Abstract
Pancreatic cancer is one of the most fatal malignancies with increasing incidence and high mortality. Possibilities for early diagnosis are limited and there is currently no efficient therapy. Molecular markers that have been introduced into diagnosis and treatment of other solid tumors remain unreciprocated in this disease. Recent discoveries have shown that certain microRNAs (miRNAs) take part in fundamental molecular processes associated with pancreatic cancer initiation and progression including cell cycle, DNA repair, apoptosis, invasivity, and metastasis. The mechanism involves both positive and negative regulation of expression of protooncogenes and tumor suppressor genes. Various miRNAs are expressed at different levels among normal pancreatic tissue, chronic pancreatitis, and pancreatic cancer and may therefore serve as a tool to differentiate chronic pancreatitis from early stages of cancer. Other miRNAs can indicate the probable course of the disease or determine the survival prognosis. In addition, there is a growing interest directed at the understanding of miRNA-induced molecular mechanisms. The possibility of intervention in the molecular mechanisms of miRNAs regulation could begin a new generation of pancreatic cancer therapies. This review summarizes the recent reports describing functions of miRNAs in cellular processes underlying pancreatic cancerogenesis and their utility in diagnosis, survival prognosis, and therapy.
1. Introduction
Despite the recent medical advances and new diagnostic possibilities, pancreatic cancer (PC) represents a frequent malignancy with disturbingly high mortality rates. Several histology subtypes of pancreatic tumors can be distinguished. The vast majority of them are represented by pancreatic ductal adenocarcinoma (PDAC) occurring at 96.3% of cases followed by less common cystic tumors, lymphomas, and metastases from other primary tumors [1].
The development of pancreatic cancer is associated with increasing cytological atypia forming precursor lesions, which can be divided into four stages of pancreatic intraepithelial neoplasia (PanIN I–PanIN IV) [2]. On a molecular level PanIN stages are characterized by gradual accumulation of DNA aberrations such as somatic point mutations within regulatory and coding sequences, gene amplifications, abnormal gene expressions, and allelic deletions (typically at 9p, 18q, 17p, and 6q) [3, 4]. However unlike other cancers, the detailed knowledge of molecular processes accompanying the pathogenesis of PC has not so far led to identification of a reliable biomarker for early detection of the disease. Patients occasionally exhibit elevated sedimentation of direct or indirect bilirubin, erythrocytes, and alkaline phosphatase and about a third of patients exhibit pathological glucose curve and/or anemia in case of protruding tumor [5]. Diagnostic utility of standard tumor markers is also very limited. The main markers are mucin antigens CA 19-9, CA 242, CA 50, and CA 72-4, but due to their relatively low specificity and sensitivity they are used in monitoring of disease progression rather than diagnosis [6]. Thus the standard diagnosis of PC is based on imaging techniques including an initial ultrasound followed by computed tomography (CT) or magnetic resonance imaging (MRI) and finally endoscopic ultrasound (EUS) which can definitively confirm the diagnosis, especially if the examination is supplemented by a fine needle biopsy (FNB). Unfortunately, due to the absence of specific manifestation, PC is usually diagnosed at the time of nonspecific symptoms such as fatigue, weight loss, dysorexia, abdominal pain, or jaundice (caused by compression of the duodenum) when the disease has already advanced and the prognosis of patients is very poor [5]. Only about 15% of all PC patients are diagnosed at an early stage of the disease when the tumor is operable. In these cases the tumor is surgically resected, which at present provides the only chance for cure. The chemotherapy (most often by gemcitabine) is administered after the pancreatic resection as well as in advanced inoperable stages; alternatively it is also administered in combination with radiotherapy or targeted biological therapy by erlotinib. Suppression of symptoms associated with the disease, such as biliary drainage in the duodenum or use of analgesics, is key in palliative therapy of advanced stages [7, 8]. Despite development in the management of the disease, the five-year survival is only about 5% [9].
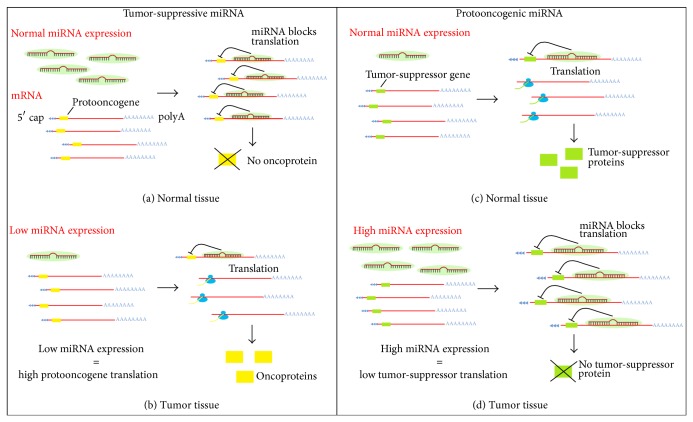
Efforts towards finding a highly sensitive and specific tool for early diagnosis of pancreatic cancer are currently leading the clinical study of this fatal disease. The role of microRNAs in malignant transformation is gradually becoming more evident [10–12] and increased emphasis is placed on finding and testing microRNAs participating in the development of pancreatic cancer in order to improve diagnosis, assess prognosis, and design new treatment options [13, 14]. MicroRNAs (miRNAs) are endogenous noncoding short RNAs (length of 21–23 nucleotides) encoded by nuclear DNA and their main function is posttranscriptional regulation of gene expression. They bind complementarily to specific sequences of messenger RNA (mRNA), which usually leads to gene silencing via translational repression or target degradation [15, 16]. miRNAs play a crucial role in various developmental, metabolic, and cellular processes including apoptosis, cell proliferation, and differentiation. Some miRNAs regulate levels of protooncogenes or tumor-suppressor genes; therefore their expression is often altered in various tumor tissues including PC (see Figure 1). These miRNAs could serve as useful tumor biomarkers [17].
Figure 1.
Effect of miRNAs expression on the regulation of protooncogenes and tumor-suppressor genes. Some miRNAs act as negative regulators of protooncogenes expression and therefore their role is tumor-suppression (a). In cancerous tissue, reduced levels of these tumor-suppressive miRNAs lead to increased target oncogenes promoting further tumor development (b). Other miRNAs negatively regulate expression of tumor-suppressor genes; hence, their function is (proto) oncogenic (c). In tumor tissue their increased expression results in blockage of translation of tumor-suppressors further assisting the malignant process (d).
2. Role of miRNAs in Processes of Malignant Transformation of Pancreatic Tissue
The development of malignant transformation consists of many steps that are characterized by the disruption of various cellular processes through the damage of their control mechanisms. These are mainly faulty DNA repair system, dysfunctional cell cycle checkpoints leading to excessive cell proliferation, the failure of apoptosis, loss of contact inhibition, and cellular migration into other tissues to form distant metastases. Current reports on involvement of miRNA in pathogenesis of pancreatic cancer are mostly based on in vitro studies of cell lines derived from malignant cells and therefore some of the results still have to be confirmed using in vivo models.
