Abstract
We have cloned the his7+ gene of the fission yeast Schizosaccharomyces pombe by complementation of the recessive mutant allele his7-366. The his7+ gene is able to complement a mutation of the Escherichia coli hisI gene, suggesting that his7+ encodes a phosphoribosyl-AMP cyclohydrase. Subcloning experiments localize the gene to a 1.9-kb XbaI-BglII fragment. We describe the construction of plasmids to facilitate the use of his7+ as a selectable marker in S. pombe studies. Plasmid pEA2 carries his7+ cloned into the pUC18 polylinker. From either pEA2 or the original his7+ clone, pMN1, fragments carrying his7+ can be isolated using a variety of restriction enzymes for the construction of gene disruptions. Plasmid pEA500 is a cloning vector that carries his7+ and ars1, yet retains the ability to use the blue/white color screen to identify recombinants.
Keywords: Fission yeast, Selectable marker, Gene disruptions, Cloning vector
Introduction
Selectable markers are a required part of both autonomously-replicating and integrating plasmids, allowing the identification and exclusive maintenance of cells that have taken up the plasmid. In yeast, selectable markers are also used to mark gene disruptions that are constructed in vitro and then transferred to the chromosome by homologous recombination (Rothstein 1983). Such integrations can also be used for the introduction of a specifically-altered copy of a gene to the genome; however, since gene disruptions are more commonly performed, we refer only to that aspect of these integrations here. Initially, the selectable marker is used to identify cells that have integrated the disrupted copy of the gene of interest. Subsequently, the selectable marker identifies progeny of a cross that carry the disrupted gene.
In prokaryotic studies, selectable markers have typically encoded proteins that confer drug resistance upon the recipient cell. In the yeasts Schizosaccharomyces pombe and Saccharomyces cerevisiae, selectable markers have more commonly encoded components of a biosynthetic pathway (Botstein et al. 1979). Host strains carry a mutation in the chromosomal copy of the gene that encodes the same step of the pathway as does the selectable marker, thus providing an auxotrophy that is complemented by introduction of the DNA carrying the selectable marker. Therefore, the selective medium lacks the required amino acid or base (such as uracil or adenine), restricting growth to cells that have received and express the selectable marker.
Most S. pombe molecular genetic studies depend upon only three selectable markers. The S. cerevisiae LEU2 gene is able to complement mutations in the S. pombe leu1 gene (Beach and Nurse 1981), while either the S. cerevisiae URA3 gene or the S. pombe ura4 gene complement mutations in the S. pombe ura4 gene (Losson and Lacroute 1983; Bach 1987). However, a single copy of the URA3 gene generally fails to complement a ura4 mutation, and cannot be used for homologous integrations of disrupted genes. While a single copy of the LEU2 gene can complement a leu1 mutation, such strains are only weakly Leu+ (Russell and Nurse 1987). Therefore, Leu+ transformants isolated from integration attempts often contain multiple copies of LEU2.
Strains that carry plasmids or marked disruptions no longer display the original auxotrophy and thus cannot be transformed with plasmids or disruptions carrying the same selectable marker. Therefore, the availability of selectable markers in only two biosynthetic pathways limits our ability to manipulate S. pombe. We describe here the cloning of the S. pombe his7+ gene and the construction of plasmids to facilitate its use as a selectable marker for molecular genetic studies. The his7+ gene should become a useful tool in S. pombe studies as a selectable marker for both autonomously-replicating plasmids and homologous integrations.
