Abstract
Objective
For decades it has been known that the HLA–DRB1 shared epitope (SE) alleles are associated with an increased risk of development and progression of rheumatoid arthritis (RA). Recently, the following variations in the peptide-binding grooves of HLA molecules that predispose to RA development have been identified: Val and Leu at HLA–DRB1 position 11, Asp at HLA–B position 9, and Phe at HLA–DPB1 position 9. This study was undertaken to investigate whether these variants are also associated with radiographic progression in RA, independent of SE and anti–citrullinated protein antibody (ACPA) status.
Methods
A total of 4,911 radiograph sets from 1,878 RA patients included in the Leiden Early Arthritis Clinic (The Netherlands), Umeå (Sweden), Hospital Clinico San Carlos–Rheumatoid Arthritis (Spain), and National Data Bank for Rheumatic Diseases (US) cohorts were studied. HLA was imputed using single-nucleotide polymorphism data from an Immunochip, and the amino acids listed above were tested in relation to radiographic progression per cohort using an additive model. Results from the 4 cohorts were combined in inverse-variance weighted meta-analyses using a fixed-effects model. Analyses were conditioned on SE and ACPA status.
Results
Val and Leu at HLA–DRB1 position 11 were associated with more radiographic progression (meta-analysis P = 5.11 × 10−7); this effect was independent of SE status (meta-analysis P = 0.022) but not independent of ACPA status. Phe at HLA–DPB1 position 9 was associated with more severe radiographic progression (meta-analysis P = 0.024), though not independent of SE status. Asp at HLA–B position 9 was not associated with radiographic progression.
Conclusion
Val and Leu at HLA–DRB1 position 11 conferred a risk of a higher rate of radiographic progression independent of SE status but not independent of ACPA status. These findings support the relevance of these amino acids at position 11.
The development and course of rheumatoid arthritis (RA) are in part determined by genetic factors. Although the genetic risk factors underlying RA development and progression of joint destruction are largely nonoverlapping (1), the genetic variants encoding the so-called HLA–DRB1 shared epitope (SE) alleles are associated with both the risk of RA development and the severity of the disease course (2–4).
The association of HLA class II with RA has been known for decades. The association between HLA–DR and RA was first reported in 1976 (5). Subsequent identification of risk HLA–DRB1 alleles that all shared a similar amino acid sequence at positions 70–74 in the peptide-binding groove of the HLA–DRB1 molecule led to the formulation of the SE hypothesis (6). This hypothesis postulates that the SE motif itself may be directly involved in the pathogenesis of RA by allowing the presentation of an arthritogenic peptide to T cells. Thus far, these peptides have not been identified. With the identification of anti–citrullinated protein antibodies (ACPAs) in the late 1990s, it became clear that SE alleles mainly predispose to ACPA-positive RA (3,7). The relevance of HLA–DRB1 for ACPA-negative RA was set by the identification of HLA–DRB1*03 (part of the conserved ancestral A1-B8-DRB1*03 haplotype) as a risk factor for ACPA-negative RA (8,9).
Recently, a further refinement of the association between HLA and RA was proposed by Raychaudhuri et al (10). Using a case–control design with 5,018 ACPA-positive RA patients and 14,974 controls, the class I and class II HLA regions were explored. The strongest association was reported for HLA–DRB1 positions 11 and 13 (which are in high linkage disequilibrium). The amino acids Val and Leu at position 11 conferred a high risk, and Ser was protective. These associations were independent of the SE status. Furthermore, performing further conditional analyses, independent associations were observed for variants in HLA–B position 9 (Asp predisposed to RA) and HLA–DPB1 position 9 (Phe predisposed to RA). In a subsequent study using a similar approach, the authors also investigated 2,406 ACPA-negative RA patients and 13,930 controls (11) and observed that Leu and Ser at HLA–DRB1 position 11 and Asp in HLA–B position 9 were associated with an increased risk of ACPA-negative RA.
These risk positions are located in the peptide-binding grooves of the HLA molecules. Studies of MHC class I and class II in mice have shown that a difference of only one or a few amino acids at such a crucial place may result in the presentation of totally different peptides (12,13). Therefore, the finding that additional amino acids located in the antigen-presenting binding grooves associate with RA development is relevant and hypothetically may fuel further studies to detect arthritogenic peptides involved in RA susceptibility (10).
Because the HLA–DRB1 SE alleles are among the strongest genetic factors for a progressive disease course, the recent findings of Raychaudhuri et al prompted us to determine the relevance of the newly identified risk factors for the severity of the course of RA, measured using radiographic joint damage progression. More specifically, first, we aimed to investigate whether Val, Leu, and Ser at HLA–DRB1 position 11, Asp at HLA–B position 9, and Phe at HLA–DPB1 position 9 are associated with radiographic progression in the total RA population and, if so, whether these effects are independent of the well-known SE effect (HLA–DRB1 positions 70–74). Second, since the SE alleles predispose to ACPAs and the SE alleles are not associated with radiographic progression independent of ACPAs (7,14), we aimed to analyze whether the newly identified associations are independent of ACPAs. Third, we aimed to evaluate whether Leu and Ser at HLA–DRB1 position 11 and Asp at HLA–B position 9, identified as risk factors for ACPA-negative RA, are associated with joint damage progression in ACPA-negative RA. To this end, a total of 4,911 sets of radiographs of 1,878 patients with RA in 4 different cohorts were studied.
PATIENTS AND METHODS
Patients
Patients were included from the following 4 cohorts: the Leiden Early Arthritis Clinic (EAC), the Umeå cohort, the Hospital Clinico San Carlos–Rheumatoid Arthritis Cohort (HCSC-RAC), and the National Data Bank for Rheumatic Diseases (NDB). In all cohorts, RA was defined according to the American College of Rheumatology 1987 criteria (15). The characteristics of the patients in each cohort are presented in Supplementary Table 1, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39018/abstract. Informed consent was obtained from all patients, and approval was obtained from the local ethics committee of each study.
Leiden EAC
This cohort consisted of 594 Dutch patients with early RA who were enrolled between 1993 and 2006 (16). The mean ± SD age at diagnosis was 57.0 ± 15.6 years, 67.0% of the patients were women, and 52.8% were ACPA positive (as determined by anti–cyclic citrullinated peptide 2 [anti–CCP-2] antibody test). Radiographs of the hands and feet were obtained at baseline and during yearly followup visits. In total 3,121 sets of radiographs obtained over 7 years of followup were chronologically scored by 1 experienced reader, who was blinded with regard to any clinical or genetic data, using the Sharp/van der Heijde score (SHS) (within reader intraclass correlation coefficient [ICC] 0.91). Initial treatment strategies changed over time. Patients enrolled in 1993–1995 were initially treated with nonsteroidal antiinflammatory drugs (NSAIDs), patients enrolled in 1996–1998 were initially treated with hydroxychloroquine or sulfasalazine, and patients enrolled in 1999–2006 were initially treated with methotrexate.
Umeå cohort
This Swedish cohort comprised 365 patients with early RA enrolled between 1995 and 2010. The mean ± SD age at diagnosis was 54.3 ± 14.5 years, 69.6% of the patients were women, and 71.8% were ACPA positive (as determined by anti–CCP-2 antibody test). At baseline and after 2 years, a total of 687 radiographs of the hands and feet were obtained and scored using the Larsen score by 2 trained readers as previously described (17). Treatment strategies differed between 1995–2000, 2000–2005, and 2006–2010, resulting in less severe radiographic progression in the subsequent treatment periods.
