Abstract
Epoxygenases belong to the cytochrome P450 family and they generate epoxyeicosatrienoic acids (EETs) known to have anti-inflammatory effects but little is known about their role in macrophage function. By high-throughput sequencing of RNA (RNA-seq) in primary macrophages derived from rodents and humans, we establish the relative expression of epoxygenases in these cells. Zinc-finger nuclease-mediated targeted gene deletion of the major rat macrophage epoxygenase Cyp2j4 (orthologue of human CYP2J2), resulted in reduced EET synthesis. Cyp2j4−/− macrophages have relatively increased PPARγ levels and show a pro-fibrotic transcriptome, displaying over-expression of a specific subset of genes (260 transcripts) primarily involved in extracellular matrix, with fibronectin being the most abundantly expressed transcript. Fibronectin expression is under the control of epoxygenase activity in human and rat primary macrophages. In keeping with the in vitro findings, Cyp2j4−/− rats show up-regulation of type I collagen following unilateral ureter obstruction (UUO) of the kidney and quantitative proteomics analysis (LC-MS/MS) showed increased renal type I collagen and fibronectin protein abundance resulting from experimentally induced crescentic glomerulonephritis in these rats. Taken together, these results identify the rat epoxygenase Cyp2j4 as a determinant of a pro-fibrotic macrophage transcriptome that could have implications in various inflammatory conditions depending on macrophage function.
Introduction
Macrophage-mediated innate immune response is associated with the synthesis of lipid mediators such as prostaglandins and leukotrienes through the metabolism of arachidonic acid (AA) (1-4). The latter is metabolised by various enzymes such as cyclooxygenases and lipoxygenases where the activity/expression of these is tightly associated with macrophage function (3, 5).
AA is also metabolised by cytochrome P450 enzyme isoforms to four cis-epoxyeicosatrienoic acids (EETs) regioisomers with CYP2 family (CYP2J2, CYP2C8 CYP2C9 and CYP2S1 in humans) being the main epoxygenases (6-8). Although considerable attention has been afforded to cyclooxygenase and lipoxygenase expression in macrophage activation, little is known about the modulatory effects of endogenous epoxygenase-derived EETs in the immune system and especially the specific role of macrophage epoxygenases and their bioactive lipid products in macrophage-driven inflammatory disease.
The anti-inflammatory effects of endogenous eicosanoids were first described by showing that EETs inhibit NF-κB in endothelial cells (9). Over the last decade, numerous studies have shown the beneficial effects of EETs in cardiovascular homeostasis and specifically within the vasculature (10). Given the relatively abundant expression of CYP2J2 in endothelial cells and cardiomyocytes, the role of CYP2J2-derived EETs were largely studied in heart failure (11), diabetic cardiomyopathy (12), hyperhomocytenemia (13) and abdominal aortic aneurysm (14). In this latter aortic over-expression of CYP2J2 resulted in reduced inflammation characterised by reduced macrophage infiltration. More recently, an EET analogue administrated to salt-sensitive hypertensive rats has resulted in reduced inflammation in the kidney and diminished renal and cardiac fibrotic response (15). Although these studies establish clearly the anti-inflammatory effect of EETs associated with reduced macrophage infiltration, they focus primarily on the effect of either systemic delivery of EETs or endothelial-cell derived EET function. Macrophages equally express epoxygenases (6, 16, 17) and a recent genome-wide association mapping using macrophage infiltrates in the bronchoalveolar lavage fluid from 36 inbred mice strains exposed to hyperoxia identified Cyp2j locus within a significant quantitative trait locus on chromosome 4 (18). This is in accordance with the soluble epoxide hydroxylase (i.e. the enzyme metabolising EETs to their corresponding diols) inhibition resulting in significantly decreased total bronchoalveolar lavage cell number by 37% in tobacco smoke-exposed rats with significant reductions in alveolar macrophages (19). These studies suggest a role for macrophage infiltration as a result of an increase in endogenous EET production. The specific role of macrophage-derived EET has been recently investigated with the identification of CYP2S1 as a novel macrophage-derived epoxygenase localised in atherosclerotic plaques in humans (6, 20).
Allelic expansion of the Cyp2j locus in rodents has made targeted gene deletion experiments challenging and a recent study generated mice deleted for the entire locus (21). Although multiple isoforms of epoxygenases may contribute to EET biosynthesis, tissue expression of specific isoforms may account for the majority of epoxygenase activity (22). Here we provide a comprehensive genome-wide expression profiling in rodent and human primary macrophages by RNA-sequencing (RNA-seq). We show that rodent macrophages express predominantly the human CYP2J2 orthologue (Cyp2j4 in rats; Cyp2j6 in mice). By using zinc-finger nuclease technology in the rat, we generated an inbred rat strain deficient for functional Cyp2j4. We show that Cyp2j4−/− bone marrow derived macrophages have reduced EET production (11,12 and 14,15 EETs) and adopt a pro-fibrotic phenotype with transcriptional activation of ECM-related transcripts amongst which fibronectin is the most abundant. Epoxygenase-derived EETs regulate fibronectin production in rat and human macrophages. Cyp2j4−/− rats show enhanced renal Col1a1, Col1a2 and Col3a1 expression and increased interstitial Col1a1 following the unilateral ureter obstruction (UUO) of the left kidney. In addition quantitative proteomics (LC-MS/MS) in control and nephrotoxic nephritis (NTN)-induced kidneys further confirmed the relative increase in renal type I collagen and fibronectin protein abundance in Cyp2j4−/− animals when compared with WT controls in a macrophage-dependent model of glomerular inflammation.
Material and Methods
ZFN-mediated gene targeting and generation of Cyp2j4−/− rats
ZFN constructs specific for rat Cyp2j4 were designed, assembled and validated by Sigma-Aldrich. Selected ZFNs were targeted to exon 4 of Cyp2j4 (target sequence GGAAAGACTGGAA; See also Supplementary Figure 1). ZFN mRNAs were micro-injected into single cell Wistar-Kyoto (WKY/NCrl) rat embryos and the injected embryos were transferred to the oviduct of day-0.5 pseudopregnant rats. The resulting offspring were screened for ZFN-induced gene alterations at the target site of Cyp2j4 by PCR on purified DNA samples derived from tail snips using specific primers flanking the ZFN target site (+/− 1kb). PCR products were identified by agarose gel electrophoresis and the ZFN-mediated 25bp deletion was further confirmed by direct automated fluorescent DNA sequencing in an ABI 3730XL (Applied Biosystems). Heterozygous animals carrying the 25bp deletion in rat Cyp2j4 exon 4 (Cyp2j4+/−) were inter-crossed and phenotypic characterisation was performed on wild-type (WT) Cyp2j4 knock-out (Cyp2j4−/−) animals.
Measurement of cellular oxygen consumption by XF Extracellular Flux Analyzer
BMDMs from WT and Cyp2j4−/− rats were plated in XF24 V7 cell culture plates (Seahorse Bioscience, North Billerica, MA, USA) at 105 cells/well and incubated for 24 hours in a 37 °C/5% CO2 incubator. Cells were equilibrated with bicarbonate-free low buffered DMEM medium without any supplement or supplemented with glucose and/or glutamine as indicated in each experiment and incubated in a 37 °C incubator without CO2 for 45 minutes immediately before XF assay. 11,12-EETs (Cayman Chemical)were prepared in the identical assay medium (1μM) as in the corresponding well and were injected from the reagent ports automatically to the wells containing Cyp2j4−/− BMDMs. OCR measurements were then taken every 10 min for a total duration of 3h.
Evaluation of 11,12- and 14,15-EETs in macrophage lysates and supernatants by ELISA
To assess 11,12- and 14,15-EETS production in macrophages, an enzyme-linked immunosorbent assay (ELISA) kit (Detroit R&D, Detroit, MI) was used to determine concentrations of the stable 11,12- and 14,15-EETs EET metabolites in the supernatants and bone marrow derived macrophage lysates of wild type and Cyp2j4−/− rats (n=4 rats per group). Eicosanoids were extracted from the cells lysates and supernatants with ethyl acetate after acidification with acetic acid.
