Abstract
MCPH1, also known as BRIT1, has recently been identified as a novel key regulatory gene of the DNA damage response pathway. MCPH1 is located on human chromosome 8p23.1, where human cancers frequently show loss of heterozygosity. As such, MCPH1 is aberrantly expressed in many malignancies, including breast and ovarian cancers, and the function of MCPH1 has been implicated in tumor suppression. However, it remains poorly understood whether MCPH1 deficiency leads to tumorigenesis. Here, we generated and studied both Mcph1−/− and Mcph1−/−p53−/− mice; we showed that Mcph1−/− mice developed tumors with long latency, and that primary lymphoma developed significantly earlier in Mcph1−/−p53−/− mice than in Mcph11+/+p53−/− and Mcph1+/−p53−/− mice. The Mcph1−/−p53−/− lymphomas and derived murine embryonic fibroblasts (MEFs) were both more sensitive to irradiation. Mcph1 deficiency resulted in remarkably increased chromosome and chromatid breaks in Mcph1−/− p53−/− lymphomas and MEFs, as determined by metaphase spread assay and spectral karyotyping analysis. Additionally, Mcph1 deficiency significantly enhanced aneuploidy as well as abnormal centrosome multiplication in Mcph1−/−p53−/− cells. Meanwhile, Mcph1 deficiency impaired double strand break (DSB) repair in Mcph1−/−p53−/− MEFs as demonstrated by neutral Comet assay. Compared with Mcph1+/+p53−/− MEFs, homologous recombination and non-homologous end joining activities were significantly decreased in Mcph1−/−p53−/− MEFs. Notably, reconstituted MCPH1 rescued the defects of DSB repair and alleviated chromosomal aberrations in Mcph1−/−p53−/− MEFs. Taken together, our data demonstrate MCPH1 deficiency promotes genomic instability and increases cancer susceptibility. Our study using knockout mouse models provides convincing genetic evidence that MCPH1 is a bona fide tumor suppressor gene. Its deficiency leading to defective DNA repair in tumors can be utilized to develop novel targeted cancer therapies in the future.
Keywords: Mcph1/Brit1, tumor suppressor gene, chromosomal instability, homologous recombination, non-homologous end joining, DNA double-strand breaks
INTRODUCTION
MCPH1 (also known as BRIT1) has been recently identified as a novel key regulator of the DNA damage response (DDR), DNA repair, and genomic instability.1-3 It belongs to the BRCA1 C-terminal domain (BRCT) protein family, and contains three BRCT domains. 1,2,4 These domains are conserved protein-protein interaction modules involved in DDR.5-8 MCPH1 interacts with key partners (such as γ-H2AX, SWI/SNF, and SET), participating into DDR, chromosomal relaxation, and cell cycle checkpoint control.9-16 Importantly, our report demonstrates that Mcph1 is essential for DNA homologous recombination repair (HR) in the Mcph1−/− mouse model.17 Moreover, the cells derived from microcephaly patients (with MCPH1 inactivation) or Mcph1−/− mice exhibit defective DNA repair and genomic instability, which are the hallmarks of many malignancies.1,17-19 Although these studies suggest that MCPH1 may act as a caretaker of the genome, one of the surveillance mechanisms of cancer, it remains to be determined whether MCPH1 plays a critical role in tumor suppression.
Recently, several lines of evidence have indicated that MCPH1 is aberrantly expressed in human cancers.1,3,20-24 For example, low expression of MCPH1 exists iñ30% of breast cancers and positively correlates with higher grade tumors;20 this suggests that MCPH1 deficiency may contribute to breast cancer. To further elucidate the tumor suppressive role of MCPH1, in this study we assessed tumor formation in both single Mcph1−/− and Mcph1−/−p53−/− double knockout mice. Our data revealed that Mcph1 deficiency leads to tumor formation and accelerates the development of malignant lymphomas in p53−/− mice. More importantly, we demonstrate that Mcph1 deficiency promotes aneuploidy and defective DNA repairs, which is confirmed by rescue experiments. Therefore, our study using a knockout mouse model strongly supports that Mcph1 deficiency plays a critical role in tumor formation and development.
RESULTS
Mcph1 deficiency promotes long-latency tumor formation in mice
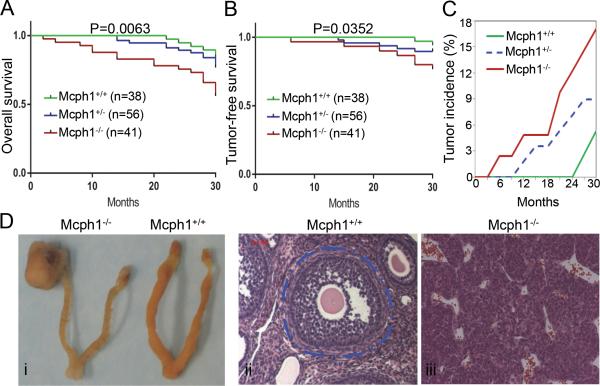
We have previously generated single Mcph1−/− mice that have an increased propensity to malignancies.17 To assess whether Mcph1 loss contributes to tumor susceptibility, we analyzed the viability of a cohort of Mcph1+/+ (n = 38), Mcph1+/− (n = 56), and Mcph1−/− (n = 41) mice over a period of 2.5 years. The survival of Mcph1−/− mice is significantly lower than that of the wild-type (WT) mice (Figure 1A, 1B). The results also showed that 17.1% of Mcph1−/− mice succumbed to malignant tumors while only 5.3% of Mcph1+/+ mice died from tumors (Figure 1C). Interestingly, Mcph1+/− mice were approximately two times more likely to develop spontaneous tumors (8.9%) than WT (Figure 1C). Although most of Mcph1−/− mice developed tumors after 1-2 years of age, a few tumors occurred as early as 6 months of age (Figure 1C). All tumors that evolved in Mcph1−/− mice originated from lymph nodes (lymphoma) or ovaries (granuloma cell tumors) (Figure 1D). Therefore, this result demonstrates that Mcph1 loss in mice promotes tumor formation with long latency.
Figure 1. Mcph1 deficiency promotes tumor formation in mice.
(A) Overall survival of a cohort of Mcph1+/+ (n=38), Mcph1+/− (n=56), and Mcph1−/− (n=41) mice monitored for 2.5 years. (B) Tumor-free survival of the cohort described in (A). (C) The cumulative tumor incidence in the cohort described in (A). Percent cumulative tumor incidence for each genotype plotted as a function of time, as indicated. (D) Representative ovarian tumors occurred in Mcph1−/− mice. (i) Gross images of ovarian tumor and ovaries. (ii and iii) H&E staining of normal ovaries (ii) and ovarian tumors (iii). The structure circled with dashed blue line is an example of a complete mature normal ovarian follicle in Mcph1+/+ mice (in ii), compare to panel iii showing the structure of ovaries with tumors in Mcph1−/− mice.
Mcph1−/−p53−/− mice display accelerated tumorigenesis
As we have shown, Mcph1−/− mice developed tumors with long latency (Figure 1). We suspected that Mcph1 deficiency may not be very effective in promoting tumors because intact p53, a guardian of the genome, may suppress propagation of tumorigenesis.25-27 To test this hypothesis, we crossed Mcph1−/− into p53−/− to generate Mcph1−/−p53−/− mice, which were confirmed by Southern blot analysis or PCR-based method (Figure 2A, 2B). Typically, Mcph1+/+p53−/− (n = 33) and Mcph1+/−p53−/− (n = 49) had an average life span of 16-18 weeks (Figure 2C). However, the medium survival time of Mcph1−/−p53−/− mice (n = 15) was only about 10 weeks (Figure 2C, 2D, and Supplementary Table S1). The significant increase in tumor predisposition observed in Mcph1−/−p53−/− mice indicates that Mcph1 deficiency and p53 loss synergize to promote tumorigenesis.