HCSC-RAC
This Spanish data set involved 380 patients with early RA, diagnosed between 1976 and 2011 (18). The mean ± SD age at diagnosis was 53.8 ± 14.2 years, 76.3% of the patients were women, and 48.6% were ACPA positive (as determined by anti–CCP-2 antibody test). During the first 10 years after disease onset, 564 radiographs of the hands were obtained and scored with known time-order using the SHS (ICC 0.99). Initial treatment strategies differed for different inclusion periods. Prior to 1990, patients were initially treated with NSAIDs, from 1990 to 1999 patients received initial monotherapy with conventional disease-modifying antirheumatic drugs, from 2000 to 2004 patients received initial monotherapy regularly and combination therapy rarely, from 2005 to 2009 patients received initial combination therapy regularly as well as biologic agents, and from 2010 to 2011 patients received tailored treatment.
NDB
This data set included 539 patients from the US and Canada who were diagnosed between 1980 and 1999 (19). The mean ± SD age at diagnosis was 48.6 ± 12.6, 78.5% of the patients were women, and 79.6% were ACPA positive (as determined by anti–CCP-2 antibody test). One set of hand radiographs was available per patient after a mean disease duration of 10.1 years (range 1–20 years) and scored according to the SHS (ICC 0.98). No treatment effects were observed for different periods of diagnosis since all patients developed RA in eras when early, tailored treatment and the use of biologic agents were uncommon.
Genotyping
For all cohorts, we used SNP2HLA to impute classic alleles and corresponding amino acid polymorphisms for HLA class I loci (HLA–A, –B, and –C) and class II loci (HLA–DPA1, –DPB1, –DQA1, –DQB1, and –DRB1) (20). We obtained patient genotypes using an Illumina Immunochip platform (1,21) and used data collected by the Type 1 Diabetes Genetics Consortium as a reference (22). The HLA imputation has been described in detail previously (10,11,20). For the present study, the classic HLA–DRB1 alleles and the amino acids at HLA–DRB1 position 11, HLA–B position 9, and HLA–DPB1 position 9 were extracted.
Analyses of the associations between variants and radiographic progression
First, the SE alleles, defined based on HLA–DRB1 positions 70–74 according to the SE hypothesis (6) (DRB1*0101, *0102, *0104, *0401, *0404, *0405, *0408, *0413, *0416, *1001, and *1402), were tested in relation to radiographic progression. Subsequently, the amino acids that were shown to confer risk to develop RA in the study by Raychaudhuri et al (10) (Val and Leu at HLA–DRB1 position 11, Asp at HLA–B position 9, and Phe at HLA–DPB1 position 9) were studied in relation to radiographic progression. Analyses were done per cohort using an additive model (0, 1, or 2 risk amino acids). Since Val and Leu at position 11 both conferred increased risk, these were combined (i.e., radiographic progression in the presence of 0, 1, or 2 Val and/or Leu was studied). Because the SE alleles and Val or Leu on position 11 are often present together, conditional analyses were performed allowing us to investigate whether Val and Leu were associated with radiographic progression independent of the known association between HLA–DRB1 SE status and radiographic progression. Furthermore, for the SE alleles it is known that the alleles are not associated with radiographic progression independent of ACPAs; in other words, the SE alleles are not associated with radiographic progression as such but this effect is mediated by ACPAs (7,14). To determine whether the same is valid for the newly identified risk factors, analyses were subsequently adjusted for ACPA status.
Previously, the amino acid Ser at HLA–DRB1 position 11 was found to protect against RA development (10). In this study, the association of Ser (0, 1, or 2 amino acids) with radiographic progression was analyzed.
The above-mentioned analyses of Val and Leu and of Ser at HLA–DRB1 position 11 do not take into account the polymorphic nature of HLA–DRB1 in which different amino acids encoded by the same position may have a predisposing or protective effect. It is crucial to ascertain that the predisposing effect identified is not actually due to the absence of a protective effect and vice versa. Stratification is the most thorough method to differentiate between these effects and has been used before to distinguish true effects from the effect of the absence of other alleles (2,23,24). Therefore, patients were stratified into one of the following 6 groups: susceptible/susceptible (S/S), susceptible/neutral (S/N), susceptible/protective (S/P), neutral/neutral (N/N), neutral/protective (N/P), and protective/protective (P/P). Val and Leu at HLA–DRB1 position 11 were the susceptibility amino acids, Ser at this position was the protective amino acid, and Asp, Pro, and Gly were the neutral amino acids. Then the groups of S/S, S/N, and N/N were compared to determine whether the effect of Val and Leu was truly predisposing and not the result of the concomitant absence of Ser. Similarly, the groups of P/P, P/N, and N/N were compared to determine whether the effect of Ser was truly protective and not the result of the concomitant absence of the susceptibility amino acids Val and Leu.
Finally, we evaluated whether the variants that were reported to predispose to ACPA-negative RA (11) (Leu and Ser at HLA–DRB1 position 11 and Asp at HLA–B position 9) were associated with radiographic progression in ACPA-negative RA.
Statistical analysis
In all data sets, radiographic scores were log-transformed (log(score+1)) to approximate a normal distribution. The residuals of the used models were normally distributed around the 0-line in all cohorts, indicating a good fit of the models (see Supplementary Figure 1, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39018/abstract).
In the cohorts in which multiple sets of radiographs were obtained over time (EAC, Umeå, and HCSC-RAC), a multivariate normal regression model for longitudinal data was used with radiographic scores as outcome (1,25,26). This method takes advantage of the within-person correlation between repeated measurements; as such the radiographic progression rates were estimated more precisely in the cohorts with serial radiographs compared to data sets with one radiograph per patient (for a detailed description, see ref. 26). In the cohort in which a radiograph set was obtained at a single time point (NDB), the estimated yearly progression rate was calculated (total SHS/disease duration in years at time of radiograph) to make the estimates of the progression rates comparable to those in the other data sets. Linear regression analysis was used with estimated yearly progression as outcome variable. In both models used, the effect sizes obtained were back-transformed and indicated the fold rate of joint destruction per year per risk amino acid compared to the reference.
In all data sets, adjustments were made for age and sex. In the cohorts that included patients in periods with different treatment strategies (EAC, Umeå, and HCSC-RAC), analyses were also adjusted for the inclusion period as proxies for differences in treatment strategies.
The individual data sets studied were estimated to be insufficiently powered to find statistically significant associations when performing conditional analyses. Therefore, the effect sizes and standard errors of the results from the individual cohorts were combined in inverse-weighted variance meta-analyses with fixed effects to test the overall association. This was allowed because the effect sizes obtained for the individual data sets, though different methods were used to score joint destruction (SHS and Larsen), all represented the relative increase (without units) of progression in joint destruction per year. The meta-analysis weights the results with a low standard error stronger than the results with a high standard error, preventing an overrepresentation of less precise data in the outcome. Subsequently, data sets with smaller 95% confidence intervals had a larger weight in the meta-analysis. P values less than 0.05 were considered significant. Statistical analysis was performed using SPSS version 20 and Stata 12.0.
