
| 108 / 158 | V | M | Reduced thermostability [17, 29–32] |
| 149 / 199 | P | L | Unknown (unpublished) |
| 153 / 203 | L | L* | No functional change expected |
These nucleic acid mutations will not alter the amino acids that are encoded by the altered codons.
In the present study, we identified a new triplet-point-mutation form of the human COMT gene. In addition, we have selectively expressed both the wild-type and mutant human S- and MB-COMTs in Escherichia coli (E. coli) for comparing their biochemical characteristics for the O-methylation of endogenous catechol estrogens. We have also conducted computational molecular modeling studies to compare the structural and catalytic differences between the mutant S-COMT and the wild-type enzyme.
Materials and Methods
Chemicals
2-OH-E2, 4-OH-E2, isopropylthio-β-D-galactoside, 1,4-dithiothreitol and phenylmethylsulfonyl fluoride were purchased from Sigma-Aldrich (St. Louis, MO). [Methyl-3H]AdoMet (specific activity = 11.2–13.5 Ci/mmol) was obtained from Perkin Elmer (Waltham, MA). All solvents used in this study were of HPLC grade or better and were obtained from Fisher Scientific Co. (Springfield, NJ).
Cloning of human S-COMT and MB-COMT cDNAs
The human liver cDNA library (obtained from Stratagene, La Jolla, CA) was used as template for cloning the human S-COMT and MB-COMT cDNAs. For PCR, the 5′ complementary forward primers (5′-CAA CAT ATG CCG GAG GCC CCG-3′ and 5′-GCA TAT GCC GGA GGC CCC GCC TC-3′) were used for S-COMT and MB-COMT, respectively, along with a common 3′ reverse primer (5′-CAG GAT CCT CAG GGC CCT GCT-3′). These primers were specifically designed to append the sequence that contains a 5′ NdeI restriction site and a 3′ BamHI restriction site (Fig. 1, lower panel). The resulting 655-bp fragment for S-COMT and the 815-bp fragment for MB-COMT amplified by PCR were eluted using a gel extract kit (QIAGEN, Valenica, CA), and they were then ligated to the pGEM T vector (Promega, Madison, WI) using T4 ligase (Invitrogen, Carlsbad, CA). The products of the ligation reactions were transformed into the chemically competent E. coli TOP-10F′ cells (Invitrogen, Carlsbad, CA), and the transformed bacteria were then selected with ampicillin (50 µg/mL) on LB agar plates. The plasmids were purified using a Miniprep purification kit (QIAGEN, Valencia, CA). The entire S-COMT and MB-COMT cDNA sequences were determined for verification. The plasmids were restriction-digested with NdeI and BamHI and ligated into NdeI and BamHI-digested pET12a vector (Novagen, Madison, WI). The recombinant DNAs were introduced into chemically competent E. coli BL21 (DE3) (Novagen, Madison, WI) according to the procedures recommended by the manufacturer, and the transformed cells were selected with ampicillin (50 µg/mL) on LB agar plates.
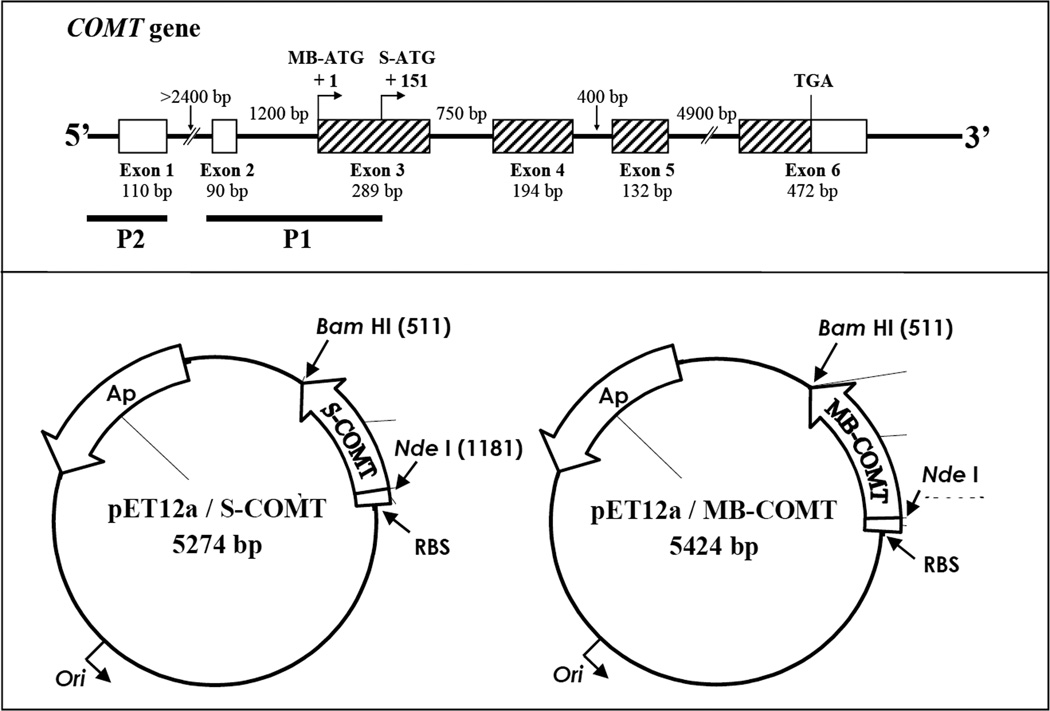
Figure 1.
Upper panel: Structure of the human COMT gene. The boxes represent exons and the thin lines between the boxes represent introns. The hatched boxes indicate protein-coding regions. The size of each exon and intron is as indicated. The positions of the initiation codons for transcription of S-COMT and MB-COMT mRNAs are indicated as S-ATG and MB-ATG. The two known promoters, P1 and P2, are shown by black bars. Note that the P1 promoter for transcritpion of S-COMT overlaps with the initiation codon and part of the coding sequence for MB-COMT. Lower panel: Construction of the pET12a/S-COMT and pET12a/MB-COMT expression vectors based on the vector pET12a. The S-COMT or MB-COMT cDNA was cloned into the NdeI and BamH I sites of pET12a to form the pET12a/S-COMT or pET12a/MB-COMT expression vectors. Each of the expression vectors was under the control of the T7 promoter and lacO-operator. Expression was induced by addition of 0.5 mM isopropylthio-β-D-galactoside.
In vitro site-directed mutagenesis
The cDNAs of the cloned mutant human S-COMT (D51G, S60F and K162R) and MB-COMT (D101G, S110F and K212R) were used as templates to generate the corresponding wild-type cDNAs. The mutations were corrected by using a PCR-based site-directed mutagenesis using the QuikChange multisite-directed mutagenesis kit (Stratagene, La Jolla, CA). The site-directed mutagenesis was carried out according to the procedures recommended by the manufacturer with the following primers: 5’-GGCAAGATCGTGGACGCCGTGATTC-3’ for D51/101G, 5’-CACCAGCCCTCCGTGCTGCTGGA-GC-3’ for S60/110F, and 5’-CTGCTGCGGAAGGGGACAGTGCTAC-3’ for K162/212R. The sequences of all reconstructed plasmid DNAs were confirmed by DNA sequencing.
