Abstract
Objective
To investigate whether a standard dental prophylaxis followed by tooth brushing with an antibacterial dentifrice will affect the oral bacterial community, as determined by denaturing gradient gel electrophoresis (DGGE) combined with 16S rRNA gene sequence analysis.
Methods
Twenty-four healthy adults were instructed to brush their teeth using commercial dentifrice for 1 week during a washout period. An initial set of pooled supragingival plaque samples was collected from each participant at baseline (0 h) before prophylaxis treatment. The subjects were given a clinical examination and dental prophylaxis and asked to brush for 1 min with a dentifrice containing 0.3% triclosan/2.0% PVM/MA copolymer/0.243% sodium fluoride (Colgate Total). On the following day, a second set of pooled supragingival plaque samples (24 h) was collected. Total bacterial genomic DNA was isolated from the samples. Differences in the microbial composition before and after the prophylactic procedure and tooth brushing were assessed by comparing the DGGE profiles of PCR-amplified and 16S rRNA gene segments sequence analysis.
Results
Two distinct clusters of DGGE profiles were found, suggesting that a shift in the microbial composition had occurred 24 h after the prophylaxis and brushing. A detailed sequencing analysis of 16S rRNA gene segments further identified six phyla and 29 genera, including known and unknown bacterial species. Importantly, an increase in bacterial diversity was observed after 24 h, including members of the Streptococcaceae family, Prevotella, Corynebacterium, TM7 and other commensal bacteria.
Conclusion
The results suggest that the use of a standard prophylaxis followed by the use of the dentifrice containing 0.3% triclosan/2.0% PVM/MA copolymer/0.243% sodium fluoride may promote a healthier composition within the oral bacterial community.
Keywords: dental plaque, microbial diversity, PCR-DGGE, 16S rDNA clone library
The bacterial community in the human oral cavity is extremely diverse. More than 700 common oral bacterial species or phylotypes have been identified from the oral cavity1,2. Many microorganisms still remain to be elucidated (http://www.homd.org), along with their behaviour and role in the dynamic oral environment. It is believed that most microorganisms in the oral cavity are host-beneficial microflora forming a commensal community, but they also include bacterial species known to cause a range of oral diseases, including dental caries, gingivitis and periodontitis.
Dental caries and chronic periodontal diseases are known to be associated with polymicrobial colonisation. Both acidogenic and aciduric bacteria, mainly the mutans streptococci (Streptococcus mutans and S. sobrinus) and lactobacilli, are known to be the primary aetiological agents of dental caries. Thus, a caries-free healthy dentition usually has low levels of mutans streptococci and lactobacilli in saliva and dental plaque. This level, however, can increase if the host frequently ingests sugar or other fermentable carbohydrates, thereby altering the bacterial composition from one that is mutually symbiotic to one associated with dental caries3. Periodontal diseases, on the other hand, are associated with the alteration from more Gram-positive facultative anaerobes found in healthy gingiva to more Gram-negative species that result from plaque accumulation3. Thus, it appears that oral health depends on maintaining a highly diverse, but balanced, bacterial composition. As such, both understanding and monitoring the interactive and dynamic changes in the microbial community are essential for developing preventive measures and promoting oral health.
In past decades, scientists have relied on conventional cultivation methods to evaluate and quantify changes in bacterial composition. However, based on in vitro culture limitations and technical difficulties, it has been estimated that over half of bacterial species in the oral cavity cannot be cultivated4. Thus, culture-independent molecular techniques, such as denaturing gradient gel electrophoresis (DGGE), terminal restriction fragment length polymorphisms (TRFLP) and 16S rRNA gene sequencing analysis, have become more favourable tools to assess both cultivable and culture-independent microbiota and to perform epidemiological analyses of the oral microbial community.
Recently, we and others demonstrated a great degree of variation in bacterial diversity and composition associated with oral health and diseases1,5,6. Studies by Goodson et al reported that professional dental prophy-laxis significantly reduced total plaque score (clinical evaluation) and, hence, bacterial level, as determined by DNA probe analysis7. Using DGGE, oral microbial changes were able to be categorised following dental prophylaxis8. Taking this procedure a step further, we decided to combine the use of DGGE with 16S rRNA gene sequence analysis to evaluate the effect of standard prophylactic procedure followed by tooth brushing using a therapeutic dentifrice on oral bacterial composition, as determined from supragingival plaque samples taken at baseline (0 h) and 24 h after the procedure.