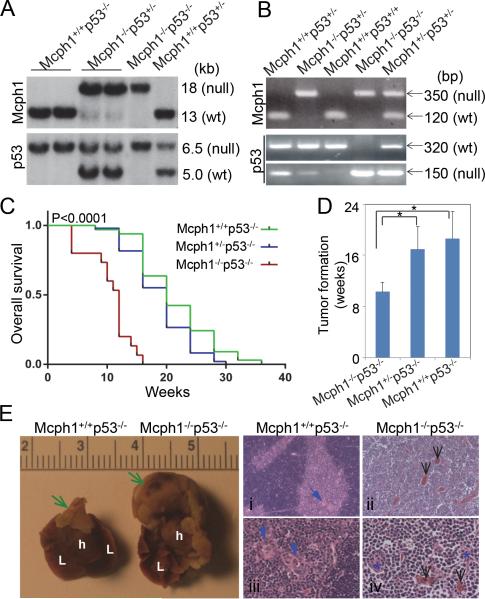
Figure 2. Mcph1 deficiency accelerates tumorigenesis in the absence of p53.
(A) Southern blot analysis of mice genotyping. For Mcph1, the sizes of the WT and null alleles are 13 kb and 18 kb, respectively; for p53, the sizes are 6.5 kb (WT) and 5.0 kb (null). (B) PCR-based genotyping of mice. For Mcph1, the sizes of the PCR products are 350 bp (null) and 120 bp (WT); for p53, the sizes are 320 bp (WT) and 150 bp (null). (C) Kaplan-Meier survival analysis of Mcph1+/+p53−/− (n=33), Mcph1+/−p53−/− (n=49), and Mcph1−/−p53−/− (n=15) mice. (D) Average tumor onset in Mcph1+/+p53−/−, Mcph1+/−p53−/−, and Mcph1−/−p53−/− mice. * P<0.05. (E) Representative thymic lymphoma in Mcph1−/−p53−/− mice. (Left) Gross images of normal thymus and thymic lymphoma (scale=cm). Green arrows indicate normal thymus (in Mcph1+/+p53−/−) or thymic lymphoma (in Mcph1−/−p53−/−). L, lung; h, heart. (Right) Histological characterization of thymus or thymic lymphoma. Tissues were cut from normal thymus (Mcph1+/+p53−/−) or thymic lymphoma (Mcph1−/−p53−/−), and stained with H&E. Normal thymus is composed of a dark-staining basophilic cortex and a light-staining eosinophilic medulla (i, iii). Normal cortex contains numerous densely packed lymphocytes, and the medulla has fewer lymphocytes and more thymic corpuscles (blue arrows). However, there is no definite cortex and medulla structure in thymic lymphoma from Mcph1−/− p53−/− mice (ii, iv), and the entire region is composed of densely packed lymphocytes and infiltrated with more blood vessels (black arrows). Thymic corpuscles are almost disrupted and infiltrated with more lymphocytes (blue asterisks in panel iv).
Of all Mcph1−/−p53−/− mice analyzed, 80% died of lymphomas and three other mice of unidentified causes (Figure 2E, Table S1). However, tumor spectra in Mcph1+/+p53−/− and Mcph1+/−p53−/− mice were broader than those in Mcph1−/−p53−/−. Mcph1+/+p53−/− mice presented 60.6% of lymphomas and 15.2% of solid tumors, including 9.1% of sarcomas. Mcph1+/−p53−/− mice developed 59.2% of lymphomas and 18.4% of solid tumors, including 10.2% of sarcomas (Table S1). These data indicate that Mcph1 deficiency mainly enhances susceptibility of malignant lymphomas in p53−/− mice.
Mcph1 deficiency in the absence of p53 induces chromosomal aberrations and aneuploidy
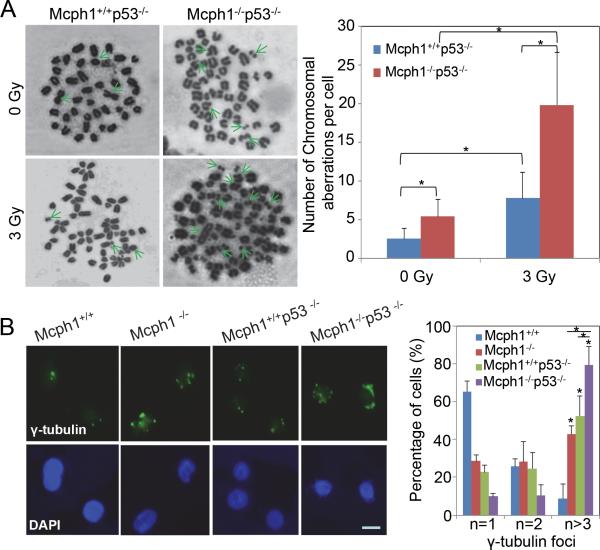
To understand how Mcph1 deficiency contributes to tumorigenesis, we used a metaphase spread assay to examine chromosomal aberrations in primary lymphomas originated in mice. As shown in Figure 3A, irradiated Mcph1−/−p53−/− lymphoma cells harbored more structural aberrations, including chromosomal breaks and translocations, than the irradiated Mcph1+/+p53−/− cells. Importantly, Mcph1−/−p53−/− cells exhibited more aneuploidy than Mcph1+/+p53−/− (Figure 3A), which was further confirmed by spectral karyotyping (SKY) analysis (Table 1). In addition, centrosome abnormalities were observed in Mcph1−/−, Mcph1+/+p53−/−, and Mcph1−/−p53−/− MEFs, as indicated by staining of γ-tubulin, a marker of centrosomes (Figure 3B). The percentage of cells with supernumerary centrosomes in Mcph1−/−p53−/− was significantly higher than that in Mcph1−/− or p53−/− MEFs (Figure 3B), indicating Mcph1 deficiency in the absence of p53 promotes abnormal multiplication of centrosomes, which may contribute to aneuploidy in Mcph1−/−p53−/− tumors. Therefore, these findings show that Mcph1 deficiency enhances chromosomal aberrations and aneuploidy in p53−/− cells.
Figure 3. Mcph1 deficiency induces chromosomal aberrations and centrosome abnormalities in the absence of p53.
(A) Chromosomal aberrations are differentially induced in IR-treated Mcph1+/+p53−/− and Mcph1−/−p53−/− primary lymphoma cells. Cells were treated with or without IR (3 Gy), and subjected to the metaphase spread assay to examine chromosomal aberrations 3 h later. (Left) Representative images of metaphase spreads. Green arrows indicate chromosomal aberrations: chromosomal breaks, fusions, and ring chromosomes. (Right) Quantitative analysis of chromosomal aberrations per cell for each genotype. The number of chromosomal aberrations was counted per spread from at least 30 spreads for each sample, and at least two pairs of primary lymphomas were used for each genotype. These experiments were repeated in duplicate, and the data represented as mean ± s.d. (* P<0.05). (B) Centrosome multiplication is induced in Mcph1−/−p53−/− MEFs. Immunofluorescent staining of centrosome marker γ-tubulin was used to detect the number of centrosomes in a cell. (Left) Representative images of γ-tubulin staining. Scale bar, 25 nM. (Right) Quantitative analysis of γ-tubulin foci per cell for each genotype. The number of γ-tubulin foci was counted per cell from at least 300 cells per sample. These experiments were repeated twice in duplicate for each MEF (* P<0.05). n, number of γ-tubulin foci (representing the number of centrosomes) in one cell.
Table 1.