Bacterial expression of recombinant human COMTs
For expression of the recombinant human S- and MB-COMT proteins in E. coli BL21 (DE3, expressing T7 polymerase), positive clones were first cultured in the LB medium supplemented with ampicillin (50 µg/mL) overnight at 37°C. The culture broth was then inoculated into 300 mL fresh LB medium supplemented with ampicillin and incubated at 37°C with vigorous shaking until the optical density reading of the bacterial culture mixture reached ~0.6 (at λ = 600 nm). The culture was then induced with isopropylthio-β-D-galactoside (at a final concentration of 0.5 mM) and cultured for another 3 hours. The cells were collected by centrifugation and were then sonicated in ice-cold lysis buffer (50 mM Tris-HCl, pH 7.5 + 200 mM NaCl). After addition of 5 mM 1,4-dithiothreitol and 1 mM phenylmethylsulfonyl fluoride to the crude homogenates, they were centrifuged at 10,000 × g for 10 minutes at 4°C. The supernatants were then subjected to column purification or directly stored at −80°C.
Assay of the enzyme activity and thermostability of recombinant human COMTs in vitro
The catalytic activity of the wild-type and mutant human recombinant S- and MB-COMTs was determined at 37°C as described earlier [34]. The reaction mixtures were consisted of the recombinant COMT protein (at 16.2 µg/mL for S-COMT or 17.1 µg/mL for MB-COMT), 1.2 mM MgCl2, 100 µM AdoMet (containing 0.5 mCi [methyl-3H]AdoMet), 1 mM 1,4-dithiothreitol, and 2-OH-E2 or 4-OH-E2 as substrate (at 10 µM or as indicated) in Tris-HCl buffer (50 mM, pH 7.4). The final volume of the reaction mixture was usually 300 µL. The reaction was initiated by addition of recombinant human COMT protein and carried out at 37°C for 15 minutes. To test the thermostability of the mutant and wild-type COMTs, the enzymes were first preincubated at 37°C for the indicated length of time immediately before testing their catalytic activity for the O-methylation of 2-OH-E2 (at 10 µM). The reaction was arrested by immediately placing the tubes on ice and followed by addition of 500 µL ice-cold saline. The reaction mixtures were extracted with 5 mL ethyl acetate for the methylated catechol products. After centrifugation at 1000 g for 10 minutes, portions of the organic extracts were measured for radioactivity content with a liquid scintillation analyzer (Packard Tri-CARB 2900TR; Downers Grove, IL). The rate of methylation of a substrate was expressed as “nmol of methylated product formed/mg of human COMT protein/minute” (abbreviated as “nmol/mg protein/min”). The kinetic parameters (KM and VMAX values) were calculated by using the curve regression method of the SigmaPlot program.
Construction of the homology models for human wild-type and mutant S-COMTs
Homology modeling was performed using the InsightII modeling program (Version 2005, Accelrys Inc., San Diego, CA) on a Dell Precision 690 workstation installed with Red Hat Enterprise Linux WS4.0 operating system (Red Hat Inc., Raleigh, NC). The energy minimization and molecular dynamics simulation were performed with Discovery Studio modeling program (Version 1.7, Accelrys Inc. San Diego, CA). The CHARMm force field was used for energy minimization and dynamics simulation.
The primary sequences of human S-COMT (GI 6466450) and rat S-COMT (GI 1633081) were obtained from the NCBI database. Sequence alignments were done by using the Homology Modeling module of InsightII, which showed a greater than 80% sequence similarity. The homology model of the human S-COMT was constructed according to the rat S-COMT (PDB code: 1VID) by using the Modeler in the Homology Modeling module of InsightII. The substrate 3,5-dinitrocatechol, AdoMet, Mg2+ ion, and the crystallographic H2O that coordinates with Mg2+ were included in the homology model. The simulation was carried out using the Standard Dynamics Cascade protocol in the Discovery Studio. For energy minimizations, the steepest descent method was employed first to a 10 kcal/(molÅ) root mean square gradient and followed by the Polak and Ribiere conjugate gradient method until the final convergence criterion reached 0.01 kcal/(molÅ) gradient. Then the whole system was heated from 50 to 300 K in 2 ps and equilibrated in 300 K for 100 ps. One hundred conformations were collected in 20-ps production phase at 300 K. The conformation with the lowest potential energy was further minimized and used for binding energy analysis. 3,5-Dinitrocatechol, Mg2+, H2O, AdoMet and key residues in the catalytic site (D141, K144, D169, N170 and E199) were constrained during the simulation process.
It should be noted that the structure of the D51G/S60F/K162R mutant human S-COMT was built with the Build Mutant function of the Homology Modeling module by using two different methods: one was based on the homology model of the wild-type human S-COMT that was built in the present study, and the other was based on the crystallographic structure of the rat S-COMT (PDB code: 1VID). The same simulation process was carried out when we built the structures of the mutant or wild-type S-COMT.
Calculation of the binding energy values (ΔEbinding) of S-COMTs with substrates
The structure of 2-OH-E2 and 4-OH-E2 were built with the Builder module of InsightII based on the X-ray structure of E2 (PDB code: 1ERE) and minimized with CHARMm force field. The catechol ring of the substrates was superimposed onto the catechol ring of 3,5-dinitrocatechol. The simulation was carried out with the Standard Dynamics Cascade protocol in Discovery Studio. The same simulation cascade was carried out as the cascade that was used for building the human COMT in complex with 3,5-dinitrocatechol as ligand. One hundred conformations were collected in the 20-ps production phase at 300 K. The conformation with the lowest potential energy was further minimized and used for the Binding Energy Calculation protocol. The backbone of the protein, key residues in the catalytic site (D141, K144, D169, N170 and E199), the catechol ring of the substrate, AdoMet, Mg2+, and H2O were constrained during the whole simulation process. ΔEbinding was calculated with the following equation: ΔEbinding = Ecomplex − (ECOMT + Esubstrate), where Ecomplex is the potential energy for the complex of human COMT with its substrate, ECOMT is the potential energy of the enzyme itself and Esubstrate is the potential energy for the substrate itself.
Results
Cloning of the mutant human S- and MB-COMT cDNAs
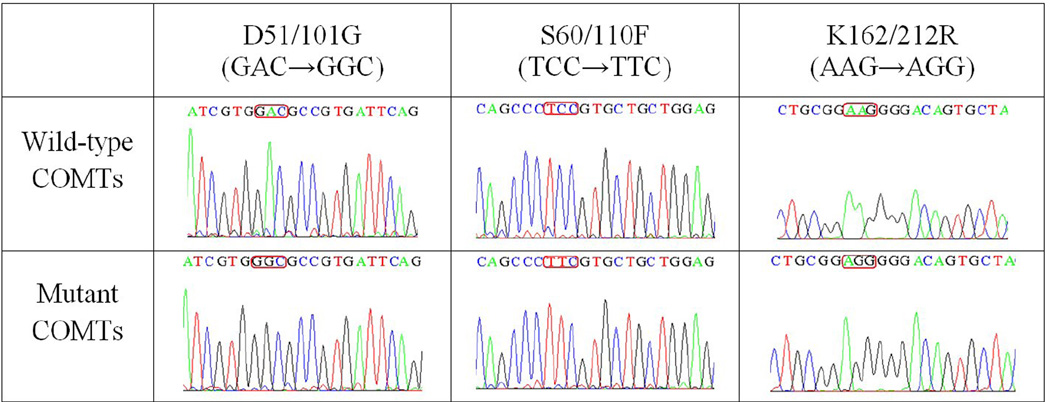
In the present study, the human S- and MB-COMT cDNAs were selectively amplified with PCR using human liver cDNA library as template. After the recombinant plasmids containing the cDNA for human S-COMT or MB-COMT were enzymatically digested, gel electrophoresis of the digests revealed that the DNA fragment bands matched the expected sizes of 655 and 815 bp, respectively (data not shown). To determine their sequences, the full-length sequence analysis of the cloned cDNAs was performed. We found that the sequences were changed at three places for both S-COMT and MB-COMT cDNAs, and the changes were exactly at the same positions for both of them. The sequence analyses were performed multiple times by two companies, and the same sequence data were obtained. Notably, since it is known that a single gene (localized to chromosome 22, band q11.2 [8, 9]) encodes both human S- and MB-COMT proteins by using two separate promoters [6], we believe that the triplet point mutations of the human S- and MB-COMT cDNAs were not artificial errors produced during the cloning procedures, because the same exact triplet point mutations were seen in the cDNAs of both S-COMT (D51G/S60F/K162R) and MB-COMT (D101G/S110F/K212R) (Fig. 2). The probability of their occurrence as a result of experimental errors was extremely low (nearly improbable).