Materials and methods
Strains and media
S. pombe strains used in this study are listed in Table 1 with full genotypes according to the nomenclature rules proposed by Kohli (1987). The ura4∷fbp1-lacZ allele is a disruption of the ura4 gene by an fbp1-lacZ translational fusion, while the fbp1∷ura4 allele is a translational fusion integrated at the fbp1 locus (Hoffman and Winston 1990). E. coli strains used include HB101 (for rescue of plasmids from S. pombe; Boyer and Roulland-Dussoix 1969), MC1061 (for small-scale plasmid preparations; Casadaban and Cohen 1980), and I903 (for plasmid complementation studies; a hisI903 mutant derivative of UTH653; Goldschmidt et al. 1970). Cells were grown at 30°C on rich medium YEA and YEL (Gutz et al. 1974) supplemented with 0.2% casamino acids, or on PM minimal medium (Beach et al. 1985) supplemented with 75 mg/l of amino acids, adenine or uracil, as needed. The inclusion of casamino acids in rich medium is not standard for S. pombe studies; however, we have found that this supplement greatly improves the growth of strains that possess multiple auxotrophies. Crosses were performed on YPD (Sherman et al. 1986) at room temperature.
Table 1.
S. pombe strain list
| Name | Genotype |
|---|---|
| CHP201 | h− leu1-32 his7-366 ade6-M216 fbp1∷ura4 ura4∷fbp1-lacZ git10-201 |
| CHP377 | h+ leu1-32 [pEA11] |
| CHP386 | h− leu1-32 his7-366 ade6-M216 fbp1∷ura4 ura4∷fbp1-lacZ git2-2∷his7+ |
| FWP5 | h+ leu1-32 |
| FWP60 | h+ leu1-32 his7-366 |
| FWP94 | h− leu1-32 his7-366 ura4∷fbp1-lacZ |
| FWP112 | h− leu1-32 his7-366 ade6-M216 fbp1∷ura4 ura4∷fbp1-lacZ |
Recombinant DNA methodology
Restriction endonucleases and T4 DNA polymerase were purchased from New England Biolabs and used according to manufacturer’s instructions. Bacterial transformations were done according to the electroporation protocol of Ausubel et al. (1987). Small-scale plasmid preparations were performed using Promega Magic Minipreps according to manufacturer’s instructions. Yeast transformations were done according to the modified LiOAc protocol of Elble (1992). Small-scale yeast DNA preparations for Southern hybridization analysis (Southern 1975) were done as previously described (Hoffman and Winston 1987), with the following modification. DNA from 10-ml cultures was resuspended in a final volume of 100 µl TE buffer, with 5 µ1 of this DNA used per digest.
Cloning of the his7+ gene
The his7+ gene was cloned by complementation of the his7-366 mutant allele. Strain CHP201 (see Table 1 for a complete genotype) was transformed to Leu+ with an S. pombe genomic library [partial HindIII-digested DNA cloned into the HindIII site of plasmid pWH5 (Wright et al. 1986); P. Young and D. Beach, unpublished]. Transformants were replica-plated to PM-leu-his medium to screen for His+ transformants. Plasmid pMN1 was rescued from one such transformant into E. coli (Hoffman and Winston 1987). Restriction mapping of pMN1 indicated that a 2-kb deletion had occurred in the region of the LEU2 gene in the vector. Plasmids recovered from other His+ S. pombe transformants were even more severely rearranged and were not studied further.
Integration and linkage analysis of his7+
Plasmid pEA11 was constructed to test whether the cloned region could direct plasmid integration to the his7 locus. Plasmid pMN1 was digested with BglII and the 3.15-kb fragment was inserted by ligation into plasmid pWH5 at the BclI site creating plasmid pEA11. We targeted the integration of plasmid pEA11 (carrying LEU2 and his7+) by digesting with SacI, which cuts in the putative his7 region, and transforming strain FWP5 to Leu+. Strain CHP377 is a transformant in which the plasmid integrated by homologous recombination as judged by Southern hybridization (data not shown). Strain CHP377 was crossed with strain FWP94 and the progeny were analyzed by tetrad dissection. In 24 tetrads with four viable progeny, the His and Leu phenotypes cosegrated, suggesting that the plasmid (carrying the LEU2 gene) had integrated at the his7 locus.