2.1. Cell Cycle and Proliferation
Cell cycle checkpoints and kinetics are important regulators of cell proliferation. Various studies conducted in connection with PC have shown several oncogenic miRNA negatively affecting tumor-suppressor genes that act as regulators of the cell cycle progression. One of the most frequently studied miRNAs, the miR-21, affects a tumor-suppressor PTEN (phosphatase and tensin homologue) whose protein product prevents the proliferation of tumor cells and controls the frequency of cell division [18]. The overexpressed miR-21 attaches to the mRNA of PTEN, thereby reducing its tumor-suppressive function. Another potential cell cycle regulator overexpressed in PC, miR-221, affects translation of p27 (CDKN1B gene), a major cyclin-dependent kinase inhibitor [18]. The p27 protein binds and prevents activation of cyclin E complex with cyclin-dependent kinase 2 (CDK 2) or cyclin D complex with cyclin-dependent kinase 4 (CDK 4), therefore having a control function in progression of cell cycle in G1 phase. Also showing increased expression, miR-192 facilitates progression from G0/G1 to S phase by regulating the expression of genes involved in cell cycle control [19]. Another oncogenic miRNA, which is often overexpressed in pancreatic cancer, is miR-424-5p. It enhances the ability of cells to proliferate and migrate through downregulation of SOCS6 protein (suppressor of cytokine signaling 6) which leads to elevated ERK pathway activity [20, 21].
In an analogy to the above, in PC a lowered expression of tumor-suppressive miRNAs that regulate the efficiency of important protooncogenes can also be detected. A significant downregulation can be observed for miR-124 (miR-124-1, miR-124-2, and miR-124-3) muted as a result of promoter hypermethylation. miR-124 inhibits proliferation, invasion, and metastasis by direct interaction with the Rac1 transcript [22]. Rac1 (Ras-related C3 botulinum toxin substrate 1) is GTPase protein which is involved not only in cell cycle control, but also in cytoskeletal reorganization, activation of protein kinases, cell adhesion, epithelial differentiation, and motility [23]. Another known miRNA that contributes to tumour cell proliferation through cell cycle deregulation in PC is miR-203, whose lowered expression leads to advancement from the G1 phase [24]. Several other miRNAs, including tumor-suppressive miR-143 [25], miR-126 [26], and let-7-d [26], regulate expression of a KRAS oncogene, which plays a crucial role by inducing abnormal cellular proliferation through mitogen-activated protein kinase (MAPK) pathways [27].
2.2. DNA Repair and Apoptosis
In a normal tissue, DNA damage triggers a wide range of cellular processes resulting in either repair of the damaged sections or a programmed cell death, apoptosis. In case of abnormal function of tumor-suppressor genes or protooncogenes, however, the DNA repair pathway as well as apoptotic cascade may be completely disrupted and the cells acquire a malignant potential.
A tumor-suppressor gene TP53 is often studied due to its major role in apoptosis and DNA repair, but it is also heavily involved in regulation of angiogenesis and cellular senescence [28]. Expression analyses have identified a number of miRNAs that contribute to TP53 regulation. One of them, miR-34a, positively regulates apoptosis and DNA repair, while negatively altering cell cycle and angiogenesis. Levels of miR-34a are reduced to less than half or are missing completely in the tissue of pancreatic cancer compared to normal ductal epithelial tissue due to a deletion of its coding region [29–31]. It was also revealed that oncogenic miR-155 represses proapoptotic gene TP53INP1 (tumor protein p53-inducible nuclear protein 1) which gets activated by p53. When present at high levels in PC, miR-155 increasingly prevents expression of TP53INP1, hence inhibiting apoptosis and allowing cell survival [32].
Another miRNA which negatively affects apoptosis appears to be miR-203, whose main function is inhibition of the apoptotic regulator survivin (baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5)). Downregulation of miR-203 results in increased expression of survivin, which inhibits apoptosis [24].
A recent study indicates that several miRNAs may also induce apoptosis. It can be triggered by elevated levels of miR-150* and miR-630, both causing the decreased expression of transmembrane tyrosine kinase receptor IGF-1R (insulin-like growth factor 1 receptor), which has antiapoptotic properties [33].
2.3. Invasivity and Metastasis
The tumor cell invasivity and ability to form metastasis are an important factor that affects cancer progression. An essential step in invasivity represents the differentiation of cells through epithelial-mesenchymal transition (EMT). This means that epithelial tumor cells undergo transition to mesenchymal type, which, among others, is capable of crossing the basement membrane and entering the bloodstream. The main feature of EMT is a loss of intercellular contacts which correlates with decreased expression of the transmembrane protein E-cadherin (epithelial cadherin), an epithelial cell marker playing a key role in cell adhesion. E-cadherin (CDH1 gene) is suppressed in several cancer types including pancreatic cancer by its repressors ZEB1 (zinc finger E-box-binding homeobox 1) and SIP1 (Smad-interacting protein 1, ZEB2, and SMADIP1) which thus act as EMT-activators [34–37]. It has been shown that members of miR-200 family (miR-200a, b, c, miR-141, and miR-429) and proteins ZEB1 and SIP1 reciprocally negatively regulate each other in a feedback loop mechanism which controls the EMT [38, 39]. Generally, the decrease in expression of miR-200 family members can trigger the epitelial to mesenchymal transition [38–43] and similar findings were published also for miR-203 [44, 45]. Most recently another miRNA, miR-208, was found to be directly involved in EMT. After overexpressing miR-208, expression of E-cadherin was decreased suggesting that miR-208 can promote the EMT [46].
Previously the involvement of miR-143 in pancreatic cancer cell invasivity was tested and its key role in regulation of Rho GTPases signaling was demonstrated [25]. Rho GTPases are G-proteins that control many processes associated with cancer metastasis formation such as cell-cell contact or cell movement. Increased activity of Rho GTPases enhances cellular invasivity and migration. miR-143 has been shown to lower Rho GTPases activity and therefore decrease levels of miR-143, which are observed frequently in pancreatic cancer cells, leading to metastatic phenotype [25].
Another miRNA involved in the formation of metastasis is miR-146a. Lower levels of mir-146a in pancreatic cancer cells were found to increase invasive behaviour [47]. Moreover it was determined that the reexpression of miR-146a can inhibit the invasivity. The molecular mechanism of this process remains unclear, but it seems to be associated with regulation of EGFR (epidermal growth factor receptor) and NF-κB transcription factor signaling [47].