Materials and methods
Subjects and bacterial sample collection
This study was carried out under the protocol for human subjects approved by the safety and regulatory authorities of Colgate-Palmolive Company (New York, NY, USA). Twenty-four healthy adults voluntarily participated in this study. All subjects were free of periodontal disease and had taken no antibiotics for a period of 3 months preceding the study. Informed consent was obtained from each individual. Subjects were then instructed to brush their teeth with the same toothpaste for 1 week during a washout period, after which a clinical examination was performed and bacterial sample collections were collected.
After the 1-week washout period, an initial set of baseline pooled plaque samples (0 h) was collected from the interproximal sites of all molars of each individual in the morning when a clinical examination was performed. The participants were then given a standard dental prophylaxis by a dental hygienist and asked to brush for 1 min with a commercial dentifrice containing 0.3% triclosan, 2.0% PVM/MA copolymer and 0.243% sodium fluoride (Colgate Total), using the same brand of toothbrush (Colgate Navigator). All participants were asked to refrain from performing any type of oral hygiene practice for 24 h and then to report back to the clinic the following day. On the morning of the next day, a second set of pooled supragingival plaque samples (24 h) was collected from the same interproximal sites of the molars of each participant. All plaque samples were collected with a sterile sickle scaling instrument and stored in DNase- and RNase-free polyethylene tubes. The plaque samples were immediately frozen at 20°C and shipped on dry ice to the microbiology laboratory at the New York University College of Dentistry (New York, NY).
PCR-DGGE of bacterial 16S rRNA genes
Bacterial samples were dissolved at 4°C and then washed in 1 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5). Total bacterial genomic DNA was extracted from the sample using the MaterPure™ DNA purification kit (Epicentre, Madison, WI), as previously described5,6,8,9. For all PCR applications, the final concentration of each DNA sample was adjusted to 10 ng/ml. PCR was performed with the GeneAmp®PCR System 9700 (PE Applied Biosystems, Foster City, CA). A set of universal bacterial 16S rDNA primers, forward prbac1 (5′-CGCCCGGGGC-GCGCCCCG-GGCGGGGCGGGGGCACGGGGGGACTACGT-GCCAGCAGCC-3′) and reverse prbac2 (5′-GGAC-TACCAGGGTATCT-ACTAATCC-3′)10, which target the hypervariable V4 V5 regions of the Escherichia coli 16S rDNA ribosomal locus, was used with a 40-nucleotide GC-clamp11 added to the 5′ end of prbac1. Each standardised PCR mixture (a total volume of 50 ml) contained 100 ng of total genomic DNA, 0.8 mM of dNTP, 40 pmol of each primer, 4.0 mM of MgCl, 2, 5 ml of 10X PCR buffer II and 2.5 U of Taq DNA polymerase (PE Applied Biosystems). The PCR conditions were the same as previously described5,6,8,9. The PCR products were evaluated by electrophoresis in 1.0% agarose gels run at 60 V for 60 min, and the sizes of all amplicons (300 base pairs) were confirmed according to a molecular size standard. A standardised 20 ml of each PCR-amplified product was loaded on the DGGE gel and separated with the Bio-Rad DcodeTM System (Bio-Rad, Hercules, CA). A 40% to 60% linear DNA denaturing gradient was formed in 8% (w/v) polyacrylamidegels. PCR products were directly loaded into each lane. Electrophoresis was performed at a constant 60 V at 58°C for 16 h in 1X Tris-acetate-EDTA (TAE) buffer (pH 8.5), as previously described5,6,8,9. After electrophoresis, gels were rinsed and stained with ethidium bromide (0.5 mg/ml) for 15 min, followed by 15 min destaining in water. DGGE images were digitally captured with the AlphaImager 3300 System (Alpha Innotech Corporation, San Leandro, CA) and analysed with Fingerprinting II Informatix™ Software (Bio-Rad). Levels of similarity between fingerprints were calculated based on the Dice coefficient of pairwise comparisons. A dendrogram was constructed based on Ward’s method and algorithm for cluster analysis12. Differences in the microbial composition before and after the prophylactic procedure and tooth brushing were assessed by comparing the DGGE profiles of the amplified 16S rRNA gene segments. Significant differences in the number of detected PCR amplicons in the DGGE gels were determined using analysis of variance (ANOVA) and paired t-test. Statistical analyses were performed using SPSS software (version 17.0, SPSS, Chicago, IL). All P-values < 0.05 were two-tailed and considered significant.