Figure 2.
Sequencing results of the wild-type and mutant human S- and MB-COMTs.
To obtain the wild-type human S- and MB-COMT cDNAs, we carried out PCR-based site-directed mutagenesis based on the mutant human S- and MB-COMT cDNAs. The full-length DNA sequences were determined twice and compared with the known wild-type human COMT gene. The DNA sequencing results confirmed their right sequences (Fig. 2).
Expression of the mutant and wild-type COMT proteins in E. coli
The E. coli BL21 (DE3) cells were transformed with the recombinant expression vector pET12a containing the gene encoding either the mutant or wild-type human S-COMT or MB-COMT. Following induction with 0.5 mM isopropylthio-β-D-galactoside, the bacteria that abundantly expressed the desired protein were harvested by centrifugation. The resulting cell pellets were lysed and analyzed using 12% SDS-PAGE, followed by Western blot analysis using polyclonal rabbit antibodies against the human wild-type COMT and donkey anti-rabbit IgG antiserum (conjugated to horseradish peroxidase).
A band with a molecular mass of approximately 24 kD was detected for both mutant and wild-type S-COMT proteins, and a band of approximately 30 kD was detected for the mutant and wild-type MB-COMTs (data not shown). The sizes of the expressed proteins matched the expected sizes for the recombinant human S- and MB-COMT proteins.
Kinetic parameters for the O-methylation of 2-OH-E2 and 4-OH-E2
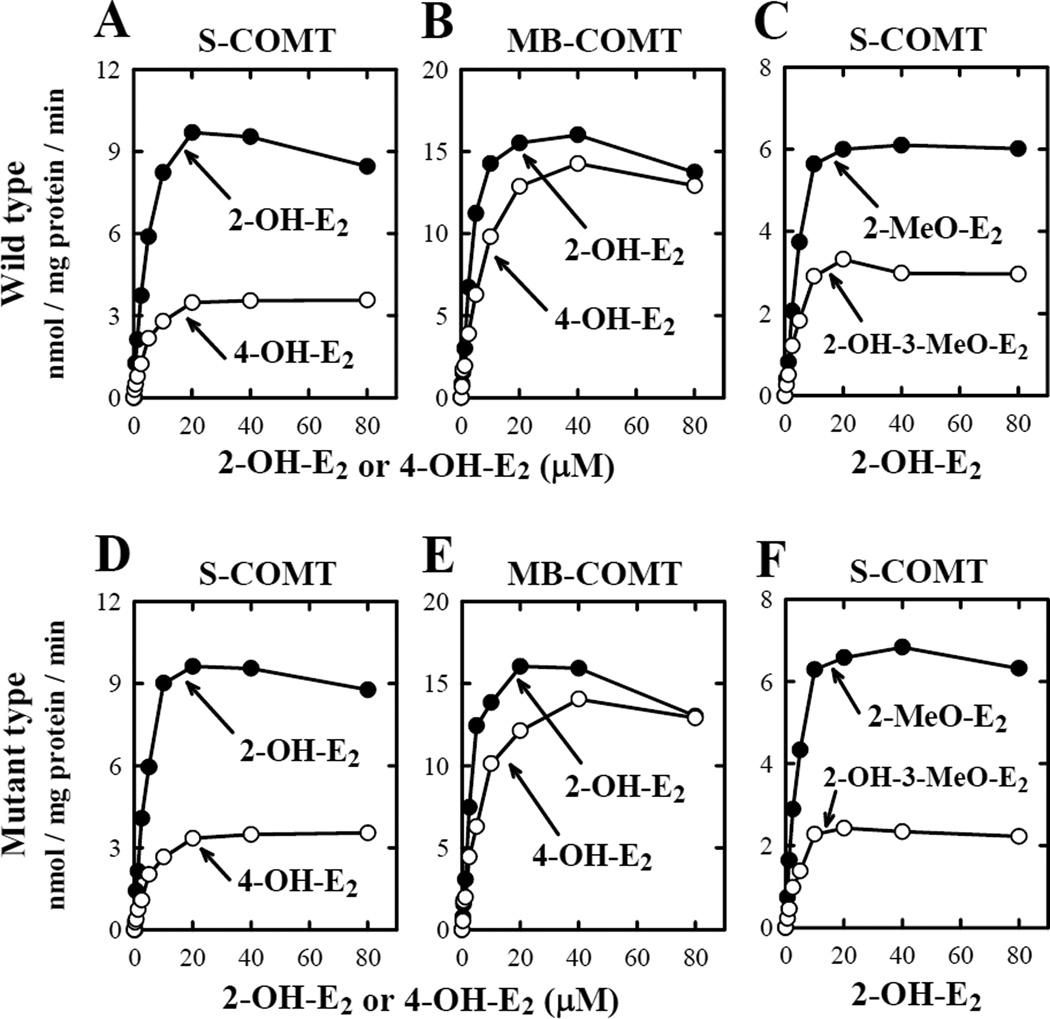
To characterize the kinetic parameters (KM, VMAX and VMAX/KM) of the mutant human S- and MB-COMTs, we selectively expressed them and also the wild-type S- and MB-COMTs in E.coli. The enzyme activity assays were performed by measuring the COMT-mediated O-methylation of 2-OH-E2 and 4-OH-E2, two representative endogenous catechol estrogen substrates. No significant difference was observed between the wild-type S-COMT and its D51G/S60F/K162R mutant form, or between the wild-type MB-COMT and its D101G/S110F/K212R mutant form (Fig. 3 and Table 2).
Figure 3.
Relationship between catechol estrogen concentrations and their rate of O-methylation by wild-type human COMTs (A, B and C) and mutant human COMTs (D, E, and F). The upper right panel (C) showed the rate of 2-O-methylation and 3-O-methylation of 2-OH-E2 catalyzed by the wild-type S-COMT, and the lower right panel (F) showed the rate of 2-O-methylation and 3-O-methylation of 2-OH-E2 by mutant S-COMT. The incubation mixture consisted of 10 different concentrations (0, 0.31, 0.63, 1.25, 2.5, 5, 10, 20, 40, and 80 µM) of each substrate, 1.2 mM MgCl2, 100 µM AdoMet (containing 0.5 mCi [methyl-3H]AdoMet), 1 mM 1,4-dithiothreitol, and the recombinant COMT protein (at 16.2 µg/mL for S-COMT or 17.1 µg/mL for MB-COMT). The incubations were carried out at 37°C for 15 minutes. Note that the rate of its total methylation was based on liquid scintillation counting of the radioactivity extracted with ethyl acetate. The rates for its 2-O- and 3-O-methylation (C and F) were determined by using HPLC that separately quantified the amount of 2-methoxyestradiol (2-MeO-E2) and 2-OH-E2 3-methyl ether (2-OH-3-MeO-E2) formed. Each value is the mean of duplicate measurements.
