Abstract
As human population density continues to increase exponentially, speeding the reduction and fragmentation of primate habitat, greater human-primate contact is inevitable, making higher rates of pathogen transmission likely. Anthropogenic effects are particularly evident in Madagascar, where a diversity of endemic lemur species are threatened by rapid habitat loss. Despite these risks, knowledge of how anthropogenic activities affect lemur exposure to pathogens is limited. To improve our understanding of this interplay, we non-invasively examined six species of wild lemurs in Ranomafana National Park for enteric bacterial pathogens commonly associated with diarrheal disease in human populations in Madagascar. Patterns of infection with Enterotoxigenic Escherichia coli, Shigella spp., Salmonella enterica, Vibrio cholerae, and Yersinia spp. (enterocolitica and pseudotuberculosis) were compared between lemurs inhabiting intact forest and lemurs inhabiting degraded habitat with frequent exposure to tourism and other human activity. Fecal samples acquired from humans, livestock, and rodents living near the degraded habitat were also screened for these bacteria. Remarkably, only lemurs living in disturbed areas of the park tested positive for these pathogens. Moreover, all of these pathogens were present in the human, livestock, and/or rodent populations. These data suggest that lemurs residing in forests altered or frequented by people, livestock, or peridomestic rodents, are at risk for infection by these diarrhea-causing enterobacteria and other similarly transmitted pathogens.
Keywords: Escherichia coli, health, Madagascar, Salmonella, Shigella, Vibrio, Yersinia
INTRODUCTION
Recent evidence of pathogen transmission to humans from wild primates [Calvignac-Spencer et al., 2012] and a greater recognition of the risk of human pathogen spread to free-ranging primates [Kondgen et al., 2008; Palacios et al., 2011; Schaumburg et al., 2012] have raised awareness of the potential impact of zoonoses on primate conservation and the health of both nonhuman primates and humans. As human population density continues to increase exponentially, speeding the fragmentation and reduction of primate habitat, greater human-primate contact is inevitable; making higher rates of pathogen transmission likely [Gillespie and Chapman, 2006; Gillespie et al., 2008; Young et al., 2013]. Despite this potential for zoonotic transmission, few studies have examined how anthropogenic disturbance affects the incidence of enteric bacterial pathogens in wild primates. However, a series of studies examining bacterial spread using non-pathogenic Escherichia coli as a model in Uganda suggest high potential for transmission. In these studies, wild primates from Kibale and Bwindi Impenetrable National Parks that had routine exposure to humans or were from fragmented forest regions, were colonized by bacteria that were genetically more similar to those carried by humans than the bacteria found in primates with minimal exposure to humans [Goldberg et al., 2007, 2008; Rwego et al., 2008].
Human disruption of natural ecosystems is particularly evident in Madagascar. This island country is home to 22 million people and at least 2% of global endemic plants and animals, while making up less than 0.4% of the Earth’s land surface [Myers et al., 2000]. Moreover, 90% of all species found on this island live entirely within forest ecosystems [Ganzhorn et al., 2001; Harper et al., 2007; IUCN, 2009]. Over the years this country has experienced widespread and dramatic changes to the original environment for agricultural purposes and resource extraction. These alterations have led to the fragmentation or clearing of nearly 90% of the native forest [Harper et al., 2007; Junge et al., 2011]. Consequently, the majority of Madagascar’s native species are at risk of extinction due to anthropogenic changes to the environment.
Lemurs, which are endemic to this island nation, are of particular importance as 91% of the known species are considered endangered; six of which are among the 25 most endangered primates in the world [IUCN, 2012; Mittermeier et al., 2012]. It has become imperative to understand if and how zoonotic pathogens may be moving between hosts, especially lemurs, which as non-human primates are susceptible to human pathogens [Nizeyi et al., 2001; Salyer et al., 2012]. Helminths, enteric protozoa, and enteric bacteria have been detected in wild and captive lemurs; some of these cases have been linked to human or rodent exposure [Junge and Louis, 2005; de Camps et al., 2008; Junge et al., 2008, 2011; Wright et al., 2009; Rasambainarivo et al., 2013].
We non-invasively examined six species of wild lemurs within Ranomafana National Park (RNP) as well as humans, livestock, and rodents from a village in close proximity to the disturbed habitat within the park. Lemurs inhabiting intact forest and lemurs exposed to humans or human-related activities (e.g., free-ranging cattle) living in degraded habitat were compared. These data were collected in order to improve our understanding of the natural occurrence of enteric bacterial pathogens, as well as the potential for anthropogenic introduction of these bacteria, commonly associated with diarrheal disease in human populations in Madagascar to rainforest-dwelling lemurs.