Analysis of 16S rRNA gene sequence libraries
To further investigate changes in bacterial composition at the bacterial gene level, a pilot study was conducted. Bacterial DNA samples, including samples from baseline (0 h) and post-procedure (24 h), were randomly selected from two individuals (no. 2 and no. 11). The targeted 16S rRNA gene, positioning at 509 to 805 of the E. coli 16S rRNA gene, was amplified using the same universal bacterial primers, prbac1 without the GC-clamp and prbac2. In order to establish 16S rRNA gene libraries, the PCR products were ligated into pCR®4-TOPO® (Invitrogen, Carlsbad, CA) and transformed into OneShot®Top10 chemically competent E. coli (Invitrogen). After culture selection (with addition of kanamycin, 50 mg/ml, to the culture medium) and blue/white colony screening, 100 colonies per sample, a total of 400 colonies, were picked randomly, including the two time points (0 h and 24 h).
The vector-specific universal primer set of M13F and M13R was used to amplify the plasmid DNA to determine all colonies containing inserts of correct size (300 bases). The purified PCR product was then sequenced in an automated ABI Prism 3730xl DNA Sequencer (Applied Biosystems). A standard nucleotide-nucleotide BLAST search was conducted to find all 16S rRNA gene sequences in the Ribosomal Database Project II (RDP-II, release 9.39)13 and NCBI GenBank (www.ncbi.nlm.gov) databases. They were first examined for chimerism by using the Chimera Detection tool available through RDP-II and further categorised into various phylotypes using Classifier analysis (95% confidence threshold)14 and the furthest-neighbour assignment algorithm in DOTUR (distance-based operational taxonomic unit and richness), a computer program which assigns sequences to phylotypes15. The coverage and the total number of sequences for the clone library analysed for microbial diversity were calculated according to Good’s coverage estimation16. Estimates of phylotype richness were calculated according to the abundance-based coverage estimator (ACE)17. Collector’s curves of observed and estimated richness were calculated in DOTUR15. The Shannon-Weaver diversity index18 was also calculated using DOTUR. Furthermore, ∫-LIBSHUFF19 was used to determine whether observed differences in 16S rDNA sequence libraries were the result of underlying variability in the microbial populations or an artefact resulting from insufficient bacterial population sampling. Pairwise comparisons were performed to analyse inter-experiment (0 h vs. 24 h) and intra-subject (subject 2 and subject 11) variability.