RNA-seq library preparation and data analysis in rodent and human macrophages
Total RNA was extracted from rat (Wild type Wistar-Kyoto strain, n=3; Cyp2j4−/−, n=3) and mice (commercially outbred mice; n=4) bone marrow derived macrophages (BMDMs) and human monocytes-derived macrophages (n=3 healthy donors) using Trizol (Invitrogen) according to manufacturer’s instructions with an additional purification step by on-column DNase treatment using the RNase-free DNase Kit (Qiagen) to ensure elimination of any genomic DNA. The integrity and quantity of total RNA was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific) and Agilent 2100 Bioanalyzer (Agilent Technologies). 1μg of total RNA from each species was used to generate RNA-seq libraries using TruSeq RNA sample preparation kit (Illumina) according to the manufacturer’s instructions. Briefly, RNA was purified and fragmented using poly-T oligo-attached magnetic beads using two rounds of purification followed by the first and second cDNA strand synthesis. Next, cDNA 3′ ends were adenylated and adapters ligated followed by 10 cycles of library amplification. Finally, the libraries were size selected using AMPue XP Beads (Beckman Coulter) purified and their quality was checked using Agilent 2100 Bioanalyzer. Samples were randomized to avoid batch effects and libraries were run on a single lane per sample of the HiSeq 2500 platform (Illumina) to generate 100bp paired-end reads. An average of 50M, 81M and 60M reads coverage per rat, mouse and human samples was achieved respectively. RNA-seq reads were aligned to the rat (rn4), mouse (mm9) and human (hg19) reference genomes using tophat 2. The average mapping percentage for all species was > 80%. Sequencing and mapping were quality controlled using standard tools provided in the fastQC software. Gene level read counts were computed using HT-Seq-count with ‘union’ mode and clustering of samples corresponding to different conditions was done using ward’s methods based on Euclidian distance of scaled sample gene expression profile. Differential expression analyses between WT and Cyp2j4−/− rat BMDMs were conducted using edgeR (23) with an FDR cut-off of 5%. PPARγ target enrichment on the differentially expressed genes (5% FDR) was performed on the targets identified by chromatin immunoprecipitation coupled with whole genome tiling arrays in 3T3-L1 adipocytes (24). The coordinates were converted to the mouse reference genome mm10 using liftOver utility from UCSC genome browser and the identified peaks (+/− 50kb of the transcription start site as recommended in the original study (24)) resulted in 2109 mouse genes (1889 rat orthologues). The RNA-seq dataset in rat BMDMs showed ~16000 expressed transcripts with 1117 PPARg targets and the enrichment was calculated using the Fisher’s exact test.
Cell Culture and RNAi
Rat bone marrow-derived macrophages were isolated from femurs and differentiated using the L929 conditioned media (25). Rat fibroblasts were isolated from skin and lungs as previously detailed (26). Primary adult human dermal fibroblasts (Cellworks) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) and F-12 medium containing 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin supplemented with 10% fetal calf serum (FCS). The cells were maintained at 37 °C in a humidified incubator in an atmosphere of 5% CO2, and fresh growth medium was added to the cells every 3–4 days until confluence. Cells were growth-arrested in serum-free medium for 24 h before use in experiments, and all experiments were performed under serum-free conditions unless otherwise stated. Human monocyte derived macrophages were differentiated from buffy coats from healthy donors using gradient separation (Histopaque 1077, Sigma) and adhesion purification. Following Histopaque separation, peripheral blood mononuclear cells were re-suspended in RPMI (Life Technologies) and monocytes were purified by adherence for 1h at 37°C, 5% CO2. The monolayer was washed 6 times with HBSS to remove non-adherent cells and monocytes were matured for 5 days in RPMI containing 100 ng/ml M-CSF (PeproTech, UK). Macrophage purity was confirmed by immunohistochemical assessment of CD68 and > 99% cells were CD68+.
For RNAi, human MDMs were replated in six-well plates (1×106 cells per well) in RPMI (Invitrogen) overnight and transfected for 48 hours with ON-TARGET PLUS for human CYP2S1 (100 nM, Dharmacon SMART pool) or non-targeting siRNA pool as the scrambled control siRNA using Dharmafect 1 (1:50, Dharmacon) as a transfection reagent in OPTIMEM medium (Invitrogen). In some experiments, following 24h incubation with CYP2S1 or non-targetting siRNA, the culture media was washed and cells were further cultured for 24h in presence or absence of 11,12 EETs (1μM). The siRNA sequences used in the siGENOME SMARTpool for all transcripts are available upon request.
Nephrotoxic nephritis (NTN) and unilateral ureter obstruction (UUO)
NTN was induced in ten week old male WT and Cyp2j4−/− rats by intravenous injection of 0.1 ml of NTS (25). The animals were then left for either 10 or 28 days to evaluate the potential effect of targeted deletion of Cyp2j4 on the progression of disease. 9, 13 and 27 days later, urine was collected by placing rats in metabolic cages for 24 hours with free access to food and water. Proteinuria was determined by the sulphosalicylic acid test. 10 and 28 days following NTN induction, rats were culled by asphyxiation with CO2 and cervical dislocation, kidneys from all rats were formalin fixed and paraffin embedded for histology (PAS, H&E, Sirius Red)
For induction of UUO, animals were anaesthetized with a mixture of isoflurane and oxygen under sterile conditions. Two ties were knotted around the mid-portion of the left ureter, using a thin non-absorbable suture (5/0, Mersilk) and the ureter was sectioned between the ligatures. The abdomen was closed with running sutures and the skin was closed with interrupted sutures. The animals were sacrificed at 7 days following UUO.
Formalin-fixed paraffin-embedded kidney sections were stained with mouse monoclonal antibody to ED-1 (anti-CD68, AbD Serotec, Oxford, UK) and fibronectin (AbD Serotec, Oxford, UK), followed by an HRP-labelled anti-mouse polymer development system (Dako Ltd, UK) for ED1 and fibronectin staining. The ED1+ cellular infiltrate in 20 consecutive glomeruli was quantified using automated image analysis software (ImagePro Plus, Media Cybernetics, Bethesda, MD) and expressed as a percentage of total glomerular cross sectional area. For immunofluorescence, kidneys were embedded in OCT (Cellpath, Newtown, UK), snap frozen in isopentane cooled with liquid nitrogen, and stored at −80°C. Sections (5-μm thick) were fixed in acetone for 10 minutes. For Col1a1 immunofluorescence, sections were blocked with BSA, incubated with the Col1a1 primary antibody raised in goat (Southern Biotech, 1/40 dilution), washed and re-incubated with secondary antibody (donkey anti-goat-FITC, 1/100 dilution, Abcam).
Quantitative proteomics by LC-MS/MS
Kidneys from untreated and NTN-treated rats (n=3 per group) were grounded to a powder by using a liquid nitrogen-cooled mini mortar and pestle set (Fisher Scientific). The samples were then lysed (8M Urea in 20mM HEPES), sonicated and 200μg of protein extracts were reduced with DTT (10mM) before trypsin digestion (TPCK trypsin beads slurry, Thermo Scientific) for 18h at 37°C. Peptides from each biological replicate were desalted by reversed-phase chromatographic cartridges (Oasis HLB, Waters Corporation), washed, dried and analysed by LC-MS/MS in technical duplicates. Samples were loaded in to a temperature-controlled autosampler (4°C) of an UltiMate 3000 RSLC nanoLC instrument coupled online to a Q-Exactive mass spectrometer (Thermo Scientific). Sample loading and elution details are available upon request. The Q-Exactive system acquired full scan survey spectra in profile (m/z 400 to 2000) with 70,000 resolution at m/z 400 using an underfill ratio of 10%. A maximum of the 12 most abundant multiply-charged ions registered in each survey spectrum were selected in a data-dependent (DDA) manner, fragmented by higher-energy collision induced dissociation (HCD) with a normalized collision energy of 28% and scanned in the orbitrap mass analyzer (m/z 200 to 2000). In DDA, a dynamic exclusion was enabled (30 second duration) as well as charge exclusion for singly charged ions. Raw data files were uploaded onto Progenesis QI for Proteomics software (Nonlinear Dynamics, 2014). Chromatographic alignment (with additional manual manipulation), data normalisation and peak picking were performed by Progenesis QI. An in-house Mascot server (version 2.5.0) was used to peptide/protein identification as searched against the Uniprot Swissprot rattus norvegicus FASTA which contained 28,864 sequences. File searching was automated using Mascot Daemon v2.5 with MS1 (precursor ions) mass tolerance set to 3ppm and MS2 (product ions) mass tolerance set to 25mmu. Data was exported from Mascot as an xml file at identity threshold with p=0.05 threshold applied. These xml files were imported into Progenesis QI and label-free relative quantitation applied using non-conflicting (unique) features or peptides only. For differential peptide abundance specific to Cyp24−/−-NTN, we have considered the following statistical workflow as we previously described elsewhere (27):
-
(i)
peptides that are differentially expressed between Cyp2j4−/− and Cyp2j4−/−-NTN (5% FDR)
-
(ii)
peptides that are not differentially expressed between WT and WT-NTN at 5% nominal P-value
-
(iii)
peptides that are differentially expressed between Cyp2j4−/−-NTN and WT-NTN at 5% nominal P-value
Western Blot, ELISA and qRT-PCR
The cells were lysed in Laemmli sample buffer supplemented with protease inhibitors and resolved by SDS-PAGE, transferred to PVDF membranes, and subjected to immunoblotting with appropriate primary and secondary antibodies. The probed proteins were detected using SuperSignal West Femto Cheluminescent Substrate (Thermo Fisher Scientific Inc., Rockford, IL). The primary antibodies used were as follows: rabbit polyclonal anti fibronectin (Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit polyclonal anti PPARγ, (Cell Signalling) and mouse monoclonal anti-β-Actin (C4, Santa Cruz Biotechnology). TNFα levels were quantified by sandwich ELISA (R&D Systems, Abingdon, UK), according to the manufacturer’s instructions. All qRT-PCRs were performed with a Viaa 7 Real-Time PCR system (Life technologies). A two-step protocol was used beginning with cDNA synthesis with iScript select (Bio-Rad) followed by PCR using Brilliant II SYBR® Green QPCR Master Mix (Agilent). A total of 10ng of cDNA per sample was used. Viia 7 RUO Software was used for the determination of Ct values. Results were analysed using the comparative Ct method and each sample was normalised to the reference gene Hprt, to account for any cDNA loading differences. The primer sequences are available upon request
Statistical Analyses
Data are presented as Mean ± S.E.M and were analysed using GraphPad Prism software (version 6.0; GraphPad). The Mann–Whitney test (two-tailed) was used for comparison of two groups. One-way ANOVA followed by Tukey’s multiple comparisons test was used for comparison of three or more groups. Differences in relative cytokine quantities (TNFα) were tested for significance using the nonparametric Wilcoxon signed-rank test.