RESULTS
Frequencies of variants
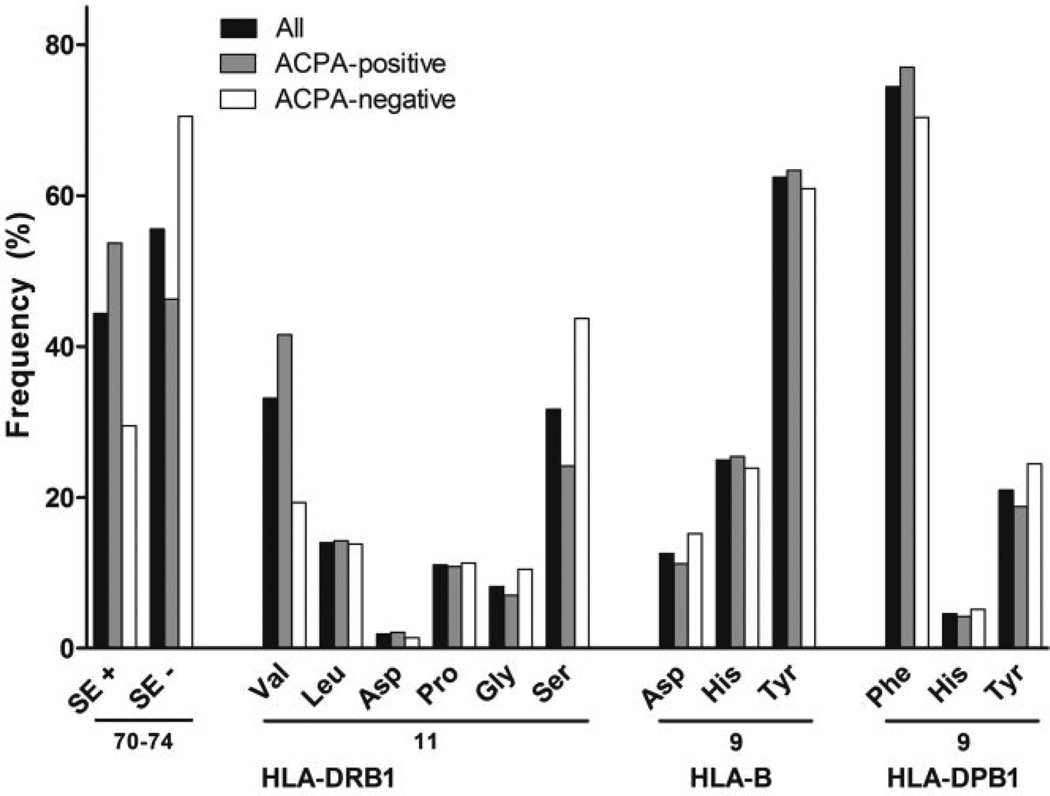
The allele frequencies of HLA–DRB1 SE and the amino acids at HLA–DRB1 position 11, HLA–B position 9, and HLA–DPB1 position 9 in the total population studied are presented in Figure 1. Of the 6 amino acids possible at HLA–DRB1 position 11, Val, Ser, and Leu were most prevalent in the total RA population (prevalence of 33.2%, 31.7%, and 14.0%, respectively). Within the ACPA-positive group of RA patients, Val was the most prevalent amino acid (41.6%), and within the ACPA-negative group, Ser was the most prevalent (43.7%). At HLA–B position 9, Tyr was the most prevalent and Asp the least prevalent amino acid (prevalence of 62.4% and 12.6%, respectively). At HLA–DPB1 position 9, Phe was the most common amino acid with a prevalence of 74.4%. For HLA–B and HLA–DPB1, the frequencies of amino acids were similar for ACPA-positive and ACPA-negative RA.
Figure 1.
Frequencies of HLA–DRB1 shared epitope (SE) alleles and the amino acids at HLA–DRB1 position 11, HLA–B position 9, and HLA–DPB1 position 9. Frequencies are expressed as the percentage of a total of 3,756 alleles/amino acids. Of the 1,878 patients, 69.0% were positive for the HLA–DRB1 SE (at least 1 SE allele), 71.2% were positive for Val/Leu at HLA–DRB1 position 11 (at least 1 Val or Leu amino acid), 23.8% were positive for Asp at HLA–B position 9 (at least 1 Asp), and 93.0% were positive for Phe at HLA–DPB1 position 9 (at least 1 Phe). ACPA = anti–citrullinated protein antibody.
HLA–DRB1 SE alleles
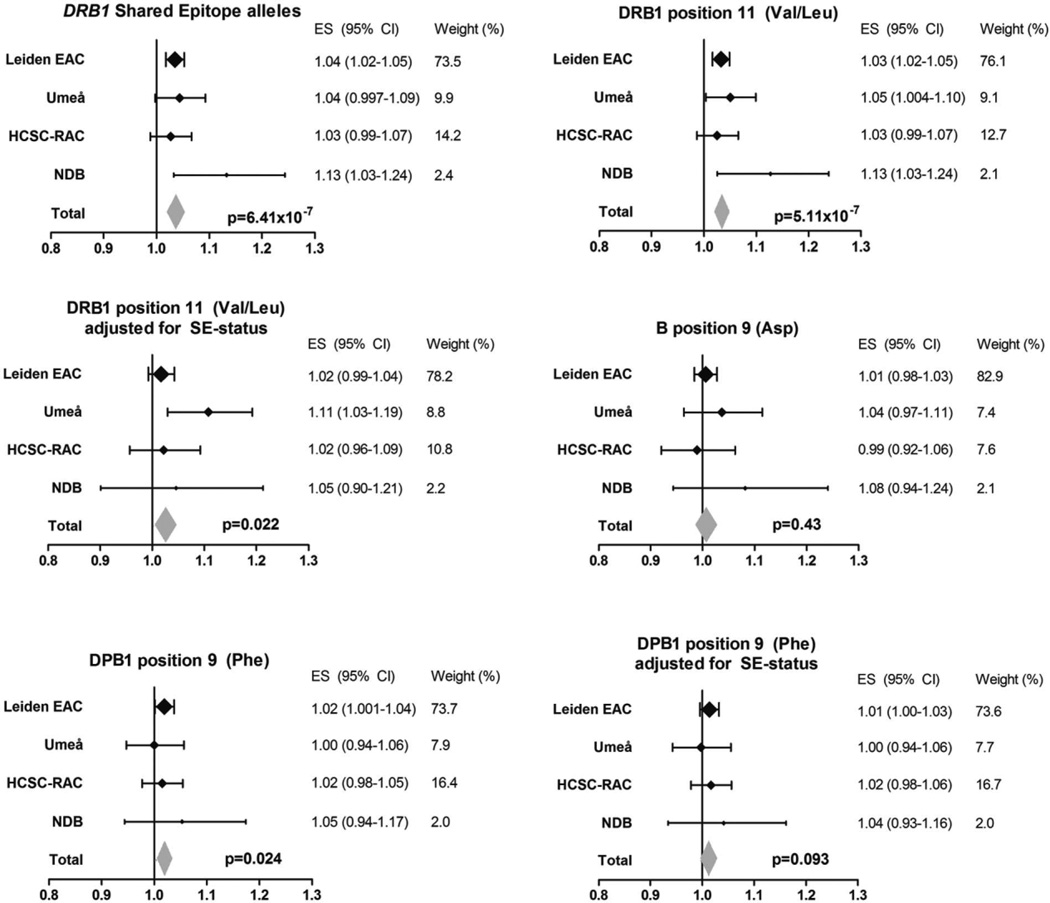
First, the association between the HLA–DRB1 SE alleles and radiographic progression was studied. As expected, the SE alleles were associated with more severe radiographic progression (P = 6.41 × 10−7 in the fixed-effects meta-analysis of the 4 cohorts) (Figure 2). When SE and ACPAs were analyzed concomitantly, the significance of the SE alleles was lost (meta-analysis P = 0.20) and ACPA was still significant (meta-analysis P = 2.22 × 10−16). The association between the SE alleles and radiographic progression was thus not independent of ACPAs, suggesting that ACPAs play a role in the causal path of the SE alleles and radiographic progression.
Figure 2.