Results
DGGE profile analysis
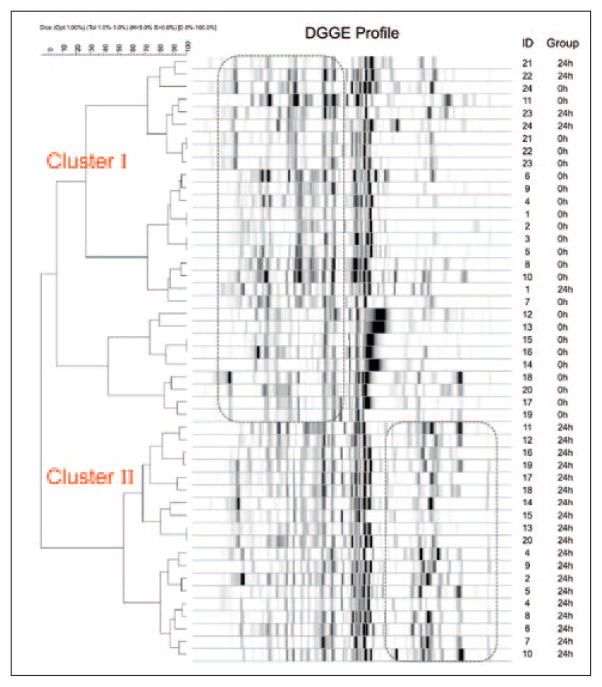
PCR amplification was performed for all 48 pooled plaque samples (0 h and 24 h) of the 24 participants to obtain the targeted 16S rDNA fragments (300 base pairs). The PCR products were separated by DGGE (Fig 1). The Fingerprinting II Informatix™ program (Bio-Rad) was used to perform identification of the DGGE banding positions and comparison of the fingerprints between baseline (Fig 1a) and the 24-h (Fig 1b) samples. The mean numbers of detected PCR amplicons were 29.5 ± 2.9 for the 0-h group and 33.1 ± 2.6 for the 24-h group. The differences were statistically significant (P < 0.001). The cluster analysis revealed two distinct clusters of DGGE profiles (Fig 2). Nineteen (79.2%) of the baseline profiles were grouped in cluster I, and all 24-h profiles (100%) were in cluster II. The different distribution patterns suggested that the overall microbial composition was changed within 24 h after prophylaxis.
Fig 1.
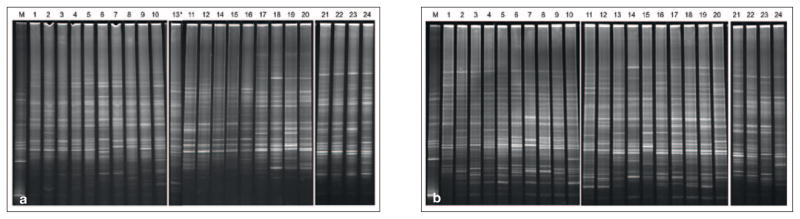
DGGE profiles for: a) baseline (0 h) samples, b) post-prophylaxis (24 h) samples. *The sample from subject 13 was misplaced before the sample from subject 11.
Fig 2.
Dendrogram of DGGE profiles. There are two distinct clusters for the baseline samples (0 h) and the post-proph-ylaxis samples (24 h), suggesting a difference in the overall bacterial profile between the two groups.
Bacterial phylogenetic analysis
A total of 400 clones were randomly selected, and 381 clones with an insert of the correct size, approximately 300 bases, were analysed. Sequence examination showed that 10 of the 381 clones were found to be chimeras. Thus, 371 sequences were included in the final phyloge-netic analysis. Good’s coverage estimates ranged from 77.4% to 89.5% (Table 1). All 16S rRNA gene sequences were carefully aligned with Near Alignment Space Termination (NAST) at Greengenes (http://greengenes.lbl.gov/cgi-bin/nph-index.cgi)20 and subjected to RDP-II Classifier analysis. A total of 110 distinct phylotypes were identified among the 371 sequences, varying from 24 to 42 per library. Most clones were assigned to six phyla, including Firmicutes (30.2% of all sequences), Bacteroidetes (25.3%), Proteobacteria (25.1%), Fuso-bacteria (8.9%), Actinobacteria (6.2%) and TM7 (2.4%), for which there are no cultivable representatives, and eight sequences remained unclassified (2.2%). The distribution of bacterial phylotypes showed slight increases in Actinobacteria and Bacteroidetes and decreases in Fusobacteria and Proteobacteria for the 24-h samples (Fig 3). The difference, however, was not statistically significant (chi-square statistics = 6.11; P > 0.05). Moreover, only 3.5% (13/371) of clones had less than 94% sequence similarity to existing database entries.
Table 1.
Phylotype richness and calculated coverage and diversity for each library
| Clone library | Subject | Sample type | No. clones in library | No. of phylotypes identified | Good’s coverage (%) | Shannon-Weaver diversity index* |
|---|---|---|---|---|---|---|
| 1 | 2 | 0 h | 93 | 24 | 77.42 | 2.65 ± 0.22 |
| 2 | 2 | 24 h | 93 | 31 | 83.87 | 3.06 ± 0.19 |
| 3 | 11 | 0 h | 90 | 39 | 88.89 | 3.35 ± 0.19 |
| 4 | 11 | 24 h | 95 | 42 | 89.47 | 3.37 ± 0.20 |
A slight increase in the diversity index in the 24-h samples indicated a higher degree of microbial diversity. Also, more bacterial phylotypes were identified in the 24-h samples.