Construction of plasmids pEA2 and pEA500
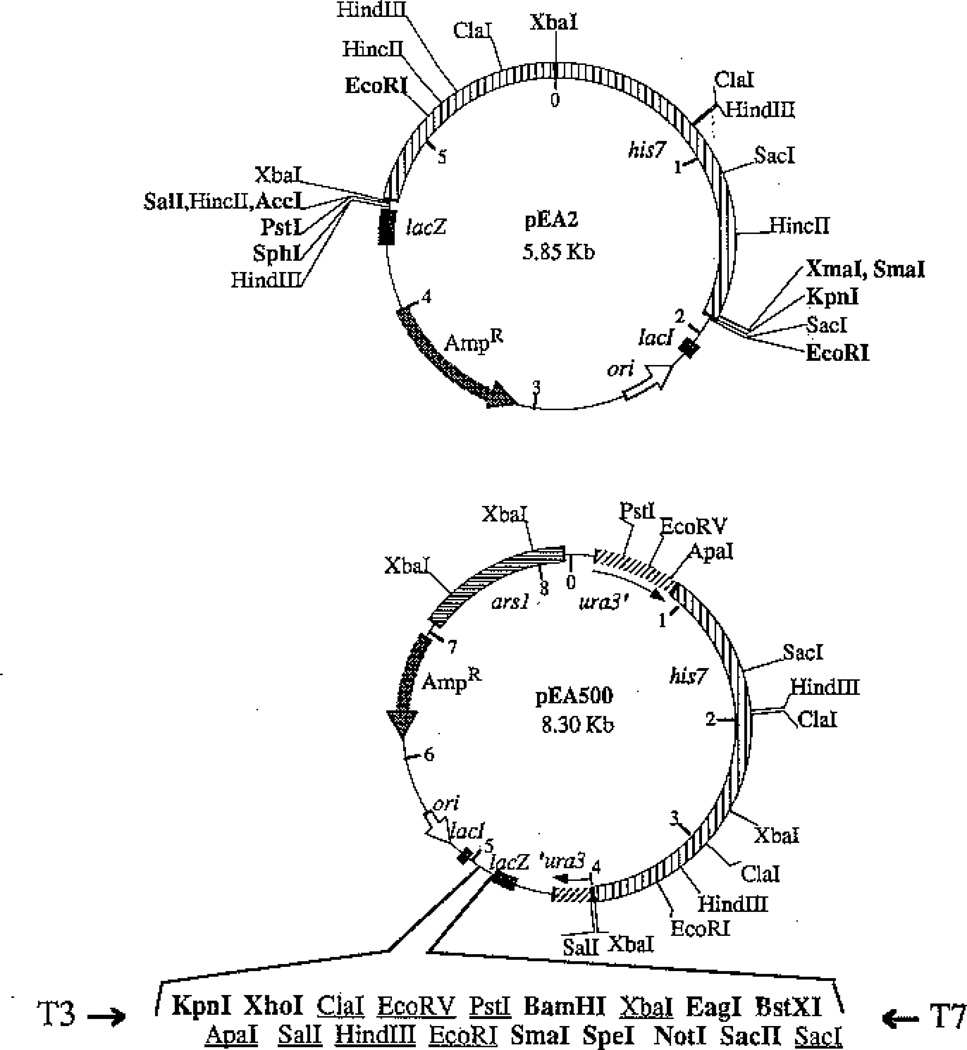
Plasmid pEA2 was constructed by inserting the 3.15-kb BglII fragment from plasmid pMN1 into the BamHI site of pUC18 (Norrander et al. 1983). Plasmid pEA500 was constructed by inserting the PstI-Smal fragment of pEA2 (see Figs. 1, 2) into plasmid pSP2 (Cottarel et al. 1993) digested with NsiI and StuI (both recognize unique sites within the URA3 gene). Both pEA2 and pEA500 were analyzed by restriction digestion analysis with all enzymes that recognize sites within the two polylinkers. All relevant sites are shown in Fig. 1.
Fig. 1.