Results
Macrophage epoxygenase expression in rodent and humans by RNA-seq
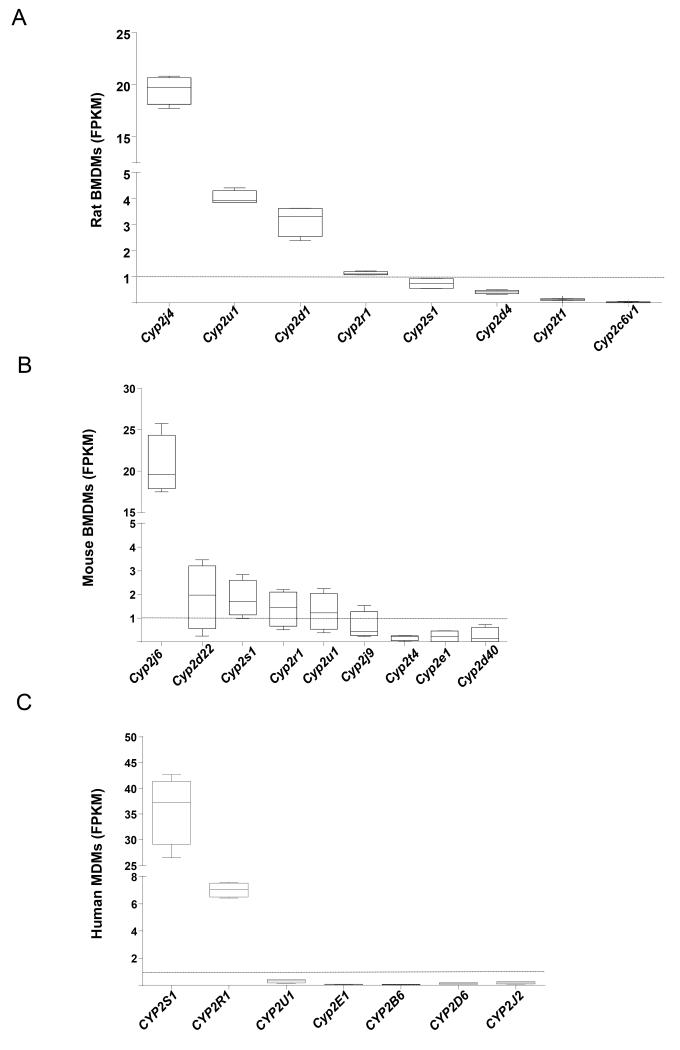
In order to establish a comprehensive expression profile of all possibly expressed epoxygenase isoforms in primary macrophages, we have performed unbiased high throughput RNA-sequencing (RNA-seq) in rodent bone marrow-derived macrophages (BMDMs) and human monocyte-derived macrophages (MDMs). The RNA-seq approach allowed robust coverage and quantification of all the transcribed elements in primary macrophages of three different species (> 40 million paired-end reads and > 90% mapped to the respective reference genomes, Figure 1). Rodent primary macrophages express primarily the CYP2J2 orthologue Cyp2j4 in rats and Cyp2j6 in mice (Figure 1A and B). Human monocytes-derived macrophages (MDMs) express primarily CYP2S1 and to a lesser extent CYP2R1 but the expression of all other CYP2 family genes were < 1 Fragments Per Kilobase of transcript per Million fragments mapped (FPKM), suggesting their relatively low levels of transcription in these cells (Figure 1C).
Figure 1.
RNA-seq in primary rodent and human macrophages establishes the mRNA expression levels from all main CYP epoxygenases (CYP2 family genes). RNA-seq FPKM values for CYP2 genes in BMDMs from rats (A), mice (B) and human monocyte-derived macrophages (MDM) from healthy donors (C). Macrophages from at least n=3 individuals/species were analysed. Genes with FPKM=0 were not plotted. The dotted line is FPKM=1 represents the threshold for low expression.
Targeted gene deletion of Cyp2j4 in the rat reduced macrophage EET production
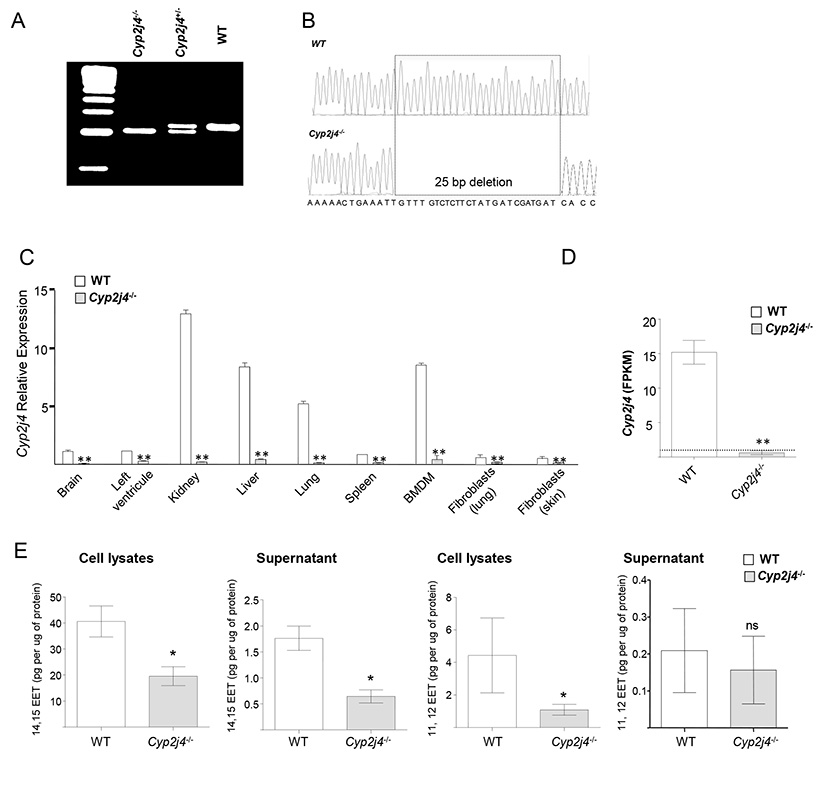
Zinc finger-mediated genome targeting of rat Cyp2j4 led to a deletion of 25bp in its exon 4. Genomic PCR of the ZFN target region of rat Cyp2j4 exon 4 showing the deletion in Cyp2j4+/− and Cyp2j4−/− animals (Figure 2A) was further confirmed with direct sequencing (Figure 2B). The 25 bp frameshift deletion in rat Cyp2j4 causes a premature stop codon upstream the putative heme binding site (Supplementary Figure 1A and B). Cyp2j4 deletion in the rat has resulted in a markedly reduced expression of its transcript levels in different tissues and primary cells (Figure 2C), suggesting a non-sense mediated decay of the Cyp2j4 mRNA resulting from the ZFN-mediated frameshift deletion. In order to establish accurately Cyp2j4 transcript copies in macrophages, RNA-seq analysis was performed in WT and Cyp2j4−/− BMDMs (Figure 2D). While WT BMDMs show average FPKM values of 15, Cyp2j4−/− BMDMs FPKM values were < 1, indicating the markedly reduced expression due to the frameshift deletion (Figure 2D). We next measured the consequence of the disrupted rat Cyp2j4 on EET production in macrophage lysates and supernatants by EET ELISA. Targeted genomic deletion in Cyp2j4 has resulted in reduced 11,12 and 14,15 EET in bone marrow derived macrophages (Figure 2E).
Figure 2.