METHODS
Study Site
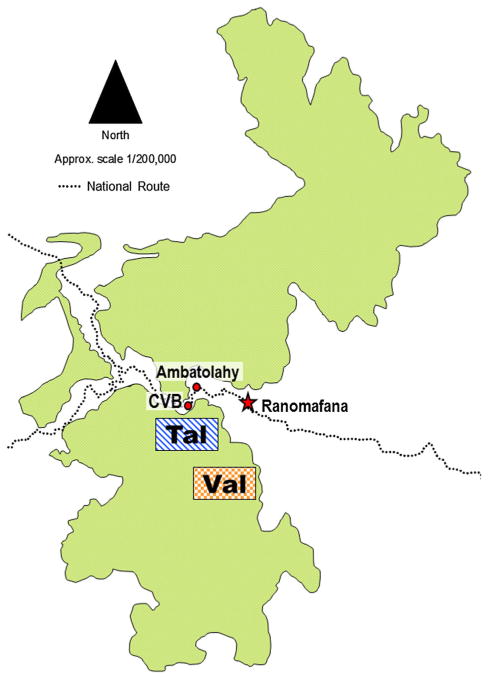
Ranomafana National Park (RNP) in Southeast Madagascar (21°02′–21°25′S and 47°18′–47°37′) is a 43,500 ha World Heritage Site and one of the primary long-term ecological research sites in Madagascar. This study focuses on two of the park’s established trail systems, located 8.5 km from each other: Talatakely (Tal) and Valohoaka (Val) (Fig. 1). These sites differ substantially in regards to both historical and current human use. Historically, Tal was subjected to a high degree of habitat disturbance, including the presence of a small settlement and market in the 1930s and 1940s and intensive “selective” logging by timber exploiters from 1986 to 1989 [Wright, 1997]. Today, Tal is the major tourist thoroughfare in RNP, containing a trail network of over 30 km and hosting over 12,000 tourists per year. In contrast, Val has historically been subject to neither human settlement nor commercial logging [Balko & Underwood, 2005] and remains a minimally traveled portion of intact rainforest today [Wright et al., 2009].
Fig. 1.
Ranomafana National Park, with study sites labeled. Tal is the disturbed forest site, Val is undisturbed forest. CVB indicates the Centre ValBio research station. The town of Ranomafana is indicated to enhance spatial perspective.
The Centre ValBio (CVB) is a permanent research station just outside RNP and within 1 km of Tal (Fig. 1). At the time of this study, the CVB complex consisted of a multipurpose building (office, research, and cafeteria space) and a campsite, regularly used by students and scientists, located in an expanse of secondary forest adjacent to the CVB property.
The village of Ambatolahy (Fig. 1), population 256, is located directly across the Namorona River from Tal and is the village where many of the park’s tourist guides and research technicians live [Bublitz et al., 2014]. Residents of Ambatolahy keep livestock included peridomestic rodents (Mus musculus and Rattus rattus), cattle (Bos indicus), pigs (Sus domesticus), and humans.
Sample Collection
In June and July 2011, fecal samples were collected noninvasive from lemurs living in RNP and the forest surrounding CVB. Twenty-four samples from distinct individual lemurs were collected: 11 from Val and 13 from Tal or CVB. The species represented included: Avahi laniger, Eulemur reinvented, Hap lemur aurous, Microcopies Rufus, Propithecus edwardsi, and Prolemur simus (Table I). Field assistants collected only fecal matter they witnessed being passed from known animals that were tracked using devices placed on the animals from prior research projects. These samples were placed in sterile tubes with gloved hands, taking care not to collect portions that had touched the ground.
TABLE I.
Species and Numbers of Lemurs Sampled by Location to Examine Enteric Infections in Ranomafana National Park, Madagascar
| Species | Total (n) | No. animals by site
|
Positive PCR (n) | |
|---|---|---|---|---|
| Tal/CVB | Val | |||
| Avahi laniger | 1 | 1 | 0 | 0 |
| Eulemur reinvented | 5 | 0 | 5 | 0 |
| Hap lemur aurous | 2 | 2 | 0 | 2/2 |
| Microcopies Rufus | 3 | 3 | 0 | 2/3 |
| Propithecus edwardsi | 9 | 3 | 6 | 0 |
| Prolemur simus | 4 | 4 | 0 | 4/4 |
| 24 | 13 | 11 | 8/24 | |
A cluster sampling method was used for the village: 10 households were chosen and every person inhabiting those households was asked to provide a fecal specimen (n = 47). Participants were chosen independent of age, sex, or symptoms, but some households were selected based on livestock ownership. Concurrently, domesticated animals of participants were sampled (n = 18) and baited rodent live-traps were set inside participant homes overnight. The next morning, fecal samples were collected from trapped rodents (n = 33). Human volunteers were instructed to collect fecal samples in sealable plastic bags that were retrieved the same or next day. Livestock (cattle, Bos indicus and pigs, Sus domesticus) were sampled by rectal fecal collection using a non-sterile latex glove or from the ground if defecation was observed. For the latter, only fecal material that had not contacted the ground was collected. All samples were preserved immediately upon collection in the following fashion: approximately 1.0 ml of feces from each specimen was homogenized with an equal volume of RNAlater® (Ambion, Life Technologies, Grand Island, NY). Samples were then transported within 12 hr to the Centre ValBio where they were stored at −20°C until transport to the United States.