Plasmid maps for pEA2 and pEA500. Plasmid pEA2 was constructed by insertion of the 3.15-kb BglII fragment (indicated by vertical stripes) from pMN1 into the BamHI site of pUC18 (Norrander et al. 1983). See Fig. 2 for subcloning information with respect to sites useful for gene disruptions (shown in bold). Plasmid pEA500 was constructed by inserting the PstI-SmaI fragment from pEA2 into plasmid pSP2 (Cottarel et al. 1993) which was digested within the URA3 gene with NsiI and StuI. Unique sites in the polylinker are in bold. Sites in the polylinker that are also present elsewhere in the plasmid are underlined, and the additional sites are displayed
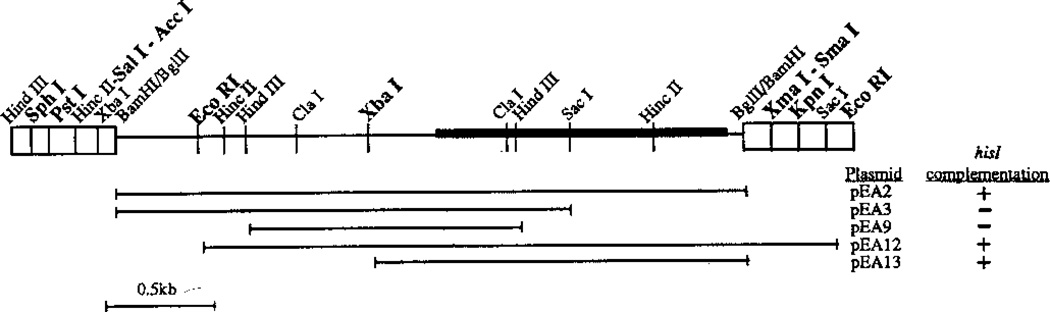
Fig. 2.
Complementation analysis of his7 subclones. The polylinker and insert from pEA2 (Fig. 1) are shown. The sites in the polylinker are indicated in the boxed regions. The plasmids carrying various subclones, described in Materials and methods, were tested for their ability to transform E. coli strain I903 (hisI903) to His+. Sites that cut outside of the his7+ gene, and are therefore useful for constructing gene disruptions, are in bold (SphI, PstI, SalI, AccI, EcoRI and XbaI to the left of his7+; XmaI, SmaI, KpnI and EcoRI to the right of his7+). The sites generated by the ligation that created pEA2 (BglII/BamHI and BamHI/BglII) can be cut with BstYI; however, other subcloning experiments indicate that there is a BstYI site within his7+ (data not shown)
Subcloning analysis of his7+ by complementation in E. coli
Portions of the 3.15-kb BglII fragment from pMN1 were subcloned and tested for their ability to complement the hisI903 mutation in E. coli strain I903. Plasmid pEA3 was constructed by digestion of pEA2 with SacI and recircularization of the vector with T4 DNA ligase, thus excising an 0.8-kb SacI fragment (Fig. 2). Plasmid pEA9 was constructed by ligating the 1.4-kb HindIII fragment from pEA2 into the HindIII site of pWH5. Plasmid pEA12 was constructed by ligating the 2.6-kb EcoRI fragment from pEA2 into the EcoRI site of pUC19 (Norrander et al. 1983). Plasmid pEA13 was constructed by digestion of pEA2 with XbaI and recircularization of the vector with T4 DNA ligase, thus excising a 1.25-kb XbaI fragment. Complementation of the hisI903 mutation was performed by transforming strain I903 to ampicillin resistance on LB Amp (Ausubel et al. 1987) and then testing for His+ by replica plating to M9-his (Maniatis et al. 1982).