Table 2.
A comparison of the kinetic parameters of the mutant forms of the human S- and MB-COMTs with those of wild-type human S- and MB-COMTs
| Genotype | Enzyme | Substrate (µM) |
AdoMet (µM) |
KM (µM) |
VMAX (nmol/mg/min) |
VMAX/KM |
|---|---|---|---|---|---|---|
| Wild-type COMT | S-COMT | 2-OH-E2 (0–80) |
100 | 3.6 | 9.7 | 2.7 |
| 4-OH-E2 (0–80 µM) |
100 | 4.5 | 3.5 | 0.8 | ||
| MB-COMT | 2-OH-E2 (0–80 µM) |
100 | 3.2 | 16.3 | 5.1 | |
| 4-OH-E2 (0–80 µM) |
100 | 6.4 | 14.3 | 2.2 | ||
| Mutant COMT (D51G/S60F/K162R) | S-COMT | 2-OH-E2 (0–80 µM) |
100 | 3.3 | 8.7 | 2.6 |
| 4-OH-E2 (0–80 µM) |
100 | 4.5 | 3.5 | 0.8 | ||
| MB-COMT | 2-OH-E2 (0–80 µM) |
100 | 2.6 | 16.1 | 6.2 | |
| 4-OH-E2 (0–80 µM) |
100 | 5.0 | 14.1 | 2.8 | ||
| Wild-type COMT | S-COMT | 2-OH-E2 (10 µM) |
0–400 | 113.6a | 18.3a | – |
| 4-OH-E2 (10 µM) |
0–400 | 145.3a | 17.1a | – | ||
| Mutant COMT (D51G/S60F/K162R) | S-COMT | 2-OH-E2 (10 µM) |
0–400 | 79.5a | 18.1a | – |
| 4-OH-E2 (10 µM) |
0–400 | 146.7a | 9.6a | – |
To determine the apparent KM and VMAX values of AdoMet for the O-methylation catalyzed by S-COMT and MB-COMT, a fixed concentration (at 10 µM) of 2-OH-E2 or 4-OH-E2 was used as substrate, and different concentrations of AdoMed (at 0, 12.5, 25, 50, 100, 200, and 400 µM) were used as the methyl donor.
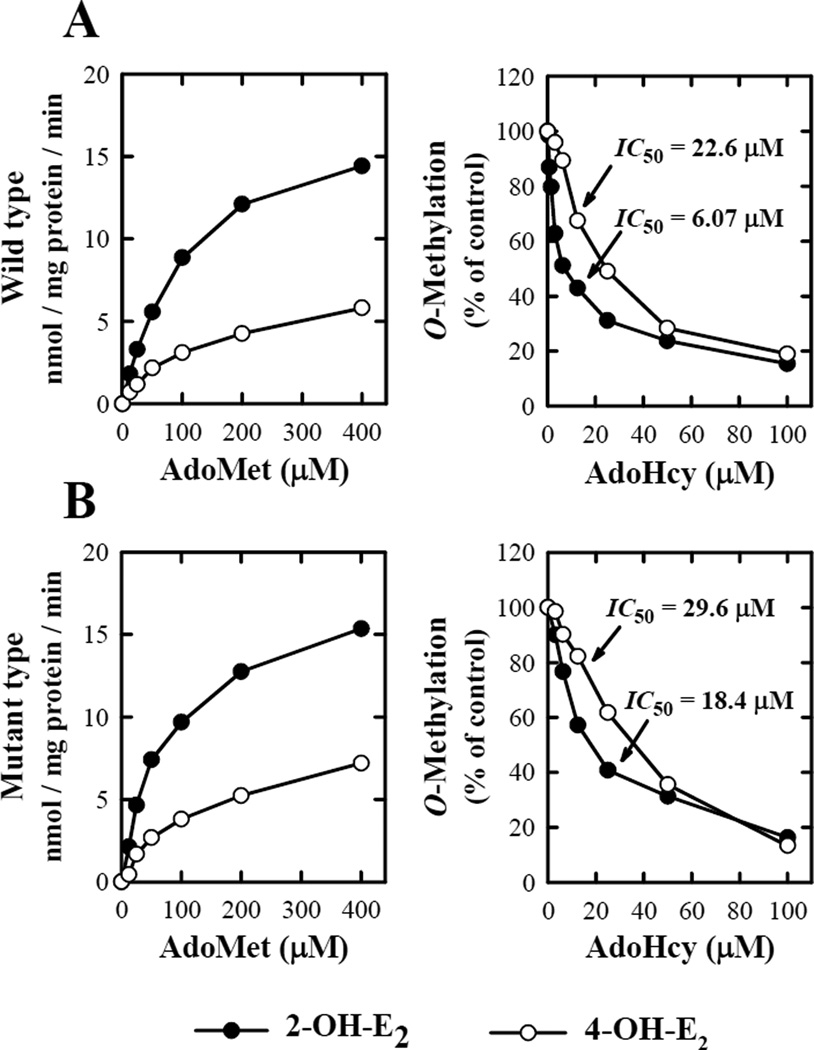
When a fixed concentration of 2-OH-E2 or 4-OH-E2 (at 10 µM) was used as substrate, the apparent KM values of the wild-type S-COMT for AdoMet were 113.6 or 145.3 µM, respectively (Fig. 4). Under the same conditions, the apparent KM values of the D51G/S60F/K162R mutant of human S-COMT for AdoMet were 79.5 and 146.7 µM. Notably, the KM value of the mutant S-COMT for AdoMet in the O-methylation of 2-OH-E2 (but not 4-OH-E2) was approximately 50% lower than the corresponding KM value for the wild-type S-COMT (Table 2).
Figure 4.
Biochemical properties of the in vitro O-methylation of catechol substrates with respect to AdoMet and AdoHcy concentrations. The representative substrates used in these assays were 2-OH-E2 and 4-OH-E2 at 10 µM concentration. The incubation mixture consisted of the substrate, 100 µM [methyl-3H]AdoMet (containing 0.2 µCi or as indicated), 16.2 µg/mL of wild-type S-COMT (A) or mutant S-COMT (B), 1 mM 1,4-dithiothreitol, and 1.2 mM MgCl2 in a final volume of 300 µL Tris-HCl buffer (50 mM) at pH 7.4. The incubations were carried out at 37°C for 15 minutes. Each value is the mean of duplicate measurements.
We have also compared the sensitivity of the mutant and wild-type human S-COMTs to inhibition by AdoHcy (a physiological feedback inhibitor of COMTs) on the O-methylation of 2-OH-E2 and 4-OH-E2. The mutant D51G/S60F/K162R S-COMT was less sensitive to inhibition by AdoHcy for the O-methylation of 2-OH-E2 as compared to the wild-type S-COMT (IC50 values of 18.4 and 6.07 µM, respectively; Fig. 5). However, AdoHcy had a comparable potency for inhibiting the O-methylation of 4-OH-E2 by the wild-type S-COMT and its D51G/S60F/K162R mutant (the IC50 values of 22.6 and 29.6 µM, respectively) (Fig. 4).
Figure 5.