Fig 3.

Comparison of the taxonomic diversity between the two library sets, 0 h vs. 24 h. The distribution of bacterial phy-lotypes showed slight increases in Actinobacteria and Bacter-oidetes and decreases in Fusobacteria and Proteobacteria for the 24-h samples.
More specific distributions of bacterial taxa in the clinical samples are summarised and listed in Table 2. (1) Within the Firmicutes phylum, 24% of sequenced clones fell into the Bacilli class. The most abundant order was Lactobacillales, which was dominated by Streptococcaceae, mainly including S. oralis, S. gon-donii, S. cristatus and S. sanguinis. (2) The phylum Bacteroidetes was found in all four libraries, but varied in abundance. The most abundant group within this phylum was the Prevotellaceae family, mainly observed in the 24-h group from subject 11 (74.29%). (3) Twenty-five percent of all clones were in the Proteobacteria phylum. The most abundant class was b-proteobacteria, as well as g-proteobacteria and e-proteobacteria. (4) Only 32 Fusobacteria clones were identified in both 24-h and 0-h samples from subject 11. These included Fusobacterium and Leptotrichia. (5) Actinobacteria found in all libraries, albeit in low abundance, included the genera Corynebacterium, Rothia and Actinomyces. (6) A total of 9 TM7 clones were obtained from both 24-h and 0-h samples, indicating that the prevalence of this group of uncultivated bacteria was as low as 2.4%.
Table 2.
Comparison of the distribution of 16S rRNA gene sequences and phylotypes before and after the prophylaxis and tooth brushing
| Phylum | Bacterial taxa | Total N = 371 (%) |
0 h N = 183 (%) |
24 h N = 188 (%) |
|---|---|---|---|---|
| Firmicutes | 112 (30.2) | 58 (31.7) | 54 (28.7) | |
| Bacilli | 89 (24.0) | 39 (21.3) | 50 (26.6) | |
| Streptococcaceae | 74 (19.9) | 31 (16.9) | 43 (22.9) | |
| Clostridia | 19 (5.1) | 16 (8.7) | 3 (1.6) | |
| Veillonellaceae | 17 (4.6) | 15 (8.2) | 2 (1.1) | |
| Aerococcaceae | 3 (0.8) | 1 (0.6) | 2 (1.1) | |
| Incertae Sedis XI | 2 (0.5) | 1 (0.6) | 1(0.5) | |
| Lachnospiraceae | 1 (0.3) | 0 | 1 (0.5) | |
| Bacteroidetes | 94 (25.3) | 41 (22.4) | 53 (28.2) | |
| Flavobacteriaceae | 43 (11.6) | 22 (12.0) | 21 (11.2) | |
| Prevotellaceae | 35 (9.4) | 9 (4.9) | 26 (13.8) | |
| Porphyromonadaceae | 12 (3.2) | 8 (4.4) | 4 (2.1) | |
| Proteobacteria | 93 (25.1) | 51 (27.9) | 42 (22.3) | |
| β-Proteobacteria | 73 (19.7) | 40 (21.9) | 33 (17.6) | |
| Order Neisseriales | 53 (14.3) | 28 (15.3) | 25 (13.3) | |
| Order Burkholderiales | 15 (4.0) | 10 (5.5) | 5 (2.7) | |
| Pasteurellaceae | 14 (3.8) | 8 (4.4) | 6 (3.2) | |
| γ-Proteobacteria | 14 (3.8) | 8 (4.4) | 6 (3.2) | |
| Campylobacteraceae | 6 (1.6) | 3 (1.6) | 3 (1.6) | |
| ε-Proteobacteria | 6 (1.6) | 3 (1.6) | 3(1.6) | |
| Fusobacteria | 33 (8.9) | 18 (9.8) | 14 (7.5) | |
| Fusobacterium | 19 (5.1) | 11 (6.0) | 8 (4.3) | |
| Leptotrichia | 13 (3.5) | 7 (3.8) | 6 (3.2) | |
| Actinobacteria | 23 (6.2) | 8 (4.4) | 15 (8.0) | |
| Corynebacteriaceae | 6 (1.6) | 2 (1.1) | 4 (2.1) | |
| Micrococcaceae | 6 (1.6) | 1 (0.6) | 5 (2.7) | |
| Actinomycetaceae | 9 (2.4) | 4 (2.2) | 5 (2.7) | |
| TM7 | ||||
| TM7 genera incertae sedi | 9 (2.4) | 3 (1.6) | 6 (3.2) |