Disruption of git2 gene with his7+
The disruption allele of git2, git2-2∷his7+, was constructed on plasmid pEA19 such that the his7+ gene replaced most of the coding region of git2 including that of the entire catalytic domain. Before constructing pEA19, we had to first remove an XbaI site in the pWH5 vector portion of plasmid pCHY26 (Hoffman and Winston 1991). Plasmid pCHY26 was digested with ClaI and then recircularized by ligation, creating pCHY26ΔClaI. The his7+ gene, carried on a 1.9-kb XbaI-BglII fragment from plasmid pMN1, was then inserted by ligation into pCHY26ΔClaI, also digested with XbaI and BglII creating plasmid pEA19. The git2-2∷his7+ DNA was isolated by digestion with XhoI and PstI and used to transform FWP112 to His+. Southern hybridization analysis (Southern 1975) was performed on HindIII-digested DNA from purified transformants, using pCHY26 as the probe. Strain CHP386 was determined to carry the appropriate single-copy disruption.
Results
Cloning of the his7+ gene
The his7+ gene was cloned by screening transformants of strain CHP201 (his7-366; see Table 1 for full genotype) for the ability to grow in the absence of histidine. Of approximately 6000 Leu+ transformants, transformed with an S. pombe genomic library (see Materials and methods for details), four were His+. Plasmid pMN1 (Fig. 1) was rescued from one of these transformants into E. coli (Hoffman and Winston 1987). Restriction analysis of the plasmid revealed an internal deletion in the vector in the region of the LEU2 gene (data not shown). Indeed, plasmid pMN1 was unable to transform strain FWP60 (leu1-32 his7-366) to Leu+, although His+ transformants could be obtained. A 3.15-kb BglII fragment was sub-cloned from pMN1 into the BamHI site of YEpl3 (Broach et al. 1979), creating plasmid pMN3. When strain FWP60 was transformed to Leu+ with pMN3, the transformants concurrently became His+.
While high-copy suppression by a gene other than his7+ seems unlikely, since this is a biosynthetic pathway, complementation of the his7-366 mutation by plasmids pMN1 and pMN3 does not prove that these plasmids carry the his7+ gene. To determine this, we tested whether the cloned region could direct the integration of a plasmid by homologous recombination to the his7 locus (see Materials and methods). Plasmid pEA11 was constructed by inserting the 3.15-kb BglII fragment from pMN1 into the BclI site of pWH5. Plasmid pEA11 was linearized by digestion with SacI, which cuts uniquely within the BgtII fragment, and used to transform FWP5 (h+ leui-32) to Leu+. Southern hybridization analysis identified a Leu+ transformant in which the plasmid integrated by homologous recombination (data not shown). This strain, CHP377, was then crossed with FWP94 (h− leu1-32 his7-366 ura4∷fbp1-lacZ). In 24 tetrads, the His and Leu phenotypes cosegregated (all progeny were either His− Leu− or His+ Leu+), suggesting that pEA11 had integrated at the his7 locus.
The S. pombe his7+ gene complements a mutation in the E. coli hisI gene
We tested whether the his7+ gene could complement any mutations in the E. coli his operon in order to determine its biological function and to provide a useful tool in the construction of plasmids carrying the his7+ gene. Complementation of E. coli mutations by yeast genes that function in the same biosynthetic pathway has previously been observed. The S. cerevisiae LEU2 gene and the S. pombe leu1 gene complement mutations in the E. coli leuB gene (Ratzkin and Carbon 1977; Kikuchi et al. 1988). The S. cerevisiae URA3 gene and the S. pombe ura4 gene complement mutations in the E. coli pyrF gene (Bach et al. 1979; Bach 1987). The practical use of such complementation is that E. coli strains carrying these mutations can be used as host strains for transformation by ligations to screen for the insertion of the yeast selectable marker into the coding region of another yeast gene carried on a plasmid, thus disrupting the gene of interest. This region of the plasmid can then be used to replace the normal copy of the yeast gene in the yeast chromosome by homologous recombination (Rothstein 1983).
Plasmids carrying the his7+ gene transform an E. coli hisI mutant to histidine prototrophy (data not shown). Such complementation did not occur with E. coli strains carrying mutations in any of the genes hisA, hisB, hisC, hisD, hisF, hisG, or hisH. The hisI enzymatic activity, encoded by the bifunctional enzyme hisIE (Chiariotti et al. 1986), is a phosphoribosyl-AMP cyclohydrase. Therefore, the S. pombe his7+ gene appears to also encode a phosphoribosyl-AMP cyclohydrase.