The opposite effect was observed for miR-10a. The expression of miR-10a promotes metastatic formation, whereas its repression leads to inhibition of invasive behavior. miR-10a supports the ability of cell to metastasize through the suppression of homeobox transcription factors HOXA1, HOXB1, and HOXB3, which, as demonstrated, may function as metastatic suppressors [48, 49].
For a summary of miRNAs involved in pancreatic carcinogenesis, which are listed in the previous text, see Table 1.
Table 1.
miRNAs involved in processes of malignant transformation of pancreatic tissue (↓ = reduced, ↑ = increased).
| miRNA | Role of miRNA | Expression in PC cells | Impact of the aberrant expression on PC cells | Target gene | Respective references |
|---|---|---|---|---|---|
| miR-21 | Oncogenic | ↑ | ↑ proliferation and frequency of cell division | PTEN | [18] |
| miR-221 | Oncogenic | ↑ | ↑ cell cycle progression | CDKN1B | [18] |
| miR-192 | Oncogenic | ↑ | ↑ cell cycle progression | SIP1 and cell cycle regulatory genes | [19] |
| miR-424-5p | Oncogenic | ↑ | ↑ cell proliferation and migration | SOCS6 | [20, 21] |
| miR-124 | Tumor-suppressive | ↓ | ↑ proliferation, invasion, and metastasis | RAC1 | [22] |
| miR-203 | Tumor-suppressive | ↓ | ↑ cell cycle progression, ↓ apoptosis | BIRC5 | [24] |
| ↑ epithelial to mesenchymal transition (EMT) | CAV1 | [45] | |||
| miR-143 | Tumor-suppressive | ↓ | ↑ cell proliferation, cellular invasivity, and migration | GET1, GET2, and KRAS | [25] |
| miR-126, let-7d | Tumor-suppressive | ↓ | ↑ cell proliferation | KRAS | [26] |
| miR-34a | Tumor-suppressive | ↓ | ↓ apoptosis and DNA repair, ↑ cell cycle progression and angiogenesis | TP53 | [29–31] |
| miR-155 | Oncogenic | ↑ | ↓ apoptosis | TP53INP1 | [32] |
| miR-200 family | Tumor-suppressive | ↓ | ↑ EMT | ZEB1, SIP1 | [43] |
| miR-208 | Oncogenic | ↑ | ↑ EMT | CDH1 | [46] |
| miR-146a | Tumor-suppressive | ↓ | ↑ invasivity | IRAK-1, EGFR | [47] |
| miR-10a | Oncogenic | ↑ | ↑ invasivity and metastatic behavior | HOXA1, HOXB1, and HOXB3 | [47] |
3. Impact of Polymorphisms and Mutations in miRNA Genes
Although changes in the coding sequences of some miRNAs such as single nucleotide polymorphisms (SNPs) and mutations may play an important role in susceptibility and development of pancreatic cancer, there are only a few studies directed at this topic. In very recent study it was shown that specific SNPs in the genes coding for precursors of miR196a2 and miR-146a may play a role in pancreatic tumorigenesis. Although significant difference between genotypes of healthy individuals and PC patients was not found, certain genetic variants of these miRNAs have been expressed more frequently in T1 and T2 stages of PC compared to T3 and T4 stages. Molecular mechanism of the different expression based on the subtle differences in sequences is currently under further study [50]. In another work a connection between mutations in miR-21 gene and pancreatic cancer was suggested [51].
4. miRNAs Differently Expressed in Pancreatic Tissue
4.1. Normal Pancreas versus Chronic Pancreatitis versus Cancer
Many research groups are engaged in analysis of aberrant expression of miRNAs in normal pancreatic tissue and pancreatic cancer. The differences are also studied and compared to data from chronic pancreatitis, which represents a major precancerous condition and an endogenous factor causing up to 16-fold increase in risk of pancreatic cancer [52]. Over the years hundreds of publications with varying levels of scientific impact have disclosed a number of miRNAs differentially expressed in all the above types of pancreatic tissues [53–75]. The most frequently reported miRNAs to be expressed at elevated levels in PDAC include miR-21 [53–57, 59, 61, 62], miR-155 [55, 58, 59, 61, 70, 72], miR-196a [58–60, 68, 72–74], miR-221 [53, 59, 75], and miR-222 [53, 59, 61, 70, 72].
4.2. Correlation of miRNAs Levels to PanINs
More recent papers have reported a number of miRNAs whose levels sharply change among different PanIN stages. This is typical for more advanced PanIN-2 and PanIN-3. There are currently no reports comprehensively describing specific miRNAs that correlate with each of the PanINs.
Significantly increased levels of miR-155 correlate directly with the advanced PanIN-2 and PanIN-3 [76, 77]. As mentioned earlier, the target of miR-155 is proapoptotic TP53INP1 gene. TP53INP1 is expressed in normal as well as in the early PanIN tissues but it is lost in PanIN-3. This could be explained by the rapid growth of miR-155 in the 2nd and 3rd PanIN stages [32]. The PanIN-2 and PanIN-3 stages are supplemented by a significant increase of miR-10b [78], miR-221 [79], miR-222 [77, 79], let-7a [79], and miR-196b [77, 78] and a significant decrease of miR-217 [77, 78]. Overexpression was also observed for miR-21 [76–79] with the most notable change in PanIN-3 [76]. Also increased expression levels of miR-146a [77, 78] and miR-196a-2 [78] were found to correlate with advanced PanINs. In contrast miR-148a, whose gene is epigenetically silenced resulting in its low expression levels, correlates with early PanIN stages [80]. All miRNAs which are significantly associated with PanIN progression are listed in Table 2. In addition, a recent extensive study has revealed more than 100 miRNAs which are aberrantly expressed in different PanIN stages [77]. From them, miRNAs whose expression increases or decreases with advanced panINs are included in Table 2.
Table 2.
miRNAs whose expression increase (↑) or decrease (↓) with increasing PanIN stages.
| let-7a [79] | ↑ | miR-155 [32, 76, 77] | ↑ | miR-200a/b/c [77] | ↑ |
| miR-10b [78] | ↑ | miR-182 [77] | ↑ | miR-217 [78, 79] | ↓ |
| miR-21 [76–79] | ↑ | miR-183* [77] | ↑ | miR-221 [79] | ↑ |
| miR-146a [77, 78] | ↑ | miR-196a-2 [78] | ↑ | miR-222 [77, 79] | ↑ |
| miR-148a [80] | ↓ | miR-196b [77, 78] | ↑ | miR-296-5p [77] | ↓ |
*Passenger strand of pre-miRNA.