Karyotype analysis of MCPH1+/+p53−/− and MCPH1−/−p53−/− lymphomas
| Genotype | Phenotype | Composite Karyotype |
|---|---|---|
| MCPH1+/+p53−/− | Thymic lymphoma | 39(2n),X,−X,+5,+Der(10)T(10B2;17B),−13,−17[4] 40(2n),X,−X,+5,+Del(10B2),−13[4] 39(2n),XX,−13[2] 40(2n),XX[1] |
| MCPH1−/−p53−/− | Thymic lymphoma | 48−53,X,−Y +2,+3,+4, +6,+8,+9,+11,DEL(12?),+14,+14,DEL(14?),+15,+15 ,+16,−17, [cp12] 89,X,−Y,+1,+2,+2,+4, +5,+n,+12+13,+H+15+15,−17+18+18,− 19,+X,[1] 82,X,− Y,+ROB(2;),ROB(3;3),ROB(4;4),DER(4)T(3?;4E2), +DER(5)T(5?D;10?),+ROB(5;5),+DER(6)T(6A1;12 ?),+ROB(6;6),+DER(8)T(8C?1;10?),−10,− 11,DEL(12C1),+13,+13,+DEL(14D3)×2,+15,−16, −16,−18,−19,−19,+X,[1] (~19 fragments) |
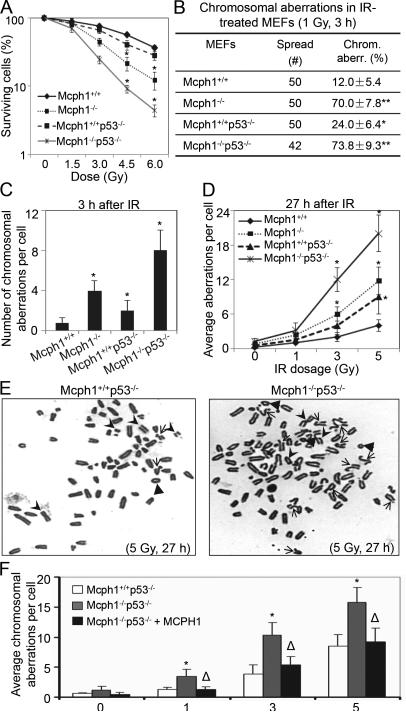
To further investigate whether Mcph1 depletion contributes to genomic instability in cells with a p53−/− background, we examined the radiosensitivity of Mcph1+/+, Mcph1−/−, Mcph1+/+p53−/−, and Mcph1−/−p53−/− MEFs. As shown in Figure 4A, Mcph1−/− and Mcph1−/−p53−/− MEFs were more sensitive to IR than Mcph1+/+ or Mcph1+/+p53−/− MEFs. We next performed a metaphase spread assay and examined chromosomal aberrations shortly (3 h) after IR (1 Gy). Mcph1−/− or Mcph1−/−p53−/− MEFs exhibited more chromosomal aberrations (70.0% and 73.8%, respectively) than Mcph1+/+ (12.0%) and Mcph1+/+p53−/− (24.0%) MEFs (Figure 4B). Also, the average number of chromosomal aberrations per cell was significantly higher in IR-treated Mcph1−/− and Mcph1−/−p53−/− MEFs 3 h after IR (Figure 4C). In parallel, we evaluated chromosomal aberrations in MEFs 27 h after IR. Compared with that in Mcph1+/+ MEFs, the number of chromosomal aberrations in Mcph1−/− and Mcph1−/−p53−/− MEFs was still notably higher 27 h after IR (Figure 4D). After IR treatment, there were three major types of structural aberrations observed in Mcph1-deficient MEFs: chromatid breaks, chromosome breaks, and ring chromosomes (Table S2). Chromatid breaks were prominent in Mcph1−/− and Mcph1−/−p53−/− MEFs (Table S2). Moreover, asymmetric chromatid-type exchanges, which are the result of fusions between two or more broken chromatids and represent defective HR, were frequently detected in Mcph1−/−p53−/−, but rarely found in Mcph1+/+p53−/− MEFs (Figure 4E). This suggests that HR is severely impaired in IR-treated, Mcph1-deficient MEFs.
Figure 4. Mcph1 deficiency enhances genomic instability in p53 null background.
(A) Mcph1−/−p53−/− MEFs are more sensitive to IR. Mcph1+/+, Mcph1−/−, Mcph1+/+p53−/−, and Mcph1−/−p53−/− MEFs were irradiated with increasing doses of IR (0, 1.5, 3, 4.5, and 6 Gy). Cells were counted 6 days later and the counts were normalized to the number of cells from unirradiated controls for each genotype. These experiments were triplicated, and the data represented as mean ± s.d. (* P<0.05, compared with Mcph1+/+ MEFs at the indicated doses). (B) Percentage of cells with chromosomal aberrations in Mcph1+/+, Mcph1−/−, Mcph1+/+p53−/−, and Mcph1−/−p53−/− MEFs treated with IR (1 Gy, 3 h). (* P<0.05, compared with Mcph1+/+ MEFs). (C) The average number of chromosomal aberrations per cell in Mcph1+/+, Mcph1−/−, Mcph1+/+p53−/−, and Mcph1−/−p53−/− MEFs treated with IR (1 Gy, 3 h). (* P<0.05, compared with Mcph1+/+ MEFs). (D) The average number of chromosomal aberrations per cell 27 h after IR at the indicated doses. Mcph1+/+, Mcph1−/−, Mcph1+/+p53−/−, and Mcph1−/−p53−/− MEFs were irradiated with increasing doses of IR (0, 1, 3, and 5 Gy), and then analyzed with the metaphase spread assay. (* P<0.05, compared with Mcph1+/+ MEFs at the indicated doses). (E) Representative metaphase spread of Mcph1+/+p53−/− and Mcph1−/−p53−/− MEFs 27 h after IR (5 Gy). Arrows, chromatid breaks or translocations; arrowheads, chromosomal breaks or fusions; triangles, ring chromosomes. (F) Ectopic MCPH1 can largely reduce chromosomal aberrations in Mcph1−/−p53−/− MEFs. MCPH1 was stably transfected into Mcph1−/−p53−/− MEFs, namely Mcph1−/−p53−/−+MCPH1, and the average number of chromosomal aberrations per cell was counted as described in (D). (* P<0.05, compared with either Mcph1+/+p53−/− or Mcph1−/− p53−/−+MCPH1 MEFs at the indicated doses; Δ, P>0.05, compared with Mcph1+/+p53−/− MEFs at the indicated doses).
To confirm that Mcph1 deficiency is indeed the cause of genomic instability in Mcph1−/−p53−/− MEFs, we performed rescue experiments in which we restored human MCPH1 in Mcph1−/−p53−/− MEFs and then analyzed genomic stability. We first assessed the effect of ectopic MCPH1 on cell proliferation with both BrdU incorporation assay and flow cytometry. Compared with Mcph1+/+ MEFs, Mcph1−/− exhibited proliferation inhibition (Figure S1AS1D). When lacking p53, Mcph1−/−p53−/− MEFs exhibited more rapid growth (Figure S1E) and a larger S phase cell population than Mcph1+/+p53−/− (Figures S1F, S1G). This phenomenon was rescued when we reconstituted Mcph1−/−p53−/− MEFs with ectopic MCPH1 (Figure S1ES1G). Moreover, ectopic MCPH1 significantly reduced the radiosensitivity of Mcph1−/−p53−/− MEFs to IR (Figure S1H). Next, we determined whether ectopic MCPH1 could restore and maintain genomic stability in Mcph1−/−p53−/− MEFs 27 h after IR. We irradiated Mcph1+/+p53−/−, Mcph1−/−p53−/−, and MCPH1-ectopic MEFs (i.e. Mcph1−/−p53−/−+MCPH1) with increasing doses of IR, and performed the metaphase spread assay 27 h later. Mcph1−/−p53−/−+MCPH1 MEFs presented an average of chromosomal aberrations that was significantly lower than that in Mcph1−/−p53−/−, but was comparable to that in Mcph1+/+p53−/− (Figure 4F). This shows that ectopic MCPH1 can restore genomic stability to a large extent in Mcph1−/−p53−/− MEFs. Taken together, these results support our hypothesis that Mcph1 deficiency indeed leads to genomic instability in p53−/− tumors.