Activities in this project complied with protocols approved by Stony Brook University’s Animal Care and Use Committee, and also adhered to all research requirements of Madagascar and the American Society of Primatologists principles for the ethical treatment of primates. All human-related protocols were reviewed and approved by the Ministry of Health of the government of Madagascar and the Stony Brook University Internal Review Board. Informed consent of human participants was obtained before specimen collection.
Molecular Methods
DNA was extracted from fecal specimens preserved in RNAlater® using the FastDNA® SPIN Kit for Soil (MP Biomedicals, LLC, Solon, OH), following the kit-recommended procedures. Using PCR, we screened the fecal samples for species-specific genes in the case of Enterotoxigenic E. coli, Salmonella enterica, and Vibrio cholerae, and genus-specific genes for Yersinia spp. (enterocolitica and pseduotuberculosis), Shigella spp. (flexneri and dysenteriae), and S. enterica (Table II). So as to maximize our small DNA samples we probed for genes encoding YadA, IpaH, and InvA for Yersinia, Shigella, and Salmonella respectively, in order to detect various pathogenic forms of these bacteria. In the case of Yersinia, we chose the gene yadA as it is similar in enterocolitica, and pseduotuberculosis species and could be used to screen for both simultaneously [Thoerner et al., 2003]. The yadA primers should generate a product of 849 bp with Y. enterocolitica serogroup 03/09 strains, a product of 759 bp with Y. enterocolitica serogroup 08 strains, and 681 bp product with Yersinia pseudotuberculosis (including the positive control strain used). Likewise, the invA gene in S. enterica was used for its conserved nature across serovars [Rahn et al., 1992]. A portion of the ipaH gene was amplified to detect Shigella flexneri and dysenteriae [Wang et al., 1997].
TABLE II.
Positive Control Bacteria Strains, Target Genes, and Primers* Used to Examine Infections in Lemurs, Humans, Livestock, and Rodents in and Around Ranomafana National Park, Madagascar
| Genus and species (ATCC #) | Target gene | PCR primers (5′–3′) | Product size |
|---|---|---|---|
| Enterotoxigenic E. coli serotype O78:H11 (35401) | Enterotoxin (LT) gene | F: GAGACCGGTATTACAGAAATC R: GAGGTGCATGATGAATCCAG |
117 bp |
| Shigella flexneri serotype 2b (12022) | ipah | F: CTTGACCGCCTTTCCGATAC R: CAGCCACCCTCTGAGAGTA |
610 bp |
| Salmonella enterica serovar Typhimurium (14028) | invA | F: TATCGCCACGTTCGGGCAA R: TCGCACCGTCAAAGGAACC |
275 bp |
| Vibrio cholerae (14035) | ctxA | F: GGCAGATTCTAGACCTCCT R: TCGATGATCTTGGAGCATTC |
563 bp |
| Yersinia pseudotuburculosis serogroup O1 (32777) | yadA | F: CTTCAGATACTGGTGTCGCTGT R: ATGCCTGACTAGAGCGATATCC |
681 bp (849a, 751b) |
All primer sequences obtained from Wang et al. [1997] except Yersinia obtained from Thoerner et al. [2003].
Product size with Y. enterocolitica serogroup 03 or 09 strains.
Product size with Y. enterocolitica serogroup 08 strains.
V. cholerae, E. coli, and S. flexneri strains were obtained from American Type Culture Collection (ATCC, Manassas, VA) for use as positive controls. The 32777 strain in the Bliska Laboratory collection was used as a positive control for Y. pseudotuberculosis. The S. enterica serovar 14028 positive control strain was obtained from the laboratory of Dr. Adrianus van der Velden (Both: Stony Brook University, Stony Brook, NY). All positive control strains are listed in Table II. Genomic DNA was isolated from each of the control strains using a DNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. PCR was conducted on 2.0 μl of DNA sample using 0.5 μmol of each primer (Table II) in 25 μl of Platinum PCR mix (Invitrogen, Life Technologies, Grand Island, NY). The amplification setting was as previously described less the “Transition S-9” setting [Wang et al., 1997].
Statistical Methods
Fisher’s exact text using Prism 6 (Graphpad, La Jolla, CA) was used to analyze statistical significance of associations, relative risk, and confidence intervals (CI). Fisher’s exact is a non-parametric test for independence, useful for small sample sizes. Fisher’s exact test assesses whether there is a significant association between a treatment and an outcome using a 2 × 2 contingency table and a two-tailed P-value. The relative risk is calculated as a difference of proportions for two treatments and a given outcome from the 2 × 2 contingency table. The relative risk describes whether a significant proportion of a given population is at risk for a defined outcome.