5. Diagnostic miRNAs
The study of miRNAs as cancer biomarkers is not restricted to tumor tissues only. More recently it was shown that almost all body fluids contain miRNA [81] as a result of either passive release from necrotic or apoptotic cells or due to an active secretion by microvesicles [82]. In comparison to mRNA, whose detection in body fluids is somewhat challenging, miRNAs are stable as they are resistant to cleavage by ribonucleases and survive extreme pH and temperature conditions [83]. With the lack of reliable approaches based on imagining techniques and/or routine tumor markers, the option of detecting miRNA in peripheral body fluids, especially blood serum, has currently a considerable potential for use in clinical practice.
Among others, miR-192 is very promising showing increased levels in serum of PDAC patients compared to healthy controls with sensitivity towards cancer at 76% and specificity at 55% [19]. Another potential biomarker is miR-18a, which occurs at high levels in tumor tissue as well as in plasma of cancer patients. In addition, miR-18a levels were found to be significantly reduced after tumor resection [84]. Other studies are focused on using combination of several circulating miRNAs to discern various stages of PC from cancer-free controls and CP [57, 85]. Recently another study introducing a promising panel of circulating miRNAs for early blood-based diagnosis of pancreatic cancer has been presented [86].
In order to increase diagnostic accuracy of early stage pancreatic cancer a combination of serum CA19-9 and quantification of miR-16 [87] or combination of CA19-9, miR-16, and miR-196a [60] may be of clinical use. The resulting sensitivity and specificity of the combined markers detected in peripheral blood for discrimination of chronic pancreatitis and pancreatic cancer are reportedly higher compared to the values for individual markers [60, 87]. Another study indicates that a combination of serum CA19-9 with the detection of the expression of miR-27a-3p from peripheral blood mononuclear cells (PBMC) can be used to diagnose pancreatic cancer with a sensitivity of 85.3% and specificity of 81.6% [69]. These tests could supposedly be applied also for screening of peripheral blood of high-risk groups. Indeed, a potential use for diagnostic purposes can be attributed to all miRNAs that are differentially expressed in PDAC compared to healthy tissue and/or chronic pancreatitis [88].
6. Prognostic miRNAs
Estimation of prognosis in terms of survival probability has a great significance in clinical management of pancreatic cancer. Patients often show a poor performance status and the effect of treatment is only minor. Systemic therapy or chemotherapy should therefore carefully be considered with regard to the quality of life, especially for unresectable tumors. Finding prognostic markers to assess probable course of the disease prior to treatment is therefore highly desirable. A number of literature reports are devoted to the use of miRNAs as prognostic markers. Many have demonstrated prognostic utility for miRNAs exhibiting aberrant expression in serum or in tumor tissue of PC patients. The sometimes contradicting findings are summarized in Table 3 showing the expression levels in normal tissue, in chronic pancreatitis (CP), and in cancer.
Table 3.
Prognostic miRNAs expression (↓ reduced, ↑ increased, and ↕ comparable expression).
| miRNA | miRNA level correlated with poor prognosis | Tumor versus normal tissue | Tumor versus CP tissue | CP versus normal tissue |
|---|---|---|---|---|
| let-7g* | ↓ [96] | |||
| miR-7 | ↓ [97] | |||
| miR-10b | ↑ [62, 98] | ↑ [59, 62, 98] | ↑ [59] | ↑ [59] |
| miR-21 | ↑ [61, 91, 92, 99] | ↑ [53–56, 59–62] | ↑ [59, 60] | |
| miR-30d | ↓ [61] | |||
| miR-31 | ↓ [96] | |||
| miR-34a | ↓ [61] | |||
| miR-122 | ↓ [96] | |||
| miR-124 | ↓ [55] | |||
| miR-142-5p | ↑ [90] | |||
| miR-145 | ↓ [99] | ↑ [61] | ||
| miR-146 | ↓ [99] ↑ [95] | ↑ [59] | ||
| miR-148a* | ↓ [96] | |||
| miR-155 | ↑ [70, 99] | ↑ [55, 58–61, 70, 72] | ↑ [59, 60] | ↑ [61] |
| miR-187 | ↓ [96] | |||
| miR-196a | ↑ [58, 59] | ↑ [55, 58–60, 72–74] | ↕ [59] ↑ [58, 60] | ↑ [59] |
| miR-200c | ↓ [100] | |||
| miR-203 | ↑ [70, 71] | ↑ [70, 71] | ↕ [59, 71] | ↕ [71] |
| miR-205 | ↓ [97] | ↑ [59] | ||
| miR-210 | ↑ [70, 99] | ↑ [59, 60, 70] | ↑ [60] | |
| miR-212 | ↑ [96] | |||
| miR-218 | ↓ [101] | ↓ [101] | ||
| miR-219 | ↑ [59] | |||
| miR-221 | ↑ [99] | ↑ [59] | ↑ [59] | |
| miR-222 | ↑ [70, 99] | ↑ [53, 59, 61, 70, 72] | ||
| miR-675 | ↑ [96] |
*Passenger strand of pre-miRNA.
7. Methods for Detection of miRNA in Pancreatic Cancer
The key to a successful analysis of miRNA in pancreatic cancer is in the type, quality, and quantity of the studied sample material. As mentioned already, serum samples are best suited for clinical diagnosis and determination of prognosis. The main problem in this case, however, is in limiting amounts of miRNA as well as somewhat lower specificity. The main focus of molecular analyses, however, remains on samples acquired directly from pancreatic tissue.
Samples from surgical resections processed into frozen or paraffin sections usually provide a great amount of material for both histology as well as molecular evaluations. By the initial histology inspection presence of malignant cells is ensured prior to their subsequent molecular analysis. High content of malignant cells in the examined material is necessary for representative quantification of expression of cancer-associated genes. Many patients, however, do not undergo surgery. In such a case pancreatic tissue is only available in a form of biopsy acquired by fine needle (FNB) during an endoscopic ultrasound (EUS) examination. If the patient status permits, EUS-FNB may be performed also over the course of the disease development, to monitor the effects of therapy. The disadvantage of utilizing EUS-FNB samples for molecular examination is relatively limited amount of material, often only in a form of a cytology smear. More importantly, for EUS-FNB samples there is always an uncertainty about the actual malignant content with respect to possible contamination by blood or surrounding nonmalignant cells. Only a few of the studies directed at miRNAs rely on EUS-FNB samples [62, 67, 74, 89], while the majority use resections. An overview of the samples types used in recent literature reports on miRNAs in pancreatic cancer is summarized in Table 4.
Table 4.