Mcph1 deficiency reduces DNA DSB repair
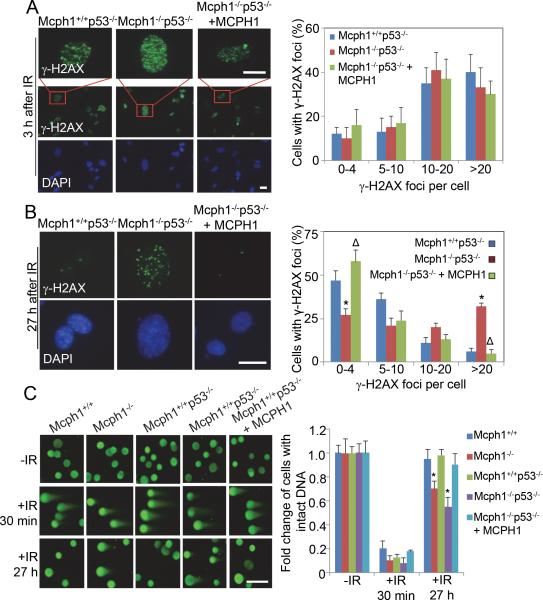
To determine how Mcph1 deficiency results in genomic instability, we evaluated DNA DSB by IR-induced foci formation (IRIF) of γ-H2AX and neutral-pH Comet assay.28-29 γ-H2AX foci were detectable in both Mcph1-intact (Mcph1+/+ and Mcph1+/+p53−/−) and Mcph1-deficient (Mcph1−/− and Mcph1−/−p53−/−) MEFs shortly (3h) after IR (Figures 5A, S2A), although Mcph1 foci is diminished in Mcph1-deficient MEFs (e.g. Mcph1−/−p53−/− MEFs, Figure S3). IRIF of γ-H2AX were also evaluated 27 h after IR, at which time point the damaged DNA are regarded to be nearly repaired in cells with intact DDR.30 At 27 h after IR, γ-H2AX foci still retained in Mcph1−/− MEFs and Mcph1−/−p53−/− MEFs, while γ-H2AX foci almost completely disappeared in Mcph1+/+ MEFs and Mcph1+/+p53−/− MEFs (Figures 5B, S2B). We then performed neutral-pH Comet assay to further assess DSB repair in Mcph1−/− p53−/− MEFs. As shown in Figure 5C and Figure S4, the percentage of cells with intact DNA was significantly reduced in Mcph1-deficienct MEFs 27 h after IR as compared Mcph1-intact MEFs, indicating IR-induced DSBs were not completely repaired at this time point in Mcph1−/−p53−/− and Mcph1−/− MEFs. This result was consistent with the finding described in Figure 4D, where chromosomal aberrations in Mcph1−/−p53−/− were more prominent than in Mcph1+/+p53−/− MEFs. Meanwhile, when ectopic MCPH1 was reintroduced into Mcph1−/−p53−/− MEFs (namely Mcph1−/−p53−/−+MCPH1), γ-H2AX foci (Figure 5B) and DSBs (revealed by Comet assay, Figure 5C) were significantly decreased 27 h after IR to the levels comparable to those observed in Mcph1+/+p53−/− MEFs (Figures 5B, 5C), indicating reconstituted MCPH1 can largely rescue DSB repair defects in Mcph1−/−p53−/− MEFs. Collectively, these results reveal that Mcph1 deficiency indeed impairs DNA DSB repairs in cells derived from Mcph1−/− mice.
Figure 5. Mcph1 deficiency impairs DNA DSB repair.
(A) Mcph1 deficiency does not affect γ-H2AX foci formation. An immunofluorescence staining assay was used to examine foci formation of γ-H2AX in the indicated MEFs 3 h after IR (2 Gy). (Left) Representative images of γ-H2AX foci. A large field of cells with γ-H2AX staining is also shown here. Scale bar, 25 μM. (Right) Quantitative analysis of γ-H2AX foci per cell in each genotype from three independent experiments. (B) Immunofluorescence analysis of residual γ-H2AX foci in the indicated MEFs 27 h after IR (2 Gy). (Left) Representative images of γ-H2AX foci. Scale bar, 25 μM. (Right) The graph represents the number of γ-H2AX foci per cell in the indicated MEFs. (* P<0.05, compared with Mcph1+/+p53−/− MEFs; Δ, P>0.05, compared with Mcph1+/+p53−/− MEFs). (C) DSB repair is impaired in Mcph1-deficienct MEFs as determined by Neutral-pH Comet assay. (Left) Representative images. A large file of these images is provided in Supplementary Figure S4. Scale bar is 50 μM. (Right) Quantitative analysis of three independent experiments. More than 96% of the untreated cells contained tail moments less than 2, which was set as the parameter for cells with intact DNA. Percentage of no-IR cells with intact DNA (tail moment less than 2) was set as 1 for each genotype. At least 75 cells were scored in each sample and each value represents the mean ± s.d. of three independent experiments. (* P<0.05, compared with Mcph1+/+ MEFs).
DSB repair by HR is defective in Mcph1−/−p53−/− MEFs
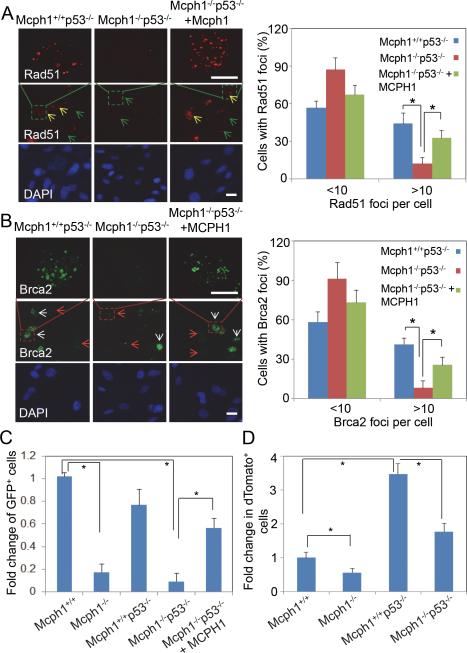
In mammalian cells, two conserved pathways are involved in DSB repair, namely HR and non-homologous end joining (NHEJ).31, 32 To determine the role of Mcph1 in HR repair in Mcph1−/−p53−/− MEFs, we examined IRIF of Rad51, a marker of HR competence, in IR-treated MEFs. As shown in Figure 6A, Rad51 foci were formed and detected 3 h after IR in Mcph1+/+p53−/− MEFs, but dramatically suppressed in Mcph1−/−p53−/− MEFs. The decrease of Rad51 foci in Mcph1−/−p53−/− MEFs was restored to a large extent by ectopic MCPH1 (Figure 6A). Additionally, we evaluated foci formation of Brca2, another key player in HR after IR. Brca2 foci exhibit a similar pattern of Rad51 (Figure 6B), further demonstrating HR activity is potentially impaired in Mcph1−/−p53−/− MEFs. The decreased foci formation of Rad51 and Brca2 in Mcph1−/−p53−/− MEFs is not due to decreased population of the cycling cells (i.e. S or G2 population); the cycling cell population was not decreased in Mcph1−/−p53−/− as compared to Mcph1+/+p53−/− MEFs (Figures S1F, S1G, and S5). To further confirm the defective HR DNA repair in Mcph1-deficienct MEFs, we analyzed their HR repair capacity by using an in vivo HR repair assay. Mcph1-deficienct MEFs showed a significant decrease of HR-repaired GFP+ population. Meanwhile, ectopic MCPH1 significantly increased the number of HR-repaired GFP+ cells (Figures 6C, S6). Together, these results show that Mcph1 deficiency impedes HR activity in Mcph1-deficient cells, which can be largely rescued by ectopic MCPH1.