Targeted gene deletion of Cyp2j4 in the rat. (A) ZFN-mediated gene targeting resulted in a deletion of 25bp in rat exon 4. (B) Sanger sequencing chromatograms of rat Cyp2j4 exon 4 shows the 25 bp deletion that creates a frameshift in the genomic sequence of Cyp2j4. The coding sequence resulting from the frameshift deletion (see Supplementary Figure 1) generates a premature stop codon at exon 6 of Cyp2j4. (C). ZFN-mediated genetic deletion in rat Cyp2j4 results in a marked reduction in Cyp2j4 expression in major tissues and bone marrow derived macrophages when assessed by qRT-PCR. Error bars indicate SEM; *, P<0.001 (D) RNA-sequencing in bone marrow derived macrophages (BMDMs) of Cyp2j4−/− and WT rats confirms the markedly reduced Cyp2j4 expression in Cyp2j4−/− BMDMs. FPKM values are shown for Cyp2j4. **, P<0.001. FPKM = 1 is shown with a dotted line. (E) 11,12- and 14,15-EETs measurements in the supernatants and cell lysates of WT and Cyp2j4−/− BMDMs. *, P<0.05; ns, non-significant.
Cyp2j4 deletion in the rat generates a pro-fibrotic macrophage transcriptome
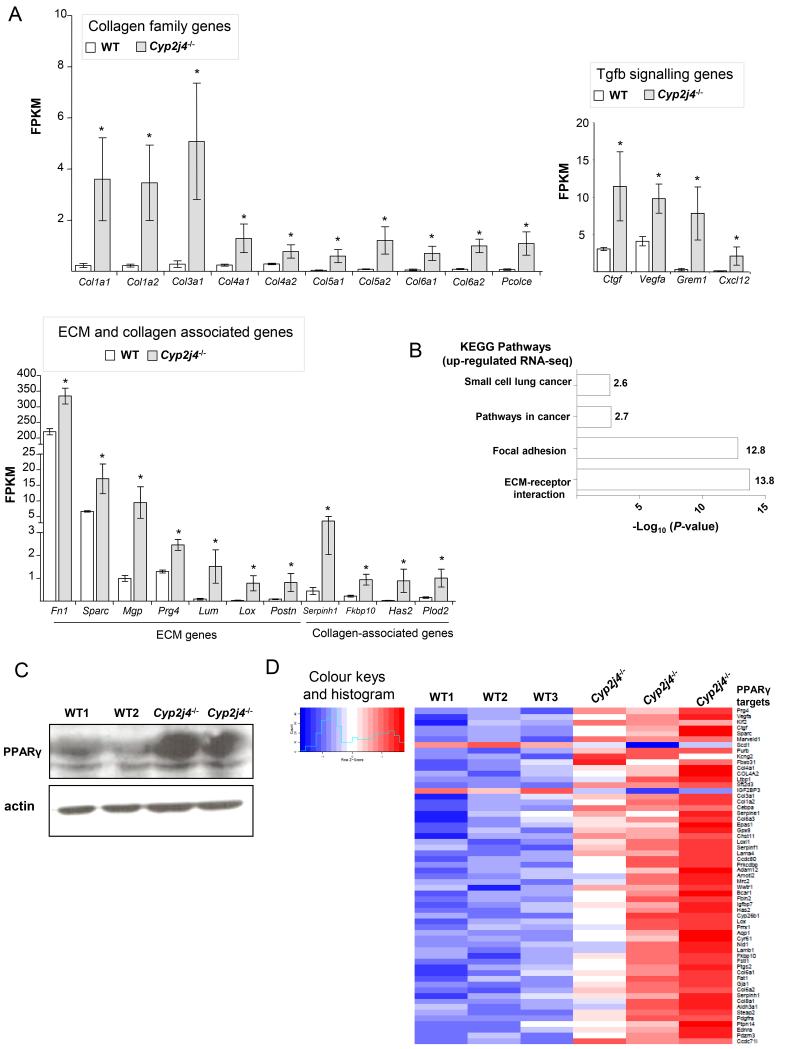
BMDMs from Cyp2j4−/− and WT animals were subjected to RNA-sequencing (RNA-seq). A total number of 260 transcripts were found to be differentially expressed (5% FDR) between Cyp2j4−/− and WT BMDMs, and interestingly 92% of those (238 in 260) were up-regulated in Cyp2j4−/− macrophages. Notably, the majority of the up-regulated genes belong to collagen, extracellular matrix (ECM), collagen-associated gene families together with genes involved in TGFβ signalling pathway (Ctgf, Vegfa, Grem1, Cxcl12, Figure 3A). The up-regulated transcripts were significantly enriched for KEGG pathways for ECM-receptor interaction (P = 1.7×10−14) and Focal adhesion (P = 1.5×10−13; Figure 3B). Because of the previously described role of peroxisome proliferator-activated receptor-gamma (PPARγ) as an effector for epoxygenase-derived EETs (28), we have investigated PPARγ levels in WT and Cyp2j4−/− BMDMs and found relatively increased PPARγ protein levels in the absence of Cyp2j4 in primary macrophages (Figure 3C). In keeping with the relatively increased PPARγ protein levels, we show that there was a significant enrichment for PPARγ targets among the differentially expressed genes between WT and Cyp2j4−/− BMDMs in the RNA-seq dataset (enrichment P = 2.5×10−17, Figure 3D). Taken together these results suggest that Cyp2j4−/− BMDMs show a distinct pro-fibrotic transcriptome signature associated with high PPARg levels. Since fibroblasts are also primarily involved in ECM production, we tested in parallel whether the pro-fibrotic transcriptome signature is a hallmark of Cyp2j4−/− fibroblasts. To this aim, we have isolated lung and skin-derived primary fibroblasts from WT and Cyp2j4−/− rats and measured the mRNA levels of Col1a1, Col1a2, Col3a1 and Fn1 in basal and TGFβ-stimulated cells (Supplementary Figure 2). These results further demonstrated the enhanced ECM gene expression in Cyp2j4−/− dermal and lung-derived fibroblasts (Supplementary Figure 2).
Figure 3.
Cyp2j4 deletion results in a pro-fibrotic macrophage transcriptome enriched for PPARγ targets. (A) Differential expression analysis of RNA-sequencing of WT and Cyp2j4−/− BMDMs (FDR<0.05) revealed an up-regulation of collagen family genes (upper-left panel), TGFβ signalling genes (upper-right panel) and ECM/collagen-associated genes (lower-left panel). *, P < 0.01 compared to WT. (B) KEGG pathway analysis showed markedly significant representation of ECM-receptor interaction and Focal adhesion pathways. (C) Western Blot analysis showing enhanced PPARγ protein levels in Cyp2j4−/− BMDMs. (D) Heat map showing the enrichment for PPARγ targets (59 out of 259 differentially expressed genes, P=2.5 10−17) among the 259 differentially expressed genes (FDR < 0.05) between WT and Cyp2j4−/− BMDMs.
Epoxygenase-derived EETs regulate fibronectin levels in macrophages
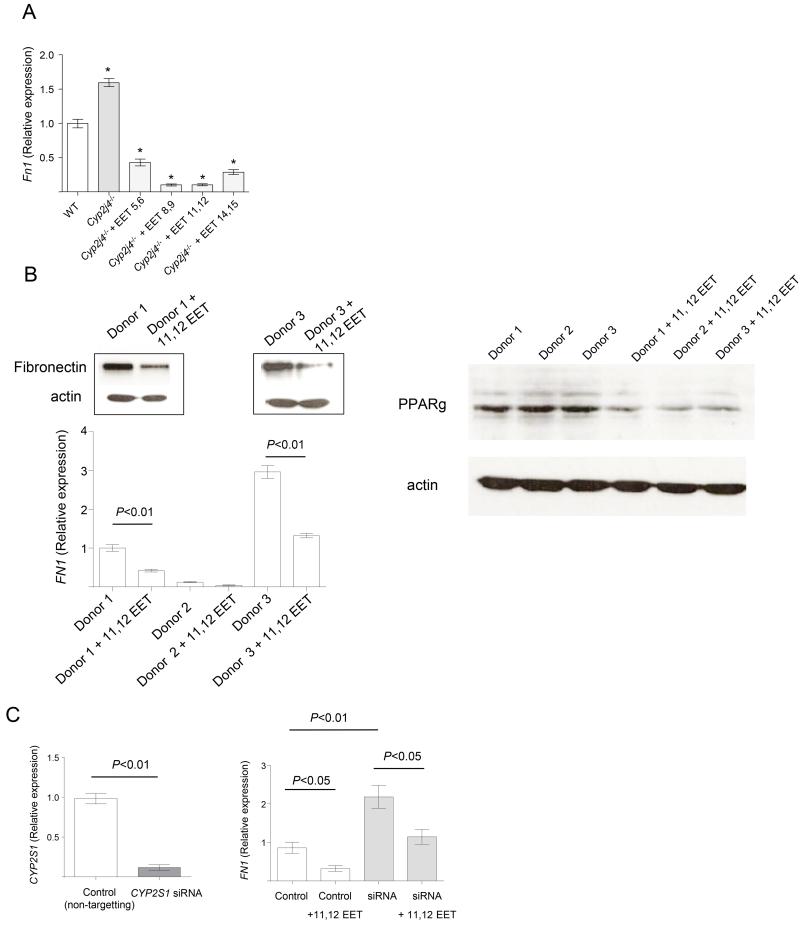
Among the significantly up-regulated transcripts in Cyp2j4−/− BMDMs, the must abundent is fibronectin (Fn1, FPKM=334). The relatively increased Fn1 expression levels are rescued and reversed by the addition of exogenous EETs into Cyp2j4−/− BMDMs (Figure 4A) and human monocyte-derived macrophages (Figure 4B) down-regulate Fn1 expression and protein levels in response to incubation with 11,12 EETs. In primary human monocyte-derived macrophages, the addition of 11,12 EETs resulted in decreased PPARγ levels suggesting that epoxygenase-mediated FN1 expression is associated with PPARγ levels in the macrophages. We next performed siRNA-mediated knock down of the most abundantly expressed human monocyte/macrophage CYP2 epoxygenase (Figure 1 and (6)), CYP2S1. This resulted in a significant increase FN1 expression (Figure 4C) which was lost following addition of 11,12 EETs (Figure 4C), indicating that epoxygenase activity and intracellular EETs levels are partly responsible for the FN1 transcription in human MDMs.