Associations of amino acids at HLA–DRB1, HLA–B, and HLA–DPB1 that are known to predispose to rheumatoid arthritis (RA) development with radiographic progression in RA. The yearly radiographic progression rates per risk amino acid in each individual cohort are shown. P values are from the fixed-effects meta-analyses evaluating the 4 cohorts, which consisted of a total of 1,878 patients and 4,911 sets of radiographs. Risk amino acids were defined according to the findings of Raychaudhuri et al (10). For the DRB1 shared epitope (SE), I2 = 22.9%, P = 0.27, fixed-effects P = 6.41 × 10−7, and random-effects P = 2.01 × 10−4. For Val/Leu at DRB1 position 11, I2 = 23.0%, P = 0.27, fixed-effects P = 5.11 × 10−7, and random-effects P = 2.19 × 10−4. For Val/Leu at DRB1 position 11 adjusted for SE status, I2 = 37.3%, P = 0.19, fixed-effects P = 0.022, and random-effects P = 0.066. For Asp at B position 9, I2 = 0.0%, P = 0.59, fixed-effects P = 0.43, and random-effects P = 0.43. For Phe at DPB1 position 9, I2 = 0.0%, P = 0.85, fixed-effects P = 0.024, and random-effects P = 0.024. For Phe at DPB1 position 9 adjusted for SE status, I2 = 0.0%, P = 0.90, fixed-effects P = 0.093, and random-effects P = 0.093. EAC = Early Arthritis Clinic; HCSC-RAC = Hospital Clinico San Carlos–Rheumatoid Arthritis Cohort; NDB = National Data Bank for Rheumatic Diseases; ES = effect size; 95% CI = 95% confidence interval.
HLA–DRB1 position 11
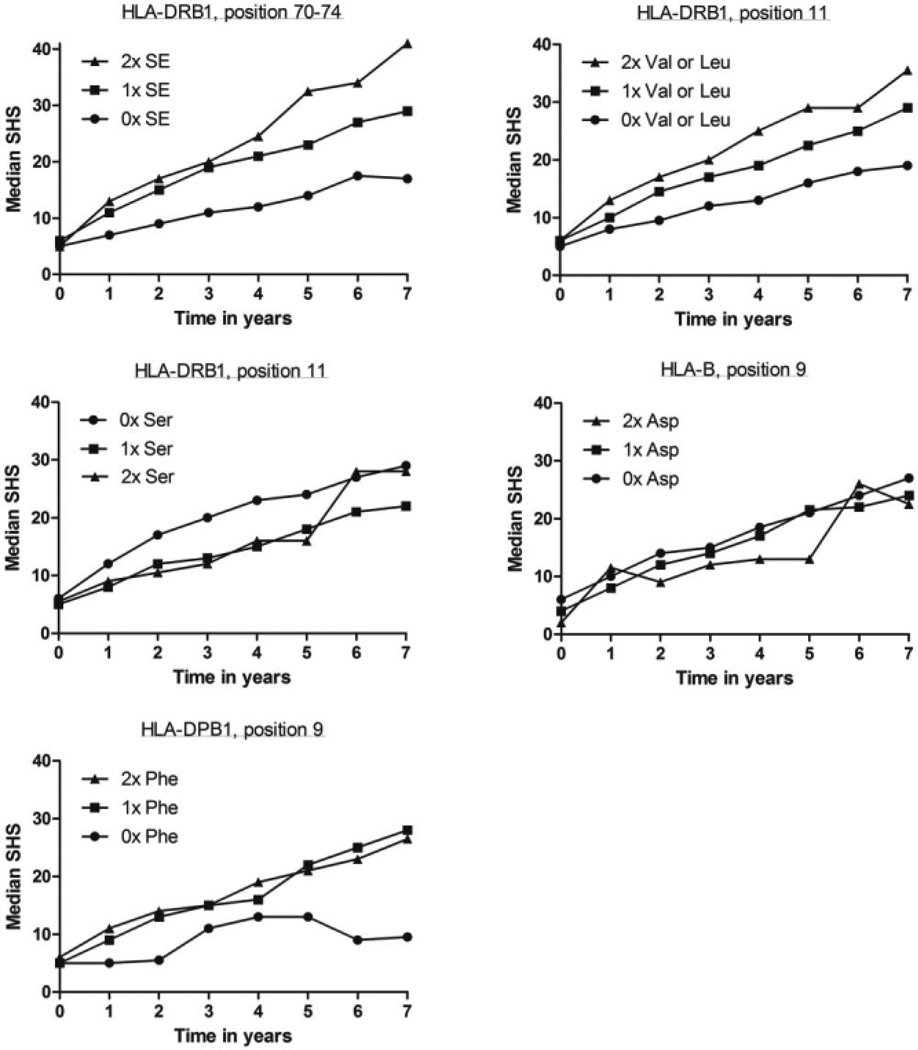
Patients with the risk amino acids Val and Leu had a higher rate of joint damage progression; this reached statistical significance in 3 individual cohorts (EAC P = 4.94 × 10−5, Umeå P = 0.032, and NDB P = 0.013) and in the meta-analysis (P = 5.11 × 10−7) (Figure 2). To illustrate, the raw radiographic data for RA patients included in the Leiden EAC are presented in Figure 3. In these patients, the presence of one Val or Leu amino acid was associated with a 1.033-fold rate of joint destruction per year compared to patients without these amino acids; this equals a 26% (1.033 to the power 7) higher rate of joint destruction over 7 years. To further illustrate the effects, RA patients with 1 and 2 Val or Leu amino acids, respectively, had SHS scores that were 10 and 16.5 points higher than patients without any Val or Leu amino acids (Figure 3). Conditioning on SE status revealed an independent association of position 11 with radiographic progression (meta-analysis P = 0.022) (Figure 2). This indicates that Val or Leu on HLA–DRB1 position 11 was associated with radiographic progression independent of the HLA–DRB1 SE status.
Figure 3.
Radiographic progression in rheumatoid arthritis patients in the Leiden Early Arthritis Clinic cohort per number of risk alleles or amino acids at HLA–DRB1, HLA–B, and HLA–DPB1. The median raw and unmodeled Sharp/van der Heijde scores (SHS) during 7 years of followup of 594 RA patients are shown. The P values obtained by multivariate normal regression analyses comparing the genotypes in relation to radiographic progression were P = 5.33 × 10−5 for the DRB1 shared epitope (SE) alleles, P = 4.94 × 10−5 for Val/Leu at DRB1 position 11, P = 0.044 for Ser at DRB1 position 11, P = 0.62 for Asp at HLA–B position 9, and P = 0.036 for Phe at DPB1 position 9.
In order to determine whether the effect of Val and Leu at position 11 on radiographic progression was independent of the effect of ACPAs on radiographic progression, analyses were also adjusted for ACPA status. Similar to the SE alleles, the effect of Val and Leu on radiographic progression was not independent of ACPA status (meta-analysis P = 0.12).