Types of samples used for miRNA analysis in pancreatic cancer.
| Serum | Fine needle biopsy | Resected tissue | |
|---|---|---|---|
| Invasiveness | Low | Medium | High |
| Possible sampling frequency | Days | Days-weeks | Mostly no or only one sampling |
| The amount of sample/target miRNA | Sufficient/very small | Small/small | Sufficient/sufficient |
| The possibility of obtaining pure tumor tissue (cells) | No | Yes (microdissection) | Yes (macrodissection, microdissection) |
| Respective references | [54, 58, 68, 84–86] | Fixation [62, 67, 89] RNA stabilization solution [67, 74] |
Frozen tissue [53, 56, 61, 63, 70, 72, 76, 97] Fixation [59, 67, 78, 84, 90, 96] |
Once the tissue sample is received, total RNA (including short RNA or miRNA) is extracted and reverse-transcribed into cDNA. Then, two alternative approaches are used (see Table 5). The first approach is based on microarray technology using hundreds to thousands of oligonucleotide hybridization probes. This approach allows screening a vast number of miRNAs in a single experiment. On the other hand miRNA array experiments require considerable amount of RNA as well as a rather sophisticated data evaluation. It is mostly used to screen for new potential miRNA biomarkers. A second approach is examination of a panel (usually up to 100) of preselected miRNAs using quantitative RT-PCR (qRT-PCR). This method is suitable, for example, to monitor aberrant levels of selected miRNAs in order to verify the context of prognosis. The qRT-PCR data are then processed by standard normalization using a set of housekeeping genes, typically including U6 [61, 62, 76, 78, 84], U44 [90], and 18S [53, 63, 70]. Normalization to other miRNAs has also been applied with miR-16 [55] and miR-54 [68].
Table 5.
Comparison of principal approaches for study of miRNA [102].
| qRT-PCR | miRNA microarray | |
|---|---|---|
| Principle | PCR amplification | Hybridization |
| The recommended amount of RNA | 10–700 ng | 100–10 000 ng |
| Limit of detection | 10–22 mol | 10–18 mol |
| Data processing | 1 day | More than 2 days |
8. Role of miRNA in Treatment Response and Potential for miRNA Therapy
Some of prognostic miRNAs also play a role in the efficacy of anticancer therapy and thus present themselves with new therapeutic possibilities. For example, it was found that nanomolar concentrations of antisense miR-21 and miR-221 oligonucleotides effectively inhibit their targets (oncogenic miR-21 and miR-221) and thus reduce proliferation of pancreatic cancer cell lines and, along with gemcitabine, prevent their growth [18, 91].
PDAC cells expressing elevated levels of miR-21 are chemoresistant to gemcitabine and reduce the efficiency of apoptosis induction [91, 92]. Addition of phosphoinositide 3-kinase inhibitors (PI3K inhibitors) and mTOR (mammalian target of rapamycin) serine/threonine protein kinase prevented the miR-21 (namely, the pre-miR-21) resistance, thus opening a way to gemcitabine-induced apoptosis [92]. miRNAs can be targeted, for example, by lentiviral vectors (a type of retroviruses) as recently demonstrated for miR-21, wherein PDA-derived cell lines were transduced by lentiviral vector for expression of miR-21 antagonist. Inhibition of mir-21 by its antagonist led to the cessation of tumor growth and the induction of apoptosis in vitro and in vivo (animal model) [93].
Potential drug triptolide acts on pancreatic tumor tissue as an inhibitor of cell proliferation and reduces the levels of the molecular chaperone HSP70. Rather than directly affecting HSP70 it causes increase of the levels of miR-142-3p. Ectopic expression of miR-142-3p in pancreatic tumors caused by the effect of water-soluble precursor triptolide (minnelide) in vivo reduces the expression of HSP70 by direct binding to the 3′UTR region of its transcript. Therefore the miR-142-3p reduces proliferation, induces cell death, and is useable as a proper target for pancreatic cancer therapies [94]. Another study has revealed a relation of miR-142-5p to the therapeutic response to gemcitabine and further states that this miRNA is an important predictive marker in patients treated with gemcitabine after tumor resection, when its higher levels indicate a longer survival [90]. Yet another therapy option comes from a possibility of recovery of function of miR-34a, a potent pro-apoptotic component involved in p53 mediated apoptosis, whose expression is reduced or lost in PDAC cells [29–31]. As shown by Ji et al. [95], restoration of miR-34a may substitute function of inactivated TP53 gene.
miR-10a and miR-146a play important roles in pancreatic cancer invasivity and metastasis and represent potential targets for antimetastatic therapies [47, 48]. It has been shown that miR-10a promotes the metastatic behavior of PC and that its expression is regulated by retinoids [48]. The use of retinoic acid receptor antagonists inhibits miR-10a expression and stops metastasis of PDAC cells [48]. In contrast, miR-146a suppresses invasion of pancreatic cancer cells but its expression is lowered in PC compared with normal pancreatic tissue. Finally, use of isoflavones or DIM (3,3′-diinodolylmethane), both nontoxic natural compounds increasing the expression of miR-146a, also presents a promising approach to block the invasivity and metastases [47].
9. Conclusion
Small noncoding RNA (microRNA and miRNA) is a new hope for improvement of poor prognosis of pancreatic cancer patients. The broad involvement in cellular mechanisms of cell cycle regulation, proper functioning of DNA repair, and apoptotic control as well as mechanisms of invasivity and metastasis positions miRNAs as potential biomarkers for clinical management of pancreatic cancer. Many of the above described miRNAs are now being tested as diagnostic or prognostic markers for use in routine clinical practice. The new instrumental development in the genomic analysis facilitates their further discovery and validation. Continuing research and better understanding of the principles and complex mechanisms of miRNA-associated gene expression control may bring new possibilities for anticancer therapy of this fatal disease.
Acknowledgment
This paper is supported by the Ministry of Health of the Czech Republic, Project no. NT 13638 and institutional support grant MO 1012.
Conflict of Interests
The authors of this paper declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Tereza Halkova has written the majority of the paper and has revised further versions according to reviewer's suggestions. Romana Cuperkova has prepared the first draft and did initial literature search. Lucie Benesova has revised further versions and contributed to detailed descriptions of the molecular pathways. Marek Minarik assisted in literature search and final wording of the paper.