Figure 4.
Epoxygenase-derived EETs regulate fibronectin levels in macrophages. (A) Measurement of Fn1 expression levels by qRT-PCR in WT (basal), Cyp2j4−/− BMDMs (basal and following incubation with different EETs, 1μM). *, P < 0.01 compared to WT. (B) Human monocyte-derived macrophages (MDMs) from different healthy donors were incubated with 11,12 EETs (1μM) and fibronectin levels were measured by Western Blot (upper panel; donor 1 and donor 3) and qRT-PCR (lower panel; all donors). PPARγ protein levels were reduced when human MDMs from different donors were incubated with 11,12 EETs (1μM, right panel). (C) siRNA knock-down of human CYP2S1 and the expression levels of CYP2S1 (left panel) and FN1 (right panel). Control samples were incubated with non-targeting scrambled siRNA. FN1 expression was measured in control and siRNA conditions, with or without addition of 11,12 EETs (1μM).
In addition to macrophage Fn1 expression, we assessed whether targeted gene deletion of Cyp2j4 affected macrophage function in general by measuring oxygen consumption and LPS-mediated TNFα secretion. The oxygen consumption rate (OCR) measured by extracellular flux analyser showed reduced OCR in Cyp2j4−/− BMDMs which was rescued by the addition of 11,12 EETs (Supplementary Figure 3A). Cyp2j4 deletion has also resulted in diminished TNFα secretion by BMDMs where exogenous addition of 11,12 EETs had no effect (Supplementary Figure 3B).
Cyp2j4 deletion and susceptibility to macrophage-dependent glomerulonephritis and unilateral ureter obstruction (UUO)
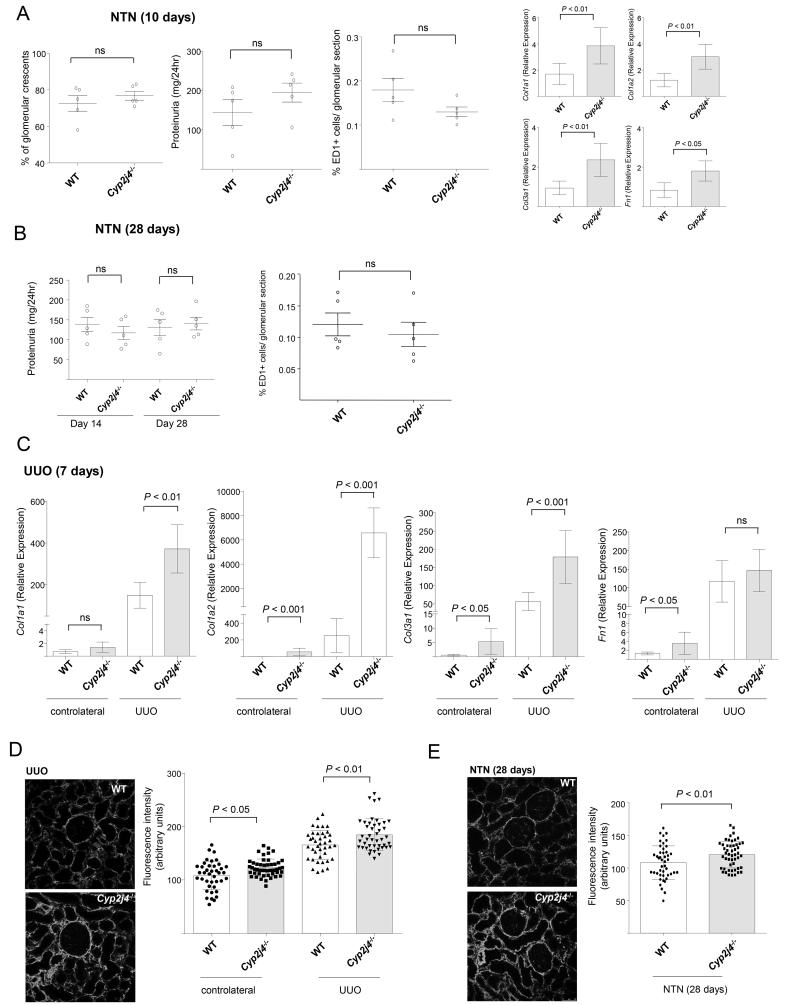
We next investigated whether Cyp2j4 deletion has resulted in altered disease outcome in nephrotoxic nephritis (NTN) model in the rat. WT and Cyp2j4−/− rats were injected with nephrotoxic serum and glomerular crescents/scarring, proteinuria and macrophage infiltration were measured at day 10 (the inflammatory phase) and at day 28 (the fibrotic phase of the disease). Targeted genetic deletion of Cyp2j4 did not result in a significant change in the inflammatory NTN phenotypes except increased Col1a1, Col1a2, Col3a1 and Fn1 expression in the nephritic kidneys at day 10. (Figure 5A and B). In addition, we have induced unilateral ureter obstruction (UUO) as a more fibrogenic model of kidney interstitial inflammation and observed that Col1a1, Col1a2 and Col3a1 expression levels were significantly increased in obstructed kidneys of Cyp2j4−/− animals when compared with WT controls (Figure 5C). Col1a2 was found to be markedly up-regulated in Cyp2j4−/− UUO kidneys (Figure 5C). Quantitative immunofluorescence for Cola1 in UUO and nephritic kidneys (day 28) showed a significant up-regulation of Col1a1 in Cyp2j4−/− rats, confirming the up-regulation of Col1a1 mRNA levels in NTN and UUO (Figure 5D and E).
Figure 5.
Cyp2j4−/− rats show increased collagen expression in UUO. (A) Nephrotoxic nephritis (NTN) was induced by injection of nephrotoxic serum (NTS) and glomerular crescents, proteinuria and glomerular ED1(CD68)+ macrophage infiltration were measured at day 10 in wild-type (WT) and Cyp2j4−/− rats. Col1a1, Col1a1, Col3a1 and Fn1 mRNA levels were measured in nephritic renal cortex of WT and Cyp2j4−/− animals. (B) NTN at day 28 following injection of NTS and measurement of proteinuria (day 14 and day 28) and glomerular ED1(CD68)+ macrophage infiltration. (C) UUO was induced in WT and Cyp2j4−/− rats and the expression of Col1a1, Col1a2, Col3a1 and Fn1 in the obstructed kidneys, were measured by qRT-PCR seven days afterwards. At least 5 animals were used in each group in NTN and UUO experiments. Representative Col1a1 immunofluorescence images (left panel) and the quantification (right panel) for UUO (D) and NTN at day 28 (E). Original magnification, ×20.