In the study, collector’s curves at a pseudo-phylum level using a distance value of 0.01 (Fig 4) were also constructed. Estimates of phylotype richness were calculated according to the ACE17 and the bias-corrected Chao1 estimator21. By randomly selecting 95 100 clones per library, we found that the gap between observed and estimated richness of bacterial phylotypes was smaller at 0 h compared with 24 h, suggesting that additional sampling at 24 h would be necessary to obtain constant estimates of the number of unobserved phylotypes in the clinical sample. Furthermore, ∫-LIBSHUFF pairwise comparisons of pooled libraries (0 h versus 24 h) revealed that all the libraries were statistically different from one another (P < 0.001) (Table 3). In addition, the statistical test results demonstrated significant differences both in subject-to-subject libraries and 0-h versus 24-h libraries (P < 0.001) (Table 3), suggesting that the observed differences between the paired libraries could have resulted from underlying differences in the dental plaque from which they were derived.
Fig 4.
Collector’s curves of the observed and estimated (ACE and Chao1) phylo-type richness of the bacterial samples per subject at different time points. Each curve reflects the series of observed or estimated richness values obtained as clones are added to the dataset in an arbitrary order. The gap between observed and estimated richness when sampling stopped suggested that more clones with more sequences will increase the unobserved phylotypes.
Table 3.
∫-LIBSHUFF comparisons of 16S rRNA gene sequence libraries from four saliva samples of two subjects
| Library X | Library Y | ||||
|---|---|---|---|---|---|
| Subject 2 | Subject 11 | ||||
| 0 h | 24 h | 0 h | 24 h | ||
| Subject 2 | 0 h | - | 0.0632 | 0.0000 | 0.0000 |
| 24 h | 0.0002 | - | 0.0000 | 0.0000 | |
| Subject 11 | 0 h | 0.0000 | 0.0000 | - | 0.0000 |
| 24 h | 0.0000 | 0.0004 | 0.0000 | - | |
The values in the table represent p values for ΔCXY of homologous library X and heterologous library Y (lower triangle) and ΔCYX of homologous library Y and heterologous library X (upper triangle). Libraries are distinct if both pairwise comparisons (ΔCXY and ΔCYX) show statistically significant difference (P < 0.05). The study found that 11 out of 12 pairwise comparisons were statistically different, except 0 h vs. 24 h of subject 2.
Discussion
The application of professional dental prophylaxis can significantly reduce plaque accumulation and total bacterial level in saliva and dental plaque7,22,23. In a previous study, a significant reduction in the number of detected 16S rRNA gene amplicons after a dental pro-phylactic treatment was also shown8. Here we employed two different molecular-based PCR techniques, DGGE profile and 16S rRNA gene sequence analysis, which demonstrated a shift in bacterial phylotype distribution, confirming changes in bacterial composition observed in the earlier study8. For example, compared with the post-prophylaxis samples, more high-density bands (DGGE) and bacterial phyla of lower G+C content were observed for Firmicutes and Fusobacteria groups in the baseline plaque samples. Conversely, compared with the baseline plaque samples, more high-density bands (DGGE) and bacterial phyla of higher G+C content were observed for the Bacteroidetes groups in the post-prophylaxis samples.