References
- 1.Jemal A., Murray T., Samuels A., Ghafoor A., Ward E., Thun M. J. Cancer statistics 2003. CA Cancer Journal for Clinicians. 2003;53(1):5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 2.Hruban R. H., Adsay N. V., Albores-Saavedra J., et al. Pancreatic intraepithelial neoplasia (PanIN): a new nomenclature and classification system for pancreatic duct lesions. The American Journal of Surgical Pathology. 2001;25(5):579–586. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Zavoral M., Minarikova P., Zavada F., Salek C., Minarik M. Molecular biology of pancreatic cancer. World Journal of Gastroenterology. 2011;17(24):2897–2908. doi: 10.3748/wjg.v17.i24.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin L.-J., Asaoka Y., Tada M., et al. Integrated analysis of copy number alterations and loss of heterozygosity in human pancreatic cancer using a high-resolution, single nucleotide polymorphism array. Oncology. 2008;75(1-2):102–112. doi: 10.1159/000155813. [DOI] [PubMed] [Google Scholar]
- 5.Bakkevold K. E., Arnesjo B., Kambestad B. Carcinoma of the pancreas and papilla of Vater: presenting symptoms, signs, and diagnosis related to stage and tumour site. A prospective multicentre trial in 472 patients. Scandinavian Journal of Gastroenterology. 1992;27(4):317–325. doi: 10.3109/00365529209000081. [DOI] [PubMed] [Google Scholar]
- 6.Jiang X.-T., Tao H.-Q., Zou S.-C. Detection of serum tumor markers in the diagnosis and treatment of patients with pancreatic cancer. Hepatobiliary and Pancreatic Diseases International. 2004;3(3):464–468. [PubMed] [Google Scholar]
- 7.Tempero M. A., Behrman S., Ben-Josef E., et al. Pancreatic adenocarcinoma: Clinical Practice Guidelines in Oncology. Journal of the National Comprehensive Cancer Network. 2005;3(5):598–626. doi: 10.6004/jnccn.2005.0035. [DOI] [PubMed] [Google Scholar]
- 8.Burris H. A., III, Moore M. J., Andersen J., et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. Journal of Clinical Oncology. 1997;15(6):2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 9.Jemal A., Siegel R., Xu J., Ward E. Cancer statistics, 2010. CA: Cancer Journal for Clinicians. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 10.Croce C. M. Causes and consequences of microRNA dysregulation in cancer. Nature Reviews Genetics. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Negrini M., Nicoloso M. S., Calin G. A. MicroRNAs and cancer—new paradigms in molecular oncology. Current Opinion in Cell Biology. 2009;21(3):470–479. doi: 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Dalmay T., Edwards D. R. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25(46):6170–6175. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- 13.Salek C., Benesova L., Zavoral M., et al. Evaluation of clinical relevance of examining K-ras, p16 and p53 mutations along with allelic losses at 9p and 18q in EUS-guided fine needle aspiration samples of patients with chronic pancreatitis and pancreatic cancer. World Journal of Gastroenterology. 2007;13(27):3714–3720. doi: 10.3748/wjg.v13.i27.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salek C., Minarikova P., Benesova L., et al. Mutation status of K-ras, p53 and allelic losses at 9p and 18q are not prognostic markers in patients with pancreatic cancer. Anticancer Research. 2009;29(5):1803–1810. [PubMed] [Google Scholar]
- 15.Ambros V., Bartel B., Bartel D. P., et al. A uniform system for microRNA annotation. RNA. 2003;9(3):277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nana-Sinkam S. P., Fabbri M., Croce C. M. MicroRNAs in cancer: personalizing diagnosis and therapy. Annals of the New York Academy of Sciences. 2010;1210(1):25–33. doi: 10.1111/j.1749-6632.2010.05822.x. [DOI] [PubMed] [Google Scholar]
- 18.Park J.-K., Lee E. J., Esau C., Schmittgen T. D. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. 2009;38(7):e190–e199. doi: 10.1097/MPA.0b013e3181ba82e1. [DOI] [PubMed] [Google Scholar]
- 19.Zhao C., Zhang J., Zhang S., et al. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncology Reports. 2013;30(1):276–284. doi: 10.3892/or.2013.2420. [DOI] [PubMed] [Google Scholar]
- 20.Wu K., Hu G., He X., et al. MicroRNA-424-5p suppresses the expression of socs6 in pancreatic cancer. Pathology and Oncology Research. 2013;19(4):739–748. doi: 10.1007/s12253-013-9637-x. [DOI] [PubMed] [Google Scholar]
- 21.Bayle J., Letard S., Frank R., Dubreuil P., de Sepulveda P. Suppressor of cytokine signaling 6 associates with KIT and regulates KIT receptor signaling. The Journal of Biological Chemistry. 2004;279(13):12249–12259. doi: 10.1074/jbc.m313381200. [DOI] [PubMed] [Google Scholar]
- 22.Wang P., Chen L., Zhang J., et al. Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene. 2014;33(4):514–524. doi: 10.1038/onc.2012.598. [DOI] [PubMed] [Google Scholar]
- 23.Ridley A. J. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends in Cell Biology. 2006;16(10):522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Xu D., Wang Q., An Y., Xu L. MiR-203 regulates the proliferation, apoptosis and cell cycle progression of pancreatic cancer cells by targeting Survivin. Molecular Medicine Reports. 2013;8(2):379–384. doi: 10.3892/mmr.2013.1504. [DOI] [PubMed] [Google Scholar]
- 25.Hu Y., Ou Y., Wu K., Chen Y., Sun W. MiR-143 inhibits the metastasis of pancreatic cancer and an associated signaling pathway. Tumour Biology. 2012;33(6):1863–1870. doi: 10.1007/s13277-012-0446-8. [DOI] [PubMed] [Google Scholar]
- 26.Jiao L. R., Frampton A. E., Jacob J., et al. Micrornas targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0032068.e32068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins M. A., di Magliano M. P. Kras as a key oncogene and therapeutic target in pancreatic cancer. Frontiers in Physiology. 2014;4, article 407 doi: 10.3389/fphys.2013.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raver-Shapira N., Marciano E., Meiri E., et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Molecular Cell. 2007;26(5):731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 29.Chang T.-C., Wentzel E. A., Kent O. A., et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Molecular Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent O. A., Mullendore M., Wentzel E. A., et al. A resource for analysis of microRNA expression and function in pancreatic ductal adenocarcinoma cells. Cancer Biology and Therapy. 2009;8(21):2013–2024. doi: 10.4161/cbt.8.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habbe N., Koorstra J.-B. M., Mendell J. T., et al. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biology and Therapy. 2009;8(4):340–346. doi: 10.4161/cbt.8.4.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gironella M., Seux M., Xie M.-J., et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(41):16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farhana L., Dawson M. I., Murshed F., Das J. K., Rishi A. K., Fontana J. A. Upregulation of miR-150∗ and miR630 induces apoptosis in pancreatic cancer cells by targeting IGF-1R. PLoS ONE. 2013;8(5) doi: 10.1371/journal.pone.0061015.e61015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li A., Omura N., Hong S.-M., et al. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Research. 2010;70(13):5226–5237. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aigner K., Dampier B., Descovich L., et al. The transcription factor ZEB1 (δEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26(49):6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spaderna S., Schmalhofer O., Wahlbuhl M., et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Research. 2008;68(2):537–544. doi: 10.1158/0008-5472.can-07-5682. [DOI] [PubMed] [Google Scholar]