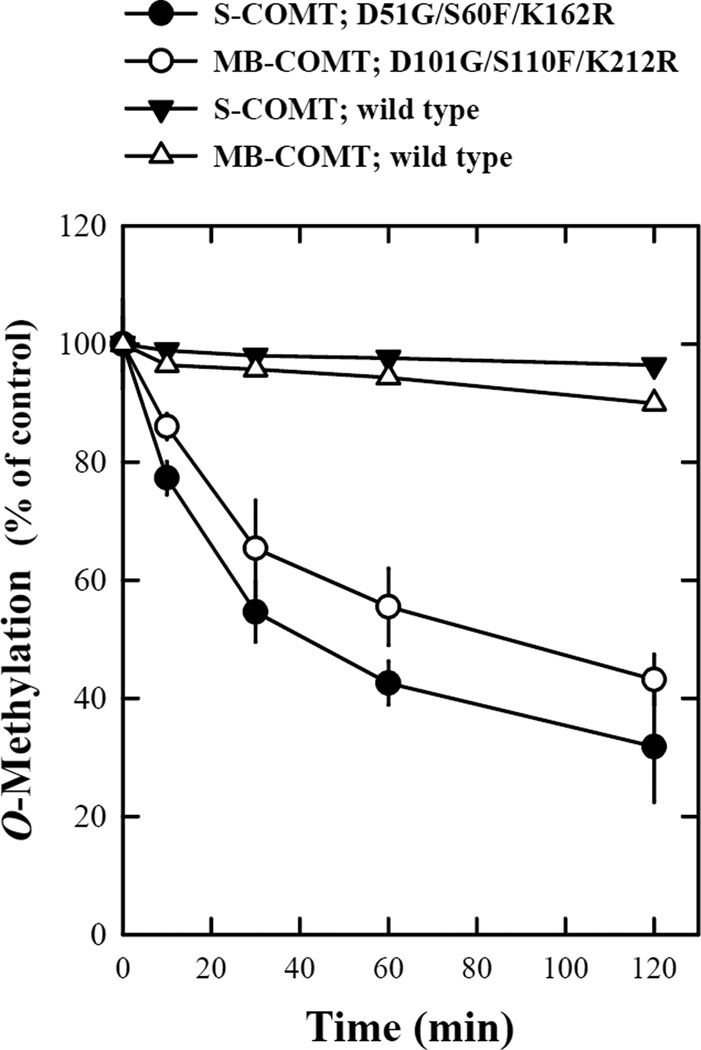
Stabilities of the wild-type and mutant human recombinant S- and MB-COMTs. The reaction mixtures consisted of the recombinant COMT protein (at 16.2 µg/mL for S-COMT or 17.1 µg/mL for MB-COMT), 1.2 mM MgCl2, 100 µM AdoMet (containing 0.5–1 mCi [methyl-3H]AdoMet), 2-OH-E2 as a substrate and 1 mM 1,4-dithiothreitol in Tris-HCl buffer (50 mM, pH 7.4). The final volume of the reaction mixture was usually 300 µL. The reaction was initiated by addition of the recombinant human COMT protein and carried out at 37°C for 15 minutes. To test the thermostability of the mutant and wild-type COMTs, the enzymes were first preincubated at 37°C for the indicated length of time immediately before testing their catalytic activity for the O-methylation of 2-OH-E2 (at 10 µM). Each value is the mean ± S.D. of triplicate measurements.
Thermostability test
The thermostability of human COMTs has often been used as an indicator of the biophysical properties of the mutant COMT proteins. In the present study, we have also compared the thermostability of the mutant human S- and MB-COMTs with the wild-type COMTs. The D51G/S60F/K162R mutant S-COMT and the D101G/S110F/K212R mutant MB-COMT were more sensitive to inactivation after pre-incubation at 37°C. The mutant S-COMT and MB-COMT lost approximately 60% of their catalytic activity after pre-incubation at 37°C for 120 minutes, while the wild-type enzymes were mostly stable after up to 120 minutes of pre-incubation at 37°C (Fig. 5).
Interestingly, the thermostability of the S60F/K162R mutant S-COMT, which has one of the three point mutations corrected by site-directed mutagenesis, was completely restored to that of the wild-type COMT (data not shown). These data showed that the aspartate residue at position 51 (D51) of the human S-COMT plays a crucial role in determining its thermostability.
Molecular modeling
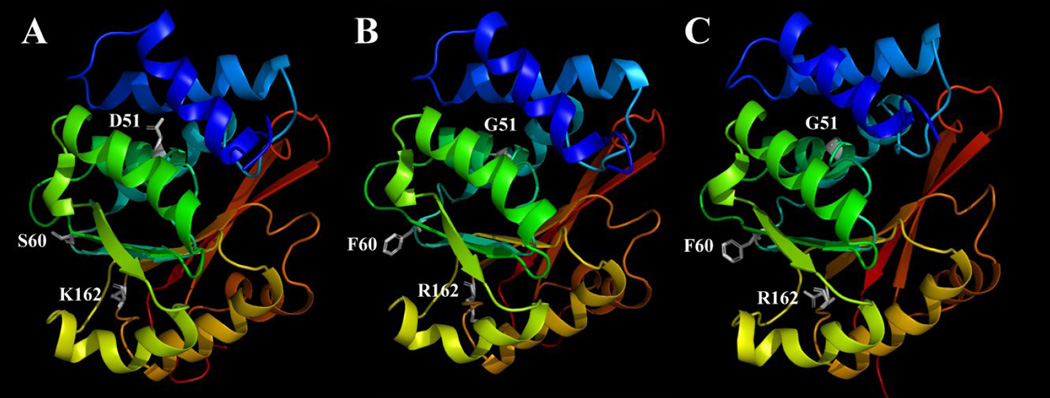
To better understand the mechanism of the triplet point mutations in affecting the structure and catalytic activity of human COMTs, we constructed the homology models for both wild-type and mutant human S-COMTs. Two different methods were used to build the mutant human S-COMT. One of them used the human S-COMT homology model as a template to build the structure of the mutant S-COMT. This method has been commonly used in many other similar studies. In addition, we have also built de novo the structure of the mutant human S-COMT according to the known crystallographic structure of the rat S-COMT. The use of two different methods was hoped to provide a more complete view of the possible structures of this new mutant human COMT. Our homology models showed that the two mutant S-COMT homology models built with two different templates are nearly the same in their backbone structures as well as their catalytic sites (Fig.6B and 6C), with only minor differences in the side-chain orientations. Therefore, for further structural comparison with the wild-type human S-COMT as well as for docking calculations using representative substrates, the mutant S-COMT built according to the wild-type human S-COMT was used.
Figure 6.
The wild-type human S-COMT (A), the D51G/S60F/K162R mutant S-COMT built according to the homology model of the wild-type human S-COMT (B) or according to the crystallographic structure of the rat S-COMT (PDB code: 1VID) (C). The figure is drawn with the PyMOL software. Secondary structures are shown with colored ribbons with blue for N-terminus and red for C-terminus. AdoMet, Mg2+ and substrates are not included in this model. The amino acids at mutation sites (D51, S60 and K162 for wild-type S-COMT and G51, F60 and R162 for mutant S-COMT) are shown in white sticks. Hydrogens are omitted from the amino acids.