Construction of a plasmid for generating marked gene disruptions
To facilitate the use of the his7+ gene as a selectable marker for gene disruptions or other integrations, we have inserted the 3.15-kb BglII fragment from plasmid pMN1 into the BamHI site in the pUC18 polylinker, creating plasmid pEA2 (Fig. 1).
From plasmids pEA2 and pMN1, we have carried out a combination of restriction mapping and subcloning analyses of the his7+ gene. Together, these studies identify the restriction sites useful for the production of restriction fragments carrying a functional copy of his7+ (Fig. 2). Function was scored on the basis of complementation of the hisI903 mutation in E. coli strain I903. [This strain is the hisI903 mutant derivative of strain UTH653 isolated by Goldschmidt et al. (1970), but was not given a strain name in that study]. The smallest functional subclone, a 1.9-kb XbaI-BglII fragment from pMN1, has also been shown to complement the his7-366 mutation in S. pombe (see below). The his7+ gene can be isolated by digestion with a variety of enzymes to be used in the construction of gene disruptions. From plasmid pEA2, the restriction enzymes SphI, PstI, SalI, AccI (not all AccI-generated ends are compatible with each other for ligation), EcoRI, and XbaI all cut to the left of the his7+ gene as depicted in Fig. 2. Restriction enzymes XmaI, SmaI, KpnI, and EcoRI (note the presence of an EcoRI site within the 3.15-kb insert) cut to the right of his7+. Plasmid pMN1 offers the additional ability to retrieve his7+ on a 3.15-kb BglII fragment, a 2.65-kb BglII-EcoRI fragment, or a 1.9-kb BglII-XbaI fragment.
Prior to performing a single-step gene disruption in yeast (Rothstein 1983) one must insert a selectable marker by ligation into the coding region of the target gene present on a plasmid. Since the E. coli hisI mutant strain I903 becomes His+ when transformed by plasmids carrying the his7+ gene, this strain can be used to identify plasmids from a ligation in which the gene disruption has occurred. Unfortunately, strain I903 does not become highly competent for transformation. Our highest transformation frequency for this strain has been 1.4 × 103 transformants/µg DNA using an electroporation protocol (Ausubel et al. 1987). This low transformation frequency makes it impractical to directly transform I903 and screen for His+ colonies. However, we have been successful at screening a ligation for his7+-carrying plasmids by transforming strain MC1061 (Casadaban and Cohen 1980) with the ligation, preparing plasmid DNA from the pooled transformants, and then transforming I903 with this amplified DNA. Ampicillin-resistant transformants of I903 were replica plated to M9-his medium to identify His+ transformants. This screen allowed the detection of his7+-carrying plasmids that represented approximately 1% of the total transformants. Such a screen is especially useful in ligations where the vector is likely to recircularize.
his7+ can function as a selectable marker for gene disruptions
In order to determine the usefulness of this system, we tested whether the 1.9-kb XbaI-BglII fragment from pMN1 could be used to mark a gene disruption in S. pombe. The his7+ fragment was used to replace most of the git2 gene (Hoffman and Winston 1991), encoding adenylate cyclase (identical to cyr1; Yamawaki-Kataoka et al. 1989; Young et al. 1989). A fragment from the resulting plasmid was used to transform strain FWP112 to His+. Of 38 transformants, six have a single copy of the git2 gene disrupted by his7+ as determined by Southern hybridization analysis. Most of the other His+ strains appear to be gene conversions of the his7-366 mutation to his7+, as opposed to nonhomologous integration events. The appearance of the His+ colonies was dependent upon transformation with the his7+ DNA and thus does not represent a reversion event. In a previous effort to mark a disruption of git2 with LEU2 (Hoffman and Winston 1991), only 1 of 88 Leu+ transformants carried the single-copy homologous replacement of git2 (Hoffman, unpublished). Therefore, his7+ appears to be distinctly better suited than LEU2 for use in gene disruptions in S. pombe.