Figure 6. HR and NHEJ are reduced in Mcph1−/−p53−/− MEFs.
(A, B) Rad51 and Brac2 foci formation are impaired in Mcph1−/−p53−/− MEFs and partially rescued by ectopic MCPH1. Three different types of MEFs were treated with IR (8 Gy), and stained for immunofluorescent analysis of Rad51 (A) and Brca2 (B) 3 h after IR. The large fields of Rad51 and Brca2 foci staining are also shown here. Yellow arrows, Rad51-positive cells; green arrows, Rad51-negative cells; white arrows, Brca2-positive cells; red arrows, Brca2-negative cells. Scale bar, 50 μM. * P<0.05. (C) HR activity is significantly decreased in MCPH1-deficienct MEFs according to the analysis of the results of the HR repair assay. The HR activity in each MEF was represented by the relative percentage of GFP+ cells. The relative percentage of GFP+ cells in each MEF was calculated as the percentage of GFP+ cells in I-SceI-transfected MEFs, which was first subtracted by the percentage of GFP+ cells in control plasmid (pCAGGS)-transfected MEFs, and then normalized by transfection efficiency determined by pEGFP-C1. Each value in the graph is relative to the percentage of GFP+ cells in Mcph1+/+ MEFs, which was set at 1, and is shown as the mean ± s.d. of three independent experiments (* P<0.05). (D) NHEJ activity is reduced in Mcph1-deficient MEFs. NHEJ activity was reported as the ratio of cells that were double positive for red (dTomato+) and green (GFP+) fluorescence to the total cells that were only positive for green fluorescence (GFP+). The level of NHEJ activity in Mcph1+/+ MEFs was set as “1”. The values for all other MEFs were normalized to that in Mcph1+/+ MEFs. This experiment was performed twice in duplicates for each MEFs (* P<0.05).
NHEJ activity is decreased in Mcph1-deficient MEFs
We further investigated whether NHEJ, another major repair pathway for DSBs, is impaired in Mcph1−/−p53−/− MEFs. To this end, we performed in vivo NHEJ assay. In line with previous studies, 1, 11 NHEJ activity in Mcph1−/− MEFs was notably decreased as compared to Mcph1+/+ MEFs (Figures 6D, S7). The level of NHEJ activity in Mcph1−/−p53−/− MEFs was also decreased as compared to Mcph1+/+p53−/− MEFs although it remained similar level to Mcph1+/+ MEFs. To further determine whether NHEJ-associated V(D)J recombination process is involved in lymphomagenesis, we examined developmental stages at which thymic lymphomas developed in Mcph1−/−p53−/− mice. We performed immunofluorescent staining of lymphoma using antibodies against CD4, CD8, CD44, and CD25, the molecular markers of thymocytes development. 33, 34 The analyses showed that lymphomas from Mcph1+/+p53−/−, Mcph1+/−p53−/−, and Mcph1−/−p53−/− mice stained positive for CD4 or CD8 (Figure S8). Most of lymphoma cells were CD44+, indicating that these cells are premature for lymphocyte development. Compared to Mcph1+/+p53−/− lymphomas, Mcph1−/−p53−/− exhibited much less CD25+ cells, suggesting lymphomas in Mcph1−/−p53−/− mice were originally formed around double negative stages (DN) 1/2, the stages slightly before the NHEJ-directed V(D)J recombination processed. Collectively, these data support that the decreased NHEJ activity in Mcph1−/−p53−/− cells may not be directly involved in lymphomagenesis in Mcph1−/−p53−/− mice.
DISCUSSION
In this study, we have generated and analyzed both Mcph1−/− and Mcph1−/−p53−/− mice. By using our unique germline Mcph1−/− mouse model, we have found that Mcph1 loss itself triggers long-latency tumor formation in mice. This phenomenon may be attributed to the fact that Mcph1 deficiency needs to cooperate with other aberrant tumor suppressors or oncoproteins to overcome the barrier of “guardian of the genome” to form a tumor.35 Indeed, when we crossed Mcph1−/− into p53−/− mice, Mcph1 deficiency enhanced tumor susceptibility in p53-null background. Deficiency of other DDR proteins (e.g. BRCA1, 53BP1, H2AX, and NBS1) in p53-null background has also accelerated lymphomagenesis in mice,25,30,36,37 supporting our observation in Mcph1−/−p53−/− mice. Meanwhile, human MCPH1 is located on chromosome 8p23.1, a locus on which loss of heterozygosity is frequently observed in human cancers. A number of previous studies have shown that MCPH1 is aberrantly downregulated in breast and ovarian cancer, particularly in higher grade tumors. 1,20,21 Moreover, we have analyzed a large cohort of Oncomine datasets including TCGA data, which revealed that MCPH1's copy number and mRNA level are notably reduced in solid tumors (Figure S9).38-40 Thus, we provide convincing genetic evidence that Mcph1 may act as a novel tumor suppressor.
Mechanistically, our study demonstrates that Mcph1 deficiency promotes aneuploidy and causes defective DSB repairs, which together may contribute to increased tumor susceptibility in Mcph1−/−p53−/− mice. First, our data reveal that loss of Mcph1 leads to substantially increased aneuploidy in Mcph1−/−p53−/− lymphomas. Aneuploidy is a well-known hallmark in cancers and it may arise from centrosome malfunction or defects in mitotic segregation.41-44 In fact, centrosome amplification has been reported in MCPH1-knockdown U2OS cells45 and Mcph1−/− DT40 cells19. Consistently, we observed centrosome hyper-amplification in Mcph1−/−p53−/− MEFs. Interestingly, the staining pattern of γ-tubulin, a centrosome marker, appears to be heterogeneous in Mcph1−/−p53−/− MEFs, which may be caused by increased centriole number associated with Mcph1 deficiency. Previous studies have also shown that loss of Mcph1 dysregulated Chk1 or Aurora A/plk1-associated mitotic pathways. 14, 45, 46 Therefore, Mcph1 deficiency causes abnormal communications between mitosis and centriole duplication, and thereby leads to centrosome hyper-amplification, which, in turn, promotes aneuploidy and ultimately tumorigenesis. Second, our studies reveal that Mcph1 deficiency leads to defective DNA repairs and severe chromosomal aberrations in p53-null background, as indicated by foci formation assay, comet assay, metaphase spread assay and DNA repair assays. In consistent with previous reports,1,9,11,12,17 we observed Mcph1 deficiency substantially decreased HR activity in Mcph1−/−p53−/− MEFs. Thus, it is possible this HR defects causes genomic instability and as a result, may contribute to tumorigenesis for the Mcph1−/−p53−/− mice. In fact, lines of evidence support the role of defective HR repair in promoting tumor formation; deletion/mutations of HR repair genes commonly occur in many cancer types, e.g. BRCA1/2 in breast and ovarian cancer,47, 48 RAD54 /CtIP in lymphoma and colon cancer,49,50 RAD51B in lipoma,51 and RECQL4 in osteosarcoma52. In addition to defective HR repair, NHEJ repair is largely impaired in Mcph1−/−p53−/− MEFs as compared to p53-null MEFs, which is in line with previous studies.1,11 However, NHEJ may be not directly involved in lymphomagenesis of Mcph1−/−p53−/− mice since lymphoma in these mice originally formed around DN1/2 stage, slightly before the stage of NHEJ-directed V(D)J process. Consistent with our study, lymphomas developed in mice deleted for both NHEJ genes and p53 (ref. 53-55) are believed to be mainly induced by a failure of apoptosis due to absence of p53, leading to the inappropriate survival of potentially defective cells that were lymphomagenic, supporting NHEJ-associated abnormalities may be not involved in lymphomagenesis in p53-null mice.