Quantitative proteomics shows relatively increased ECM peptide abundance in the nephritic kidneys of Cyp2j4−/− rats
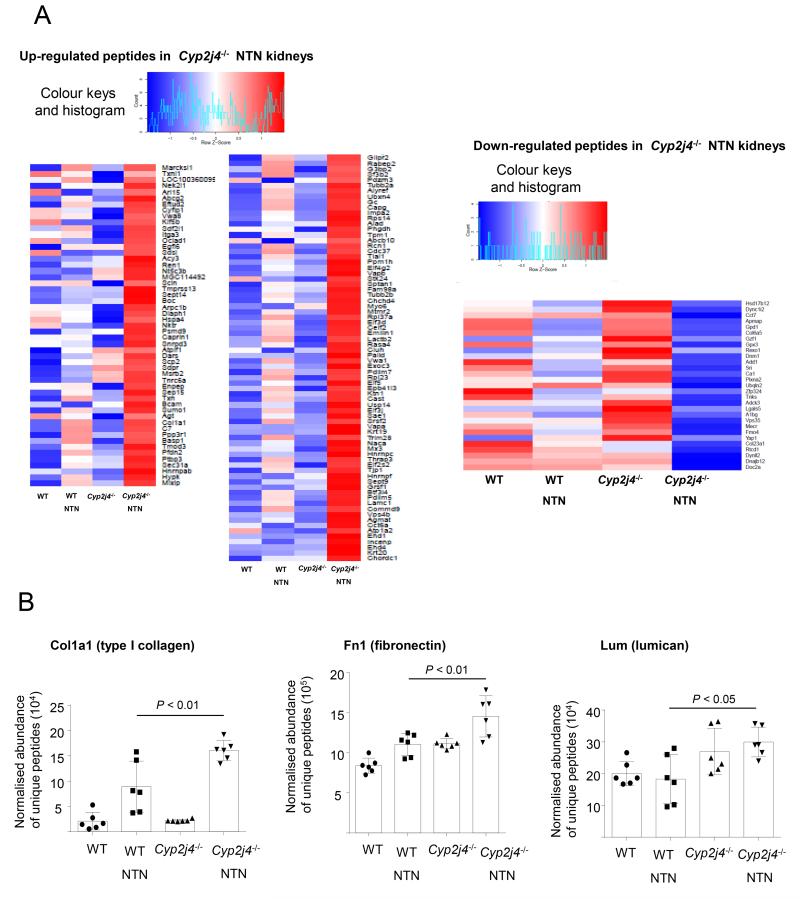
To further confirm the relative increase in ECM peptides in macrophage-dependent kidney inflammation, we have performed quantitative proteomics (LC-MS/MS) in control and NTN (day 10) WT and Cyp2j4−/− rats. Day 10 NTN corresponds to the peak of macrophage infiltration in the glomerulus of the WKY rat (25, 29). This analysis led to the identification of a total of 1,902 proteins reliably detected (≥ 2 peptides detected per protein) in all conditions for quantitative analysis of differential peptide abundance. At 5% FDR, we identified 170 annotated proteins where their abundance was differentially detected in Cyp2j4−/− kidneys following NTN induction (peptides that are differentially expressed between Cyp2j4−/− and Cyp2j4−/−-NTN but not between WT and WT-NTN; peptides that are significantly expressed between Cyp2j4−/−-NTN and WT-NTN; Figure 6). Gene Ontology analysis on the 170 differentially detected proteins showed the most significant enrichment for extracellular matrix organization (Benjamini corrected- P- value < 0.01) and confirmed the major effect of Cyp2j4 deletion on ECM production in nephritic kidneys. The results have also confirmed that the expression of the extracellular matrix genes up-regulated by Cyp2j4−/− macrophages (i.e. Fn1 Col1a1 and Lum, Figure 3A) is mirrored by an increased abundance in protein levels in Cyp2j4−/− kidneys when compared with WT controls following NTN induction (Figure 6).
Figure 6.
Quantitative proteomics (LC-MS/MS) confirms increased ECM peptide abundance in Cyp2j4−/− nephritic kidneys. (A) Heat map showing up-regulated (left panel) and down-regulated (right panel) peptides in Cyp2j4−/− specifically under NTN (day 10) conditions (i. differentially expressed between Cyp2j4−/−-basal and -NTN but not differentially expressed between WT and WT-NTN, ii. differentially expressed between Cyp2j4−/−-NTN and WT-NTN). (B) Normalised abundance of unique peptides for Col1a1, Fn1 and Lum in WT and Cyp2j4−/− rat kidneys in basal (untreated) and under NTN conditions. Note that these 3 genes’ expression is up-regulated in Cyp2j4−/− BMDMs (Figure 3A).
Discussion
Cytochrome P450 (CYP) epoxygenases can metabolise endogenous substrates such as arachidonic acid to generate bioactive lipid mediators. The arachidonic epoxides or epoxyeicosatrienoic acids (EETs) have been mainly described as anti-inflammatory mediators with attenuating effects on inflammatory cell infiltration into different tissues (19, 30-32). Macrophages are effector innate immune cells in various inflammatory reactions and although the role of cyclooxygenase and lipoxygenase pathways were largely studied in relation to their activation levels, the effect of expoygenase-derived EETs on macrophage function is an emerging field of research. In a previous comprehensive study, Fromel et al., have described human CYP2S1 as a novel macrophage epoxygenase regulating phagocytosis and co-localising with CD68+ cells in human atherosclerotic plaques (6).
In this study, we have established by RNA-sequencing in rat, mouse and human primary macrophages, the relative expression levels of CYP2 epoxygenases. We show that the most abundant CYP2 epoxygeases are Cyp2j4, Cyp2j6 and CYP2S1 in primary rat, mouse and human macrophages, respectively. This is in accordance with the previous report describing the identification of human CYPS1 by mass spectrometry as the main monocyte/macrophage epoxygenase (6). We have then generated a Cyp2j4−/− rat strain by ZFN-mediated gene targeting to study the effect of targeted genomic deletion of Cyp2j4 on macrophage phenotype, and macrophage-dependent inflammatory kidney disease in comparison with Cyp2j4+/+ animals. Bone marrow derived macrophages isolated from Cyp2j4−/− rats showed a specific transcriptome signature defined by up-regulation of 260 genes encoding primarily proteins of extracellular matrix, collagen and mediators of TGFß signalling. Cyp2j4−/− BMDMs showed increased PPARγ levels and there was a significant enrichment for known PPARγ targets among the up-regulated transcripts, suggesting that the deletion of Cyp2j4 may have resulted in a PPARγ-mediated pro-fibrotic macrophage phenotype. The nuclear orphan receptor peroxisome proliferator-activated receptor-gamma (PPARγ) is known to regulate adipogenesis (33) and inflammatory responses (34) but previous studies have also established a novel role for PPARγ signalling in the regulation of TGFß-dependent fibrogenesis (35, 36). Importantly, a recent study showed the importance of endothelial-derived EETs in organ regeneration and wound healing (37) and an orally active EET analogue have been shown to attenuate kidney fibrosis in the rat (15), but to our knowledge, our study is the first demonstration of a pro-fibrotic macrophage transcriptome depending on epoxygenase level/activity. It should be noted that epoxygenase products other than EETs (i.e. linoleic, eicosapentaenoic and docosahexenoic acid products) could also be affected in Cyp2j4−/− macrophages. This is the case for human macrophage epoxygenase CYP2S1-containing microsomes generating epoxides of linoleic and eicosapentaenoic acid (6) so further studies must focus on their potential pro-fibrotic effect.
Tissue fibrosis is characterised by activated mesenchymal fibroblasts synthesizing elevated levels of matrix proteins, including collagen and fibronectin (38). In order to measure the effects of Cyp2j4 deletion in fibroblasts, we have cultured dermal and lung fibroblasts from Cyp2j4−/− and WT animals. We found that despite Cyp2j4 expression being 10 fold less in dermal and lung fibroblasts when compared with bone marrow-derived macrophages, Cyp2j4−/− fibroblasts show relatively increased collagen and fibronectin expression levels suggesting that epoxygenases determine a pro-fibrotic transcriptomic signature in major ECM producing cells.
Among the up-regulated transcripts in the Cyp2j4−/− BMDMs, fibronectin (Fn1) had the highest expression levels (FPKM ~200). We have also confirmed the relatively elevated levels of FN1 in human macrophages (FPKM > 200, data not shown). Macrophages are known as producers of fibronectin (39, 40) and our results show that endogenous EETs have an effect on the production of FN1 in human macrophages but a more generalised effect of epoxygenase-derived EETs on other ECM genes including collagens cannot be ruled out. Macrophage activation associated with collagen production has been previously described (41) and in breast cancer, macrophages regulate collagen fibrillogenesis (42). Our study underlines the importance of epoxygenase-mediated ECM production in pathologies where macrophages are likely to adopt a pro-fibrotic phenotype. Given that epoxygenase mRNA levels are enhanced following M1-type activation of macrophages in vitro (6, 16), intracellular levels of EETs or other epoxgenase products could be responsible for a plastic macrophage phenotype between pro-inflammatory and pro-fibrotic activation states depending on the in vivo ‘immunological context’ (43).
In order to test whether the epoxygenase-mediated pro-fibrotic macrophage transcriptome contribute to enhanced ECM production in vivo, we have measured the susceptibility of Cyp2j4−/− rats to nephrotoxic nephritis (NTN) and unilateral ureter obstruction (UUO). The latter is characterised by interstitial fibrosis where depletion studies have previously shown the importance of macrophage function in renal fibrosis (44). Our results have shown increased Col1a1, Col1a2 and Col3a1 in the UUO kidney of Cyp2j4−/− animals suggesting that pro-fibrotic macrophage phenotype observed in vitro could translate into enhanced collagen expression in the fibrotic kidney. In keeping with this, type I collagen and fibronectin peptides measured by LC-MS/MS are increased in Cyp2j4−/− rat kidneys following NTN induction. Despite glomerular macrophage infiltration being unchanged between WT and Cyp2j4−/− kidneys following NTN, Cyp2j4−/− nephritic kidneys were characterised by enhanced ECM production, suggesting that the effect of Cyp2j4 is more likely to be on the fibrotic phase of experimental crescentic glomerulonephritis.