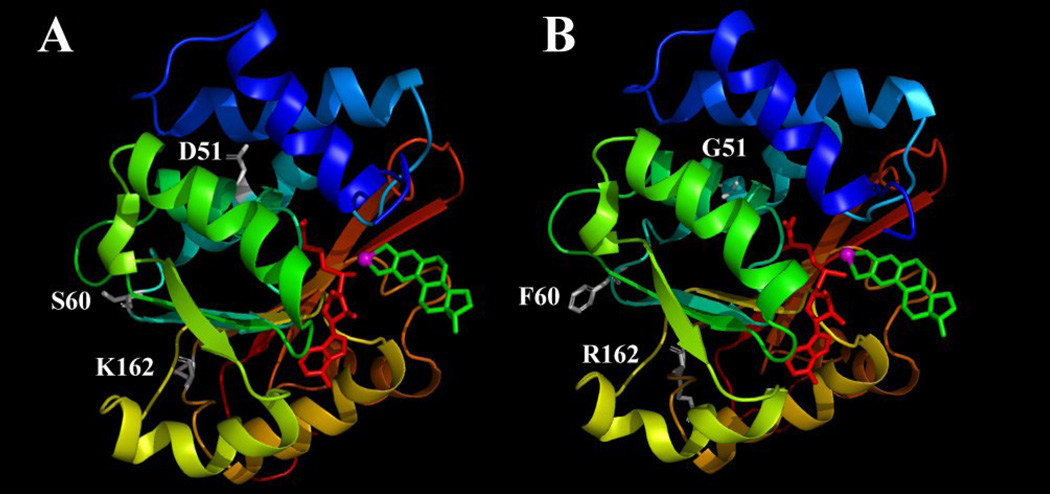
When the enzyme is complexed with a representative substrate and also the methyl donor AdoMet, the secondary structures of the wild-type and mutant S-COMTs were nearly identical (Fig. 7). Although the S60F mutation caused some minor changes in loop 5 which connects α-helix 4 and β-sheet 2, the overall structure and configuration of all nine α-helixes and seven β-sheets remained mostly the same. The amino acid residue D51 in the wild-type S-COMT was located in α-helix 3, and S60 and K162 were located in two different loop regions (loops 3 and 11, respectively). A comparison of homology-modeled structures of the wild type and mutant form showed that the D51G mutation did not noticeably interrupt the structure of α-helix 3 of the enzyme, and similarly, the S60K and K162R mutations also did not significantly affect the overall structure of the enzyme. Notably, these three mutations are mostly located in the outer surface regions of the COMT protein, and they are on the opposite side of the catalytic pocket. The distances between the α-carbon of D51, S60 and K162 to Mg2+ are 13.8, 23.2 and 25.2 Å, respectively, which are beyond the defined catalytic site (within the 7-Å reach of the Mg2+ ion). The structures of the catalytic sites in the wild-type and mutant COMTs are nearly identical when they are in complex with 2-OH-E2 or 4-OH-E2. Also, the orientations of the two substrates are very similar when they are bound as a substrate in the catalytic pocket.
Figure 7.
The wild-type human S-COMT (A) and the D51G/S60F/K162R mutant S-COMT built according to the homology model of the wild-type human S-COMT (B). The enzymes are complexed with 2-OH-E2 in the catalytic pocket in its geometry for 2-O-methylation. The figure is drawn with the PyMOL software. Secondary structures are shown with colored ribbons with blue for N-terminus and red for C-terminus. AdoMet is colored red, 2-OH-E2 is colored green and Mg2+ is colored magenta. The amino acids at mutation sites (D51, S60 and K162 for wild-type S-COMT and G51, F60 and R162 for mutant S-COMT) are shown in white sticks. Hydrogens are omitted from AdoMet, 2-OH-E2 and the amino acids.
Using the homology models we have developed, we have also computed the relative binding energy values (ΔEbinding) of the wild-type and mutant human COMTs for 2-OH-E2 and 4-OH-E2. We found that 2-OH-E2 and 4-OH-E2 have similar binding energy values for the wild-type and mutant S-COMTs (data summarized in Table 3). When 2-OH-E2 was the substrate, the ΔEbinding values for its interactions with the wild-type and mutant S-COMTs for its 2-O-methylation were quite comparable, −124.5 and −135.4 kcal/mol, respectively (Table 3), suggesting that the wild-type and mutant S-COMTs have a similar binding affinity for 2-OH-E2, which agreed well with our experimental data (Table 2). Similarly, the computed binding energy values for the binding interactions of 4-OH-E2 with the wild-type and mutant S-COMTs for its 4-O-methylation were − 122.9 and −138.5 kcal/mol, respectively (Table 3), which are also in agreement with our experimental data (Table 2).
Table 3.
The binding energy of 2-OH-E2 and 4-OH-E2 with the wild-type human S-COMT and the mutant S-COMT (D51G/S60F/K162R).
| Genotype | Substrate | Site of O-methylation | ΔEbinding |
|---|---|---|---|
| Wild-type | 2-OH-E2 | 2-O-methylation | −124.5 |
| 3-O-methylation | −121.9 | ||
| 4-OH-E2 | 3-O-methylation | −122.9 | |
| 4-O-methylation | −123.2 | ||
| Mutant (D51G/S60F/K162R) | 2-OH-E2 | 2-O-methylation | −135.4 |
| 3-O-methylation | −148.7 | ||
| 4-OH-E2 | 3-O-methylation | −138.5 | |
| 4-O-methylation | −143.4 |
ΔEbinding was calculated with this equation: ΔEbinding = Ecomplex − (ECOMT + Esubstrate), where Ecomplex is the potentical energy for the complex of COMT with substrate, ECOMT is the potential energy of the enzyme itself and Esubstrate is the potential energy for the substrate itself.
DISCUSSION
Earlier familial studies based on measurement of COMT activity in red blood cells revealed that COMT is a polymorphic enzyme, with a bimodal/trimodel distribution pattern [35]. Moreover, the trait of the low COMT activity was found to be associated with a decreased thermostability of the enzyme preparation [19–21, 30]. Following the subsequent cloning of the human COMT gene [4, 36, 37], a number of single point mutations of this gene have been reported (listed in Table 1). Among them, the V108/158M polymorphism has been most extensively studied. Many epidemiological studies have shown that women, homozygous with the V108/158M mutant, have an increased risk of developing estrogen-associated cancers [18–24]. In addition, this polymorphism has also been suggested to be associated with an elevated risk of schizophrenia, obsessive-compulsive disorder, bipolar disorder, and Parkinson’s disease in both genders [19–21, 25–29]. These studies provide the basis for further studies to identify other mutations/polymorphisms of the human COMT gene and particularly those that alter the catalytic functions of the enzymes.
In the present study, we identified a new mutant human COMT gene with triplet point mutations (D51G/S60F/K162R for S-COMT and D101G/S110F/K212R for MB-COMT), and we have also selectively expressed the mutant enzymes in E. coli for characterization of their catalytic properties. We found that the kinetic parameters (KM and VMAX) of the mutant S- and MB-COMTs did not differ significantly from those of the wild-type enzymes when 2-OH-E2 and 4-OH-E2 were used as substrates. For instance, the mutant S- and MB-COMTs have similar KM values (3.3 and 2.6 µM, respectively) as the wild-type S- and MB-COMTs (3.6 and 3.2 µM, respectively) for the O-methylation of 2-OH-E2. In addition, the mutant S-COMT had almost the same regio-preference for the 2-O-methylation of 2-OH-E2 over its 3-O-methylation as that of the wild-type S-COMT.
One of the notable differences between the mutant and wild-type human COMTs was their thermostability. While the wild-type human COMTs (both S- and MB-COMT) are very stable after 2 hours of pre-incubation at 37°C, the mutant COMTs lost approximately 60% of their catalytic activity under the same pre-incubation conditions. Notably, earlier biochemical analyses of the V108M mutant and wild-type human S-COMTs also showed that whereas they had similar catalytic activity for the O-methylation of catechol estrogens in vitro, the mutant protein was more susceptible to heat inactivation [31, 32]. In both cases, since the mutant enzymes are more unstable under prolonged incubation at 37°C (the physiological temperature), it is suggested that this will eventually result in decreased total levels of catalytically-active COMTs in a given tissue or cell.