Analysis of our single Mcph1−/− mice shows that Mcph1 deficiency itself triggers long-latency tumor formation; it is possible that those cells with genomic instability induced by Mcph1 deficiency is eliminated by p53 or other pathway-mediated apoptosis and few tumorigenesis occurs. When we cross Mcph1−/− into p53−/− mice, Mcph1 deficiency greatly enhances tumor susceptibility in p53-null background with increased aneuploidy and chromosomal structural aberrations. Here, p53 loss may attenuate apoptosis, allowing cells with severe chromosomal aberrations to survive and transform to tumor cells in the absence of Mcph1. These data together demonstrate that Mcph1 can cooperate with p53, a master tumor suppressor, to prevent tumor formation. In addition to its critical role in DDR signaling and DNA repair, recently, MCPH1 has been shown to be a novel regulator for p53 protein stability using human breast cancer cell lines.56A MCPH1 can interact with MDM2 (murine double minute 2) and p53, and control p53 stability by regulating MDM2-mediated p53 ubiquitination. This post-translational modification of p53 is likely to be another mechanism by which MCPH1 functions as a tumor suppressor.
Of note, Mcph1−/−p53−/− mice mainly develop lymphoma, but not carcinomas (i.e. epithelial tumors), although both MCPH1 and p53 aberrations have been frequently observed in human carcinomas (including breast and ovarian cancer).20, 57 Similarly, other studies have shown that carcinomas cannot be developed in p53−/− mice or p53−/− mice with deficiency of other DDR proteins (such as BRCA1, 53BP1, H2AX, and NBS1). 25,30,36,37 This preferential bias of tumorigenesis may occur at least partially because of p53-null mutation in mice. In humans, p53-associated carcinomas mainly harbor point mutations, but not the large deletions (null status) of p53 (ref. 58). In fact, two p53 gain-of-function mutant mouse models, such as p53R172H/− and p53R270H/− , have been observed to frequently develop carcinomas.57,59 This demonstrates that gain of function of p53 mutants (regarding as “oncogenes”), but not the null mutation of p53, plays a key role in the development of carcinomas. In our current Mcph1−/−p53−/− mouse model, p53 is in a null background, lacking gain of function of p53 mutants, therefore, carcinomas have not been developed in Mcph1−/− p53−/− mice. Since MCPH1 aberrations are common events in several types of human carcinomas, it will be critical to evaluate Mcph1's role in carcinomas such as breast cancer using mouse models. In the future, we will exploit our epithelia-specific Mcph1−/− mice crossed with p53R270H/- , Ptenfloxed/floxed, or K-RasG12D mutant mice to elucidate pathological role of Mcph1 in the development of epithelial tumors.
Together, our data clearly demonstrate that Mcph1 deficiency can promote tumor initiation in Mcph1−/− mice and synergize with p53−/− mice in lymphomagenesis in Mcph1−/−p53−/− mouse model. We further reveal that Mcph1 deficiency can induce chromosomal structural and numeric aberrations in tumors developed in Mcph1−/−p53−/− mice. Our data also demonstrate that Mcph1 deficiency causes defective DNA DSB repair. These mechanistic studies will provide new insights into development of novel targeted therapy for MCPH1-deficient cancer patients in the future.
MATERIALS AND METHODS
Mice and genotyping
All mice were bred and maintained in a conventional animal facility under protocol AN-3575 approved by Baylor College of Medicine. Previously established Mcph1−/− mice17 were bred with p53−/− mice 60 to generate Mcph1−/−p53−/− mice. Mice were monitored every other day for the development of tumors and other signs of poor health. Those that had difficulty breathing and a hunched back, or signs of thoracic congestion because of an enlarged thymus, were sacrificed within 1 day of detection of the problem and screened for tumors.36
Genotyping was performed with Southern blot or a PCR-based method. For Southern blot, genomic DNA was digested with BamHI (New England BioLabs, Ipswich, MA, USA), and then hybridized with probes appropriate for Mcph1 or p53. For the PCR-based method, the primers used for Mcph1 and p53 were provided upon request.
Tumor histology and developmental staging
Tissues from solid tumors were stained with hematoxylin and eosin (H&E) according to a standard histological protocol, and examined under the microscope by experienced pathologists.
Lymphoma staging was determined according to the immunofluorescent staining of lymphocyte markers, as described previously with modifications. 61, 62 Before staining, Fc II/III receptors were blocked first by preincubation with anti-CD16/32 (Fc-Block, BD Biosciences, San Jose, CA, USA). Cell markers were then stained either directly (CD4-FITC, CD8-FITC; Sigma, St. Louis, MO, USA), or indirectly (CD44, CD25; Novus Biologicals, Littleton, CO, USA) in 1% BSA/PBS for 2 h at 4°C. The indirectly-stained slides were further incubated with goat-anti-mouse (or rabbit) IgG-Alexa Fluor 594 or Alexa Fluor 488 (Life Technologies, Carlsbad, CA, USA) for 2 h at 4°C. The slides were counterstained with DAPI, and finally observed microscopically. About ~1000 cells were counted in each slide from each lymphoma, and at least two lymphomas were used for each genotype. The experiments were duplicated for analysis.
Primary thymic lymphoma cells
Thymic lymphoma cells were isolated from Mcph1+/+, Mcph1−/−, Mcph1+/+p53−/−, and Mcph1−/−p53−/− mice and maintained in RPMI 1640 complete medium with 10 ng/ml of IL-2 (BioLegend, San Diego, CA, USA) at low passages (P<5 passages) for future use.
Metaphase spread assay
Metaphase spreads were performed as previously described.63 Primary thymic lymphoma cells and MEFs were irradiated at the indicated doses. Cells were then treated with KaryoMax Colcemid (Life Technologies, Grand Island, NY, USA) for 2 h and metaphase spreads were prepared 3 h and/or 27 h after IR, according to a standard protocol. At least 30-50 metaphase spreads were analyzed for each case, and at least two pairs of lymphomas or MEFs from each genotype were used to repeat the experiments.
Spectral karyotyping (SKY) analysis
SKY analysis was carried out according to the manufacturer's protocol.64 The cocktail of mouse chromosome paints was obtained from Applied Spectral Imaging (ASI, Vista, CA, USA). Chromosomes were counterstained with DAPI. For each case, a minimum of 10 metaphase cells was analyzed with SKY, and at least two lymphomas were used for each genotype.
Neutral-pH Comet assay
Neutral Comet assay was performed with the Trevigen's CometAssay kit (Trevigen, Gaithersburg, MD, USA) according to the manufacturer's instructions. MEFs were treated with or without IR (4Gy), and then subjected to Comet assay at 30 min or 27 h after IR. SYBR Green I-stained Comet images were captured with fluorescence microscopy. Tail moments were determined in at least 75 cells/slide with the Comet Assay IV software (Perceptive Instruments, Bury St Edmunds, UK). Data were collected from three independent experiments.
HR repair assay
DR-GFP/I-SceI-based HR repair assay was performed with transient transfection. Briefly, MEFs (5 x 106 cells) were seeded into 6-well plates and transfected with both DR-GFP and either of pCAGGS (mock plasmid), pEGFP-C1 (GFP-expressing plasmid), or pCBASce (I-SceI-expressing plasmid) (1.25 μg each plasmid, totally 2.5μg/well). Forty-eight hours later, GFP+ cells were detected with LSR II flow cytometry (Becton Dickinson, San Jose, CA). Data were analyzed with Flowjo Version 10 (Tree Star, Ashland, OR, USA). This experiment was repeated at least three times.