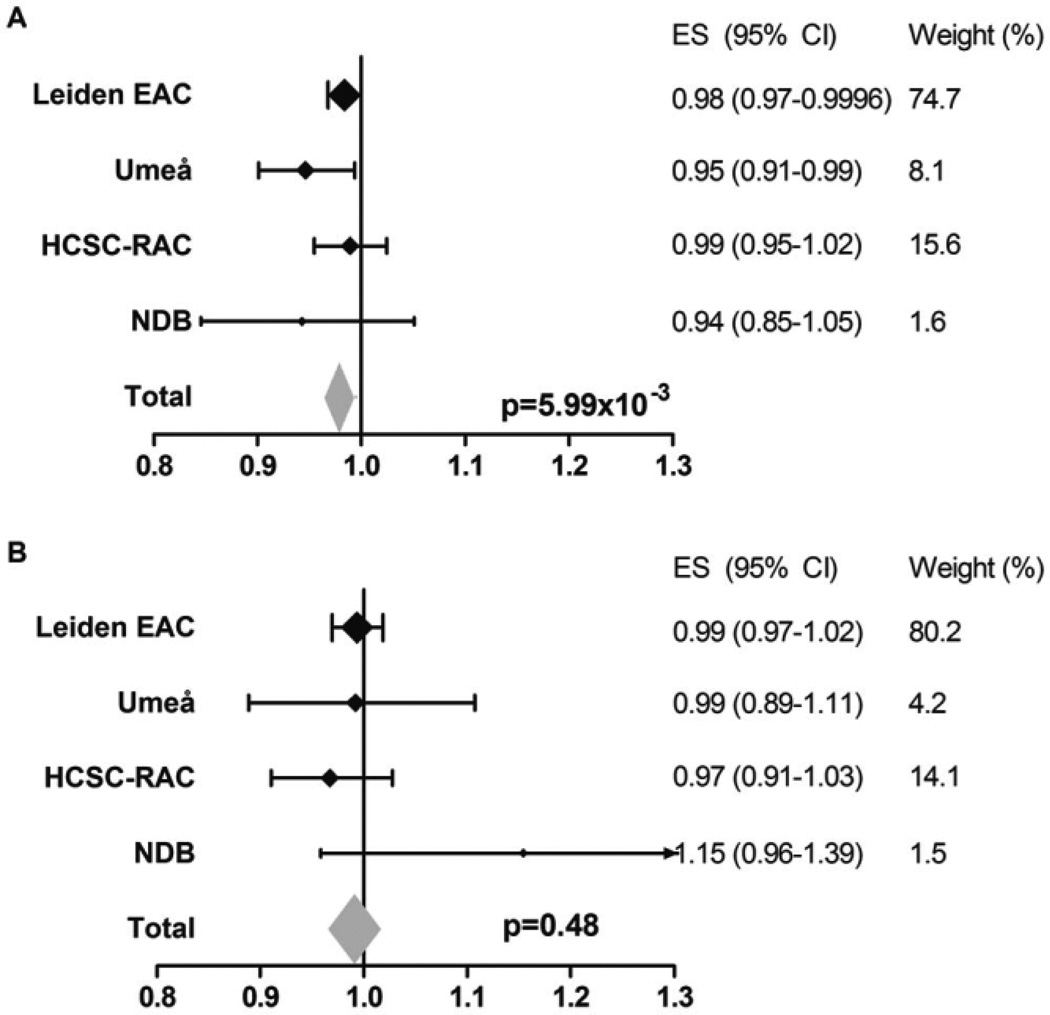
We next studied amino acid Ser at HLA–DRB1 position 11 in more detail, since this variant was reported to protect against RA (10). Analysis showed that Ser was associated with a lower rate of joint destruction (meta-analysis P = 5.99 × 10−3) (Figure 4A).
Figure 4.
Association of Ser at HLA–DRB1 position 11 with radiographic progression of rheumatoid arthritis in the total population and in the subgroup of patients who did not carry any predisposing Val or Leu amino acids. A, Yearly radiographic progression rates per Ser amino acid at HLA–DRB1 in the total population. Data are shown for each individual cohort and for the fixed-effects meta-analyses evaluating the 4 cohorts, consisting of a total of 1,878 patients and 4,911 sets of radiographs. I2 = 0.0%, P = 0.40, fixed-effects P = 5.99 × 10−3, and random-effects P = 5.99 × 10−3. B, Yearly radiographic progression rates per Ser amino acid at HLA–DRB1 in the patients who did not carry any predisposing Val or Leu amino acids. To determine whether the observed association of Ser with radiographic progression was truly protective and not due to the concomitant absence of the predisposing amino acids Val and Leu, analyses were performed within the subgroup of patients not carrying Val and Leu (thus, patients with 2 copies of Ser, patients with 1 Ser and 1 neutral amino acid, and patients with 2 neutral amino acids were compared). Data are shown for each individual cohort and for the fixed-effects meta-analyses evaluating the 4 cohorts, consisting of a total of 541 patients and 1,477 sets of radiographs. I2 = 9.5%, P = 0.35, fixed-effects P = 0.48, and random-effects P = 0.56. See Figure 2 for definitions.
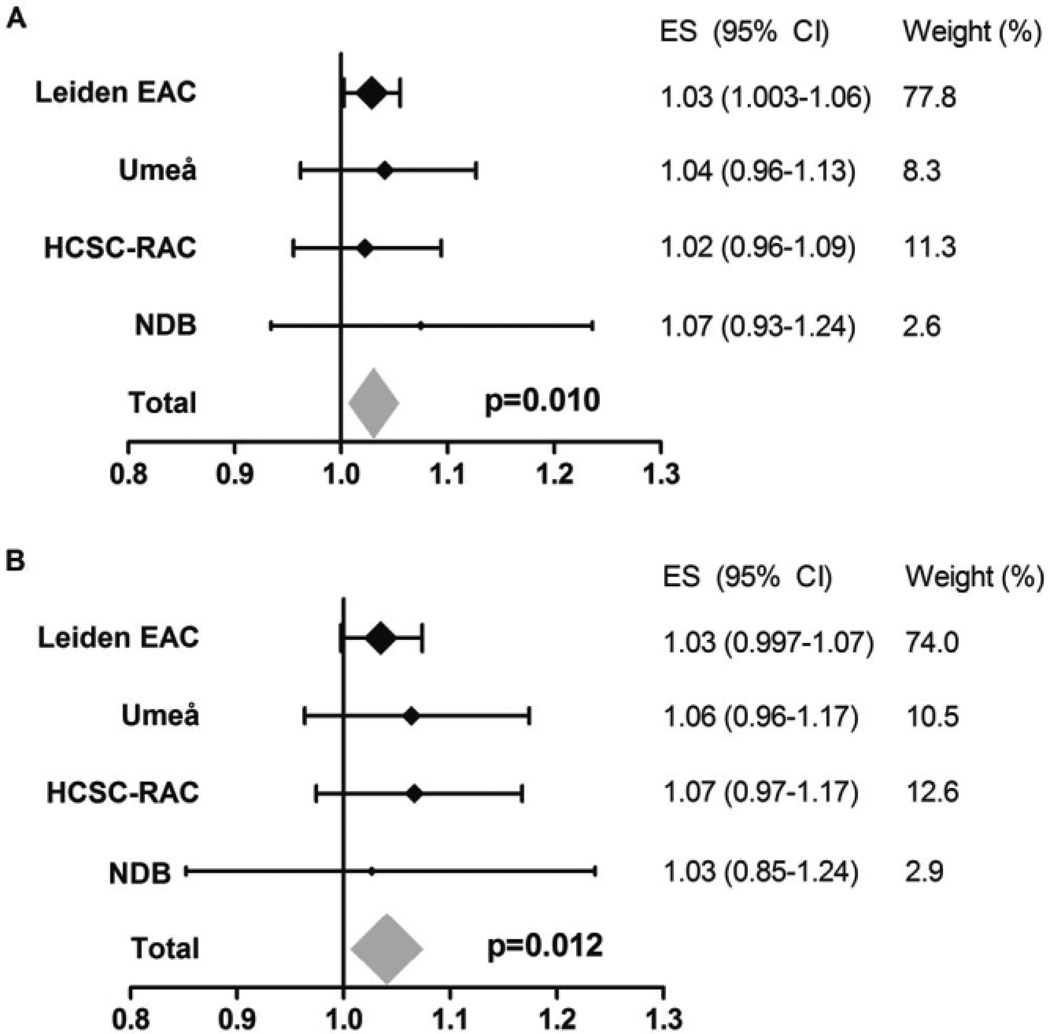
Because of the polymorphic nature of HLA–DRB1 and because predisposing as well as protective associations of variants encoded by the same position with radiographic progression were observed, we subsequently performed stratified analyses to distinguish between true-positive signals (i.e., a truly predisposing variant) and false-positive signals (i.e., seemingly predisposing but actually caused by the absence of protective variants). First, the association of Val and Leu was studied when comparing patient groups with S/S, S/N, and N/N, thus excluding patients with the protective amino acid Ser. Carrying Val and Leu was still associated with more radiographic progression (meta-analysis P = 0.010) (Figure 5A), and this association remained after additionally conditioning on SE status (meta-analysis P = 0.012) (Figure 5B), indicating a truly predisposing association for Val and Leu with radiographic progression. Furthermore, when performing stratified analysis to evaluate the effect of Ser in patients not carrying the predisposing variants Val and Leu (thus comparing 3 groups of patients: P/P, P/N, and N/N) the protective effect of Ser was lost (meta-analysis P = 0.48) (Figure 4B). This might suggest that the protective effect of Ser observed in the total RA population was the consequence of the absence of the predisposing amino acids Val and Leu rather than a truly protective effect.