RESULTS
Of the 24 lemurs sampled, 33% of the individuals tested positive for at least one of the enterobacteria. Of the lemurs testing positive for at least one of the bacteria, E. coli was the most common infectious agent detected (33%) followed by V. cholerae (11%), Salmonella and Yersinia spp. (7.4%), and Shigella spp. (3.7%) (Table III). Importantly, there was a striking difference in the prevalence of enteric infection between lemur samples collected from intact portions of RNP (0%, P = 0.0011, CI = 0.00–0.26) and those collected in disturbed and/or heavily trafficked region of the park (60%, P = 0.0011, CI = 0.32–0.84). Incidence rates for each individual pathogen nearly doubled when looking at only lemurs from CVB and the disturbed region of RNP (60%) versus all of the individuals (33%). As such, there was a significant negative association between lemurs found in intact habitat and these pathogens (P <0.0001, Relative Risk = 0, CI = Infinity).
TABLE III.
Incidence of Entererobacterium in Lemurs Found in Disturbed Versus Intact Habitat in Ranomafana National Park, Madagascar
| Pathogen | Disturbed habitat
|
Intact habitat
|
Total
|
|||
|---|---|---|---|---|---|---|
| +/Total | Prevalence | +/Total | Prevalence | +/Total | Prevalence | |
| Enterotoxigenic E. coli | 8/13 | 0.61 | 0/11 | 0.00 | 8/24 | 0.33 |
| Shigella spp. | 1/13 | 0.08 | 0/11 | 0.00 | 1/24 | 0.04 |
| S. enterica | 2/13 | 0.15 | 0/11 | 0.00 | 2/24 | 0.08 |
| V. cholerae | 2/13 | 0.15 | 0/11 | 0.00 | 2/24 | 0.08 |
| Yersinia spp. | 2/13 | 0.15 | 0/11 | 0.00 | 2/24 | 0.08 |
Of the humans screened, 91% carried at least one of the target pathogens. Individually, Enterotoxigenic E. coli and Shigella spp. were the most prevalent in the human population near RNP at 43% and 64%, respectively (Table IV). Rodents trapped in these villages were also found to carry all target pathogens with the exceptions of Yersinia spp. and V. cholerae (Table IV). S. enterica and Enterotoxigenic E. coli were the most prevalent in rodents sampled at 18% and 30%, respectively. Lastly, there was a relatively low prevalence of these bacteria in the livestock tested (as compared to the sampled humans and rodents). Only Enterotoxigenic E. coli was found in livestock with a prevalence of 14% (Table IV). When combined, Enterotoxigenic E. coli was the most prevalent pathogen detected in the Ambatolahy with a 32% carriage rate.
TABLE IV.
Incidence of Each Enterobacterium Species in Humans, Livestock, and Rodents Near Disturbed Habitat Within Ranomafana National Park, Madagascar
| Pathogen | Human
|
Livestock
|
Rodent
|
|||
|---|---|---|---|---|---|---|
| +/Total | Prevalence | +/Total | Prevalence | +/Total | Prevalence | |
| Enterotoxigenic E. coli | 20/47 | 0.43 | 4/29 | 0.14 | 10/33 | 0.30 |
| Shigella spp. | 30/47 | 0.64 | 0/29 | 0.00 | 4/33 | 0.12 |
| S. enterica | 15/47 | 0.32 | 0/29 | 0.00 | 6/33 | 0.18 |
| V. cholerae | 1/47 | 0.02 | 0/29 | 0.00 | 0/33 | 0.00 |
| Yersinia spp. | 13/47 | 0.28 | 0/29 | 0.00 | 0/33 | 0.00 |
| Total | 43/47 | 0.81 | 4/18 | 0.22 | 12/33 | 0.36 |
DISCUSSION
Enteric bacterial diseases are a leading cause of illness and death in the developing world. A recent 3-year study evaluating over 9,000 children with moderate to severe diarrhea in Africa and Asia found that Shigella and Enterotoxigenic E. coli were two of the four most common causes of infection while V. cholerae was also identified as a significant cause of diarrhea at some sites [Kotloff et al., 2013]. In Madagascar, diarrheal disease is of exceptional concern for both children and adults [Johansson & Wardlaw, 2009; Randremanana et al., 2012; WHO/DFID-AHP, 2013; WHO/UNICEF, 2000]. Humans near RNP demonstrate high prevalence of enteric bacterial pathogens at 77% [Bublitz et al., 2014]. The close phylogenetic relationship between humans and lemurs, as well as all non-human primates, enhances their susceptibility to many human infectious diseases [Junge et al., 2011; Nunn, 2012; Rasambainarivo et al., 2013; Salyer et al., 2012]. This relatedness, coupled with the exponential expansion of human populations and human activities within lemur habitats, has resulted in a high potential for pathogen exchange [Ganzhorn et al., 2001; Harper et al., 2007; Junge et al., 2011]. Moreover, recent work highlights enhanced risk for lemurs due to climate-mediated shifts in pathogen distribution [Barrett et al., 2013]. Using current climate shifts in Madagascar and projected climate data, Barrett et al. predicted an expansion in the range of parasites typically associated with lemurs and detrimental to their health. If global climate change results in shifting the range of populations from microbes to mammals, habitat overlaps that did not previously exist have the potential to expose any number of plants and animals to novel pathogens.