- 37.Vandewalle C., Comijn J., de Craene B., et al. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Research. 2005;33(20):6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bracken C. P., Gregory P. A., Kolesnikoff N., et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Research. 2008;68(19):7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 39.Burk U., Schubert J., Wellner U., et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Reports. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregory P. A., Bert A. G., Paterson E. L., et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biology. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 41.Park S.-M., Gaur A. B., Lengyel E., Peter M. E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & Development. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korpal M., Lee E. S., Hu G., Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. The Journal of Biological Chemistry. 2008;283(22):14910–14914. doi: 10.1074/jbc.c800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Y., Lu J., Li X., et al. MiR-200a inhibits epithelial-mesenchymal transition of pancreatic cancer stem cell. BMC Cancer. 2014;14(1, article 85) doi: 10.1186/1471-2407-14-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wellner U., Schubert J., Burk U. C., et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nature Cell Biology. 2009;11(12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 45.Miao L., Xiong X., Lin Y., et al. miR-203 inhibits tumor cell migration and invasion via caveolin-1 in pancreatic cancer cells. Oncology Letters. 2014;7(3):658–662. doi: 10.3892/ol.2014.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu A., Shao C., Jin G., et al. miR-208-induced epithelial to mesenchymal transition of pancreatic cancer cells promotes cell metastasis and invasion. Cell Biochemistry and Biophysics. 2014;69(2):341–346. doi: 10.1007/s12013-013-9805-3. [DOI] [PubMed] [Google Scholar]
- 47.Li Y., VandenBoom T. G., II, Wang Z., et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Research. 2010;70(4):1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss F. U., Marques I. J., Woltering J. M., et al. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137(6):2136.e7–2145.e7. doi: 10.1053/j.gastro.2009.08.065. [DOI] [PubMed] [Google Scholar]
- 49.Ohuchida K., Mizumoto K., Lin C., et al. MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Annals of Surgical Oncology. 2012;19(7):2394–2402. doi: 10.1245/s10434-012-2252-3. [DOI] [PubMed] [Google Scholar]
- 50.Pavlakis E., Papaconstantinou I., Gazouli M., et al. MicroRNA gene polymorphisms in pancreatic cancer. Pancreatology. 2013;13(3):273–278. doi: 10.1016/j.pan.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 51.Zhu Z., Gao W., Qian Z., Miao Y. Genetic variation of miRNA sequence in pancreatic cancer. Acta Biochimica et Biophysica Sinica (Shanghai) 2009;41(5):407–413. doi: 10.1093/abbs/gmp023. [DOI] [PubMed] [Google Scholar]
- 52.Lowenfels A. B., Maisonneuve P., Lankisch P. G. Chronic pancreatitis and other risk factors for pancreatic cancer. Gastroenterology Clinics of North America. 1999;28(3):673–685. doi: 10.1016/S0889-8553(05)70080-7. [DOI] [PubMed] [Google Scholar]
- 53.Eun J. L., Gusev Y., Jiang J., et al. Expression profiling identifies microRNA signature in pancreatic cancer. International Journal of Cancer. 2007;120(5):1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dillhoff M., Liu J., Frankel W., Croce C., Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. Journal of Gastrointestinal Surgery. 2008;12(12):2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J., Chen J., Chang P., et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prevention Research. 2009;2(9):807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bauer A. S., Keller A., Costello E., et al. Diagnosis of pancreatic ductal adenocarcinoma and chronic pancreatitis by measurement of microRNA abundance in blood and tissue. PloS ONE. 2012;7(4) doi: 10.1371/journal.pone.0034151.e34151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu R., Chen X., Du Y., et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clinical Chemistry. 2012;58(3):610–618. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- 58.Kong X., Du Y., Wang G., et al. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR196a could be a potential marker for poor prognosis. Digestive Diseases and Sciences. 2011;56(2):602–609. doi: 10.1007/s10620-010-1285-3. [DOI] [PubMed] [Google Scholar]
- 59.Bloomston M., Frankel W. L., Petrocca F., et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. Journal of the American Medical Association. 2007;297(17):1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 60.Liu J., Gao J., Du Y., et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. International Journal of Cancer. 2012;131(3):683–691. doi: 10.1002/ijc.26422. [DOI] [PubMed] [Google Scholar]
- 61.Jamieson N. B., Morran D. C., Morton J. P., et al. MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clinical Cancer Research. 2012;18(2):534–545. doi: 10.1158/1078-0432.CCR-11-0679. [DOI] [PubMed] [Google Scholar]
- 62.Preis M., Gardner T. B., Gordon S. R., et al. MicroRNA-10b expression correlates with response to neoadjuvant therapy and survival in pancreatic ductal adenocarcinoma. Clinical Cancer Research. 2011;17(17):5812–5821. doi: 10.1158/1078-0432.CCR-11-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park J. K., Henry J. C., Jiang J., et al. MiR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochemical and Biophysical Research Communications. 2011;406(4):518–523. doi: 10.1016/j.bbrc.2011.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang S., Hao J., Xie F., et al. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32(8):1183–1189. doi: 10.1093/carcin/bgr105. [DOI] [PubMed] [Google Scholar]
- 65.Yu S., Lu Z., Liu C., et al. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Research. 2010;70(14):6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 66.Vogt M., Munding J., Grüner M., et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Archiv. 2011;458(3):313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 67.Torrisani J., Bournet B., Du Rieu M. C., et al. Let-7 microRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression. Human Gene Therapy. 2009;20(8):831–844. doi: 10.1089/hum.2008.134. [DOI] [PubMed] [Google Scholar]
- 68.Ho A. S., Huang X., Cao H., et al. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Translational Oncology. 2010;3(2):109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang W.-S., Liu L.-X., Li G.-P., et al. Combined serum CA19-9 and miR-27a-3p in peripheral blood mononuclear cells to diagnose pancreatic cancer. Cancer Prevention Research. 2013;6(4):331–338. doi: 10.1158/1940-6207.CAPR-12-0307. [DOI] [PubMed] [Google Scholar]
- 70.Greither T., Grochola L. F., Udelnow A., Lautenschläger C., Würl P., Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. International Journal of Cancer. 2010;126(1):73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 71.Ikenaga N., Ohuchida K., Mizumoto K., et al. MicroRNA-203 expression as a new prognostic marker of pancreatic adenocarcinoma. Annals of Surgical Oncology. 2010;17(12):3120–3128. doi: 10.1245/s10434-010-1188-8. [DOI] [PubMed] [Google Scholar]