Construction of a cloning vector with his7+
We have constructed plasmid pEA500 to create a cloning vector that employs the his7+ selectable marker, yet retains its ability to screen for recombinant plasmids using the blue/white color screen. Plasmid pSP2 (Cottarel et al. 1993) carries the lacZα region and cloning polylinker from pBluescript (Stratagene) along with the S. pombe ars1 region. Plasmids carrying the ars1 region transform S. pombe more efficiently and are less likely to undergo rearrangements in S. pombe than plasmids lacking this region (Maundrell et al. 1985). Plasmid pEA500 was constructed by replacing a region of the URA3 gene (from NsiI to StuI) on plasmid pSP2 (Cottarel et al. 1993) with the PstI-SmaI fragment from pEA2. Restriction mapping of plasmid pEA500 reveals that the KpnI, XhoI, SmaI, BamHI, SpeI, NotI, EagI, and SacII sites in the polylinker are unique, and thus suitable for cloning (Fig. 1). The BstXI site is also unique; however, the overhang generated by this enzyme varies among BstXI sites.
Discussion
We have described the cloning of the S. pombe his7+ gene and the construction of plasmids that will facilitate its use as a selectable marker for both plasmids and integrations. We believe that the major use of the his7+ gene will be for marking integrations. As mentioned above, a single copy of the LEU2 gene does not fully complement a leu1 mutation. Therefore, this marker is not convenient for constructing integrations. Furthermore, the ura4 gene is commonly employed as a target site for integrations since the loss of its activity results in resistance to the pyrimidine analog 5-fluoro-orotic acid (5FOA; Boeke et al. 1984; Grimm et al. 1988). If ura4 is used to mark a gene disruption, this strain cannot be used later for an integration event at ura4. The ura4 gene can also be used as a reporter gene (Hoffman and Winston 1990), in which case ura4 cannot also be used to mark a gene disruption. For these reasons, his7+ is likely to become the marker of choice for integrations.
One molecular technique that is commonly used in S. cerevisiae, but rarely used in S. pombe, is the plasmid shuffle (Boeke et al. 1987). This technique can be employed to isolate conditional lethal mutations in a gene in cases where the gene disruption is a lethal event. In it, a strain is constructed carrying a chromosomal deletion of a gene whose function is essential for viability, as well as two distinct plasmids carrying the gene of interest. The URA3-based (or ura4 for S, pombe) plasmid carries a wild-type copy of the gene while the other plasmid carries a copy of the gene that has been subjected to some form of mutagenesis. By plating the cells on medium containing 5FOA, one can select for cells that have lost the wild-type copy of the gene, as 5FOA will kill the cells carrying the URA3-based plasmid. One can then determine if a phenotype is associated with the expression of the altered copy. In general, the plasmid shuffle requires three selectable markers, one for the chromosomal deletion and two for the two plasmids. (This technique can be employed with only two selectable markers by constructing an unmarked chromosomal deletion.) The his7+ gene should be useful for marking chromosomal deletions as part of the plasmid shuffle technique. Alternatively, plasmid pEA500 can be used to carry the mutagenized copy of the gene in studies where LEU2-marked disruptions already exist.
Plasmid pEA500 and other vectors that employ the his7+ gene as a selectable marker will be useful in situations requiring an additional or alternate selectable marker to LEU2, URA3, or ura4. Such situations arise in studies in which strains carry integrations marked with LEU2 or ura4, when ura4 is used as a reporter gene, or when multiple plasmids are required.
Acknowledgements
We thank Philip E. Hartman for his gift of E. coli his mutants and Guillaume Cottarel for his gift of plasmid pSP2. We thank Linda Lepnis Hoffman and Maureen McLeod for critical reading of the manuscript. This work was supported by Boston College Research Expense Grants and National Institutes of Health grant GM46226 to C.S.H..