NHEJ activity assay
NHEJ activity assay was performed as described previously with minor modifications.65 In this study, BamHI-cleaved pCSCMV-tdTomato was used as a target for NHEJ, and pEGFPC1 as a control plasmid. Briefly, MEFs were co-electroporated with pCSCMV-tdTomato plasmid (Plasmid 30530, Addgene, Cambridge, MA, USA) digested with BamHI, which cleaves between the promoter and the tdTomato open reading frame, and pEGFP-C1 at 2.5 μg of DNA per 106 cells each. Cells were then cultured for 10-12 h. The counts of tdTomato-expressing and GFP-expressing cells were estimated with LSRFortessa (BD Biosciences, San Jose, CA, USA), and data were analyzed with FlowJo Version 10 (Tree Star, Ashland, OR, USA). Two pairs of MEFs for each genotype were used, and the experiments were duplicated for analysis.
Immunofluorescent staining
IR-induced foci formation assay was performed as described previously.1 The primary antibodies used here were anti-GST-Mcph1 (gift from Dr. Shiaw-Yih Lin at MD Anderson Cancer Center, Houston, Texas), anti-γ-H2AX (Bethyl Laboratories, Montgomery, TX, USA; Cat# A300-081), anti-Rad51 (Santa Cruz Biotechnology, Santa Cruz, CA, USA; Cat# sc-8349), and anti-Brca2 (Santa Cruz Biotech, Santa Cruz, CA, USA; Cat# sc-28235). Centrosome analysis was performed as described elsewhere,66 with primary antibody anti-γ-Tubulin (Sigma, St. Louis, MO, USA; Cat# T6557). Centrosome numbers were counted in approximately 300 cells from at least 10 fields per type of cells. These experiments were repeated at least three times.
Flow Cytometry
MEFs were treated with or without the indicated doses of IR, collected by trypsinization, fixed by 75% ethanol, and kept at −20°C for at least 2 h. After centrifugation, MEFs were stained with propiduim iodide (PI, 2 μg/mL in PBS) and RNase A (10 μg/mL). FACS analysis was performed with FACScan (BD, San Jose, CA, USA) and the data analyzed with Flowjo software (Ashland, OR, USA).
BrdU incorporation assay
The BrdU incorporation assay was performed with the BrdU labeling and detection kit (Roche, Pleasanton, CA, USA), according to the manufacturer's instructions. BrdU-positive cells were stained intensely brown, and BrdU incorporation rate of the entire population was then scored. About 500 cells were counted for each case, and the data were analyzed from three independent experiments.
Statistical analysis
Cultured cell-based assays were repeated at least two or three times in triplicate and the significance was assessed with unpaired Student t-test. Kaplan-Meier survival curve and log-rank test were used for survival analysis. Error bars show s.d. P< 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health to K. Li (R01-CA155151 and R21-CA161513), a grant from the St. Luke's Episcopal Hospital McDonald Research Foundation to K. Li, and a grant from National Natural Science Foundation of China (to Y. Liang, 81372589). We thank BCM Cytometry and Cell Sorting Core (NCRR S10RR024574, NIAID AI036211, NCI P30CA125123) and Molecular Morphology Core Laboratory (PHS DK56338) for technical assistance. We also thank Drs. Zhijie Li, Han Lin, Ana Rodríguez (all from Baylor College of Medicine, Houston, TX, USA), and Guang Peng (UT MD Anderson Cancer Center, Houston, TX, USA) for providing anti-CD4/CD8 antibodies (ZL), assisting in NHEJ analysis (HL), editing of this manuscript (AR), and providing HR-related plasmids (GP).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Rai R, Dai H, Multani AS, Li K, Chin K, Gray J, et al. BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell. 2006;10:145–157. doi: 10.1016/j.ccr.2006.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin SY, Liang Y, Li K. Multiple roles of MCPH1/MCPH1 in DNA damage response, DNA repair, and cancer suppression. Yonsei Med J. 2010;51:295–301. doi: 10.3349/ymj.2010.51.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venkatesh T, Suresh PS. Emerging roles of MCPH1: expedition from primary microcephaly to cancer. Eur J Cell Biol. 2014;93:98–105. doi: 10.1016/j.ejcb.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Mohammad DH, Yaffe MB. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst) 2009;8:1009–1017. doi: 10.1016/j.dnarep.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 6.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 7.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 8.Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, et al. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- 9.Wood JL, Singh N, Mer G, Chen J. MCPH1 functions in an H2AX-dependent but MDC1-independent pathway in response to DNA damage. J Biol Chem. 2007;282:35416–35423. doi: 10.1074/jbc.M705245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang SZ, Lin FT, Lin WC. MCPH1/MCPH1 cooperates with E2F1 in the activation of checkpoint, DNA repair and apoptosis. EMBO Rep. 2008;9:907–915. doi: 10.1038/embor.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng G, Yim EK, Dai H, Jackson AP, Burgt I, Pan MR, et al. MCPH1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol. 2009;11:865–872. doi: 10.1038/ncb1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu X, Mondal G, Wang X, Wu J, Yang L, Pankratz VS, et al. Microcephalin regulates BRCA2 and Rad51-associated DNA double-strand break repair. Cancer Res. 2009;69:5531–5536. doi: 10.1158/0008-5472.CAN-08-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung JW, Leitch A, Wood JL, Shaw-Smith C, Metcalfe K, Bicknell LS, et al. SET nuclear oncogene associates with microcephalin/MCPH1 and regulates chromosome condensation. J Biol Chem. 2011;286:21393–21400. doi: 10.1074/jbc.M110.208793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruber R, Zhou Z, Sukchev M, Joerss T, Frappart PO, Wang ZQ. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat Cell Biol. 2011;13:1325–1334. doi: 10.1038/ncb2342. [DOI] [PubMed] [Google Scholar]
- 15.Singh N, Wiltshire TD, Thompson JR, Mer G, Couch FJ. Molecular basis for the association of microcephalin (MCPH1) protein with the cell division cycle protein 27 (Cdc27) subunit of the anaphase-promoting complex. J Biol Chem. 2012;287:2854–2862. doi: 10.1074/jbc.M111.307868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi L, Li M, Su B. MCPH1/MCPH1 represses transcription of the human telomerase reverse transcriptase gene. Gene. 2012;495:1–9. doi: 10.1016/j.gene.2011.12.053. [DOI] [PubMed] [Google Scholar]
- 17.Liang Y, Gao H, Lin SY, Peng G, Huang X, Zhang P, et al. MCPH1/MCPH1 is essential for mitotic and meiotic recombination DNA repair and maintaining genomic stability in mice. PLoS Genet. 2010;6:e1000826. doi: 10.1371/journal.pgen.1000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alderton GK, Galbiati L, Griffith E, Surinya KH, Neitzel H, Jackson AP, et al. Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat Cell Biol. 2006;8:725–733. doi: 10.1038/ncb1431. [DOI] [PubMed] [Google Scholar]
- 19.Brown JA, Bourke E, Liptrot C, Dockery P, Morrison CG. MCPH1/MCPH1 limits ionizing radiation-induced centrosome amplification. Oncogene. 2010;29:5537–5544. doi: 10.1038/onc.2010.302. [DOI] [PubMed] [Google Scholar]
- 20.Richardson J, Shaaban AM, Kamal M, Alisary R, Walker C, Ellis IO, et al. Microcephalin is a new novel prognostic indicator in breast cancer associated with BRCA1 inactivation. Breast Cancer Res Treat. 2011;127:639–648. doi: 10.1007/s10549-010-1019-4. [DOI] [PubMed] [Google Scholar]