- 72.Szafranska A. E., Davison T. S., John J., et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26(30):4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 73.Schultz N. A., Werner J., Willenbrock H., et al. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Modern Pathology. 2012;25(12):1609–1622. doi: 10.1038/modpathol.2012.122. [DOI] [PubMed] [Google Scholar]
- 74.Szafranska A. E., Doleshal M., Edmunds H. S., et al. Analysis of microRNAs in pancreatic fine-needle aspirates can classify benign and malignant tissues. Clinical Chemistry. 2008;54(10):1716–1724. doi: 10.1373/clinchem.2008.109603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kawaguchi T., Komatsu S., Ichikawa D., et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. British Journal of Cancer. 2013;108(2):361–369. doi: 10.1038/bjc.2012.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ryu J. K., Hong S. M., Karikari C. A., Hruban R. H., Goggins M. G., Maitra A. Aberrant microRNA-155 expression is an early event in the multistep progression of pancreatic adenocarcinoma. Pancreatology. 2010;10(1):66–73. doi: 10.1159/000231984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu J., Li A., Hong S.-M., Hruban R. H., Goggins M. MicroRNA alterations of pancreatic intraepithelial neoplasias. Clinical Cancer Research. 2012;18(4):981–992. doi: 10.1158/1078-0432.CCR-11-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xue Y., Abou Tayoun A. N., Abo K. M., et al. MicroRNAs as diagnostic markers for pancreatic ductal adenocarcinoma and its precursor, pancreatic intraepithelial neoplasm. Cancer Genetics. 2013;206(6):217–221. doi: 10.1016/j.cancergen.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 79.يu Rieu M. C., Torrisani J., Selves J., et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clinical Chemistry. 2010;56(4):603–612. doi: 10.1373/clinchem.2009.137364. [DOI] [PubMed] [Google Scholar]
- 80.Hanoun N., Delpu Y., Suriawinata A. A., et al. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clinical Chemistry. 2010;56(7):1107–1118. doi: 10.1373/clinchem.2010.144709. [DOI] [PubMed] [Google Scholar]
- 81.Weber J. A., Baxter D. H., Zhang S., et al. The microRNA spectrum in 12 body fluids. Clinical Chemistry. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brase J. C., Wuttig D., Kuner R., Sültmann H. Serum microRNAs as non-invasive biomarkers for cancer. Molecular Cancer. 2010;9, article 306 doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mitchell P. S., Parkin R. K., Kroh E. M., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morimura R., Komatsu S., Ichikawa D., et al. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. British Journal of Cancer. 2011;105(11):1733–1740. doi: 10.1038/bjc.2011.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schultz N. A., Dehlendorff C., Jensen B. V., et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. Journal of the American Medical Association. 2014;311(4):392–404. doi: 10.1001/jama.2013.284664. [DOI] [PubMed] [Google Scholar]
- 86.Ganepola G. A., Rutledge J. R., Suman P., Yiengpruksawan A., Chang D. H. Novel blood-based microRNA biomarker panel for early diagnosis of pancreatic cancer. World Journal of Gastrointestinal Oncology. 2014;6(1):22–33. doi: 10.4251/wjgo.v6.i1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gao L., He S.-B., Li D.-C. Effects of miR-16 plus CA19-9 detections on pancreatic cancer diagnostic performance. Clinical Laboratory. 2014;60(1):73–77. doi: 10.7754/Clin.Lab.2013.121210. [DOI] [PubMed] [Google Scholar]
- 88.Humeau M., Torrisani J., Cordelier P. MiRNA in clinical practice: pancreatic cancer. Clinical Biochemistry. 2013;46(10-11):933–936. doi: 10.1016/j.clinbiochem.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 89.Ali S., Saleh H., Sethi S., Sarkar F. H., Philip P. A. MicroRNA profiling of diagnostic needle aspirates from patients with pancreatic cancer. British Journal of Cancer. 2012;107(8):1354–1360. doi: 10.1038/bjc.2012.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ohuchida K., Mizumoto K., Kayashima T., et al. MicroRNA expression as a predictive marker for gemcitabine response after surgical resection of pancreatic cancer. Annals of Surgical Oncology. 2011;18(8):2381–2387. doi: 10.1245/s10434-011-1602-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hwang J. H., Voortman J., Giovannetti E., et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS ONE. 2010;5(5) doi: 10.1371/journal.pone.0010630.e10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giovannetti E., Funel N., Peters G. J., et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Research. 2010;70(11):4528–4538. doi: 10.1158/0008-5472.can-09-4467. [DOI] [PubMed] [Google Scholar]
- 93.Sicard F., Gayral M., Lulka H., Buscail L., Cordelier P. Targeting miR-21 for the therapy of pancreatic cancer. Molecular Therapy. 2013;21(5):986–994. doi: 10.1038/mt.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.MacKenzie T. N., Mujumdar N., Banerjee S., et al. Triptolide induces the expression of miR-142-3p: a negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Molecular Cancer Therapeutics. 2013;12(7):1266–1275. doi: 10.1158/1535-7163.mct-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ji Q., Hao X., Zhang M., et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS ONE. 2009;4(8) doi: 10.1371/journal.pone.0006816.e6816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schultz N. A., Andersen K. K., Roslind A., Willenbrock H., Wøjdemann M., Johansen J. S. Prognostic microRNAs in cancer tissue from patients operated for pancreatic cancer—five microRNAs in a prognostic index. World Journal of Surgery. 2012;36(11):2699–2707. doi: 10.1007/s00268-012-1705-y. [DOI] [PubMed] [Google Scholar]
- 97.Singh S., Chitkara D., Kumar V., Behrman S. W., Mahato R. I. MiRNA profiling in pancreatic cancer and restoration of chemosensitivity. Cancer Letters. 2013;334(2):211–220. doi: 10.1016/j.canlet.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 98.Nakata K., Ohuchida K., Mizumoto K., et al. MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery. 2011;150(5):916–922. doi: 10.1016/j.surg.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 99.Papaconstantinou I. G., Manta A., Gazouli M., et al. Expression of micrornas in patients with pancreatic cancer and its prognostic significance. Pancreas. 2013;42(1):67–71. doi: 10.1097/mpa.0b013e3182592ba7. [DOI] [PubMed] [Google Scholar]
- 100.Yu J., Ohuchida K., Mizumoto K., et al. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Molecular Cancer. 2010;9, article 169 doi: 10.1186/1476-4598-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu Z., Xu Y., Du J., Tan J., Jiao H. Expression of microRNA-218 in human pancreatic ductal adenocarcinoma and its correlation with tumor progression and patient survival. Journal of Surgical Oncology. 2014;109(2):89–94. doi: 10.1002/jso.23475. [DOI] [PubMed] [Google Scholar]
- 102.Meyer S. U., Pfaffl M. W., Ulbrich S. E. Normalization strategies for microRNA profiling experiments: a ‘normal’ way to a hidden layer of complexity? Biotechnology Letters. 2010;32(12):1777–1788. doi: 10.1007/s10529-010-0380-z. [DOI] [PubMed] [Google Scholar]