Although these in vivo models of kidney inflammation do not establish the cellular origin of enhanced type I collagen and fibronectin after UUO and NTN, our data shows that macrophages are not the only cell type responsible of an enhanced ECM expression in the absence of Cyp2j4. It is noteworthy that NTN is characterised by a peak in macrophage infiltration at day 10 following the injection of the nephrotoxic serum, the time point where the total kidney protein extracts were subjected to LC-MS/MS. Considered together, these findings suggest that the increased ECM production seen in NTN and UUO could have multiple cellular sources, with a combined contribution from stromal (fibroblasts) and innate immune cells (macrophages). Since we have previously established the importance of genes expressed by macrophages in the pathophysiology of crescentic glomerulonephritis (45-47), the current study emphasises the importance of factors influencing glomerular scarring. The effect of CYP2J2 over-expression on chronic kidney failure was previously investigated in a nephrectomy model where adenovirus-mediated CYP2J2 gene delivery significantly lowered collagen I and collagen IV deposition in rats (48).
Recently, the role of endothelial-derived EETs in wound healing as well as normal organ and tissue growth was established (49), and our results, taken together with others (6), suggest that epoxygenase-mediated macrophage function could also contribute to pathophysiological conditions depending on the activity of these cells. Inter-individual variations were reported in P450-dependent AA metabolism (reviewed in (50)). In the kidney, inter-individual variations in the cytochrome P450-dependent arachidonic acid metabolism (51) suggest a genetic predisposition for the regulation of tissue (or cell)-specific EET levels. In keeping with this, we found Cyp2j4 expression to be under cis-genetic control (47) responsible for differential expression between two inbred rat strain BMDMs (52). Hence epoxygenase-mediated pro-fibrotic macrophage phenotype is likely to be genetically determined.
In summary, our results show that macrophage epoxygenase determines a pro-fibrotic transcriptomic signature. Macrophage EET levels and epoxygenase activity must be taken into account on the activity of these cells during inflammatory reactions. These findings could have important implications for regulating macrophage activity in fibrotic disorders with an inflammatory component.
Supplementary Material
Acknowledgements
We thank Zelpha D’Souza for her excellent technical assistance.
This work was supported by the Kidney Research UK (RP9/2013 to J.B.) and the Medical Research Council (MR/M004716/1 to J.B.). We acknowledge funding of Advanced grant ERC-2010-AdG_20100317 (ELABORATE) from the European Research Council to T.J.A.
Footnotes
Accession number
The RNA-seq results in rat macrophages presented in this article have been submitted to the National Center for Biotechnology Information’s Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE65715.
References
- 1.Barrios-Rodiles M, Chadee K. Novel regulation of cyclooxygenase-2 expression and prostaglandin E2 production by IFN-gamma in human macrophages. J Immunol. 1998;161:2441–2448. [PubMed] [Google Scholar]
- 2.Dalli J, Serhan CN. Specific lipid mediator signatures of human phagocytes: microparticles stimulate macrophage efferocytosis and pro-resolving mediators. Blood. 2012;120:e60–72. doi: 10.1182/blood-2012-04-423525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 4.Peters-Golden M, Henderson WR., Jr. Leukotrienes. N Engl J Med. 2007;357:1841–1854. doi: 10.1056/NEJMra071371. [DOI] [PubMed] [Google Scholar]
- 5.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fromel T, Kohlstedt K, Popp R, Yin X, Awwad K, Barbosa-Sicard E, Thomas AC, Lieberz R, Mayr M, Fleming I. Cytochrome P4502S1: a novel monocyte/macrophage fatty acid epoxygenase in human atherosclerotic plaques. Basic Res Cardiol. 2013;108:319. doi: 10.1007/s00395-012-0319-8. [DOI] [PubMed] [Google Scholar]
- 7.Imig JD. Epoxides and soluble epoxide hydrolase in cardiovascular physiology. Physiological reviews. 2012;92:101–130. doi: 10.1152/physrev.00021.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeldin DC. Epoxygenase pathways of arachidonic acid metabolism. J Biol Chem. 2001;276:36059–36062. doi: 10.1074/jbc.R100030200. [DOI] [PubMed] [Google Scholar]
- 9.Node K, Huo Y, Ruan X, Yang B, Spiecker M, Ley K, Zeldin DC, Liao JK. Anti-inflammatory properties of cytochrome P450 epoxygenase-derived eicosanoids. Science. 1999;285:1276–1279. doi: 10.1126/science.285.5431.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imig JD, Hammock BD. Soluble epoxide hydrolase as a therapeutic target for cardiovascular diseases. Nat Rev Drug Discov. 2009;8:794–805. doi: 10.1038/nrd2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Ni L, Yang L, Duan Q, Chen C, Edin ML, Zeldin DC, Wang DW. CYP2J2-derived epoxyeicosatrienoic acids suppress endoplasmic reticulum stress in heart failure. Mol Pharmacol. 2014;85:105–115. doi: 10.1124/mol.113.087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma B, Xiong X, Chen C, Li H, Xu X, Li X, Li R, Chen G, Dackor RT, Zeldin DC, Wang DW. Cardiac-specific overexpression of CYP2J2 attenuates diabetic cardiomyopathy in male streptozotocin-induced diabetic mice. Endocrinology. 2013;154:2843–2856. doi: 10.1210/en.2012-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moshal KS, Zeldin DC, Sithu SD, Sen U, Tyagi N, Kumar M, Hughes WM, Jr., Metreveli N, Rosenberger DS, Singh M, Vacek TP, Rodriguez WE, Ayotunde A, Tyagi SC. Cytochrome P450 (CYP) 2J2 gene transfection attenuates MMP-9 via inhibition of NF-kappabeta in hyperhomocysteinemia. J Cell Physiol. 2008;215:771–781. doi: 10.1002/jcp.21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Z, Zhao G, Yan J, Liu W, Feng W, Ma B, Yang L, Wang JA, Tu L, Wang DW. CYP2J2 overexpression increases EETs and protects against angiotensin II-induced abdominal aortic aneurysm in mice. J Lipid Res. 2013;54:1448–1456. doi: 10.1194/jlr.M036533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hye Khan MA, Neckar J, Manthati V, Errabelli R, Pavlov TS, Staruschenko A, Falck JR, Imig JD. Orally active epoxyeicosatrienoic acid analog attenuates kidney injury in hypertensive Dahl salt-sensitive rat. Hypertension. 2013;62:905–913. doi: 10.1161/HYPERTENSIONAHA.113.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bystrom J, Thomson SJ, Johansson J, Edin ML, Zeldin DC, Gilroy DW, Smith AM, Bishop-Bailey D. Inducible CYP2J2 and its product 11,12-EET promotes bacterial phagocytosis: a role for CYP2J2 deficiency in the pathogenesis of Crohn’s disease? PLoS One. 2013;8:e75107. doi: 10.1371/journal.pone.0075107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakayama K, Nitto T, Inoue T, Node K. Expression of the cytochrome P450 epoxygenase CYP2J2 in human monocytic leukocytes. Life Sci. 2008;83:339–345. doi: 10.1016/j.lfs.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 18.Nichols JL, Gladwell W, Verhein KC, Cho HY, Wess J, Suzuki O, Wiltshire T, Kleeberger SR. Genome-wide association mapping of acute lung injury in neonatal inbred mice. Faseb J. 2014;28:2538–2550. doi: 10.1096/fj.13-247221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith KR, Pinkerton KE, Watanabe T, Pedersen TL, Ma SJ, Hammock BD. Attenuation of tobacco smoke-induced lung inflammation by treatment with a soluble epoxide hydrolase inhibitor. Proc Natl Acad Sci U S A. 2005;102:2186–2191. doi: 10.1073/pnas.0409591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleming I. The pharmacology of the cytochrome P450 epoxygenase/soluble epoxide hydrolase axis in the vasculature and cardiovascular disease. Pharmacological reviews. 2014;66:1106–1140. doi: 10.1124/pr.113.007781. [DOI] [PubMed] [Google Scholar]
- 21.Zhou GL, Beloiartsev A, Yu B, Baron DM, Zhou W, Niedra R, Lu N, Tainsh LT, Zapol WM, Seed B, Bloch KD. Deletion of the murine cytochrome P450 Cyp2j locus by fused BAC-mediated recombination identifies a role for Cyp2j in the pulmonary vascular response to hypoxia. PLoS genetics. 2013;9:e1003950. doi: 10.1371/journal.pgen.1003950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu S, Moomaw CR, Tomer KB, Falck JR, Zeldin DC. Molecular cloning and expression of CYP2J2, a human cytochrome P450 arachidonic acid epoxygenase highly expressed in heart. J Biol Chem. 1996;271:3460–3468. doi: 10.1074/jbc.271.7.3460. [DOI] [PubMed] [Google Scholar]