- 21.Brüning-Richardson A, Bond J, Alsiary R, Richardson J, Cairns DA, McCormack L, et al. ASPM and microcephalin expression in epithelial ovarian cancer correlates with tumour grade and survival. Br J Cancer. 2011;104:1602–1610. doi: 10.1038/bjc.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venkatesh T, Nagashri MN, Swamy SS, Mohiyuddin SM, Gopinath KS, Kumar A. Primary microcephaly gene MCPH1 shows signatures of tumor suppressors and is regulated by miR-27a in oral squamous cell carcinoma. PLoS One. 2013;8:e54643. doi: 10.1371/journal.pone.0054643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Wu XB, Fan JJ, Mai L, Cai W, Li D, et al. MCPH1 Protein Expression in Normal and Neoplastic Lung Tissues. Asian Pac J Cancer Prev. 2013;14:7295–7300. doi: 10.7314/apjcp.2013.14.12.7295. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharya N, Mukherjee N, Singh RK, Sinha S, Alam N, Roy A, et al. Frequent alterations of MCPH1 and ATM are associated with primary breast carcinoma: clinical and prognostic implications. Ann Surg Oncol. 2013;20(Suppl 3):S424–432. doi: 10.1245/s10434-012-2715-6. [DOI] [PubMed] [Google Scholar]
- 25.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 28.Olive PL, Banáth JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 29.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward IM, Difilippantonio S, Minn K, Mueller MD, Molina JR, Yu X, et al. 53BP1 cooperates with p53 and functions as a haploinsufficient tumor suppressor in mice. Mol Cell Biol. 2005;25:10079–10086. doi: 10.1128/MCB.25.22.10079-10086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 32.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human nonhomologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 33.Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto K, Yoshikai Y, Moroi Y, Asano T, Ando T, Nomoto K. Two differential pathways from double-negative to double-positive thymocytes. Immunology. 1991;72:20–26. [PMC free article] [PubMed] [Google Scholar]
- 35.Efeyan A, Serrano M. p53: guardian of the genome and policeman of the oncogenes. Cell Cycle. 2007;6:1006–1010. doi: 10.4161/cc.6.9.4211. [DOI] [PubMed] [Google Scholar]
- 36.Bachelier R, Xu X, Wang X, Li W, Naramura M, Gu H, et al. Normal lymphocyte development and thymic lymphoma formation in Brca1 exon-11-deficient mice. Oncogene. 2003;22:528–537. doi: 10.1038/sj.onc.1206208. [DOI] [PubMed] [Google Scholar]
- 37.Kang J, Ferguson D, Song H, Bassing C, Eckersdorff M, Alt FW, et al. Functional interaction of H2AX, NBS1, and p53 in ATM-dependent DNA damage responses and tumor suppression. Mol Cell Biol. 2005;25:661–670. doi: 10.1128/MCB.25.2.661-670.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 40.Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 41.Duesberg P, Fabarius A, Hehlmann R. Aneuploidy, the primary cause of the multilateral genomic instability of neoplastic and preneoplastic cells. IUBMB Life. 2004;56:65–81. doi: 10.1080/15216540410001667902. [DOI] [PubMed] [Google Scholar]
- 42.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 43.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, et al. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 44.Tarapore P, Fukasawa K. Loss of p53 and centrosome hyperamplification. Oncogene. 2002;21:6234–6240. doi: 10.1038/sj.onc.1205707. [DOI] [PubMed] [Google Scholar]
- 45.Rai R, Phadnis A, Haralkar S, Badwe RA, Dai H, Li K, et al. Differential regulation of centrosome integrity by DNA damage response proteins. Cell Cycle. 2008;7:2225–2233. doi: 10.4161/cc.7.14.6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tibelius A, Marhold J, Zentgraf H, Heilig CE, Neitzel H, Ducommun B, et al. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J Cell Biol. 2009;185:1149–1157. doi: 10.1083/jcb.200810159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 48.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 49.Wong AK, Ormonde PA, Pero R, Chen Y, Lian L, Salada G, et al. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene. 1998;17:2279–2285. doi: 10.1038/sj.onc.1202150. [DOI] [PubMed] [Google Scholar]
- 50.Hiramoto T, Nakanishi T, Sumiyoshi T, Fukuda T, Matsuura S, Tauchi H, et al. Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene. 1999;18:3422–3426. doi: 10.1038/sj.onc.1202691. [DOI] [PubMed] [Google Scholar]
- 51.Schoenmakers EF, Huysmans C, Van de Ven WJ. Allelic knockout of novel splice variants of human recombination repair gene RAD51B in t(12;14) uterine leiomyomas. Cancer Res. 1999;59:19–23. [PubMed] [Google Scholar]
- 52.Mohaghegh P, Hickson ID. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum Mol Genet. 2001;10:741–746. doi: 10.1093/hmg/10.7.741. [DOI] [PubMed] [Google Scholar]
- 53.Guidos CJ, Williams CJ, Grandal I, Knowles G, Huang MT, Danska JS. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes Dev. 1996;10:2038–2054. doi: 10.1101/gad.10.16.2038. [DOI] [PubMed] [Google Scholar]
- 54.Nacht M, Strasser A, Chan YR, Harris AW, Schlissel M, Bronson RT, et al. Mutations in the p53 and SCID genes cooperate in tumorigenesis. Genes Dev. 1996;10:2055–2066. doi: 10.1101/gad.10.16.2055. [DOI] [PubMed] [Google Scholar]
- 55.Gurley KE, Vo K, Kemp CJ. DNA double-strand breaks, p53, and apoptosis during lymphomagenesis in scid/scid mice. Cancer Res. 1998;58:3111–3115. [PubMed] [Google Scholar]
- 56.Zhang B, Wang E, Dai H, Hu R, Liang Y, Li K, et al. BRIT1 regulates p53 stability and functions as a tumor suppressor in breast cancer. Carcinogenesis. 2013;34:2271–2280. doi: 10.1093/carcin/bgt190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 58.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 59.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 60.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 61.Mailloux AW, Young MR. NK-dependent increases in CCL22 secretion selectively recruits regulatory T cells to the tumor microenvironment. J Immunol. 2009;182:2753–2765. doi: 10.4049/jimmunol.0801124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grundy S, Plumb J, Lea S, Kaur M, Ray D, Singh D. Down regulation of T cell receptor expression in COPD pulmonary CD8 cells. PLoS One. 2013;8:e71629. doi: 10.1371/journal.pone.0071629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mei J, Huang X, Zhang P. Securin is not required for cellular viability, but is required for normal growth of mouse embryonic fibroblasts. Curr Biol. 2001;11:1197–1201. doi: 10.1016/s0960-9822(01)00325-6. [DOI] [PubMed] [Google Scholar]
- 64.Singh B, Stoffel A, Gogineni SK, Poluri A, Pfister DG, Shaha AR, et al. Amplification of the 3q26.3 locus is associated with progression to invasive cancer and is a negative prognostic factor in head and neck squamous cell carcinomas. Am J Pathol. 2002;161:365–371. doi: 10.1016/S0002-9440(10)64191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sotiropoulou PA, Candi A, Mascré G, De Clercq S, Youssef KK, Lapouge G, et al. Bcl-2 and accelerated DNA repair mediates resistance of hair follicle bulge stem cells to DNA-damage-induced cell death. Nat Cell Biol. 2010;12:572–582. doi: 10.1038/ncb2059. [DOI] [PubMed] [Google Scholar]
- 66.Milliken EL, Lozada KL, Johnson E, Landis MD, Seachrist DD, Whitten I, et al. Ovarian hyperstimulation induces centrosome amplification and aneuploid mammary tumors independently of alterations in p53 in a transgenic mouse model of breast cancer. Oncogene. 2008;27:1759–1766. doi: 10.1038/sj.onc.1210815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.