References
- Ausubel F, Brent R, Kingston R, Moore D, Smith J, Scidman J, Struhl K, editors. Current protocols in molecular biology. New York: Wiley; 1987. [Google Scholar]
- Bach ML. Curr Genet. 1987;12:527–534. doi: 10.1007/BF00419562. [DOI] [PubMed] [Google Scholar]
- Bach ML, Lacroute F, Botstein D. Proc Natl Acad Sci USA. 1979;76:386–390. doi: 10.1073/pnas.76.1.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D, Nurse P. Nature. 1981;290:140–142. doi: 10.1038/290140a0. [DOI] [PubMed] [Google Scholar]
- Beach D, Rogers L, Gould J. Curr Genet. 1985;10:297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Lacroute F, Fink GR. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Botstein D, Falco SC, Stewart S, Brennan M, Scherer S, Stinch-comb DT, Struhl K, Davis R. Gene. 1979;8:17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Boyer HW, Roulland-Dussiox D. J Mol Biol. 1969;41:458–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Broach JR, Strathern JN, Hicks JB. Gene. 1979;8:121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Casadaban MJ, Cohen SN. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chiariotti L, Alifano P, Carlomagno MS, Bruni CB. Mol Gen Genet. 1986;203:382–388. doi: 10.1007/BF00422061. [DOI] [PubMed] [Google Scholar]
- Cottarel G, Beach D, Deuschle U. Curr Genet. 1993;23:547–578. doi: 10.1007/BF00312650. [DOI] [PubMed] [Google Scholar]
- Elble R. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Goldschmidt EP, Cater MS, Matney TS, Butler MA, Greene A. Genetics. 1970;66:219–229. doi: 10.1093/genetics/66.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Kohli J, Murray J, Maundrell K. Mol Gen Genet. 1988;215:81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- Gutz H, Heslot H, Leupold U, Loprieno N. In: Handbook of genetics. King RC, editor. New York: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- Hoffman CS, Winston F. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. Genetics. 1990;124:807–816. doi: 10.1093/genetics/124.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. Genes Dev. 1991;5:561–571. doi: 10.1101/gad.5.4.561. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Kitazawa Y, Shimatake H, Yamamoto Y. Curr Genet. 1988;14:375–379. doi: 10.1007/BF00419995. [DOI] [PubMed] [Google Scholar]
- Kohli J. Curr Genet. 1987;11:575–589. doi: 10.1007/BF00393919. [DOI] [PubMed] [Google Scholar]
- Losson R, Lacroute F. Cell. 1983;32:371–377. doi: 10.1016/0092-8674(83)90456-7. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Cold Spring Harbor Laboratory. New York: Cold Spring Harbor; 1982. Molecular cloning: a laboratory manual. [Google Scholar]
- Maundrell K, Wright APH, Piper M, Shall S. Nucleic Acids Res. 1985;13:3711–3722. doi: 10.1093/nar/13.10.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J, Kempe T, Messing J. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ratzkin B, Carbon J. Proc Natl Acad Sci USA. 1977;74:487–491. doi: 10.1073/pnas.74.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein RJ. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Cold Spring Harbor Laboratory. New York: Cold Spring Harbor; 1986. Methods in yeast genetics. [Google Scholar]
- Southern EM. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wright A, Maundrell K, Heyer HD, Beach D, Nurse P. Plasmid. 1986;15:156–158. doi: 10.1016/0147-619x(86)90051-x. [DOI] [PubMed] [Google Scholar]
- Yamawaki-Kataoka Y, Tamaoki T, Choe HR, Tanaka H, Kataoka T. Proc Natl Acad Sci USA. 1989;86:5693–5697. doi: 10.1073/pnas.86.15.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D, Riggs M, Fields J, Vojtek A, Broek D, Wigler M. Proc Natl Acad Sci USA. 1989;86:7989–7993. doi: 10.1073/pnas.86.20.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]