- 23.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr., Liu XS, Lazar MA. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes & development. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’Souza Z, McAdoo SP, Smith J, Pusey CD, Cook HT, Behmoaras J, Aitman TJ. Experimental crescentic glomerulonephritis: a new bicongenic rat model. Disease models & mechanisms. 2013;6:1477–1486. doi: 10.1242/dmm.012328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seluanov A, Vaidya A, Gorbunova V. Establishing primary adult fibroblast cultures from rodents. Journal of visualized experiments: JoVE. 2010 doi: 10.3791/2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rotival M, Ko JH, Srivastava PK, Kerloc’h A, Montoya A, Mauro C, Faull P, Cutillas PR, Petretto E, Behmoaras J. Integrating phosphoproteome and transcriptome reveals new determinants of macrophage multinucleation. Molecular & cellular proteomics: MCP. 2014 doi: 10.1074/mcp.M114.043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Zhang Y, Schmelzer K, Lee TS, Fang X, Zhu Y, Spector AA, Gill S, Morisseau C, Hammock BD, Shyy JY. The antiinflammatory effect of laminar flow: the role of PPARgamma, epoxyeicosatrienoic acids, and soluble epoxide hydrolase. Proc Natl Acad Sci U S A. 2005;102:16747–16752. doi: 10.1073/pnas.0508081102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tam FW, Smith J, Morel D, Karkar AM, Thompson EM, Cook HT, Pusey CD. Development of scarring and renal failure in a rat model of crescentic glomerulonephritis. Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association. 1999;14:1658–1666. doi: 10.1093/ndt/14.7.1658. [DOI] [PubMed] [Google Scholar]
- 30.Abraham NG, Sodhi K, Silvis AM, Vanella L, Favero G, Rezzani R, Lee C, Zeldin DC, Schwartzman ML. CYP2J2 Targeting to Endothelial Cells Attenuates Adiposity and Vascular Dysfunction in Mice Fed a High-Fat Diet by Reprogramming Adipocyte Phenotype. Hypertension. 2014 doi: 10.1161/HYPERTENSIONAHA.114.03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng Y, Edin ML, Theken KN, Schuck RN, Flake GP, Kannon MA, DeGraff LM, Lih FB, Foley J, Bradbury JA, Graves JP, Tomer KB, Falck JR, Zeldin DC, Lee CR. Endothelial CYP epoxygenase overexpression and soluble epoxide hydrolase disruption attenuate acute vascular inflammatory responses in mice. Faseb J. 2011;25:703–713. doi: 10.1096/fj.10-171488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manhiani M, Quigley JE, Knight SF, Tasoobshirazi S, Moore T, Brands MW, Hammock BD, Imig JD. Soluble epoxide hydrolase gene deletion attenuates renal injury and inflammation with DOCA-salt hypertension. Am J Physiol Renal Physiol. 2009;297:F740–748. doi: 10.1152/ajprenal.00098.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, Spiegelman BM. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes & development. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehrke M, Lazar MA. The many faces of PPARgamma. Cell. 2005;123:993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 35.Ghosh AK, Bhattacharyya S, Lakos G, Chen SJ, Mori Y, Varga J. Disruption of transforming growth factor beta signaling and profibrotic responses in normal skin fibroblasts by peroxisome proliferator-activated receptor gamma. Arthritis and rheumatism. 2004;50:1305–1318. doi: 10.1002/art.20104. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh AK, Bhattacharyya S, Wei J, Kim S, Barak Y, Mori Y, Varga J. Peroxisome proliferator-activated receptor-gamma abrogates Smad-dependent collagen stimulation by targeting the p300 transcriptional coactivator. Faseb J. 2009;23:2968–2977. doi: 10.1096/fj.08-128736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panigrahy D, Kalish BT, Huang S, Bielenberg DR, Le HD, Yang J, Edin ML, Lee CR, Benny O, Mudge DK, Butterfield CE, Mammoto A, Mammoto T, Inceoglu B, Jenkins RL, Simpson MA, Akino T, Lih FB, Tomer KB, Ingber DE, Hammock BD, Falck JR, Manthati VL, Kaipainen A, D’Amore PA, Puder M, Zeldin DC, Kieran MW. Epoxyeicosanoids promote organ and tissue regeneration. Proc Natl Acad Sci U S A. 2013;110:13528–13533. doi: 10.1073/pnas.1311565110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leask A, Abraham DJ. TGF-beta signaling and the fibrotic response. Faseb J. 2004;18:816–827. doi: 10.1096/fj.03-1273rev. [DOI] [PubMed] [Google Scholar]
- 39.Rennard SI, Hunninghake GW, Bitterman PB, Crystal RG. Production of fibronectin by the human alveolar macrophage: mechanism for the recruitment of fibroblasts to sites of tissue injury in interstitial lung diseases. Proc Natl Acad Sci U S A. 1981;78:7147–7151. doi: 10.1073/pnas.78.11.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamauchi K, Martinet Y, Crystal RG. Modulation of fibronectin gene expression in human mononuclear phagocytes. J Clin Invest. 1987;80:1720–1727. doi: 10.1172/JCI113263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnoor M, Cullen P, Lorkowski J, Stolle K, Robenek H, Troyer D, Rauterberg J, Lorkowski S. Production of type VI collagen by human macrophages: a new dimension in macrophage functional heterogeneity. J Immunol. 2008;180:5707–5719. doi: 10.4049/jimmunol.180.8.5707. [DOI] [PubMed] [Google Scholar]
- 42.Pollard JW. Macrophages define the invasive microenvironment in breast cancer. J Leukocyte Biol. 2008;84:623–630. doi: 10.1189/jlb.1107762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000prime reports. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng XM, Nikolic-Paterson DJ, Lan HY. Inflammatory processes in renal fibrosis. Nature reviews. Nephrology. 2014;10:493–503. doi: 10.1038/nrneph.2014.114. [DOI] [PubMed] [Google Scholar]
- 45.Behmoaras J, Bhangal G, Smith J, McDonald K, Mutch B, Lai PC, Domin J, Game L, Salama A, Foxwell BM, Pusey CD, Cook HT, Aitman TJ. Jund is a determinant of macrophage activation and is associated with glomerulonephritis susceptibility. Nature genetics. 2008;40:553–559. doi: 10.1038/ng.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Behmoaras J, Smith J, D’Souza Z, Bhangal G, Chawanasuntoropoj R, Tam FW, Pusey CD, Aitman TJ, Cook HT. Genetic loci modulate macrophage activity and glomerular damage in experimental glomerulonephritis. Journal of the American Society of Nephrology: JASN. 2010;21:1136–1144. doi: 10.1681/ASN.2009090968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang H, Kerloc’h A, Rotival M, Xu X, Zhang Q, D’Souza Z, Kim M, Scholz JC, Ko JH, Srivastava PK, Genzen JR, Cui W, Aitman TJ, Game L, Melvin JE, Hanidu A, Dimock J, Zheng J, Souza D, Behera AK, Nabozny G, Cook HT, Bassett JH, Williams GR, Li J, Vignery A, Petretto E, Behmoaras J. Kcnn4 is a regulator of macrophage multinucleation in bone homeostasis and inflammatory disease. Cell reports. 2014;8:1210–1224. doi: 10.1016/j.celrep.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao G, Tu L, Li X, Yang S, Chen C, Xu X, Wang P, Wang DW. Delivery of AAV2-CYP2J2 protects remnant kidney in the 5/6-nephrectomized rat via inhibition of apoptosis and fibrosis. Hum Gene Ther. 2012;23:688–699. doi: 10.1089/hum.2011.135. [DOI] [PubMed] [Google Scholar]
- 49.Panigrahy D, Kalish BT, Huang S, Bielenberg DR, Le HD, Yang J, Edin ML, Lee CR, Benny O, Mudge DK, Butterfield CE, Mammoto A, Mammoto T, Inceoglu B, Jenkins RL, Simpson MA, Akino T, Lih FB, Tomer KB, Ingber DE, Hammock BD, Falck JR, Manthati VL, Kaipainen A, D’Amore PA, Puder M, Zeldin DC, Kieran MW. Epoxyeicosanoids promote organ and tissue regeneration. Proc Natl Acad Sci U S A. 2013;110:13528–13533. doi: 10.1073/pnas.1311565110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shahabi P, Siest G, Meyer UA, Visvikis-Siest S. Human cytochrome P450 epoxygenases: Variability in expression and role in inflammation-related disorders. Pharmacol Ther. 2014 doi: 10.1016/j.pharmthera.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 51.Schwartzman ML, Martasek P, Rios AR, Levere RD, Solangi K, Goodman AI, Abraham NG. Cytochrome P450-dependent arachidonic acid metabolism in human kidney. Kidney Int. 1990;37:94–99. doi: 10.1038/ki.1990.13. [DOI] [PubMed] [Google Scholar]
- 52.Hull RP, Srivastava PK, D’Souza Z, Atanur SS, Mechta-Grigoriou F, Game L, Petretto E, Cook HT, Aitman TJ, Behmoaras J. Combined ChIP-Seq and transcriptome analysis identifies AP-1/JunD as a primary regulator of oxidative stress and IL- 1beta synthesis in macrophages. BMC genomics. 2013;14:92. doi: 10.1186/1471-2164-14-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.