Previous studies had demonstrated DGGE analysis to be a powerful tool for microbial 16S rRNA gene characterisation, as well as for assessing the overall microbial profile in the oral cavity5,6,8,9. The cluster analysis of 16S rRNA gene amplicons revealed two distinct clusters of DGGE profiles, suggesting that the overall microbial composition had changed within 24 h. Since all subjects in this study were instructed to brush their teeth with the dentifrice containing 0.3% triclosan/2.0% PVM/MA copolymer/0.243% sodium fluoride after receiving dental prophylaxis, the changes in bacterial composition could not be attributed either to dental prophylaxis or the use of the dentifrice alone. In spite of that, a body of evidence based on clinical studies has shown the 0.3% triclosan (a broad-spectrum antibacterial agent), 2.0% PVM/MA copolymer and 0.243% sodium fluoride dentifrice has therapeutic effect against bacterial colonisation, gingivitis and the progression of periodontal disease24–26. The observed changes in the present study, therefore, could be from the combined effect of dental prophylaxis and the use of the 0.3% triclosan/2.0% PVM/MA copolymer/0.243% sodium fluoride dentifrice, suggesting that their combined effect could provide a potentially beneficial effect on balancing the bacterial community in the oral cavity.
Extensive 16S rRNA-based sequence analysis has played a pivotal role in studies of microbial identification27,28. Based on the analysis of 36,043 16S rRNA gene sequences, Dewhirst and others reported six major phyla, including Firmicutes (36.7%), Bacteroidetes (17.3%), Proteobacteria (17.1%), Actinobacteria (11.6%), Spirochaetes (7.9%), Fusobacteria (5.2%) and TM7 (1.9%)29. The phylogenetic distribution observed in this study was similar to their findings, except for spirochaetes. The difference in the findings compared with those of other investigators was not unexpected1,29 given the relatively low prevalence of spirochaete species in saliva, and the fact that only four 16S rRNA gene libraries containing 371 sequences were included in this pilot study. Furthermore, the DNA sequences were based on an average molecular size of 300 base pair PCR products; therefore, interpreting the results of a 16S rRNA gene similarity search based on ‘first hit’ or ‘closest match’ may not necessarily represent the actual identity of a bacterial isolate27. Consequently, the sequences of 16S rRNA gene similarity identified in our study can only be confidently identified at the genus, not the species, level, especially the sequences which show <94% similarity. Interestingly, this study showed a relatively moderate value for library sample coverage and high value for diversity index. Since only 100 clones per library of 16S rRNA genes were sufficient to provide valuable insight into the primary membership of microbial communities, we anticipate that additional sampling would increase the phylotype richness in each subject.
In summary, the current study demonstrated two distinct clusters of DGGE profiles of bacterial 16S rRNA genes in the two microbial communities tested, both before and after prophylaxis and tooth brushing. Second, the results from 16S rRNA-based molecular analysis indicated that these changes occurred within three phyla: Firmicutes, Fusobacteria and Bacteroidetes. It is well known that dental caries and chronic periodontal diseases are associated with polymicrobial colonisation. Although significant variation in oral bacterial community composition has been reported, a greater degree of diversity is associated with good oral health5,6,8. The findings of this study suggest that application of the standard prophylaxis plus brushing with the 0.3% triclosan/2.0% PVM/MA copolymer/0.243% sodium fluoride dentifrice may promote a healthier microbial composition in dental plaque. A full-scale clinical trial is needed to further determine the potential effect of the dentifrice, as well as good oral health practices, on microbiota shifts. Understanding the nature of the microbial composition and its response to perturbation, such as prophylaxis, brushing, flossing and other oral health practices, could provide valuable insight for the development of novel preventive dental care programmes and treatment.
Acknowledgments
Supported by NIH/NIDCR Grant DE13937 and the Col-gate-Palmolive Company.