We tested lemurs for the presence of five enterobacteria that are known to cause severe disease in humans. We found that 33% of the lemurs sampled were positive for at least one enterobacterium; the most common being Enterotoxigenic E. coli, followed by V. cholerae, Salmonella and Yersinia spp., and S. flexneri (Table II). Strikingly, none of the lemurs found in Val, the undisturbed portion of RNP, carried any of the pathogens, meaning that the prevalence rate was 60% among lemurs in the disturbed sites, Tal and CVB. A similar pattern was seen when examining gastrointestinal helminthes and protozoa of lemurs in RNP [Rasambainarivo et al., 2013; Wright et al., 2009]. In these previous studies, lemurs in intact zones of RNP were not infected, while those in the disturbed regions were. These data complement work in Northeast Madagascar where Indri indri in degraded forest had significantly compromised health compared to those living in more intact areas, suggesting that lemurs in disturbed habitats may be prone to disease due to increased physiological stress [Junge et al., 2011].
Chronic physiological stress in humans is associated with a suppressed immune response and thus, increased susceptibility to infection and cancer [Dhabhar et al., 2012; Schneiderman et al., 2005]. Likewise, little red flying foxes experiencing nutritional stress are more likely to be seropositive for Hendra virus [Plowright et al., 2008]. If lemurs are under constant stress from food scarcity, human interaction, or increased pressure from predators due to loss of intact habitat, their immune systems could be compromised, thus allowing a greater frequency of successful infection. Of the six lemur species examined here, only one occurs in both the disturbed and intact study sites (P. edwardsi). In response to habitat disturbance, sifakas living in the disturbed forest have altered their diet; decreasing dietary diversity and relying more heavily on non-tree plant foods [Arrigo-Nelson, 2006]. Furthermore, disturbance has limited the ability of sifakas to consume preferred food types, forcing them to consume leaves instead of fruits and seeds and creating a discrepancy in the nutrient intake of sifakas within the pristine and disturbed forest habitats that peaks during the austral winter—the most climatologically and reproductively harsh season of the year. A between-site comparison revealed that adult females living in intact forest weighed significantly more than females living in the disturbed forest [Arrigo-Nelson, 2006]. If habitat is further fragmented, it is probable that population density will increase, creating more opportunities for interaction among species, more dynamic dietary and nutritional disparities, and a greater chance for the transmission of pathogens [Debinski and Holt, 2001].
Interestingly, we found a 32% carriage of Enterotoxigenic E. coli among humans, livestock, and rodents from the same region. In fact, E. coli was the most prevalent bacterium detected in the livestock and rodent populations and the second most prevalent in the human population (Table IV). These data suggest that local human population and associated livestock or peridomestic rodents could be a source of infection for lemurs. Moreover, all of the bacteria species found in the lemur samples were also detected in samples from humans, livestock, and/or rodents from the surrounding area (Table IV). Recent work in Kirindy Forest Reserve in Madagascar found that 72% of wild lemurs sampled carry a lineage of Staphylococcus auerus found to be prevalent in humans in other parts of Africa, indicating the possibility for transmission of these strains both to, and from, humans [Schaumburg et al., 2013]. Future studies should include sequencing of these strains to determine if those detected in these various populations are the same.
Another point to consider is the fitness of individual lemur species. As only one of the six species examined (P. edwardsi) was sampled at both forest sites, we are currently unable to discern whether certain species are simply more susceptible to these pathogens or are simply not exposed to these bacteria at the frequency of the lemurs found in Tal and CVB. These are important areas of study to be pursued in the future. Likewise, we sampled only one lemur species from CVB and thus cannot draw conclusions about researchers versus tourists as a more likely source of these bacteria. However, the aforementioned sequencing work would shed light on this point.
Combined with our results, these studies all suggest a link between pathogen transmission to lemurs and exposure to humans or human practices (e.g. agriculture). It remains to be determined if the prevalence of these pathogens is due to direct contact with humans or if habitat fragmentation due to resource extraction and agriculture are allowing greater exposure of lemurs to other infected reservoirs (i.e., rodents or livestock). Previous studies found that pathogen diversity was higher for primates in disturbed versus intact habitats [Gillespie and Chapman, 2006, 2008; Gillespie et al., 2005; Sa et al., 2013]. Furthermore, as forests are fragmented, lemur population densities will likely increase, at least initially [Debinski and Holt, 2001]. Host density is of central importance to infection rates in directly transmitted pathogens, and intraspecific studies have demonstrated that host density is positively associated with pathogen prevalence and diversity [Gillespie and Chapman, 2006]. Thus, interactive effects of disturbance and altered host densities may facilitate pathogen transmission.