In addition, we have also noticed a difference in the apparent KM values of the mutant and wild-type S-COMTs for the methyl donor AdoMet. Whereas the mutant S-COMT had a lower KM value than did the wild-type S-COMT when 2-OH-E2 was used as substrate, this difference disappeared when 4-OH-E2 was the substrate. As expected, the mutant and wild-type S-COMTs also have a different sensitivity to inhibition by AdoHcy, a demethylated AdoMet which binds to same pocket in the COMTs. The IC50 value (18.4 µM) of AdoHcy for the mutant S-COMT was 3-fold higher than the IC50 value (6.1 µM) for the wild-type S-COMT when 2-OH-E2 was the substrate, but when 4-OH-E2 was the substrate, the sensitivity of inhibition by AdoHcy was comparable (26.6 and 29.6 µM). Taken together, these data suggest that the mutant S-COMT has a higher binding affinity for AdoMet than does the wild-type S-COMT when 2-OH-E2 is the substrate, and because of its higher binding affinity, the concentration of AdoHcy needed to compete for its binding site is also proportionally higher.
To better understand the precise structural and functional differences between the D51G/S60F/K162R mutant of S-COMT and the wild-type protein, we have built the homology models of the mutant and wild-type S-COMTs for comparison. It is apparent that the overall secondary structure of the mutant enzyme and especially the structure of its catalytic site (built in two different ways) are very similar to those of the wild-type enzyme. Consistent with this observation, the calculated binding energy values (ΔEbinding) for the interactions of the mutant and wild-type enzymes with substrates were also comparable, which agreed well with the experimental data for the O-methylation of 2-OH-E2 and 4-OH-E2. The overall structural similarities between the mutant and wild-type S-COMTs likely are attributable to the following two factors: (i) All three mutant amino acid residues are found to be located at the surface regions of the protein and are relatively away from the catalytic site. (ii) Two of the three mutations are located in the loop regions of the protein sequence (S60F in loop 3 and K162R in loop 11), and their presence did not alter any of the existing α-helical structures. Although D51 is located in the middle of α-helix 3, this mutation also did not cause a disruption of the original α-helical structure.
In summary, we have identified a new mutant human COMT gene with triple point mutations, i.e., D51G/S60F/K162R for S-COMT and D101G/S110F/K212R for MB-COMT. The selectively-expressed S- and MB-COMT proteins have been biochemically characterized for their catalytic and kinetic properties. Whereas the catalytic activity of the mutant S- and MB-COMTs does not differ significantly from that of the wild-type enzymes when 2-OH-E2 or 4-OH-E2 is the substrate, the mutant COMTs have a lower thermostability compared to the wild-type proteins. Also, the mutant and wild-type S-COMTs have different binding affinities for AdoMet and AdoHcy, depending on the substrate used.
Footnotes
This study was supported, in part, by a grant from the NIH (CA97109).
Abbreviations used: COMT, catechol-O-methyltransferase; E2, 17β-estradiol; 2-OH-E2 and 4-OH-E2, 2- and 4-hydroxyestradiol, respectively; AdoMet, S-adenosyl-L-methionine; AdoHcy, S-adenosyl-L-homocysteine.
References
- 1.Axelrod J, Tomchick R. Enzymatic O-methylation of epinephrine and other catechols. J Biol Chem. 1958;233:702–705. [PubMed] [Google Scholar]
- 2.Axelrod J. Methylation reactions in the formation and metabolism of catecholamines and other biogenic amines. Pharmacol Rev. 1996;18:95–113. [PubMed] [Google Scholar]
- 3.Zhu BT. Catechol-O-Methyltransferase (COMT)-mediated methylation metabolism of endogenous bioactive catechols and modulation by endobiotics and xenobiotics: importance in pathophysiology and pathogenesis. Curr Drug Metab. 2002;3:321–349. doi: 10.2174/1389200023337586. [DOI] [PubMed] [Google Scholar]
- 4.Tenhunen J, Salminen M, Jalanko A, Ukkonen S, Ulmanen I. Structure of the rat catechol-O-methyltransferase gene: separate promoters are used to produce mRNAs for soluble and membrane-bound forms of the enzyme. DNA Cell Biol. 1993;12:253–263. doi: 10.1089/dna.1993.12.253. [DOI] [PubMed] [Google Scholar]
- 5.Zhu BT, Liehr JG. Quercetin increases the severity of estradiol-induced tumorigenesis in hamster kidney. Toxicol Appl Pharmacol. 1994;125:149–158. doi: 10.1006/taap.1994.1059. [DOI] [PubMed] [Google Scholar]
- 6.Zhu BT, Patel UK, Cai MX, Conney AH, et al. O-Methylation of tea polyphenols catalyzed by human placental cytosolic catechol-O-methyltransferase. Drug Metab Dispos. 2000;28:1024–1030. [PubMed] [Google Scholar]
- 7.Zhu BT, Ezell EL, Liehr JG. Catechol-O-methyltransferase-catalyzed rapid O-methylation of mutagenic flavonoids. Metabolic inactivation as a possible reason for their lack of carcinogenicity in vivo. J Biol Chem. 1994;269:292–299. [PubMed] [Google Scholar]
- 8.Zhu BT, Liehr JG. Inhibition of catechol O-methyltransferase-catalyzed O-methylation of 2- and 4-hydroxyestradiol by quercetin. Possible role in estradiol-induced tumorigenesis. J Biol Chem. 1996;271:1357–1363. doi: 10.1074/jbc.271.3.1357. [DOI] [PubMed] [Google Scholar]
- 9.Zhu BT. CNS dopamine oxidation and catechol-O-methyltransferase: importance in the etiology, pharmacotherapy, and dietary prevention of Parkinson's disease. Int J Mol Med. 2004;13:343–353. [PubMed] [Google Scholar]
- 10.Karayiorgou M, Altemus M, Galke BL, Goldman D, Murphy DL, Ott J, et al. Genotype determining low catechol-O-methyltransferase activity as a risk factor for obsessive-compulsive disorder. Proc Natl Acad Sci USA. 1997;94:4572–4575. doi: 10.1073/pnas.94.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirov G, Murphy KC, Arranz MJ, Jones I, McCandles F, Kunugi H, et al. Low activity allele of catechol-O-methyltransferase gene associated with rapid cycling bipolar disorder. Mol Psychiatry. 1998;3:342–345. doi: 10.1038/sj.mp.4000385. [DOI] [PubMed] [Google Scholar]
- 12.Papolos DF, Veit S, Faedda GL, Saito T, Lachman HM. Ultra-ultra rapid cycling bipolar disorder is associated with the low activity catecholamine-O-methyltransferase allele. Mol Psychiatry. 1998;3:346–349. doi: 10.1038/sj.mp.4000410. [DOI] [PubMed] [Google Scholar]
- 13.Strous RD, Bark N, Parsia SS, Volavka J, Lachman HM. Analysis of a functional catechol-O-methyltransferase gene polymorphism in schizophrenia: evidence for association with aggressive and antisocial behavior. Psychiatry Res. 1997;69:71–77. doi: 10.1016/s0165-1781(96)03111-3. [DOI] [PubMed] [Google Scholar]