References
- 1.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paster BJ, Boches SK, Galvin JL, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruby J, Goldner M. Nature of symbiosis in oral disease. J Dent Res. 2007;86:8–11. doi: 10.1177/154405910708600102. [DOI] [PubMed] [Google Scholar]
- 4.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Peri-odontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Ge Y, Saxena D, Caufield PW. Genetic profiling of the oral microbiota associated with severe early-childhood caries. J Clin Microbiol. 2007;45:81–87. doi: 10.1128/JCM.01622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Ismail AI, Ge Y, Tellez M, Sohn W. Similarity of bacterial populations in saliva from African-American mother-child dyads. J Clin Microbiol. 2007;45:3082–3085. doi: 10.1128/JCM.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodson JM, Palys MD, Carpino E, et al. Microbiological changes associated with dental prophylaxis. J Am Dent Assoc. 2004;135:1559–1564. doi: 10.14219/jada.archive.2004.0082. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Saxena D, Barnes V, et al. PCR-based denaturing gradient gel electrophoresis in the evaluation of oral microbiota. Oral Microbiol Immunol. 2006;21:333–339. doi: 10.1111/j.1399-302X.2006.00301.x. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Ku CY, Xu J, Saxena D, Caufield PW. Survey of oral microbial diversity using PCR-based denaturing gradient gel electrophoresis. J Dent Res. 2005;84:559–564. doi: 10.1177/154405910508400614. [DOI] [PubMed] [Google Scholar]
- 10.Rupf S, Kneist S, Merte K, et al. Quantitative determination of Streptococcus mutans by using competitive polymerase chain reaction. Eur J Oral Sci. 1999;107:75–81. doi: 10.1046/j.0909-8836.1999.eos107201.x. [DOI] [PubMed] [Google Scholar]
- 11.Sheffield VC, Cox DR, Lerman LS, et al. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci U S A. 1989;86:232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ward JH. Hierarchical Grouping to optimize an objective function. J Am Stat Assoc. 1963;58:236–244. [Google Scholar]
- 13.Cole JR, Chai B, Farris RJ, et al. The ribosomal database project (RDP-II): Introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- 17.Chao A, Lee S-M. Estimating the number of classes via sample coverage. J Am Stat Assoc. 1992;87:210–217. [Google Scholar]
- 18.Magurran A. Ecological Diversity and Its Measurement. Princeton, NJ: Princeton University Press; 1988. [Google Scholar]
- 19.Schloss PD, Larget BR, Handelsman J. Integration of microbial ecology and statistics: A test to compare gene libraries. Appl Environ Microbiol. 2004;70:5485–5492. doi: 10.1128/AEM.70.9.5485-5492.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeSantis TZ, Jr, Hugenholtz P, Keller K, et al. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chao A. Nonparametric estimation of the number of classes in a population. Scand J Statist. 1984;11:265–270. [Google Scholar]
- 22.Handleman SL, Hess C. Effect of dental prophylaxis on tooth-surface flora. J Dent Res. 1970;49:340–345. doi: 10.1177/00220345700490022401. [DOI] [PubMed] [Google Scholar]
- 23.al-Yahfoufi Z, Mombelli A, Wicki A, Lang NP. The effect of plaque control in subjects with shallow pockets and high prevalence of periodontal pathogens. J Clin Periodontol. 1995;22:78–84. doi: 10.1111/j.1600-051x.1995.tb01774.x. [DOI] [PubMed] [Google Scholar]
- 24.Panagakos FS, Volpe AR, Petrone ME, et al. Advanced oral antibacterial/anti-inflammatory technology: A comprehensive review of the clinical benefits of a triclosan/copolymer/fluoride dentifrice. J Clin Dent. 2005;16(suppl):S1–S19. [PubMed] [Google Scholar]
- 25.Davies RM. The clinical efficacy of triclosan/copolymer and other common therapeutic approaches to periodontal health. Clin Microbiol Infect. 2007;13(Suppl 4):25–29. doi: 10.1111/j.1469-0691.2007.01801.x. [DOI] [PubMed] [Google Scholar]
- 26.Blinkhorn A, Bartold PM, Cullinan MP, et al. Is there a role for tri-closan/copolymer toothpaste in the management of periodontal disease? Br Dent J. 2009;207:117–125. doi: 10.1038/sj.bdj.2009.669. [DOI] [PubMed] [Google Scholar]
- 27.Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect. 2008;14:908–934. doi: 10.1111/j.1469-0691.2008.02070.x. [DOI] [PubMed] [Google Scholar]
- 28.Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies Interactions within Oral Microbial Communities. Microbiol Mol Biol Rev. 2007;71:653–670. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]