Figure 5.
Association of Val and Leu at HLA–DRB1 position 11 with radiographic progression in the subgroup of rheumatoid arthritis patients who did not carry any Ser at position 11 and with additional adjustment for shared epitope (SE) status. A, Yearly radiographic progression rates per Val or Leu amino acid at HLA–DRB1 position 11 in patients who did not carry any Ser at position 11. These analyses were performed to determine whether the observed association of Val and Leu with radiographic progression was truly predisposing and not due to the concomitant absence of Ser (thus, patients carrying 2 copies of Val or Leu, patients with 1 Val or Leu and 1 neutral amino acid, and patients with 2 neutral amino acids were compared). Data are shown for each individual cohort and for the fixed-effects meta-analyses evaluating a total of 781 patients and 1,747 sets of radiographs. I2 = 0.0%, P = 0.92, fixed-effects P = 0.010, and random-effects P = 0.010. B, Yearly radiographic progression rates per Val or Leu amino acid at HLA–DRB1 position 11 in patients who did not carry any Ser at position 11, adjusted for SE status. I2 = 0.0%, P = 0.90, fixed-effects P = 0.012, and random-effects P = 0.012. See Figure 2 for definitions.
HLA–B position 9
Because risk factors for RA located outside the HLA–DRB1 region were also identified, we subsequently studied the association of 2 amino acids that are located elsewhere in the HLA region. First, the association between amino acid Asp at HLA–B position 9 and radiographic progression was assessed. No association was observed, neither in the individual cohorts nor in a meta-analysis (meta-analysis P = 0.43). Also, no tendency for association was observed (Figure 2) (Figure 3 illustrates the raw radiographic data for HLA–B position 9 in the Dutch RA patients).
HLA–DPB1 position 9
The other variant that is not located within the HLA–DRB1 region was HLA–DPB1. Amino acid Phe at position 9 in HLA–DPB1 was significantly associated with more severe radiographic progression (meta-analysis P = 0.024) (Figure 2) (Figure 3 illustrates raw radiographic data for RA patients in the EAC). However, after conditioning on SE status, no significance was obtained (meta-analysis P = 0.093) (Figure 2). Furthermore, when conditioning the effect of Phe for ACPA significance was lost (meta-analysis P = 0.27).
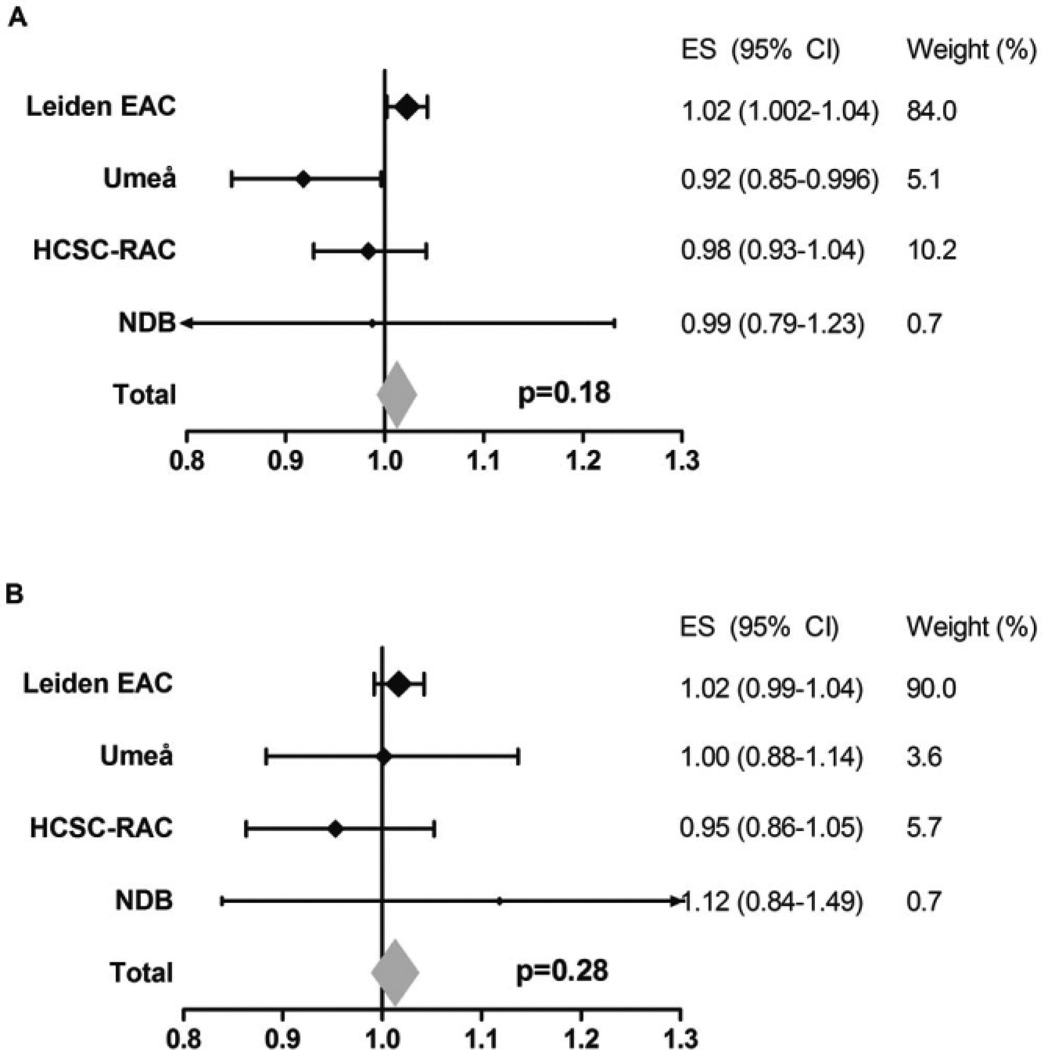
Predisposing variants within ACPA-negative RA
Recently, the amino acids Leu and Ser on HLA–DRB1 position 11 and Asp on B position 9 were identified as risk factors for ACPA-negative RA (11). Associations between these amino acids and radiographic progression were also studied within the ACPA-negative subgroup, consisting of 657 patients with 1,877 radiograph sets. No significant results were obtained for Leu/Ser at HLA–DRB1 position 11 or Asp at HLA–B position 9 (meta-analysis P = 0.18 and P = 0.28, respectively). Also, the directionality of the effects was diverse (Figure 6) (see Supplementary Figure 2, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39018/abstract).
Figure 6.
Association of Leu and Ser at HLA–DRB1 position 11 and Asp at HLA–B position 9 with radiographic progression in anti–citrullinated protein antibody (ACPA)–negative RA. Data are shown for each individual cohort and for the fixed-effects meta-analyses evaluating the 4 cohorts, consisting of a total of 657 patients and 1,877 sets of radiographs. Risk amino acids were defined according to the findings of Han et al (11). A, Yearly radiographic progression rates per risk amino acid Leu or Ser at HLA–DRB1 position 11 in ACPA-negative patients. I2 = 60.8%, P = 0.054, fixed-effects P = 0.18, and random-effects P = 0.59. B, Yearly radiographic progression rates per risk amino acid Asp at HLA–B position 9 in ACPA-negative patients. I2 = 0.0%, P = 0.56, fixed-effects P = 0.28, random-effects P = 0.28. See Figure 2 for other definitions.