These findings are of special importance given that 91% of lemur species are endangered, six of which are among the 25 most endangered primates in the world [IUCN, 2012; Mittermeier et al., 2012]. Previous studies modeling disease transmission among humans, livestock, and non-human primates indicate that there could be major population declines of non- human primates, such as lemurs, resulting from gastrointestinal infections [Nunn, 2012]. Lemur species that regularly consume soil may be particularly at risk for infection via environmental contamination [Wright et al., 2009]. The fragmented habitats of Madagascar coupled with small population sizes for many lemur species leaves them particularly susceptible to disease events [Daszak et al., 2000; Junge and Sauther, 2007]. The interplay between humans and their environment is complicated. As was highlighted in a recent study of western lowland gorillas, it is difficult to determine if pathogens have a long history in wild populations or have been recently introduced by humans [Sak et al., 2013]. However, that does not diminish the need to understand and evaluate where we may be able to mitigate the transference of infectious agents to, and from, wildlife.
Emerging infectious diseases continue to be a critical threat to public health across the globe. With ease of travel as well as increased populations and demands for resources, the spaces for humans and wildlife are increasingly overlapping. A situation like this creates opportunity for novel pathogens to be spread both to, and from, wildlife. While the “One Health” approach in predicting and controlling disease outbreaks is increasingly gaining credence [Rabinowitz et al., 2013], there remains a pressing need for further understanding of how anthropogenic changes to the environment influence zoonotic disease transmission. Only then will we be able to implement proper measures for conservation and public health.
Acknowledgments
We are grateful for logistical and infrastructural support from MICET, particularly director Benjamin Andriamihaja, the administration and support personnel of the Centre ValBio, Madagascar National Parks, Ian Fried, and Emilie Redwood. We would also like to thank Dr. Martha Furie and Dr. Jorge Benach for the generous use of their laboratory space at Stony Brook University and for helping to make this work possible. This study was supported by the Jim and Robin Herrnstein Foundation, Stony Brook University, and the Emory University Global Health Institute. Research reported in this publication that was performed by DeAnna Bublitz was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number T32AI007539.
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Arrigo-Nelson SJ. PhD Thesis. Stony Brook, NY: Stony Brook University; 2006. The Impact of Habitat Disturbance on the Feeding Ecology of the Milne-Edwards’ Sifaka (Propithecus edwardsi) in Ranomafana National Park, Madagascar. [Google Scholar]
- Balko EA, Underwood HB. Effects of forest structure and composition on food availability for Varecia variegata at Ranomafana National Park, Madagascar. American Journal of Primatology. 2005;66:45–70. doi: 10.1002/ajp.20127. [DOI] [PubMed] [Google Scholar]
- Barrett MA, Brown JL, Junge RE, Yoder AD. Climate change, predictive modeling and lemur health: assessing impacts of changing climate on health and conservation in Madagascar. Biological Consesrvation. 2013;157:409–422. [Google Scholar]
- Bublitz DC, Wright PC, Bodager JR, et al. Epidemiology of pathogenic enterobacteria in humans, livestock, and peridomestic rodents in rural Madagascar. PLoS ONE. 2014;9:e101456. doi: 10.1371/journal.pone.0101456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvignac-Spencer S, Leendertz SA, Gillespie TR, Leendertz FH. Wild great apes as sentinels and sources of infectious disease. Clinical Microbiology and Infection. 2012;18:521–527. doi: 10.1111/j.1469-0691.2012.03816.x. [DOI] [PubMed] [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- de Camps S, Dubey JP, Saville WJ. Seroepidemiology of Toxoplasma gondii in zoo animals in selected zoos in the midwestern United States. Journal of Parasitology. 2008;94:648–653. doi: 10.1645/GE-1453.1. [DOI] [PubMed] [Google Scholar]
- Debinski DM, Holt RD. A survey and overview of habitat fragmentation experiments. Conservation Biology. 2001;14:342–355. [Google Scholar]
- Dhabhar FS, Saul AN, Holmes TH, et al. High-anxious individuals show increased chronic stress burden, decreased protective immunity, and increased cancer progression in a mouse model of squamous cell carcinoma. PLoS ONE. 2012;7:e33069. doi: 10.1371/journal.pone.0033069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganzhorn JU, Lowery PP, II, Schatz GE, Sommer S. The biodiversity of Madagascar: one of the world’s hottest hotspots on its way out. Oryx. 2001;35:346–348. [Google Scholar]
- Gillespie TR, Chapman CA. Prediction of parasite infection dynamics in primate metapopulations based on attributes of forest fragmentation. Conservation Biology. 2006;20:441–448. doi: 10.1111/j.1523-1739.2006.00290.x. [DOI] [PubMed] [Google Scholar]
- Gillespie TR, Chapman CA, Greiner EC. Effects of logging on gastrointestinal parasite infections and infection risk in African primates. Journal of Applied Ecology. 2005;42:699–707. [Google Scholar]
- Gillespie TR, Chapman CA. Forest fragmentation, the decline of an endangered primate, and changes in host- parasite interactions relative to an unfragmented forest. American Journal of Primatology. 2008;70:222–230. doi: 10.1002/ajp.20475. [DOI] [PubMed] [Google Scholar]
- Gillespie TR, Nunn CL, Leendertz FH. Integrative approaches to the study of primate infectious disease: implications for biodiversity conservation and global health. Yearbook of Physical Anthropology. 2008;51:53–69. doi: 10.1002/ajpa.20949. [DOI] [PubMed] [Google Scholar]
- Goldberg TL, Gillespie TR, Rwego IB, Estoff EL, Chapman CA. Anthropogenic disturbance promotes bacterial trasnimission among primates, humans, and livestock across a fragmented forest landscape. Emerging Infectious Diseases. 2008;14:1375–1382. doi: 10.3201/eid1409.071196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg TL, Gillespie TR, Rwego IB, et al. Patterns of gastrointestinal bacterial exchange between chimpanzees and humans involved in research and tourism in western Uganda. Biological Conservation. 2007;135:511–517. [Google Scholar]
- Harper GJ, Steininger MK, Tucker CJ, Juhn D, Hawkins F. Fifty years of deforestation and forest fragmentation in Madagascar. Environmental Conservation. 2007;34:325–333. [Google Scholar]
- IUCN. IUCN statement on Madagascar. 2009 http://www.iucn.org/?2995/IUCN-statement-on-Madagascar. Downloaded on 15 July 2013.