- 14.Tiihonen J, Hallikainen T, Lachman H, Saito T, Volavka J, Kauhanen J, et al. Association between the functional variant of the catechol-O-methyltransferase (COMT) gene and type 1 alcoholism. Mol Psychiatry. 1999;4:286–289. doi: 10.1038/sj.mp.4000509. [DOI] [PubMed] [Google Scholar]
- 15.Enoch MA, Xu K, Ferro E, Harris CR, Goldman D. Psychiatr. Genetic origins of anxiety in women: a role for a functional catechol-O-methyltransferase polymorphism. Genet. 2003;13:33–41. doi: 10.1097/00041444-200303000-00006. [DOI] [PubMed] [Google Scholar]
- 16.Rosa A, Peralta V, Cuesta MJ, Zarzuela A, Serrano F, Martinez-Larrea A, et al. New evidence of association between COMT gene and prefrontal neurocognitive function in healthy individuals from sibling pairs discordant for psychosis. Am J Psychiatry. 2004;161:1110–1112. doi: 10.1176/appi.ajp.161.6.1110. [DOI] [PubMed] [Google Scholar]
- 17.Nolan KA, Bilder RM, Lachman HM, Volavka J. Catechol O-methyltransferase Val158Met polymorphism in schizophrenia: differential effects of Val and Met alleles on cognitive stability and flexibility. Am J Psychiatry. 2004;161:359–361. doi: 10.1176/appi.ajp.161.2.359. [DOI] [PubMed] [Google Scholar]
- 18.Illi A, Mattila KM, Kampman O, Anttila S, Roivas M, Lehtimaki T. Catechol-O-methyltransferase and monoamine oxidase A genotypes and drug response to conventional neuroleptics in schizophrenia. J Clin Psychopharmacol. 2003;23:429–434. doi: 10.1097/01.jcp.0000088916.02635.33. [DOI] [PubMed] [Google Scholar]
- 19.Zhu BT, Conney AH. Functional role of estrogen metabolism in target cells: review and perspectives. Carcinogenesis. 1998;19:1–27. doi: 10.1093/carcin/19.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Lee AJ, Cai MX, Tomas PE, Conney AH, Zhu BT. Characterization of the oxidative metabolites of 17β-estradiol and estrone formed by 15 selectively expressed human cytochrome p450 isoforms. Endocrinology. 2003;144:3382–3398. doi: 10.1210/en.2003-0192. [DOI] [PubMed] [Google Scholar]
- 21.Zhu BT, Conney AH. Is 2-methoxyestradiol an endogenous estrogen metabolite that inhibits mammary carcinogenesis? Cancer Res. 1998;58:2269–2277. [PubMed] [Google Scholar]
- 22.Huang CS, Chern HD, Chang KJ, Cheng CW, Hsu SM, Shen CY. Breast cancer risk associated with genotype polymorphism of the estrogen-metabolizing genes CYP17, CYP1A1, and COMT: A multigenic study on cancer susceptibility. Cancer Res. 1999;59:4870–4875. [PubMed] [Google Scholar]
- 23.Mitrunen K, Jourenkova N, Kataja V, Eskelinen M, Kosma VM, Benhamou S, et al. Glutathione S-transferase M1, M3, P1, and T1 genetic polymorphisms and susceptibility to breast cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:635–640. [PubMed] [Google Scholar]
- 24.Mitrunen K, Kataja V, Eskelinen M, Kosma VM, Kang D, Benhamou S, et al. Combined COMT and GST genotypes and hormone replacement therapy associated breast cancer risk. Pharmacogenetics. 2002;12:67–72. doi: 10.1097/00008571-200201000-00009. [DOI] [PubMed] [Google Scholar]
- 25.Thompson PA, Shields PG, Freudenheim JL, Stone A, Vena JE, Marshall JR, et al. Genetic polymorphisms in catechol-O-methyltransferase, menopausal status, and breast cancer risk. Cancer Res. 1998;58:2107–2110. [PubMed] [Google Scholar]
- 26.Suzuki K, Nakazato H, Matsui H, Koike H, Okugi H, Kashiwagi B, et al. Genetic polymorphisms of estrogen receptor alpha, CYP19, catechol-O-methyltransferase are associated with familial prostate carcinoma risk in a Japanese population. Cancer. 2003;98:1411–1416. doi: 10.1002/cncr.11639. [DOI] [PubMed] [Google Scholar]
- 27.Yim DS, Parkb SK, Yoo KY, Yoon KS, Chung HH, Kang HL, et al. Relationship between the Val158Met polymorphism of catechol O-methyl transferase and breast cancer. Pharmacogenetics. 2001;11:279–286. doi: 10.1097/00008571-200106000-00001. [DOI] [PubMed] [Google Scholar]
- 28.Goodman MT, McDuffie K, Kolonel LN, Terada K, Donlon TA, Wilkens LR, et al. Case-control study of ovarian cancer and polymorphisms in genes involved in catecholestrogen formation and metabolism. Cancer Epidemiol Biomarkers Prev. 2001;10:209–216. [PubMed] [Google Scholar]
- 29.Lavigne JA, Helzlsouer KJ, Huang HY, Strickland PT, Bell DA, Selmin O, et al. An association between the allele coding for a low activity variant of catechol-O-methyltransferase and the risk for breast cancer. Cancer Res. 1997;57:5493–5497. [PubMed] [Google Scholar]
- 30.Lotta T, Vidgren J, Tilgman C, Ulmanen I, Melen K, Julkunen I, et al. Kinetics of human soluble and membrane-bound catechol O-methyltransferase: a revised mechanism and description of the therolabile variant of the enzyme. Biochemistry. 1995;34:4202–4210. doi: 10.1021/bi00013a008. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Yao J, Chang M, Nikolic D, Yu L, Yager JD. Equine catechol estrogen 4-hydroxyequilenin is a more potent inhibitor of the variant form of catechol O-methyltransferase. Chem Res Toxicol. 2004;17:512–520. doi: 10.1021/tx0342464. [DOI] [PubMed] [Google Scholar]
- 32.Goodman JE, Jensen LT, He, Yager JD. Characterization of human soluble high and low activity catechol O-methyltrasferase catalyzed catechol estrogen methylation. Pharmacogenetics. 2002;12:517–528. doi: 10.1097/00008571-200210000-00003. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Yang X, Breemen RB, Bolton JL. Characterization of two new variants of human catechol O-methyltransferase in vitro. Cancer Letter. 2005;230:81–89. doi: 10.1016/j.canlet.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 34.Zhu BT, Roy D, Liehr JG. The carcinogenic activity of ethinyl estrogens is determined by both their hormonal characteristics and their conversion to catechol metabolites. Endocrinology. 1993;132:577–583. doi: 10.1210/endo.132.2.8381068. [DOI] [PubMed] [Google Scholar]
- 35.Cohn CC, Dunner DL, Axelrod J. Reduced catechol-O-methyltransfease activity in red blood cells of women with primary affective disorder. Science. 1970;170:125–135. doi: 10.1126/science.170.3964.1323. [DOI] [PubMed] [Google Scholar]
- 36.Tenhunen J, Ulmanen I. Production of rat soluble and membrane-bound catechol O-methyltransferase forms from bifunctional mRNAs. Biochem J. 1993;296(Pt 3):595–600. doi: 10.1042/bj2960595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winqvist R, Lundstrom K, Salminen M, Laatikainen M, Ulmanen I. The human catechol-O-methyltransferase (COMT) gene maps to band q11.2 of chromosome 22 and shows a frequent RFLP with BglI. Cytogenet Cell Genet. 1992;59:253–257. doi: 10.1159/000133262. [DOI] [PubMed] [Google Scholar]