DISCUSSION
This study was undertaken to investigate whether genetic variants at HLA–DRB1 position 11, HLA–B position 9, and HLA–DPB1 position 9, which were recently identified as risk factors for RA, were also associated with radiographic progression in RA, and whether the associations were independent of HLA–DRB1 SE status and ACPA status. We observed that RA patients carrying the amino acids Val or Leu at HLA–DRB1 position 11 had more severe radiographic progression. Despite linkage between the variants at position 11 and the HLA–DRB1 SE alleles, the effect of Val and Leu was independent of the known SE effect. In addition, similar to the SE alleles, the effect of Val and Leu at HLA–DRB1 position 11 was not independent of ACPA. Therefore, the findings of the present study validate the relevance of these amino acids at position 11 in HLA–DRB1.
Identifying which variants in the HLA region are associated with the development or course of RA provides the opportunity to increase the understanding of the mechanisms underlying the progression of RA. The recent observation that the genetic variants that are associated with risk of RA development and those that are associated with risk of a progressive course of RA are largely different suggests that the processes driving disease development and disease severity are to a large extent dissimilar (1). However, a few genetic variants are risk factors for both RA development and RA severity, highlighting the importance of the pathway indicated by these risk factors throughout the disease. One such risk factor is the presence of the SE alleles encoded by HLA–DRB1 positions 70–74, which act via ACPAs on disease development and outcome. The findings of the present study demonstrate that Val and Leu at HLA–DRB1 position 11 are also associated with both the development and severity of RA as measured by radiographic progression.
The hope of the formulation of the SE hypothesis was that it would allow the identification of the auto-antigens of RA (6). However, thus far, arthritogenic peptides have not been found. Mouse studies have revealed that small changes in amino acids located within the antigen-presenting binding site may have large influences on the antigens that are presented (12,13). The amino acids at position 11 and position 13 are observed to be in very tight linkage disequilibrium (10); therefore, only the amino acids of position 11 were analyzed. Future studies will reveal whether taking the amino acids at position 11 and 13 into account will be helpful in identifying the pathogenic antigens that result in immune activation and autoantibody production, thereby stimulating disease development or progression.
Investigation of the HLA region is challenging due to high linkage disequilibrium. This is illustrated by the fact that 91.8% of all patients evaluated in the present study that had 0, 1, or 2 SE alleles also had, respectively, 0, 1, or 2 Val/Leu amino acids at HLA–DRB1 position 11, implying that the observation of an additive value of position 11 was actually based on a relatively small proportion of patients. In addition, the association between SE and radiographic progression is strong, providing another reason why large data sets are required to determine whether other HLA variants have additional effects that are independent of the SE effect. Thus far, to the best of our knowledge, no studies have been published providing replication of the results regarding RA susceptibility published by Raychaudhuri et al in completely independent data sets (10). Also in this respect, the findings of the present study, though evaluating a different outcome, support the relevance of position 11 on HLA–DRB1.
In addition to linkage and power issues, the polymorphic nature of the HLA–DRB1 region in which the same position can encode for both predisposing and protective variants adds another level of complexity. To unravel these effects, adjusted multivariable analyses and stratified analyses can be done. The advantage of adjusting in multivariable analyses is that it is more powerful than subgroup analyses; however, this is at the cost of possible disturbing effects due to the assumptions underlying the model and eventual disturbances due to multicollinearity. In our view, in the present setting, stratified analyses yield the most conclusive results. We observed that Val and Leu at HLA–DRB1 position 11 were truly predisposing to more severe disease. In contrast, our data suggested that carrying the amino acid Ser at the same position was not protective. Although a protective effect was observed initially, in stratified analyses (excluding the effect of the absence of predisposing effects of Val and Leu), Ser was not significantly associated with less severe radiographic progression. These data might suggest that Ser does not have a protective effect on the severity of radiographic progression in RA. However, it is also possible that the present study was insufficiently powered to detect a protective effect of Ser in the subgroup of patients studied. We cannot discriminate between these two possible explanations, and further studies are needed to determine the association of Ser with the severity of radiographic progression.
Raychaudhuri et al also defined 16 haplotypes, based on positions 11, 71, and 74 in HLA–DRB1 (10). We did not perform haplotype analyses, since in our view stratification is needed to distinguish predisposing or protective haplotypes from the absence of protective and predisposing haplotypes, respectively, and then subgroups became too small.
Within the field of RA susceptibility, it has been notable that Ser has been reported to be protective against ACPA-positive RA but predisposing to ACPA-negative RA (10,11). Because of this finding, we also evaluated the association of Ser with progression in the ACPA-negative subgroup. The presence of Ser in these patients was not associated with the severity of radiographic progression.
We could not identify an association between Asp at HLA–B9 and radiographic progression. This position, which is highly correlated with HLA–B*08, is part of the conserved ancestral A1–B8–DRB1*03 (8.1) haplotype and has been associated with susceptibility to ACPA-negative disease (8,9). In contrast to the undisputed association of DRB1*03 or this haplotype with susceptibility to ACPA-negative RA, we did not detect an association with the severity of the disease in RA or in ACPA-negative RA.
The presence of Phe at HLA–DPB1 position 9 was associated with the severity of radiographic progression, but significance was lost (P = 0.093) when conditioning for the HLA–DRB1 SE status. This might suggest that Phe at HLA–DPB1 is not associated with radiographic progression independent of the association between SE status and radiographic progression. Notably, the directionality of the effect of Phe at HLA–DPB1 was similar in all cohorts, with more severe progression in the patients carrying Phe. Although the linkage between this position with the HLA–DRB1 SE alleles is less than for HLA–DRB1 position 11 with SE (in our study 32.7% of the patients who had 0, 1, or 2 SE alleles also had, respectively, 0, 1, or 2 Phe amino acids at HLA–DPB1 position 9), it is possible that our data were insufficiently powered to find an independent association of Phe with radiographic progression. The present data do not allow making definite conclusions regarding HLA–DPB1 and radiographic progression; though if an effect is present, this effect seems to act in the path of ACPAs, similar to HLA–DRB1.
In order to get some indication of the variance in joint destruction explained by the genetic factors studied, the interindividual variance in joint destruction in the Leiden RA patients at year 7 was evaluated. The SE alleles explained 4.2% of the variance in joint destruction and the combination of SE alleles, HLA–DRB1 position 11, HLA–DPB1 position 9, and HLA–B position 9 explained 4.5%. This is lower than the variance in the risk of ACPA-positive RA explained by a combination of HLA–DRB1 positions 71, 74, and 11, HLA–DPB1 position 9, and HLA–B position 9, which was reported to be 12.7% (10).
To conclude, Val and Leu at position 11 in the HLA–DRB1 locus were recently identified as additional susceptibility factors for ACPA-positive RA. We observed that these amino acids were also associated with a more severe disease course, an effect that was not independent of ACPA status. Further 3-dimensional modeling studies or binding assays, such as described previously by Scally et al (27) are needed to determine whether the present findings result in novel insights in the consequences of the HLA–DRB1 motif on antigen presentation and its function as an immune response gene.
Supplementary Material
Acknowledgments
Supported by the Netherlands Organization for Scientific Research (Vidi grant), the Innovative Medicines Initiative (BTCure grant), and the European Union Framework Programme (project Masterswitch).
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. van Steenbergen had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Van Steenbergen, Rantapää-Dahlqvist, Huizinga, van der Helm-van Mil.
Acquisition of data. Van Steenbergen, Rodríguez-Rodríguez, Rantapää-Dahlqvist, Berglin, Toes, Fernández-Gutiérrez, Gregersen, van der Helm-van Mil.
Analysis and interpretation of data. Van Steenbergen, Raychaudhuri, Huizinga, van der Helm-van Mil.