- IUCN. Primates in peril - conservationists reveal the world’s 25 most endanhgered primates. 2012 http://www.iucn.org/?11259/Primates-in-peril-conservationists-reveal-the-worlds-25-most-endangered-primates. Downloaded on 20 August 2013.
- Johansson EW, Wardlaw T. Diarrhoea: why children are still dying and what can be done. 2009 doi: 10.1016/S0140-6736(09)61798-0. http://whqlibdoc.who.int/publications/2009/9789241598415_eng.pdf. Downloaded on 20 December 2013. [DOI] [PubMed]
- Junge RE, Barrett MA, Yoder AD. Effects of anthropogenic disturbance on indri (Indri indri) health in Madagascar. American Journal of Primatology. 2011;73:632–642. doi: 10.1002/ajp.20938. [DOI] [PubMed] [Google Scholar]
- Junge RE, Dutton CJ, Knightly F, et al. Comparison of biomedical evaluation for white-fronted brown lemurs (Eulemur fulvus albifrons) from four sites in Madagascar. Journal of Zoo and Wildlife Medicine. 2008;39:567–575. doi: 10.1638/2007-0137.1. [DOI] [PubMed] [Google Scholar]
- Junge RE, Louis EE. Biomedical evaluation of two sympatric lemur species (Propithecus verreauxi decking and Eulemur fulvus Rufus) in Tsiombokibo Classified Forest, Madagascar. Journal of Zoo and Wildlife Medicine. 2005;36:581–589. doi: 10.1638/05-025.1. [DOI] [PubMed] [Google Scholar]
- Junge RE, Sauther ML. Overview on the health and disease ecology of wild lemurs: conservation implications. In: Gould L, Sauther ML, editors. Lemurs. New York, NY: Springer; 2007. pp. 423–440. [Google Scholar]
- Kondgen S, Kuhl H, N’Goran PK, et al. Pandemic human viruses cause decline of endangered great apes. Current Biology. 2008;18:260–264. doi: 10.1016/j.cub.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Mittermeier RA, Schwitzer C, Rylands AB, et al. Primates in Peril: The World’s 25 Most Endangered Primates 2012–2014. Arlington, VA: IUCN/SSC Primate Specialist Group International Primatological Society, Conservation International, Bristol Conservation and Science Foundation; 2012. [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GA, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Nizeyi JB, Innocent RB, Erume J, et al. Campylobacteriosis, salmonellosis, and shigellosis in free-ranging human-habituated mountain gorillas of Uganda. Journal of Wildlife Diseases. 2001;37:239–244. doi: 10.7589/0090-3558-37.2.239. [DOI] [PubMed] [Google Scholar]
- Nunn CL. Primate disease ecology in comparative and theoretical perspective. American Journal of Primatology. 2012;74:497–509. doi: 10.1002/ajp.21986. [DOI] [PubMed] [Google Scholar]
- Palacios G, Lowenstine LJ, Cranfield MR, et al. Human metapneumovirus infection in wild mountain gorillas, Rwanda. Emerging Infectious Diseases. 2011;17:711–713. doi: 10.3201/eid1704.100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, Field HE, Smith C, et al. Reproduction and nutritional stress are risk factors for Hendra virus infection in little red flying foxes (Pteropus scapulatus) Proceedings of the Royal Society of B. 2008;275:861–869. doi: 10.1098/rspb.2007.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz PM, Kock R, Kachani M, et al. Toward proof of concept of a One Health approach to disease prediction and control. Emerging Infectious Pisease. 2013 doi: 10.3201/eid1912.130265. http://dx.doi.org/10.3201/eid1912.130265. Downloaded on 25 November 2013. [DOI] [PMC free article] [PubMed]
- Rahn K, De Grandis SA, Clarke RC, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Molecular and Cellular Probes. 1992;6:271–279. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- Randremanana R, Randrianirina F, Gousseff M, et al. Case–control study of the etiology of infant diarrheal disease in 14 districts in Madagascar. PLoS ONE. 2012;7:e44533. doi: 10.1371/journal.pone.0044533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasambainarivo FT, Gillespie TR, Wright PC, et al. Survey of Giardia and Cryptosporidium in lemurs from the Ranomafana National Park, Madagascar. Journal of Wildlife Diseases. 2013;49:741–743. doi: 10.7589/2012-10-264. [DOI] [PubMed] [Google Scholar]