REFERENCES
- 1.De Rooy DP, Zhernakova A, Tsonaka R, Willemze A, Kurreeman BA, Trynka G, et al. A genetic variant in the region of MMP-9 is associated with serum levels and progression of joint damage in rheumatoid arthritis. Ann Rheum Dis. 2014;73:1163–1169. doi: 10.1136/annrheumdis-2013-203375. [DOI] [PubMed] [Google Scholar]
- 2.Van der Helm-van Mil AH, Huizinga TW, Schreuder GM, Breedveld FC, de Vries RR, Toes RE. An independent role of protective HLA class II alleles in rheumatoid arthritis severity and susceptibility. Arthritis Rheum. 2005;52:2637–2644. doi: 10.1002/art.21272. [DOI] [PubMed] [Google Scholar]
- 3.Van der Woude D, Lie BA, Lundstrom E, Balsa A, Feitsma AL, Houwing-Duistermaat JJ, et al. Protection against anti–citrullinated protein antibody–positive rheumatoid arthritis is predominantly associated with HLA–DRB1*1301: a meta-analysis of HLA–DRB1 associations with anti–citrullinated protein antibody–positive and anti–citrullinated protein antibody–negative rheumatoid arthritis in four European populations. Arthritis Rheum. 2010;62:1236–1245. doi: 10.1002/art.27366. [DOI] [PubMed] [Google Scholar]
- 4.Lindqvist E, Jonsson K, Saxne T, Eberhardt K. Course of radiographic damage over 10 years in a cohort with early rheumatoid arthritis. Ann Rheum Dis. 2003;62:611–616. doi: 10.1136/ard.62.7.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stastny P. Mixed lymphocyte cultures in rheumatoid arthritis. J Clin Invest. 1976;57:1148–1157. doi: 10.1172/JCI108382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis: an approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 7.Huizinga TW, Amos CI, van der Helm-van Mil AH, Chen W, van Gaalen FA, Jawaheer D, et al. Refining the complex rheumatoid arthritis phenotype based on specificity of the HLA–DRB1 shared epitope for antibodies to citrullinated proteins. Arthritis Rheum. 2005;52:3433–3438. doi: 10.1002/art.21385. [DOI] [PubMed] [Google Scholar]
- 8.Verpoort KN, van Gaalen FA, van der Helm-van Mil AH, Schreuder GM, Breedveld FC, Huizinga TW, et al. Association of HLA–DR3 with anti–cyclic citrullinated peptide antibody–negative rheumatoid arthritis. Arthritis Rheum. 2005;52:3058–3062. doi: 10.1002/art.21302. [DOI] [PubMed] [Google Scholar]
- 9.Jawaheer D, Li W, Graham RR, Chen W, Damle A, Xiao X, et al. Dissecting the genetic complexity of the association between human leukocyte antigens and rheumatoid arthritis. Am J Hum Genet. 2002;71:585–594. doi: 10.1086/342407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han B, Diogo D, Eyre S, Kallberg H, Zhernakova A, Bowes J, et al. Fine mapping seronegative and seropositive rheumatoid arthritis to shared and distinct HLA alleles by adjusting for the effects of heterogeneity. Am J Hum Genet. 2014;94:522–532. doi: 10.1016/j.ajhg.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kast WM, de Waal LP, Melief CJ. Thymus dictates major histo-compatibility complex (MHC) specificity and immune response gene phenotype of class II MHC-restricted T cells but not of class I MHC-restricted T cells. J Exp Med. 1984;160:1752–1766. doi: 10.1084/jem.160.6.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumacher TN, Van Bleek GM, Heemels MT, Deres K, Li KW, Imarai M, et al. Synthetic peptide libraries in the determination of T cell epitopes and peptide binding specificity of class I molecules. Eur J Immunol. 1992;22:1405–1412. doi: 10.1002/eji.1830220612. [DOI] [PubMed] [Google Scholar]
- 14.Van der Helm-van Mil AH, Verpoort KN, Breedveld FC, Huizinga TW, Toes RE, de Vries RR. The HLA–DRB1 shared epitope alleles are primarily a risk factor for anti–cyclic citrullinated peptide antibodies and are not an independent risk factor for development of rheumatoid arthritis. Arthritis Rheum. 2006;54:1117–1121. doi: 10.1002/art.21739. [DOI] [PubMed] [Google Scholar]
- 15.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 16.De Rooy DP, van der Linden MP, Knevel R, Huizinga TW, van der Helm-van Mil AH. Predicting arthritis outcomes–what can be learned from the Leiden Early Arthritis Clinic? Rheumatology (Oxford) 2011;50:93–100. doi: 10.1093/rheumatology/keq230. [DOI] [PubMed] [Google Scholar]
- 17.Innala L, Kokkonen H, Eriksson C, Jidell E, Berglin E, Rantapaa-Dahlqvist S. Antibodies against mutated citrullinated vimentin are a better predictor of disease activity at 24 months in early rheumatoid arthritis than antibodies against cyclic citrullinated peptides. J Rheumatol. 2008;35:1002–1008. [PubMed] [Google Scholar]
- 18.Rodriguez-Rodriguez L, Jover-Jover J, Fontsere O, Pena-Blanco R, Leon L, Fernandez-Gutierrez B, et al. Leflunomide discontinuation in rheumatoid arthritis and influence of associated disease-modifying anti-rheumatic drugs: a survival analysis. Scand J Rheumatol. 2013;42:433–436. doi: 10.3109/03009742.2013.785590. [DOI] [PubMed] [Google Scholar]
- 19.Wolfe F, Michaud K. The National Data Bank for rheumatic diseases: a multi-registry rheumatic disease data bank. Rheumatology (Oxford) 2011;50:16–24. doi: 10.1093/rheumatology/keq155. [DOI] [PubMed] [Google Scholar]
- 20.Jia X, Han B, Onengut-Gumuscu S, Chen WM, Concannon PJ, Rich SS, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown WM, Pierce J, Hilner JE, Perdue LH, Lohman K, Li L, et al. Overview of the MHC fine mapping data. Diabetes Obes Metab. 2009;11:2–7. doi: 10.1111/j.1463-1326.2008.00997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reviron D, Perdriger A, Toussirot E, Wendling D, Balandraud N, Guis S, et al. Influence of shared epitope–negative HLA–DRB1 alleles on genetic susceptibility to rheumatoid arthritis. Arthritis Rheum. 2001;44:535–540. doi: 10.1002/1529-0131(200103)44:3<535::AID-ANR101>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 24.De Vries N, Tijssen H, van Riel PL, van de Putte LB. Reshaping the shared epitope hypothesis: HLA-associated risk for rheumatoid arthritis is encoded by amino acid substitutions at positions 67–74 of the HLA–DRB1 molecule. Arthritis Rheum. 2002;46:921–928. doi: 10.1002/art.10210. [DOI] [PubMed] [Google Scholar]
- 25.Knevel R, de Rooy DP, Zhernakova A, Grondal G, Krabben A, Steinsson K, et al. Association of variants in IL2RA with progression of joint destruction in rheumatoid arthritis. Arthritis Rheum. 2013;65:1684–1693. doi: 10.1002/art.37938. [DOI] [PubMed] [Google Scholar]
- 26.Knevel R, Tsonaka R, le Cessie S, van der Linden MP, Huizinga TW, van der Heijde DM, et al. Comparison of methodologies for analysing the progression of joint destruction in rheumatoid arthritis. Scand J Rheumatol. 2013;42:182–189. doi: 10.3109/03009742.2012.728618. [DOI] [PubMed] [Google Scholar]
- 27.Scally SW, Petersen J, Law SC, Dudek NL, Nel HJ, Loh KL, et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J Exp Med. 2013;210:2569–2582. doi: 10.1084/jem.20131241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.