- Rwego IB, Isabirye-Basuta G, Gillespie TR, Goldberg TL. Gastrointestinal bacterial transmission among humans, mountain gorillas, and domestic livestock in Bwindi Impenetrable National Park, Uganda. Conservation Biology. 2008;22:1600–1607. doi: 10.1111/j.1523-1739.2008.01018.x. [DOI] [PubMed] [Google Scholar]
- Sa RM, Petrasova J, Pomajbikova K, et al. Gastrointestinal symbionts of chimpanzees in Cantanhez National Park, Guinea-Bissau with respect to habitat fragmentation. American Journal of Primatology. 2013;75:1032–1041. doi: 10.1002/ajp.22170. [DOI] [PubMed] [Google Scholar]
- Sak B, Petrzelkova KJ, Kvetonova D, et al. Long-term monitoring of Microsporidia, Cryptosporidium and Giardia infections in western lowland gorillas (Gorilla gorilla gorilla) at different stages of habituation in Dzanga Sangha protected areas, Central African Republic. PLoS ONE. 2013;8:e71840. doi: 10.1371/journal.pone.0071840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyer SJ, Gillespie TR, Rwego IB, Chapman CA, Goldberg TL. Epidemiology and molecular relationships of Cryptosporidium spp. in people, primates, and livestock from Western Uganda. PLoS Neglected Tropical Diseases. 2012;6:e1597. doi: 10.1371/journal.pntd.0001597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumburg F, Mugisha L, Kappeller P, et al. Evaluation of non-invasive biological samples to monitor Staphylococcus aurous colonization in great apes and lemurs. PLoS ONE. 2013;8:e78046. doi: 10.1371/journal.pone.0078046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumburg F, Mugisha L, Peck B, et al. Drug-resistant human Staphylococcus aurous in sanctuary apes pose a threat to endangered wild ape populations. American Journal of Primatology. 2012;74:1071–1075. doi: 10.1002/ajp.22067. [DOI] [PubMed] [Google Scholar]
- Schneiderman N, Ironson G, Siegel SD. Stress and health: psychological, behavioral, and biological determinants. Annual Review of Clinical Psychology. 2005;1:607–628. doi: 10.1146/annurev.clinpsy.1.102803.144141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoerner P, Bin Kingombe CI, Bogli-Stuber K, et al. PCR detection of virulence genes in Yersinia enterocolitica and Yersinia pseudotuberculosis and investigation of virulence gene distribution. Applied Environmental Microbiology. 2003;69:1810–1816. doi: 10.1128/AEM.69.3.1810-1816.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RF, Cao WW, Cerniglia CE. A universal protocol for PCR detection of 13 species of foodborne pathogens in foods. Journal of Applied Microbiology. 1997;83:727–736. doi: 10.1046/j.1365-2672.1997.00300.x. [DOI] [PubMed] [Google Scholar]
- WHO/DFID-AHP. Madagascar: health profile. 2013 http://www.who.int/gho/countries/mdg.pdf. Downloaded on 06 July 2013.
- WHO/UNICEF. Global water supply and sanitation assessment 2000 report. Switzerland: WHO/UNICEF; 2000. www.who.int/water_sanitation_health/monitoring/jmp2000.pdf. Downloaded on 06 July 2013. [Google Scholar]
- Wright PC. The future of biodiversity in Madagascar: a view from Ranomafana National Park. In: Patterson BD, Goodman SM, editors. Natural change and human impact in Madagascar. Washington, DC: Smithsonian University Press; 1997. pp. 381–405. [Google Scholar]
- Wright PC, Arrigo-Nelson SJ, Hogg KL, et al. Habitat distrubance and seasonal fluctuations of lemur parasites in the rain forest of Ranomafana National Park, Madagascar. In: Huffman MA, Chapman CA, editors. Primate parasite ecology; the dynamics of host-parasite relationships. Cambridge, UK: Cambridge University Press; 2009. pp. 311–330. [Google Scholar]
- Young H, Griffin RH, Wood CL, Nunn CL. Does habitat disturbance increase infectious disease risk for primates? Ecology Letters. 2013;16:656–663. doi: 10.1111/ele.12094. [DOI] [PubMed] [Google Scholar]